Abstract
Background:
Ductal carcinoma is one of the most common breast cancer (BrC) among the women in the world. Several factors may involve in establishment of breast cancer. The role of viral infections have been investigated in BrC, Among them the association of Epstein Barr virus have been reported in the patients with breast cancer type ductal carcinoma. Thus this study was conducted to evaluate the rate of Epstein Barr virus in women with breast cancer type ductal carcinoma.
Material and methods:
A total of 72 formalin-fixed paraffin-embedded tissue blocks samples were collected from 37 (51.38%) women with breast cancer type ductal carcinoma and 35 (48.61%) samples of breast with fibro adenoma as control group. The DNA was extracted for all the samples. The detection of EBNA 3C EBV DNA was done by nested PCR. The results of positive were sequenced to confirm PCR product and determine EBV genotypes.
Results:
About 10/37 (27.02%) samples of ductal breast carcinoma were showed positive for EBNA 3C EBV DNA while 4/35 (11.42%) of fibro adenoma were positive for EBNA 3C EBV DNA (p= 0.095). Randomly 7 PCR products were sequenced and the results of sequencing EBNA 3C shows, the detected EBVDNA were type 1 EBV type.
Conclusion:
This study shows high prevalence of 27.02% EBV DNA type 1 was found in formalin-fixed paraffin-embedded tissue of Patients with ductal breast carcinoma. The outcomes of this study suggesting that EBV might have a significant role in breast cancer in Ahvaz city, south west region of Iran. However the expression of EBV oncoproteins, EBNA1, LMP1, and LMP2 require to be determined with ductal carcinoma cells. About 72.97% breast samples showed negative for EBVDNA. The role other viruses including Human cytomegalovirus, papilloma viruses and Merkel viruses are required to be investigated in further studies.
Keywords: Epstein Barr virus, breast cancer, ductal carcinoma, nested PCR
Introduction
Breast cancers, are the main cause of the death among the women worldwide (Murata et al., 2014). It is estimated about 1000,000 cases of breast cancers occur in different regions of the world (Parkin et al., 2001). Breast cancer ranked first among all tumors and presented about 17.5% of all the diagnosed cancer cases (Ok et al., 2015). Detection of Epstein-Barr virus (EBV) DNA in the neoplastic tissues of breast cancer cases have been reported by some authors (Khan et al., 2014; Jha et al., 2016; Aboulkassim et al., 2015; Mazouni et al., 2015; Joshi et al., 2009; Fawzy et al., 2008; Mohammadizadeh et al., 2014; Zekri et al., 2012). The etiology of breast cancers remains unknown but risk factors including female gender family history obesity, physical inactivity old age nulliparity and alcohol consumption have been described (Reza et al., 2015). EBV is the common cause of infectious mononucleosis among the children and infects over 90% of the world’s population (Murata et al., 2014). The association of EBV have been reported with other disease including hepatitis, encephalitis and Posttransplant Lymphoproliferative Disease (PTLD) (Ok et al., 2015). EBV genome comprises a linear double-stranded DNA molecule with 172 kb in length and encodes 100 genes (Parkin et al., 2001). EBV belong to γ herpes and two EBV types A and B circulate in the most populations. EBV can establish a latent status and its genome can persist in the nucleus of infected cells in the form of non-integrated circular episomes with a nucleosomal pattern similar to the host chromatin. Through histone modification and DNA methylation, viral promoters are regulated for the lytic cell cycle and reactivation. The EBV latent episomes have a capacity to influence the epigenetic state of the host DNA and can express several types of oncoproteins which associated with different type of malignant tumors (Ok et al., 2015). EBV accounts for 0.5–2% cancers in different geographical regions of the world, The recent analysis indicates that 1.8% of all cancer deaths in 2010 were associated with EBV. EBV is closely associated with human malignancies, including Burkitt’s lymphoma (BL), Hodgkin’s disease (HD), T-cell lymphoma, nasopharyngeal carcinoma (NPC), a subset of gastric carcinoma and lung carcinoma with lymphoepithelioma-like histology (Ishtiaq et al., 2013., Leila et al., 2016; Monabati et al.,2016; Khan et al., 2014; Jha et al., 2016). Several mechanisms and hypotheses have been described regarding the association between EBV infection and breast carcinoma (Zekri et al., 2012; Reza et al., 2015; Tempera et al., 2014; Glenn et ., 2012). The oncoprotein or EBNA-1 (Epstein Barr Nuclear Antigen) gene have been expressed in Burkitt’s lymphoma, The oncoprotein or EBNA-1, LMP1 (Latent Membrane protein -1) and LMP2 genes are expressed in NPC (Mesri et al., 2014). Epstein Barr-Nuclear antigen EBNA-3C is expressed in latency stage and can transform primary B lymphocytes to lymphoblastoid cell lines (Whitehurst et al., 2015). In the course of transformation both EBNA3C targets several tumor suppressors like p53, p73 and pRB through a mechanism of phosphorylation and ubiquitination (Jha et al., 2015c). it was found EBNA3C has a specific interaction with IRF4 which results in enhanced stabilization of IRF4 (Banerjee et al., 2013). Additionally, the stabilizing IRF4 and at the same time enhancing ubiquitin-mediated IRF8 degradation which contributes to EBV mediated lymphomagenesis (Banerjee et al., 2013). The relationship between breast cancer and EBV could be of potential importance not only for better understanding of breast cancer etiology, but also for early detection, strategy for prevention of breast cancer and treatment. Thus this study was conducted to evaluate the presence of EBNA-3C in the tissue specimens of ductal breast carcinomas in Ahvaz city, IRAN. Ahvaz city is capital of Khozestan province, located at south west region of Iran.
Material and Methods
A total of 37 paraffin embedded tissues of the patients with ductal breast carcinoma and 35 paraffin embedded tissues of the patients with fibroadenoma as control group were collected from archive Eimamkhomaini hospital, Ahvaz, Iran during 2006-2014 (Table 1). The diagnostic accuracy of ductal breast carcinoma were confirmed by a pathologist. The patients ages were between 40 to 59 years with the mean age of 55±8 years. The sections of 10 μm thickness were prepared from each sample and stored at 4°C till performance of tests.
Table 1.
Primers Used for PCR with Thermal Cycling Conditions
| Primer | Sequence | PCR thermal cycling conditions |
|---|---|---|
| First Cycle | EBNA 3C 5′-GAGAAGGGG AGC GTG TGT TGT-3′ 5′ -GCT CGT TTT TGA CGT CGG C -3′ |
95°C for 10 min, 94°C for 45 s, 54.5°C for 45 s, 72°C for 45 s: 35 cycles 72°C for 10 min; final extension |
| Nested Cycle | EBNA 3C ′-TCA TAG AGG TGA TTG ATG TT-3′ 5′-ATG TTT CCG ATG TGG CTT AT-3′ |
95°C for 10 min, 94°C for 45 s, 54.5°C for 45 s, 72°C for 45 s: 35 cycles 72°C for 10 min; final extension |
Deparaffinization
Deparaffinization was done by xylene and ethanol (Germany, Merk). Initially, all the specimens were placed in microtubes then xylene was added and kept at 45°C for 15 min followed by centrifuge at 14,000 rpm. This stage was repeated again. The supernatant was discarded and 1ml absolute ethanol was added to precipitate and stored at the room temperature for 10 min and centrifuged again at 14,000 rpm for 1 minute. The supernatant was discarded. This process was repeated by adding 70% ethanol, followed the same condition. Finally supernatant was discarded and all microtubes were placed at 65°C for 5 min to vaporize the ethanol residue and the pellet was used in DNA extraction (Habibian et al., 2013).
DNA extraction
High pure PCR template preparation kit (Roche, Germany, code No: 11796828001) was applied for the extraction of DNA, according to the manufacturer’s instruction. The extracted DNA was stored at -70°C until PCR amplification.
PCR amplification
All the extracted DNA samples were initially subjected to PCR with consensus primers PCO3/PCO4 (β-globin) to confirm the quality of the extracted DNA (used as an internal control). The following primers (PCO3: 5’ACA CAA CTG TGT TCA CTA GC /PCO4: 5’CAA CTTC AT CCACGT TCA CC with PCR product of 110 bp (Shahab et al., 2015).
Two oligonucleotide primers of the EBNA-C was used to discriminate type 1 and type 2 EBV (Table 1). The first round of PCR was performed in 25μl mixture, containing 7μl of extracted DNA, 2.5μl PCR buffer 10X (Roche), 0.5μl dNTP 10 mM (Roche), 1U Taq Polymerase (Roche),20μM of each primer sequence was subjected to thermocycler (Techne TC-5000, UK). The second round was carried out with 4μl of the first round product, under the same condition described previously with the set of Nested primers and different annealing temperature which are reported in Table 1. The products of nested PCR were 75 bp for type 1 and 168bp for type2 EBV (Kim et al., 2002). Cell line B95.8 was used as a positive control to detect EBV type 1 (Kim et al., 2002).
Gel electrophoresis
The second round PCR product was separated on a 2% agarose gel and developed by Safe Stain under voltage at 100V. The result was seen under ultra violet in transilluminator. The sizes of bands are compared with 50bp Ladder (Fermentas) which was placed on the well as an indicator.
Sequencing
To confirm the results of PCR and to determine genotyping randomly 7 positive PCR products were selected and sequenced (Bioneer company, South Korea). The sequences were blasted using available databases.
Statistical analysis
The obtained results were analyzed by the version 17 of SPSS software and the role of age and sex on positive cases were surveyed by the Fisher`s exact and Chi square test.
Results
Among of 37 ductal breast carcinoma 10 (27.02%) cases showed positive for EBVDNA (EBNA 3C) by the Nested PCR. While among the 35 control group (fibroadenoma) 4 (11.42%) cases were positive for EBVDNA (P > 0.095). The ages of the patients positive for EBVDNA EBNA 3C was between 50 to 58 years. Table 2 shows the frequency of positive EBVDNA among the ductal breast carcinoma. The results of blast of sequencing of EBVDNA EBNA 3C displayed all the 7 positive samples were showed belonged to EBV type 1.
Table 2.
Shows the Number of Positive and Negative EBNA3C in Tumors
| Category | EBNA3C positive | EBNA3C negative | p - value |
|---|---|---|---|
| Ages | |||
| 40-49 | 3/15 (20%) | 12 (80%) | |
| <50 | 7/22 (31.8%) | 15 (68.18%) | 0.427 |
| Ductal | |||
| carcinoma | 10/37 (27.02%) | 27/37 (72.97%) | |
| Fibroma | 4/35 (11.42%) | 31/35 (88.57%) | 0.095 |
| Tumor grade | |||
| Grade I | 1/12 (8.33%) | 11 (91.16%) | |
| Grade II | 2/13 (15.38%) | 11 (84.61%) | 0.007 |
| Grade III | 7/12 (58.33%) | 5 (41.66%) | |
| Breast cancer | |||
| Invasive | |||
| N=34 | 9/33 (27.27%) | 0.798 | |
| In situ | |||
| N=3 | 1/3 (33.33%) | ||
Table 2, shows the distributions of EBNA 3C detection among the age groups (p=0.427), in Ductal carcinoma and fibroma (p>0.095). While the distributions of EBNA3C in histology grade III with histology grade I and II was found significant (p<0.003). The frequency of EBNA 3C in invasive and in situ ductal carcinoma was not found significant (p=0.798).
Discussion
Several factors including status of host genetic, immunodeficiency, geographically endemic patterns of EBV infection, co-infection of EBV and HIV, long persistence of EBV infection in B cells and epithelial cells may lead to lymphoma and some carcinoma tumors (Joseph, 2010). The prevalence of EBV association with lymphoma such as Burkitt’ s Endemic (>99%), sporadic (~15%), B cell, immunoblastic 100%, AIDS, PTLD, congenital immunodeficiency Hodgkin’ s ~40% – 50% and with association with Carcinomas such as Nasopharyngeal 100% and Gastric ~15% (Joseph, 2010). It have been revealed that the few EBV oncoproteins are expressed in different types of cancers, i.e. in Burkett’s lymphoma expressed EBNA1 and EBBRs, Nasopharyngeal carcinoma expressed EBNA1, LMP1, LMP2, EBERs, Gastric Carcinoma expressed EBNA 1, LMP2, EBERs, T/NK cell Lymphoma expressed EBNA1, LMP1, LMP2, EBERs, Hodgkin’s Lymphoma EBNA1, LMP1, LMP2, EBERs, posttransplant lymphoproliferative disorder (PTLD) (Hsin-Pai, 2010).
Figure 1.
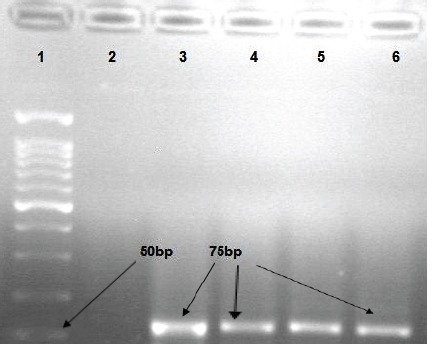
The Results of PCR for Detection of EBV EBNA 3C. Line 1, DNA Molecular Marker (50bp size), Line 2, Negative control, Line 3 positive control B95-8 cell line, Lines 4-6 positive samples.
Breast cancer is a dominant cause of death among women worldwide. The rate of breast cancer is rising about 3% per year (Forouzanfar et al., 2011). Using PCR-based techniques, a number of studies have reported a positive correlation between EBV and breast cancer (Fawzy et al., 2008; Preciado et al., 2005). The frequency of EBV detection in breast cancer is varied in different region of world. In the present study About 10/37 (27.02%) samples of ductal breast carcinoma showed positive for EBV DNA and 4/35 (11.42%) of fibro adenoma as the control group were positive for EBV DNA (p=0.095). Our finding is in agreement with Abdel-Rahman et al, that have detected EBV DNA in 14/50 (28%) of Iraqi women with breast cancer type ductal carcinoma (Abdel-Rahman et al., 2012). Our finding was in accordance with Naushad et al results that have described, EBV DNA was detected in 24.4% of Pakistani women with Breast cancer (Naushad et al., 2017).
The low frequency of 7.3% EBV DNA have been reported among the breast cancer in Iran (Torabizadeh et al., 2014). While high frequency of 55.5% of EBV DNA detection in women with breast cancer have been reported in Sudan (Yahia et al., 2014). Although the role of EBV in breast cancer is a controversial , since some investigators failed to identify EBV DNA in the breast cancer sample (Kadivar et al., 2011).
Virus related breast cancer has been exhibited a poor prognosis, particularly when multiple viruses are detected in breast specimens (Tsai et al., 2007). It was revealed that EBV positive breast cancer displays more aggressive characteristics (Mazouni et al., 2011). In our present study the high frequency of 58.33% EBVDNA have been found among the ductal carcinoma specimens with grade III.
The role of EBV latent antigens EBNA1, EBNA2, EBNA3C, and LMP1 were found essential for transformation and immortalization of EBV-infected cells which leading to cancers (Jha et al., 2016). Some investigators have demonstrated that onco-proteins of EBV, LMP1 and EBNA1 enhance cancer progression and metastasis of nasopharyngeal malignancy and can induces an Epithelial–Mesenchymal Transition (EMT) and increases the Stem-like Cancer Cells in NPC which is a crucial event during cancer metastasis progression, (Kong et al., 2010; Wang et al., 2014). The presence of EBNA1 gene have been detected in the breast cancers tissue. The detection of EBV LMP1 and LMP2 genes have been found in the breast cancer tissues (Yahia et al., 2014). In the detection of EBVLMP1, LMP2 and EBNA1 genes have not been investigated but it requires further investigation.
Current anti-herpes virus drugs, including nucleoside analogs ganciclovir GCV) and acyclovir are used for acute or reactivation of EBV, but these drugs are inefficient to eliminate the EBV virus in a latent stage (Ghosh et al., 2012). Recently combination of betulinic acid BA which is a natural product, derived from plant sources, and Chidamide CDM synergistically inhibits EBV replication with ROS over-generation and subsequent DNA damage and apoptosis in. An in vivo xenograft tumor development study with the tail vein injection of EBV-transformed LCL cells (lymphoblastoid cell line) in nude mice proves that the combination of BA and CDM synergistically increases superoxide anion release in tumor tissues and suppresses EBV replication and tumor growth, and significantly prolongs mouse survival. The combination of BA and CDM may be an efficient strategy for clinical EBV removal in the latent stage (Haibing et al., 2017). Formalin-fixed and paraffin-embedded (FFPE) tissues support the tissue structure for histopathological diagnosis of diseases like cancer are used routinely world-wide (Rogier et al., 2015). Detection and characterization of novel viruses is often barricade the lack of adequately stored materials. Formalin-fixed paraffin embedded (FFPE) tissues can be used to detect known viral sequences (Bodewes et al., 2014; Cimino et al., 2014). The viral RNA was extracted from a FFPE lung tissue sample of a victim of the 1918 pandemic, ‘Spanish’ influenzaA/H1N1 virus, this virus was characterized and subsequently recovered by reverse genetics (Tumpey et al., 2005;Taubenberger et al., 1997).
In the present study about 27 (72.97%) breast samples showed negative for EBVDNA. The role other viruses including papilloma viruses and Merkel viruses have been investigated (Reza et al., 2015; Haibing et al., 2017; Glenn et al., 2011). The role of mentioned viruses requires for the further investigation.
With the aforementioned data it is suggested to manage and control the spreading of EBV infection, the screening of EBV DNA should be implemented for women with breast cancer.
In summary, this study shows high prevalence of 27.02% EBV DNA type 1 was found in formalin-fixed paraffin-embedded tissue of Patients with ductal breast carcinoma. The outcomes of this study suggesting that EBV might have a significant role in breast cancer in Ahvaz city, south west region of Iran However the expression of EBNA1, LMP1, and LMP2 is required to determine association of EBV oncoproteins with invasive ductal carcinoma cells. About 72.97% breast samples showed negative for EBVDNA. The role other viruses including Human cytomegalovirus, papilloma viruses and Merkel viruses are required to be investigated in future studies.
Acknowledgements
This study was done as a research project with registration number 92,114 has been carried out by Chia sharifpour MSc (Virology), which financially supported by Infectious and Tropical Disease Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
References
- 1.Aboulkassim T, Yasmeen A, Akil N, et al. Incidence of Epstein-Barr virus in Syrian women with breast cancer: A tissue microarray study. Hum Hum Vaccin Immunother. 2015;11:951–5. doi: 10.1080/21645515.2015.1009342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Rahman N Z, Abeer AB, Waleed S. Epstein-Barr virus and breast cancer: Epidemiological and Molecular study on Egyptian and Iraqi women. J Egypt Natl Canc Inst. 2012;24:123–31. doi: 10.1016/j.jnci.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee S, Lu J, Cai Q, et al. The EBV latent antigen3C inhibits apoptosis through targeted regulation of interfere on regulatory factors4 and 8. PLoS Pathog. 2013;9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodewes R, Sánchez CGJ, Rubio GA, et al. Identification of DNA sequences that imply a novel gammaherpesvirus in seals. J Gen Virol. 2014. http://dx.doi.org/10.1099/vir.0.000029 . pii: vir.0.000029. [DOI] [PubMed]
- 5.Cimino PJ, Zhao GWD, Sehn JK, et al. Detection of viral pathogens in high grade gliomas from unmapped next-generationsequencing data. Exp Mol Pathol. 2014;96:310–15. doi: 10.1016/j.yexmp.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Fawzy S, Sallam M, Awad NM. Detection of Epstein- Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–92. doi: 10.1016/j.clinbiochem.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–84. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 8.Glenn WK, Heng B, Delprado W, et al. Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One. 2012;7:e48788. doi: 10.1371/journal.pone.0048788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh SK, Perrine SP, Williams RM, et al. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood. 2012;119:1008–17. doi: 10.1182/blood-2011-06-362434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn WK, Heng B, Delprado W, et al. Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One. 2012;7:e48788. doi: 10.1371/journal.pone.0048788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habibian A, Makvandi M, Samarbaf-zadeh AR, et al. Epstein-Barr virus DNAfrequency in paraffin embedded tissues of Non-Hodgkin lymphoma patients from Ahvaz, Iran. Jentashapir J Health Res. 2013;4:315–20. [Google Scholar]
- 12.Haibing Yu, Hongyu Z, Zhigang C, et al. Combination of betulinic acid and chidamide synergistically inhibits Epstein-Barr virus replication through over-generation of reactive oxygen species. Oncotarget. 2017;8:61646–61. doi: 10.18632/oncotarget.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsin-Pai Li, Mei Chao, Shu-Jen Chenand, et al. Yu-Sun Chang, Epstein-Barr virus and its oncogenesis. In: James J.-H, Ou T.S, Benedict Yen, editors. Human oncogenic Viruses. 1th edition. Singapore: World Scientific Publishing; 2010. pp. 208–67. [Google Scholar]
- 14.Ishtiaq S, Hassan U, Mushtaq S, et al. Determination of frequency of Epstein-barr virus in non- Hodgkin lymphomas Using EBV latent membrane protein 1 (EBV-LMP1) immunohistochemical staining. Asian Pac J Cancer Prev. 2013;14:3963–7. doi: 10.7314/apjcp.2013.14.6.3963. [DOI] [PubMed] [Google Scholar]
- 15.Jha HC, Yang K, El-Naccache EBNA3C regulates p53 through induction of Aurorakinase B. Oncotarget. 2015c;6:5788–5803. doi: 10.18632/oncotarget.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha HC, Pei Y, Robertson ES. Epstein–Barr virus: Diseases linked to infection and transformation. Front Microbiol. 2016;7:1602. doi: 10.3389/fmicb.2016.01602. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi D, Quadri M, Gangane N, et al. Association of Epstein Barr virus infection (EBV) with breast cancer in rural Indian women. PLoS One. 2009;4:e8180. doi: 10.1371/journal.pone.0008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph SP. Molecular pathobiology of EBV infection. In: Kamel Khalili, Kuan-Teh Jeang., editors. Basic science and clinical applications. 1th edition. Hoboken, New Jersey: John Wiley &Sons, Inc, 2010; 2010. pp. 409–23. [Google Scholar]
- 19.Kadivar M, Monabati A, Joulaee A, et al. Epstein-Barr virus and breast cancer: lack of evidence for an association in Iranian women. Pathol Oncol Res. 2011;17:489–92. doi: 10.1007/s12253-010-9325-z. [DOI] [PubMed] [Google Scholar]
- 20.Khan G, Hashim MJ. Global burden of deaths from Epstein Barr virus attributable malignancies 1990–2010. Infect Agent Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim I, Park ER, Park SH, et al. Characteristics of Epstein Barr virus isolated from the malignant lymphoma in Korea. J Med Virol. 2002;67:59–66. doi: 10.1002/jmv.2193. [DOI] [PubMed] [Google Scholar]
- 22.Kong QL, Hu LJ, Cao JY, et al. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010;6:e1000940. doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leila Z, Arabzadeh SA, Afshar RM, et al. Detection of Epstein-Barr virus and cytomegalovirus in gastric cancers in Kerman. Iran Asian Pac J Cancer Prev. 2016;17:2423–8. [PubMed] [Google Scholar]
- 24.Monabati A, Vahedi A, Safaei A, et al. Epstein-Barr virus-positive diffuse large B-cell lymphoma: Is it different between over and under 50 years of age? Asian Pac J Cancer Prev. 2016;17:2285–9. doi: 10.7314/apjcp.2016.17.4.2285. [DOI] [PubMed] [Google Scholar]
- 25.Mazouni C, Fina F, Romain S, et al. Epstein Barr virus as a marker of biological aggressiveness in breast cancer. Br J Cancer. 2011;104:332–7. doi: 10.1038/sj.bjc.6606048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazouni C, Fina F, Romain S. Outcome of Epstein-Barr virus-associated primary breast cancer. Mol Clin Oncol. 2015;3:295–8. doi: 10.3892/mco.2014.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–82. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadizadeh F, Zarean M, Abbasi M. Association of Epstein-Barr virus with invasive breast carcinoma and its impact on well-known clinicopathologic parameters in Iranian women. Adv Biomed Res. 2014;3:141. doi: 10.4103/2277-9175.135158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata T, Tsurumi T. Switching of EBV cycles between latent and lytic states. Rev Med Virol. 2014;24:142–53. doi: 10.1002/rmv.1780. [DOI] [PubMed] [Google Scholar]
- 30.Naushad W, Surriya O, Sadia H. Prevalence of EBV, HPV and MMTV in Pakistani breast cancer patients: A possible etiological role of viruses in breast cancer. Infect Genet Evol. 2017;10:230–7. doi: 10.1016/j.meegid.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Ok CY, Li L, Young KH. EBV-driven B-cell lymphoproliferative disorders: from biology, classification and differential diagnosis to clinical management. Exp Mol Med. 2015;23:e132. doi: 10.1038/emm.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37:4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 33.Preciado MV, Chabay PA, De Matteo EN, et al. Epstein-Barr virus in breast carcinoma in Argentina. Arch Pathol Lab Med. 2005;129:377–81. doi: 10.5858/2005-129-377-EVIBCI. [DOI] [PubMed] [Google Scholar]
- 34.Reza MA, Reza MH, Mahdiyeh L, et al. Evaluation frequency of Merkel cell polyoma, Epstein-Barr and mouse mammary tumor viruses in patients with breast cancer in Kerman, Southeast of Iran. Asian Pac J Cancer Prev. 2015;16:7351–7. doi: 10.7314/apjcp.2015.16.16.7351. [DOI] [PubMed] [Google Scholar]
- 35.Rogier B, Peter RWAVR, Anita CS, et al. Virus characterization and discovery in formalin-fixed paraffin-embedded tissues. Virol Methods. 2015;214:54–59. doi: 10.1016/j.jviromet.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahab M, Akbar S, Nasrollah E, et al. Presence of human Papillomavirus DNA in colorectal cancer tissues in Shiraz, Southwest Iran. Asian Pac J Cancer Prev. 2015;16:7883–7. doi: 10.7314/apjcp.2015.16.17.7883. [DOI] [PubMed] [Google Scholar]
- 37.Taubenberger JK, Reid AH, Krafft AE, et al. Initial genetic characterization of the 1918 Spanish influenza virus. Science. 1997;275:1793–6. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 38.Tumpey TM, Basler CF, Aguilar PV, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 39.Tsai JH, Hsu CS, Tsai CH, et al. Relationship between viral factors, axillary lymph node status and survival in breast cancer. J Cancer Res Clin Oncol. 2007;133:13–21. doi: 10.1007/s00432-006-0141-5. [DOI] [PubMed] [Google Scholar]
- 40.Tsai MH, Lin X, Shumilov A, et al. The biological properties of different EpsteinBarr virus strains explain their association with various types of cancers. Oncotarget. 2017;8:10238–54. doi: 10.18632/oncotarget.14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tempera I, Lieberman PM. Epigenetic regulation of EBV persistence and oncogenesis. Semin Cancer Biol. 2014;26:22–9. doi: 10.1016/j.semcancer.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torabizadeh Z, Nadji A, Naghshvar F, et al. Association between Epstein-Barr Virus (EBV) and Breast Cancer. Res Mol Med. 2014;2:24–9. [Google Scholar]
- 43.Wang L, Tian WD, Xu X. Epstein-Barr virus nuclear antigen 1 (EBNA1) protein induction of epithelial-mesenchymal transition in nasopharyngeal carcinoma cells. Cancer. 2014;120:363–72. doi: 10.1002/cncr.28418. [DOI] [PubMed] [Google Scholar]
- 44.Whitehurst CB, Li G, Montgomery SA, et al. Knockout of Epstein-Barrvirus BPLF1 retards B-cell transformation and lymphoma formation in humanized mice. MBio. 2015;6:e01574–15. doi: 10.1128/mBio.01574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yahia ZA, Adam AA, Elgizouli M, et al. Epstein Barr virus: a prime candidate of breast cancer aetiology in Sudanese patients. Infect Agent Cancer. 2014;9:9. doi: 10.1186/1750-9378-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zekri A-RN, Bahnassy AA, Mohamed WS, et al. Epstein-Barr virus and breast cancer: epidemiological and molecular study on Egyptian and Iraqi women. J Egypt Natl Cancer Inst. 2012;24:123–31. doi: 10.1016/j.jnci.2012.06.001. [DOI] [PubMed] [Google Scholar]


