Abstract
Global outbreaks of drug resistant fungi such as Candida auris are thought to be due at least in part to excessive use of antifungal drugs. Baker’s yeast Saccharomyces cerevisiae has gained importance as an emerging opportunistic fungal pathogen that can cause infections in immune-compromised patients. Analyses of over 1000 S. cerevisiae isolates are providing rich resources to better understand how fungi can grow in human environments. A large percentage of clinical S. cerevisiae isolates are heterozygous across many nucleotide sites, and a significant proportion are of mixed ancestry and/or are aneuploid or polyploid. Such features potentially facilitate adaptation to new environments. These observations provide strong impetus for expanding genomic and molecular studies on clinical and wild isolates to understand the prevalence of genetic diversity and instability generating mechanisms and how they are selected for and maintained. Such work can also lead to the identification of new targets for anti-fungal drugs.
Keywords: Saccharomyces cerevisiae, baker’s yeast, clinical isolates, adaptation to stress, genome instability
Saccharomyces cerevisiae, a model for studying pathogenic yeast
Saccharomyces cerevisiae, or baker’s yeast, is the best studied single cell model eukaryote. It is found in many natural environments including trees, fruits and soil, is extensively used in industry to make bread, beer, and wine (e.g.[1, 2]), and has been found in the respiratory, gastrointestinal, and urinary tracts of healthy individuals ([3–7], reviewed in [8]). It is not thought to adapt quickly to the changing conditions that occur in the human body, and its presence does not normally cause infections because it can be cleared by the immune system and does not cross epithelial barriers [3–7]. However, recent molecular and genomic analyses of S. cerevisiae natural isolates (see Glossary) have identified loci and mechanisms that promote genetic variability and genomic instability, and consequently aid in adaptation to stressful environments [8–11]. This review provides an overview of genomic processes that can promote adaptation of baker’s yeast to changing environments and how such adaptation may lead to virulence in a human host.
S. cerevisiae, an opportunistic pathogen
Over the past 25 years, a significant effort has been made to collect and characterize S. cerevisiae isolated from patients (for examples see [11–17]). It has been designated as an emerging opportunistic pathogen that can cause infections in immune compromised patients, and is associated with virulence when the epithelial barrier is breached [5, 18]. The association of baker’s yeast with human infection has become better recognized due to improved diagnostic methods. S. cerevisiae causes roughly 1–4% of severe fungal infections, with Candida albicans (~53–58%) and Candida glabrata (~20–23%) causing the majority [19]. There are a few extreme cases where S. cerevisiae is the direct cause of mortality, usually by inducing sepsis [19]. Baker’s yeast infection is associated with several illnesses including pneumonia, peritonitis, esophagitis, and liver abscesses (reviewed in [20]). Furthermore, it can cause infections ranging from vaginitis in healthy patients, cutaneous infections, systemic bloodstream infections, and infections of essential organs in immunocompromised and critically ill patients [5, 20–22]. Based on these reports, S. cerevisiae is considered a low-virulence human pathogen [8, 22].
Challenges for baker’s yeast in the human host and clinical environment
The human body is a stressful growth environment for baker’s yeast because it is subjected to heat stress, antifungal agents, antimicrobials, and competing commensals and/or infectious microbes. These stresses are analogous to what yeast might experience in a wine fermentation environment, which is constantly fluctuating, and differs from a typical natural environment such as the bark of a tree or fruit [23]. Major factors that allow for opportunistic infection by S. cerevisiae are impaired host immune response, use of invasive and infected catheters in hospitals, antibiotic therapies to treat some infections, and probiotics that contain S. cerevisiae [20, 21]. Thus, it is not a surprise that the majority of infections caused by S. cerevisiae are hospital acquired. Apart from being a commensal in patients and health-care workers, it may be acquired from other infected patients and may appear in the hospital environment through probiotics and other food sources [5, 20, 21, 22, 24].
Can human behavior impact adaptation mechanisms?
Indiscriminate use of antimicrobials is thought to be a leading cause of outbreaks of multi-drug resistant bacterial pathogens [25]. Recent work has suggested a similar situation in fungi [26, 27]. Excessive use of antifungals in farm animals and crops is thought to be a major source of drug-resistant fungi that cause infections. This is exemplified by global outbreaks of Candida auris within the last five years [26]. This pathogen is resistant to major antifungal medications and causes infections in patients with weak immunity such as critically ill patients, infants and patients on immune-suppressive medications. The commensal fungal population is important to maintain gut health and promote anti-fungal immunity [27]. However, the use of probiotics, especially in immune-compromised patients, is also thought to provide an advantage for specific fungi to grow in a clinical environment. For example, Saccharomyces boulardii, a subtype of S. cerevisiae, is used in probiotics for the treatment of diarrheal disease such as those caused by Clostridium difficile [5, 21]. These live preparations are often consumed at high doses for long periods of time and have been associated with invasive S. cerevisiae infections in immunosuppressed individuals [5, 21].
Phenotypes associated with virulence
Some of the earliest studies analyzing baker’s yeast infections in mammals involved introducing clinical and non-clinical human isolates into immune-compromised mice [12]. Such studies showed that virulence was likely to be a dominant trait involving multiple loci [12]. Outlined below and in Table 1 are summaries of phenotypes that are important for growth in clinical conditions.
Table 1.
Phenotypes observed in fungi grown in clinical environments or conditions: a few examples.
| Phenotype | Description |
|---|---|
| Antifungal resistance |
1. Genes that act in drug efflux pathways. A. Overexpression of genes in clinical isolates of C. albicans [32]. i. CDR1 and CDR2: ABC (ATP-binding cassette) transporters ii. MDR1: Regulates intracellular protein transport B. Experimentally evolved S. cerevisiae [29]. i. PDR1: Mutations in this transcription factor cause overexpression of PDR5 and SNQ2 ii. PDR3: Mutations in this transcriptional activator alter expression of ABC transporters. iii. Overexpression increased drug efflux [30]: ICT1 (phosphatidic acid biosynthesis), YOR1 (ABC transporter), GRE2 (catabolism of some sugars), PDR16 (lipid synthesis), YGR035C and YPL088W (unknown). |
|
3. Modulation of expression of ergosterol biosynthesis pathway genes in experimentally evolved S. cerevisiae [29, 33]. A. Loss of function mutation in ERG3 causes overexpression of ERG11 which causes resistance to fluconazole B. Loss of function of ERG6 causes resistance to fluconazole | |
| 4. Aneuploidy of isochromosome 5L and trisomy of chromosome 7 in experimental evolution of C. albicans [31]. | |
| High temperature growth | 1. Linkage to NCS2, MKT1, END3 and RHO2 in S. cerevisiae [34, 35]. 2. Alleles of S. cerevisiae housekeeping genes were found to be important for high temperature growth phenotypes [36]. |
| Colony phenotype switching | Linked to clinical isolates of S. cerevisiae [13, 37]. |
| Pseudohyphal growth | Linked to clinical isolates of S. cerevisiae [39] and a large gene set [40, 41]. |
| Nutrient starvation | Determining fitness effects of genes from the yeast amplification and deletion library in nutrient limited conditions resulted in the identification of 73 genes in phosphate limited conditions, 210 in glucose limited, and 223 in sulfate limited conditions [94] |
Antifungal resistance.
The most common class of antifungals, azoles, targets enzymes in the ergosterol biosynthesis pathway. Ergosterol is the major sterol in the plasma and mitochondrial membranes of fungi. It is not present in mammals and is an established target of antifungals [28]. Resistance to antifungals can occur through the deletion or overexpression of genes that regulate membrane transporters that cause efflux of the drug, or modulate the expression of targets in the ergosterol pathway (Table 1; [29–33]). For example, in experimentally evolved populations (defined as cultures grown in a laboratory condition for a large number of generations) of S. cerevisiae, resistance to the antifungal fluconazole resulted from mutations in several targets [29]. At low drug concentrations, diploids acquired resistance more rapidly than haploids. This advantage was thought to be due to resistance resulting from dominant mutations and diploids having twice the number of mutational targets. At high drug concentrations, resistance occurred more quickly in haploids due to the acquisition of recessive mutations. In experimentally evolved populations of C. albicans, resistance to fluconazole resulted from an additional copy of isochromosome 5L, which became fixed in multiple independent populations [31].
High temperature growth.
The ability to grow at high temperature aids in virulence of S. cerevisiae. Four genes have been linked to high temperature growth in S. cerevisiae: NCS2, MKT1, END3 and RHO2 [34, 35]. They were identified through a targeted backcross mapping strategy and reciprocal hemizygosity analysis [34, 35]. More recently, S. cerevisiae specific housekeeping alleles were identified by a reciprocal hemizygosity mapping strategy involving an interspecific hybrid of S. cerevisiae and its thermosensitive relative S. paradoxus [36].
Colony phenotype switching.
Many clinical isolates display several colony phenotypes that can include variation in smoothness, color, size and colony border color [13, 37]. Clinical isolates were observed that showed different but reversible colony phenotypes that were dependent on growth media [37]. Colony phenotype switching was observed more often in clinical compared to non-clinical isolates [37]. Such properties likely facilitate colonizing different parts of the human body that provide different stress conditions and nutrient availability. It is also important to note that meiotic progeny of clinical isolates can display a wide variety of colony phenotypes in a single growth condition (for example, see [10]).
Pseudohyphal growth.
In this growth phase, cells bud and become elongated but the buds do not separate, creating chains of cells that can invade the growth substrate [38]. This unipolar growth may be important under nutrient deprived conditions to identify food sources. Such growth has been linked to over 500 genes in S. cerevisiae, with virulent isolates showing significantly higher pseudohyphal growth compared to avirulent isolates [39–41].
Genetic mechanisms that contribute to baker’s yeast virulence
Adaptation to stress can often be accomplished through immediate physiological responses involving induction or repression of stress response genes. Rapid adaptation can also occur through epigenetic mechanisms, in which changes in transcription patterns, chromatin organization through histone or DNA modifications, and protein folding (e.g. prions) can be maintained across generations in the absence of changes in DNA sequence [42–44]. Adaptive evolution experiments in S. cerevisiae indicated that levels of epigenetic gene silencing can impact, through population size expansion, the rate of acquisition of novel alleles that further enhance silencing. This observation suggests a connection between epigenetic mechanisms and accelerated adaptation [45]. The adaptive mechanisms described above occur either in parallel to, or are followed by changes at the genetic level, the focus of this section.
Studies on natural and clinical isolates have identified different genetic factors that aid in the adaptation of baker’s yeast to stress environments (Figure 1, Key Figure). Many of these factors (Figure 2A; Table 2) have been associated with exposure to human associated environments. They include: 1. Multi-site heterozygosity and 2. Mosaicism, which offer wider explorations of phenotypes in the progeny of an organism. 3. High mutation rates, which provide a source of mutations that can accelerate adaptation. 4. Aneuploidy, or change in the number of one or more chromosomes, provides a transient adaptive mechanism. 5. Polyploidy, which is also observed in natural isolates, and can generate genome instability and variability associated with rapid adaptation.
Figure 1. Factors aiding in adaptation of S. cerevisiae clinical isolates.
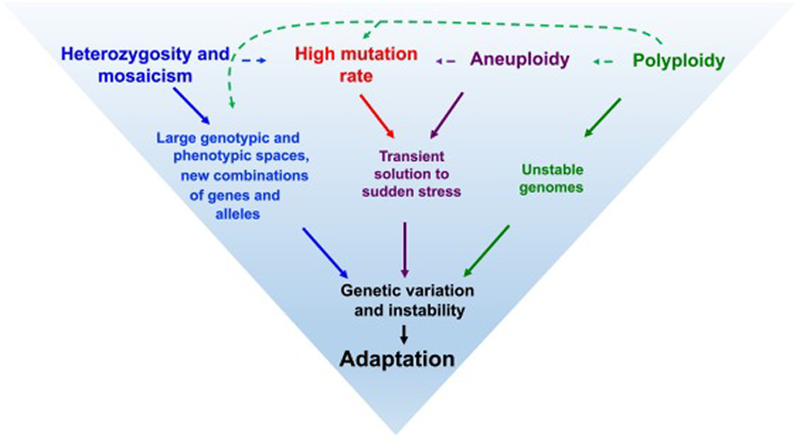
Interrelated factors that lead to genetic variation and genomic instability help baker’s yeast adapt to growth in clinical environments. Heterozygosity and mosaicism of isolates result in spore clones accessing larger genotypic and phenotypic spaces and also give rise to new combinations of genes and alleles, which aid in adaptation to new environments. The allelic variation and hybrid incompatibility resulting from heterozygosity and mosaicism can also cause variation in mutation rates. Aneuploidy and high mutation rates provide a transient advantage to sudden stress conditions and aneuploidy generates large phenotypic variations, which may also lead to high mutation rates. Polyploids show genome instability phenotypes that include an increase in the frequency of chromosome mis-segregation events that lead to aneuploidy, higher mutation rates, and larger genotypic and phenotypic spaces, all of which facilitate adaptation [82].
Figure 2. Genetic and phenotypic properties of clinical and nonclinical isolates of yeast.
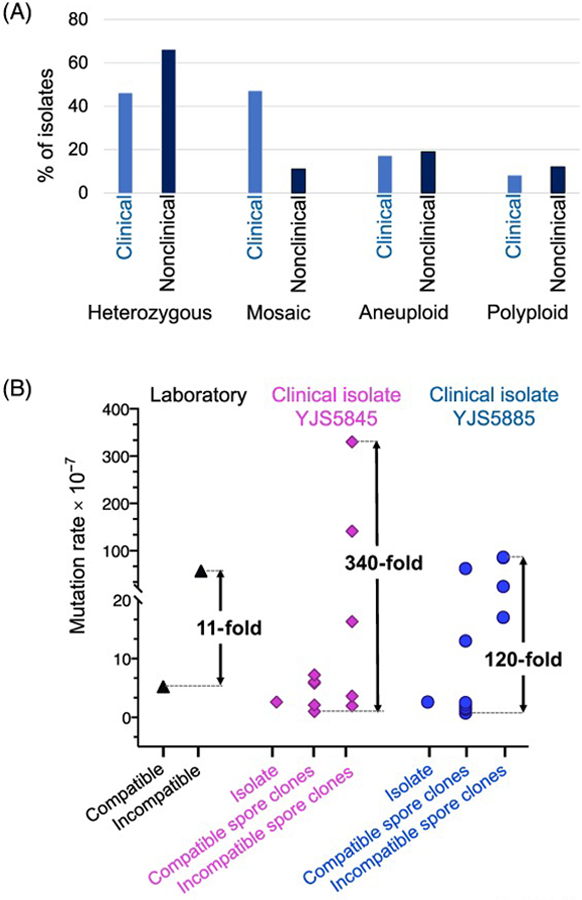
A. Proportions of 107 clinical and 904 nonclinical isolates from the 1011 S. cerevisiae genome project [9] with a given genetic property. See Table 2 for details. For the heterozygous and polyploid categories, only natural isolates (107 clinical, 687 nonclinical) were analyzed. B. Mutation rate variation observed in a clinical isolate. Mutation rates were determined using a plasmid-based frameshift reversion assay of an MLH1-PMS1 compatible laboratory strain (S288c) and an isogenic derivative containing an incompatible MLH1-PMS1 combination. This assay detects frameshift mutations that restore the reading frame in a homopolymeric run of nucleotides inserted into KanMX. In wild-type strains, the reversion rate of such events (~10−7) [10] is considerably higher than seen for base substitutions (~5 × 10−10) [101]. Mutation rates of the heterozygous diploid clinical isolates YJS5845 and YJS5885 were also determined, as well as the rates for six compatible and five incompatible spore clones of YJS5845 and eight compatible and four incompatible spore clones of YJS5885. Larger variations in mutation rate were observed between incompatible and compatible spore clones of YJS5845 (340-fold) and YJS5885 (120-fold) compared to the S288c laboratory strain (11-fold). Adapted from [10].
Table 2.
Adaptive mechanisms observed in S. cerevisiae clinical isolate studies.
| Adaptive mechanism | Description |
|---|---|
| Multi-site heterozygosity | 794 natural isolates [9] • 46% of 107 clinical origin isolates were heterozygous • 66% of 687 non-clinical origin isolates were heterozygous |
| Higher levels of heterozygosity in clinical isolates [15] • 37%−59% of total SNPs were heterozygous in clinical isolates • 0.9–15% of total SNPs were heterozygous in non-clinical isolates | |
| Mosaicism | 1011 natural isolates [9] • 47% of 107 clinical isolates belong to mosaic clades • 11% of 904 non-clinical isolates belong to mosaic clades |
| 144 natural isolates [8] • Approximately 19% of the 132 clinical isolates belong to mosaic clades • None of the 12 non-clinical isolates are mosaics | |
| 100 natural isolate segregants [11] • 63% of 43 clinical strains belong to mosaic clades • 33% of 57 non-clinical strains belong to mosaic clades | |
| High mutation rate | Clinical isolates YJS5885 and YJS5845 with a genetic incompatibility in mismatch repair genes MLH1 and PMS1 generate spore clones with a range of mutation rates [10]. |
| Aneuploidy | 1011 natural isolates [9] • 17% of 107 clinical isolates are aneuploid, 50% of which have aneuploidies in more than one chromosome (two to seven). • 19% of 904 non-clinical isolates are aneuploid, 7.2% of which have aneuploidies in more than one chromosome (two to nine). |
| 144 natural isolates [8]. 36% contained aneuploidies, 132 of which are clinical isolates. Eight clinical isolates contained multiple aneuploidies. | |
| 93 natural isolate segregants [11] • 5% of 47 clinical strains were aneuploid. • 10% of 50 non-clinical strains were aneuploid. Two contained multiple aneuploidies (one with two, the other with three). | |
| 47 natural isolates [76] • 30% of 10 clinical isolates are aneuploid. One had aneuploidy in two chromosomes. • 24% of 37 non-clinical natural isolates are aneuploid. Two of them had aneuploidies in multiple chromosomes. One had aneuploidy in two chromosomes and the other in four chromosomes. | |
| Polyploidy | 794 natural isolates [9] • 8% of 107 clinical isolates are polyploid • 12% of 687 non-clinical isolates are polyploid |
| 32% of 144 natural isolates are polyploid, 132 of which are clinical isolates [8]. | |
| 30% of both clinical and non-clinical isolates are polyploid in a total of 137 isolates [46]. |
1. Multi-site heterozygosity
Multi-site heterozygosity is defined as the presence of allelic variation at multiple loci in the same yeast strain. In 1011 S. cerevisiae genome project [9], isolates were identified that contained 2,000–78,000 single nucleotide heterozygous sites. 63% of the 794 diploid natural isolates and 46% of the 107 clinical isolates in this project displayed multi-site heterozygosity ([9]; Table 2; Figure 2A). Interestingly, higher levels of heterozygosity were associated with isolates recovered from human-associated compared to natural environments [9, 11, 15, 46]. A simple explanation for the above findings is that actions by humans, such as consuming yeast and culturing them for beer and wine, increase the possibility of outcrossing by increasing stress conditions and providing proximity to other isolates [15, 46]. Thus, by inference, outcrossing between different isolates would lead to multi-site heterozygosity.
How is heterozygosity generated?
Baker’s yeast can reproduce asexually as well as enter meiosis infrequently for sexual reproduction to generate haploid spores. Once spores are formed, S. cerevisiae haploid cells have the potential to switch mating type (if homothallic) and autodiploidize to create a homozygous diploid or mate with other haploid progeny in the vicinity, termed as outcrossing. Heterozygosity at multiple loci can result by recent outcrossing events or from the accumulation of mutations during vegetative growth. Outcrossing in baker’s yeast, which depends on growth environment and proximity to other isolates, has been estimated to occur between 1 in 100, to 1 in 50,000 vegetative divisions [23, 47], with high rates observed in the gut of social wasps [48]. A closely related yeast, Saccharomyces paradoxus, is unable to survive in the gut of social wasps, but conditions in the gut favor their sporulation and germination and most importantly, mating with S. cerevisiae to form hybrids that can survive [48]. Mixed ancestry from two or more populations due to outcrossing would result in a larger proportion of heterozygosity in the genome and is termed mosaicism. While it is difficult to distinguish heterozygosity created by outcrossing from that created by mutation accumulation, the finding that 63% of wild isolates are heterozygotes despite a prevalence of asexual reproduction suggests that the heterozygous state is advantageous [9].
Advantages of heterozygosity.
High levels of heterozygosity in baker’s yeast, coupled with rare meiotic cycles, are thought to increase genotypic and phenotypic spaces and promote rapid adaptation to novel environments and stress conditions. High levels of heterozygosity, especially in clinical isolates, also point towards a heterosis-like advantage that allows for adaptation and survival in particular environments [46, 49]. This advantage is demonstrated by the variation seen in the phenotypes of meiotic progeny of the clinical isolate YJM311 [39, 49, 50], and in the variation in mutation rate of meiotic progeny of the clinical isolates YJS5845 and YJS5885 ([10]; Table 3).
Table 3.
Heterozygosity in an isolate results in a phenotypic range in its progeny: a few examples.
| Isolate | Phenotype of spore clones |
|---|---|
| YJM311 | 1. Varied resistance to the antifungal drug fluconazole [49]. Determined by measuring MIC50 (minimum concentration of fluconazole that inhibits 50% growth) for 288 homozygous diploid spore clones. 2. Colony biofilm complexity [49, 50]. Colony morphologies of 288 spore clones were categorized visually ranging from simple, non-biofilm phenotype to highly complex biofilm colony morphology. 3. Varied invasive growth on agar [49]. Assessed by growing yeast on agar containing low ammonium, washing the surface growing cells and quantifying the levels of invasive growth. 4. Varied growth at 42ºC [39, 49]. Determined by measuring growth on agar plates at 42ºC for the 288 spores. |
| YJS5885 | 1. 120-fold variation in mutation rate in 12 spore clones [10]. Determined by a frameshift reversion assay involving a plasmid containing a frameshift mutation in the gene encoding resistance for Geneticin. 2. Variation in colony sizes in 12 spore clones as determined by growing on agar plates at 30ºC for 2 days [10]. |
| YJS5845 | 1. 340-fold variation in mutation rate in 11 spore clones [10]. 2. Variation in colony sizes in 11 spore clones [10]. |
Importantly, loss of heterozygosity (LOH) can act to fix beneficial alleles during adaptation. This has been observed in natural baker’s yeast populations as well as in experimental evolution studies [9, 51]. Studies have shown that LOH events are common and can occur over shorter evolutionary time-scales such as in a 500-generation adaptive evolution experiment [51]. Furthermore, LOH events have been shown to be beneficial during adaptation and result from direct selection of one allele over the other [51].
How is heterozygosity maintained?
Baker’s yeast is primarily homothallic and capable of self-mating, and so it seems surprising that high levels of heterozygosity are maintained in natural isolates. The frequency of heterothallism (inability to switch mating-type to form diploids) was reported to be higher in clinical isolates than non-clinical isolates [46]. For example, based on genotyping of the HO locus, it was previously observed that four of eight clinical heterozygous isolates were heterothallic, indicating they would thus support maintenance of heterozygosity [46]. The heterozygote advantage may select for the heterothallic phenotype in these isolates. For example, in a recent study, one of two clinical isolates studied was functionally heterothallic, with its meiotic spore progeny remaining haploid [10]. This isolate did not have any defects in the open reading frame of the HO gene, so there are likely to be mutations in other loci in this isolate that confer a heterothallic phenotype. Interestingly, it was reported in a set of 28 natural isolates that there was a seven-fold higher sporulation efficiency in homozygous compared to heterozygous isolates [15]. Thus, lower sporulation efficiency may also play a role in maintaining heterozygosity. It would be interesting to see if this correlation holds for the 1011 isolates [9].
2. Mosaicism
Different isolates can often colonize and cause infection in individual immune-deficient patients. Such multiple isolate infections can provide an opportunity for isolates to outcross and generate mosaics [8, 11, 16, 52]. Isolates are classified as mosaics if their genomes contain sequences derived from more than one genetically diverse ancestor. Importantly, they may also contain high levels of genomic heterozygosity if the outcrossing events occurred relatively recently. Human influence is likely to play a major role in bringing different isolates together and generating stress conditions that promote outcrossing between such isolates. Thus, it is not a surprise that a majority of the isolates that are mosaics are isolated from human-related (e.g., clinical and wine isolates) rather than natural environments ([8, 9, 11]; Table 2; Figure 2A). Additionally, a high proportion of clinical isolates belong to mosaic clades; 47% of clinical isolates compared to 11% of non-clinical isolates, belong to mosaic clades, though it is important to note that a significant number of clinical isolates are part of the wine clade ([8, 9, 11]; Table 2; Figure 2A).
Outcrossing between different isolates with varying degrees of adaptive potential will likely lead to the generation of mosaic isolates with higher adaptive potentials because they would contain new and unexplored combinations of genes and alleles that facilitate adaptation to new environments. Mosaicism may also aid in adaptation by providing a phenotypic range in the progeny as mentioned above for heterozygosity.
3. High mutation rates
High mutation rates can accelerate adaptation to stress conditions because they provide an elevated mutation supply that can more rapidly yield beneficial mutations. Bacteria that display high mutation rates are frequently found in nature [53–60]; however, modeling analyses and molecular studies indicate that bacteria prevent the long-term fitness cost of accumulating deleterious mutations through the horizontal transfer of genes that restore a low mutation rate [55, 59, 61]. Horizontal gene transfer events are rare in fungi [62, 63], and baker’s yeast active mutators have yet to be isolated in natural environments. Recently, active mutators were identified in clinical isolates of the human fungal pathogen Cryptococcus that contain mutations in the mismatch repair gene MSH2 [64, 65], demonstrating that a high mutation rate can provide beneficial mutations for adaptation in fungi despite the associated fitness costs.
Outcrossing between isolates with high sequence divergence can result in the creation of novel mutator combinations of non-mutator variants in different genes. For example, mating between two laboratory baker’s yeast strains can yield progeny that display mutator phenotypes due to the presence of an incompatible combination of the MLH1 and PMS1 mismatch repair genes which act in a highly conserved pathway to remove DNA replication errors [10, 66]. The proteins Mlh1 and Pms1 function as a heterodimer, and a specific incompatible combination of single amino-acid polymorphisms in Mlh1 and Pms1 results in elevated mutation rates [67], which can provide an adaptive advantage to stress [68]. In the heterozygous clinical isolates YJS5885 and YJS5845, which are not mutators, MLH1-PMS1 incompatibility acts as a major contributor to high mutation rates seen in spore clones derived from either isolate [10]. Interestingly, these spore clones displayed a wide range of mutation rates, indicating the presence of extragenic suppressors and enhancers of mutation rate ([10], Figure 2B).
MLH1-PMS1 incompatibility allele combinations are rare in nature, most likely due to the detrimental effects of defects in mismatch repair on fitness [66]. Analysis of the patterns of sequence polymorphisms in DNA encompassing the PMS1 locus provided evidence that recombination to generate incompatible genotypes had occurred in the past, suggesting that natural isolates of baker’s yeast do mate and occasionally produce the incompatible genotype [67]. Only one isolate among 1011 natural isolates was found to be homozygous for the incompatibility genotype, but a spore clone of this isolate had acquired suppressor mutations and was not a mutator [9, 52, 66]. However, a diploid strain containing an MLH1-PMS1 incompatibility in the heterozygous state may have an advantage because the incompatibility is recessive and the effect of incompatibility is only observed in spore progeny [10]. The presence of the MLH1-PMS1 incompatibility could thus provide a transient advantage for adaptation, with mutator spore clones adapting to a stress condition and escaping fitness costs by acquiring suppressors, mating to nearby spore clones, or outcrossing to become non-mutators [10]. Thus, transient hypermutators are likely to permit the acquisition of drug resistance to anti-fungal agents without increasing mutational load, and may also provide a supply of mutations that can promote resistance to other stresses. Consistent with this idea, in a study where diploid baker’s yeast were evolved by sequential strong selection to three drugs, clones were identified that displayed high genetic instability, including increased mutation rate, chromosome loss, and mitotic recombination [69].
4. Aneuploidy
A change in the chromosome number from the euploid set is referred to as aneuploidy; in most cases such events are deleterious to the cell. Effects of aneuploidy could be mediated by a direct change in the expression of genes on the aneuploid chromosome due to copy number variation, or be an indirect effect due to a change in expression of a gene that regulates targets located throughout the genome [70]. Additionally, there could be a general effect of aneuploidy that is not specific to a particular chromosome. For example, an extra copy of almost any yeast chromosome caused a reduction in cellular proliferation, which was attributed to altered levels of gene products encoded by genes that reside on the extra chromosome [71]. In organisms such as Drosophila, C. elegans, mice, plants and humans, most aneuploidies are lethal [72]. Aneuploidies are observed in almost all cancerous cells, and it is debated whether they are a consequence of chromosome instability and segregation defects, or they are direct drivers of cellular transformation [72].
Despite conferring negative fitness effects, aneuploidy has been commonly observed in natural baker’s yeast and may provide an important route to natural genetic variation. Aneuploidy has been suggested to help in adaptation in environments with human association such as in brewing, baking and wine strains of yeast [73]. In the 1011 genomes project [9] the highest levels of aneuploidy (40 to 60%) were observed in isolates from sake, beer, or bakery origin. Levels were lower from other origins including, but not limited to trees, wine, human clinical, fruit, dairy and industrial sources (11 to 40%). The lowest levels of aneuploidy were observed for soil isolates (5%). One possible explanation for the high level of aneuploidy in the sake, beer and bakery isolates is that they encounter a high frequency of fluctuating stress that favors aneuploidy as an adaptation mechanism (Figure 2A; Table 2, with other examples provided). Furthermore, aneuploid isolates may be more likely to generate aneuploid progeny, as seen in the clinical isolate, YJS5845 [10]. This isolate was a mix of aneuploid and euploid cells, probably generated due to mitotic chromosome segregation defects. When sporulated in the laboratory, two of sixteen spore clones displayed aneuploidies in different chromosomes [10].
In certain circumstances, the beneficial effects of aneuploidy could offset negative effects on cell survival [74, 75]. Aneuploidy appears to be one of the first lines of defense to stress that increases chances for survival under strong and abrupt selective pressures [75]. In baker’s yeast, sudden stress conditions induced in the laboratory selected for aneuploidy, with return to euploidy occurring when the stressor was removed [75]. When the stress condition was maintained, a return to euploidy occurred in about 2,000 generations, with a stable solution obtained through mutations that modified the expression of stress resistance genes, as seen for heat stress [75, 76]. Thus, aneuploidy may be a valuable mechanism for adaptation in clinical isolates, which live in an environment with variable and sudden stressors.
How is aneuploidy tolerated?
In many situations, an extra chromosomal copy provides tolerance to a particular stress condition [74]. For example, extra chromosomes confer resistance to heat (Chr. III), high pH (Chr. V), and the ultra violet light mimetic mutagen 4-Nitroquinoline 1-oxide (Chr. XIII) [74, 75]. Aneuploid isolates appear to better tolerate chromosome gains or losses due to dosage compensation mechanisms [76, 77]. In support of this, a previous study analyzed twelve isolates containing an extra chromosome [76]. They found that for 40% of genes located on such a chromosome, gene expression levels were lower than expected based on their dosage [76]. Furthermore, in contrast to artificially created aneuploids in the laboratory, natural aneuploids had growth rates similar to closely related euploids [76].
5. Polyploidy
Baker’s yeast is most commonly diploid in nature, but polyploidy has been associated with human interference in natural isolates of yeast [8, 9]. In the 2011 S. cerevisiae isolate study [9] 87% of 794 natural isolates were found to be diploid and 11.4% were polyploid. These polyploid isolates were enriched in human associated environments including beer, mixed-origin and African palm wine clades, although there was no significant enrichment in clinical isolates ([9]; Table 2; Figure 2A). Previously, a diverse natural population was analyzed [46] and it was estimated that 70–80% of their isolates were diploid, with the remaining 20–30% of isolates being triploid or tetraploid. Another group [78] found that 95% of more than 200 wine strains were diploid. Polyploidy has also been observed in clinical isolates [8, 46]. In a study of 144 isolates, the majority of which were clinical (132), 34% were polyploid [8].
In baker’s yeast, polyploid genomes are less stable than haploid and diploid genomes, and have been hypothesized to act as drivers of adaptation [46]. There is also variation in adaptation rates of haploids compared to diploids; haploids have been observed to adapt faster and more commonly by recessive mutations whereas diploids accumulate mostly dominant mutations [79]. Polyploid baker’s yeast display genetic instability phenotypes that are thought to arise as a result of their tendency to mis-segregate chromosomes [80–83]. Consistent with this idea, polyploid baker’s yeast genomes are associated with more than a two-fold increase in aneuploidy [8] and display higher mutation rates [82, 84]. Laboratory evolution experiments support these ideas, but such support depends on the stress condition used [82, 85]. For example, a group compared adaptation of tetraploid, diploid, and haploid S. cerevisiae to a low carbon environment and found that tetraploids adapted significantly faster. This more rapid adaptation was due to a higher rate of beneficial mutations as well as a higher fitness conferred by the mutations [82]. In contrast, another group showed in C. albicans that both higher and lower ploidy states can be advantageous under different stress conditions [85]. Since polyploidy can be beneficial in adaptation by multiple mechanisms, it may be used by clinical isolates to adapt to the unpredictable environment of the human host.
Conclusions
Adaptation to novel stress environments often requires organisms to incur genetic changes. Such changes can be accelerated in organisms by high mutation rates, acquiring specific stress resistance genes through horizontal gene transfer, or genomic instabilities that lead to chromosome gains and losses. Clinical yeast isolates have been subjected to novel and repeated stresses aimed at suppressing their growth and survival. Thus, they must be able to rapidly and repeatedly adapt to stress conditions to survive. As summarized in this review, multi-site heterozygosity, mosaicism, transient high mutation rates, aneuploidy, and polyploidy appear to be major sources of genetic raw material for adaptation in baker’s yeast.
Analyzing more extensive collections of S. cerevisiae isolates from hospitals will be critical to determine more precisely the prevalence of phenotypes and genetic mechanisms that lead to these phenotypes. To aid in this, a plasmid-based antibiotic reversion assay was developed that can rapidly analyze mutator phenotypes in natural/clinical isolates and their spore clones (Figure 2B, [10, 66]). This approach can also be used to identify loci in natural isolates that impact mutation rate [86–88].
It is critical to identify new targets that can be exploited by anti-fungal compounds, recognizing that such drugs need to selectively act on eukaryotic pathogens without disrupting essential cellular processes conserved in humans. Promising areas of research include identifying targets in ergosterol biosynthesis [33], calcium homeostasis [89], and the cyclic AMP/Protein Kinase A (cAMP/PKA) nutrient sensing pathway [90–92]. The cAMP/PKA pathway has received a lot of attention because disrupting it in Cryptococcus neoformans resulted in reduced virulence of the pathogen in a mouse model [90], and alterations in cAMP signaling affect pseudohyphal growth in S. cerevisiae, a response important for cell adhesion and invasion, phenotypes linked to virulence [92]. Phage therapies have also been proposed to target fungal infections; recent studies identified bacteriophage present in isolates of the gram-negative bacteria P. aeruginosa that sequester resources such as iron that are important for the growth of fungal pathogens (reviewed in [93]). Finally, methods to rapidly detect probiotic fungi have been developed [24] which will help identify sources of infection in humans, and provide information on whether probiotic yeast found in infections should still be available for consumption.
New gene targets can be identified by evolving fungi under conditions that mimic a human environment; these include high temperature, nutrient limitation, and the presence of antifungal agents. Using systematic and experimental evolution approaches, researchers have identified mutations in diploid and haploid yeast that confer adaptation to such conditions (see examples from [69, 79, 94]). Interestingly systematic screens, which involved deleting or overexpressing genes, provided predictive power for mutations identified by experimental evolution [94]. Also, computational approaches have been developed to identify interactions between potential driver mutations based on the prediction that genes that display genetic or physical interactions are more likely to be seen in evolved genotypes [95]. Such experiments can generate candidate target genes and determine if they are mutated at a higher frequency in clinical versus non-clinical isolates, with the goal of focusing on adaptive loci enriched in clinical isolates. Furthermore, molecular evolution studies performed with baker’s yeast grown under different stress conditions have identified beneficial mutations that drive adaptation and propagate the population [96, 97]. These studies were performed by barcoding individual cells and tracking their lineages through whole genome sequencing to identify adaptive mutations. Such methods will be helpful to identify adaptive mutations in clinical yeast isolates grown in a mouse model. Lastly, CRISPR technologies have been developed to rapidly test candidate target genes by reconstructing genotypes in naive strains [98, 99].
It is clear that excessive use of antifungals plays an important role in the generation of fungal infections. Their use can result in the development of multi-drug resistant fungi [26, 27], and also affect the protective commensal fungal population that aids in immunity [27]. The use of live cultures of S. cerevisiae and S. boulardii in probiotics has also resulted in infections in immunocompromised patients [5, 21, 100]. It is unlikely that restricting doses of antifungals will be effective in controlling the spread of fungal pathogens that have already become resistant to these compounds (e.g. C. auris; [26]). Thus, it will also be important to control anti-fungal resistance by developing measures that reduce human influence in the promotion of outcrossing [49]. Recently outcrossed isolates, identified as mosaics, appear to have a major advantage in the clinical environment [8, 9, 11]. It seems reasonable to take precautions to restrict activities that cause intensive outcrossing; these could include strict regulations in the transport of live isolates by the public, research institutes, breweries and bakeries. We also need to rethink the inclusion of S. cerevisiae in probiotic preparations. Additionally, learning more about adaptation mechanisms and genetic signatures of clinical isolates will help in generation of anti-fungal drugs to fight these infections.
Outstanding questions.
How prevalent are phenotypes such as high temperature growth, resistance to antifungal drugs, colony phenotype switching, and pseudohyphal growth in clinical yeast isolates? Have all loci been identified that are responsible for these phenotypes?
How frequently are heterozygosity, mosaicism, high mutation rates, aneuploidy and polyploidy seen in clinical isolates that facilitate adaptation?
Can loci be mapped that enhance or suppress mutation rates in natural isolates and their spore clones?
Adaptive evolution experiments have yet to be performed for clinical yeast isolates grown in an immune-compromised mouse model. What are the evolutionary landscapes and mutational signatures that accompany adaptation of clinical yeast isolates in a mammalian host environment? Can these approaches efficiently identify new targets for anti-fungal drugs?
Highlights.
Excessive use of antifungals in hospitals and agriculture is thought to promote the origin and spread of drug-resistant fungi that are pathogenic in humans.
Hospital patients provide a unique and stressful environment for baker’s yeast. Clinical yeast isolates are subjected to novel and repeated stresses such as high temperature and exposure to antifungals aimed at suppressing their growth and survival.
Genomic heterozygosity, mosaicism, high mutation rates, aneuploidy, and polyploidy appear to be major sources of genetic raw material for adaptation in baker’s yeast.
Recent studies of natural baker’s yeast isolate collections, including the sequencing of a set of 1011 isolates which contain over 100 clinical samples, are providing useful models from which to gain insights into adaptation to stressful environments, identify anti-fungal targets, and develop strategies to limit the spread of drug-resistant fungi.
Acknowledgements
We are grateful to Joseph Schacherer for discussions and comments on the manuscript, Michael McGurk for discussions, and anonymous reviewers for critical comments. V.R. and E.A. were supported by National Institutes of Health (NIH) grant GM-053085. C.F.A. was supported by NIH grant GM-095793. The authors have declared that no competing interests exist.
Glossary
- Aneuploidy
Loss or gain of one or few chromosomes that causes a change in the chromosome number from a multiple of the haploid set (euploidy).
- Autodiploidization
Mating between mother and daughter cells after mating type switching.
- Epigenetic mechanisms
Inheritance of phenotypes resulting from alterations in cellular mechanisms, such as changes in transcription patterns, chromatin organization, and protein folding, that occur in the absence of genetic changes.
- Heterosis
Higher fitness of the hybrid resulting from mating between different strains/isolates. Also referred to as hybrid vigor.
- Heterothallic
Baker’s yeast cells that cannot undergo mating type switching and can be maintained as stable haploids.
- Homothallic
Baker’s yeast cells that have the ability to undergo mating type switching. This allows mating (auto-diploidization) between mother and daughter cells.
- Isolate
An isolate is found in nature, whereas a strain has been manipulated in the laboratory.
- Loss of heterozygosity
Loss of genetic information derived from one parent. Such events can be caused by chromosomal deletions and loss, gene conversion, and mitotic recombination.
- Mosaic
Mixed genotype derived from two or more genetically different populations. An isolate is classified as mosaic when it has multiple sources of ancestry and when it has less than 60% ancestry from any one population [11]. Mosaic isolates are polymorphic for a majority of segregating sites and are derived from outbreeding [9,14]. They frequently manifest as isolated branches on a phylogenetic tree [9].
- Multi-site heterozygosity
Allelic variation seen in the same individual at multiple genetic loci. Heterozygous isolates were classified as having more than 5% heterozygous sites out of a total number of SNPs (single nucleotide polymorphisms) compared to the reference genome S288c [9]. In 1011 isolates, heterozygosity ranged from 0.63–6.56 SNP sites per kilobase in heterozygous isolates [9]. There were 2,000–78,000 total heterozygous SNPs in the 1011 isolates [9].
- Outcrossing
Mating between genetically different strains/isolates.
- Polyploidy
Having more than two complete sets of chromosomes, e.g.: triploid has three copies of each chromosome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavalieri D et al. (2003) Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol 57, S226–232. [DOI] [PubMed] [Google Scholar]
- 2.McGovern PE et al. (2004) Fermented beverages of pre- and proto-historic China. Proc Natl Acad Sci U S A 101, 17593–17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackerman AL and Underhill DM (2017) The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann Transl Med 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui L et al. (2015) Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 191, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Llanos R et al. (2011) In vivo virulence of commercial Saccharomyces cerevisiae strains with pathogenicity-associated phenotypical traits. Int J Food Microbiol 144, 393–399. [DOI] [PubMed] [Google Scholar]
- 6.Nash AK et al. (2017) The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai U et al. (2014) Invasive Saccharomyces cerevisiae infection: a friend turning foe? Saudi J Kidney Dis Transpl 25, 1266–1269. [DOI] [PubMed] [Google Scholar]
- 8.Zhu YO et al. (2016) Whole Genome Analysis of 132 Clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 6, 2421–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peter J et al. (2018) Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghavan V et al. (2018) Incompatibilities in mismatch repair genes MLH1-PMS1 contribute to a wide range of mutation rates in human isolates of baker’s yeast. Genetics 210, 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strope PK et al. (2015) The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res 25, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemons KV et al. (1994) Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis 169, 859–867. [DOI] [PubMed] [Google Scholar]
- 13.Diezmann S and Dietrich FS (2009) Saccharomyces cerevisiae: population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLoS One 4, e5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liti G et al. (2009) Population genomics of domestic and wild yeasts. Nature 458, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magwene PM et al. (2011) Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 108, 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schacherer J et al. (2009) Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458, 342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller LA et al. (2011) Genome-wide association analysis of clinical vs. nonclinical origin provides insights into Saccharomyces cerevisiae pathogenesis. Mol Ecol 20, 4085–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Torrado R et al. (2012) Clinical Saccharomyces cerevisiae isolates cannot cross the epithelial barrier in vitro. Int J Food Microbiol 157, 59–64. [DOI] [PubMed] [Google Scholar]
- 19.Piarroux R et al. (1999) Are live Saccharomyces yeasts harmful to patients? Lancet 353, 1851–1852. [DOI] [PubMed] [Google Scholar]
- 20.Munoz P et al. (2005) Saccharomyces cerevisiae fungemia: an emerging infectious disease. Clin Infect Dis 40, 1625–1634. [DOI] [PubMed] [Google Scholar]
- 21.Enache-Angoulvant A and Hennequin C (2005) Invasive Saccharomyces infection: a comprehensive review. Clin Infect Dis 41, 1559–1568. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Torrado R and Querol A (2015) Opportunistic strains of Saccharomyces cerevisiae: A potential risk sold in food products. Front Microbiol 6, 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsit S and Dequin S (2015) Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res 15, pii: fov067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imre A, et al. (2019) A new, rapid multiplex PCR method identifies frequent probiotic origin among clinical Saccharomyces isolates. Microbiol Res 227, 126298. [DOI] [PubMed] [Google Scholar]
- 25.Davies J and Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhary A et al. (2017) Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13, e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonardi I et al. (2018) CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kodedova M and Sychrova H (2015) Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10, e0139306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson JB et al. (2004) Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 168, 1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson JB et al. (2009) Gene expression and evolution of antifungal drug resistance. Antimicrob Agents Chemother 53, 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selmecki AM et al. (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5, e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White TC et al. (2002) Resistance mechanisms in clinical isolates of Candida albicans. Antimicrobial Agents and Chemotherapy 46, 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharya S et al. (2018) Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha H et al. (2008) Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmetz LM et al. (2002) Dissecting the architecture of a quantitative trait locus in yeast. Nature 416, 326–330. [DOI] [PubMed] [Google Scholar]
- 36.Weiss CV, et al. (2018) Genetic dissection of interspecific differences in yeast thermotolerance. Nat Genet 50, 1501–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clemons KV et al. (1996) Colony phenotype switching in clinical and non-clinical isolates of Saccharomyces cerevisiae. J Med Vet Mycol 34, 259–264. [DOI] [PubMed] [Google Scholar]
- 38.Halme A et al. (2004) Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116, 405–415. [DOI] [PubMed] [Google Scholar]
- 39.McCusker JH et al. (1994) Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 degrees C and form pseudohyphae. Infect Immun 62, 5447–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shively CA, et al. (2013) Genetic networks inducing invasive growth in Saccharomyces cerevisiae identified through systematic genome-wide overexpression. Genetics 193, 1297–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Q et al. (2014) Pooled segregant sequencing reveals genetic determinants of yeast pseudohyphal growth. PLoS Genet 10, e1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabrizio P, et al. (2019) Histone methylation and memory of environmental stress. Cells 8, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westergard L, and True HL (2014) Wild yeast harbour a variety of distinct amyloid structures with strong prion-inducing capabilities Mol Microbiol 92, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yona AH, et al. (2015) A relay race on the evolutionary adaptation spectrum. Cell 163, 549–559. [DOI] [PubMed] [Google Scholar]
- 45.Stajic D et al. (2019) Epigenetic gene silencing alters the mechanisms and rate of evolutionary adaptation. Nat Ecol Evol 3, 491–498. [DOI] [PubMed] [Google Scholar]
- 46.Muller LA and McCusker JH (2009) Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol Ecol 18, 2779–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruderfer DM et al. (2006) Population genomic analysis of outcrossing and recombination in yeast. Nat Genet 38, 1077–1081. [DOI] [PubMed] [Google Scholar]
- 48.Stefanini I et al. (2016) Social wasps are a Saccharomyces mating nest. Proc Natl Acad Sci U S A 113, 2247–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magwene PM (2014) Revisiting Mortimer’s genome renewal hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast. Adv Exp Med Biol 781, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Granek JA et al. (2013) The genetic architecture of biofilm formation in a clinical isolate of Saccharomyces cerevisiae. Genetics 193, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smukowski Heil CS et al. (2017) Loss of Heterozygosity Drives Adaptation in Hybrid Yeast. Mol Biol Evol 34, 1596–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skelly DA et al. (2017) Known mutator alleles do not markedly increase mutation rate in clinical Saccharomyces cerevisiae strains. Proc Biol Sci 284, pii: 20162672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boe L et al. (2000) The frequency of mutators in populations of Escherichia coli. Mutat Res 448, 47–55. [DOI] [PubMed] [Google Scholar]
- 54.Chao L and Cox EC (1983) Competition between high and low mutating strains of Escherichia coli. Evolution 37, 125–134. [DOI] [PubMed] [Google Scholar]
- 55.Denamur E et al. (2000) Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103, 711–721. [DOI] [PubMed] [Google Scholar]
- 56.LeClerc JE et al. (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- 57.Taddei F et al. (1997) Role of mutator alleles in adaptive evolution. Nature 387, 700–702. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka MM et al. (2003) The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Townsend JP et al. (2003) Horizontal acquisition of divergent chromosomal DNA in bacteria: effects of mutator phenotypes. Genetics 164, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giraud A et al. (2001) The rise and fall of mutator bacteria. Curr Opin Microbiol 4, 582–585. [DOI] [PubMed] [Google Scholar]
- 61.Giraud A et al. (2001) Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291, 2606–2608. [DOI] [PubMed] [Google Scholar]
- 62.Fitzpatrick DA (2012) Horizontal gene transfer in fungi. FEMS Microbiol Lett 329, 1–8. [DOI] [PubMed] [Google Scholar]
- 63.Hall C et al. (2005) Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell 4, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Billmyre RB et al. (2017) Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. Elife 6, e28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyce KJ et al. (2017) Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. MBio 8, e00595–00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bui DT et al. (2017) Mismatch repair incompatibilities in diverse yeast populations. Genetics 205, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heck JA et al. (2006) Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc Natl Acad Sci U S A 103, 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bui DT et al. (2015) A genetic incompatibility accelerates adaptation in yeast. PLoS Genet 11, e1005407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coehlo MC, et al. (2019) Heterozygous mutations cause genetic instability in a yeast model of cancer evolution. Nature 566, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cromie GA et al. (2017) Dissecting gene expression changes accompanying a ploidy-based phenotypic switch. G3 7, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres EM et al. (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924. [DOI] [PubMed] [Google Scholar]
- 72.Gordon DJ et al. (2012) Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13, 189–203. [DOI] [PubMed] [Google Scholar]
- 73.Rancati G and Pavelka N (2013) Karyotypic changes as drivers and catalyzers of cellular evolvability: a perspective from non-pathogenic yeasts. Semin Cell Dev Biol 24, 332–338. [DOI] [PubMed] [Google Scholar]
- 74.Pavelka N et al. (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yona AH et al. (2012) Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci U S A 109, 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hose J et al. (2015) Dosage compensation can buffer copy-number variation in wild yeast. Elife 4, e05462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cromie GA and Dudley AM (2015) Aneuploidy: Tolerating Tolerance. Curr Biol 25, R771–773. [DOI] [PubMed] [Google Scholar]
- 78.Cubillos FA et al. (2009) Self-fertilization is the main sexual reproduction mechanism in native wine yeast populations. FEMS Microbiol Ecol 67, 162–170. [DOI] [PubMed] [Google Scholar]
- 79.Marad DA, et al. (2018) Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids. Nat Ecol Evol 2, 882–889. [DOI] [PubMed] [Google Scholar]
- 80.Mayer VW and Aguilera A (1990) High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat Res 231, 177–186. [DOI] [PubMed] [Google Scholar]
- 81.Rancati G et al. (2008) Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Selmecki AM et al. (2015) Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Storchova Z et al. (2006) Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547. [DOI] [PubMed] [Google Scholar]
- 84.Scott AL et al. (2017) The Influence of polyploidy on the evolution of yeast grown in a sub-optimal carbon source. Mol Biol Evol 34, 2690–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gerstein AC et al. (2017) Ploidy tug-of-war: Evolutionary and genetic environments influence the rate of ploidy drive in a human fungal pathogen. Evolution 71, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Demogines A et al. (2008) Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet 4, e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Demogines A et al. (2008) Identification and dissection of a complex DNA repair sensitivity phenotype in Baker’s yeast. PLoS Genet 4, e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gou L et al. (2019) The genetic basis of mutation rate variation in Yeast. Genetics 211, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Odom AR (2014) The triphenylethylenes, a novel class of antifungals. mBio 5, e01126–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caza M, and Kronstad JW (2019) cAMP/Protein kinase A pathway regulates virulence and adaptation to host conditions in Cryptococcus neoformans. Front Cell Infect Microbiol 9, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang G et al. (2019) Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol Microbiol 111, 6–16. [DOI] [PubMed] [Google Scholar]
- 92.Kayikci O, and Magwene PM (2018) Divergent Roles for cAMP–PKA signaling in the regulation of filamentous growth in Saccharomyces cerevisiae and Saccharomyces bayanus. G3 8, 3529–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gorski A et al. (2019) Perspectives of phage therapy in non-bacterial infections. Front. Microbiol 9, 3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Payen C, et al. (2016) High-throughput identification of adaptive mutations in experimentally evolved yeast populations. PLoS Genet 12, e1006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fisher KJ, et al. (2019) Detecting genetic interactions using parallel evolution in experimental populations. Phil Trans R Soc B 374, 20180237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blundell JR et al. (2019) The dynamics of adaptive genetic diversity during the early stages of clonal evolution. Nat Ecol Evol 3, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy SF et al. (2015) Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanchez JC, et al. (2019) Phenotypic and Genotypic Consequences of CRISPR/Cas9 Editing of the Replication Origins in the rDNA of Saccharomyces cerevisiae. Genetics pii: genetics.302351.2019. doi: 10.1534/genetics.119.302351. [DOI] [PMC free article] [PubMed]
- 99.Xie Z-X (2017) “Perfect” designer chromosome V and behavior of a ring derivative. Science 355, eaaf4704. [DOI] [PubMed] [Google Scholar]
- 100.Herbrecht R and Nivoix Y (2005) Saccharomyces cerevisiae fungemia: an adverse effect of Saccharomyces boulardii probiotic administration. Clin Infect Dis 40, 1635–1637. [DOI] [PubMed] [Google Scholar]
- 101.Lang GI and Murray AW (2008) Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


