Abstract
An efficient genetic transformation system and suitable promoters are essential prerequisites for gene expression studies and genetic engineering in streptomycetes. In this study, firstly, a genetic transformation system based on intergeneric conjugation was developed in Streptomyces rimosus M527, a bacterial strain which exhibits strong antagonistic activity against a broad range of plant-pathogenic fungi. Some experimental parameters involved in this procedure were optimized, including the conjugative media, ratio of donor to recipient, heat shock temperature, and incubation time of mixed culture. Under the optimal conditions, a maximal conjugation frequency of 3.05×10−5 per recipient was obtained. Subsequently, based on the above developed and optimized transformation system, the synthetic promoters SPL-21 and SPL-57, a native promoter potrB, and a constitutive promoter permE * commonly used for gene expression in streptomycetes were selected and their activity was analyzed using gusA as a reporter gene in S. rimosus M527. Among the four tested promoters, SPL-21 exhibited the strongest expression activity and gave rise to a 2.2-fold increase in β-glucuronidase (GUS) activity compared with the control promoter permE *. Promoter SPL-57 showed activity comparable to that of permE *. Promoter potrB, which showed the lowest activity, showed a 50% decrease in GUS activity compared with the control permE *. The transformation system developed in this study and the tested promotors provide a basis for the further modification of S. rimosus M527.
Keywords: Streptomyces rimosus M527, Intergeneric conjugation, Promoter, β-Glucuronidase (GUS)
1. Introduction
Streptomycetes are soil-dwelling Gram-positive filamentous bacteria well known for their ability to produce several bioactive secondary metabolites, including therapeutic molecules such as antibiotic (Schlatter and Kinkel, 2015), antiparasitic (Cao et al., 2018), anticancer (Noomnual et al., 2016), and immunosuppressive agents (Yoo et al., 2017). Therefore, Streptomyces species have important commercial value in agriculture and medicine (Rey and Dumas, 2017; Kemung et al., 2018). Although Streptomyces attract a great deal of attention in genetic engineering and synthetic biology due to their outstanding ability to produce various secondary metabolites, the limited production and long fermentation period remain a bottleneck for their further application.
A strain of antagonistic microorganism named Streptomyces rimosus M527 (China Center for Type Culture Collection (CCTCC) M2013270) has been isolated from the plant rhizosphere of wetlands. This strain has been shown to produce rimocidin, a 28-membered tetraene macrolide, comprising a large lactone ring with a sugar moiety. Rimocidin exhibits strong antifungal activity and is recognized as a promising fungicide that can be used to control plant-pathogenic fungi (Jeon et al., 2016). However, systems for genetic manipulation of S. rimosus M527, such as transformation of plasmids or gene over-expression, have not been developed. This situation hampers the genetic improvement of S. rimosus M527 using molecular biotechnological approaches. The development of an efficient genetic transformation method and expression system for this special strain is essential for enhancing the production of rimocidin.
Intergeneric conjugation between Escherichia coli and Streptomyces, has been proven to be a reliable and efficient transformation method (Du et al., 2012) and has been widely used as a means of transferring plasmids into Streptomyces strains (Zhou et al., 2012). However, there is still no universal protocol applicable to all Streptomyces strains.
Promoter elements are of indisputable importance in gene expression systems because they are responsible for efficient transcription, which is the first stage of gene expression (Myronovskyi and Luzhetskyy, 2016). In Streptomyces, only a small number of native promoters have been described (Luo et al., 2015; Li et al., 2018). Promoter engineering, which is a strategy for the design and construction of synthetic promoters (Seghezzi et al., 2011; Siegl et al., 2013; Sohoni et al., 2014; Ji et al., 2018) and engineered native promoters (Wang et al., 2013; Kakule et al., 2015; Yi et al., 2017), has been successfully employed to modulate and optimize gene expression in Streptomyces. However, the transcriptional activity of a promoter can change in different Streptomyces hosts.
In this study, firstly, a reliable intergeneric conjugation system for S. rimosus M527 was developed by optimization of experimental parameters including the conjugative medium, heat-shock temperature, ratio of donor to recipient cell number, and incubation time of the conjugation plates. Subsequently, based on the developed transformation system, plasmids harboring different native promoters (permE * and potrB) and synthetic promoters (SPL-21 and SPL-57) were separately transferred into S. rimosus M527, and their activity was determined using gusA encoding β-glucuronidase (GUS) as a reporter gene.
2. Materials and methods
2.1. Strains, plasmids, and primers
The strains, plasmids, and primers used in this study are described and listed in Table 1. The rimocidin producer S. rimosus M527 has been deposited at the CCTCC (M2013270), Wuhan, China. E. coli JM109 was used as a general host for gene cloning and expression. E. coli ET12567 harboring plasmid pUZ8002 was used as the donor for intergeneric conjugation.
Table 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, primer | Description | Source or reference |
| Strain | ||
| E. coli JM109 | General cloning host | Our lab |
| E. coli ET12567/pUZ8002 | Cm r, Km r, donor strain for conjugation | Our lab |
| S. rimosus M527 | Parental strain, rimocidin producer | CCTCC M2013270 |
| M527-21S | M527 with integrative vector pGUS-SPL-21 | This work |
| M527-57S | M527 with integrative vector pGUS-SPL-57 | This work |
| M527-ES | M527 with integrative vector pGUS-ermE* | This work |
| M527-BS | M527 with integrative vector potrB-GUS | This work |
| Plasmid | ||
| pGUS-SPL-21 | gusA under the control of synthetic promoter SPL-21 | Siegl et al., 2013 |
| pGUS-SPL-57 | gusA under the control of synthetic promoter SPL-57 | Siegl et al., 2013 |
| pGUS-ermE* | gusA under the control of promoter permE * | Siegl et al., 2013 |
| potrB-GUS | gusA under the control of promoter potrB | This work |
| Primer | ||
| P1 | 5'-ACGTCTAGAAGGCGGCCGTTGACCGCGAA-3' (XbaI) | This work |
| P2 | 5'-ACGGGTACCGGCCGTCAGGATCTGCCGAT-3' (KpnI) | This work |
Primers P1 and P2 were used for amplification of otrB promoter. XbaI and KpnI restriction enzyme sites are underlined
2.2. Media and culture conditions
E. coli strains were cultured in liquid or on solid Luria-Bertani (LB) medium (Wang et al., 2018) containing appropriate antibiotics at 37 °C. Apramycin (50 µg/mL), chloramphenicol (25 µg/mL), ampicillin (100 µg/mL), kanamycin (50 µg/mL), and nalidixic acid (50 µg/mL) were added as needed. S. rimosus M527 and its derivates were incubated according to the method described by Zhao et al. (2019). S. rimosus M527 was incubated at 28 °C and grown in solid mannitol soya flour (MS) medium (soya flour 20 g/L, mannitol 20 g/L, agar 20 g/L, tap water) for sporulation. MS, tryptic soy broth (TSB), and 2CMC solid media were used for conjugation. TSB medium contains tryptone 17 g/L, soytone 3 g/L, D-glucose 2.5 g/L, NaCl 5 g/L, and K2HPO4 2.5 g/L (adjusted to pH 7.5 by NaOH). 2CMC solid medium, which is optimized based on 2CM medium (Wang, 2007), contains starch 10 g/L, tryptone 2 g/L, NaCl 1 g/L, (NH4)2SO4 2 g/L, MgSO4·7H2O 2 g/L, CaCO3 2 g/L, casamino acids 2 g/L, K2HPO4·3H2O 1 g/L, FeSO4· 7H2O 1 g/L, MgCl2·6H2O 1 g/L, ZnSO4·7H2O 1 g/L, and agar 20 g/L (adjusted to pH 7.2 by NaOH).
S. rimosus M527 spores (1×106 mL−1) were inoculated into a 250-mL Erlenmeyer flask containing 50 mL seed medium, and shaken at 28 °C and 180 r/min. The CP liquid medium used as seed medium had the same composition as that described in an earlier study (Zhao et al., 2018). Five percent of the seed culture was inoculated into 50 mL fermentation medium containing 1 g soya flour, 1 g D-mannitol, and 0.03 g K2HPO4·3H2O.
2.3. Construction of plasmids
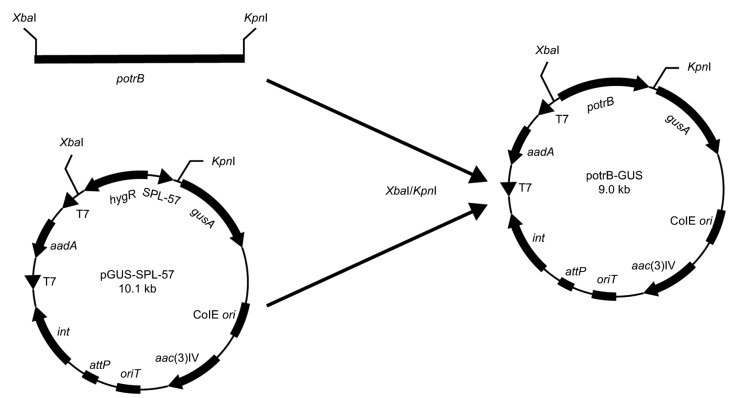
Vectors pGUS-SPL-21, pGUS-SPL-57 and pGUS-ermE* were gifts from Prof. LUZHETSKYY (Siegl et al., 2013). The 300-bp otrB promoter fragment (potrB) (nucleotides −300 to −1 upstream from gene otrB translational start codon) was amplified from genomic DNA of S. rimosus M527 using primers P1 and P2 (Table 1). The PCR amplification procedure was performed as described by Xu et al. (2017). The synthetic promoter SPL-57 in vector pGUS-SPL-57 was removed by digesting pGUS-SPL-57 with XbaI/KpnI, and was replaced by XbaI/KpnI potrB to create the plasmid potrB-GUS. Sequencing of the inserted fragment containing potrB confirmed that the gene contained no mutations.
2.4. Intergeneric conjugation procedure and analysis of exconjugants
Intergeneric conjugation between E. coli and Streptomyces was conducted as described by Phornphisutthimas et al. (2010) and Ma et al. (2014) with minor modifications. S. rimosus M527 spores (1×108 mL−1) were collected from solid medium, filtered by sterile absorbent cotton to remove mycelia and then incubated at different test temperatures for 10 min to induce germination. A culture of the spores was cultured in 2×yeast extract tryptone (YT) broth (Ma et al., 2014) at 28 °C for 2 h. Then the spores, serving as recipients, were washed twice with an equal volume of LB broth and resuspended in 500 μL fresh LB medium. The donor strain, E. coli ET12567/pUZ8002 harboring the constructed plasmids, was grown in LB with appropriate antibiotics until an optical density at 600 nm (OD600) of 0.4–0.6 was reached. The cells were washed twice with LB and resuspended in a final volume of 500 µL of LB. E. coli donor cells and the recipients were mixed and spread on 2CMC, MS, and TSB agar plates containing 10 mmol/L MgCl2. The plates were incubated at 28 °C for 10–20 h and overlaid with 500 µL fresh LB medium containing 100 µg/mL nalidixic acid and 300 µg/mL apramycin. The plates were further incubated at 28 °C for 3–4 d, and the exconjugants were then counted. For conjugation with mycelia, S. rimosus M527 was incubated in CP liquid medium for 48 h at 28 °C. Mycelia were collected and mixed with donor cells, and spread on the agar plates.
All the putative exconjugants were purified on solid 2CMC plates supplemented with 300 μg/mL apramycin to confirm the integration of the target plasmids. After transferring into S. rimosus M527, a constructed plasmid can be maintained only in a chromosomally integrated state. Total exconjugant DNA was isolated according to the methods described by Kieser et al. (2000). The existence of the apramycin resistance (apr) gene from the constructed plasmids in the chromosome of exconjugants was verified by PCR amplification. The average value of the conjugation frequency of three independent experiments was calculated. Each conjugation experiment was carried out three times.
2.5. GUS assay
GUS activity was measured according to a method described previously (Myronovskyi et al., 2011; Siegl et al., 2013; Xu et al., 2017). Spores of recombinant strains were inoculated into 100 mL of TSB liquid medium and grown in shaking flasks for 48 h at 28 °C. A 40-mL sample was collected into an Eppendorf (EP) tube and dried at 60 °C. Another 40-mL sample was used for the GUS assay. The culture was centrifuged, and the deposited cells were collected and resuspended in 20 mL of GUS buffer 2 (50 mmol/L phosphate-buffered saline (PBS; pH 7.0), 5 mmol/L dithiothreitol (DTT), 0.1% Triton X-100, 1 mg/mL lysozyme). After incubating at 37 °C for 30 min, the lysates were subsequently diluted with GUS buffer 1 (50 mmol/L PBS (pH 7.0), 5 mmol/L DTT, 0.1% Triton X-100) to 45 mL and centrifuged at 11 000 r/min at 4 °C for 10 min. A 500-μL sample of the supernatant was mixed with 500 μL of GUS buffer 3 (50 mmol/L PBS (pH 7.0), 5 mmol/L DTT, 0.1% Triton X-100, supplemented with 2 mmol/L p-nitrophenyl-β-D-glucuronide), and the absorption was spectrophotometrically measured at 415 nm for 30 min (UV-1800, Shimadzu, Japan). As a control, 500 μL of GUS buffer 1 was mixed with 500 μL of the sample.
Specific enzyme activity is indicated as unit per gram of protein. The following equation was used to calculate the enzymatic activity (EA): EA=2V×ΔA 415/14×DCW, where V equals the total end volume of the sample (45 mL), ΔA 415 equals the absorbance which was spectrophotometrically measured at 415 nm per minute, and DCW equals the dry weight in grams of the original sample size (40 mL). All experiments were performed in triplicate, and the reported values are the average from three assays with their calculated standard deviations (SDs).
2.6. Statistical analysis
All experiments were carried out at least three times, and the results were expressed as mean±SD. Statistical analysis was performed using Student’s t-test (Zhou et al., 2017).
3. Results
3.1. Development and optimization of intergeneric conjugation transformation for S. rimosus M527
It has been reported that both mycelia and spores of streptomycetes can be used as recipients in intergeneric conjugation (Phornphisutthimas et al., 2010). To determine the appropriate status of the recipient for transformation efficiency, mycelia and spores of S. rimosus M527 were assessed. When mycelia were used as the recipient in conjugation, almost no exconjugants appeared when using MS or TSB as the conjugation medium. Exconjugants were obtained only when 2CMC medium was used, and the conjugation efficiency was about 1×10−8–1×10−7. After using spores as the recipient, exconjugants were generated at a frequency of 1×10−7–1×10−5, an almost 100-fold improvement compared with using mycelia as the recipient. Our results confirmed that both spores and mycelia of S. rimosus M527 could be used as recipient, but the conjugation efficiency achieved with spores was much higher than that obtained with mycelia.
The selection of an appropriate conjugation medium has an obvious influence on the conjugation efficiency in streptomycetes (Du et al., 2012; Sun et al., 2014), so three applicable solid media, MS, TSB, and 2CMC, were tested to determine the appropriate medium. The 2CMC medium proved to be optimal for the intergeneric conjugation of E. coli-S. rimosus M527, whether mycelia or spores were chosen as recipient (Table 2). Therefore, 2CMC was used as the conjugation medium in follow-up experiments.
Table 2.
Effects of conjugation media and the number of recipient spores on the efficiency of intergeneric conjugation
| Conjugation medium | Number of recipient spores (mL−1)b | Number of exconjugants | Conjugation frequencyc |
| MS | 1×109 | 330 | 3.30×10−7 |
| 1×108 | 168 | 1.68×10−6 | |
| 1×107 | 64 | 6.40×10−6 | |
| 1×106 | 5 | 5.00×10−6 | |
| TSB | 1×109 | 264 | 2.64×10−7 |
| 1×108 | 106 | 1.06×10−6 | |
| 1×107 | 28 | 2.80×10−6 | |
| 1×106 | |||
| 2CMCa | 1×109 | 583 | 5.83×10−7 |
| 1×108 | 397 | 3.97×10−6 | |
| 1×107 | 167 | 1.67×10−5 | |
| 1×106 | 13 | 1.30×10−5 |
Different kinds of medium were tested to determine the appropriate medium. 2CMC medium was proven to be optimal for the conjugation of E. coli-S. rimosus M527.
1×108 mL−1 E. coli ET12567 donor cells were used, and a range of S. rimosus M527 spores (1×106–1×109) were tested as recipients.
Values are presented as the number of conjugations per colony forming units of the recipient, and each value represents the average efficiency from three independent experiments
It has been reported that an appropriate ratio between the number of donor cells and recipient spores is an important parameter in the case of Streptomyces species (Enríquez et al., 2006; Ma et al., 2014). In this study, 1×108 mL−1 E. coli ET12567 as donor cells and a range of S. rimosus M527 spores (1×106–1×109 mL−1) as recipients were tested. When the number of the spores reached 1×107 mL−1, the conjugation efficiency reached the highest value of 1.67×10−5 (Table 2), indicating that maximum efficiency was achieved with a donor-to-recipient ratio of 10:1.
Heat-shock treatment of Streptomyces spores has generally been carried out prior to mixing with donor E. coli (Sun et al., 2014; Zhang et al., 2018). To determine the optimal temperature for heat treatment and incubation time of heat-shocked spores, the effects of temperature and different incubation time on conjugation efficiency were investigated. In this study, high conjugation efficiencies were achieved when spores were treated at 45–55 °C for 10 min (Table 3), and the efficiencies rapidly decreased when spores were treated above 55 °C or below 45 °C.
Table 3.
Effects of heat-shock temperature on transformation frequency
| Heat-shock temperature (°C) | Average transformation frequency |
| 35 | 4.89×10−6 |
| 40 | 7.66×10−6 |
| 45 | 1.08×10−5 |
| 50 | 2.23×10−5 |
| 55 | 1.67×10−5 |
| 60 | 8.39×10−6 |
S. rimosus M527 spores (1×107 mL−1) were collected and heat-treated at various temperatures for 10 min to induce germination
In addition, before the plates are flooded with fresh LB medium containing antibiotics, the incubation time of the mixed culture of Streptomyces and E. coli plays an important role in conjugation efficiency (Zhang et al., 2018). If the time of mixed culture is too short, Streptomyces mycelia grow weakly and are easily scraped off, leading to a lower conjugation efficiency. Conversely, the longer the incubation time of mixed culture, the greater the possibility of false positives, which hinders the selection of the positive exconjugants. Our results demonstrated that the optimal incubation time of mixed culture plates was 12–18 h (Table 4).
Table 4.
Effect of different incubation time before the plates were flooded with antibiotics on transformation frequency
| Incubation time (h) | Average transformation frequency |
| 0–6 | 2.74×10−6 |
| 6–12 | 7.07×10−6 |
| 12–18 | 3.05×10−5 |
| 18–24 | 1.16×10−5 |
| 24–30 | 5.13×10−6 |
S. rimosus M527 spores (1×107 mL−1) in mixed culture with the donor cells E. coli ET12567 (1×108 mL−1) were incubated for various lengths of time at 28 °C before the plates were flooded with fresh LB medium containing antibiotics
The optimal conjugation conditions were determined: pre-incubated S. rimosus M527 spores were selected as recipients, the appropriate donor-to-recipient ratio was 10:1, and spores were heat-treated at 50 °C for 10 min prior to mixing with donor E. coli ET12567/pUZ8002. After incubating at 28 °C for 12–18 h, the plates were overlaid with 500 µL fresh LB medium containing nalidixic acid and apramycin (100 and 300 µg/mL, respectively). Incubation was continued at 28 °C until exconjugant cells appeared, and the highest efficiency of conjugation was 3.05×10−5 when 2CMC medium was used as the conjugation medium.
3.2. Comparison of promoter activity in S. rimosus M527 using GUS as reporter
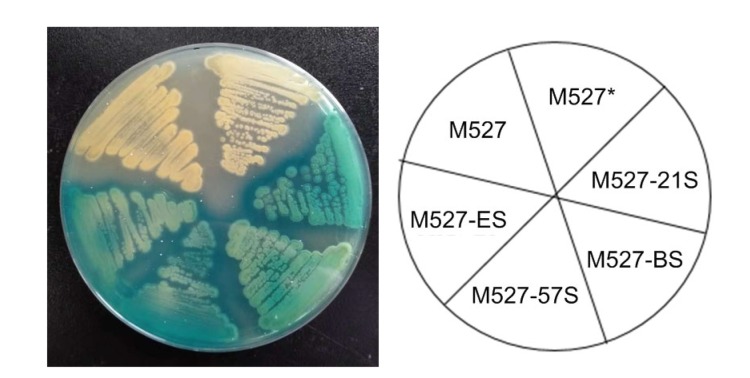
To test the transcriptional activity of four promoters (synthetic promoters SPL-21 and SPL-57, native promoter potrB, and constitutive promoter permE *), gene gusA was used as a reporter gene and was placed under the control of the tested promoters. The construction of plasmid potrB-GUS was performed as follows: the 300-bp otrB promoter fragment was amplified from genomic DNA of S. rimosus M527 and replaced the synthetic promoter SPL-57 in vector pGUS-SPL-57 to create plasmid potrB-GUS (Fig. 1). Plasmids potrB-GUS, pGUS-SPL-21, pGUS-SPL-57, and pGUS-ermE* were separately introduced into E. coli ET12567/pUZ8002 and then transferred into S. rimosus M527 using the optimized intergeneric conjugation conditions described above to generate exconjugants M527-BS, M527-21S, M527-57S, and M527-ES, respectively. GUS activity was visible in all resulting recombinant strains and compared with that of the wild-type strain S. rimosus M527 (Fig. 2). Strains M527-21S and M527-BS exhibited darker and lighter blue color than the other strains, respectively. No significant difference in the intensity of blue color was detectable in the recombinant strain M527-57S or M527-ES.
Fig. 1.
A map of plasmid potrB-GUS constructed in this study
Fig. 2.
Visual observation of GUS activity in solid medium of wild-type strain S. rimosus M527 and different recombinant strains
Colonies of wild-type strain M527, vector control strain M527* (containing the empty plasmid pSET152), and recombinant strains (M527-ES, M527-BS, M527-21S, M527-57S) containing different promoters were covered with X-Gluc (1 mmol/L) on 2CMC medium and incubated at 28 °C for 3–4 d. Blue halos are 5,5'-dibromo-4,4'-dichloro-indigo, formed by β-glucuronidase (GUS) activity
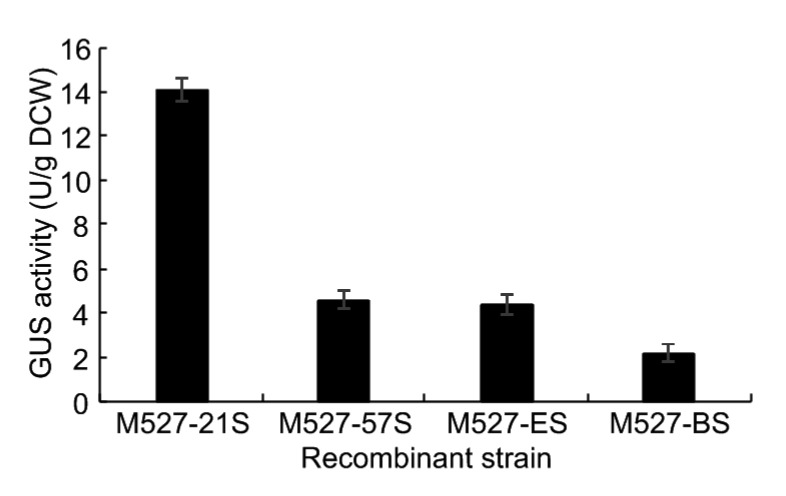
The GUS enzymatic assay, which precisely reflected the promoter strength, was performed to determine the expression of these four promoters in detail. In consideration of variation among different exconjugants, three independent exconjugants were selected for screening in the GUS assay. Promoter SPL-21 exhibited the strongest expression activity among the four tested promoters (a 2.2-fold increase in GUS activity compared with permE *) and promoter potrB the weakest (a 50% decrease in GUS activity compared with permE *). Promoter SPL-57 showed the activity comparable to that of permE * (Fig. 3). The data were in accordance with aforementioned intuitive observations.
Fig. 3.
Detection of GUS activity in different recombinant strains
β-Glucuronidase (GUS) was used as a reporter gene for examining protein synthesis. GUS activity was measured in cell lysates of strains M527-21S, M527-57S, M527-ES, and M527-BS from fermentation broth. Data are expressed as mean±SD
4. Discussion
There are three main genetic transformation methods for Streptomyces: polyethylene glycol (PEG)-mediated protoplast transformation, electroporation, and intergeneric conjugation. To genetically engineer S. rimosus M527, the most suitable transformation system needs to be identified. In this study, the three transformation methods were performed to introduce plasmids into S. rimosus M527. The results showed that the transformation efficiency obtained from intergeneric conjugation was significantly higher than those from the other two methods (data not shown).
Intergeneric conjugation between E. coli cells and Streptomyces spores was initially reported by Mazodier et al. (1989). The conjugation takes advantage of the donor E. coli ET12567 host with a transfer function (tra gene in pUZ8002), which allows for mobilization of a plasmid harboring an origin of transfer (oriT). Conjugation is advantageous over other Streptomyces transformation protocols for two reasons: (1) it can readily transform large plasmids with an oriT into a Streptomyces recipient; and (2) many oriT-harboring plasmids allow direct integration into a chromosomal specific attachment site (attB) (e.g., via int-attP on pSET152).
Intergeneric conjugation allows the transfer of plasmids manipulated in E. coli into Streptomyces spores. Thus, higher conjugation efficiencies have been reported in several Streptomyces when spores rather than mycelia were used. Mycelia are the vegetative and propagative forms of actinobacteria. Recently, mycelia have shown potency in intergeneric conjugation. Some research results concerning intergeneric conjugation using mycelia as recipient have been reported for several Streptomyces (Du et al., 2012; Rocha et al., 2018; Zhang et al., 2018). The conjugation conditions were highly strain-specific and usually ineffective for different actinobacteria. Due to interspecific variation, appropriate conditions for intergeneric conjugation have to be developed and optimized for each particular strain (Sun et al., 2014; Wang and Jin, 2014; Zhang et al., 2018).
In this study, both the mycelia and spores of S. rimosus M527 were chosen as recipients for establishment of intergeneric conjugation. The results showed that conjugation efficiencies achieved with spores were higher than those achieved with mycelia. From the point of morphology, spores are more suitable for the introduction or acceptance of plasmid DNA than mycelia. In particular, plasmid DNA can be effectively transferred from the donor to the recipient through the germ tube when the spores are in a germination state after heat-shock treatment, leading to high conjugation efficiency.
MS medium is usually used for sporulation and conjugation. Unfortunately, using the mycelia as the recipient for conjugation failed despite considerable effort. On TSB medium S. rimosus M527 grows well but does not produce spores. This hampers subsequent conjugation. Whether spores or mycelia were used as the recipient, the conjugation efficiency was lower on TSB medium than on the other two media. In this study, 2CMC medium was shown to be optimal for the intergeneric conjugation of E. coli-S. rimosus M527, whether mycelia or spores were chosen as recipient. The components of 2CMC medium may be more effective for conjugation. Some other experimental parameters involved in conjugation were further optimized. Under optimal conditions, a maximal conjugation frequency of 3.05×10−5 per recipient was obtained.
The conjugation transfer system developed in this study provides a basis for the introduction of genes into S. rimosus M527. However, the promoter is a crucial element for gene expression. Selection of a suitable promoter for gene expression is also important for further improving rimocidin production in S. rimosus M527 using genetic engineering technology. Promoter permE * is widely used for heterologous expression of genes in streptomycetes. Ji et al. (2018) reported that the native promoter potrB was stronger than permE * in S. rimosus. Also, two artificial synthetic promoters, SPL-21 and SPL-57, were shown to be much stronger than permE * in different Streptomyces. In Streptomyces diastatochromogenes 1628, SPL-57 showed activity comparable to that of permE *. In contrast, promoter SPL-21 showed a 5.2-fold increase in GUS activity compared with permE * (Xu et al., 2017). One promoter may show different transcriptional activity due to differences in the genetic background of the host strain. In this study, two synthetic promoters, SPL-21 and SPL-57, a native promoter potrB, and a constitutive promoter permE * were compared and analyzed for GUS activity. Similar results were obtained from S. diastatochromogenes 1628, in which the promoter SPL-21 showed the strongest activity among the tested promoters, and SPL-57 showed activity comparable to that of permE *. Surprisingly, the promoter potrB showed the lowest activity compared with that of promoter permE *, which is not consistent with the results reported by Ji et al. (2018). This inconsistency may have been caused by the different backgrounds or regulatory mechanisms of the host strains.
Note that the constructed plasmids used for detection of GUS activity of promoters were derived from the pSET152 backbone. The effects of the different plasmids on GUS activity driven by the tested promoters would have been excluded. In addition, the whole genome was sequenced. The results of scanning the genome sequence revealed that S. rimosus M527 contained a single copy of pSET152 integrated at a unique chromosomal attachment site (attB). The copy number of plasmid integration had no effect on the gusA expression driven by the tested promoters.
5. Conclusions
In conclusion, in this study a genetic transfer system based on intergeneric conjugation was developed and optimized for S. rimosus M527. The activity of four promoters using gusA as reporter gene was determined and analyzed. These four promoters exhibited different expression activity and may be classified as strong, medium, and weak, which will facilitate fine-tuning of the expression of different genes in S. rimosus M527. This work will provide a basis for further modification of S. rimosus M527.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31772213 and 31972320) and the Excellent Youth Fund of Zhejiang Province, China (No. LR17C140002)
Contributors: Zhang-qing SONG, Zhi-jun LIAO, and Ye-feng HU participated in the design. Zheng MA and Zhang-qing SONG wrote this article. Andreas BECHTHOLD revised the manuscript. Zheng MA and Xiao-ping YU checked the final version. All authors approved the final manuscript and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Zhang-qing SONG, Zhi-jun LIAO, Ye-feng HU, Zheng MA, Andreas BECHTHOLD, and Xiao-ping YU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Cao XM, Luo ZS, Zeng WZ, et al. Enhanced avermectin production by Streptomyces avermitilis ATCC 31267 using high-throughput screening aided by fluorescence-activated cell sorting. Appl Microbiol Biotechnol. 2018;102(2):703–712. doi: 10.1007/s00253-017-8658-x. [DOI] [PubMed] [Google Scholar]
- 2.Du L, Liu RH, Ying L, et al. An efficient intergeneric conjugation of DNA from Escherichia coli to mycelia of the lincomycin-producer Streptomyces lincolnensis . Int J Mol Sci. 2012;13(4):4797–4806. doi: 10.3390/ijms13044797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enríquez LL, Mendes MV, Antón N, et al. An efficient gene transfer system for the pimaricin producer Streptomyces natalensis . FEMS Microbiol Lett. 2006;257(2):312–318. doi: 10.1111/j.1574-6968.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 4.Jeon BJ, Kim JD, Han JW, et al. Antifungal activity of rimocidin and a new rimocidin derivative BU16 produced by Streptomyces mauvecolor BU16 and their effects on pepper anthracnose. J Appl Microbiol. 2016;120(5):1219–1228. doi: 10.1111/jam.13071. [DOI] [PubMed] [Google Scholar]
- 5.Ji CH, Kim JP, Kang HS. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth Biol. 2018;7(8):1946–1955. doi: 10.1021/acssynbio.8b00175. [DOI] [PubMed] [Google Scholar]
- 6.Kakule TB, Jadulco RC, Koch M, et al. Native promoter strategy for high-yielding synthesis and engineering of fungal secondary metabolites. ACS Synth Biol. 2015;4(5):625–633. doi: 10.1021/sb500296p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemung HM, Tan LTH, Khan TM, et al. Streptomyces as a prominent resource of future anti-MRSA drugs. Front Microbiol, 9:2221. 2018 doi: 10.3389/fmicb.2018.02221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieser T, Bibb MJ, Buttner MJ, et al. Practical Streptomyces Genetics. John Innes Foundation, Norwich, UK; 2000. [Google Scholar]
- 9.Li SS, Wang JY, Xiang WS, et al. An autoregulated fine-tuning strategy for titer improvement of secondary metabolites using native promoters in Streptomyces . ACS Synth Biol. 2018;7(2):522–530. doi: 10.1021/acssynbio.7b00318. [DOI] [PubMed] [Google Scholar]
- 10.Luo YZ, Zhang L, Barton KW, et al. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus . ACS Synth Biol. 2015;4(9):1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Liu JX, Bechthold A, et al. Development of intergeneric conjugal gene transfer system in Streptomyces diastatochromogenes 1628 and its application for improvement of toyocamycin production. Curr Microbiol. 2014;68(2):180–185. doi: 10.1007/s00284-013-0461-z. [DOI] [PubMed] [Google Scholar]
- 12.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myronovskyi M, Luzhetskyy A. Native and engineered promoters in natural product discovery. Nat Prod Rep. 2016;33(8):1006–1019. doi: 10.1039/c6np00002a. [DOI] [PubMed] [Google Scholar]
- 14.Myronovskyi M, Welle E, Fedorenko V, et al. Β-Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol. 2011;77(15):5370–5383. doi: 10.1128/AEM.00434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noomnual S, Thasana N, Sungkeeree P, et al. Streptanoate, a new anticancer butanoate from Streptomyces sp. DC3. J Antibiot (Tokyo) 2016;69(2):124–127. doi: 10.1038/ja.2015.95. [DOI] [PubMed] [Google Scholar]
- 16.Phornphisutthimas S, Sudtachat N, Bunyoo C, et al. Development of an intergeneric conjugal transfer system for rimocidin-producing Streptomyces rimosus . Lett Appl Microbiol. 2010;50(5):530–536. doi: 10.1111/j.1472-765X.2010.02835.x. [DOI] [PubMed] [Google Scholar]
- 17.Rey T, Dumas B. Plenty is no plague: Streptomyces symbiosis with crops. Trends Plant Sci. 2017;22(1):30–37. doi: 10.1016/j.tplants.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Rocha D, Ruiz-Villafán B, Manzo M, et al. Development of an efficient conjugal DNA transfer system between Escherichia coli and a non-sporulating Streptomyces strain. J Microbiol Methods. 2018;144:60–66. doi: 10.1016/j.mimet.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Schlatter DC, Kinkel LL. Do tradeoffs structure antibiotic inhibition, resistance, and resource use among soil-borne Streptomyces? BMC Evol Biol, 15:186. 2015 doi: 10.1186/s12862-015-0470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seghezzi N, Amar P, Koebmann B, et al. The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Appl Microbiol Biotechnol. 2011;90(2):615–623. doi: 10.1007/s00253-010-3018-0. [DOI] [PubMed] [Google Scholar]
- 21.Siegl T, Tokovenko B, Myronovskyi M, et al. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng. 2013;19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Sohoni SV, Fazio A, Workman CT, et al. Synthetic promoter library for modulation of actinorhodin production in Streptomyces coelicolor A3(2) PLoS ONE. 2014;9(6):e99701. doi: 10.1371/journal.pone.0099701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun FH, Luo D, Shu D, et al. Development of an intergeneric conjugal transfer system for xinaomycins-producing Streptomyces noursei xinao-4. Int J Mol Sci. 2014;15(7):12217–12230. doi: 10.3390/ijms150712217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HX. Studies on strain breeding and fermentation process of flavomycin. MS T. Heilongjiang Bayi Agricultural University, Daqing, China; 2007. (in Chinese) [Google Scholar]
- 25.Wang WD, Zhang NN, Chanda W, et al. Antibacterial and anti-biofilm activity of the lipid extract from Mantidis ootheca on Pseudomonas aeruginosa . J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(5):364–371. doi: 10.1631/jzus.B1700356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang WS, Li X, Wang J, et al. An engineered strong promoter for streptomycetes. Appl Environ Microbiol. 2013;79(14):4484–4492. doi: 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang XK, Jin JL. Crucial factor for increasing the conjugation frequency in Streptomyces netropsis SD-07 and other strains. FEMS Microbiol Lett. 2014;357(1):99–103. doi: 10.1111/1574-6968.12507. [DOI] [PubMed] [Google Scholar]
- 28.Xu XH, Wang J, Bechthold A, et al. Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol. 2017;33(2):30. doi: 10.1007/s11274-016-2194-1. [DOI] [PubMed] [Google Scholar]
- 29.Yi JS, Kim MW, Kim M, et al. A novel approach for gene expression optimization through native promoter and 5' UTR combinations based on RNA-seq, Ribo-seq, and TSS-seq of Streptomyces coelicolor . ACS Synth Biol. 2017;6(3):555–565. doi: 10.1021/acssynbio.6b00263. [DOI] [PubMed] [Google Scholar]
- 30.Yoo YJ, Kim H, Park SR, et al. An overview of rapamycin: from discovery to future perspectives. J Ind Microbiol Biotechnol. 2017;44(4-5):537–553. doi: 10.1007/s10295-016-1834-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhang SM, Chen TS, Jia J, et al. Establishment of a highly efficient conjugation protocol for Streptomyces kanamyceticus ATCC12853. MicrobiologyOpen. 2018;8(6):e00747. doi: 10.1002/mbo3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao YF, Lu DD, Bechthold A, et al. Impact of otrA expression on morphological differentiation, actinorhodin production, and resistance to aminoglycosides in Streptomyces coelicolor M145. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(9):708–717. doi: 10.1631/jzus.B1800046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao YF, Song ZQ, Ma Z, et al. Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J Ind Microbiol Biotechnol. 2019;46(5):697–708. doi: 10.1007/s10295-019-02146-w. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Wang YM, Yu YF, et al. A non-restricting and non-methylating Escherichia coli strain for DNA cloning and high-throughput conjugation to Streptomyces coelicolor . Curr Microbiol. 2012;64(2):185–190. doi: 10.1007/s00284-011-0048-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhou YS, Gu YX, Qi BZ, et al. Porcine circovirus type 2 capsid protein induces unfolded protein response with subsequent activation of apoptosis. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2017;18(4):316–323. doi: 10.1631/jzus.B1600208. [DOI] [PMC free article] [PubMed] [Google Scholar]