Abstract
Background
Coronary heart disease (CHD) is one of the most severe cardiovascular diseases. Cyclin‐dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B‐AS1) is a significant susceptibility locus for cardiovascular disease by regulating inflammation response and cell cycle. The aim of this study was to assess whether CDKN2B‐AS1 polymorphisms are associated with CHD risk in the Chinese Han population.
Methods
A total of 501 CHD patients and 496 healthy controls were recruited from Central South University Xiangya School of Medicine Affiliated Haikou Hospital, five CDKN2B‐AS1 polymorphisms (rs10115049, rs75227345, rs2383205, rs10738606, and rs1333049) were analyzed by the Agena MassARRAY platform. The association of CDKN2B‐AS1 polymorphisms and CHD risk was determined by odd ratios (OR) and 95% confidence intervals (CI) using logistic regression.
Results
CDKN2B‐AS1 rs10738606 was significantly associated with CHD under codominant (p = .03), dominant (p = .019), recessive (p = .010), additive (p = .003), and allele (p = .003) models. Gender‐based subgroup tests showed that four polymorphisms (rs75227345, rs2383205, rs10738606 and rs1333049) were associated with CHD in males (p < .05). And age‐based subgroup tests indicated that rs2383205 and rs10738606 were associated with CHD among individuals, respectively (p < .05). For CHD patients, rs1333049 decreased the risk of diabetes under heterozygote (p = .014) and dominant (p = .024) models.
Conclusions
In conclusion, CDKN2B‐AS1 polymorphisms were associated with CHD risk in the combined or subgroup tests, suggesting an important role of CDKN2B‐AS1 in CHD susceptibility.
Keywords: case–control study, CDKN2B‐AS1, coronary heart disease, polymorphism, subgroups analysis
1. BACKGROUND
Coronary heart disease (CHD) is one of the most common cause of morbidity and mortality in cardiovascular diseases (CVD) worldwide, especially in the developed countries (Gaunt & Davey, 2015). CHD is characterized by the deposition of excessive cholesterol in the arterial intima (Lusk et al., 2014). The interaction of genetic and environmental factors can explain the majority of CHD cases (Peyser, 1997). Genetic factors play a vital role in the occurrence and development of CHD (Cunnington, Koref, Mayosi, Burn, & Keavney, 2010; Roberts, 2014). Single nucleotide polymorphisms (SNPs) are the most frequent genetic variation. Therefore, further exploration of the gene SNPs is much more significant and helpful for specific diagnosis on CHD.
Cyclin‐dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B‐AS1), also called ANRIL, is located within the CDKN2A‐CDKN2B cluster. Its product is a functional RNA molecule that interacts with polycomb repressive complex‐1 (PRC1) and ‐2 (PRC2), leading to epigenetic silencing of other genes in this cluster (Jing et al., 2018). CDKN2B‐AS1 is expressed in vascular endothelial cells and coronary smooth muscle cells (Broadbent et al., 2008). The regulation of CDKN2B‐AS1 expression level affects vascular cell proliferation and senescence. Genome‐wide association studies (GWAS) have reported that CDKN2B‐AS1 contains multiple genetic markers for CHD (Kunnas, Piesanen, & Nikkari, 2018). Genetic variants of CDKN2B‐AS1 are related to CVD by mediating the response to inflammatory signalling (Harismendy et al., 2011). However, the definite polymorphisms of CDKN2B‐AS1 affect CHD risk remain unclear, especially in the subgroups of age, gender, CHD patients with hypertension, and diabetes. Previous studies have shown that CDKN2B‐AS1 polymorphisms are associated with susceptibility to many diseases, including brain diseases (Sun et al., 2017), gout (Hsu et al., 2012), myocardial infarction (MI; Ivanova et al., 2017), and cancers (Gong et al., 2017). Although there have been several studies about the relationship between rs1333049 and CHD risk, the conclusions were not entirely consistent (Foroughmand, Nikkhah, Galehdari, & Jadbabaee, 2015; Lian et al., 2014; Pignataro et al., 2017; Pinós et al., 2014). Moreover, there were no studies regarding the association of rs10115049, rs75227345, rs2383205, rs10738606, and CHD susceptibility.
Hence, we conducted a case–control study to investigate the association of CDKN2B‐AS1 polymorphisms (rs10115049, rs75227345, rs2383205, rs10738606, and rs1333049) and CHD risk in the Chinese Han population.
2. METHODS
2.1. Study subjects
This study included 501 CHD patients (320 males and 181 females) and 496 healthy controls (318 males and 178 females) enrolled from Central South University Xiangya School of Medicine Affiliated Haikou Hospital, China. Patients were diagnosed with CHD according to standardized coronary angiography. The healthy controls were healthy individuals determined by medical history and clinical examinations. The healthy controls who had congenital heart disease, family history of CVD or known disease, were excluded in this study. The demographic and clinical characteristics of the participants are recorded in Table 1. Our study protocol was approved by the Medical Ethics Committees of Central South University Xiangya School of Medicine Affiliated Haikou Hospital. And written informed consent was obtained from all study objects.
Table 1.
Demographic and clinical characteristics of the study objects
| Characteristics |
CHD patients (N = 501) |
Healthy controls (N = 496) |
p |
|---|---|---|---|
| Age, years | 61.32 ± 11.70 | 60.69 ± 6.43 | .289 |
| >61 | 250 (49.9%) | 233 (47.0%) | |
| ≤61 | 251 (50.1%) | 263 (53.0%) | |
| Sex | |||
| Male | 320 (63.9%) | 318 (64.1%) | .895 |
| Female | 181 (36.1%) | 178 (35.9%) | |
| HDL (mmol/L) | 1.13 ± 0.25 | 1.09 ± 0.23 | <.001 |
| LDL (mmol/L) | 1.92 ± 0.82 | 2.55 ± 0.71 | <.001 |
| PLT (109/L) | 169.37 ± 75.18 | 211.10 ± 55.50 | <.001 |
| PDW (%) | 14.30 ± 2.87 | 13.74 ± 2.87 | .010 |
| MPV (FL) | 13.01 ± 7.14 | 10.91 ± 1.23 | <.001 |
| PCT (%) | 1.08 ± 3.151 | 0.30 ± 0.89 | <.001 |
| WBC | 11.68 ± 15.45 | 5.93 ± 1.50 | <.001 |
| RBC | 14.72 ± 35.86 | 4.84 ± 0.45 | <.001 |
| HGB | 132.67 ± 31.56 | 148.48 ± 14.77 | <.001 |
| Urea | 5.81 ± 6.51 | 7.01 ± 21.29 | .241 |
| UA (μmol/L) | 292.30 ± 88.75 | 330.55 ± 82.58 | <.001 |
| TG (mmol/L) | 1.78 ± 1.48 | 1.77 ± 1.13 | .947 |
| TC (mmol/L) | 4.09 ± 1.16 | 4.82 ± 5.47 | .006 |
| Hypertension | 296 (60%) | ||
| Diabetes | 101 (20%) | ||
| Gastritis | 59 (12%) |
Numbers in bold mean statistical significance.
Abbreviations: CHD, coronary heart disease; HDL, high‐density lipoprotein; HGB, hemoglobin; LDL, low‐density lipoprotein; MPV, mean platelet volume; PCT, plateletcrit; PDW, platelet distribution width; PLT, platelet; RBC, red blood cells; TC, total cholesterol; TG, triglyceride; UA, uric acid; WBC, white blood cells.
2.2. SNP selection and genotyping
Genomic DNA was extracted from peripheral blood stored with EDTA using blood DNA kit (GoldMag Co. Ltd.). The concentration of the DNA samples was measured with Nanodrop 2000 (Thermo Scientific). In this study, five SNPs in CDKN2B‐AS1 were selected from UCSC database and each candidate SNP had larger than 5% minor allele frequency in Chinese Han population. The primers used in this study were designed using MassARRAY Assay Design 3.0 software (Table S1), and the genotyping was performed on the MassARRAY iPLEX platform (Agena Bioscience) (Sun et al., 2017). We checked the quality of the genotype determination by the same method. We predicted functions of selected polymorphisms by HaployReg v4.1. Agena Bioscience TYPER version 4.0 software was used to perform data management and analyses.
2.3. Statistical analysis
Differences in categorical and continuous variables between cases and controls were assessed using the chi‐squared test and t test, respectively. Hardy–Weinberg equilibrium (HWE) was conducted for each SNP in controls using Fisher's exact test. Genotype and allele distributions were compared using the chi‐squared test. The relationships between CDKN2B‐AS1 polymorphisms and CHD risk were evaluated in multiple genetic models using PLINK software. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using logistic regression analysis after adjusting with gender and age. In addition, haplotype analysis and linkage disequilibrium (LD) were conducted by PLINK software and Haploview software (version 4.2; Kaushal et al., 2007). All statistical analyses were performed using the SPSS 17.0 (IBM®). The p‐values were two‐sided in our study, and p < .05 was considered statistical significant.
3. RESULTS
3.1. Characteristics of the study objects
A total of 501 CHD cases (mean age: 61.32 ± 11.70) and 496 healthy controls (mean age: 60.69 ± 6.43) were included in this study. Demographic and clinical characteristics of the study objects that include age, sex, high‐density lipoprotein (HDL), low‐density lipoprotein (HDL), platelet (PLT), platelet distribution width (PDW), mean platelet volume (MPV), plateletcrit (PCT), white blood cells (WBC), red blood cells (RBC), hemoglobin (HGB), urea, uric acid (UA), triglyceride (TG), total cholesterol (TC) are shown in Table 1. There were no significant differences in the age and sex distribution between two groups (p > .05). Among the CHD patients, 296 (60%) individuals with hypertension, 101 (20%) individuals with diabetes, and 59 (12%) individuals had gastritis.
3.2. Associations between CDKN2B‐AS1 polymorphisms and CHD risk
As shown in Table 2, the genotype distributions of all the five SNPs in controls met HWE (p > .05). Genotype and allele frequencies of CDKN2B‐AS1 polymorphisms are listed in Table 2. In Table S2, all CDKN2B‐AS1 polymorphisms are located in intronic region, and these SNPs are related to the regulation of DNAse, Motifs changed, Selected eQTL hits, Enhancer histone marks and NHGRI/EBI GWAS hits. In Table 3, logistic regression analyses revealed that rs10738606 conferred a decreased risk of CHD in codominant (OR = 0.54, 95% CI = 0.36–0.81, p = .003), dominant (OR = 0.74, 95% CI = 0.57–0.95, p = .019), recessive (OR = 0.60, 95% CI = 0.41–0.89, p = .010) and additive (OR = 0.75, 95% CI = 0.63–0.91, p = .003) models. A allele carriers of rs10738606 significantly decreased CHD risk (OR = 0.76, 95% CI = 0.63–0.91, p = .003). No significant associations were observed in the other CDKN2B‐AS1 polymorphisms and CHD risk (p > .05).
Table 2.
Comparison of genotype and allele frequencies between cases and controls
| SNP | Location: Position | Groups | Genotype (counts) | p | Allele (counts) | p | MAF | HWE p | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10115049 | Chr9: 22032120 | AA | GA | GG | A | G | |||||
| Cases | 73 (14.6%) | 208 (41.5%) | 220 (43.9%) | 354 (35.3%) | 648 (64.7%) | 0.353 | |||||
| Controls | 63 (12.7%) | 227 (45.8%) | 206 (41.5%) | .370 | 353 (35.6%) | 639 (64.4%) | .905 | 0.356 | 1.000 | ||
| rs75227345 | Chr9: 22042298 | TT | TC | CC | T | C | |||||
| Cases | 26 (5.2%) | 139 (27.7%) | 336 (67.1%) | 191 (19.1%) | 811 (80.9%) | 0.191 | |||||
| Controls | 17 (3.4%) | 127 (25.6%) | 352 (71.0%) | .257 | 161 (16.2%) | 831 (83.8%) | .097 | 0.162 | 0.188 | ||
| rs2383205 | Chr9: 22060936 | AA | GA | GG | A | G | |||||
| Cases | 10 (2.0%) | 115 (23.0%) | 376 (75.0%) | 135 (13.5%) | 867 (86.5%) | 0.134 | |||||
| Controls | 13 (2.6%) | 133 (26.8%) | 350 (50.4%) | .265 | 159 (16.0%) | 833 (84.0%) | .108 | 0.160 | 0.869 | ||
| rs10738606 | Chr9: 22088091 | AA | AT | TT | A | T | |||||
| Cases | 48 (9.6%) | 224 (45.0%) | 226 (45.4%) | 320 (32.1%) | 606 (67.9%) | 0.321 | |||||
| Controls | 74 (15.0%) | 230 (46.7%) | 188 (38.3%) | .011 | 378 (38.4%) | 676 (61.6%) | .003 | 0.384 | 0.776 | ||
| rs1333049 | Chr9: 22125504 | CC | CG | GG | C | G | |||||
| Cases | 110 (22.0%) | 263 (52.5%) | 128 (25.5%) | 483 (48.2%) | 519 (51.8%) | 0.482 | |||||
| Controls | 94 (19.0%) | 254 (51.2%) | 148 (29.8%) | .241 | 442 (44.6%) | 550 (55.4%) | .103 | 0.446 | 0.467 | ||
Numbers in bold mean statistical significance.
Abbreviations: HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Table 3.
The association between CDKN28‐AS1 polymorphisms and CHD risk
| SNP | Model | Genotype/Allele | OR (95%CI) | p |
|---|---|---|---|---|
| rs10115049 | Codominant | AA/GG | 1.08 (0.73–1.59) | .690 |
| GA/GG | 0.86 (0.66–1.12) | .257 | ||
| Dominant | AA‐AG/GG | 0.91 (0.70–1.16) | .438 | |
| Recessive | AA/AG‐GG | 1.17 (0.81–1.68) | .396 | |
| Additive | 0.99 (0.83–1.18) | .894 | ||
| Allele | A/G | 0.99 (0.82–1.19) | .905 | |
| rs75227345 | Codominant | TT/CC | 1.59 (0.85–2.99) | .147 |
| TC/CC | 1.15 (0.87–1.53) | .332 | ||
| Dominant | TT‐CT/CC | 1.20 (0.92–1.58) | .178 | |
| Recessive | TT/CT‐CC | 1.53 (0.82–2.86) | .181 | |
| Additive | 1.20 (0.96–1.50) | .110 | ||
| Allele | T/C | 1.22 (0.96–1.53) | .097 | |
| rs2383205 | Codominant | AA/GG | 0.72 (0.31–1.66) | .435 |
| GA/GG | 0.80 (0.60–1.07) | .130 | ||
| Dominant | AA‐AG/GG | 0.79 (0.60–1.05) | .104 | |
| Recessive | AA/AG‐GG | 0.76 (0.33–1.75) | .515 | |
| Additive | 0.81 (0.64–1.04) | .103 | ||
| Allele | A/G | 0.82 (0.64–1.05) | .108 | |
| rs10738606 | Codominant | AA/TT | 0.54 (0.36–0.81) | .003 |
| TA/TT | 0.80 (0.61–1.05) | .108 | ||
| Dominant | AA‐AT/TT | 0.74 (0.57–0.95) | .019 | |
| Recessive | AA/AT‐TT | 0.60 (0.41–0.89) | .010 | |
| Additive | 0.75 (0.63–0.91) | .003 | ||
| Allele | A/T | 0.76 (0.63–0.91) | .003 | |
| rs1333049 | Codominant | CC/GG | 1.36 (0.95–1.96) | .095 |
| GC/GG | 1.20 (0.90–1.61) | .218 | ||
| Dominant | CC‐CG/GG | 1.25 (0.94–1.65) | .122 | |
| Recessive | CC/GC‐GG | 1.21 (0.89–1.65) | .231 | |
| Additive | 1.17 (0.98–1.40) | .089 | ||
| Allele | C/G | 1.16 (0.97–1.38) | .103 |
Numbers in bold mean statistical significance.
Abbreviations: 95% CI, 95% confidence interval; CHD, coronary heart disease; OR, odds ratio; SNP, single nucleotide polymorphism.
3.3. Associations between CDKN2B‐AS1 polymorphisms and CHD risk in subgroups
The relationships between CDKN2B‐AS1 polymorphisms and CHD risk were further assessed in four subgroups (age, gender, hypertension, and diabetes). The significant associations were presented in Table 4. Rs75227345, rs2383205, rs10738606, and rs1333049 were associated with CHD risk in males. Rs75227345 and rs1333049 increased the risk of CHD (rs75227345: homozygote, OR = 2.62, 95% CI = 1.07–6.44, p = .036; recessive: OR = 2.49, 95% CI = 1.02–6.10, p = .045; additive: OR = 1.35, 95% CI = 1.01–1.81, p = .040; allele: OR = 1.38, 95% CI = 1.03–1.86, p = .033; rs1333049: homozygote, OR = 1.67, 95% CI = 1.06–2.64, p = .027; recessive: OR = 1.59, 95% CI = 1.08–2.37, p = .018; additive: OR = 1.28, 95% CI = 1.02–1.60, p = .034; allele: OR = 1.26, 95% CI = 1.01–1.58, p = .037), whereas rs2383205 and rs10738606 decreased CHD risk (rs2383205: dominant, OR = 0.67, 95% CI = 0.47–0.97, p = .035; additive, OR = 0.67, 95% CI = 0.48–0.92, p = .014; allele, OR = 0.67, 95% CI = 0.49–0.93, p = .014; rs10738606, homozygote, OR = 0.44, 95% CI = 0.27–0.73, p = .001; dominant, OR = 0.64, 95% CI = 0.47–0.88, p = .006; recessive, OR = 0.52, 95% CI = 0.33–0.84, p = .007; additive, OR = 0.68, 95% CI = 0.54–0.85, p = .001; allele, OR = 0.67, 95% CI = 0.53–0.85, p < .001). For the individuals equal or younger than 61 years old, rs2383205 had a lower risk of CHD in heterozygote (OR = 0.61, 95% CI = 0.39–0.95, p = .030) and dominant (OR = 0.63, 95% CI = 0.41–0.97, p = .035) models. Among the elderly group (age > 61), rs10738606‐A allele was a protective factor of CHD (OR = 0.74, 95% CI = 0.57–0.96, p = .023). In addition, we found that rs1333049 decreased the risk of diabetes for CHD patients (heterozygote, OR = 0.53, 95% CI = 0.32–0.88, p = .014; dominant: OR = 0.58, 95% CI = 0.36–0.93, p = .024).
Table 4.
The association between CDKN28‐AS1 polymorphisms and CHD risk in the subgroups
| SNP | Subgroup | Homozygote | Heterozygote | Dominant | Recessive | Additive | Allele | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | ||
| rs75227345 | Male | 2.62 (1.07–6.44) | .036 | 1.21 (0.84–1.73) | .309 | 1.32 (0.94–1.86) | .109 | 2.49 (1.02–6.10) | .045 | 1.35 (1.01–1.81) | .040 | 1.38 (1.03–1.86) | .033 |
| rs2383205 | Age ≤ 61 | 0.89 (0.23–3.45) | .864 | 0.61 (0.39–0.95) | .030 | 0.63 (0.41–0.97) | .035 | 1.01 (0.26–3.91) | .987 | 1.01 (0.26–3.91) | .987 | 0.72 (0.51–1.03) | .071 |
| Male | 0.30 (0.08–1.12) | 0.072 | 0.71 (0.49–1.02) | .065 | 0.67 (0.47–0.96) | .028 | 0.32 (0.09–1.21) | .094 | 0.67 (0.48–0.92) | .014 | 0.67 (0.49–0.93) | .014 | |
| rs10738606 | Age > 61 | 0.60 (0.33–1.08) | .089 | 0.86 (0.58–1.28) | .453 | 0.79 (0.54–1.15) | .217 | 0.64 (0.37–1.13) | .124 | 0.80 (0.61–1.04) | .100 | 0.74 (0.57–0.96) | .023 |
| Male | 0.44 (0.27–0.73) | .001 | 0.72 (0.51–1.00) | .051 | 0.64 (0.47–0.88) | .006 | 0.52 (0.33–0.84) | .007 | 0.68 (0.54–0.85) | .001 | 0.67 (0.53–0.85) | <.001 | |
| rs1333049 | Male | 1.67 (1.06–2.64) | .027 | 1.08 (0.74–1.56) | .698 | 1.22 (0.86–1.73) | .264 | 1.59 (1.08–2.34) | .018 | 1.28 (1.02–1.60) | .034 | 1.26 (1.01–1.58) | .037 |
| Diabetes | 0.72 (0.39–1.32) | .284 | 0.53 (0.32–0.88) | .014 | 0.58 (0.36–0.93) | .024 | 1.07 (0.63–1.82) | .791 | 0.81 (0.59–1.12) | .199 | 0.81 (0.60–1.11) | .187 | |
Numbers in bold mean statistical significance.
Abbreviations: 95% CI, 95% confidence interval; CHD, coronary heart disease; OR, odds ratio; SNP, single nucleotide polymorphism.
3.4. Genotypes and clinical characteristics
We evaluated the association between genotypes of CDKN2B‐AS1 polymorphisms and clinical characteristics of patients, including HDL, LDL, PLT, PDW, MPV, PCT, WBC, RBC, HGB, urea, UA, TG, and TC (Table S3 and Table 5). We observed that patients carried different genotypes of CDKN2B‐rs75227345 had significant differences in MPV, PCT, WBC, RBC, and HGB (p < .05).
Table 5.
Clinical characteristics of rs75227345 on CHD patients
| Characteristics | CDKN2B‐rs75227345 | |||
|---|---|---|---|---|
| TT | CT | CC | p | |
| HDL (mmol/L) | 1.10 ± 0.29 | 1.13 ± 0.24 | 1.14 ± 0.26 | .786 |
| LDL (mmol/L) | 1.84 ± 0.60 | 1.96 ± 0.82 | 1.91 ± 0.84 | .753 |
| PLT (109/L) | 170.77 ± 37.87 | 157.71 ± 80.58 | 174.15 ± 74.70 | .097 |
| PDW (%) | 14.08 ± 2.61 | 14.41 ± 3.13 | 14.28 ± 2.80 | .846 |
| MPV (FL) | 11.08 ± 0.93 | 14.18 ± 8.67 | 12.67 ± 6.64 | .044 |
| PCT (%) | 0.19 ± 0.04 | 1.60 ± 3.72 | 0.93 ± 2.98 | .038 |
| WBC | 7.03 ± 2.65 | 14.83 ± 20.20 | 10.72 ± 13.42 | .009 |
| RBC | 4.82 ± 0.75 | 22.02 ± 46.70 | 12.44 ± 31.35 | .011 |
| HGB | 138.00 ± 20.16 | 125.37 ± 36.47 | 135.23 ± 29.61 | .006 |
| Urea | 5.43 ± 2.53 | 5.75 ± 2.11 | 5.86 ± 7.80 | .942 |
| UA (μmol/L) | 309.72 ± 81.47 | 297.77 ± 82.16 | 288.74 ± 91.79 | .372 |
| TG (mmol/L) | 1.69 ± 1.10 | 1.88 ± 1.67 | 1.75 ± 1.43 | .644 |
| TC (mmol/L) | 3.93 ± 1.01 | 4.27 ± 1.31 | 4.03 ± 1.09 | .110 |
Numbers in bold mean statistical significance.
Abbreviations: CHD, coronary heart disease; HDL, high‐density lipoprotein; HGB, hemoglobin; LDL, low‐density lipoprotein; MPV, mean platelet volume; PCT, plateletcrit; PDW, platelet distribution width; PLT, platelet; RBC, red blood cells; TC, total cholesterol; TG, triglyceride; UA, uric acid; WBC, white blood cells.
3.5. Haplotype analysis of CDKN2B‐AS1 polymorphisms and CHD risk
We also performed haplotype analysis of CDKN2B‐AS1 polymorphisms and CHD risk. We found one block including rs10115049 and rs75227345 (Figure 1). As shown in Table S4, there was no significantly association between haplotypes of CDKN2B‐AS1 polymorphisms and CHD risk (p > .05).
Figure 1.
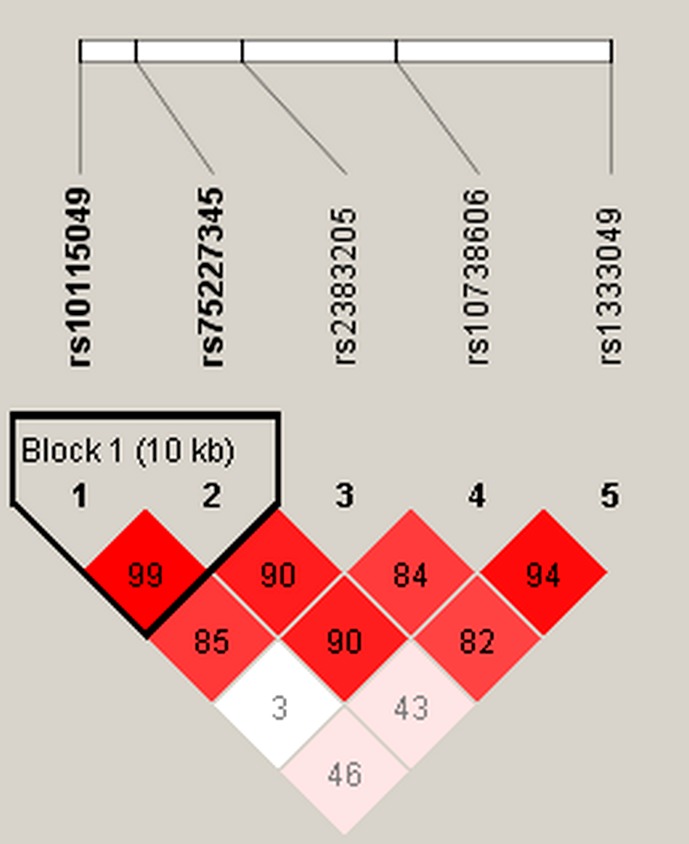
Linkage disequilibrium plots containing five polymorphisms from CDKN2B‐AS1. Block 1 includes rs10115049 and rs75227345. The numbers inside the diamonds indicate the D′ for pairwise analyses
4. DISCUSSION
In this study, we genotyped five SNPs of CDKN2B‐AS1 and evaluated the association between these SNPs and CHD risk in the Chinese Han population. We found that CDKN2B‐AS1 polymorphisms had strong relationships with CHD risk, especially rs10738606 could protect the Chinese Han population from CHD. Age, sex, and complications of CHD significantly influenced the association of CDKN2B‐AS1 polymorphisms and CHD risk. Our results gave a clue in the prevention, diagnosis, and individual treatment of CHD.
CDKN2B‐AS1 encodes a 3.8 kb lnc RNA which consists of 19 exons, and is located at chromosome 9p21 (Holdt et al., 2011; Kong, Sharma, Nwosu, & Alonso, 2016). The 9p21.3 locus was first identified by GWAS to be strongly associated with CHD and MI (Glinsky, 2008; Ruth et al., 2007). It then reported that this locus was associated with PLT reactivity and polymorphisms at 9p21 influence inflammatory signaling and vascular cell proliferation (Harismendy et al., 2011; Musunuru et al., 2010; Visel et al., 2010). We observed significant difference in PLT between CHD patients and healthy controls, and genotypes of rs75227345 also associated with PLT, it may explain the association of polymorphism with CHD. According to several published studies, rs2383206, rs10757274, and rs10757278 may serve as genetic biomarkers of CHD in Caucasians, East Asians, and West Asian (Wang, Dong, & Yang, 2016). The expression of CDKN2B‐AS1 genetic variants could influence CHD susceptibility in the Iranian patients (Bochenek et al., 2013). Among the selected SNPs, rs1333049 is the frequently studied polymorphism. Rs1333049 was found to be associated with CHD in the Turkish (Cakmak et al., 2015), Indians (Kashyap et al., 2018), Japanese (Pinós et al., 2014) population, but no studies focused on the Chinese population. As shown in our study, rs1333049 was remarkably associated with CHD susceptibility in subgroups. In addition, we found rs10738606 was a protective factor for CHD.
Age and gender disparities widely existed in the prevalence of CHD (Chen et al., 2016; Cline & Beckie, 2013). The incidence of CHD is 0.6% for the people younger than 40 years old, it will increase twofold or more with aging (Yan et al., 2013). In our study, rs2383205 was associated with a decreased CHD risk among younger people (age ≤ 61), whereas rs10738606 decreased CHD risk among elderly people (age > 61). It revealed an age‐based mechanism on the genetic variations. Previous studies have indicated that gender differences could influence gene expression and then affect disease progression (Coban et al., 2014; Xu et al., 2008). We found rs75227345, rs2383205, rs10738606, and rs1333049 had relationships with CHD risk in males, no associations were found in females. It confirmed previous results. Additionally, hypertension and diabetes are considered as traditional risk factors of CHD. In this study, we studied the relationship of CDKN2B‐AS1 polymorphisms and complications (hypertension and diabetes) among CHD cases. Rs1333049 could protect CHD patients from diabetes under heterozygote and dominant models. Nevertheless, larger sample size and well‐designed studies are required to validate our results.
Some limitations could not be ignored in this study. First, all samples were collected from hospital, which inevitably exist the choosing bias. Second, we did not evaluate some factor that could have effects on CHD risk, because of a lack of data from both CHD patients and healthy controls. Third, this study did not analysis the mechanisms of CDKN2B‐AS1 polymorphisms influence CHD risk. Further experiments on cell or animal level are required to explain the detailed molecular mechanism.
5. CONCLUSION
Our results indicate that CDKN2B‐AS1 polymorphisms are associated with CHD risk in the Chinese Han population. These SNPs may serve as biomarkers for CHD in the Chinese Han population.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
Supporting information
ACKNOWLEDGMENTS
We sincerely thank all the participators in this study.
Huang K, Zhong J, Li Q, et al. Effects of CDKN2B‐AS1 polymorphisms on the susceptibility to coronary heart disease. Mol Genet Genomic Med. 2019;7:e955 10.1002/mgg3.955
Contributor Information
Shijuan Lu, Email: 1157416676@qq.com.
Shufang Zhang, Email: zhangshufang0898@126.com.
REFERENCES
- Bochenek, G. , Häsler, R. , El Mokhtari, N.‐E. , König, I. R. , Loos, B. G. , Jepsen, S. , … Schaefer, A. S. (2013). The large non‐coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Human Molecular Genetics., 22(22), 4516–4527. 10.1093/hmg/ddt299 [DOI] [PubMed] [Google Scholar]
- Broadbent, H. M. , Peden, J. F. , Lorkowski, S. , Goel, A. , Ongen, H. , Green, F. , … Watkins, H. (2008). Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human Molecular Genetics, 17(6), 806–814. 10.1093/hmg/ddm352 [DOI] [PubMed] [Google Scholar]
- Cakmak, H. A. , Bayoglu, B. , Durmaz, E. , Can, G. , Karadag, B. , Cengiz, M. , … Yuksel, H. (2015). Evaluation of association between common genetic variants on chromosome 9p21 and coronary artery disease in Turkish population. Anatolian Journal of Cardiology., 15(3), 196–203. 10.5152/akd.2014.5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Chen, X. , Xu, Y. , Yang, W. , Wu, N. , Ye, H. , … Duan, S. (2016). Association of six CpG‐SNPs in the inflammation‐related genes with coronary heart disease. Human Genomics, 10(Suppl 2), 21 10.1186/s40246-016-0067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, J. L. , & Beckie, T. M. (2013). The relationships between FAM5C SNP (rs10920501) variability and metabolic syndrome and inflammation in women with coronary heart disease. Biological Research for Nursing, 15(2), 160–166. 10.1177/1099800411424487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coban, N. , Onat, A. , Kömürcü, B. E. , Güleç, C. , Can, G. , & Erginel, Ü. N. (2014). Gender specific association of ABCA1 gene R219K variant in coronary disease risk through interactions with serum triglyceride elevation in Turkish adults. Anadolu Kardiyol Derg, 14(1), 18–25. 10.5152/akd.2013.234 [DOI] [PubMed] [Google Scholar]
- Cunnington, M. S. , Koref, M. S. , Mayosi, B. M. , Burn, J. , & Keavney, B. (2010). Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genetics, 6(4), e1000899 10.1371/journal.pgen.1000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroughmand, A. M. , Nikkhah, E. , Galehdari, H. , & Jadbabaee, M. H. (2015). Association study between coronary artery disease and rs1333049 and rs10757274 polymorphisms at 9p21 locus in South‐West Iran. Cell Journal (Yakhteh), 17(1), 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, T. R. , & Davey, S. G. (2015). eNOS and coronary artery disease: Publication bias and the eclipse of hypothesis‐driven meta‐analysis in genetic association studies. Gene, 556(2), 257–258. 10.1016/j.gene.2014.11.052 [DOI] [PubMed] [Google Scholar]
- Glinsky, G. V. (2008). An SNP‐guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle, 7(16), 2570–2583. 10.4161/cc.7.16.6524 [DOI] [PubMed] [Google Scholar]
- Gong, W.‐J. , Peng, J.‐B. , Yin, J.‐Y. , Li, X.‐P. , Zheng, W. , Xiao, L. , … Liu, Z.‐Q. (2017). Association between well‐characterized lung cancer lncRNA polymorphisms and platinum‐based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacologica Sinica, 38(4), 581–590. 10.1038/aps.2016.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy, O. , Notani, D. , Song, X. , Rahim, N. G. , Tanasa, B. , Heintzman, N. , … Frazer, K. A. (2011). 9p21 DNA variants associated with coronary artery disease impair interferon‐γ signalling response. Nature, 470(7333), 264–268. 10.1038/nature09753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt, L. M. , Sass, K. , Gäbel, G. , Bergert, H. , Thiery, J. , & Teupser, D. (2011). Expression of Chr9p21 genes CDKN2B (p15(INK4b)), CDKN2A (p16(INK4a), p14(ARF)) and MTAP in human atherosclerotic plaque. Atherosclerosis, 214(2), 264–270. 10.1016/j.atherosclerosis.2010.06.029 [DOI] [PubMed] [Google Scholar]
- Hsu, A. , Dalbeth, N. , Gow, P. , Harrison, A. , Highton, J. , Jones, P. B. , … Merriman, T. R. (2012). No evidence for association of Chr 9p21 variant rs1333049 with gout in New Zealand case‐control sample sets. Rheumatology, 51(6), 1129–1130. 10.1093/rheumatology/kes029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova, A. A. , Maksimov, V. N. , Orlov, P. S. , & Ivanoshchuk, D. E. , Savchenko S. V., Voevoda M. I. Association of the genetic markers for myocardial infarction with sudden cardiac death. Indian Heart Journal. 2017;69(Suppl 1), S8–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, X. J. , Lin, J. , Jun, X. L. , Zhan, G. , Yan, Z. X. , Yin, Z. , et al. (2018). Association of CDKN2B‐AS1 polymorphisms with premature triple‐vessel coronary disease and their sex specificity in the Chinese population. Biomedical and Environmental Sciences, 31(11), 787–796. 10.3967/bes2018.106 [DOI] [PubMed] [Google Scholar]
- Kashyap, S. , Kumar, S. , Agarwal, V. , Misra, D. P. , Rai, M. K. , & Kapoor, A. (2018). The association of polymorphic variants, rs2267788, rs1333049 and rs2383207 with coronary artery disease, its severity and presentation in North Indian population. Gene, 648, S0378111918300283 10.1016/j.gene.2018.01.021 [DOI] [PubMed] [Google Scholar]
- Kaushal, R. , Pal, P. , Alwell, K. , Haverbusch, M. , Flaherty, M. , Moomaw, C. , … Woo, D. (2007). Association of ALOX5AP with ischemic stroke: A population‐based case‐control study. Human Genetics, 121(5), 601–607. 10.1007/s00439-007-0338-y [DOI] [PubMed] [Google Scholar]
- Kong, Y. , Sharma, R. B. , Nwosu, B. U. , & Alonso, L. C. (2016). Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia, 59(8), 1579–1593. 10.1007/s00125-016-3967-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnas, T. , Piesanen, J. , & Nikkari, S. T. (2018). Association of a Chromosome Locus 9p21.3 CDKN2B‐AS1 Variant rs4977574 with Hypertension: The TAMRISK Study. Genet Test Mol Biomarkers., 22(5), 327–330. 10.1089/gtmb.2017.0249 [DOI] [PubMed] [Google Scholar]
- Lian, J. , Ba, Y. , Dai, D. , Chen, Z. , Lou, Y. , Jiang, Q. , … Duan, S. (2014). A replication study and a meta‐analysis of the association between the CDKN2A rs1333049 polymorphism and coronary heart disease. Journal of Atherosclerosis and Thrombosis, 21(11), 1109–1120. 10.5551/jat.23507 [DOI] [PubMed] [Google Scholar]
- Lusk, C. M. , Dyson, G. , Clark, A. G. , Ballantyne, C. M. , Frikke‐Schmidt, R. , Tybjærg‐Hansen, A. , … Sing, C. F. (2014). Validated context‐dependent associations of coronary heart disease risk with genotype variation in the chromosome 9p21 region: The Atherosclerosis Risk in Communities study. Human Genetics., 133(9), 1105–1116. 10.1007/s00439-014-1451-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru, K. , Post, W. S. , Herzog, W. , Shen, H. , O'Connell, J. R. , McArdle, P. F. , … Mitchell, B. D. (2010). Association of single nucleotide polymorphisms on chromosome 9p21.3 with platelet reactivity: A potential mechanism for increased vascular disease. Circulation: Cardiovascular Genetics, 3(5), 445–453. 10.1161/CIRCGENETICS.109.923508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyser, P. A. (1997). Genetic epidemiology of coronary artery disease. Epidemiologic Reviews, 19(1), 80 10.1093/oxfordjournals.epirev.a017949 [DOI] [PubMed] [Google Scholar]
- Pignataro, P. , Pezone, L. , Di Gioia, G. , Franco, D. , Iaccarino, G. , Iolascon, A. , … Capasso, M. (2017). Association study between coronary artery disease and rs1333049 polymorphism at 9p21. 3 locus in Italian population. Journal of Cardiovascular Translational Research, 10(5–6), 455–458. 10.1007/s12265-017-9758-9 [DOI] [PubMed] [Google Scholar]
- Pinós, T. , Fuku, N. , Cámara, Y. , Arai, Y. , Abe, Y. , Rodríguez‐Romo, G. , … Lucia, A. (2014). The rs1333049 polymorphism on locus 9p21.3 and extreme longevity in Spanish and Japanese cohorts. AGE, 36(2), 933–943. 10.1007/s11357-013-9593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. (2014). Genetics of coronary artery disease: An update. Methodist Debakey Cardiovascular Journal, 10(1), 7–12. 10.14797/mdcj-10-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruth, M. P. , Alexander, P. , Nihan, K. , Alexandre, S. , Robert, R. , Cox, D. R. , et al. (2007). A common allele on chromosome 9 associated with coronary heart disease. Science, 316(5830), 1488–1491, 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. X. , Gao, C. Y. , Lu, Y. , Fu, X. , Jia, J. G. , Zhao, Y. J. , et al. (2017). Association between PPAP2B gene polymorphisms and coronary heart disease susceptibility in Chinese Han males and females. Oncotarget, 8(8), 10.18632/oncotarget.14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel, A. , Zhu, Y. , May, D. , Afzal, V. , Gong, E. , Attanasio, C. , … Pennacchio, L. A. (2010). Targeted deletion of the 9p21 non‐coding coronary artery disease risk interval in mice. Nature, 464(7287), 409–412. 10.1038/nature08801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Dong, P. , & Yang, X. (2016). ANRIL rs2383207 polymorphism and coronary artery disease (CAD) risk: A meta‐analysis with observational studies. Cellular and Molecular Biology, 62(12), 6–10. 10.14715/cmb/2016.62.12.2 [DOI] [PubMed] [Google Scholar]
- Xu, H. , Hou, X. , Wang, N. , Hui, B. O. , Jin, J. , Yun, S. , … Han, Y. (2008). Gender‐specific effect of estrogen receptor‐1 gene polymorphisms in coronary artery disease and its angiographic severity in Chinese population. Clinica Chimica Acta, 395(1), 130–133. 10.1016/j.cca.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Yan, Y. L. , Qiu, B. , Hu, L. J. , Jing, X. D. , Liu, Y. J. , Deng, S. B. , … She, Q. (2013). Efficacy and safety evaluation of intensive statin therapy in older patients with coronary heart disease: A systematic review and meta‐analysis. European Journal of Clinical Pharmacology, 69(12), 2001–2009. 10.1007/s00228-013-1570-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


