Abstract
Background
PH domain and leucine‐rich repeat protein phosphatase 2 (PHLPP2) has been reported to be a potent tumor suppressor in many human cancers. However, PHLPP2 has not been fully researched as a putative clinical prognostic biomarker of lung cancer.
Methods
The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases including data on 1383 non‐small cell lung cancer (NSCLC) patients were used to determine PHLPP2 expression. PHLPP2 expression was then examined by immunohistochemistry, and its clinical significance analyzed in 134 NSCLC patients, including 73 patients with adenocarcinoma and 81 with squamous cell carcinoma.
Results
We found PHLPP2 expression to be less pronounced in NSCLC tissue samples than that in nontumoral lung tissues according to data taken from TCGA and GEO datasets; this outcome was further validated by immunohistochemistry assay. The low PHLPP2 expression level was found to be associated with the presence of lymph node metastasis (P = 0.003). Importantly, PHLPP2 was found to be an independent indicator of prognosis for overall (hazard ratio [HR] = 0.520, 95% confidence interval [Cl] = 0.327–0.827; P = 0.006) and disease‐free survival (HR = 0.489, 95% Cl = 0.308–0.775; P = 0.002) in patients with surgically‐resected NSCLC by multivariate analysis.
Conclusion
Taken together, our findings show that PHLPP2 is a robust clinical marker for NSCLC survival and could serve as a potential therapeutic target.
Keywords: Metastasis, non‐small cell lung cancer, PHLPP2, prognosis
Introduction
Lung cancer, one of the most commonly diagnosed malignancies, has an incidence of 11.6% among all new cancer cases and currently accounts for 18.4% of cancer‐related mortality worldwide.1 Non‐small cell lung cancer (NSCLC) accounts for a striking 87% of all lung cancer cases with a five‐year survival rate of 21% after diagnosis.2 While various treatments such as surgery, chemotherapy, radiotherapy and immunotherapy have improved the prognosis of NSCLC to some extent, the risk of relapse remains high, highlighting the need to identify oncogenic and tumor suppressive genes related to patient prognosis as molecular biomarkers and to develop therapeutic targets.3
PHLPP2, a pleckstrin homology domain leucine‐rich repeat protein phosphatases (PHLPP) isozyme, catalyzes the dephosphorylation of AGC kinases including Akt.4 Maintaining balanced levels of PHLPP2 expression is essential in preventing pathologies, as changes in steady‐state levels of PHLPP2 are correlated with many diseases including diabetes, hepatic steatosis and cancer.5, 6 Recently, many studies have shown that PHLPP2 expression is ubiquitously lost in multiple cancers and plays a key role in a wide range of biological processes such as cancer cell proliferation, metastasis, autophagy and apoptosis.6, 7, 8, 9, 10, 11, 12
PHLPP2 regulates several cellular pathways in cancer.13 Notably, direct dephosphorylation by PHLPP2 inactivates pro‐oncogenic AGC kinases such as Akt, S6K, and PKC and activates pro‐apoptotic kinases such as Mst1.14, 15, 16, 17 In addition, PHLPP2 can regulate the epigenome by suppressing histone acetylation and phosphorylation to reduce the gene expression of epidermal growth factor receptor (EGFR), an oncogenic driver of tumor progression.18 The essential role of PHLPP2 in promoting the autophagic degradation of MMP2 protein and bladder cancer invasion was recently identified.19 In reference to NSCLC, studies of the tumor suppressive effects and functional mechanisms of PHLPP2 are ongoing.13 PHLPP2 downregulation was reported to promote lung carcinogenesis by inducing inflammatory tumor necrosis factor alpha (TNFα).20 Contactin‐1 contributes to the malignant behavior of lung cancer cells by reducing PHLPP2 expression.21 By targeting the 3′ UTR of PHLPP2, miR‐205 and miR‐141 frequently were upregulated and drive malignant phenotypes in NSCLC.22, 23 However, the diagnostic and prognostic significance of PHLPP2 in NSCLC has not been fully studied.
Therefore, this study aimed to assess protein levels of PHLPP2 in NSCLC tissue samples and found a correlation between PHLPP2 and clinicopathological indicators of NSCLC. In conclusion, our data show that elevated PHLPP2 levels may predict negative lymph node metastasis and the favorable survival of patients with NSCLC.
Methods
Patients and tissue specimens
In this study, samples of 134 human NSCLC tissues and 30 normal lung tissues were fixed in formalin and then embedded in paraffin for immunohistochemistry after surgical removal between January 2009 and December 2009 at the Harbin Medical University Cancer Hospital. Patients who had received neoadjuvant therapy or radiotherapy prior to surgery were not enrolled in this work. Follow‐up data for NSCLC patients expired on 30 March 2019 or on the date of death. Clinicopathological information for these patients is given in Table 1. A pathologist provided diagnosis and executed histological typing and tumor staging following the World Health Organization Consensus Classification and Staging System for lung cancer. Our study was approved by the Harbin Medical University Institute Research Medical Ethics Committee.
Table 1.
Association between PHLPP2 expression and clinicopathological characteristics in 134 NSCLC patients
| PHLPP2 expression | ||||
|---|---|---|---|---|
| All patients | High (%) | Low (%) | ||
| Variable | (n = 134) | (n = 47) | (n = 87) | P‐value |
| Gender | ||||
| Male | 88 | 34 (72.3) | 54 (62.1) | 0.232 |
| Female | 46 | 13 (27.7) | 33 (37.9) | – |
| Age (years) | ||||
| <60 | 90 | 28 (59.6) | 62 (71.3) | 0.169 |
| ≥60 | 44 | 19 (40.4) | 25 (28.7) | – |
| Differentiation | ||||
| Well | 4 | 2(44.7) | 2(44.8) | 0.838† |
| Moderate | 65 | 23(48.9) | 42(48.3) | – |
| Poor | 65 | 22(6.4) | 43(6.9) | – |
| pTNM stage | ||||
| I | 72 | 31 (66.0) | 41 (47.1) | 0.096 |
| II | 23 | 7 (14.9) | 16 (18.4) | – |
| III | 39 | 9 (19.1) | 30 (34.5) | – |
| pT classification | ||||
| T1 | 42 | 17 (36.2) | 25 (28.7) | 0.215† |
| T2 | 83 | 25 (53.2) | 58 (66.7) | – |
| T3/4 | 9 | 5 (10.6) | 4 (4.6) | – |
| Lymph node metastasis | ||||
| Present | 83 | 37 (78.7) | 46 (52.9) | 0.003* |
| Absent | 51 | 10 (21.3) | 41 (47.1) | – |
P < 0.05 was considered statistically significant.
The Fisher's exact test.
NSCLC, non‐small cell lung cancer; pT, pathological tumor; pTNM, pathological tumor node metastasis.
Immunohistochemistry (IHC)
IHC experimental procedures were performed as previously described.24 The paraffin‐embedded tissue blocks were sectioned into 4 μm thick portions for IHC. These sections were then deparaffinized and rehydrated. The samples were immersed in 3% H2O2 solution to block endogenous peroxidase activity before antigen retrieval with citrate buffer (PH 6.0). The sections were then incubated with an anti‐PHLPP2 antibody (1:100 dilution, 25 244‐1‐AP, ProteinTech) at 4°C overnight, after which the slides were incubated with a corresponding rabbit secondary antibody at 37°C. The anti‐PHLPP2 antibody is specific to the peptide sequence of PHLPP2 protein “LDLQHNALTRLPDTLFSKALNLRYLNASANSLESLPSACTGEESLSMLQLLYLTNNLLTDQCIPVLVGHLHLRILHLANNQLQTFPASKLNKLEQLEELNLSGNKLKTIPTTIANCKRLHTLVAHSNNISIFPEILQLPQIQFVDLSCNDLTEILIPEALPATLQDLDLTGNTNLVLEHKTLDIFSHITTLKIDQKPLPTTDSTVTS” encoded by BC129927 (https://www.ncbi.nlm.nih.gov/nuccore/BC129927). Finally, PHLPP2 expression was scored by two pathologists using the following criteria: (i) The percentage of immunopositive cells: 0 (0%), one (0%–10%), two (11%–50%), three (51%–70%), and four (≥71%) and (ii) the intensity of staining: 0 (negative staining), 1 (mild staining), two (moderate staining), three (intense staining). Staining scores collected from the two scoring systems were summed to obtain a final point score of 0 to 7 where 0–3 points denoted low PHLPP2 expression while 4–7 points denoted high PHLPP2 expression.
Bioinformatics analysis
We used web‐based tools available through GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/) based on the TCGA database to detect PHLPP2 mRNA levels in various tumors. NSCLC genome‐wide gene profiles were downloaded from TCGA (https://tcga-data.nci.nih.gov/). Gene expression datasets GSE81089, GSE40419 and GSE19804 were obtained from the publicly accessible GEO (http://www.ncbi.nlm.nih.gov/geo/) database. The Kaplan‐Meier Plotter (http://kmplot.com/analysis/) online tool was used to test the predictive significance of PHLPP2 expression.
Statistical analysis
We used the SPSS 21.0 statistical software program (Chicago, IL, USA) to analyze the data. Distinctions between the two groups were analyzed with a χ2 test. A Kaplan‐Meier analysis was performed to analyze the survival time using a log‐rank test. Using the Cox proportional hazards model, the independent prognostic factors of OS and DFS were analyzed by multivariate analysis. P‐value <0.05 was regarded as statistically significant.
Results
PHLPP2 exhibited downregulated expression in human NSCLC tissues
GEPIA was used to detect PHLPP2 mRNA levels in different carcinomas.25 We first determined that PHLPP2 was significantly downregulated in kidney renal clear cell carcinoma and thyroid carcinoma tissues (Fig 1a). Consistent with previous findings for bladder urothelial carcinoma, breast invasive carcinoma, colon adenocarcinoma and prostate adenocarcinoma, PHLPP2 levels were significantly lower in the tumor tissues than those in non‐tumor tissues (Fig 1a).
Figure 1.
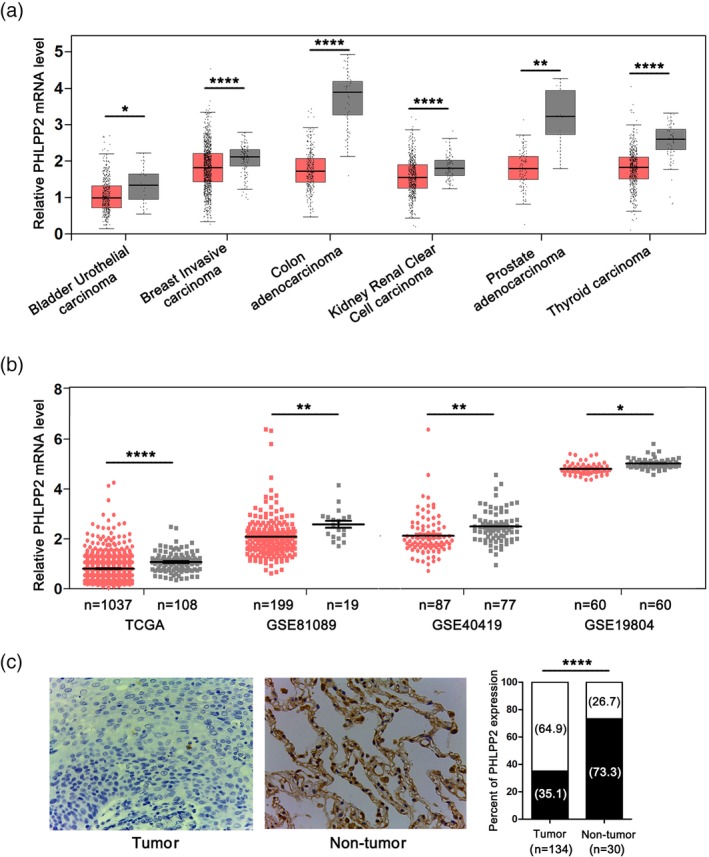
PHLPP2 expression in human NSCLC tissues. (a) Relative mRNA levels of PHLPP2 in different tumors were analyzed by GEPIA. (b) The expression levels of PHLPP2 mRNA in NSCLC and nontumoral lung tissues were analyzed using TCGA and GEO databases. ( ) Tumor and (
) Tumor and ( ) Nontumor. (c) Representative immunostaining images for PHLPP2 expression in 134 human NSCLC tissues (Tumor) and 30 normal lung tissues (Nontumor). (
) Nontumor. (c) Representative immunostaining images for PHLPP2 expression in 134 human NSCLC tissues (Tumor) and 30 normal lung tissues (Nontumor). ( ) High and (
) High and ( ) low. Histogram showing percentages of PHLPP2 expression for tumor and nontumor tissues. ****P < 0.0001, **P < 0.01, *P < 0.05.
) low. Histogram showing percentages of PHLPP2 expression for tumor and nontumor tissues. ****P < 0.0001, **P < 0.01, *P < 0.05.
In NSCLC, PHLPP2 was also found to be downregulated in tumor as opposed to what was observed in nontumor from TCGA and GSE81089, GSE40419, and GSE19804 datasets (Fig 1b). The results demonstrated that PHLPP2 expression levels in the NSCLC tissues were lower than those observed in the nontumoral lung tissues (Fig 1c).
Correlation between PHLPP2 expression and clinicopathologic characteristics in NSCLC tissue samples
Negative control and positive control of PHLPP2 staining was shown in Figure S1. Negative or weak levels of PHLPP2 immunoreactivity were observed in NSCLC tissue samples (64.9%), and PHLPP2 was clearly localized in the cytoplasmic and nuclear compartments of tumor cells (Figs 1c, 2). High and low PHLPP2 expression levels in ADC and SCC tissues by immunohistochemical staining were shown in Figure 2.
Figure 2.
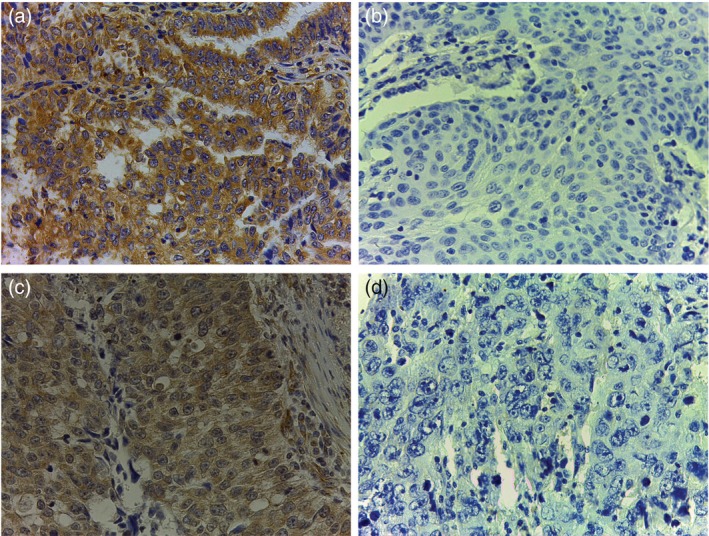
Representative immunohistochemical staining photographs of PHLPP2. (a) High and (b) low levels of PHLPP2 protein expression in adenocarcinoma. (c) High and (d) low levels of PHLPP2 expression in squamous cell carcinoma.
The association between PHLPP2 expression and diverse clinicopathological factors are illustrated in Table 1. Low PHLPP2 expression was found to be significantly correlated with the presence of lymph node metastasis (P = 0.003). However, no strong associations were observed between PHLPP2 and gender, age, differentiation, pTNM stage and pT classification.
Low PHLPP2 protein levels predicted poor survival of NSCLC
The survival curves of OS (HR 0.508, 95% CI 0.327–0.790; P = 0.002) and DFS (HR 0.481, 95% CI 0.311–0.745; P = 0.001) are shown in Figure 3a. Furthermore, the Kaplan‐Meier plotter online tool was used to validate the effect of PHLPP2 on lung cancer survival (n = 1926).26 The results consistently show that patients with elevated levels of PHLPP2 expression had longer OS (P = 0.00013) and post progression survival (PPS) (P = 0.012) than those with low PHLPP2 expression levels (Fig 3b). Using the web‐based tools in The Human Protein Atlas (https://www.proteinatlas.org) based on TCGA, low PHLPP2 expression predicts a poor prognosis in ADC (Fig S2).
Figure 3.
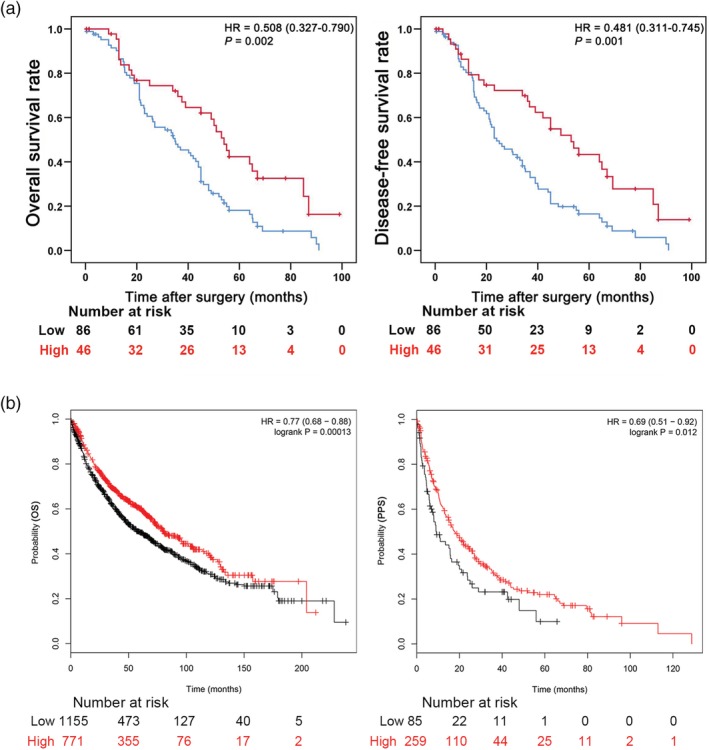
High PHLPP2 expression correlated with favorable survival outcomes in NSCLC patients. (a) Kaplan‐Meier analysis of overall (left) and disease‐free survival (right) for high and low PHLPP2 expression levels. ( ) Low‐PHLPP2 and (
) Low‐PHLPP2 and ( ) high‐PHLPP2. (b) Kaplan‐Meier survival curves of OS and PPS comparing high and low PHLPP2 expression estimates of NSCLC patient survival probability determined from the Kaplan‐Meier plotter database (213407_at). Expression (
) high‐PHLPP2. (b) Kaplan‐Meier survival curves of OS and PPS comparing high and low PHLPP2 expression estimates of NSCLC patient survival probability determined from the Kaplan‐Meier plotter database (213407_at). Expression ( ) low and (
) low and ( ) high. * P < 0.05 was considered statistically significant.
) high. * P < 0.05 was considered statistically significant.
Furthermore, the results of univariate analysis demonstrated that advanced pTNM stage, positive lymph node metastasis and low PHLPP2 expression were the key factors leading to poorer OS in patients with NSCLC. Multivariate analysis results showed that pTNM stage (HR 1.953, 95% CI 1.368–2.789; P < 0.001) and PHLPP2 expression (HR 0.520, 95% CI 0.327–0.827; P = 0.006) were independent prognostic factors for OS. The results of univariate analysis of DFS were similar to the findings observed in OS. In our multivariate analysis, pTNM stage (HR 1.892, 95% CI 1.326–2.700; P < 0.001) and PHLPP2 expression (HR 0.489, 95% CI 0.308–0.775; P = 0.002) were independent predictors for DFS, as shown in Table 2.
Table 2.
Univariate and multivariate analyses of OS and DFS
| OS | DFS | |||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||
| Variable | P‐value | HR (95% CI) | P‐value | P‐value | HR (95% CI) | P‐value |
| Age | – | – | – | – | – | |
| < 60 | – | – | – | – | – | |
| ≥ 60 | 0.453 | – | – | 0.610 | – | – |
| Gender | – | – | – | – | – | |
| Female | – | – | – | – | – | |
| Male | 0.401 | – | – | 0.358 | – | – |
| Differentiation | – | – | – | – | – | |
| Good | – | – | – | – | – | |
| Moderate | – | – | – | – | – | |
| Poor | 0.053 | – | – | 0.073 | – | – |
| pT stage | – | – | – | – | – | |
| I | – | – | – | – | – | |
| II | – | – | – | – | – | |
| III/IV | 0.109 | – | – | 0.067 | – | – |
| pTNM stage | – | – | – | – | – | |
| I | – | – | – | – | – | |
| II | – | – | – | – | – | |
| III | <0.001* | 1.953 (1.368 to 2.789) | <0.001* | <0.001* | 1.892 (1.326 to 2.700) | <0.001* |
| Lymph node metastasis | – | – | – | – | – | |
| Present | – | – | – | – | – | |
| Absent | 0.003* | 0.646 (0.336 to 1.244) | 0.191 | 0.002* | 0.730 (0.384 to 1.390) | 0.339 |
| PHLPP2 expression | – | – | – | – | – | |
| Low | – | – | – | – | – | |
| High | 0.003* | 0.520 (0.327 to 0.827) | 0.006* | 0.001* | 0.489(0.308 to 0.775) | 0.002* |
P < 0.05 was regarded as statistically significant.
CI, confidence interval; DFS, disease‐free survival; HR, hazard ratio; NSCLC, non‐small cell lung cancer; OS, overall survival; pT, pathological tumor; pTNM, pathological tumor node metastasis.
Discussion
The present study illustrates the clinical significance of PHLPP2 in NSCLC. Our data showed that PHLPP2 expression in protein levels in NSCLC tissue specimens are downregulated relative to nontumoral lung tissues. We also found that PHLPP2 expression is deregulated in kidney renal clear cell carcinoma and thyroid carcinoma. Previous studies show decreased expressions of PHLPP2 in several types of cancer.27, 28, 29, 30, 31 In agreement with our findings, PHLPP2 was lost or decreased in most colon and pancreatic tumor tissues, strongly supporting its tumor suppressive role.30, 31
Researchers have made efforts to identify novel and important metastatic and prognostic biomarkers to guide research and provide therapeutic targets.16, 32, 33 Another interesting finding was that we found that low PHLPP2 expression levels were associated with positive lymph node metastasis. Moreover, the present study suggested that overexpression of PHLPP2 as an effective molecular biomarker is associated with favorable prognosis for NSCLC. Consistently, low PHLPP2 expression was significantly related to the advanced tumor clinical stage, poor differentiation and positive lymph node metastasis in hypopharyngeal squamous cell carcinoma.28 Although there was a trend of the correlation between low PHLPP2 expression and advanced pTNM stage in our study, there was no statistical significance. As this study of lung cancer involved a limited number of patients, it is essential to determine the tumor suppressive role of PHLPP2 as a promising prognostic biomarker based on investigations of larger samples.
However, for 75 advanced lung adenocarcinoma patients receiving EGFR tyrosine kinase inhibitor (EGFR‐TKI) treatment, there was no significant correlation between PHLPP2 protein expression and any clinicopathological characteristics reported by Xie et al.34 The inconsistent results might be due to discrepancies in different techniques of detection, scoring methods of PHLPP2 expression, unequal cutoff points, sample size, and the treatment of EGFR‐TKI for patients after surgery between these two studies. Xie et al. showed that the high expression rate of PHLPP2 reached 61.3% based on the intensity of staining, while in the present study we found 35% of cases with high PHLPP2 expression based on the percentage of immunopositive cells and the intensity of staining.34 How to standardize the technology and criteria on PHLPP2 detection is the key point to evaluate its clinical significance. Some miRNAs reported to target the 3′‐untranslated region (3′‐UTR) of PHLPP2, including miR‐27a, miR‐205 and miR‐93, were found to be significantly and positively correlated with lymph node metastasis and poor survival, indicating that PHLPP2 can be defined as an important molecular marker for cancer patients to prognosticate spreading to lymph nodes and prognosis.23, 35, 36, 37, 38
Our IHC analysis results show that PHLPP2 plays a critical role in tumor cell metastasis. Numerous findings have provided supporting evidence of the tumor suppressive roles of PHLPP2.17, 19, 35, 39, 40 Peng et al. revealed that PHLPP2 promotes MMP2 degradation to inhibit bladder cancer invasion via p62‐mediated autophagy.19 PHLPP2 has been reported to induce epithelial‐mesenchymal transition (EMT), an important step of tumor metastasis, through an Akt/GSK3β dependent pathway.35 Moreover, PHLPP2 suppresses the invasive growth and migration of colorectal cancer cells by negatively regulating EMT and RAF1.17
Researchers have made efforts to identify several important cancer biomarkers to guide research and provide therapeutic targets.32 The present study suggested that PHLPP2 is an effective molecular biomarker for NSCLC diagnosis. It has consistently been reported that lower PHLPP2 expression levels might predict poor prognosis in hypopharyngeal squamous cell carcinomas.28 However, for 75 advanced lung adenocarcinoma patients receiving EGFR tyrosine kinase inhibitor (EGFR‐TKI) treatment, it was PHLPP1 rather than PHLPP2 protein expression levels that were found to be negatively associated with poor outcomes, showing that PHLPP2 has no effect on acquired EGFR‐TKI resistance.34 As previous works and our studies of lung cancer involved a limited number of patients, it is essential to determine the tumor suppressive role of PHLPP2 as a promising prognostic biomarker based on investigations of larger samples.
As an identified tumor suppressive gene, multiple lines of evidence reveal the role of PHLPP2 in tumor progression.7, 8, 9, 13, 23, 29, 41, 42, 43 PHLPP2 is involved in the inhibition of malignant behaviors of cancer attributed to the suppression of prosurvival signaling such as through the PI3K/Akt signaling pathway.13, 29 In addition, PHLPP2 can dephosphorylate and stabilize the Myc oncogene to suppress the progression of prostate cancer.42 A recent study reported on a novel molecular cascade of PHLPP2/CREB/miR‐302d that mediates cell cycle progression and anchorage‐independent growth.41 Therefore, the identification of PHLPP2 as an effective prognostic biomarker is reasonable.
Strenuous efforts have been made to understand how expression of PHLPP2 itself is regulated. PHLPP2 mRNA translation is regulated by miRNA, the mammalian target of rapamycin complex 1 (mTORC1) and ubiquitinating enzymes.13 Many different miRNAs target the 3′‐UTR of PHLPP2 in various cancers.23, 35, 38 Knockdown of key components of the mTORC1 complex results in decreased translation of PHLPP mRNA.44 Intriguingly, a recent study suggests a role of free Raptor, which is normally associated with the mTORC1 complex, interacts and binds PHLPP2, resulting in PHLPP2 protein stabilization and reduced signaling through Akt.45 Moreover, potassium channel tetramerization domain containing 17 (KCTD17) plays a role in the regulation of PHLPP2 protein stability by targeting to PHLPP2 for ubiquitin‐mediated degradation.5
In summary, this study shows that high levels of PHLPP2 expression predict better survival outcomes and that PHLPP2 serves as a promising candidate target for NSCLC. Further studies will continue to shed light on the functions and mechanisms of PHLPP2 as a crucial tumor suppressor in NSCLC. Our study presents knowledge that will prove central to the development of novel therapies targeting PHLPP2.
Disclosure
The authors have no competing interests to declare.
Supporting information
Figure S1 Negative control and positive control of PHLPP2 staining. (a) Negative staining of lung cancer. (b) Positive control for osteosarcoma.
Figure S2 Kaplan–Meier curves showing survival for ADC patients with high and low PHLPP2 expression from TCGA database.
Acknowledgments
This study was supported by the Certificate of China Postdoctoral Science Foundation Grant (2017M621307), Hei Long Jiang Postdoctoral Foundation (LBH‐Z17182), Hai Yan Youth Fund and Top‐Notch Youth Fund from Harbin Medical University Cancer Hospital (JJQN2018‐11 and BJQN2019‐07), and the NSFC (Grant Nos. 81803023, 81772474, 81572276).
Contributor Information
Li Cai, Email: caili@ems.hrbmu.edu.cn.
Ying Xing, Email: xingying0618@163.com.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Miller KD, Siegel RL, Lin CC et al Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016; 66: 271–89. [DOI] [PubMed] [Google Scholar]
- 3. Mathur A, Pandey VK, Kakkar P. PHLPP: A putative cellular target during insulin resistance and type 2 diabetes. J Endocrinol 2017; 233: R185–r98. [DOI] [PubMed] [Google Scholar]
- 4. Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol 2014; 54: 537–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim K, Ryu D, Dongiovanni P et al Degradation of PHLPP2 by KCTD17, via a glucagon‐dependent pathway, promotes hepatic steatosis. Gastroenterology 2017; 153: 1568–80.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeh ST, Zambrano CM, Koch WJ, Purcell NH. PH domain leucine‐rich repeat protein phosphatase 2 (PHLPP2) regulates G‐protein‐coupled receptor kinase 5 (GRK5)‐induced cardiac hypertrophy in vitro. J Biol Chem 2018; 293: 8056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li CF, Li YC, Jin JP, Yan ZK, Li DD. miR‐938 promotes colorectal cancer cell proliferation via targeting tumor suppressor PHLPP2. Eur J Pharmacol 2017; 807: 168–73. [DOI] [PubMed] [Google Scholar]
- 8. Strotbek M, Schmid S, Sanchez‐Gonzalez I, Boerries M, Busch H, Olayioye MA. miR‐181 elevates Akt signaling by co‐targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int J Cancer 2017; 140: 2310–20. [DOI] [PubMed] [Google Scholar]
- 9. Liao Y, Deng Y, Liu J et al MiR‐760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol 2016; 143: 655–63. [DOI] [PubMed] [Google Scholar]
- 10. Molina JR, Agarwal NK, Morales FC et al PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 2012; 31: 1264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu Y, Dai M, Lu A, Yu E, Merlino G. PHLPP1 mediates melanoma metastasis suppression through repressing AKT2 activation. Oncogene 2018; 37: 2225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toivanen R, Furic L. A balancing act: PHLPP2 fine tunes AKT activity and MYC stability in prostate cancer. J Cell Biol 2019; 218: 1771–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grzechnik AT, Newton AC. PHLPPing through history: A decade in the life of PHLPP phosphatases. Biochem Soc Trans 2016; 44: 1675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu J, Stevens PD, Li X, Schmidt MD, Gao T. PHLPP‐mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol Cell Biol 2011; 31: 4917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiao M, Wang Y, Xu X et al Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol Cell 2010; 38: 512–23. [DOI] [PubMed] [Google Scholar]
- 16. Yan Y, Hanse EA, Stedman K et al Transcription factor C/EBP‐beta induces tumor‐suppressor phosphatase PHLPP2 through repression of the miR‐17‐92 cluster in differentiating AML cells. Cell Death Differ 2016; 23: 1232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Stevens PD, Liu J et al PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology 2014; 146: 1301–12.e1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reyes G, Niederst M, Cohen‐Katsenelson K et al Pleckstrin homology domain leucine‐rich repeat protein phosphatases set the amplitude of receptor tyrosine kinase output. Proc Natl Acad Sci U S A 2014; 111: E3957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng M, Wang J, Zhang D et al PHLPP2 stabilization by p27 mediates its inhibition of bladder cancer invasion by promoting autophagic degradation of MMP2 protein. Oncogene 2018; 37: 5735–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H, Pan X, Jin H et al PHLPP2 downregulation contributes to lung carcinogenesis following B[a]P/B[a]PDE exposure. Clin Cancer Res 2015; 21: 3783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan J, Wong N, Hung C, Chen WX, Tang D. Contactin‐1 reduces E‐cadherin expression via activating AKT in lung cancer. PLOS One 2013; 8: e65463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mei Z, He Y, Feng J et al MicroRNA‐141 promotes the proliferation of non‐small cell lung cancer cells by regulating expression of PHLPP1 and PHLPP2. FEBS Lett 2014; 588: 3055–61. [DOI] [PubMed] [Google Scholar]
- 23. Cai J, Fang L, Huang Y et al miR‐205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non‐small cell lung cancer. Cancer Res 2013; 73: 5402–15. [DOI] [PubMed] [Google Scholar]
- 24. Xing Y, Liu Y, Liu T et al TNFAIP8 promotes the proliferation and cisplatin chemoresistance of non‐small cell lung cancer through MDM2/p53 pathway. Cell Commun Signal 2018; 16: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei CY, Wang L, Zhu MX et al TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J Exp Clin Cancer Res 2019; 38: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018; 8: 9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He X, Zhang Z, Li M et al Expression and role of oncogenic miRNA‐224 in esophageal squamous cell carcinoma. BMC Cancer 2015; 15: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou J, Yu X, Wang J et al Aberrant expression of PHLPP1 and PHLPP2 correlates with poor prognosis in patients with hypopharyngeal squamous cell carcinoma. PLOS One 2015; 10: e0119405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishibashi O, Akagi I, Ogawa Y, Inui T. MiR‐141‐3p is upregulated in esophageal squamous cell carcinoma and targets pleckstrin homology domain leucine‐rich repeat protein phosphatase‐2, a negative regulator of the PI3K/AKT pathway. Biochem Biophys Res Commun 2018; 501: 507–13. [DOI] [PubMed] [Google Scholar]
- 30. Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: Role in proliferation and tumorigenesis. Oncogene 2009; 28: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nitsche C, Edderkaoui M, Moore RM et al The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhibits tumor formation. Gastroenterology 2012; 142: 377–87.e1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hemingway H, Croft P, Perel P et al Prognosis research strategy (PROGRESS) 1: A framework for researching clinical outcomes. BMJ 2013; 346: e5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu B, Yang JR, Jia Y et al Overexpression of NR4A1 is associated with tumor recurrence and poor survival in non‐small‐cell lung carcinoma. Oncotarget 2017; 8: 113977–86. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Xie Y, Lv D, Wang W, Ye M, Chen X, Yang H. High PHLPP1 expression levels predicts longer time of acquired resistance to EGFR tyrosine kinase inhibitors in patients with lung adenocarcinoma. Oncotarget 2017; 8: 59000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding L, Zhang S, Xu M, Zhang R, Sui P, Yang Q. MicroRNA‐27a contributes to the malignant behavior of gastric cancer cells by directly targeting PH domain and leucine‐rich repeat protein phosphatase 2. J Exp Clin Cancer Res 2017; 36: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma YJ, Ha CF, Bai ZM, Li HN, Xiong Y, Jiang J. Overexpression of microRNA‐205 predicts lymph node metastasis and indicates an unfavorable prognosis in endometrial cancer. Oncol Lett 2016; 12: 4403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li G, Ren S, Su Z et al Increased expression of miR‐93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumour Biol 2015; 36: 3949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang L, Wang C, Lei F et al miR‐93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget 2015; 6: 8286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nowak DG, Cho H, Herzka T et al MYC drives Pten/Trp53‐deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer Discov 2015; 5: 636–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang C, Liao X, Jin H et al MEG3, as a competing endogenous RNA, binds with miR‐27a to promote PHLPP2 protein translation and impairs bladder cancer invasion. Mol Ther Nucleic Acids 2019; 16: 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu J, Wang Y, Hua X et al Inhibition of PHLPP2/cyclin D1 protein translation contributes to the tumor suppressive effect of NFkappaB2 (p100). Oncotarget 2016; 7: 34112–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nowak DG, Katsenelson KC, Watrud KE et al The PHLPP2 phosphatase is a druggable driver of prostate cancer progression. J Cell Biol 2019; 218: 1943–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warfel NA, Newton AC. Pleckstrin homology domain leucine‐rich repeat protein phosphatase (PHLPP): A new player in cell signaling. J Biol Chem 2012; 287: 3610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Stevens PD, Gao T. mTOR‐dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem 2011; 286: 6510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim K, Qiang L, Hayden MS, Sparling DP, Purcell NH, Pajvani UB. mTORC1‐independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun 2016; 7: 10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Negative control and positive control of PHLPP2 staining. (a) Negative staining of lung cancer. (b) Positive control for osteosarcoma.
Figure S2 Kaplan–Meier curves showing survival for ADC patients with high and low PHLPP2 expression from TCGA database.


