Abstract
Objective:
To date, The Cancer Genome Atlas (TCGA) has provided the most extensive molecular characterization of invasive cervical cancer (ICC). Analysis of reverse phase protein array (RPPA) data from TCGA samples showed that cervical cancers could be stratified into 3 clusters exhibiting significant differences in survival outcome: hormone, EMT, and PI3K/AKT. The goals of the current study were to: 1) validate the TCGA RPPA results in an independent cohort of ICC patients and 2) to develop and validate an algorithm encompassing a small antibody set for clinical utility.
Methods:
Subjects consisted of 2 ICC patient cohorts with accompanying RPPA and clinical-pathologic data: 155 samples from TCGA (TCGA-155) and 61 additional, unique samples (MCW-61). Using data from 173 common RPPA antibodies, we replicated Silhouette clustering analysis in both ICC cohorts. Further, an index score for each patient was calculated from the survival-associated antibodies (SAAs) identified using Random survival forests (RSF) and the Cox proportional hazard regression model. Kaplan-Meier survival analysis and the log-rank test were performed to assess and compare cluster or risk group survival outcome.
Results:
In addition to validating the prognostic ability of the proteomic clusters reported by TCGA, we developed an algorithm based on 22 unique antibodies (SAAs) that stratified women with ICC into low-, medium-, or high-risk survival groups.
Conclusions:
We provide a signature of 22 antibodies which accurately predicted survival outcome in 2 separate groups of ICC patients. Future studies examining these candidate biomarkers in additional ICC cohorts is warranted to fully determine their clinical potential.
Keywords: Cervical cancer, reverse phase protein array, survival risk, prognostic biomarkers, The Cancer Genome Atlas (TCGA)
Introduction
In March 2017, The Cancer Genome Atlas (TCGA) published data on the largest molecular characterization of invasive cervical cancer (ICC; [1]). The study included resampling-based consensus clustering analysis of protein expression data from 192 antibodies on a reverse phase protein analysis (RPPA) platform and identified 3 distinct clusters defined as EMT (epithelial-mesenchymal transition), PI3K/AKT, and hormone (see Figure 3 of [1]). In their analysis, consensus clusters of 155 samples were found and a silhouette clustering approach identified the core members of each RPPA cluster with a silhouette width >0.02 (n=115) associated with 5-year survival. Patients falling within the EMT cluster had the poorest survival, while those within the hormone cluster had the best. Although very informative, these results have not yet been validated in other datasets and need further refinement to translate to a clinical approach.
Protein expression arrays offer complementary information to RNA sequencing data. Frequently, however, proteomic data do not correlate directly with those obtained from RNA sequencing, suggesting that protein content might reflect a tissue’s dynamic state more accurately than its nucleic acid content. mRNA expression studies also do not reflect posttranslational modifications that can heavily influence a protein’s structure and/or function and thus its potential role in tumorigenesis. Moreover, a protein signature that predicts poor clinical outcome in a patient population can also provide important information for steering therapeutic decisions. The main goal of the current study was to validate RPPA-based prediction of survival results in an independent cohort of subjects with ICC. We also sought to build a prediction model with a reduced number of proteins in the prognostic signature that could be assessed by simpler technologies than RPPA, thus making it more applicable to clinical management. Indeed, we identified a novel signature that associates with patient outcome using a the TCGA samples and datasets developed in this project. Studies like these that focus on prognostic markers with clinical utility are greatly needed to improve our understanding of cervical cancer and to provide useful data for generating targeted therapies for this disease which has limited options for cure in metastatic cases.
Material and methods
Study Population
Subjects included 155 women with ICC and associated TCGA RPPA data [1], as well as 61 additional ICC samples with associated clinical-pathologic features recruited through the CerGE study (MCW-61 cohort; Table 1) [2]. All samples were obtained at the time of cancer diagnosis and prior to treatment. Procedures for the collection, shipping, processing, and quality management of samples used in the current study were generated according to the guidelines provided in The NCI Best Practices for Biospecimen Resources (https://biospecimens.cancer.gov/bestpractices/). ICC biopsies from the operating room or clinic were flash frozen in liquid nitrogen within 1 hour of their removal and stored at −80°C. Human papillomavirus (HPV) typing was performed as previously published for each study [1–3]. There were 33 ICC tumors with HPV typing data from both MCW and TCGA, and the results showed excellent concordance except for 1 discrepant case for which MCW typing identified 2 HPV types, while only 1 was found by TCGA. The study protocol was approved by the institutional review board at the Medical College of Wisconsin and all patients consented.
Table 1.
Patient characteristics.
| TCGA-155 | MCW-61 | ||
|---|---|---|---|
| Age (years) | 47 (38, 56) | 45 (37, 60) | |
| Race | White | 112 (72) | 43 (70) |
| African American | 11 (7) | 18 (30) | |
| Asian | 15 (10) | 0 | |
| American Indian or Alaska Native | 2 (1) | 0 | |
| Other | 15 (10) | 0 | |
| Ethnicity | |||
| Hispanic or Latino | 12 (8) | 2 (3) | |
| Not Hispanic or Latino | 99 (64) | 40 (66) | |
| Not specified | 44 (28) | 19 (31) | |
| Stage | |||
| 1 (Ib, Ib1, Ib2) | 97 (63) | 40 (66) | |
| 2 (IIa, IIb) | 31 (20) | 8 (13) | |
| 3 (IIIb, IVa, IVb) | 24 (15) | 13 (21) | |
| Unstaged | 3 (2) | 0 | |
| Histology | |||
| Adenocarcinoma | 22 (14) | 9 (15) | |
| Adenosquamous | 3 (2) | 5 (8) | |
| Squamous cell carcinoma | 130 (84) | 47 (77) | |
| HPV clade | A7 | 43 (28) | 25 (41) |
| A9 | 100 (64) | 32 (52) | |
| Other | 4 (3) | 1 (2) | |
| Negative | 8 (5) | 3 (5) |
Median age (interquartile range) or n (%) are presented.
RPPA
Fresh-frozen ICC tumors (10 mg) from the MCW-61 cohort were shipped to The Functional Proteomics RPPA Core Facility at The University of Texas MD Anderson Cancer Center (https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core.html) for protein extraction and RPPA identical to that employed by TCGA [1]. Tumor samples contained ≥60% tumor nuclei and ≤20% necrosis. Protein was extracted as previously described [1] and specific protein expression was examined using 282 unique antibodies. Of note, TCGA RPPA analysis employed 192 antibodies, 173 of which were shared across both RPPA platforms (Table S1). The 19 antibodies used in TCGA but subsequently excluded from the RPPA platform are: 4EBP1PT70, ACETYLATUBULIN_LYS40, ALPHACATENIN, ASNS, BRCA2, CD20, CIAP, CYCLINE2, ERALPHAPS118, ERK2, KU80, LKB1, MRE11, P62LCKLIGAND, P90RSK, P90RSKPT359S363, PRDX1, S6, and VHL. These 19 antibodies were excluded from the RPPA platform for 2 reasons: 1) the antibody used was discontinued in the market and/or 2) there was a change in antibody validation status due to antibody lot/batch variation.
Statistical Analysis
There were 173 common proteins across the 2 RPPA platforms (see Table S1) in 2014 (TCGA-155) and 2015 (MCW-61) used by the RPPA Core Facility. Following the methodology published by the TCGA [1], we performed consensus clustering and silhouette clustering [4] on the expression of 173 common proteins in the TCGA-155 cohort and validated them in the MCW-61 cohort (Figure 1). Silhouette width values (defined as the ratio of each sample’s average distance to patients in the same cluster to the smallest distance to patients not in the same cluster) were calculated for the TCGA-155 cohort and patients with a silhouette cluster width >0.02 were identified as core patients. MCW-61 patients were assigned to a cluster with minimum Euclidean distance to the centroids, which were calculated using the silhouette core patients of TCGA-155 cohort. Then, patients with a silhouette cluster width >0.02 were identified as core patients of MCW-61 cohort. Kaplan-Meier survival analysis was then performed to assess 5-year survival and the log-rank test was used to compare survivals of the different clusters in the silhouette core patients. Raw RPPA expression data from the 173 common antibodies across RPPA platforms is provided in Table S2.
Figure 1. Cluster data survival analysis methods.
See Statistical analysis section of Materials and Methods for detailed description of analyses.
To further explore RPPA’s ability to identify small protein groups that could accurately predict survival, we performed a machine learning technique called Random forests [5]. Random Forests are collections of decision trees that are grown by recursive binary partitioning on a different, random subsample of the training data. Random survival forests (RSF) [6], an extension of Random forests, was employed using the TCGA-155 data and a classification tree was generated using survival status as the outcome variable. The predictors were the 173 common antibodies across the 2 RPPA platforms. The optimal candidate variables were chosen using a log-rank splitting rule. The resulting parameters used were: (1) Number of trees: 10000, (2) Forest terminal node size: 3, and (3) Number of variables tried at each split: 14.
RSF and a Cox proportional hazard regression model were further used to identify the antibodies with expression data that associated with survival status (survival-associated antibodies: SAAs). Cox proportional hazard regression models examined 1 protein at a time, while RSF was used to account for interdependent relationships of many proteins. RSF variable importance (VIMP) and minimal depth were used for variable selection. VIMP ranks the proteins by their impact on a forest’s predictive ability, while minimum depth is based on the forest construction [7]. To ensure a more robust selection of informative proteins, we used a combination of VIMP and minimal depth or VIMP and Cox proportional hazard regression models.
Graphic assessment of standardized score process versus follow-up time and Kolmogorov-type supremum test were used to check the proportional hazards assumption for each predictor for Cox proportional hazard regression model. If the test were rejected implying the assumption of proportionality was rejected, then to see if the assumption could hold, with modification, we included an interaction of the predictor and a function of survival time (log) in the final model. An interaction term was generated and included in the final model. The proteins that remained significant using the selection criteria that we have described, namely using RSF VIMP & COX, were chosen for further examination.
An index score for each patient was calculated from the significant SAAs using RSF and the Cox proportional hazard regression model. The direction of the hazard for each protein was determined by Cox proportional hazard regression. An index score was calculated as the sum of genes with increased hazard minus the sum of genes with reduced hazard. The terciles of the index score were used to divide patients into 3 survival risk groups (LR – low risk; MR – medium risk; HR – high risk). We then used Kaplan-Meier survival analysis and the log-rank test to assess risk group survival and to compare risk group survivals, respectively.
Ancillary clinical-pathologic associations of SAA protein expression were performed on the combined TCGA-155 and MCW-61 cases. Patients’ characteristics were summarized as median and interquartile range (IQR) or n (%). Non-parametric Mann-Whitney-Wilcoxon tests were used to compare differences between continuous variables, while a Chi-square test or Fisher’s exact test was employed to examine associations between categorical variables. A p<0.05 was considered significant. No correction for multiple testing was done since RSF and Cox proportional hazard were used for selection of candidates. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC) and R version 3.4.2 (Vienna, Austria).
Results
Validation of TCGA cluster survival results in the MCW-61 cohort
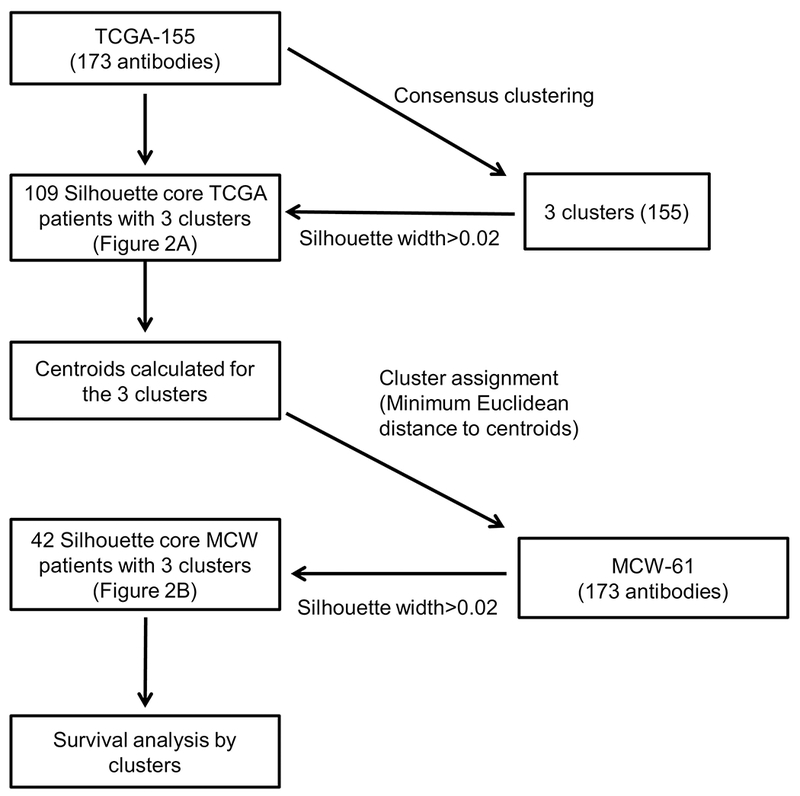
We completed consensus clustering in the TCGA-155 using expression data from the 173 common antibodies across both RPPA platforms in the same manner as the TCGA study performed with 192 antibodies [1]. From that analysis, we classified subjects with a silhouette width of >0.02 as core patients (n=109; Figure 1). As with the original TCGA survival data, we found that overall survival in these core patients was significantly better for patients harboring tumors with hormone-classified RPPA pathway scores and significantly worse for those falling within the EMT cluster (Figure 2A). Further, using the 42 core patients identified from the MCW-61 cohort, we validated the significant difference in survival probability between the EMT and hormone groups (p=0.038; Figure 2B). Somewhat unexpectedly, we found that the prognostic ability of the PI3K signature was not consistent across the 2 patient cohorts as the MCW-61 PI3K group exhibited significantly worse survival than its hormone group (p=0.030; Figure 2B).
Figure 2. Validation of TCGA cluster survival results in the TCGA-155 and MCW-61 cohorts using 173 common antibodies.
Tumors from the TCGA-155 (A) and MCW-61 (B) cohorts were assigned to survival-associated clusters that were previously identified by TCGA (EMT, Hormone, and PI3K/AKT). In agreement with the original TCGA data, we found that overall survival was significantly better for patients assigned to the hormone cluster and significantly worse for those within the EMT cluster in both TCGA-155 and MCW-61 cohorts in our analysis of expression data from the 173 common antibodies across RPPA platforms.
Identification of 22 antibodies that associate with survival outcome and risk
RSF and Cox proportional hazard models were used to further explore the ability of the RPPA data to identify small protein groups that could accurately predict survival. RSF analysis, using minimum depth and VIMP, identified 11 antibodies that associated significantly with survival status (Table 2). In addition, 52 proteins given an importance score of >0 by VIMP but not identified by minimum depth were further tested using Cox proportional hazard regression. For those proteins that violated the proportional hazard assumption, the protein was selected if it remained significant in the final model. From these analyses, we found that expression data from 11 of the 52 antibodies significantly associated with survival (p<0.05), bringing our total number of candidates for further study to 22. Table 2 lists these 22 antibodies and their hazard ratios for risk of death. Of note, the 22 antibodies represent 21 proteins as Her3 was represented twice.
Table 2.
SAA candidates: Proteins selected using RSF and a Cox proportional hazards model.
| Proteins | Gene | Selection algorithm | Hazard*** | Hazard ratio and 95% CI | p-value**** |
|---|---|---|---|---|---|
| her3 | ERBB3 | RSF Minimum Depth & VIMP* | Reduced | 0.825 (0.337, 2.022) | 0.674 |
| hsp70 | HSPA1A | RSF Minimum Depth & VIMP | Increased | ***** | 0.238 |
| acc1 | ACACA | RSF Minimum Depth & VIMP | Increased | 1.779 (0.838, 3.778) | 0.134 |
| chk1_pS345 | CHEK1 | RSF Minimum Depth & VIMP | Reduced | 0.041 (0.005, 0.371) | 0.004 |
| myosin-Ila pS1943 | MYH9 | RSF Minimum Depth & VIMP | Increased | 1.939 (0.521, 7.222) | 0.323 |
| rad51 | RAD51 | RSF Minimum Depth & VIMP | Increased | ***** | 0.029 |
| paxillin | PXN | RSF Minimum Depth & VIMP | Increased | 1.758 (0.780, 3.962) | 0.173 |
| p70S6K_pT389 | RPS6KB1 | RSF Minimum Depth & VIMP | Increased | ***** | 0.024 |
| fasn | FASN | RSF Minimum Depth & VIMP | Increased | 1.991 (0.962, 4.121) | 0.064 |
| p27_pT198 | CDKN1B | RSF Minimum Depth & VIMP | Reduced | 0.066 (0.004, 1.156) | 0.063 |
| cyclin D1 | CCND1 | RSF Minimum Depth & VIMP | Increased | 6.091 (2.145, 17.294) |
0.0007 |
| bak | BAK1 | RSF VIMP & Cox** | Reduced | 0.050 (0.005, 0.490) | 0.010 |
| src_pY416 | SRC | RSF VIMP & Cox | Reduced | 0.386 (0.191, 0.780) | 0.008 |
| pai1 | SERPINE1 | RSF VIMP & Cox | Increased | 1.382 (1.012, 1.886) | 0.042 |
| cjun_pS73 | JUN | RSF VIMP & Cox | Reduced | 0.327 (0.111, 0.959) | 0.042 |
| raptor | RPTOR | RSF VIMP & Cox | Increased | 15.710 (1.783, 138.390) | 0.013 |
| her3_pY1298 | ERBB3 | RSF VIMP & Cox | Increased | 6.245 (1.107, 35.244) | 0.038 |
| C-raf_S338 | RAF1 | RSF VIMP & Cox | Increased | ***** | 0.037 |
| E-cadherin | CDH1 | RSF VIMP & Cox | Reduced | ***** | 0.035 |
| N-cadherin | CDH2 | RSF VIMP & Cox | Increased | ***** | 0.044 |
| yb1 | YBX1 | RSF VIMP & Cox | Increased | 5.554 (1.048, 29.422) | 0.044 |
| bcl2 | BCL2 | RSF VIMP & Cox | Reduced | 0 319 (0.141, 0.720) | 0.0059 |
Proteins selected by both RSF minimum depth & VIMP: RSF minimum depth & RSF VIMP >0
Proteins selected by RSF VIMP & Cox: RSF VIMP >0 & Cox proportional hazards model p<0.05
Increased or reduced risk of death determined by Cox proportional hazards model
p value from Cox proportional hazards model
Proportional hazard assumption was violated and an interaction of the predictor and survival time (log) was included in the final model; only direction and p value were reported
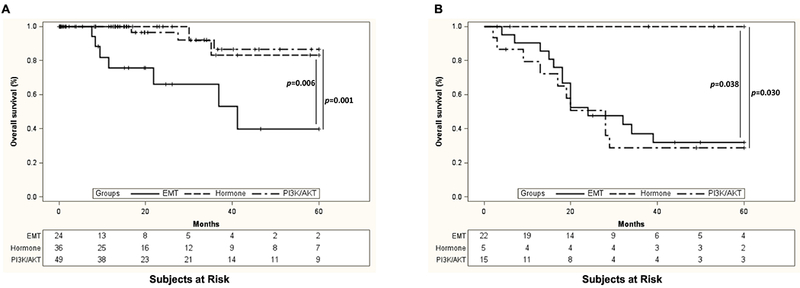
We next calculated an index score for each patient to investigate whether these 22 SAAs could accurately stratify patients into survival risk groups. Indeed, an index score from the protein hazard ratios predicted 3 significantly different survival risk groups (low risk=LR, medium risk=MR, and high risk=HR) from the TCGA-155 cohort (Figure 3A), which we then tested in the MCW-61 dataset (Figure 3B). We found a significant difference in survival between the MR and HR groups (p=0.039 for TCGA-155; p=0.045 for MCW-61) and the HR and LR groups (p<0.0001 for TCGA-155; p=0.001 for MCW-61). While outcomes between the MR and LR groups in the MCW-61 cohort were not statistically significant (p=0.077; Figure 3B), the TCGA-155 cohort’s LR and MR groups survival was significantly different (p=0.007; Figure 3A). A linear-by-linear association test showed that the correlation between the SAA risk groups and the TCGA clusters was significant (p=0.0028; Table S3).
Figure 3. Expression data from 22 antibodies can stratify cervical cancer patients into risk groups with significant differences in survival outcome.
Using RSF and Cox proportional hazard models, we determined that expression data from 22 specific antibodies (SAAs) could accurately predict survival. An index score from the protein hazard rations predicted 3 significantly different survival risk groups (low, medium, and high) in both the TCGA-155 cohort (A) and MCW-61 cohort (B).
A subgroup of 22 SAAs associate with clinical and pathologic features
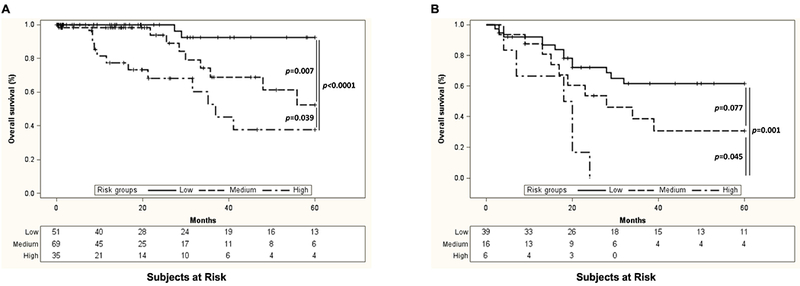
Finally, we determined whether protein expression detected by the 22 SAAs associated with clinical and/or pathologic data from the combined TCGA and MCW cohorts (n=216; Figure 4). Expression of the phosphorylated forms of both checkpoint kinase 1 (Chk1_pS345) and jun proto-oncogene (cJun_pS73) was significantly lower in HPV clade A7 than in HPV clade A9 and HPV-negative cancers (p=0.0036 and p=0.048, respectively; Figure 4A and 4B). On the other hand, N-cadherin expression was significantly lower in HPV clade A9-positive cancers compared to HPV clade A7 and HPV-negative cancers (p=0.029; Figure 4C). Further, we found that HPV-negative cancers expressed significantly higher levels of the cell cycle-related protein cyclin D1 (p=0.008; Figure 4D) and significantly lower levels of both the multifunctional enzyme acetyl-coA carboxylase alpha (Acc1; p=0.021; Figure 4E) and phosphorylated Erb-B2 receptor tyrosine kinase 3 (Her3_pY1298; p=0.012; Figure 4F). Finally, expression of the DNA repair protein, RAD51 recombinase (Rad51), was significantly higher in stage 3 cancer patients than in those with either stage 1 or 2 disease (p=0.043; Figure 4G).
Figure 4. A subgroup of the 22 SAAs associate with clinical and pathological features of cervical cancer.
Expression of chk1_pS345 (A), cjun_pS73 (B), N-cadherin (C), cyclin D1 (D), accl (E), and her3_pY1298 (F) associated with HPV clade, while expression of rad51 (G) associated with cervical cancer stage.
Discussion
We began this study by validating TCGA [1] results that characterized patient survival risk clusters based on protein expression data from RPPA. Due to the number of antibodies analyzed in RPPA, we reanalyzed the TCGA data along with the new MCW cohort using the 173 common antibodies across both datasets. Through this work, we were able to again demonstrate that the hormone cluster exhibited the best survival, while the EMT cluster exhibited the worst. The survival outcome of PI3K groups, however, were noticeably different across the 2 patient cohorts studied here. As stated before, the original TCGA manuscript used RPPA expression data from 192 antibodies, while our TCGA reanalysis and MCW validation used data from 173 common antibodies. Thus, the expression data examined here are different than that in the original TCGA paper and could have contributed to the observed differences. Additionally, it could simply be that the PI3K signature is not as robust as the EMT and hormone signatures, leading to inconsistencies in its prognostic ability across different antibody platforms and sample cohorts.
Building on the above results, we developed and validated an SAA index score using Cox proportional hazard and RSF, which integrates gene interaction information when selecting meaningful protein biomarkers to predict survival outcome. Using this methodology, we were able to stratify women with cervical tumors into 3 survival risk groups based solely on expression data obtained from 22 unique antibodies (SAA index) making this assay more clinically applicable. Fourteen of the 22 antibodies (Table 2) indicated an increased hazard ratio for death while 8 indicated a reduced hazard ratio. The increased hazard ratio group included oncogenic drivers such as Rad51, PAI1, cyclin D1, FASN, and phosphorylated isoforms of Her3 and Raf-1. Finally, when we further analyzed data from the 22 antibodies, we found significant associations between their expression and patients’ HPV status and clinical stage (Figure 4).
Several of the 22 antibodies highlight the prognostic biomarker results obtained from previous mRNA gene expression studies in cervical cancer. Rad51 is central to the homologous recombination DNA repair pathway, and its overexpression associates with treatment resistance and worse outcome in multiple cancer types [8]. Importantly, Rad51 has been identified as a prognostic biomarker and therapeutic target for cervical cancer. Inhibition of Rad51 significantly suppresses proliferation of cervical cancer cells in vitro and tumor growth in vivo, and its downregulation sensitizes cervical cancer cells to chemotherapy and radiation [9]. Gene expression microarrays used to examine pretreatment cervical tumor biopsies have indicated that the most significantly altered canonical pathway associated with intrinsic resistance is the BRCA-related DNA damage response [10]. Rad51 is a key member of this pathway, and this RNA-based result extends to immunohistochemistry experiments demonstrating that Rad51 protein expression is significantly higher in non-responsive cervical tumors compared to responsive tumors [10]. Finally, high Rad51 expression significantly predicts poor outcome and is associated with a lower likelihood of response to chemoradiation in women with locally advanced cervical cancer [11].
In support of previous findings, we found that PAI1 protein expression was associated with an increased hazard ratio (Table 2). PAI1 is encoded by the Serpin Family E Member 1 (SERPINE1) gene, which is a member of the serine proteinase inhibitor superfamily and the principal inhibitor of tissue plasminogen activator and urokinase. The plasminogen activator system contributes to tumor progression and metastasis by initiating a series of proteolytic cascades to degrade components of the extracellular matrix [12]. High expression levels and activity of PAI1 associate with prognostic factors in cervical cancer such as advanced stage, positive lymph nodes, tumor size, and lymph-vascular invasion [13–15]. Additionally, PAI1 expression is a strong independent prognostic factor for overall and disease-free survival in a group of 108 women with cervical cancer [15]. Phosphorylation of serine 1943 of MYH9 enhances invadopodia function and is critical for matrix degradation in vitro and experimental metastasis in vivo [16]. These data suggest that extracellular matrix degradation and tumor cell invasion and metastasis networks warrant additional work to further define their potential as prognostic markers and therapeutic targets.
FASN (fatty acid synthase) is involved in fatty acid synthesis, with its main function being catalyzation of palmitate synthesis from acetyl-CoA and malonyl-CoA. High expression of FASN associates with significantly shorter overall survival in women treated by surgical exenteration for recurrent cervical cancer [17]. cBioPortal [18, 19] mutual exclusivity analysis of genomic alterations in cervical cancer suggests a strong co-occurrence of alterations in FASN and ACC1 (acetyl-CoA carboxylase alpha), which encodes the rate-limiting enzyme in fatty acid synthesis. A recent study by Stoiber et al. [20] targeted lipogenesis using Acc1 and FASN inhibitors and found that they induce cell membrane alterations, which result in the inhibition of cancer cell migration and invasion, proliferative potential, and tumor growth in vivo [20]. FASN and ACC1 are also target genes of sterol regulatory element-binding proteins (SREBPs), which are key transcription factors in lipid biosynthesis that can be targeted for degradation using clinically-approved heat shock protein 90 (Hsp90) inhibitors [21]. In addition to the effects on SREBPs, Hsp90 inhibitors have also been shown to result in degradation of Rad51 [22] and loss of Her3-associated PI3K activity due to Her2 degradation [23]. Together, these results and ours suggest that targeting lipogenesis, potentially with these existing Hsp90 inhibitors, may be beneficial for a subset of cervical cancer patients with poor prognosis.
The work here complements available genomic-based studies, provides important information on prognostic biomarkers, and identifies tumor protein signatures that may be examined for identification of more efficacious combinatorial drug therapies for invasive cervical cancer. Examination and analysis of SAA expression data in additional datasets could help to further support and refine this set of protein prognostic biomarkers.
Supplementary Material
Highlights.
The prognostic ability of TCGA RPPA signatures was validated
Expression data from 22 antibodies predicts cervical cancer survival risk
A subgroup of the 22 antibodies associate with clinical features of cervical cancer
Acknowledgements
This study was supported in part by the Women’s Health Research Program in the Department of Obstetrics and Gynecology, Medical College of Wisconsin (JSR), NCI CA 16672 (YL), NIH CA 221675 (YL), and NIH/NCI CA 210950 and CA 210949 (RA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
Dr. Gordon B. Mills is a member of the following scientific advisory boards: AstraZeneca, ImmunoMET, Ionis, Symphogen (honorarium), and Tarveda (honorarium). He has acted as a consultant for Chrysalis (received travel reimbursement monies only), Nuevolution (received travel reimbursement monies only), PDX Pharm (receives consultant fees), and Signalchem Lifesciences (honorarium). Dr. Mills has research sponsored by AstraZeneca, Ionis, Karus Therapeutics, Takeda/Millenium Pharmaceuticals, and Pfizer and reports stock options in ImmunoMET, Signalchem Lifesciences, Tarveda, Catena Pharmaceuticals, and Spindletop Ventures. He has received travel reimbursement monies from Mills Institute for Personalized Care Center and reports licensed technology with Myriad Genetics and Nanostring.
All other authors have no conflicts of interest to report.
References
- [1].Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang Z, Borecki I, Nguyen L, Ma D, Smith K, Huettner PC, et al. CD83 gene polymorphisms increase susceptibility to human invasive cervical cancer. Cancer Res. 2007;67:11202–8. [DOI] [PubMed] [Google Scholar]
- [3].Hu X, Zhang Z, Ma D, Huettner PC, Massad LS, Nguyen L, et al. TP53, MDM2, NG01, and susceptibility to cervical cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(3):755–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathmatics. 1987;20:53–65. [Google Scholar]
- [5].Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ishwaran H, Kogalur UB. Random survival forests for R. R News. 2007;7(2):25–31. [Google Scholar]
- [7].Ehrlinger J ggRandomForests: Exploring Random Forest Survival2016. Available from: https://arxiv.org/abs/1612.08974.
- [8].Ward A, Khanna KK, Wiegmans AP. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treat Rev. 2015;41(1):35–45. [DOI] [PubMed] [Google Scholar]
- [9].Chen Q, Cai D, Li M, Wu X. The homologous recombination protein RAD51 is a promising therapeutic target for cervical carcinoma. Oncol Rep. 2017;38(2):767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Balacescu O, Balacescu L, Tudoran O, Todor N, Rus M, Buiga R, et al. Gene expression profiling reveals activation of the FA/BRCA pathway in advanced squamous cervical cancer with intrinsic resistance and therapy failure. BMC Cancer. 2014;14:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leonardi S, Buttarelli M, De Stefano I, Ferrandina G, Petrillo M, Babini G, et al. The relevance of prelamin A and RAD51 as molecular biomarkers in cervical cancer. Oncotarget. 2017;8(55):94247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front Oncol. 2018;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Daneri-Navarro A, Macias-Lopez G, Oceguera-Villanueva A, Del Toro-Arreola S, Bravo-Cuellar A, Perez-Montfort R, et al. Urokinase-type plasminogen activator and plasminogen activator inhibitors (PAI-1 and PAI-2) in extracts of invasive cervical carcinoma and precursor lesions. Eur J Cancer. 1998;34(4):566–9. [DOI] [PubMed] [Google Scholar]
- [14].Kobayashi H, Fujishiro S, Terao T. Impact of urokinase-type plasminogen activator and its inhibitor type 1 on prognosis in cervical cancer of the uterus. Cancer Res. 1994;54(24):6539–48. [PubMed] [Google Scholar]
- [15].Hazelbag S, Kenter GG, Gorter A, Fleuren GJ. Prognostic relevance of TGF-beta1 and PAI-1 in cervical cancer. Int J Cancer. 2004;112(6):1020–8. [DOI] [PubMed] [Google Scholar]
- [16].Norwood Toro LE, Wang Y, Condeelis JS, Jones JG, Backer JM, Bresnick AR. Myosin-IIA heavy chain phosphorylation on S1943 regulates tumor metastasis. Exp Cell Res. 2018;370(2):273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Roszik J, Ring KL, Wani KM, Lazar AJ, Yemelyanova AV, Soliman PT, et al. Gene Expression Analysis Identifies Novel Targets for Cervical Cancer Therapy. Front Immunol. 2018;9:2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stoiber K, Naglo O, Pernpeintner C, Zhang S, Koeberle A, Ulrich M, et al. Targeting de novo lipogenesis as a novel approach in anti-cancer therapy. Br J Cancer. 2018;118(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kuan YC, Hashidume T, Shibata T, Uchida K, Shimizu M, Inoue J, et al. Heat Shock Protein 90 Modulates Lipid Homeostasis by Regulating the Stability and Function of Sterol Regulatory Element-binding Protein (SREBP) and SREBP Cleavage-activating Protein. J Biol Chem. 2017;292(7):3016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sottile ML, Nadin SB. Heat shock proteins and DNA repair mechanisms: an updated overview. Cell Stress Chaperones. 2018;23(3):303–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Basso AD, Solit DB, Munster PN, Rosen N. Ansamycin antibiotics inhibit Akt activation and cyclin D expression in breast cancer cells that overexpress HER2. Oncogene. 2002;21(8):1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


