Abstract
Background
Despite recent advances in treating non‐small cell lung cancer (NSCLC) with immune checkpoint inhibitors (ICIs), their role in ALK‐positive NSCLC patients is unclear. We investigated the efficacy of ICIs in patients with ALK‐positive NSCLC.
Methods
Between 2011 and 2018, a total of 14 ALK‐positive NSCLC patients treated with ICIs were evaluated retrospectively. Clinicopathologic features including age, PD‐L1 expression, and treatment outcomes were analyzed. RNA expression level and cytolytic activity by ALK positivity were analyzed using The Cancer Genome Atlas (TCGA) and National Cancer Center Research Institute (NCCRI) data sets.
Results
A total of 13 patients (92.9%) received ALK inhibitors. Patients received a median of three (range 2–8) courses of therapy. The study included nine patients (64.3%) who were PD‐L1‐high (>50%) and four (28.6%) who were PD‐L1‐low (<50%). The objective response rate was 14.3% (2/14). The median progression‐free survival time was 2.18 months (95% confidence interval [CI] 1.13 months‐not reached [NR]). The median overall survival time was 5.67 months (95% CI 3.00 months‐NR). RNA expression levels of CD274 were similar between the ALK‐positive and negative groups in both TCGA and NCCRI datasets. RNA levels of CD8A in both TCGA and NCCRI data sets were nonsignificantly lower in the ALK‐positive group. Cytolytic activity scores including interferon‐γ‐related response were lower in the ALK‐positive group in the NCCRI but not TCGA dataset.
Conclusions
Despite high PD‐L1‐positive rates, ICIs show limited efficacy in ALK‐positive NSCLC. Decreased interferon‐γ‐related response may underlie these findings.
Keywords: Anaplastic lymphoma kinase, immune checkpoint inhibitors, non‐small cell lung cancer
Key points
From retrospective review of 14 ALK‐positive NSCLC patients in South Korea treated with ICIs, the role of ICI in ALK‐positive NSCLC was navigated. Despite high PD‐L1‐positive rates, ICIs show limited efficacy (ORR 14.3%; median PFS 2.18 months; median OS 5.67 months). From TCGA and NCCRI dataset analysis, decreased interferon‐γ‐related response may underlie these findings.
Introduction
Anaplastic lymphoma kinase (ALK) rearrangement has been observed in 3%–5% of non‐small cell lung cancers (NSCLCs).1, 2 ALK‐positive NSCLC is a distinct subgroup, and ALK tyrosine kinase inhibitors (TKIs) have been shown to prolong survival.3, 4, 5 Crizotinib, the first ALK inhibitor, showed superior outcomes compared to standard cytotoxic chemotherapy in advanced ALK‐positive NSCLC.3 Subsequently, ceritinib also showed its superiority to cytotoxic chemotherapy, and alectinib proved its efficacy compared to crizotinib.4, 5
Although ALK TKIs yield high response rates (>60%) in advanced ALK‐positive NSCLC,3, 4, 5 patients who initially respond to these agents eventually experience resistance and disease progression.6 For patients who progress after ALK TKI therapy, initial cytotoxic chemotherapy regimens, which are used for first‐line treatment of NSCLC (e.g., carboplatin/paclitaxel), are recommended.6
Recently, immune checkpoint inhibitors (ICIs) targeting programmed death 1 (PD‐1) and its ligand (PD‐L1) have demonstrated impressive results in NSCLC.7, 8, 9 NSCLC harboring an ALK rearrangement is reported to be significantly associated with PD‐L1 expression.10 Preclinical data suggest that ALK TKIs enhance antitumor immunity by downregulating PD‐L1, implying possibilities for anti‐PD‐1/PD‐L1 antibodies as therapeutic options for progression following ALK TKI therapy.11 However, data from the US Flatiron Health database and clinical trials show poor results of ICIs for ALK‐positive NSCLC.12 Clinical trials evaluating the combination of ICIs with ALK TKIs in advanced NSCLC are ongoing (NCT02574078, NCT01998126, NCT02393625, NCT02013219), but clinical data of patients with ALK‐positive NSCLC still remain insufficient.
Overall, the role of ICIs in treating ALK‐positive NSCLC is not yet elucidated. Therefore, in this study, we conducted a retrospective analysis to investigate the efficacy of ICIs in treating patients with ALK‐positive NSCLC.
Methods
Patients and data collection
We reviewed the records of patients diagnosed with ALK‐positive NSCLC and receiving treatments to block PD‐1/PD‐L1 at Seoul National University Hospital (SNUH) and Seoul National University Bundang Hospital (SNUBH) between 1 January 2011 and 31 August 2018. We identified 14 ALK‐positive NSCLC patients who received ICIs in their disease course. Clinicopathologic features including age, sex, smoking history, and treatment outcomes were reviewed from their medical history.
Detection of ALK rearrangement
ALK positivity was defined by the ALK break‐apart fluorescence in situ hybridization (FISH) assay as evaluated by three experienced pathologists. FISH was performed on formalin‐fixed paraffin‐embedded tumor tissues using a break‐apart probe specific to the ALK locus (Vysis LSI ALK Dual Color, break‐apart rearrangement probe; Abbott Molecular, Abbott Park, IL, USA) according to the manufacturer's instructions, as described in previous reports.13 ALK FISH was performed from biopsy specimens prior to ALK TKI treatment.
Immunohistochemistry of PD‐L1
Immunohistochemistry was performed using the following monoclonal antibodies against PD‐L1: 22C3 (Dako, Glostrup, Denmark), E1L3N (Cell Signaling Technology, Danvers, MA, USA), and SP263 (Ventana, Tucson, AZ, USA). Staining with clone 22C3 was performed with the corresponding Dako pharmDx assay. Staining with clone SP263 on the Ventana Benchmark Ultra platform was performed with the SP263 Ventana assay. In this study, a score of 50% was defined as “PD‐L1‐high” and a score of less than 50% or equal to 50% as “PD‐L1‐low”. Details of PD‐L1 immunohistochemistry profiles are described in Table S1.
Open database analyses of RNA expression
Two publicly available data sets were used; one was lung adenocarcinoma of The Cancer Genome Atlas (TCGA)14 and the other was generously provided by the authors from an earlier publication which is referred to as the National Cancer Center Research Institute (NCCRI) dataset (accession number GSE31210).15 Level 3 data of TCGA lung adenocarcinoma from the UCSC Cancer Browser (https://genome-cancer.ucsc.edu) was downloaded on 3 June 2015. Both datasets included clinical information of ALK translocation and mRNA expression data obtained by RNAseq (TCGA using the Illumina HiSeq V2 platform) and microarray (NCCRI using the Affymetrix Human Genome U133 Plus 2.0 Array). Array data from the NCCRI dataset were normalized using the limma R package and were log2 transformed.
In these datasets, we analyzed differences in the mRNA expression of CD8A and CD274 (PD‐L1) between samples with and without ALK translocation.16 We also analyzed differences in the cytolytic activity score, defined as the mean value of mRNA expression of GZMA (granzyme A) and PRF1 (perforin 1).17 Finally, to assess interferon‐γ‐responsive gene expression, we analyzed the data sets using the “Module3_IFN_Score” gene set obtained from earlier publications, and the gene set enrichment analysis method through the GenePattern website (https://genepattern.broadinstitute.org).18
Statistical analysis
Progression‐free survival (PFS) was calculated from the start date of ICI treatment to the date of disease progression by RECISTv1.1 criteria,19 as confirmed by imaging, death, or the last follow‐up date, if censored. Overall survival (OS) was measured from the initiation of ICI treatment until death or the last follow‐up date, if censored. Survival analyses were carried out according to the Kaplan‐Meier method with the log‐rank test. All tests were two‐sided and P‐values <0.05 were considered statistically significant. For survival analysis, STATA Statistical Software version 15.0 (StataCorp, College Station, TX, USA) was used for computation. Statistical analyses and image creations for RNA expression were performed using R version 3.4.3 software (R Development Core Team, https://www.r-project.org/). The Wilcoxon rank sum test was applied to analyze gene expression differences.
Results
Patient characteristics
We identified 14 patients with ALK‐positive NSCLC who were treated with ICIs between 2011 and 2018 at SNUH (n = 9) and SNUBH (n = 5). Baseline clinical and pathological features of these patients are summarized in Table 1.
Table 1.
Patient characteristics
| N = 14 | N (%) | |
|---|---|---|
| Age (years) | Median (range) | 49 (25–64) |
| Sex | Male | 10 (71.4) |
| Female | 4 (28.6) | |
| Ethnicity | Asian | 14 (100.0) |
| Smoking | Never smoker | 5 (35.7) |
| Current or ex‐smoker | 9 (64.3) | |
| Pathology | Adenocarcinoma | 13 (92.9) |
| Poorly differentiated | 1 (7.1) | |
| Prior ALK TKIs | None | 1 (7.1) |
| 1 | 1 (7.1) | |
| 2 | 8 (57.2) | |
| 3 | 4 (28.6) | |
| Prior lines of therapy | 2 | 2 (14.3) |
| 3 | 7 (50.0) | |
| ≥ 4 | 5 (35.7) | |
| PD‐1 inhibitor | Nivolumab | 8 (57.2) |
| Pembrolizumab | 5 (35.7) | |
| PD‐L1 inhibitor | Atezolizumab | 1 (7.1) |
| ALK rearrangement | FISH positive | 14 (100.0) |
| PD‐L1 expression | High (>50%) | 9 (64.3) |
| Low (≤50%) | 4 (28.6) | |
| Not checked | 1 (7.1) |
ALK, Anaplastic lymphoma kinase; ICI, immune checkpoint inhibitor; PD‐1, programmed death 1; PD‐L1, programmed death ligand; TKI, tyrosine kinase inhibitor.
A majority of patients (n = 13; 92.9%) had previously received and progressed on ALK TKI treatment. Patients received a median of three (range 2–8) courses of therapy. All patients received single‐agent PD‐1 or PD‐L1 inhibitors. Thirteen patients (92.9%) received the PD‐1 inhibitor nivolumab (n = 8; 57.2%) or pembrolizumab (n = 5, 35.7%). The study also included one patient (n = 1; 7.1%) who received atezolizumab, a PD‐L1 inhibitor. PD‐L1 expression over 50% was found in nine patients (64.3%). Four patients (28.6%) did not show PD‐L1 expression (≤50%) by immunohistochemistry.
ICI responses
Among patients with ALK‐positive NSCLC, the objective response rate to ICIs was 14.3% (2/14). Details are presented in Table 2. Two patients treated with pembrolizumab showed responses (duration: 8.2 and 4.1+ months). The median PFS among ALK‐positive NSCLC patients treated with ICIs was 2.18 months (95% confidence interval [CI] 1.13‐not reached [NR] months) (Fig 1). The median OS among ALK‐positive NSCLC patients treated with ICIs was 5.67 months (95% CI 3.00‐NR months) (Fig 2). The patients were followed for a median of 4.2 months (range 0.8–30.5 months).
Table 2.
Response rates of immune checkpoint inhibitors
| N = 14 | N (%) | |
|---|---|---|
| ICI response | CR | 0 (0.0) |
| PR | 2 (14.3) | |
| SD | 2 (14.3) | |
| PD | 9 (64.3) | |
| Not evaluable | 1 (7.1) | |
| ORR | 2 (14.3) | |
| PFS a (months) | Median (95% CI) | 2.2 (1.1–NR) |
| OS b (months) | Median (95% CI) | 5.7 (3.0–NR) |
| Follow‐up (months) | Median (range) | 4.2 (0.9–30.5) |
Progression free survival from the initiation of ICI to the date of disease progression by RECISTv1.1 criteria by imaging, death, or the last follow‐up.
Overall survival from the initiation of ICI to death or last follow‐up.
ICI, immune checkpoint inhibitor; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; ORR, objective response rate; PFS, progression‐free survival; OS, overall survival; NR, not reached; CI, confidence interval.
Figure 1.

Kaplan‐Meier survival curves showing the progression‐free survival of ALK‐positive NSCLC patients treated with immune checkpoint inhibitors. ALK, anaplastic lymphoma kinase; NSCLC, non‐small‐cell lung cancer; PD‐1, programmed death 1; PD‐L1, programmed death ligand 1.
Figure 2.
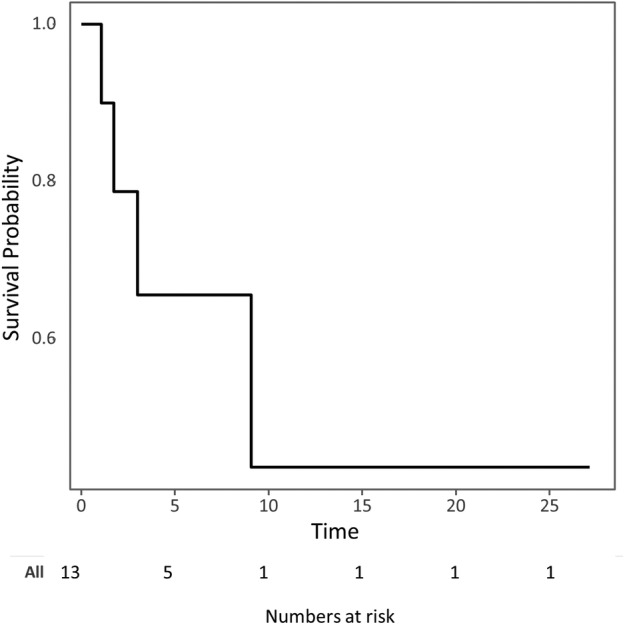
Kaplan‐Meier survival curves showing the overall survival of ALK‐positive NSCLC patients treated with immune checkpoint inhibitors. ALK, anaplastic lymphoma kinase; NSCLC, non‐small‐cell lung cancer; PD‐1, programmed death 1; PD‐L1, programmed death ligand 1.
RNA expression levels and cytolytic activity score
To investigate a possible biologic explanation of our clinical observations, we used lung adenocarcinoma of TCGA and NCCRI datasets.15 Three out of 513 cases (0.58%) had ALK translocation in TCGA dataset. In the NCCRI dataset, ALK translocation was present in 11 out of 246 cases (4.47%).
RNA levels of CD274 (PD‐L1) were similar between the ALK translocation positive and negative groups in both TCGA and NCCRI datasets (P‐values = 0.780 and 0.913, respectively, Fig 3a,b, left). RNA levels of CD8A in both TCGA and NCCRI datasets tended to be lower in the ALK translocation positive group, although the tendency was not statistically significant (P‐values = 0.37 and 0.062, respectively) (Fig 3a,b, middle).
Figure 3.

Open database analyses of RNA expression according to anaplastic lymphoma kinase translocation. Boxplots of (a) were created with lung adenocarcinoma of The Cancer Genome Atlas (TCGA) dataset. Boxplots of (b) were created with the National Cancer Center Research Institute (NCCRI) dataset. The left boxplots show RNA expression levels of CD274 (PD‐L1), and the middle boxplots show RNA expression levels of CD8A. The right boxplots show cytolytic activity scores. Statistical significance is shown with P‐values. (c) Enrichment plots were created using gene set enrichment analysis through the GenePattern website (left, TCGA dataset; right, NCCRI dataset) ( ) Enrichment profile, (
) Enrichment profile, ( ) Hits, and (
) Hits, and ( ) Ranking metric scores.
) Ranking metric scores.
Next, we determined the cytolytic activity scores, which are related to local immunogenicity of the tumor microenvironment.17 Cytolytic activity scores were significantly lower in the ALK translocation positive group in the NCCRI (P‐value = 0.040) dataset, but not TCGA dataset (P‐value = 0.160) (Fig 3a,b, right). Enrichment scores of ALK‐positive sample gene sets of interferon‐γ,18 by the gene set enrichment analysis, were −0.46 and −0.70 in TCGA and NCCRI datasets (FDR = 0.67 and 0.16, respectively, Fig 3c). Collectively, these results suggest that ALK translocation tended to be associated with decreased interferon‐γ‐related responses, underlying lower response rates.
Discussion
In this study, ALK‐positive NSCLC patients showed a low response rate of 14.3% to ICIs, and short PFS of 2.1 months and OS of 9.1 months. Although most patients exhibited high PD‐L1 expression, it did not result in higher response rates.
As a predictive biomarker for ICI treatment response, PD‐L1 expression has been widely studied in multiple clinical trials since the early time of ICI development. Both preclinical and clinical data demonstrate that ALK‐positive NSCLC is significantly associated with high expression of PD‐L1.10 Patients with high PD‐L1 expression, based on varying assays with different cutoffs, tend to have better responses to ICIs.20, 21 However, a positive correlation between PD‐L1 expression and the response to ICIs was not observed in ALK‐positive NSCLC patients. In this study, even though the majority of study patients had high expression levels of PD‐L1, it did not lead to a good response to ICIs.
A retrospective study including six ALK‐positive NSCLC patients found ALK rearrangement to be associated with low overall response rates to PD‐1/PD‐L1 blockade.22 Most recently, in the Supplementary data of a large scale phase 2 study (ATLANTIC) that included 15 ALK‐positive NSCLC patients treated with durvalumab as third‐line or later therapy, ALK‐positive NSCLC patients had a tendency of poor OS and PFS compared to EGFR‐positive patients.23 These findings are consistent with our study.
Preclinical data from Koh et al.10 showed that EML4‐ALK enhanced PD‐L1 expression in pulmonary adenocarcinoma via hypoxia‐inducible factor‐1α and STAT3. Similarly, induction of PD‐L1 expression by the EML4‐ALK oncoprotein and downstream signaling pathways has been reported in ALK‐positive NSCLC patients.24 This implies that NSCLCs harboring an ALK rearrangement are significantly associated with PD‐L1 expression.10, 11, 24
The effect of ALK‐TKIs on PD‐LI expression is controversial. Ota et al.24 showed that endogenous PD‐L1 expression in ALK‐positive NSCLC cells was attenuated by treatment with the specific ALK inhibitor, alectinib, or by ALK siRNA. Hong et al.11 reported that an ALK‐TKI downregulated PD‐L1 in tumor cells. Most recently, Kim et al.25 reported that PD‐L1 was expressed at higher levels in ALK inhibitor‐resistant cell lines than in the ALK inhibitor‐naïve parental cell line at the total protein, surface protein, and mRNA levels. Because of these diverse results, the clinical impact of PD‐L1 expression on ALK‐positive NSCLC tumor cells remains unclear.
Recently, the tumor mutational burden (TMB) has emerged as a predictive biomarker of ICI effectiveness.26 While a large‐scale clinical trial has shown that the TMB has a role in NSCLC,27 ALK‐positive patients have a relatively low TMB.28 In addition, Gainor et al.22 reported low rates of concurrent PD‐L1 expression and CD8+ tumor infiltrating lymphocytes within the tumor microenvironment of ALK‐positive NSCLC. Thus, ALK‐positive NSCLC patients may express fewer neoantigens, possibly leading to an ineffective tumor environment for ICIs. Furthermore, a decreased interferon‐γ‐related response is suggested from cytolytic activity score analysis, even though the difference in CD274 (PD‐L1) and CD8A RNA expression levels was not significant.
There are several limitations to this study. Firstly, this was a retrospective analysis. Because ALK‐positive NSCLC is rare, only a small number of patients were included in the study, resulting in limited statistical power. In addition, the small study population did not allow us to perform comparisons between different groups. Secondly, we performed immunohistochemistry of PD‐L1 utilizing three different anti‐PD‐L1 antibodies (22C3, E1L3N, and SP263). Although different antibody clones can have varying cutoff levels for tumor PD‐L1 expression, our study used a unified cutoff for all antibodies. Because of a lack of tissue, we did not repeat PD‐L1 immunohistochemistry with single PD‐L1 antibodies. However, recently, the Blueprint PD‐L1 IHC Assay Comparison Project revealed that these PD‐L1 antibodies were closely aligned on tumor cell staining,29 minimizing this concern. Thirdly, our study was not free from tumor heterogeneity and sampling variability. Even in the same patient, PD‐L1 expression can be discordant both within and between tumor specimens.30 Due to invasiveness of the procedure, we could only perform a limited number of biopsies during treatment. Moreover, the timing of assessing PD‐L1 expression varied according to individual patients. As there are preclinical data that ALK‐TKIs may affect the expression level of PD‐L1,11, 24 the diversity of examination points probably influenced the results.
Despite these limitations, to our knowledge, this retrospective study is the first report in ALK‐positive NSCLC and includes the largest number of ALK‐positive NSCLC patients treated with ICIs. We believe that this report may provide useful information to clinicians regarding ALK‐positive NSCLC patients.
In conclusion, ICIs show limited effectiveness in treating ALK‐positive NSCLC even though this cancer exhibits high PD‐L1‐positive rates. Further large‐scale prospective studies are required to confirm our findings.
Disclosure
The authors declare that there are no conflicts of interest.
Supporting information
Table S1. Survival and immunohistochemistry profile of PD‐L1.
Acknowledgments
This study was supported by a grant of the Korea Health Technology R&D Project “Strategic Center of Cell and Bio Therapy for Heart, Diabetes & Cancer” through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (MHW), Republic of Korea (grant number: HI17C2085). The authors thank Juyoun Kim, a data manager in SNUH, who managed the database. This manuscript has been edited by native English‐speaking experts. The institutional review board of SNUH and SNUBH approved the study protocol (approval number H‐1809‐045‐971). All procedures were carried out in accordance with the ethical standards of the institutional research ethics committee and the Helsinki declaration revised in 2013 by the World Medical Association.
Contributor Information
Ja Yoon Heo, Email: jayoonheo@gmail.com.
Bhumsuk Keam, Email: bhumsuk@snu.ac.kr.
References
- 1. Rikova K, Guo A, Zeng Q et al Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007; 131 (6): 1190–203. [DOI] [PubMed] [Google Scholar]
- 2. Soda M, Choi YL, Enomoto M et al Identification of the transforming EML4‐ALK fusion gene in non‐small‐cell lung cancer. Nature 2007; 448 (7153): 561–6. [DOI] [PubMed] [Google Scholar]
- 3. Solomon BJ, Mok T, Kim D‐W et al First‐line Crizotinib versus chemotherapy in ALK‐positive lung cancer. N Engl J Med 2014; 371 (23): 2167–77. [DOI] [PubMed] [Google Scholar]
- 4. Peters S, Camidge DR, Shaw AT et al Alectinib versus Crizotinib in untreated ALK‐positive non‐small‐cell lung cancer. N Engl J Med 2017; 377 (9): 829–38. [DOI] [PubMed] [Google Scholar]
- 5. Soria JC, Tan DSW, Chiari R et al First‐line ceritinib versus platinum‐based chemotherapy in advanced ALK‐rearranged non‐small‐cell lung cancer (ASCEND‐4): A randomised, open‐label, phase 3 study. Lancet 2017; 389 (10072): 917–29. [DOI] [PubMed] [Google Scholar]
- 6. Network NCC . Non‐small cell lung cancer. Version 6. 2018.
- 7. Carbone DP, Reck M, Paz‐Ares L et al First‐line Nivolumab in stage IV or recurrent non‐small‐cell lung cancer. N Engl J Med 2017; 376 (25): 2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reck M, Rodríguez‐Abreu D, Robinson AG et al Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med 2016; 375 (19): 1823–33. [DOI] [PubMed] [Google Scholar]
- 9. Socinski MA, Jotte RM, Cappuzzo F et al Atezolizumab for first‐line treatment of metastatic nonsquamous NSCLC. N Engl J Med 2018; 378 (24): 2288–301. [DOI] [PubMed] [Google Scholar]
- 10. Koh J, Jang JY, Keam B et al EML4‐ALK enhances programmed cell death‐ligand 1 expression in pulmonary adenocarcinoma via hypoxia‐inducible factor (HIF)‐1alpha and STAT3. Oncoimmunology 2016; 5 (3): e1108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong S, Chen N, Fang W et al Upregulation of PD‐L1 by EML4‐ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti‐PD‐1/PD‐L1 immune therapy for ALK‐TKIs sensitive and resistant NSCLC patients. Oncoimmunology 2016; 5 (3): e1094598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwartzberg L, Korytowsky B, Penrod JR et al Real‐world clinical impact of immune checkpoint inhibitors in patients with advanced/metastatic non–small cell lung cancer after platinum chemotherapy. Clin Lung Cancer 2019; 20: 287–96. [DOI] [PubMed] [Google Scholar]
- 13. Won JK, Keam B, Koh J et al Concomitant ALK translocation and EGFR mutation in lung cancer: A comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol 2015; 26 (2): 348–54. [DOI] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511 (7511): 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okayama H, Kohno T, Ishii Y et al Identification of genes upregulated in ALK‐positive and EGFR/KRAS/ALK‐negative lung adenocarcinomas. Cancer Res 2012; 72 (1): 100–11. [DOI] [PubMed] [Google Scholar]
- 16. Ock CY, Keam B, Kim S et al Pan‐cancer Immunogenomic perspective on the tumor microenvironment based on PD‐L1 and CD8 T‐cell infiltration. Clin Cancer Res 2016; 22 (9): 2261–70. [DOI] [PubMed] [Google Scholar]
- 17. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015; 160 (1–2): 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thorsson V, Gibbs DL, Brown SD et al The immune landscape of cancer. Cancer Immun 2018; 48 (4): 812–30.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eisenhauer EA, Therasse P, Bogaerts J et al New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45 (2): 228–47. [DOI] [PubMed] [Google Scholar]
- 20. Borghaei H, Paz‐Ares L, Horn L et al Nivolumab versus Docetaxel in advanced nonsquamous non–small‐cell lung cancer. N Engl J Med 2015; 373 (17): 1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herbst RS, Baas P, Kim DW et al Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): A randomised controlled trial. Lancet 2016; 387 (10027): 1540–50. [DOI] [PubMed] [Google Scholar]
- 22. Gainor JF, Shaw AT, Sequist LV et al EGFR mutations and ALK rearrangements are associated with low response rates to PD‐1 pathway blockade in non‐small cell lung cancer: A retrospective analysis. Clin Cancer Res 2016; 22 (18): 4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garassino MC, Cho B‐C, Kim J‐H et al Durvalumab as third‐line or later treatment for advanced non‐small‐cell lung cancer (ATLANTIC): An open‐label, single‐arm, phase 2 study. Lancet Oncol 2018; 19 (4): 521–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ota K, Azuma K, Kawahara A et al Induction of PD‐L1 expression by the EML4‐ALK Oncoprotein and downstream Signaling pathways in non‐small cell lung cancer. Clin Cancer Res 2015; 21 (17): 4014–21. [DOI] [PubMed] [Google Scholar]
- 25. Kim SJ, Kim S, Kim DW et al Alterations in PD‐L1 expression associated with acquisition of resistance to ALK inhibitors in ALK‐rearranged lung cancer. Cancer Res Treat 2019; 51 (3): 1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizvi NA, Hellmann MD, Snyder A et al Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015; 348 (6230): 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hellmann MD, Ciuleanu T‐E, Pluzanski A et al Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018; 378 (22): 2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis AA, Chae YK, Agte S et al Association of tumor mutational burden with smoking and mutation status in non‐small cell lung cancer (NSCLC). J Clin Oncol 2017; 35(Suppl. 7: 24.28034071 [Google Scholar]
- 29. Hirsch FR, McElhinny A, Stanforth D et al PD‐L1 immunohistochemistry assays for lung cancer: Results from phase 1 of the blueprint PD‐L1 IHC assay comparison project. J Thorac Oncol 2017; 12 (2): 208–22. [DOI] [PubMed] [Google Scholar]
- 30. McLaughlin J, Han G, Schalper KA et al Quantitative assessment of the heterogeneity of PD‐L1 expression in non‐small‐cell lung cancer. JAMA Oncol 2016; 2 (1): 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Survival and immunohistochemistry profile of PD‐L1.


