Abstract
Background:
Nudix Hydrolase 15 gene (NUDT15) encodes nucleotide triphosphate diphosphatase which metabolizes the purine analog drugs, such as anticancer thiopurine and anti-gout allopurinol. Genetic variants on Nudix Hydrolase 15 gene (NUDT15) gene effects the drug’s hydrolyses and hence increases the susceptibility to drug-induced toxicity. The NUDT15 gene has been genotyped in various ethnic groups, however, it has not been genotyped among the Middle Eastern Arab Jordanian population.
Aim:
The current study aimed to identify NUDT15 genetic variants among Jordanian Arab population.
Method:
The DNA samples were isolated from leukocytes of 85 unrelated Jordanian Arab volunteers. The coding regions of NUDT15 gene; Exon 1,2 and 3, in addition to some regions of intron 1,2 and 3’UTR, were amplified using polymerase chain reaction (PCR). the PCR products were then subjected to purification and sequenced using Applied Biosystems Model (ABI3730x1).
Results:
Six NUDT15 genetic variants were found among the volunteers. The results were as followed: A novel synonymous variant 36A>G on exon 1 (6%, 95%CI= 3- 9%), the intronic IVS1 +116C>T variant on intron 1 (0.6%, 95%CI= 0-2%), the non-synonymous variant on exon 3; 415C>T (0.6%, 95%CI= 0-2%), A novel non-synonymous variant on exon 3; 404C>A (0.6%, 95%CI= 0-2%) , and two novel variants on 3’UTR ;502G>A (2%, 95%CI= 0.5-4%) and 588T>C (0.6%, 95%CI= 0-2%). NUDT15 36A>G wasfound to be the most common allele among Jordanians was. In silico softwares predicted that the novel NUDT15 404C>A was harmful and affected NUDT15 enzyme’sstability and function. Furthermore, the frequency of NUDT15 IVS1 +116C>T , among Jordanians, showed to be significantly lower from what was reported in other ethnicities with ap value > 0.05 on the other hand, the frequency of 415C>T variant showed to be similar to Europeans in contrast to Asians and Indians that showed to be significantly lower (p value > 0.05).
Conclusions:
The frequency of NUDT15 genetic variants is low among the Jordanian volunteers and significantly lower than other ethnic groups. The findings of this study may increase our understanding of the inter-individual variation in the response to purine analog drugs. Further clinical studies are needed to investigate the influence of novel NUDT15 404C>A on drug metabolism and response.
Keywords: NUDT15, Jordanians, genetic variants, metabolism
Introduction
The thiopurine drugs are commonly prescribed for patients with neoplastic and autoimmune diseases (Weinshilboum, 2001). It is found, clinically, that there is an inter-individual variation in the response of thiopurine drugs among cancer patients (Goel et al., 2015). This variation in drug response can be attributed, in some parts, to the genetic variants coding for thiopurine metabolizing enzymes (Weinshilboum, 2001). It is reported that thiopurine methyl transferase (TPMT) and Nudix hydrolase 15 (NUDT15) enzymes metabolize purine analog drugs and genetic variants on TPMT and Nudix hydrolase 15 (NUDT15) gene significantly affect the pharmacokinetics, and hence the efficacy and toxicity of the purine analog drugs (Moriyama et al., 2016).
The genetic variants on TPMT are well studied and genotyped among different ethnic populations (Liang et al., 2013; Chen et al., 2014). However, limited studies investigated the role of NUDT15 genetic variants on thiopurine drugs’ response.
The NUDT15 gene is located on the long arm of chromosome 13 at position 14.2 (GenBank accession NC_000013.11). It consists of 3 exons with 495 encoding base pairs. Genetic variants on NUDT15 influence the purine analog drug’s response (Moriyama et al., 2017a). It was found that NUDT15 genetic variants resulted in decrease or loss of enzyme’s function, leading to poor metabolism of the drug and accumulation of thiopurinetoxic metabolites and subsequently causing serious side effects such as thiopurine induced leukopenia (Fei et al., 2018). A Korean study reported that pediatric patients with Cohn’s disease, who carried homozygous NUDT15 415C>T genotype, were more sensitive to thiopurine treatment and required more dose reduction than those with wild or heterozygote NUDT15 415C>T genotype (Lee et al., 2016).
Furthermore, there is an inter-ethnic variation in the frequency of NUDT15 genetic variants (Singh et al., 2017). The non-synonymous NUDT15 variants Arg139Cys (rs116855232), Arg139His (rs147390019), Val18Ile (rs186364861) and Val18_Val19insGlyVal (rs554405994) were most common in East Asians and Hispanics, rare in Europeans, and not observed in Africans (Tanaka et al., 2015; Yang et al., 2015; Shah et al., 2017). On the other hand, the deletion variant p.G17_V18del was only seen in African and European, but not in Asian population (Moriyama et al., 2017b). This inter-ethnic variation in the frequency of NUDT15 genetic variants may cause inter-ethnic variation in the response of purine analog drugs among different ethnic populations (Singh et al., 2017).
There are limited studies regarding NUDT15 variants among Middle Eastern Arab populations (Zgheib et al., 2017). In addition, no study has investigated the genetic variants of NUDT15 gene among Jordanian population yet. Therefore, the aim of this study was to identify the NUDT15 gene structure among unrelated Jordanian Arab volunteers and compare it with other major ethnic populations.
Materials and Methods
Chemical Compounds
The PCR primers were obtained from Integrated DNA Technologies, USA. The primers sequence is illustrated in Table 1. The Taq polymarse, Taq polymerase buffer, deoxyribonucleotide triphosphate (dNTPs), nuclease free water (NFW), DNA laddar, and MgCl2 were obtained from Promega, USA. Agarose powder was purchased from Bio- basic, Canada; and the buffer, used to carry out electrophoresis (TBE), was obtained from Bio- basic, Canada. The solution had to be stained with redsafe dye which was purchased from iNtRON Biotechnology, South Korea.
Table 1.
Name of Primers, Primer Sequence, and Annealing Temperature Used in PCR Amplification of NUDT15 Gene
| Primer | Primer sequence 5’-3’ | Annealing temperature (°C) | Region | Size (bp) |
|---|---|---|---|---|
| NUDT15 1st- F | CAA AGC ACA ACT GTA AGC GAC T | 57 | exon 1 | 600 |
| NUDT15 1st - R | GAA AGA CCC AGC TAG CAA AGA C | |||
| NUDT15 2nd- F | CGG CCT TCC AAA AGA TTA CA | 56 | exon 2 | 580 |
| NUDT15 2nd- R | TGA TCT AAT CAC CTC CCA AGG | |||
| NUDT15 3rd- F | AAG CAA ATG CAA AGC ATC AC | 57 | exon 3 | 480 |
| NUDT15 3rd- R | GGC TGA AAG AGT GGG GGA TA |
F, forward primer; R, reverse primer; bp, base pair
Study Participants and protocol
A total of 100 unrelated healthy Jordanian participants of both genders with 27±5 years (mean age± standard deviation)were recruited as blood donors. The participants included in this study were not taking any drugs in regular basis and had no chronic or serious diseases.
The study was approved by the ethical committee of AlZaytoonah University of Jordan, (reference number 18/04-2016), and written informed consent was obtained from each participnat in accordance with the Decleration of Helsinky (World Medical, 2013).
Sample Collection and DNA Extraction
Approximately, 3 to 5 ml of peripheral blood sample was drawn from each participant and kept in ethylenediaminetetraacetic acid (EDTA) tubes. Around 300 microliter of the Buffy coat layer, which contained the concentrated leukocytes, was isolated by centrifugation of the blood samples at 3500 rpm for 5 minutes (Jarrar et al., 2010), and then transferred to an Eppendorf tube
The Genomic DNA was isolated from the leukocytes and purified according to the Wizard® Genomic DNA purification kit manufacturer’s instructions.
Breifly, the leukocytes were lysed using cellular lysis solution and the resulting supernatent that contained cellular debris was discarded. Sequentially, lysing the nucleus was done by the additon of nucleo lysing solution followed by the addition of 100 microliters of protein lysing solution to accelate the precipitation of the proteins. In order to isolate the genomic DNA that was present in theresultant supernatent solution, we transferred it to another eppendorf tube and isopropyl alchohol was added .
The DNA was then washed by 75% ethanol, diluted in NFW to achieve final concentration of 100 ng/μl, and stored at -20°C until further use.
The total genomic DNA was quantified using the Nano Drop 2000 spectrophotometer (Quawell DNA/Protein Analyzer, USA). The 260/280 ratio was used to measure the purity of DNA samples. Only samples with 260/280 ratio of 1.8±0.1 were considered for further molecular analysis (Olson and Morrow, 2012).
PCR Reaction
DNA samples were then subjected to NUDT15 gene amplification. The coding regions of the gene, Exons1, 2, and 3, were amplified using the Bio-Rad (model, UK) thermal cycler.
The PCR reaction was first optimized by determining the optimal concentration of the PCR components and the best annealing temperatures which would result in amplification of NUDT15 gene and removal of any unwanted and non-specific amplifications.
After optimization of the concentrations, the PCR reaction of a total volume of 50 μl was carried out containing 100ng of the extracted genomic DNA,
10 pico moles of forward and reverse primers, 10µl green of 5X Taq DNA polymerase, 0.5 mM MgCl2, 0.2 mM dNTPs, and 0.15 units of Taq DNA polymerase
The PCR amplification reaction started with the initial denaturation stage at 94°C lasting for 5 min, and followed by multiple steps that were repeated for 30 cycles. These steps included the secondary denaturation phase that lasted for 1 min at temperature of 94°C , followed by the Annealing phase that took place at 56-57°C for 1 min (Table 1), and finally ending each repitition with the elongation step at 72°C for 1 min. In the last step, all samples were exposed to a final extension step at 72°C for 7 min.
Gel Preparation and Gel Electrophoresis
Gel electrophoresis was carried out after every PCR. A 1.5% agarose gel was used to separate the amplified PCR products. The preparation of a 1.5% (w/v) gel agarose was done by melting 0.75g powdered agarose in 50ml of 1X Tris-Borate-EDTA buffer. The gel was stained with 1.25µl of 20,000x Redsafe dye.
Seven µl of the PCR product was mixed with 2µl of the green loading buffer (Promega, USA). DNA loading ladder, with a standard size of 100 bp, was used to compare the size of PCR product bands. The gel was electrophoresed at 125 electrical Ampere for 30 - 45 min. Lastly, DNA bands were visualized by exposing the gel to UV light using the bench top U.V transluminator (Bio Doc-ITTM, UK).
DNA Sequencing
The PCR product samples were sent out to GENEWIZ company (South Plainfield, USA) for sequencing of exons 1,2, and 3 in NUDT15 gene using Sanger Sequencing, as prescribed previously (Jarrar et al., 2018). The PCR products were purified before sequencing. The same PCR primers (Table 1) were used for DNA sequencing. Sequence analysis was performed using Applied Biosystems Model (ABI3730x1) which is based on the dye terminator method.
The DNA sequence for the three exons of the NUDT15 gene in Homo sapiens were obtained from the GenBank databases (Benson et al., 2013). The alignment of the DNA sequences were provided using the Multialign software (LaMarche et al., 2013). The chromatograms of the sequences were seen by DNA Based v3.5.4 software (Esteves et al., 2008)
In silico prediction of novel NUDT15 variants
The current study applied two different software programs, SIFT, and Mutpred, in order to predict the effect of novel non-synonemous NUDT15 404C>A genetic variants on NUDT15 protein’s stability and function (Kumar et al., 2009; Adzhubei et al., 2010). In addition, effect of novel intronic NUDT15 IVS1+116C>T variants on splicosome formation was predicted using Berkeley Drosophila Genome Project (BDGP) software (Reese et al., 1997). These software programs use numerical scores representing the probability that genetic variants are damaging.
Statistical Analysis
Hardy-Weinberg equilibrium and Chai-Square (χ2) test were used to test the frequency of the NUDT15 alleles. Comparison between NUDT15 allele frequencies among Jordanians and allele frequencies for the same gene in other different ethicities was performed running the χ2 test in the SPSS (IBM analytics, USA). The p < 0.05 was considered as the cut-off value for significance.
Results
AA total of 100 Jordanian Arab volunteers participated in this study. However, 15 samples were excluded from this study due to unclear sequencing results, leaving a total number of 85 DNA samples. Figure 1 shows the gel electrophoresis of the 3 amplified DNA fragments, representing the total NUDT15 exons.
Figure 1.
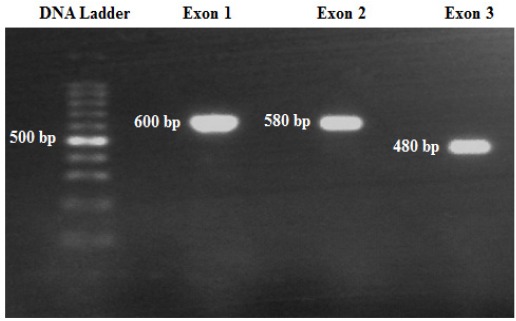
Representative Gel Electrophoresis of NUDT15 Gene’s Exons after PCR. Ten µl of PCR product was run on 1.5% agarose gel and stained with Redsafe dye. The bands were visualized by exposing the gel to UV light using the bench top U.V transluminator (Bio Doc-ITTM, UK). Further information are presented in the Materials and Methods section.
Sequencing the PCR products of the amplified NUDT15 gene for 85 volunteers showed the presence of 6 genetic variants, namely 36A>G (Figure 1b) in exon one, IVS1+116C>T (Figure 1d) located on intron one, a novel variant 404C>A (Figure 1f), the well-known variant 415C>T (Figure 1h) both located on exon 3, 2 variants 502G>A (Figure 1j), and 588T>C (Figure 1l) both found on the 3’ untranslated region (3’UTR). According to the GenBank databases (Benson et al., 2013) database, only IVS1+116C>T and 415C>T have been so far reported suggesting the novelty of the other 4 variants. Figure 2 schemes the subsequent location of the NUDT15 variants that were found in the Jordanian population.
Figure 2.
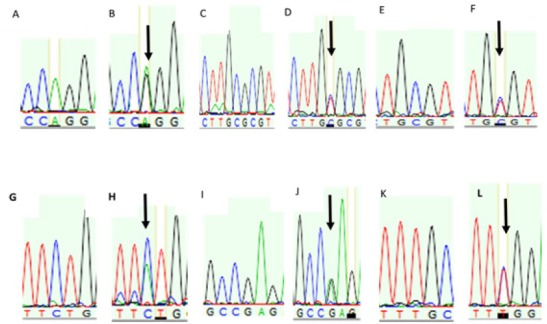
Chromatogram of DNA Sequencing of NUDT15 Variants among Jordanian Volunteers. A and B represent wild and heterozygote NUDT15 36A>G variant, respectively. The wild and heterozygote NUDT15 IVS+116C>T alleles were represented in C and D chromatogram, respectively. E and F represent wild and heterozygote NUDT15 415C>T allele, respectively. G and H are chromatograms of wild and heterozygote NUDT15 404C>A alleles, respectively. Wild and heterozygote NUDT15 502G>A are represented in I and J chromatograms, respectively . Lastly, the NUDT15 588T>C was represented in wild (K) and hetwrzygote (L) genotype.
Figure 3.
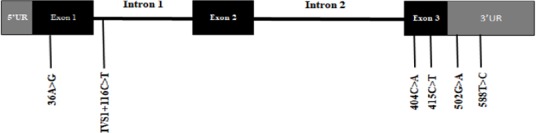
Location of NUDT15 Genetic Variants among Jordanian Volunteers. The gene structure of NUDT15 was obtained from GenBank accession NC_000013.11. UR is the abbreviation for untranslated region.
The frequency of NUDT15 genetic alleles along with the expected activity of NUDT15 enzyme among Jordanian Arab participants are represented in Table 2.
Table 2.
The Distribution of NUDT15 Genetic Variants among Healthy Unrelated Jordanian Population (n=85)
| Nucleic acid changea | Location | Reference ID | Amino Acid Change | Methylation Activity | Allele Frequency(95%CI) |
|---|---|---|---|---|---|
| 36A>Gb | Exon 1 | - | Proline12> proline | ND | 0.06 (0.03- 0.09) |
| +116C>T | Intron 1 | rs79687000 | - | ND | 0.006 (0 – 0.02) |
| 415C>T | Exon 3 | rs116855232 | Arginine 139> cysteine | Low | 0.006 (0 – 0.02) |
| 404C>Ab | Exon 3 | - | Phenylalanine 134> Laucine | ND | 0.006 (0 – 0.02) |
| 7G>Ab | 3’UR | - | - | ND | 0.2 (0.005 – 0.04) |
| 93T>Cb | 3’UR | - | - | ND | 0.006 (0 – 0.02) |
ND means that enyzme activity of genetic variant was not determined; The used reference sequence was GenBank accession NC_000013.11;
Position is indicated in relation to the start codon ATG of the NUDT15 gene; the A in ATG is +1.
A newly identified variant in the present study.
We could detect, for the first time, the NUDT15 36A>G variant which resulted in the transition of adenine to guanine at nucleotide base 36. This transition showed to be a synonymous variant that both, the wild and variant allele, coded for the same amino acid proline at the amino acid residue 12. The results of this study revealed that this variant was the most frequent NUDT15 variant among healthy Jordanian Arab participants with an allele frequency of 0.06, 95% CI=0.03 – 0.09 (Table 2). In our study, ten participants (12%), were found to carry the heterozygote NUDT15 36A>G genotype, while none of the participants carried the homozygote NUDT15 36A>G genotype (Table 3).
Table 3.
The Distribution of NUDT15 Genotype among Healthy Unrelated Jordanian Population (n=85)
| NUDT15 allele | wild genotype: frequency (proportion, 95%CI) | Heterozygote genotype: frequency (proportion, 95%CI) | homozygote genotype: frequency (proportion 95%CI) | P-value |
|---|---|---|---|---|
| 36A>G | 90 (0.88, 0.81-0.92) | 12 (0.12, 0.051-0.179) | 0 | 0.598 |
| IVS1+116C>T | 99 (0.99, 0.946-1) | 1 (0.01, 0-0.054) | 0 | 0.867 |
| 415C>T | 99 (0.99, 0.946-1) | 1 (0.01, 0-0.054) | 0 | 0.867 |
| 404C>A | 96 (0.96,0.901-0.989) | 4 (0.04,0.11-0.99 ) | 0 | 0.833 |
| 502G>A | 99 (0.99, 0.946-1) | 1 (0.01, 0-0.054) | 0 | 0.867 |
| 588T>C | 99 (0.99, 0.946-1) | 1 (0.01, 0-0.054) | 0 | 0.867 |
χ2 < 3.84 at 1 degree of freedom, p value 0.05. All of the genotype frequencies were within Hardy-Weinberg equilibrium.
The intronic variant IVS1+116C>T results in the conversion of cysteine into thymine residue. After reviewing the literature, we found that this variant held the reference number rs79687000. However, the effect of this variant on NUDT15 activity was not yet determined. The frequency of NUDT15 IVS1+116C>T allele was 0.006, 95% CI= 0.00-0.02 (Table 2). Only one participant was found to carry the NUDT15 IVS1+116C>T heterozygote genotype (Table 3).
The new NUDT15 404C>A variant was found in one participant, which was in the heterozygote form. None of the participants carried the homozygote NUDT15 404C/C genotype. This variant was confirmed by re-sequencing the DNA samples of the participants. The frequency of NUDT15 404C>A allele was 0.006, 95% CI= 0.00-0.02 (Table 2) and the heterozygote genotype frequency was 0.01, 95% CI= 0 – 0.05 (Table 3). It was also found that alteration of cysteininto adenine at nucleotide number 404, on NUDT15 coding sequence, resulted in a non-synonymous variant, causing the substitution of phenylalanine with luecine at amino acid residue 134 in the polypeptide sequence of NUDT15 protein.
Only one participant harbored the heterozygote NUDT15 415C>T (rs116855232) variant on exon 3, which is known to cause a non-synonymous variant with the conversion of arginine into cysteine at amino acid residue 139. The frequency of NUDT15 415C>T allele was 0.006, 95% CI= 0.00-0.02 (Table 2) and the heterozygote genotype frequency was 0.01, 95% CI= 0 – 0.05 (Table 3) in this study.
Furthermore, 2 novel variants were identified in the current study. Both variants; 502G>A and 588T>C, were located on the 3’UTR, and were
heterozygotes with an allelic frequency of 0.02, 95% CI= 0.005-0.04 and 0.006, 95% CI= 0.00-0.02 , respectively . (Table 2). The heterozygote genotype frequency was 0.04, 95% CI= 0.01 – 0.09 for NUDT15 502G>A and 0.01, 95% CI= 0 – 0.05 for 588T>C variant (Table 3). These novel variants were confirmed by re-sequencing of the PCR products of NUDT15 gene of the volunteers.
In the present study, we tested whether the frequency of NUDT15 genotype were deviated from Hardy-Weinberg equation by comparing the observed and the expected NUDT15 genotype frequencies among the Jordanian participants. As illustrated in Table 3, all of NUDT15 genotyped frequencies were within Hardy-Weinberg equation, with a p-value >0.05.
In silico predictions of NUDT15 404C>A and NUDT15 IVS1+116C>T are presented in Table 4. Both SIFT and Mutpred softwares predicted that NUDT15 404C>A might have harmful effects on NUDT15 protein with a probability score of 0.6 and 1, respectively. The BDGP software showed that the intronic NUDT15 variant IVS1+116C>T had no effect on the splicosome formation of NUDT15 messenger RNA.
Table 4.
In Silico Prediction of the Effect of NUDT15 Genetic Variant by Using SIFT, MutPred, and BDGP Software Programs
| NUDT15 variants | SIFT (probability score) | MutPred (probability score) | BDGP (probability score) |
|---|---|---|---|
| 404C>A | 1 | 0.596 | ND |
| IVS1+116C>T | ND | ND | 0 |
Probability of being pathogenic, 0 = lowest; 1 = highest; ND is an abbreviation for "not determined"
Table 5 compares the frequencies of NUDT15 IVS1+116C>T and NUDT15 415C>T in Jordanians with those frequencies among other major ethnic groups as reported in HapMap data (Thorisson et al., 2005). The frequency of NUDT15 415C>T (Rs116855232) in Jordanians (0.6%) was not statistically different (p value > 0.05) from those in Europeans (0.2%). In contrast, NUDT15 415C>T frequency in Jordanians showed to be significantly higher (p value < 0.05) than that in Africans (0.06%) and lower (p value < 0.05) than that in Asians (7%) and Indians (8%).
Table 5.
Comparison of Jordanian Population and Other Major Ethnic Populations in Terms of NUDT15 rs79687000 and rs116855232 Variant Proportions
| NUDT15 variants | Reference number | Jordanian (current study) | Indian allele frequency | Europeana | Asiansa | Africana |
|---|---|---|---|---|---|---|
| 415C>A | rs116855232 | 0.006 | 0.075* (Shah et al., 2018) | 0.002 | 0.066* | 0.0008* |
| IVS1+116C>T | rs79687000 | 0.006 | ND | 0.025* | 0.016* | 0.026* |
aHapmap data (Thorisson et al., 2005).The Asian, African, and European NUDT15 variant frequencies are for Han Chinese in Beijing, China(CHB), African ancestry of Yorubain Ibadan, Nigeria(YRI), and Utah residents with Northern and Western European ancestry(CEU), respectively; ND is an abbreviation for "not determined";
Significant difference (χ2 , p <0.05) in comparison with the proportion of NUDT15 genetic variant among Jordanians
On the other hand, the frequency of the intronic variant NUDT15 IVS1+116C>T (rs79687000) among healthy Jordanians (0.5%) tended to be the lowest in comparison with Africans (2.6%), Europeans (2.5%), and Asians (1.6%).
Discussion
NUDT15 gene plays a major role in drugs’ response by affecting their metabolic routes. Such drugs are purine analogues like anticancer and anti-gout drugs. During the last decade, the importance of NUDT15 is being appreciated and a strong association between thiopurine toxicity and this gene’s polymorphisms in patients with Asian ethnicity has been reported (Moriyama et al., 2016; Kim et al., 2017; Singh et al., 2017). Even though the frequency of risk alleles in the European and African individuals is low, such studies were important to be reported.
The current study was the first one .. healthy Jordanian population. In this study, the total protein coding regions and the region responsible for spliceosomal formation were sequenced. We found that the frequency of NUDT15 genetic variants was very low among Jordanians and we could identify 4 novel variants through in silico prediction of the effect of novel synonymous and intronic variant on NUDT15 mRNA and protein. These identified NUDT15 genetic variants may contribute, at least in part, in the inter-individual variation in the metabolism of purine analog drugs and susceptibility to the drug-induced toxicity among Jordanian population.
The current study genotyped NUDT15 gene by sequencing the 3 exons, which code the total protein region of NUDT15. The DNA sequencing, which was used in this study, is considered the most accurate genotyping method (Cao et al., 2013). We also confirmed the novel variants by re-sequencing the PCR products of the participants. Furthermore, unclear chromatograms of DNA sequencing results were excluded from further analysis. Therefore, the proportion of genotyping errors in this study was very low or even absent.
Several studies showed that genetic variants on exon 1 and 3 influenced the hydrolysis capacity of NUDT15 enzyme (Kakuta et al., 2018; Kojima et al., 2018). However, only few articles addressed exon 2 and its role in the enzymes activity. In this study, we sequenced exon 2 to identify the presence of any novel genetic variants. The results showed that exon 2 was not polymorphic in Arab Jordanian participants. Therefore, it seems that its role in inter-individual variation of drug response might be negligible. This finding is in line with previous studies referring that exon 2 on NUDT15 gene was not polymorphic and hence did not affect the variation of drug response (Kakuta et al., 2018).
The most common NUDT15 genetic variant among Jordanian Arab participants was NUDT15 36A>G. This variant is novel among Jordanian population and no study has reported it yet. Given that NUDT15 36A>G is a synonymous variant and does not change the amino acid proline in the polypeptide sequence of NUDT15 protein, it is expected that NUDT15 36A>G does not influence NUDT15 methylation capacity.
NUDT15 IVS1+116C>T intronic variant was identified previously among other ethnic groups (Thorisson et al., 2005). In our study, this variant was found in only one participant and it was in the heterozygote form with a low frequency (0.6%). Having compared this allelic frequency between Jordanians and other ethnic populations, it was found that it had the lowest frequency among Jordanian population (p value > 0.05) (Table 5). The effect of NUDT15 IVS1+116C>T on the metabolic capacity of NUDT15 has not been investigated yet. This variant is located on the exon 1/intron 1 boundary and might affect the spliceosome formation of NUDT15 mRNA. To investigate whether our intronic variant did affect spliceosomic formation or not, we used in silico BDGP software which showed that NUDT15 IVS1+116C>T did not affect the spliceosome formation of NUDT15 mRNA (Table 4).
Two non-synonymous genetic variants, with low frequency, were found locating on exon 3. According to literature review, the well-known variant NUDT15 415C>T (Rs116855232) clinically affects the purine anti-cancer’s response (Singh et al., 2017). It is found in high frequencies in the Asian population (>6%) such as the Koreans (Kim et al., 2017) , while it has low frequency among Caucasians (0.2%). Our study focused only on Jordanian Arab populations and found that the frequency of NUDT15 415C>T variant was low (0.6%) . This finding makes sense since Arab population is considered to be part of the Caucasians (Table 5). Therefore, there is a significant interethnic variation in the frequency of NUDT15 415C>T and this variant might play more significant role in the inter-individual variation in the metabolism of purine analog drugs among Asians than Caucasians, including Jordanians.
The other genetic variant identified on exon 3 of NUDT15 gene was a non-synonymous NUDT15 404C>T variant. This variant is novel among Jordanians which results in substation of phenylalanine at amino acid position 134 to leucine. Therefore, changing the aromatic side chain at phenylalanine to aliphatic linear chain at leucine, and hence may affect the structural stability and function of NUDT15 protein. After applying in silico tools, it was predicted that NUDT15 404C>T variant was harmful and affected the stability of NUDT15 protein. As a result, it is expected that people with NUDT15 404C>T allele have lower capacity to metabolize the purine analog drugs. However, the frequency of this genetic variant was found to be low among the Jordanian participants (0.6%). Further clinical studies are needed to identify the effect of NUDT15 404C>T genetic variant on the metabolism of purine drugs their response .
Two other genetic variants, NUDT15 502G>A and 588T>C, were identified on the 3’UR among the Jordanian participants in this study. These variants are novel and their roles on NUDT15 function have not been investigated yet. Previous studies reported that genetic variants on 3’UR influenced t he regulation of gene expression (Steri et al., 2018). Therefore, the NUDT15 592G>A and 588T>C might affect the regulation of NUDT15 transcription.
In this study, we compared the NUDT15 allele frequency between Jordanians and other major ethnic groups Caucasians, Africans, Indians and Asians. We found that there was an inter-ethnic variation in the frequency of NUDT15 alleles. The NUDT15 gene is more polymorphic among Asians than Caucasians, including Middle Eastern Arabs. This is in line with previous studies revealing that NUDT15 genetic variants played a major role in the variation of purine drug metabolism among Asians, while it had a minor role in explaining the inter-individual variation in drug metabolism among Caucasians (Moriyama et al., 2016). On the other hand, TPMT gene showed more clinical impact on the response of anticancer purine analog among Caucasians than Asians (Chouchana et al., 2012; Moriyama et al., 2016).
Arabs are considered as Caucasians and many studies showed that Middle Eastern Arabs had genetic variant frequencies which were closer to European Caucasians than other major ethnic groups (Jarrar et al., 2018). However, this study found that NUDT15 IVS1+116C>T allele frequency was significantly (p value < 0.05) lower than Europeans, indicating that Caucasian Arabs are not similar to Caucasian Europeans in the pharmacogenetics of the drugs.
In conclusion, we genotyped NUDT15 gene among healthy unrelated Arab Jordanian participants and found that the frequency of NUDT15 genetic variants was low and it was different from those reported among other ethnic populations. In addition, we identified 4 novel genetic variants and predicted, by in silico tools, that novel NUDT15 404C>A variant was harmful to NUDT15 protein and may affect the drug response. The findings of this study can help explain the inter-individual variation in purine analogue drug metabolism, and hence its response among Jordanians.
Conflict of Study
The authors declare no conflicts of interest.
Acknowledgments
This study was funded by Deanship of the Scientific Research at AlZaytoonah University of Jordan with fund number 13-10-2016.
References
- 1.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao H, Wang Y, Zhang W, et al. A short-read multiplex sequencing method for reliable, cost-effective and high-throughput genotyping in large-scale studies. Hum Mutat. 2013;34:1715–20. doi: 10.1002/humu.22439. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, Lian F, Yuan S, et al. Association of thiopurine methyltransferase status with azathioprine side effects in Chinese patients with systemic lupus erythematosus. Clin Rheumatol. 2014;33:499–503. doi: 10.1007/s10067-013-2441-x. [DOI] [PubMed] [Google Scholar]
- 5.Chouchana L, Narjoz C, Beaune P, et al. Review article: the benefits of pharmacogenetics for improving thiopurine therapy in inflammatory bowel disease. Aliment Pharmacol Ther. 2012;35:15–36. doi: 10.1111/j.1365-2036.2011.04905.x. [DOI] [PubMed] [Google Scholar]
- 6.Esteves ARF, Domingues AF, Ferreira IL, et al. Mitochondrial function in Parkinson's disease cybrids containing an nt2 neuron-like nuclear background. Mitochondrion. 2008;8:219–28. doi: 10.1016/j.mito.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Fei X, Shu Q, Zhu H, et al. NUDT15 R139C variants increase the risk of azathioprine-induced leukopenia in Chinese autoimmune patients. Front Pharmacol. 2018;9:460. doi: 10.3389/fphar.2018.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goel RM, Blaker P, Mentzer A, et al. Optimizing the use of thiopurines in inflammatory bowel disease. Ther Adv Chronic Dis. 2015;6:138–46. doi: 10.1177/2040622315579063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarrar YB, Balasmeh AA, Jarrar W. Sequence analysis of the N-acetyltransferase 2 gene (NAT2) among Jordanian volunteers. Libyan J Med. 2018;13:1408381. doi: 10.1080/19932820.2017.1408381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarrar YB, Ismail S, Irshaid YM. N-Acetyltransferase-2 (NAT2) genotype frequency among Jordanian volunteers. Int J Clin Pharmacol Ther. 2010;48:688–94. doi: 10.5414/cpp48688. [DOI] [PubMed] [Google Scholar]
- 11.Kakuta Y, Kawai Y, Okamoto D, et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: a multicenter study. J Gastroenterol. 2018;53:1065–78. doi: 10.1007/s00535-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HT, Choi R, Won HH, et al. NUDT15 genotype distributions in the Korean population. Pharmacogenet Genomics. 2017;27:197–200. doi: 10.1097/FPC.0000000000000274. [DOI] [PubMed] [Google Scholar]
- 13.Kojima Y, Hirotsu Y, Omata W, et al. Influence of NUDT15 variants on hematological pictures of patients with inflammatory bowel disease treated with thiopurines. World J Gastroenterol. 2018;24:511–8. doi: 10.3748/wjg.v24.i4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 15.LaMarche BL, Crowell KL, Jaitly N, et al. MultiAlign: a multiple LC-MS analysis tool for targeted omics analysis. BMC Bioinformatics. 2013;14:49. doi: 10.1186/1471-2105-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YJ, Hwang EH, Park JH, et al. NUDT15 variant is the most common variant associated with thiopurine-induced early leukopenia and alopecia in Korean pediatric patients with Crohn's disease. Eur J Gastroenterol Hepatol. 2016;28:475–8. doi: 10.1097/MEG.0000000000000564. [DOI] [PubMed] [Google Scholar]
- 17.Liang JJ, Geske JR, Boilson BA, et al. TPMT genetic variants are associated with increased rejection with azathioprine use in heart transplantation. Pharmacogenet Genomics. 2013;23:658–65. doi: 10.1097/FPC.0000000000000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriyama T, Nishii R, Lin TN, et al. The effects of inherited NUDT15 polymorphisms on thiopurine active metabolites in Japanese children with acute lymphoblastic leukemia. Pharmacogenet Genomics. 2017a;27:236–9. doi: 10.1097/FPC.0000000000000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriyama T, Nishii R, Perez-Andreu V, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48:367–73. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriyama T, Yang YL, Nishii R, et al. Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry. Blood. 2017b;130:1209–12. doi: 10.1182/blood-2017-05-782383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson ND, Morrow JB. DNA extract characterization process for microbial detection methods development and validation. BMC Res Notes. 2012;5:668. doi: 10.1186/1756-0500-5-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese MG, Eeckman FH, Kulp D, et al. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 23.Shah SA, Paradkar M, Desai D, et al. Nucleoside diphosphate-linked moiety X-type motif 15 C415T variant as a predictor for thiopurine-induced toxicity in Indian patients. J Gastroenterol Hepatol. 2017;32:620–4. doi: 10.1111/jgh.13494. [DOI] [PubMed] [Google Scholar]
- 24.Shah SA, Paradkar MU, Desai DC, et al. Preemptive NUDT15 genotyping: redefining the management of patients with thiopurine-induced toxicity. Drug Metab Pers Ther. 2018;33:57–60. doi: 10.1515/dmpt-2017-0038. [DOI] [PubMed] [Google Scholar]
- 25.Singh M, Bhatia P, Khera S, et al. Emerging role of NUDT15 polymorphisms in 6-mercaptopurine metabolism and dose related toxicity in acute lymphoblastic leukaemia. Leuk Res. 2017;62:17–22. doi: 10.1016/j.leukres.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Steri M, Idda ML, Whalen MB, et al. Genetic variants in mRNA untranslated regions. Wiley Interdiscip Rev RNA. 2018;9:e1474. doi: 10.1002/wrna.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Kato M, Hasegawa D, et al. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol. 2015;171:109–15. doi: 10.1111/bjh.13518. [DOI] [PubMed] [Google Scholar]
- 28.Thorisson GA, Smith AV, Krishnan L, et al. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinshilboum R. Thiopurine pharmacogenetics: clinical and molecular studies of thiopurine methyltransferase. Drug Metab Dispos. 2001;29:601–5. [PubMed] [Google Scholar]
- 30.World Medical A. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–4. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 31.Yang JJ, Landier W, Yang W, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33:1235–42. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zgheib NK, Akika R, Mahfouz R, et al. NUDT15 and TPMT genetic polymorphisms are related to 6-mercaptopurine intolerance in children treated for acute lymphoblastic leukemia at the Children's Cancer Center of Lebanon. Pediatr Blood Cancer. 2017;64:146–50. doi: 10.1002/pbc.26189. [DOI] [PubMed] [Google Scholar]


