ABSTRACT
The process of tumor cell invasion and metastasis includes assembly of invadopodia, protrusions capable of degrading the extracellular matrix (ECM). The effect of cell cycle progression on invadopodia has not been elucidated. In this study, by using invadopodia and cell cycle fluorescent markers, we show in 2D and 3D cultures, as well as in vivo, that breast carcinoma cells assemble invadopodia and invade into the surrounding ECM preferentially during the G1 phase. The expression (MT1-MMP, also known as MMP14, and cortactin) and localization (Tks5; also known as SH3PXD2A) of invadopodia components are elevated in G1 phase, and cells synchronized in G1 phase exhibit significantly higher ECM degradation compared to the cells synchronized in S phase. The cyclin-dependent kinase inhibitor (CKI) p27kip1 (also known as CDKN1B) localizes to the sites of invadopodia assembly. Overexpression and stable knockdown of p27kip1 lead to contrasting effects on invadopodia turnover and ECM degradation. Taken together, these findings suggest that expression of invadopodia components, as well as invadopodia function, are linked to cell cycle progression, and that invadopodia are controlled by cell cycle regulators. Our results caution that this coordination between invasion and cell cycle must be considered when designing effective chemotherapies.
KEY WORDS: Invasion, Invadopodia, Cell cycle, Intravital imaging, FUCCI, G1
Highlighted Article: Simultaneous time-lapse imaging of invadopodia and FUCCI markers in 2D, spheroids and tumors in vivo demonstrates that cancer cells invade preferentially during the G1 phase of the cell cycle.
INTRODUCTION
Metastasis is responsible for the majority of cancer-related mortalities (Steeg, 2016). Metastasis is a complex, multi-step process that is initiated when cancer cells in the primary tumor acquire invasive properties, including motility and the ability to breakdown the extracellular matrix (ECM) (Vanharanta and Massagué, 2013). In breast cancer, invadopodia are the main cellular structures responsible for ECM degradation. Invadopodia are small membrane protrusions, which first assemble as precursors rich in actin and actin-binding proteins, including cortactin and Tks5 (also known as SH3PXD2A) (Beaty et al., 2013; Diaz et al., 2009; Gligorijevic et al., 2012; Magalhaes et al., 2011; Moshfegh et al., 2014; Oser et al., 2010). During invadopodia maturation, matrix metalloproteinases [MMP2, MMP9 and MT1-MMP (also known as MMP14)] are delivered to the invadopodium, leading to the degradation of the surrounding extracellular matrix (Poincloux et al., 2009).
We previously demonstrated that invasive cells oscillate between two distinct states, termed the ‘invadopodia’ state and the ‘migration’ state (Esmaeili Pourfarhangi et al., 2018a). The invadopodia state is characterized by cell stasis and the presence of invadopodia, whereas the migration state is defined by cell translocation and the absence of invadopodia. This dynamic oscillatory behavior is regulated by the activity of ECM receptor integrin β1, which determines the length of time that a cell spends in each state. Within the tumor microenvironment in vivo, invadopodia assembly is stimulated in perivascular niches, which contain highly cross-linked ECM, whereas cells outside of such niches migrate along collagen fibers in an MMP-independent fashion, a motility mode referred to as contact guidance (Gligorijevic et al., 2014). Decreasing ECM cross-linking in the perivascular niche abolishes invadopodia assembly and leads to the appearance of contact guidance, demonstrating that cell motility modes can be regulated by changes in the tumor microenvironment. However, for any given ECM architecture, a maximum of 15% of tumor cells are engaged in either invadopodia-driven motility or contact guidance, while the rest of the cells are non-motile. This suggests that there must be cell intrinsic factors, in addition to the extrinsic microenvironmental factors, that determine the motility mode and the assembly of invadopodia.
A deregulated cell cycle is a hallmark of cancer (Hanahan and Weinberg, 2011). Cell cycle progression is governed by a complex network of regulator proteins that define not only the phase of the cell cycle, but also the timing of transitions between phases (Malumbres and Barbacid, 2009). In recent years, cell cycle regulators have been shown to exhibit roles in both tumor suppression and tumor promotion, particularly the CIP/KIP family of cyclin-dependent kinase inhibitors (CKI) p27kip1 and p21cip1 (also known as CDKN1B and CDKN1A, respectively) (Besson et al., 2008). There is a growing body of evidence linking the cytoplasmic mislocalization of these proteins with the acquisition of motile and invasive characteristics during tumorigenesis (Besson et al., 2004). The cytoplasmic CIP/KIP proteins have been shown to regulate actin cytoskeletal dynamics; p21cip1 via physical binding to ROCK proteins (Lee and Helfman, 2004; Tanaka et al., 2002) and p27kip1 through interaction with RhoA (Besson et al., 2008). Consistent with this role, their mislocalization to the cytoplasm is associated with a higher aggressiveness of several types of cancer, including breast cancer (Liang et al., 2002; Roninson, 2002; Shin et al., 2002; Viglietto et al., 2002; Winters et al., 2001; Zhou et al., 2001). Most recently, a study demonstrated that p27kip1 facilitates invadopodia turnover through the PAK1–cortactin axis (Jeannot et al., 2017). Thus, further investigation is required to determine the molecular mechanism by which invadopodia-driven motility is coordinated with cell cycle progression.
Transcriptional profiling of invasive cells isolated from primary tumors has revealed that, in addition to the invasion gene signature, these cells also exhibit a proliferation gene signature (Patsialou et al., 2014). However, it is unclear whether cancer cells switch between invasive and proliferative phenotypes or display both phenotypes simultaneously. The ‘go-or-grow’ hypothesis proposes that proliferation and motility are mutually exclusive. This dichotomy was tested in a number of cancer types, resulting in studies either supporting this notion, for example, in glioblastoma (Gerlee and Nelander, 2012), or challenging it, for example, in mesothelioma and lung cancer (Garay et al., 2013). Such discrepancy could be attributed to differences in cancer types, or to the fact that the measurements of ‘go’ largely focused on migration (i.e. translocation of the cell without ECM breaching), and not invasion (i.e. motility involving an active breakdown of ECM). In melanoma, cancer cells were shown to undergo a phenomenon termed phenotype switching, where they switch from a highly proliferative and less invasive state to a highly invasive, but less proliferative state, thus, alternating between proliferation and invasion (O'Connell and Weeraratna, 2013; Widmer et al., 2013). This phenotypic plasticity in melanoma is deemed to be similar to the epithelial-to-mesenchymal (EMT) transition in other cancer types and is key to developing resistance to chemotherapies (Li et al., 2015). Therefore, it is critically important to understand the relationship between proliferation and motility, and distinguish between migration and invasion.
Towards understanding how cell cycle progression is coordinated with cell migration in breast cancer cells, we recently introduced fluorescent ubiquitination-based cell cycle indicator (FUCCI)-labeled cells into different microenvironments (Esmaeili Pourfarhangi et al., 2018b). This study has shown that cell migration can occur during any of the cell cycle phases except mitosis. In both 2D and 3D environments, the speed and persistence of random migration did not vary between cells in G1 versus S, G2 or M (S/G2/M) phases. However, in the presence of contact guidance cues, such as microchannels or aligned collagen fibers in 3D, cells performed directed migration at higher velocity and persistence in G1 phase compared to S or G2 (S/G2) phases.
In this study, we investigate the coordination of cell cycle progression with invasion, specifically with invadopodia. We show that invadopodia-driven ECM degradation is cell cycle-dependent and is enhanced in the G1 phase of the cell cycle, in 2D and 3D invasion assays and in vivo. An enhancement in ECM degradation during G1 phase is achieved by upregulating the gene and protein levels of the invadopodia components MT1-MMP and cortactin, as well as by increasing the localization of Tks5 to invadopodia. ECM degradation and invasion are enhanced in cells synchronized in G1 and elimination of invadopodia through Tks5 knockdown changes the dynamics of cell cycle progression. Finally, the CKI p27kip1 is recruited to the sites of invadopodia assembly and regulates invadopodia dynamics and function. Thus, we propose that invadopodia function, and therefore the metastatic potential of tumor cells, is linked to cell cycle progression.
RESULTS
Cells degrade and invade ECM during the G1 phase of the cell cycle
To visualize both cell cycle progression and invadopodia assembly in the breast carcinoma MDA-MB-231 cell line, we stably expressed the FUCCI reporters (Sakaue-Sawano et al., 2008) with the actin reporter mCerulean3–Lifeact7 (Markwardt et al., 2011; Riedl et al., 2008) (Fig. 1A). The FUCCI system consists of two nuclear fluorescent reporters, mKO2-hCdt1 (red) and mAG-hGem (green). mKO2-hCdt1 expression peaks in G1 and drops to background levels during early S phase. Reciprocally, mAG-hGem level is high in late S, throughout G2 and in M phases, then sharply declines in late mitosis (Sakaue-Sawano et al., 2008). In early S phase, when both probes are expressed, the cell nuclei appear yellow. Time-lapse imaging of FUCCI reporters in our system confirmed the expected oscillatory expression patterns of mKO2-hCdt1 and mAG-hGem fluorescence. The FUCCI reporter trace for a representative cell progressing through the cell cycle is shown in Fig. 1B.
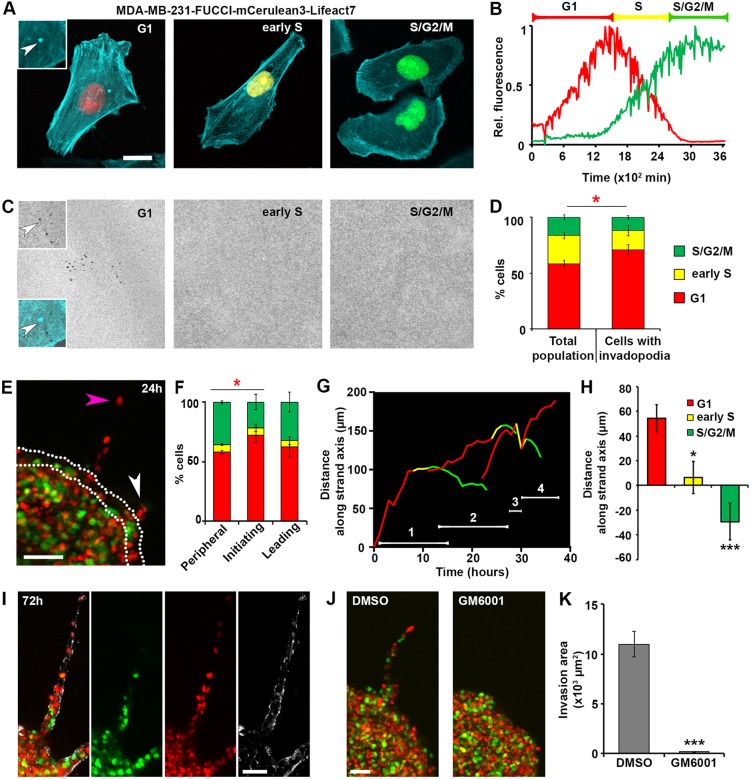
Fig. 1.
ECM degradation by invadopodia is enriched in the G1 phase of the cell cycle. (A) Representative images of MDA-MB-231 cells expressing FUCCI cell cycle reporters and the F-actin marker mCerulean3–Lifeact7 in G1, early S (middle) and S/G2/M (right) phases. Note the actin-enriched punctum in G1 cell (arrowhead, inset 1.5× magnification compared to main image). Scale bar: 10 µm. (B) Representative traces of red and green fluorescent signals in an MDA-MB-231-FUCCI cell monitored over time. Brackets indicate how the cell cycle phases are classified based on the fluorescence signal: early S phase corresponds to the period where red fluorescence has a negative slope. The ratio of the mean to maximum gray values is presented. (C) Representative images of a gelatin layer of cells in as in A during a 2D degradation assay. Note mature invadopodia in G1, where gelatin holes colocalize with actin enriched-punctum (arrow; see top inset for gelatin and bottom inset for overlay). (D) Percentage of cells in each phase of the cell cycle, either in total cell population, or in cells with invadopodia. Counts are based on 435 cells from three biological replicates. The red asterisk represents the comparison of percentage of cells in G1. Means±s.e.m. are shown. *P<0.05 (Mann–Whitney U-test). See Movies 1 and 2. (E) Representative image of a 3D spheroid at 24 h after embedding in high-density collagen I (5 mg/ml). Cells initiating and leading the invasive strands are marked by white and magenta arrowheads, respectively. Peripheral cells are marked by the white dashed outline. Scale bar: 100 µm. See Movie 3. (F) Percentage of cells in each phase of the cell cycle, either located in the periphery of the spheroid, or initiating or leading strand invasion (n=70 invasion strands, 20 spheroids, three biological replicates). Leader cells were evaluated at 70 h after embedding. The red asterisk represents the comparison of percentage of cells in G1. Mean±s.e.m. are shown. *P<0.05 (Mann–Whitney U-test). (G) Tracks of four consecutive leader cells along the strand axis, over time (Movie 4, Fig. S2). The red portion of each line corresponds to G1 phase, yellow to early S phase and green to late S/G2/M. The time period during which a cell was the leader is marked by white numbered line. (H) Quantification of the distance traveled by leader cells in G1 (red), early S (yellow) and late S/G2/M (green). The positive values indicate distance covered in the direction of the strand invasion (i.e. away from the spheroid core), while the negative values indicate backward movement, towards the spheroid core. Means±s.e.m. are shown. *P<0.05, ***P<0.001 (Mann–Whitney U-test). (I) Representative image of an invasive strand in a 3D spheroid at 72 h after embedding in high-density collagen (5 mg/ml) merged (left) and individual channels showing FUCCI nuclei (red, green) and cleaved collagen (Col1-¾C antibody, white). Scale bar: 50 µm. (J) Representative images and (K) quantification of the invasion area in DMSO control (n=25) and 25 µM GM6001-treated (n=17) spheroids from three biological replicates, taken at 72 h after embedding. Means±s.e.m. are shown. ***P<0.001 (Mann–Whitney U-test).
Using this system, we investigated whether invadopodia function correlated with any specific phase of the cell cycle. First, we surveyed the cell cycle phase distribution of total cells in an asynchronously dividing population. Through the flow cytometry analysis of the FUCCI signal, we observed that 44.54% of cells were in G1, 16% in early S and 38.2% of cells in late S/G2/M phases (Fig. S1A). Since the FUCCI signal does not distinguish between the late S and G2 phases, we employed BrdU incorporation and flow cytometry analysis to accurately determine the number of cells in S phase. The results of the BrdU/DAPI assay showed that 45.3% cells reside in G1 (2n, BrdU−), 40% in S (BrdU+) and 13.1% in G2/M (4n, BrdU−) (Fig. S1B). Importantly, the proportion of G1 cells was similar based on either method of measurement, thus, validating the use of FUCCI reporters in our model.
Next, we performed a 2D invasion assay. We plated the Lifeact and FUCCI-expressing cells (MDA-MB-231-FUCCI-mCerulean3-Lifeact7), on fluorescent gelatin for 8 h; we then quantified the cell cycle distribution for the total cell population, as well as for the cells with mature invadopodia, which were defined by the presence of the actin puncta (Fig. 1A) and colocalized degradation holes in the underlying gelatin (ECM) (Fig. 1C, see insets). By locating cells with mature invadopodia, we found that 79% of ECM-degrading cells resided in the G1 phase (Fig. 1D). Importantly, this proportion was significantly higher than the proportion of G1 cells (57%) found in the total population, suggesting that the invadopodia may be enriched in the G1 phase of the cell cycle. Live-cell imaging corroborated the results of the 2D degradation assay, and showed that cells that degrade the underlying gelatin are mostly in G1, while cells in early S and S/G2/M phases largely do not degrade gelatin (Movies 1 and 2).
To confirm our findings in a physiologically relevant setting, we used live-cell imaging and monitored the dynamics of MDA-MB-231-FUCCI-mCerulean3-Lifeact7 3D spheroid invasion into high-density rat tail collagen I, which requires MMP-driven degradation of the matrix (Wolf et al., 2013). First, we quantified the percentage of cells in each phase of the cell cycle among the cells located at the periphery of the spheroid: 57% resided in G1, 6% in early S and 35% in late S/G2/M phase. Next, we analyzed the cells that initiate strand formation, that is, invasion out of the spheroid: 72% of strand-initiating cells were in G1 phase, while only 5% and 21% were observed to be in early S and late S/G2/M phases, respectively (Fig. 1E,F, white arrowhead; Movie 3). In addition, we analyzed the ‘leader’ cells that were located at the tip of the invasion strand once the strand involved multiple cells. In contrast to cells initiating invasion, the leader cells were not enriched in G1, when measured at a fixed time (70 h) (Fig. 1E,F, magenta arrowhead; Movie 3). These findings suggest that there is a significant enrichment of G1 cells among those that initiate strand invasion, but not among leader cells.
To better understand the difference between cells initiating invasion and leader cells, we tracked the leader cells over time (Movie 4, Fig. S2). Interestingly, upon transitioning to the S phase, the ‘leader’ cells of each strand retreated back along the strand axis and were replaced by one of the cells that were previously following them. Strikingly, the new cells to assume the leader position were found to be in G1 phase, causing ‘leader switching’. To quantify this, we measured the distance that individual leader cells moved along a given strand over time (Fig. 1G), and collected results across multiple strands and spheroids (Fig. 1H). We found that the leader cells in G1 phase invaded the most (red line), moving into the collagen and away from the spheroid core. Once they transitioned to S/G2 phases, these cells were replaced by a new leader cell in G1 (Movie 4; Fig. 1G) and tend to travel backwards, towards the spheroid core (Movie 4; Fig. 1G,H). This type of leader switching in MDA-MB-231 cells was recently reported and shown to be the result of high energy requirements during invasion (Zhang et al., 2019). Here, we show that the forward invasion of the strands is driven by invadopodia-degrading cells in G1 phase, corroborating the reports of a high energy requirement for invadopodia assembly and ECM degradation (van Horssen et al., 2013). Collectively, our results indicate that the MDA-MB-231 breast cancer cells degrade ECM and invade preferentially during the G1 phase of the cell cycle, both in 2D and 3D environments.
We next asked whether the invasion we observe in 3D spheroids is truly MMP dependent. Immunostaining with an antibody that recognizes exposed collagen fragments specifically cleaved by MMPs revealed positive signal for cleaved ends in the collagen gel surrounding invasion strands (Fig. 1I). To exclude the possibility that the collagen fragments were exposed by cell mechanical remodeling of the ECM, we treated 3D spheroids with the MMP inhibitor GM6001 (25 µM). In the presence of the inhibitor, the formation of the invasion strands was effectively blocked (Fig. 1J,K), thus confirming that cells initiating invasion engage in MMP-dependent degradation of the collagen matrix.
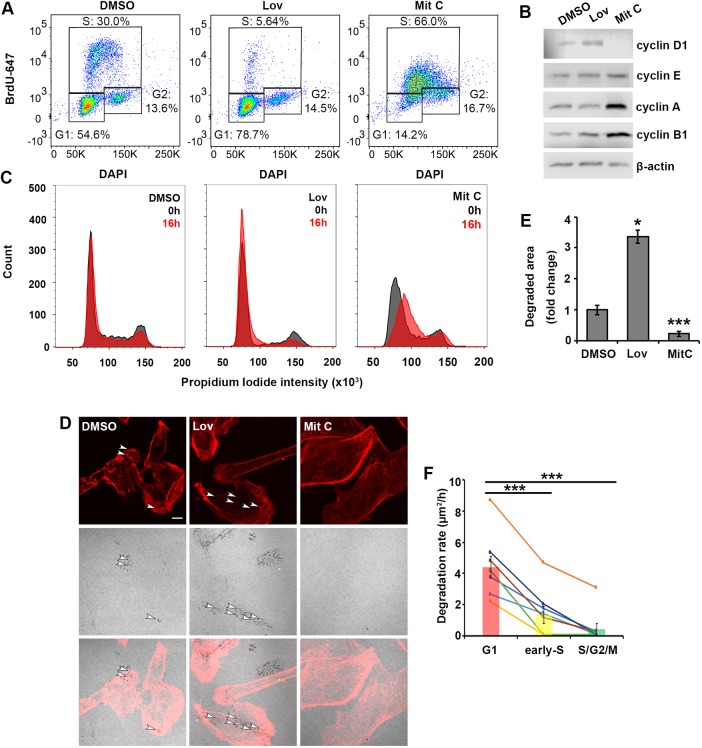
Based on our findings so far, we reasoned that ECM degradation may be conducted more efficiently in the G1 phase compared to S/G2 phases of the cell cycle. To test this notion, we synchronized cells in early G1 (treatment with lovastatin) (Jackman and O'Connor, 2001; Javanmoghadam-Kamrani and Keyomarsi, 2008) and S phase (treatment with mitomycin C) (Kang et al., 2009; Seki et al., 2005), released cells from the arrest and performed the 2D degradation assay for 16 h. The efficiency of synchronization was assessed by flow cytometry analysis of BrdU incorporation (Fig. 2A) and propidium iodide (PI) staining of cells at the time of release (0 h) (Fig. 2C, gray histograms) and at 16 h following release (Fig. 2C, red histograms). Treatment with lovastatin resulted in an increase of cells in G1 and a dramatic reduction of cells in S and G2/M. Mitomycin C, which stalls replication forks during DNA replication, induces an efficient accumulation of cells in S phase. Importantly, the overlay of PI histograms showed that at 16 h following the release from lovastatin-mediated G1 arrest, not only had the accumulation of cells in G1 persisted, but the remaining cells in G2/M had progressed to G1. Similarly, the cells synchronized in S with mitomycin C showed a shift towards G2/M, but no significant change overall. Further validation of synchronization was undertaken through the assessment of protein levels of cyclins at the time of release. The level of the G1 cyclin D1 was the highest in lovastatin-treated cells, while the G2/M cyclins, cyclin A and cyclin B1, were elevated in mitomycin C-treated conditions (Fig. 2B). Cells released from an early G1 arrest showed a 3-fold increase in ECM degradation compared to the asynchronous population (Fig. 2D,E). In contrast, cells released from an S/G2 arrest showed a marked decrease in the ECM degradation (Fig. 2D,E). To test whether this effect was not specific to the MDA-MB-231 cells only, we performed the same experiment on another breast cancer cell line, BT-549. We performed 2D degradation assay on BT-549-FUCCI cells (Fig. S3A). We observed that cells in G1 constituted 60% of total cells, while early S and S/G2/M phases comprised 15% and 25%, respectively (Fig. S3B). The quantification of degrading cells with invadopodia revealed that there was a significant enrichment of cells in G1 (84%), compared to degrading cells in early S (6%) and S/G2/M (10%). These results suggest that ECM degradation is conducted at a higher efficiency during the G1 phase in both MDA-MB-231 and BT-549 breast carcinoma cell lines.
Fig. 2.
Rate of ECM degradation decreases as cells progress from G1 to S and G2 phases. (A) BrdU–DAPI plots of DMSO-treated control cells, cells synchronized in G1 with lovastatin (Lov, 10 µM) treatment and cells synchronized in S phase with mitomycin C (Mit C, 1 µg/ml) treatment. The cells were harvested after a 24 h treatment. The percentage of cells in each phase of the cell cycle is indicated. (B) Western blot analysis of cells treated with DMSO control, lovastatin (Lov, 10 µM) and mitomycin C (Mit C, 1 µg/ml) for 24 h and blotted for cyclin D1, cyclin E, cyclin A and cyclin B1. β-actin is used as a loading control. (C) Histograms of PI staining of cells released (0 h, gray; 16 h, red) from synchronization by treatments with DMSO control, lovastatin (Lov, 10 µM) and mitomycin C (Mit C, 1 µg/ml). The cells were pre-treated with the indicated drugs for 24 h. (D) Representative images of ECM degradation and phalloidin-labeled cells after 16 h, for cells treated with DMSO (control) or released from Lov and Mit C treatments. Top panels show actin (red), middle panels show gelatin, and bottom panels show overlay images. White arrowheads point to mature invadopodia. Scale bar: 10 µm. (E) Quantification of the degraded area per cell represented as fold change relative to DMSO control. The mean±s.e.m. of the degraded area per field of view (FOV) is shown; >100 cells, NFOV=30 and three biological repeats per treatment. *P<0.05, ***P<0.001 (t-test). (F) The degradation rates of cells decrease as cells progress from G1 to early S and S/G2/M. See Movie 5. Each line represents an individual cell. Bars represent the mean degradation rate per cell cycle phase. ***P<0.001 (for the mean degradation rate; Mann–Whitney U-test). In addition, to determine the statistical significance of the degradation rate trends, we fit the data to repeated measures model and performed repeated measures analysis of variance in Matlab (Mathworks, Natick, MA). We obtained a P-value of 1.1×10−3, indicating there is a significant decreasing trend of degradation rate as the cell progresses through the cell cycle.
In the 2D degradation assay, populated by asynchronously dividing cells, ECM degradation exhibits a slow, monotonic increase over time. In the static image taken at the end of the assay, only cumulative degradation can be measured and it is unclear whether the degradation was contributed to by cells residing in the G1 phase or by all cells. To clarify this point, we performed time-lapse experiments, simultaneously monitoring the cell cycle, cell migration, actin puncta and ECM degradation over 50 h. We followed cells as they transitioned from G1 to S to G2 and mitosis, and plotted the rate at which they mediated degradation in each phase (Fig. 2F). We found that the highest rate of degradation (degraded area divided by the duration of the phase for individual cells) was achieved during the G1 phase, compared to early S and S/G2/M phases. In most cells, ECM degradation increased during G1 and then halted as the cell transitioned from G1 to S/G2 phase (Movie 5).
The expression and localization of invadopodia key components are cell cycle regulated
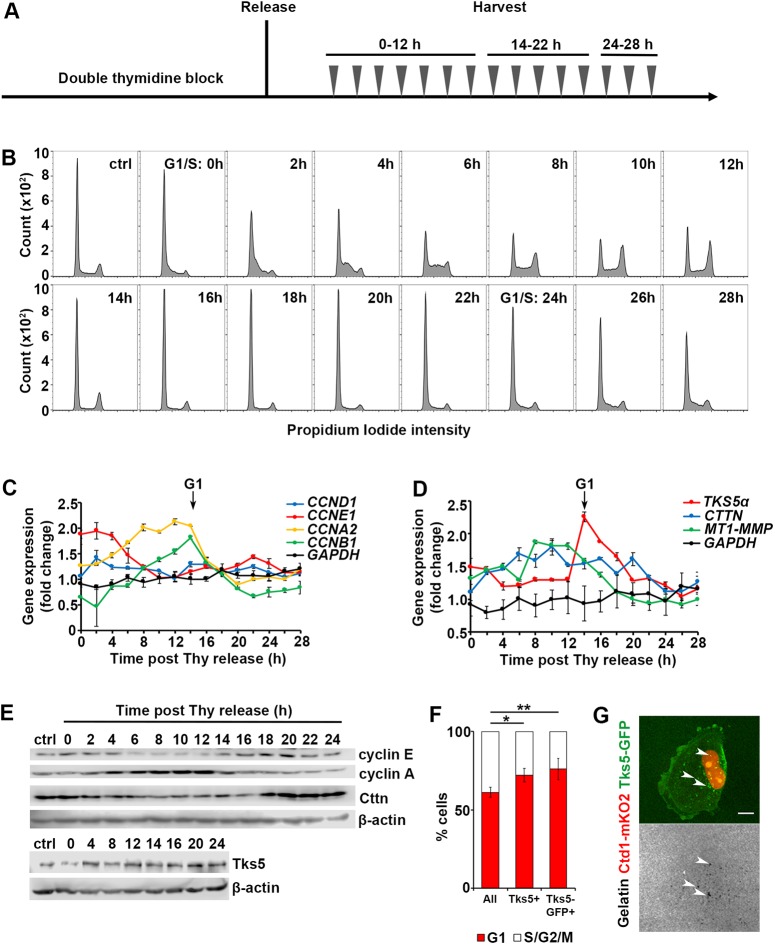
Invadopodia assembly requires an orchestration of over 50 structural and enzymatic proteins (Murphy and Courtneidge, 2011). We hypothesized that either the expression levels or the localization of core proteins would be enhanced to promote invadopodia function in G1 phase. To test this, we aimed to follow cells throughout the cell cycle, and measure the gene and protein expression levels of the key invadopodia components cortactin, Tks5 and, the main metalloproteinase, MT1-MMP. In order to follow cells on a population level and extract sufficient material for analysis, we synchronized cells at the G1/S boundary with a double thymidine block, released cells and harvested them every 2 h (Fig. 3A). Based on the PI flow cytometry data, the cells undergo a full cycle and reach the next G1/S transition ∼24–28 h following the release (Fig. 3B). Therefore, we focused on the 0–28 h time points for further analysis. The cells were efficiently arrested at the G1/S boundary, as evidenced by a sharp G1 peak at time 0 h (Fig. 3B). Following the release, the cells synchronously entered S phase and continued to accumulate in S phase until approximately the 8 h time point, where cells start progressing to the G2 phase, as indicated by an increase in the G2/M peak. Consistent with this transition, the mRNA level for cyclin E (CCNE1) drops, while the mRNA levels for the S and G2-specific cyclins, CCNA2 and CCNB1, continue to increase (Fig. 3C). At 14 h following the release from the G1/S boundary arrest, the cells start accumulating in G1 (Fig. 3B). This is accompanied by a sharp drop in mRNA levels of CCNA2 and CCNB1 (Fig. 3C). CCND1 gene expression level remained stable throughout the cell cycle, consistent with the reports that CCND1 is amplified in breast cancers (Musgrove et al., 2011).
Fig. 3.
The key components of invadopodia are cell cycle regulated. (A) The schematic of the double-thymidine block, release and harvest regimen. The cells are synchronized at the G1/S boundary with a double-thymidine block; cells are released and harvested every 2 hs over 28 h, in two batches: time points 0–12 h and 24–28 h were collected in batch 1, and 14–22 h in batch 2. (B) Histograms of PI staining of cells harvested at 0–28 h. G1/S transitions are marked at time 0 h and 24 h. (C) Expression levels of genes encoding for cyclins as measured by qRT-PCR. The data were normalized to the reference gene GAPDH (black lines), and presented as fold change relative to that in the asynchronously cycling population. Entry to G1 is marked by arrow (14 h). (D) Expression levels of genes encoding for invadopodia components measured by qRT-PCR. The data were normalized to the reference gene GAPDH (black lines), and presented as fold change relative to that in the asynchronously cycling population. Entry to G1 is marked by the arrow (14 h). (E) Protein expression levels of cyclins and invadopodia components as assessed by western blotting; β-actin is used as loading control. (F) Quantification of the percentage of cells in G1 versus S/G2/M in the total cell population, in cells assembling endogenous Tks5+ invadopodia and Tks5–GFP+ invadopodia (overexpression). The mean±s.e.m. number of cells per field of view (FOV) is shown; >100 cells, NFOV=15. *P=0.01539, **P=0.00326 (Mann–Whitney U-test). (G) Representative image of a G1 cell assembling Tks5–GFP+ invadopodia and the underlying degraded gelatin (white arrowheads). Scale bar: 10 µm.
When we evaluated the expression levels of invadopodia components, we saw that cortactin (CTTN) transcript levels remained relatively stable throughout the cell cycle (Fig. 3D). On the other hand, the MT1-MMP transcript levels peaked in G2/M and gradually dropped during G1 phase, while the TKS5 transcript (full-length, long isoform also known as TKS5α) level peaked in early G1 and also gradually dropped until approximately 20 h after the release. Surprisingly, unlike the transcript levels, the protein level of cortactin increased in G1 (18–24 h), while the Tks5 protein exhibited minimal fluctuation throughout the cell cycle (Fig. 3E). These results imply that the Tks5 protein is present in all phases of the cell cycle. This prompted us to test whether its localization to invadopodia was enriched in the G1 phase. First, we performed immunofluorescence for endogenous Tks5 and assessed the number of Tks5+ invadopodia in G1 versus S/G2/M phases. Consistent with the increased ECM degradation in G1, 71% of cells containing Tks5+ invadopodia were also in G1 phase (Fig. 3F). Furthermore, in order to amplify recruitment of Tks5 to invadopodia, we expressed Tks5–GFP in MDA-MB-231-Cdt1-mKO2 cells, which allowed us to classify cells into G1 and S/G2/M phases. The quantification of Tks5–GFP puncta confirmed the results of immunofluorescence assays, and revealed that 75% of cells containing Tks5–GFP+ puncta were in G1 (Fig. 3F,G). These proportions are significantly higher than the proportion of cells in G1 found in a total cell population, indicating that Tks5 is recruited to invadopodia primarily during the G1 phase.
Taken together, our findings suggest that invasion during the G1 phase is supported by an increase in the expression levels of the key invadopodia components cortactin and MT1-MMP and the enhanced recruitment of Tks5 to invadopodia.
Elimination of invadopodia affects cell cycle phase transitions
Considering that the assembly of invadopodia is a structurally and metabolically intensive process, requiring a large number of actin regulators, proteases and kinases, as well as both oxidative phosphorylation and glycolysis, suggesting high consumption of energy (Kropyvko, 2015; Murphy and Courtneidge, 2011; Saini and Courtneidge, 2018; van Horssen et al., 2013), we asked whether cell cycle progression may be affected in cells producing invadopodia. As Tks5 is an essential component of invadopodia assembly, its elimination effectively reduces ECM degradation (Fig. S4A). We used stable Tks5 knockdown (Tks5-KD) cells to test the effect of invadopodia assembly on cell cycle progression. Consistent with previous reports, cell proliferation assay in 2D indicated that the elimination of invadopodia has no effect on the overall timing of cell division (Fig. S4B) (Iizuka et al., 2016). However, the FUCCI reporters indicate that there is an accumulation of cells in early S phase (yellow) when invadopodia are blocked (Tks5-KD), which is accompanied by a concomitant reduction of cells in late S/G2/M phases (Fig. S4C), and no change in the proportion of cells in G1. Consistent with the flow cytometry measurements, the live imaging data showed that the duration of G1 remained the same. On the other hand, the duration of early S phase increased, while late S/G2/M duration decreased in Tks5-KD cells (Fig. S4D). To confirm these findings by methods independent of the FUCCI reporters, we pulsed the wild-type Tks5 (Tks5-WT) and Tks5-KD cells with BrdU for 1 h and chased for 4 and 7 h to monitor the transition of BrdU-positive cells (Fig. S4E). We observed a modest accumulation of cells in the early S phase bracket at 7 h chase and a reduction in the proportion of cells in late G2/M phase. These results suggest that elimination of invadopodia does not affect the length of G1 phase, but may indirectly delay progression through the S phase and accelerate the progression through G2/M.
Cyclin-dependent kinase inhibitor p27kip1 localizes to invadopodia and facilitates invadopodia turnover
The CIP/KIP family of cyclin-dependent kinase inhibitors, in addition to their roles in cell cycle regulation, have also been implicated in pro-tumorigenic roles in the cytoplasm, such as inhibition of apoptosis (Blagosklonny, 2002), and regulation of the cytoskeletal reorganization, cell migration and invasion (Besson et al., 2008). Furthermore, the cytoplasmic pool of p27kip1 was recently shown to play a novel role in invadopodia turnover through the recruitment of PAK1 in mouse embryonic fibroblasts (Jeannot et al., 2017), making it a candidate for mediating the invadopodia function in G1.
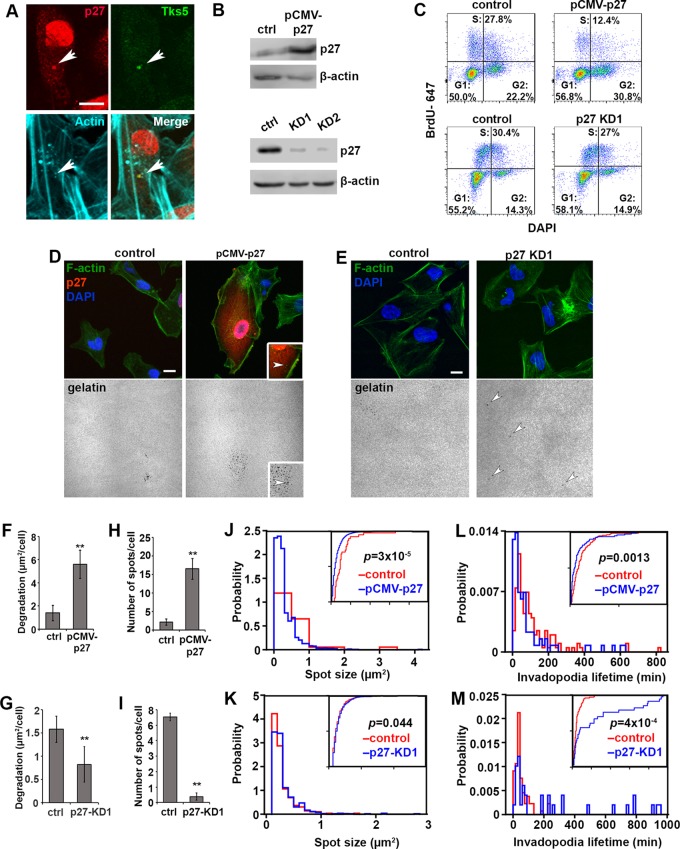
To gain insight into the molecular interplay that supports invadopodia function in G1 phase, we first sought to confirm that p27kip1 was recruited to the sites of invadopodia in MDA-MB-231 cells. We indeed observed p27kip1 protein in the cytoplasm, colocalizing with actin-rich puncta (Fig. 4A). Then, we either overexpressed or depleted p27kip1 and assessed the effect on cell cycle and invadopodia function (Fig. 4B). Consistent with its role in cell cycle progression, overexpression of p27kip1 reduced the proportion of cells entering S phase, as shown by the BrdU incorporation assay (Fig. 4C). The depletion of p27kip1, however, had a minimal effect on the cell cycle profile, similar to previous observations in MDA-MB-231 cells (Fig. 4C) (Zhao et al., 2015). To monitor the effect on invadopodia function, we performed the 2D degradation assay and measured the total degraded area per cell (Fig. 4D,E). Overexpression of p27kip1 resulted in a significant increase in the amount of degradation (Fig. 4F), while the knockdown showed a decrease (Fig. 4G), suggesting that p27kip1 is required for invadopodia-mediated degradation.
Fig. 4.
Cyclin-dependent kinase inhibitor p27kip1 localizes to invadopodia and regulates invadopodia dynamics. (A) Representative image of endogenous p27kip1 (red), Tks5 (green) and actin (cyan) in MDA-MB-231 cells. Colocalization of puncta is marked with white arrows. Scale bar: 10 µm. (B) Western blot analysis of the vector control, overexpression (pCMV-p27) and two different knockdown lines (p27-KD) of p27kip1. β-actin is used as a loading control. (C) BrdU/DAPI flow cytometric analysis of cell cycle profiles of pCMV-p27 or p27kip1 (p27)-KD1 cells and their corresponding controls. The areas show the percentage of cells in each cell cycle phase. (D) Representative immunofluorescence images of vector control and p27kip1-overexpressing cells with labeled nuclei (DAPI, blue), actin (phalloidin, green), p27kip1 (red) and gelatin (gray). Insets are a magnified area of a p27kip1-overexpressing cell. The white arrowhead points at the colocalization of actin, p27kip1 and the underlying degradation spot, indicating a mature invadopodium. Scale bar: 10 µm. (E) Representative immunofluorescence images of control and p27-KD1 cells with labeled nuclei (DAPI, blue), actin cytoskeleton (phalloidin, green) and gelatin (gray). White arrowheads point to degraded spots. Scale bar: 10 µm. (F) Quantification of the degraded area per cell in control and p27-overexpressing cells (pCMV-p27). (G) Quantification of the degraded area per cell in control and p27-KD1 cells. (H) Quantification of the number of degradation spots per cell in control and p27kip1-overexpressing cells (pCMV-p27). (I) Quantification of the number of degradation spots per cell in control and p27-KD1 cells. (J) Distribution of the degradation spot size values in control (red) and cells overexpressing p27kip1 (pCMV-p27, blue), P=3×10−5. (K) Distribution of the degradation spot size values in control (red) and p27-KD1 (blue) cells, P=0.044. (L) Distribution of the invadopodia lifetime values measured from control (red) or p27kip1-over-expressing cells (pCMV-p27, blue), P=0.0013. (M) Distribution of the invadopodia lifetime values measured from control (red) or p27-KD1 (blue), P=4×10−4. For each bar or distribution presented in F–M, >60 cells were analyzed in multiple biological replicates. For F–I, the means±s.e.m. per field of view (FOV) from three biological replicates are presented. **P<0.01, ***P<0.001 (Mann–Whitney U-test). For J–M, cumulative distribution function plots are included as insets and Kolmogorov–Smirnov tests were performed to determine the statistical significance of the difference between two distributions.
In addition to the measurements of total degraded area per cell, the size and number of degradation spots are known to reflect invadopodia dynamics and lifetime (Moshfegh et al., 2014). Invadopodia exert mechanical forces onto the ECM via protrusion–retraction oscillations (Esmaeili Pourfarhangi et al., 2018a; Magalhaes et al., 2011). These oscillations, along with the delivery of MT1-MMP (Monteiro et al., 2013), lead to the degradation of the surrounding ECM, that is, the ‘degradation spot’ in 2D invadopodia degradation assays. The number of degradation spots indicates the number of mature invadopodia assembled in a cell, and the size of the individual degradation spots positively correlates with the number of protrusion–retraction cycles an invadopodium undergoes before its disassembly. These measurements can serve as an indirect readout of invadopodia lifetime. In other words, the smaller the degradation spot, the shorter the lifetime and the higher the turnover (Moshfegh et al., 2014).
Using these indicators, we observed that the increase in total degradation in cells overexpressing p27kip1 was due to an increase in the number of degradation spots (Fig. 4H), suggesting that p27kip1 stimulates invadopodia assembly. Plotting the probability distribution functions (PDFs) of the spot sizes revealed that the overexpression of p27kip1 leads to a significant enrichment of smaller spot size compared to control [Fig. 4J, cumulative distribution functions (CDFs) in inset]. A smaller size and higher number of degradation spots suggests that p27kip1 enhances invadopodia turnover. To corroborate this, we performed live imaging and measured invadopodia lifetime. In Fig. 4L, the distribution of invadopodia lifetimes is presented. Consistent with the smaller degradation spot sizes, invadopodia with high expression of p27kip1 displayed significantly shorter lifetimes (Fig. 4L). Conversely, the reduced degraded area per cell upon stable knockdown of p27kip1 results from fewer degradation spots (Fig. 4I; Fig. S5) of larger size (Fig. 4K; Fig. S5). The lifetime measurements show that the invadopodia are more stable in the knockdown cells, further corroborating the notion that p27kip1 regulates invadopodia dynamics (Fig. 4M, CDFs in inset). These results suggest that p27kip1 regulates invadopodia dynamics and is required for the invadopodia turnover.
Tumor cells in vivo form invadopodia in G1 phase
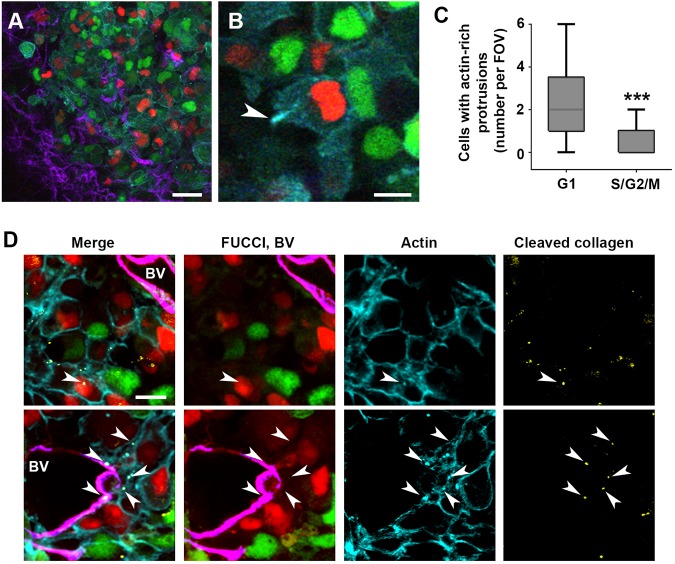
To determine whether the requirement for G1 phase is also true for invadopodia-driven invasion in vivo, we injected MDA-MB-231-FUCCI-mCerulean3-Lifeact7 cells orthotopically in the mammary fat pad of SCID mice. After 8–10 weeks, the xenograft tumors were exposed with a skin flap surgery and imaged using intravital multiphoton microscopy (Bayarmagnai et al., 2018; Harney et al., 2016). We recorded 4D stacks of tumor cells in the perivascular niche, where the invadopodia phenotype is found, and assessed the cell cycle phase of all cells and of cells forming invadopodia. We observed that the cells of each phase were evenly distributed throughout the tumor in any given field of view (Fig. 5A). Next, we identified cells forming actin-rich protrusions, indicative of invadopodia (Fig. 5B; Movie 6) and quantified the number of G1 (red nuclei) versus S/G2/M (green nuclei) cells forming such protrusions. We found that, in each field of view, G1 cells were more likely to show dynamic protrusions than S/G2/M cells (Fig. 5C).
Fig. 5.
Invadopodia are enriched in G1 cells in vivo. (A) Intravital image of MDA-MB-231-FUCCI-Lifeact7-mCerulean3 mouse xenograft. Cells in G1 (red nuclei), and S/G2/M (green nuclei) are shown, with actin cytoskeleton in cyan and collagen fibers in purple. Scale bar: 50 µm. (B) Zoom-in on a cell in G1 phase (red nucleus) extending an actin-rich protrusion (arrowhead). Also see Movie 6. Scale bar: 10 µm. (C) Quantification of cells with actin-rich protrusions per field of view, grouped per phase of the cell cycle. >50 cells with actin-rich protrusions from 12 fields of view in three mice were analyzed. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum and maximum values. ***P<0.001 (Mann–Whitney U-test). (D) Immunofluorescence images of cryosections from two different tumors, showing cells in G1 (red nuclei), cells in S/G2/M (green nuclei), the actin cytoskeleton (cyan), blood vessels (BV, magenta) and cleaved collagen I (Col1-C¾ antibody, yellow). Also see Movie 7. White arrowheads indicate mature invadopodia defined as actin puncta that colocalize with degraded/cleaved collagen. Scale bar: 20 µm.
Next, we harvested the tumor tissues and immunostained cryosections to confirm that the actin-rich protrusions were indeed invadopodia. The defining feature of an invadopodium is its ability to break down the ECM through deposition of MMPs, which are capable of cleaving the triple helix of collagen fibers. This cleavage and the subsequent untwisting of the fibers leads to the local denaturation of the triple helical structure and exposure of cleaved ends (Gligorijevic et al., 2014; Lodillinsky et al., 2016). We identified the actin-rich puncta in cryosections and asked whether they colocalized with local denaturation of the collagen, as identified by antibody staining for the type 1 collagen cleavage site. We focused on the perivascular niche identified by the presence of large blood vessels (CD31 staining for endothelial cells). We found that actin-rich puncta on top of cells in G1 indeed colocalized with cleaved collagen staining (Fig. 5D, white arrowheads). The 3D reconstruction revealed that the actin punctum is located at the surface of the cell (Movie 7). Additionally, the G1 cells forming invadopodia can also be seen degrading collagen surrounding blood vessels, consistent with their essential role for intravasation during metastasis. Thus, we show that in our in vivo model system, invadopodia also perform their ECM-degrading function in the G1 phase.
DISCUSSION
In this study, we show that invadopodia function is enhanced in the G1 phase of the cell cycle in vitro as well as in vivo. We show that the cells initiating and leading invasion strands from 3D spheroids (i.e. leader cells) largely reside in G1. Through time course experiments, we monitored cell cycle progression and found that the expression level of key invadopodia components (cortactin and MT1-MMP) is upregulated in G1 phase, while the localization to invadopodia of Tks5 is enriched in G1. The inhibition of invadopodia assembly by silencing Tks5 alters cell cycle progression by increasing the proportion of cells in early S phase. This leads to the elongation of the early S phase, while late S/G2/M phases are shortened.
Our current study suggests that cell cycle status is another determinant of the invadopodia in vivo. Previously, we demonstrated that invadopodia assemble in vivo under specific extrinsic microenvironmental conditions present in the perivascular niche, and that the invadopodia function was necessary for metastasis (Gligorijevic et al., 2012, 2014). However, even in the perivascular niche, only 15% of cells formed invadopodia at any given time, raising the possibility that additional intrinsic factors were required for invadopodia assembly. Our in vivo analysis of MDA-MB-231-FUCCI-mCerulean3-Lifeact7 tumors supports such a line of thinking and identifies cell cycle as an important determinant of invadopodia function in vivo, in conjunction with the microenvironmental conditions.
The molecular relationship between cell cycle progression and invadopodia in cancer has not been studied. Two recent studies aimed to address this gap. First, a mechanistic link implicating cell cycle regulators in invadopodia function has been revealed in Caenorhabditis elegans (Matus et al., 2015). During development, the anchor cell (AC) requires an arrest in G1 for invadopodia-driven breakdown of the basement membrane. The G1 arrest is essential for invasion, as mutations leading to a mitotic AC have been shown to abolish invadopodia and invasion (Matus et al., 2015). This arrest is mediated by the cyclin-dependent kinase inhibitor CKI-1 and histone deacetylase HDA-1, which promotes pro-invasive gene expression required for invadopodia formation and function in G1. The upregulation of pro-invasive genes is consistent with our findings that gene and protein levels of Tks5 and MT1-MMP are increased in G1 in MDA-MB-231 breast cancer cells.
Second, in a recent study using mammalian cells, p27kip1, a cyclin-dependent kinase inhibitor highly expressed in G0/G1 phases, was shown to physically interact with the structural invadopodia protein cortactin. This interaction mediates phosphorylation of cortactin by PAK1, promoting invadopodia turnover (Jeannot et al., 2017). Since the role of p27kip1 in invadopodia function is restricted to the cytoplasmic compartment, its role in invadopodia was assumed to be distinct from the cell cycle-related function of p27kip1 in the nucleus. However, in Swiss 3T3 cells, the ratio of the cytoplasmic to the nuclear pools of p27kip1 was shown to be high in G1 and drop with cell cycle progression (Shin et al., 2005; Wang et al., 1999). Careful assessment of the nuclear and cytoplasmic distributions of p27kip1 in cancer cells throughout cell cycle is required to test whether there is a direct link between the cell cycle- and invadopodia-related roles of p27kip1.
We observed that Tks5 localization to invadopodia is regulated by the cell cycle, while the Tks5 protein level remains stable throughout cell cycle (Fig. 3E). In addition to controlling Tks5 localization, the cell cycle may also be regulating its activation. Tks5 is heavily regulated by post-translational modifications, particularly by phosphorylation of its tyrosine residues. Recent large-scale phosphoproteomics studies identified novel phospho-serine/threonine sites on Tks5, the phosphorylation of which varies depending on the cell cycle (Schaffer et al., 2015). This finding is intriguing as the CDKs, which are key cell cycle regulators, are serine/threonine kinases and thus potential modulators of Tks5 activity and recruitment. In fact, a member of the CDK family, CDK5, was found to promote invadopodia and invasion through the phosphorylation of the actin regulator caldesmon (Quintavalle et al., 2011), warranting further studies on the effect of other CDKs on invadopodia function. Moreover, the altered dynamics of cell cycle progression in Tks5-KD cells suggests a reciprocal effect of invadopodia on cell cycle (Fig. S4). While we and others did not observe an effect of Tks5-KD on the rate of proliferation in 2D, there are reports demonstrating that the Tks5 proteins are required for cell proliferation in 3D cultures and tumor growth in vivo in breast cancer (Blouw et al., 2015) and melanoma (Iizuka et al., 2016). Given the potential of Tks5 as a prognostic marker of cancer progression in patients (Stylli et al., 2014), it is of utmost importance to further investigate the mechanistic link between Tks5 activity and cell cycle progression.
Studies of tumor cell motility and metastasis are motivated by the need to develop effective therapeutics that can halt the spread of cancer cells to secondary locations, which is the main cause of cancer-related deaths. On the other hand, most clinically available chemotherapies target actively cycling cells and often work by inducing a G1 arrest (Diaz-Moralli et al., 2013). It is becoming increasingly clear that both the proliferation and motility characteristics of cancer cells need to be taken into account when designing therapies. We previously showed a direct link between invadopodia in vivo and metastasis (Gligorijevic et al., 2014). Our current study indicates that breast carcinoma cells in G1 possess the highest capabilities for invadopodia function, and hence, metastasis. This suggests that anti-proliferative drugs, many of which are arresting cancer cells in G1, may result in higher invasion and metastasis. Conversely, targeting cells with invadopodia may promote the proliferative state and the growth of metastases. Taken together, our data suggest that understanding the mechanistic relationship between cell cycle progression and invadopodia is essential for effectively targeting cancer metastasis.
MATERIALS AND METHODS
Ethics statement
All experiments on mice (Mus musculus) were conducted in accordance with the NIH regulations and approved by the Temple University IACUC protocol number 4766.
Cell lines and culture conditions
The cell cultures were maintained in DMEM supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals) and 50 U penicillin/50 µg streptomycin (Corning) per ml. BT549 cells (Fox Chase Cell Culture Facility, Philadelphia PA) were maintained in RPMI medium supplemented with 2 mM glutamine, 10% FBS and 50 U penicillin/50 µg streptomycin (Corning) per ml.
MDA-MB-231-FUCCI and BT549-FUCCI stable cell lines were generated by transducing FUCCI constructs, mKO2-hCdt1 (amino acids 30/120) and mAG-hGem (amino acids 1/110), with the replication-defective, self-inactivating lentiviral expression system. To visualize cell motility, the plasmid bearing the actin-binding protein mCerulean3-Lifeact7 was introduced to MDA-MB-231-FUCCI cells by electroporation (Lonza) and selected for with 500 µg/ml G418 sulfate (cat. BP6735, Fisher Bioreagents). We performed fluorescence-activated cell sorting (FACS) after 14–20 days of selection and collected the top 20% highest fluorescing cells (after excluding the top 5% highest fluorescing population). The mCerulean3-Lifeact7 expression was maintained under 500 µg/ml G418 sulfate. The mCerulean3-Lifeact7 plasmid was Addgene plasmid #54721 (deposited by Michael Davidson). FUCCI constructs were a gift from the Miyawaki laboratory and RIKEN Tsukuba BioResource Center (Japan). The Tks5–GFP construct was a gift from Sara Courtneidge (OHSU, Portland, OR).
The stable knockdown (KD) cell lines was generated by transducing MDA-MB-231-FUCCI-mCerulean3-Lifeact7 or unlabeled MDA-MB-231 cells with lentiviral particles (5 particles/cell) containing shRNA in pLKO.1 vector targeting either Tks5 (TRCN0000136512, Sigma-Aldrich MISSION library) or p27kip1 (TRCN0000039928-KD2, and TRCN0000009856-KD1; Sigma-Aldrich MISSION library). The cells were selected with 0.5 µg/ml puromycin (cat. 0210055210, MP Biomedicals).
2D matrix degradation assay and immunofluorescence
35 mm glass-bottom MatTek (MatTek Corporation) dishes were coated with fluorescently labeled gelatin as previously described (Sharma et al., 2013). For cell cycle synchronization experiments, cells pre-treated with DMSO control, 10 µM lovastatin (G1 phase) (PHR1285, Sigma-Aldrich) or 1 µg/ml mitomycin C (S phase) (11435, Cayman Chemical) for 24 h, were then washed with PBS, trypsinized and re-plated on gelatin-coated glass-bottom dishes for the indicated number of hours. The cells were fixed for 15 min with 4% paraformaldehyde, permeabilized for 5 min in 0.1% Triton X-100, blocked for 2 h in 1% BSA with 1% FBS in PBS, and incubated with primary antibodies for 1–3 h, then with secondary antibodies, DAPI and phalloidin–Alexa Fluor 633 (1:250) for 1–2 h at room temperature. Rabbit anti-p27kip1 (Abcam, ab32034, 1:100) and mouse anti-Tks5 (clone 13H6.3, MABT336, Millipore) were used. The samples were imaged in the laser scanning confocal mode of a hybrid confocal-multiphoton microscope (Olympus FV1200). Gelatin degradation was quantified by thresholding the signal in the gelatin channel and measuring the number, size and the total area of degradation spots by using the Particle Analysis tool in FIJI (Schindelin et al., 2012). The FUCCI signal was scored to identify the cell cycle phase of cell populations. The number of cells in each phase was counted, and represented as percentage values per field of view. Cells with invadopodia were identified as cells with a presence of actin or Tks5-positive puncta located on degradation holes in the underlying gelatin.
3D spheroid invasion assay and immunofluorescence
3D spheroids were generated by the hanging drop method. For this, 5000 cells per 20 µl drop containing 4.8 mg/ml methylcellulose, 20 µg/ml Nutragen (Advanced Biomatrix), were placed on the lid of tissue culture dishes. The lids were carefully turned and placed on the bottom reservoir of the dishes filled with PBS to prevent evaporation. The spheroids were allowed to form for 48 h. Alternatively, spheroids were generated in a 96-well flat bottom dish, coated with 1.5% (w/v) agarose. Briefly, 0.3 g of agarose was added to 20 ml DMEM and autoclaved for 20 min at 121.1°C, 103 kPa. Then, 50 µl of 1.5% agarose was distributed to each well of a flat bottom 96-well plate and cooled to room temperature for ∼20 min. The hermetically sealed plates were stored for no more than 10 days. The cells were trypsinized, counted and diluted to 2.5×104 cells/ml in ice-cold medium. Matrigel (thawed on ice overnight) was added to final concentration of 2.5%. Cell mixture was distributed to the agarose-coated 96-well plate, with each well containing 5000 cells in 200 µl. All reagents were kept on ice and pre-chilled pipette tips were used. The plate was centrifuged at 4°C for 10 min at 1000 g and spheroids were allowed to form over 72 h at 37°C.
The fully formed spheroids were embedded in 50 µl of 5 mg/ml rat-tail collagen I gel (cat. 354249, Corning, the alternate gelation procedure was followed from the manufacturer's guidelines) and distributed to each well of a 24-well plate. Invasion was recorded by live imaging at 1-h intervals over 72 h, using the laser scanning confocal mode of a hybrid confocal-multiphoton microscope (Olympus FV1200). The cell cycle distribution was determined by counting cells in each phase of the cell cycle and presented as percentage values per spheroid. For the quantification of cells at the spheroid periphery, at time 0 h of time-lapse imaging, the ‘periphery’ was defined as a 20 µm-wide ring which includes two or three layers of cells most likely to initiate strand invasion. The cells ‘initiating invasion’ were identified as the first cell to emerge out of the spheroid periphery into the surrounding collagen, at a unique direction. The leader cells were identified as cells at the tip of the multicellular invasion strand.
For immunofluorescence, the spheroids were fixed in 4% paraformaldehyde for 20 min, permeabilized in 0.1% Triton X-100 for 20 min, blocked in 1% FBS with 1% BSA in PBS overnight at 4°C, incubated with primary antibody against cleaved collagen I (Col1-C¾, ImmunoGlobe, 1:100) overnight at 4°C and with secondary antibody for 3–4 h at room temperature. For the MMP inhibitor treatment, GM6001 (cat. 13533, Cayman Chemical) was added to the collagen gel mixture and the medium to give a final concentration of 25 µM. The spheroids were allowed to invade for 72 h, after which the spheroids were fixed and imaged. The invasion area was measured using FIJI as an area covered by cells outside the spheroid core.
Leader cell switching in 3D invasion strands
The image stacks (72 h, 1 frame/h; 1024 µm×1024 µm×150 µm, with 10 µm z-slices) used in the analysis were maximum-projected, and saved as composite images. Using maximum projection images, the leader cells at the tip of multicellular strands were tracked and analyzed using a semi-automated in-house algorithm written in Matlab (Mathworks, Natick, MA; available from E.T. upon request). The axis of a given invasion strand was defined as the line connecting the initial position of the first leader cell to the final position of the last leader cell (as shown in Fig. S2A). The individual leader cells were tracked through time (Fig. S2B), and their position vectors were projected onto the strand axis using a standard 2D rotation matrix (Fig. S2C). These projected distances in (x′, y′) reference frame were then plotted as a function of time, and color-coded according to their corresponding cell cycle phase (Fig. S2D). A representative combined plot of projected distances for subsequent leaders in the same invasive strand is shown in Fig. 1G; see also Fig. S2E for additional examples.
2D live-cell imaging
5×104 cells were seeded on gelatin-coated glass-bottom dishes and allowed to adhere for 2 h before imaging. Live-cell imaging was performed on a wide-field Olympus IX-81 microscope, equipped with an environmental chamber (In Vivo Scientific). The images were acquired at a single focal plane using an Olympus 60× oil immersion objective in the CFP, FITC, TRITC, Cy5 and DIC channels. Images were acquired at 10-min or 1-h intervals for up to 60 h. For FUCCI expression validation, nuclei of single cells were tracked in both red and green channels, and their mean gray values were recorded.
Invadopodia lifetime assay
MDA-MB-231-mCerulean3-Lifeact7 cells transiently transfected with vector control or pCMV-p27 plasmids, as well as control and p27-KD1 or p27-KD2 cells were plated on gelatin-coated glass-bottom dishes. Time-lapse images were acquired every 6 min for 24 h and analyzed using FIJI softwarw. The invadopodia lifetime was defined as the time between the appearance and the disappearance of actin puncta. Actin puncta present throughout the entire stack were excluded from the analysis. The lifetime PDFs and CDFs were calculated using Matlab (Mathworks, Natick, MA) and presented as histograms and CDFs. Statistical significance of the difference between distributions was calculated using Kolmogorov–Smirnov test (also see the ‘Statistical Analysis’ section).
Cell cycle synchronization methods
For cell cycle synchronization with drugs, cells were pre-treated with DMSO control, 10 µM lovastatin (G1 phase) (cat. PHR1285, Sigma-Aldrich) or 1 µg/ml mitomycin C (S phase) (cat. 11435, Cayman Chemical) for 24 h. The cells were harvested for PI staining/flow cytometry and western blot analyses.
For synchronization with a double-thymidine block, the 5×105 cells were plated per 10 cm dish per time time point. The cells were blocked with 2 mM thymidine (cat. T9250, Sigma-Aldrich) for 18 h, released for 9 h and blocked with 2 mM thymidine again for 16 h. The cells were released in two batches. One batch was released in the morning with the time points 0 h–12 h collected the same day, and with 24 h–28 h time points the following day. Another batch was released in the evening, allowed to progress through the cell cycle overnight and collected for time points 14 h–22 h the following morning. The samples at each time point were harvested for flow cytometry, gene and protein expression analyses.
Intravital imaging
106 of MDA-MB-231-FUCCI-mCerulean3-Lifeact7 cells were suspended in 100 µl of 20% collagen I in PBS and injected orthotopically into the mammary fat pad of 5–7-week-old female SCID mice. After 8–10 weeks, when the tumor diameter reached 8–10 mm, the animals were surgically prepared for continuous intravital imaging. The tumor tissue was exposed by skin flap non-survival surgery as previously described (Gligorijevic et al., 2014; Harney et al., 2016). Briefly, anesthesia in mice was induced with 5% isoflurane, the levels of which were gradually decreased to, and maintained at 1.5%. The mice were kept in a 30°C environmental chamber (In Vivo Scientific) and hydrated by intraperitoneal injection of 100 µl of warm, sterile PBS every hour.
4D stacks were collected in the multiphoton imaging mode of a hybrid confocal-multiphoton microscope (Olympus FV1200MPE), with a 30× objective with silicone oil immersion (UPLSAPO 30×, NA 1.05, Olympus). The areas were selected based on the presence of flowing macrovessels, visible through the eyepiece using the green FITC filter, as the lack of green autofluorescence. While blood flow indicates proper circulation, large capillary diameter is indicative of the perivascular niches amenable for invadopodia assembly (Gligorijevic et al., 2014). 4D stacks were collected at 3 min intervals for up to 6 h. At the end of each imaging session, the animals were euthanized and the tumor sample was collected.
The acquired time-lapse movies were analyzed using FIJI software. The frames were corrected for motion artifacts using HyperStackReg, an extension based on StackReg plug-in (Thevenaz et al., 1998). The cells extending invadopodia were identified based on the morphodynamic properties and scored for cell cycle phase based on the FUCCI signal.
Tissue immunofluorescence
At the conclusion of intravital imaging, the animals bearing the MDA-MB-231-FUCCI-mCerulean3-Lifeact7 tumors were euthanized and the tumors were excised. The tumor tissue was fixed in 4% PFA for 30 min at room temperature and shifted to 4°C overnight. The next day, the tumor tissue was washed in cold PBS and transferred to 30% sucrose, incubated overnight at 4°C until the tissue sank to the bottom of the tube. The tumor tissue was embedded in Optimal Cutting Temperature (OCT) compound and frozen in isopentane chilled on dry ice. The sample was sectioned with a cryotome at −15°C to −20°C into 8–10 µm slices and placed on positively charged glass slides. The tissue sections were stored in −80°C until staining. Prior to staining, the OCT was washed off with water. The tissue sections were permeabilized with 0.1% Triton X-100 in PBS, washed in PBS and briefly air dried. The tissue sections were outlined with a liquid-repellent marker pen to contain liquid droplets during the subsequent incubations. The samples were blocked overnight with PBS containing 5% BSA and 1% FBS at 4°C, in a humidified chamber to prevent the evaporation of the droplets and protected from light. Primary antibody for cleaved collagen I (Col1-C¾, ImmunoGlobe, 1:100) and CD31 (ab28364, Abcam, 1:100) were diluted in the blocking buffer and incubated with the tissue sections overnight at 4°C, in a humidified chamber. The secondary antibody incubation was performed at room temperature for 3–4 h. The tissue sections were then washed with PBS, briefly air dried and mounted in the anti-fade mounting medium (Fluoromount-G, Invitrogen). The slides were imaged with a 60× objective on the Olympus FV1200 confocal microscope, collecting native fluorescence from mCerulean3–Lifeact7 and FUCCI reporters mAG and mKO2. In addition, immunofluorescence signals for the anti-CD31-Alexa Fluor 546 and the anti-Col1-C¾–Alexa Fluor 647 antibodies were collected. The differential morphological features were used to decouple blood vessels and the mKO2-FUCCI-labeled nuclei in the red channel via the particle analysis plugin. The images were analyzed with FIJI software and the 3D reconstruction performed using the Imaris software (Bitplane Imaris x64 8.3.1).
Gene expression qRT-PCR
Total mRNA was extracted with Trizol (Invitrogen) according to the manufacturer's recommendations. The RNA yield was measured with NanoDrop (Thermo Fisher Scientific). 1 µg of RNA was used for reverse transcriptase PCR (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems) reactions; a 1:20 dilution of cDNA was used for quantitative real time PCR (qRT-PCR; PowerUp SYBR Green Mastermix, Applied Biosystems). For Tks5, the primers detecting the most abundantly expressed isoform in adult tissues, the full-length, long isoform TKS5α, were used (Cejudo-Martin et al., 2014; Li et al., 2013). The primer sequences are provided in Table S1.
Western blot analysis
The cells grown on a gelatin layer were harvested in RIPA lysis buffer (cat. R3792, Teknova), supplemented with protease inhibitors (Roche, complete cocktail) and phosphatase inhibitors (Sigma-Aldrich, Halt cocktail). SDS-PAGE was performed with 20–30 µg total protein per sample, transferred to a PVDF membrane (Immobilon), blocked with 5% BSA in TBST (Tris-buffered saline with 0.1% Tween-20) for 1 h at room temperature and incubated with the primary antibody in 5% BSA in TBST overnight at 4°C. Mouse anti-Tks5 (anti-SH3PXD2A, Millipore, 1:1000), mouse anti-cortactin (Abcam, ab33333, 1:1000), mouse anti-cyclin D1 (Santa Cruz Biotechnology, sc-8396, 1:500), mouse anti-cyclin E (Santa Cruz Biotechnology, sc-377100, 1:500), mouse anti-cyclin A (Santa Cruz Biotechnology, sc-271682, 1:500), mouse anti-Cyclin B1 (Santa Cruz Biotechnology, sc-245, 1:500) and mouse anti-β-actin, as loading control (Santa Cruz Biotechnology, C4, sc-47778, 1:500), antibodies were used. The membranes were then incubated with HRP-conjugated secondary antibody (Cell Signaling, 7076S, 7074S, 1:5000) in 5% non-fat milk in TBST for 1 h at room temperature. Bands were visualized using chemiluminescence detection reagents (WesternBright, Advansta) and a blot scanner (C-DiGit, LI-COR).
Cell cycle analysis via PI staining and flow cytometry
Cells subjected to the indicated treatments were harvested and re-suspended in ice-cold PBS containing 2% FBS. Cells were counted, spun at 170 g for 5 min and re-suspended in 250 µl PBS. Cells were fixed by a dropwise addition of 750 µl of ice-cold 95% ethanol, while vortexing gently at low speed. The samples were incubated at 4°C overnight or up to 1 week until ready for staining. Before staining, cells were allowed to equilibrate to room temperature (∼30 min), to loosen the cell pellet, and then centrifuged at 400 g for 5 min. The supernatant was aspirated and the cell pellet was re-suspended in 150 µl of the PI staining solution (50 µg/ml PI, 0.2 mg/ml RNase A, 0.02% Triton X-100, PBS) and incubated for 30 min at room temperature, protected from light. After the staining incubation, the samples were diluted with 150 µl of PBS in 10 mM EDTA to prevent clumps, and filtered through 40 µm mesh cell strainer. The samples were measured using BD LSRII flow cytometer (BD Biosciences). The results were analyzed using the FlowJo™ 10 software.
Cell cycle analysis with BrdU incorporation and DAPI staining
The BrdU incorporation and DAPI staining was performed according to the manufacturer's guidelines (BD Pharmingen APC BrdU/DAPI flow kit, cat. 552598). Briefly, cells were treated with 10 µM BrdU for 1 h. The cells were harvested and fixed in the Cytofix/Cytoperm buffer for 15 min at room temperature, permeabilized in the Cytoperm Buffer Plus for 10 min on ice and re-fixed with the Cytofix/Cytoperm buffer for 5 min on ice. The cells were treated with DNase for 1 h at 37°C. Then, the cells were incubated with APC-conjugated anti-BrdU antibody for 20 min at room temperature, washed and stained with DAPI to determine total DNA content. For the p27kip1-KD stable knockdown cells, Alexa Fluor-647-conjugated anti-BrdU antibody and the associated reagents were used from the Phase-Flow kit from Biolegend (cat. 370706, Biolegend). The data were acquired on BD LSRII flow cytometer and analyzed using FlowJo™ 10 software.
Cell proliferation assay
104 cells were plated in 96-well plates in triplicates per cell line per time point. Resazurin (final concentration of 15 µg/ml) was added 60 min before reading measurements. A TECAN plate reader was used to measure the fluorescence (using an excitation at 540 nm and emission at 590 nm) at 24-h intervals over 5 days from blank wells containing media only and test wells. The blank measurement values were subtracted from the test well values at each time point. The relative fluorescence units compared to values at day 0 were calculated for each time point.
Statistical analyses
All data points per condition in the manuscript were pooled from multiple biological replicates (numbers indicated in figure captions), and subjected to a one-tailed Mann–Whitney U-test, with the following exceptions. In Fig. 2F, to determine the statistical significance of the degradation rate trends, we fit the data to repeated measures model and performed repeated measures analysis of variance in Matlab (Mathworks, Natick, MA). A two-sample two-sided Kolmogorov–Smirnov test was performed to determine the statistical significance between distributions in Fig. 4J–M, Fig. S5E, once again using Matlab. For datasets with normal distributions, we used t-test, namely in Fig. 2E; Fig. S4A,D.
Supplementary Material
Acknowledgements
We are grateful to Dr Sara Courtneidge and her group (OHSU) for the help with Tks5–GFP and Tks5 immunofluorescence, Dr Asako Sakaue-Sawano and the Miyawaki lab (RIKEN) for FUCCI constructs, Ziran Zhao (Graña lab, Temple Medical School) for advice on cell synchronization strategies and Seul Jung, Theodore Tucker (Gligorijevic lab) for technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: B.B., B.G.; Methodology: B.B., X.G., E.T., B.G.; Software: E.T.; Validation: B.B., L.P., K.E.P., E.T.; Formal analysis: B.B., L.P., K.E.P., E.T.; Investigation: B.B., L.P., K.E.P., B.G.; Resources: B.G.; Writing - original draft: B.B., L.P., B.G.; Writing - review & editing: B.B., L.P., X.G., E.T., B.G.; Visualization: B.B., E.T., B.G.; Supervision: E.T., B.G.; Project administration: B.G.; Funding acquisition: X.G., E.T., B.G.
Funding
This work was supported by the National Institutes of Health [R00 CA172360 and R01 CA230777 to B.G., R01 GM117437 and R03 CA216134-01 to X.G., R01 GM121679 to E.T.]; and by a Concern Foundation Conquer Cancer Now Young Investigator Award to B.G. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.227116.supplemental
References
- Bayarmagnai B., Perrin L., Esmaeili Pourfarhangi K. and Gligorijevic B. (2018). Intravital imaging of tumor cell motility in the tumor microenvironment context. Methods Mol. Biol. 1749, 175-193. 10.1007/978-1-4939-7701-7_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B. T., Sharma V. P., Bravo-Cordero J. J., Simpson M. A., Eddy R. J., Koleske A. J. and Condeelis J. (2013). β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol. Biol. Cell 24, 1661-1675, S1-S11 10.1091/mbc.e12-12-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A., Assoian R. K. and Roberts J. M. (2004). Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors? Nat. Rev. Cancer 4, 948-955. 10.1038/nrc1501 [DOI] [PubMed] [Google Scholar]
- Besson A., Dowdy S. F. and Roberts J. M. (2008). CDK inhibitors: cell cycle regulators and beyond. Dev. Cell 14, 159-169. 10.1016/j.devcel.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M. V. (2002). Are p27 and p21 cytoplasmic oncoproteins? Cell Cycle 1, 391-393. 10.4161/cc.1.6.262 [DOI] [PubMed] [Google Scholar]
- Blouw B., Patel M., Iizuka S., Abdullah C., You W. K., Huang X., Li J.-L., Diaz B., Stallcup W. B. and Courtneidge S. A. (2015). The invadopodia scaffold protein Tks5 is required for the growth of human breast cancer cells in vitro and in vivo. PLoS ONE 10, e0121003 10.1371/journal.pone.0121003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejudo-Martin P., Yuen A., Vlahovich N., Lock P., Courtneidge S. A. and Díaz B. (2014). Genetic disruption of the Sh3pxd2a gene reveals an essential role in mouse development and the existence of a novel isoform of Tks5. PLoS ONE 9, e107674-e13. 10.1371/journal.pone.0107674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B., Shani G., Pass I., Anderson D., Quintavalle M. and Courtneidge S. A. (2009). Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2, ra53-ra53. 10.1126/scisignal.2000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Moralli S., Tarrado-Castellarnau M., Miranda A. and Cascante M. (2013). Targeting cell cycle regulation in cancer therapy. Pharmacol. Ther. 138, 255-271. 10.1016/j.pharmthera.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Esmaeili Pourfarhangi K., Bergman A. and Gligorijevic B. (2018a). ECM cross-linking regulates invadopodia dynamics. Biophys. J. 114, 1455-1466. 10.1016/j.bpj.2018.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili Pourfarhangi K., De La Hoz E. C., Cohen A. R. and Gligorijevic B. (2018b). Contact guidance is cell cycle-dependent. APL Bioeng. 2, 031904 10.1063/1.5026419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay T., Juhász É., Molnár E., Eisenbauer M., Czirók A., Dekan B., László V., Hoda M. A., Döme B., Tímár J. et al. (2013). Cell migration or cytokinesis and proliferation?--revisiting the “go or grow” hypothesis in cancer cells in vitro. Exp. Cell Res. 319, 3094-3103. 10.1016/j.yexcr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- Gerlee P. and Nelander S. (2012). The impact of phenotypic switching on glioblastoma growth and invasion. PLoS Comput. Biol. 8, e1002556 10.1371/journal.pcbi.1002556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B., Wyckoff J., Yamaguchi H., Wang Y., Roussos E. T. and Condeelis J. (2012). N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J. Cell. Sci. 125, 724-734. 10.1242/jcs.092726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B., Bergman A. and Condeelis J. (2014). Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol. 12, e1001995 10.1371/journal.pbio.1001995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harney A. S., Wang Y., Condeelis J. S. and Entenberg D. (2016). Extended time-lapse intravital imaging of real-time multicellular dynamics in the tumor microenvironment. J. Vis. Exp., 112, e54042 10.3791/54042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka S., Abdullah C., Buschman M. D., Diaz B. and Courtneidge S. A. (2016). The role of Tks adaptor proteins in invadopodia formation, growth and metastasis of melanoma. Oncotarget 7, 78473-78486. 10.18632/oncotarget.12954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. and O.Connor P. M. (2001). Methods for synchronizing cells at specific stages of the cell cycle. Curr. Protoc. Cell Biol. Chapter 8, Unit 8.3–8.3.20 10.1002/0471143030.cb0803s00 [DOI] [PubMed] [Google Scholar]
- Javanmoghadam-Kamrani S. and Keyomarsi K. (2008). Synchronization of the cell cycle using lovastatin. Cell Cycle 7, 2434-2440. 10.4161/cc.6364 [DOI] [PubMed] [Google Scholar]
- Jeannot P., Nowosad A., Perchey R. T., Callot C., Bennana E., Katsube T., Mayeux P., Guillonneau F., Manenti S. and Besson A. (2017). p27(Kip1) promotes invadopodia turnover and invasion through the regulation of the PAK1/Cortactin pathway. eLife 6, 2711 10.7554/eLife.22207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. G., Chung H., Yoo Y. D., Lee J. G., Choi Y. I. and Yu Y. S. (2009). Mechanism of growth inhibitory effect of Mitomycin-C on cultured human retinal pigment epithelial cells: Apoptosis and cell cycle arrest. Curr. Eye Res. 22, 174-181. 10.1076/ceyr.22.3.174.5513 [DOI] [PubMed] [Google Scholar]
- Kropyvko S. V. (2015). Interactome of invadopodia scaffold protein TKS5. Biopolym. Cell 31, 417-421. 10.7124/bc.0008FE [DOI] [Google Scholar]
- Lee S. and Helfman D. M. (2004). Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J. Biol. Chem. 279, 1885-1891. 10.1074/jbc.M306968200 [DOI] [PubMed] [Google Scholar]
- Li C. M.-C., Chen G., Dayton T. L., Kim-Kiselak C., Hoersch S., Whittaker C. A., Bronson R. T., Beer D. G., Winslow M. M. and Jacks T. (2013). Differential Tks5 isoform expression contributes to metastatic invasion of lung adenocarcinoma. Genes Dev. 27, 1557-1567. 10.1101/gad.222745.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Z., Dhillon A. S., Anderson R. L., McArthur G. and Ferrao P. T. (2015). Phenotype Switching in Melanoma: Implications for Progression and Therapy. Front. Oncol. 5, 1017 10.3389/fonc.2015.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Zubovitz J., Petrocelli T., Kotchetkov R., Connor M. K., Han K., Lee J.-H., Ciarallo S., Catzavelos C., Beniston R. et al. (2002). PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 8, 1153-1160. 10.1038/nm761 [DOI] [PubMed] [Google Scholar]
- Lodillinsky C., Infante E., Guichard A., Chaligné R., Fuhrmann L., Cyrta J., Irondelle M., Lagoutte E., Vacher S., Bonsang-Kitzis H. et al. (2016). p63/MT1-MMP axis is required for in situ to invasive transition in basal-like breast cancer. Oncogene 35, 344-357. 10.1038/onc.2015.87 [DOI] [PubMed] [Google Scholar]
- Magalhaes M. A. O., Larson D. R., Mader C. C., Bravo-Cordero J. J., Gil-Henn H., Oser M., Chen X., Koleske A. J. and Condeelis J. (2011). Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195, 903-920. 10.1083/jcb.201103045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. and Barbacid M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153-166. 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- Markwardt M. L., Kremers G.-J., Kraft C. A., Ray K., Cranfill P. J. C., Wilson K. A., Day R. N., Wachter R. M., Davidson M. W. and Rizzo M. A. (2011). An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS ONE 6, e17896-e17811. 10.1371/journal.pone.0017896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Lohmer L. L., Kelley L. C., Schindler A. J., Kohrman A. Q., Barkoulas M., Zhang W., Chi Q. and Sherwood D. R. (2015). Invasive cell fate requires G1 cell-cycle arrest and histone deacetylase-mediated changes in gene expression. Dev. Cell 35, 162-174. 10.1016/j.devcel.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D. et al. (2013). Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203, 1063-1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshfegh Y., Bravo-Cordero J. J., Miskolci V., Condeelis J. and Hodgson L. (2014). A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat. Cell Biol. 16, 574-586. 10.1038/ncb2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. A. and Courtneidge S. A. (2011). The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413-426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove E. A., Caldon C. E., Barraclough J., Stone A. and Sutherland R. L. (2011). Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558-572. 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- O'Connell M. P. and Weeraratna A. T. (2013). Change is in the air: the hypoxic induction of phenotype switching in melanoma. J. Invest. Dermatol. 133, 2316-2317. 10.1038/jid.2013.208 [DOI] [PubMed] [Google Scholar]
- Oser M., Mader C. C., Gil-Henn H., Magalhaes M., Bravo-Cordero J. J., Koleske A. J. and Condeelis J. (2010). Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell. Sci. 123, 3662-3673. 10.1242/jcs.068163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsialou A., Bravo-Cordero J. J., Wang Y., Entenberg D., Liu H., Clarke M. and Condeelis J. S. (2014). Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. IntraVital 2, e25294-e25229. 10.4161/intv.25294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F. and Chavrier P. (2009). Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell. Sci. 122, 3015-3024. 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Quintavalle M., Elia L., Price J. H., Heynen-Genel S. and Courtneidge S. A. (2011). A cell-based high-content screening assay reveals activators and inhibitors of cancer cell invasion. Sci. Signal. 4, ra49-ra49. 10.1126/scisignal.2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z. et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Meth 5, 605-607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson I. B. (2002). Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 179, 1-14. 10.1016/S0304-3835(01)00847-3 [DOI] [PubMed] [Google Scholar]
- Saini P. and Courtneidge S. A. (2018). Tks adaptor proteins at a glance. J. Cell. Sci. 131, jcs203661-j6. 10.1242/jcs.203661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487-498. 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Schaffer B. E., Levin R. S., Hertz N. T., Maures T. J., Schoof M. L., Hollstein P. E., Benayoun B. A., Banko M. R., Shaw R. J., Shokat K. M. et al. (2015). Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab. 22, 907-921. 10.1016/j.cmet.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y., Toba K., Fuse I., Sato N., Niwano H., Takahashi H., Tanabe N. and Aizawa Y. (2005). In vitro effect of cyclosporin A, mitomycin C and prednisolone on cell kinetics in cultured human umbilical vein endothelial cells. Thromb. Res. 115, 219-228. 10.1016/j.thromres.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Sharma V. P., Entenberg D. and Condeelis J. (2013). High-resolution live-cell imaging and time-lapse microscopy of invadopodium dynamics and tracking analysis. Methods Mol. Biol. 1046, 343-357. 10.1007/978-1-62703-538-5_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin I., Yakes F. M., Rojo F., Shin N.-Y., Bakin A. V., Baselga J. and Arteaga C. L. (2002). PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat. Med. 8, 1145-1152. 10.1038/nm759 [DOI] [PubMed] [Google Scholar]
- Shin I., Rotty J., Wu F. Y. and Arteaga C. L. (2005). Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin alpha during G1 and prevents nuclear re-entry. J. Biol. Chem. 280, 6055-6063. 10.1074/jbc.M412367200 [DOI] [PubMed] [Google Scholar]
- Steeg P. S. (2016). Targeting metastasis. Nat. Rev. Cancer 16, 201-218. 10.1038/nrc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylli S. S., Luwor R. B., Kaye A. H., I S. T., Hovens C. M. and Lock P. (2014). Expression of the adaptor protein Tks5 in human cancer: Prognostic potential. Oncol. Rep. 32, 989-1002. 10.3892/or.2014.3310 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Yamashita T., Asada M., Mizutani S., Yoshikawa H. and Tohyama M. (2002). Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J. Cell Biol. 158, 321-329. 10.1083/jcb.200202071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P., Ruttimann U. E. and Unser M. (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27-41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- van Horssen R., Buccione R., Willemse M., Cingir S., Wieringa B. and Attanasio F. (2013). Cancer cell metabolism regulates extracellular matrix degradation by invadopodia. Eur. J. Cell Biol. 92, 113-121. 10.1016/j.ejcb.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Vanharanta S. and Massagué J. (2013). Origins of metastatic traits. Cancer Cell 24, 410-421. 10.1016/j.ccr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglietto G., Motti M. L., Bruni P., Melillo R. M., D'Alessio A., Califano D., Vinci F., Chiappetta G., Tsichlis P., Bellacosa A. et al. (2002). Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 8, 1136-1144. 10.1038/nm762 [DOI] [PubMed] [Google Scholar]
- Wang G., Miskimins R. and Miskimins W. K. (1999). The cyclin-dependent kinase inhibitor p27Kip1 is localized to the cytosol in Swiss/3T3 cells. Oncogene 18, 5204-5210. 10.1038/sj.onc.1202912 [DOI] [PubMed] [Google Scholar]
- Widmer D. S., Hoek K. S., Cheng P. F., Eichhoff O. M., Biedermann T., Raaijmakers M. I. G., Hemmi S., Dummer R. and Levesque M. P. (2013). Hypoxia contributes to melanoma heterogeneity by triggering HIF1α-dependent phenotype switching. J. Invest. Dermatol. 133, 2436-2443. 10.1038/jid.2013.115 [DOI] [PubMed] [Google Scholar]
- Winters Z. E., Hunt N. C., Bradburn M. J., Royds J. A., Turley H., Harris A. L. and Norbury C. J. (2001). Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer association with prognosis. Eur. J. Cancer 37, 2405-2412. 10.1016/S0959-8049(01)00327-6 [DOI] [PubMed] [Google Scholar]
- Wolf K., te Lindert M., Krause M., Alexander S., te Riet J., Willis A. L., Hoffman R. M., Figdor C. G., Weiss S. J. and Friedl P. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069-1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Goliwas K. F., Wang W., Taufalele P. V., Bordeleau F. and Reinhart-King C. A. (2019). Energetic regulation of coordinated leader-follower dynamics during collective invasion of breast cancer cells. Proc. Natl. Acad. Sci. USA 116, 7867-7872. 10.1073/pnas.1809964116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Besser A. H., Wander S. A., Sun J., Zhou W., Wang B., Ince T., Durante M. A., Guo W., Mills G. et al. (2015). Cytoplasmic p27 promotes epithelial-mesenchymal transition and tumor metastasis via STAT3-mediated Twist1 upregulation. Oncogene 34, 5447-5459. 10.1038/onc.2014.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. P., Liao Y., Xia W., Spohn B., Lee M.-H. and Hung M.-C. (2001). Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3, 245-252. 10.1038/35060032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.