ABSTRACT
Extracellular vesicles (EVs) are a heterogeneous collection of membrane-bound vesicles released by cells that contain bioactive cargoes including proteins, lipids and nucleic acids. Multiple subpopulations of EVs have now been recognized and these include exosomes and microvesicles. EVs have been thought to facilitate intercellular and distal communication to bring about various processes that enable tumor progression and metastases. Here, we describe the current knowledge of the functional cargo contained within EVs, with a focus on tumor microvesicles, and review the emerging theory of how EVs support immune suppression in cancer.
KEY WORDS: Immune response, Intercellular communication, Microvesicles
Summary: This article reviews current literature on cargoes contained within extracellular vesicles (EVs) and the emerging theory of how they support immune suppression in cancer.
Introduction
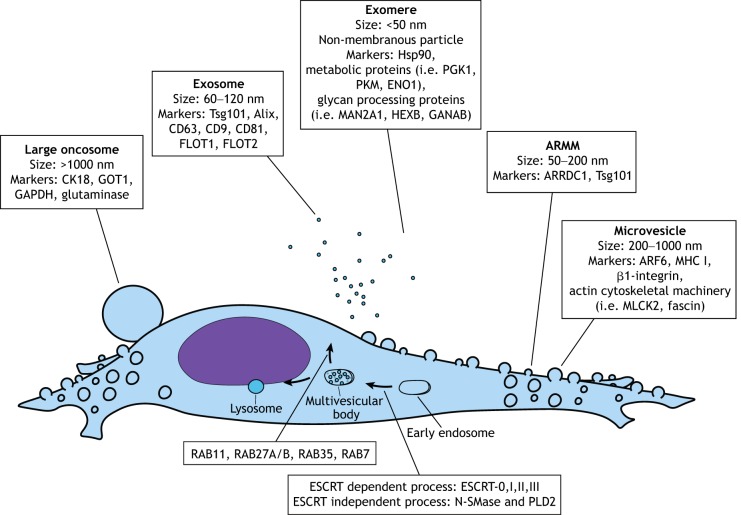
Extracellular vesicles (EVs) are a heterogeneous class of membrane-enclosed particles emitted by virtually all cell types, which function as mediators of intercellular communication (Raposo and Stoorvogel, 2013). Currently, the EV family comprises a large number of unique particles, including exomeres, nanovesicles, exosomes, microvesicles, arrestin domain-containing protein 1-mediated microvesicles (ARMMs), apoptotic bodies and large oncosomes (Sedgwick and D'Souza-Schorey, 2018; van Niel et al., 2018). These species of EVs often co-exist in biological samples and have overlapping characteristics; however, they can each be recognized by a distinct mechanism of biogenesis and source of molecular biomarkers (Fig. 1). For instance, exosomes are a type of small EV (60–120 nm) of endosomal origin, which are formed through an inward budding of late endosomes. This event produces a multivesicular body (MVB) that then may be trafficked to the cell surface to release its exosome-loaded contents or rerouted for lysosomal degradation (Colombo et al., 2014). In contrast, another less extensively studied type of EV, microvesicles, are considered large EVs (200–1000 nm) generated by outward pinching of the plasma membrane through the action of actin cytoskeletal machinery (Tricarico et al., 2017). Adding to the already complex situation, the EV field continues to grow rapidly in its identification of more distinct EV populations, and even subpopulations within the same biogenesis route have now been reported (Zhang et al., 2018).
Fig. 1.
Extracellular vesicles (EVs). Schematic representation of the different types of EVs identified in eukaryotic cells. The size range and protein markers of each type is shown.
In cancer, tumor cells often ramp up microvesicle shedding, indicating deviation from a homeostatic state (Ginestra et al., 1998). These tumor-derived microvesicles, along with other types of EVs, are released to carry out functions that support tumorigenesis, such as enabling stromal invasion, stimulating angiogenesis and promoting metastatic colonization (Broekman et al., 2018; Clancy et al., 2015; Wortzel et al., 2019). In addition to these roles, a burgeoning amount of recent literature has revealed the immunosuppressive activities of EVs, including modulating antigen presentation and evading immune surveillance (Mittal et al., 2014).
To exert these effects, EVs are enriched with a highly heterogeneous pool of biological cargo, the composition of which is continually debated and revised. Here, we describe the current knowledge of the functional cargo contained within EVs with a focus on microvesicles, and review the studies giving rise to the emerging view of how EVs modulate immune activities.
Proteomic content and cargo sorting
EVs possess a diverse collection of proteomic cargo (Greening et al., 2017; Vagner et al., 2019) that changes dynamically according to cell state and environmental conditions. For instance, Kreger et al. (2016) found that transformation of mouse embryonic fibroblasts with an oncogenic form of diffuse B cell lymphoma (onco-Dbl) both increased the rate of microvesicle production, as well as altered their proteomic contents. Further, tumor cell-derived microvesicles (TMVs) possess tissue transglutaminase (tTG, TGM2), which crosslinks external microvesicle proteins such as fibronectin – the activity of which appears to be required to impart transformative phenotypes upon recipient cells (Antonyak et al., 2011). TMVs can also contain oncogenic proteins, such as the truncated variant of the epidermal growth factor (EGF) receptor, EGFRvIII, which could contribute to transformative phenotypes in recipient cells (Al-Nedawi et al., 2009, 2008). In addition, TMVs from invasive cell types are enriched in proteases, which become utilized for extracellular matrix (ECM) degradation, including transmembrane proteins such as membrane-type 1 matrix metalloproteinase (MT1-MMP, also known as MMP14), or soluble gelatinases, such as MMP2 and MMP9 (Clancy et al., 2015; Ginestra et al., 1998; Sedgwick et al., 2015; Taraboletti et al., 2002) (see Fig. 2). It has been suggested that the acidic conditions of tumor microenvironments may promote vesicle rupture, thus releasing the protease-loaded payload and facilitating cell invasion (Giusti et al., 2008).
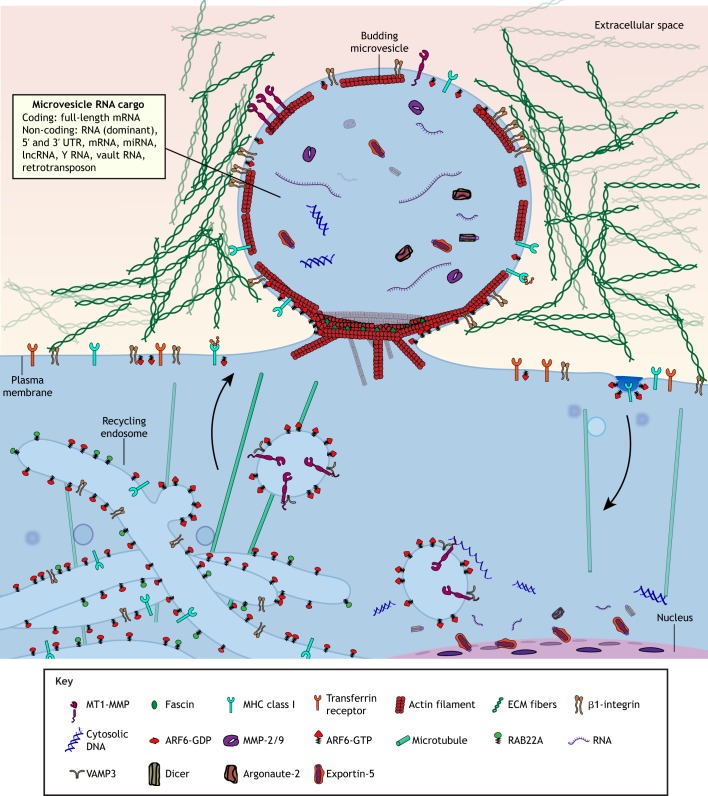
Fig. 2.
Overview over microvesicle biogenesis and cargo. Microvesicles (MVs) formed at the cell surface contain a variety of protein and nucleic acid cargo that enables them to influence recipient cell behavior. MVs receive membrane from tubular recycling endosome compartments. The cargo of specific MVs is likely further refined through other yet undescribed mechanisms, such as lipid sorting at the site of MV formation.
Mechanisms for how molecular cargo is selectively packaged into EVs remain poorly understood within the field. For exosomes, a portion of cargo is selected by direct protein interactions with components of the endosomal sorting complexes required for transport (ESCRT) complex (Frankel and Audhya, 2018). However, during microvesicle biogenesis, packaging appears to occur by regulating the traffic of specific endosome populations – carrying MV-destined cargo – to the site of budding microvesicles at the plasma membrane. In this regard, MVs are highly enriched for proteins derived from ARF6-positive recycling endosomal compartments, such as ARF6, Rab22A, major histocompatibility complex (MHC) class I protein and β1-integrin (Clancy et al., 2019a, 2015; Muralidharan-Chari et al., 2009). These endosomes may act as a junction for cellular cargo in transit to sites of microvesicle formation at the cell surface (D'Souza-Schorey and Schorey, 2018; Tricarico et al., 2017). One example of how endosomal trafficking mechanisms can allow for selective cargo delivery to microvesicles has been described for MT1-MMP. While a vesicle-specific soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor (v-SNARE) known as vesicle-associated membrane protein 7 (VAMP7) has been implicated in the delivery of MT1-MMP to invadapodia at the adherent cell surface (Steffen et al., 2008), MT1-MMP is also delivered to microvesicles along endosomes carrying a complex involving CD9 and another v-SNARE, VAMP3 (Clancy et al., 2015).
Other mechanisms for cargo sorting into extracellular vesicles are continuing to be uncovered in EV biology. While directing endosomal traffic is important in producing the proteomic composition of microvesicles, it is likely that the collective actions of multiple dynamic sorting mechanisms are responsible for creating the distinct molecular profile of extracellular vesicles. For instance, it was recently determined that HIV particles, which closely relate to ARMMs, sort membrane proteins by dynamic remodeling of the lipid composition at the particle assembly site (Sengupta et al., 2019). It is possible that similar lipid-based partitioning mechanisms contribute to generating the unique protein composition of EVs; for instance, EVs are often reported to be enriched for lipid-raft-associated proteins and studies have found that disturbance of these lipid domains can inhibit biogenesis (del Conde et al., 2005; Llorente et al., 2013; Raposo and Stoorvogel, 2013; Sengupta et al., 2019; Wei et al., 2018).
RNA cargo
Early work a decade ago suggested that EVs carry mRNA cargo that can be received and translated by recipient cells (Valadi et al., 2007). Since then, a wealth of studies have reinforced the idea that nearly all EV classes carry a diverse set of RNA species, such as messenger (m)RNA, ribosomal (r)RNA, micro (mi)RNA, piwi-interacting (pi)RNA and small nuclear (sn)RNA (Abels and Breakefield, 2016; Turchinovich et al., 2019). However, the proportional contributions of particular RNA species across EV subclasses and their functional significance remain unresolved issues. Small RNA sequencing of paired EV and whole-cell samples reveals that EV samples are more heterogeneous with regard to the proportional makeup of RNA classes compared to the whole cell they originate from (Sork et al., 2018; Wei et al., 2017). This finding corroborates with the notion of EVs acting as versatile intercellular messengers; it also sheds light on the basis of conflicting findings with respect to the RNA content. Multiple variables contribute to the differential findings among published studies, including (i) cell-type-specific effects and variation in culturing conditions, (ii) adaptor ligation biases in favor of particular RNA classes (Fuchs et al., 2015), and (iii) inconsistent methodologies of EV sample preparation. Moreover, recent studies have found that a significant proportion – if not the majority – of cell-free miRNA is not actually associated with EVs, and thus the results of many RNA studies may be confounded by the co-precipitation of ribonucleoprotein complexes with small EVs (Arroyo et al., 2011; Bettegowda et al., 2014; Jeppesen et al., 2019).
With respect to the RNA content in tumor-derived microvesicles, the current literature indicates that the majority are rRNA sequences (Fiskaa et al., 2016; Sork et al., 2018; Wei et al., 2017). Full-length mRNA cargo can also be detected and the majority of annotated mRNA cargoes are incomplete sequences derived from 3′ and 5′ untranslated regions (Jeppesen et al., 2019; Wei et al., 2017). EVs also display enrichment for transposon elements, which are capable of functional transfer into recipient cell types (Balaj et al., 2011; Li et al., 2013). Following the discovery of miRNA in EVs, a plethora of studies to date have attributed the functional effects of EVs in recipient cells to the transfer of miRNA cargo (Ismail et al., 2013; Li et al., 2013; Liang et al., 2015; Melo et al., 2014; Wei et al., 2017). However, the relative abundance of miRNA in EVs brings this hypothesis into question. Even after filtering out rRNA annotated reads, studies have found miRNA abundance is relatively small (Wei et al., 2017). Stoichiometric analyses of miRNA abundance in exosomes has found that biological samples consistently possess fewer than one copy per exosome for a given miRNA species (Akers et al., 2015; Chevillet et al., 2014). Often overlooked, EVs also display an enrichment for a variety of non-coding RNA species, such as transfer (t)RNA, vault RNA, long non-coding (lnc)RNAs and Y RNA (small non-coding RNAs that are components of the Ro60 ribonucleoprotein particle), whose functional roles in the cell continue to be uncovered (Chiou et al., 2018; Fiskaa et al., 2016; Jeppesen et al., 2019; Li et al., 2013; Wei et al., 2017). In some cases, these unique RNA species are not involved in the transcriptional or translational regulation of the recipient cell, but rather function as ligands for Toll-like receptors and other molecules, as shown in recent studies (Haderk et al., 2017; Nabet et al., 2017). Future studies will likely uncover how many of these less-studied RNA classes contribute to the phenotypic effects of TMVs.
Advancements in the field have elucidated trafficking mechanisms of RNA cargo for both exosomes and, as of very recently, microvesicles. Multiple EV studies have reported the detection of protein components involved in miRNA maturation, such as the Dicer protein family, TRBP (also known as TARBP2), the GW182 protein family and argonaute-2 (Ago2), which could potentially mediate the trafficking of pre-miRNA or a mature RISC complex through direct interactions with protein components involved in EV biogenesis (Clancy et al., 2019b; McKenzie et al., 2016; Melo et al., 2014). In particular, Ago2 directly associates with MVBs, enabling its incorporation into budding exosomes; this interaction is regulated by KRAS–MEK–ERK signaling, which directly phosphorylates the S387 site on Ago2, thereby disturbing Ago2-miRNA delivery (McKenzie et al., 2016). In addition, the tetraspanin CD43 (also known as SPN) was shown to mediate the recruitment of Dicer proteins into exosomes, which enables cell-free processing of pre-miRNA cargo into mature miRNA (Melo et al., 2014). In tumor-derived microvesicles, pre-miRNA cargo is routed to the plasma membrane via a handoff of the dsRNA-binding protein, exportin-5 (XPO5), from a nuclear export complex with Ran-GTP to a cytoplasmic shuttle involving ARF6 and an ARF guanine exchange factor (GEF), GRP1 (also known as CYTH3) (Clancy et al., 2019b).
These models suggest that EVs indiscriminately incorporate miRNA; however, the majority of studies to date have actually found that EVs exhibit enrichment for particular RNA species, adding further complexity to the means of miRNA delivery (Li et al., 2013; Shurtleff et al., 2016; Wei et al., 2017). Different theories have been proposed to explain this phenomenon, such as a bias for particular sequence motifs or post-transcriptional RNA modifications that direct trafficking (Bolukbasi et al., 2012; Lee et al., 2019; Santangelo et al., 2016; Villarroya-Beltri et al., 2013). In addition, it appears that some miRNA species are principally trafficked to EVs by proteins other than those involved in classical miRNA maturation. Ago2 knockdown was shown to promote a significant decrease in total exosomal miRNA, while levels of particular species such as miR-320a remained unaffected (McKenzie et al., 2016). Another study revealed Y box-binding protein 1 (YBX1) as a unique regulator of miRNA delivery to exosomes (Shurtleff et al., 2016) and, in a follow-up paper, expanded the role of YBX1 to having a broader function in the delivery of many small RNAs to EVs (Shurtleff et al., 2017). Similarly, another RNA-binding protein, HNRNPA2B1, was reported to direct particular miRNA species to exosomes based on affinity for particular sequence motifs (Villarroya-Beltri et al., 2013). In addition, a recent study has found that a complex with caveolin-1 directs HNRNPA2B1 incorporation into microvesicles, and a post-translational modification induced by oxidative cell stress alters its selection for miRNA (Lee et al., 2019).
DNA cargo
There is considerable interest surrounding the finding that EVs appear to carry DNA cargo, including single-stranded and double-stranded molecules, retrotransposon elements, mitochondrial DNA and genomic DNA (Balaj et al., 2011; García-Romero et al., 2016; Kalluri and LeBleu, 2016; Lee et al., 2014; Thakur et al., 2014; Vagner et al., 2018). However, this remains the least understood of the biological cargo carried by EVs. Studies involving EV DNA have reported its molecular identity to be largely single-stranded, with a smaller proportion of double-stranded DNA (Vagner et al., 2018). Further, whole-genome sequencing of EV DNA reflects the cell of origin and is representative of the entire genome, suggesting an indiscriminant loading process (Kahlert et al., 2014; Lee et al., 2014; Takahashi et al., 2017; Thakur et al., 2014; Vagner et al., 2018). Several studies to date have provided evidence for exosomes containing DNA (Diamond et al., 2018; Kahlert et al., 2014; Lázaro-Ibáñez et al., 2014; Takahashi et al., 2017; Thakur et al., 2014; Torralba et al., 2018). In one of these, electron micrograph imaging provided evidence for DNA contained in multivesicular bodies and demonstrated that histones co-fractionate with exosome markers TSG101 and CD63. Further, the authors found that disrupting regulators of exosome biogenesis results in the stimulation of DNA damage-response pathways, suggesting that tumor cells utilize exosomes for the emission of accumulating cytosolic DNA (Takahashi et al., 2017).
Although several studies report the detection of DNA in isolated EVs in vitro and in circulation in vivo, skepticism continues to hang over whether such extracellular DNA is truly enclosed within the lipid membrane of EVs (Jeppesen et al., 2019; Shurtleff et al., 2018). Indeed, only a few reports have performed a thorough biochemical analysis to demonstrate the internal location of DNA within EVs, including identifying a DNase-resistant pool of EV DNA that only becomes susceptible to degradation upon permeabilization of the lipid membrane (Torralba et al., 2018; Vagner et al., 2018). In this context, Vagner et al. (2018) reported that the majority of cell-free DNA emitted by prostate cancer cells is carried by large EVs, comprising microvesicle and large oncosome EV populations, whereas the amount of DNA isolated from small EVs was negligible. This dissimilarity in EV content is also supported by another study, which found that only the microvesicle fraction of emitted EVs from transiently transfected HEK293FT cells are capable of delivering intact plasmid DNA to recipient cell types (Kanada et al., 2015). Furthermore, a very recent report has suggested that small EVs do not contain DNA, and, rather, exosome-associated DNA is actually based on the active secretion of extracellular chromatin via multivesicular bodies (Jeppesen et al., 2019). These intriguing developments suggest a discrepancy in the molecular cargo contained in EVs, whereby DNA associated with small EVs is external, while large EVs, such as TMVs, apoptotic bodies and large oncosomes, may actually hold an internal pool of extracellular DNA.
Since the discovery of extracellular DNA, it has been suggested that cancer cells may deliver oncogenic sequences via EVs, which might result in the transformation of surrounding normal cell types akin to an infection-like process (Bergsmedh et al., 2001; Fischer et al., 2016; García-Olmo et al., 2010; Kanada et al., 2015; Rak and Guha, 2012). Indeed, it has been demonstrated that EVs can impart transformed phenotypes into recipient cell types (characterized by cellular capacity for formation of foci in culture or anchorage-independent growth) (Antonyak et al., 2011; Hallal et al., 2019; Lee et al., 2014; Oushy et al., 2018). However, cancerous phenotypes are transient in nature and donor DNA is not permanently incorporated in the recipient genome (Lee et al., 2014). In addition, further analysis has revealed that many specialized cell types, such as normal intestinal epithelia and astrocytes, are resistant to cellular transformation, bringing into question the physiological significance of this model (Lee et al., 2016).
Recent evidence shows that cancer cells accumulate cytosolic DNA as a consequence of DNA damage events and nuclear envelope rupture, resulting in an activation of intracellular signaling events that alerts surrounding cells of a compromised cell state (Mackenzie et al., 2017; Vanpouille-Box et al., 2018). Given this, the intercellular transfer of DNA as a molecular regulator of inflammatory responses may be a more reasonable explanation for the function of EV-associated DNA, but would still have important implications for many diseases like cancer. This would also provide an explanation for the apparent lack of a discriminant selection of DNA sequences as cargo. Supporting this, a recent study showed that cancer cells depleted of the cytoskeletal regulator diaphanous-related formin 3 (DIAPH3) display increased indications of nuclear instability, such as nuclear blebbing and increased irregular shape. Coinciding with these phenotypes, DIAPH3-knockdown cells secrete large EVs possessing increased levels of nuclear components, such as emerin and gDNA (Reis-Sobreiro et al., 2018). In addition, other studies have revealed that activated T-lymphoblasts and irradiated tumor cells secrete EVs enriched in dsDNA cargo, which are capable of activating the cGAS–STING pathway in recipient dendritic cells (Diamond et al., 2018; Torralba et al., 2018).
In the above sections, we discussed the world of EV cargo, focusing primarily on protein and nucleic acid content, cargo loading and release of MVs formed at the cell surface (summarized in Fig. 2). While the majority of these studies have been conducted in tumor cells, it is conceivable that other cell types may form similar structures at the cell surface. However, the functional cell-signaling roles of these EVs remains an unanswered, but alluring question. In the following sections, we focus our attention on the growing intersection between the EV field and cancer immunology, and highlight some of the currently described mechanisms by which tumor-derived EVs could significantly contribute to the evasion of immune surveillance.
Extracellular vesicles as vehicles of immune evasion
In normal cellular physiology, EVs are now appreciated as important messengers in orchestrating immune responses during processes, such as wound repair and infection with foreign organism (Laberge et al., 2018; Robbins and Morelli, 2014). Early work in the field revealed that immune cells utilize EVs, particularly exosomes, to mediate antigen presentation with other cells at a systemic level (Raposo et al., 1996; Wolfers et al., 2001; Zitvogel et al., 1998). This occurs by way of cells processing foreign antigens in late endosomal compartments and subsequently loading antigenic peptides onto MHC class I and II molecules displayed on the surface of intraluminal vesicles (Roche and Furuta, 2015). Among other mechanisms, anti-tumor immune responses in human cancers can be stimulated following the detection of unique peptides expressed by cancer cells as a consequence of genomic alterations, a term now referred to as ‘neoantigens’ (Schumacher and Schreiber, 2015). In 1998, Zitvogel et al. reported that professional antigen-presenting-cells (APCs), the dendritic cells (DC), secrete exosomes displaying MHC class I and II molecules and T-cell co-stimulatory molecules. In a subsequent study, they showed that tumor cells themselves release EVs that act as an antigen source to promote cytotoxic T-cell priming (Wolfers et al., 2001). Others have suggested that EVs may actually be a superior mechanism of antigen delivery to prime dendritic cells than soluble peptides; mechanistically, this appears to be mediated by reactive oxygen species (ROS) carried by EVs that result in early alkalization of DC phagosomal compartments (Battisti et al., 2017; Rughetti et al., 2014).
While still a subject of active investigation (Battisti et al., 2017; Dionisi et al., 2018), dendritic-cell-based immunotherapies have had very limited success in provoking anti-tumor responses in clinical trials where allogenic or autologous DCs are primed with soluble antigen or tumor lysates (Bu et al., 2011; Flörcken et al., 2013; Gu et al., 2015; Liu et al., 2017; Palucka et al., 2006; Sabado et al., 2017). Growing evidence shows that many of the interactions between tumor-derived EVs and immune cells are unfavorable for the host (Pucci et al., 2016). Hence, extending our understanding of how tumor-derived EVs manipulate the behavior of host immune cells is important for improving current therapeutic strategies and unleashing anti-tumor immune responses. Below, we describe the currently known mechanisms by which tumor-derived EVs contribute to suppressing immune responses in cancer (Fig. 3).
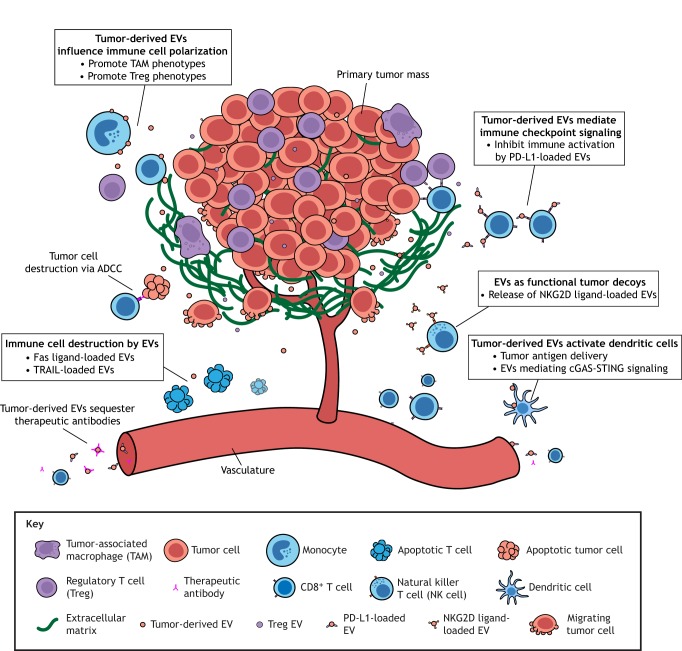
Fig. 3.
EV-mediated immune suppression. Three different mechanisms have been described thus far for how tumor-derived EVs silence anti-tumor immune responses. (1) EVs directly suppress innate and adaptive immune cell responses through interactions of cargo present at their surface, such as immune checkpoint ligands (e.g. PD-L1) and NKG2D ligands. (2) EVs can eliminate specific classes of immune cells through Fas ligand-mediated apoptosis. (3) EVs strongly promote pro-tumorigenic phenotypes in some immune cells, such as CD4+ T cells and CD14+ monocytes. In addition, coerced immune cells further support immune suppression by producing their own EVs for the secretion of immunosuppressive compounds. ADCC, antibody-dependent cell-mediated cytotoxicity.
Immune checkpoint-loaded EVs
Immune checkpoints such as PD-L1 (also known as CD274) and CTLA-4 are molecules often expressed on the plasma membrane of cancer cells or colluding cell types, which act as powerful regulators of immune tolerance. Targeted antibody-based therapies against some of these molecules have been developed as a method of relieving cytotoxic T-cells of this negative signaling and thus enable antigen activation in tumors (Fritz and Lenardo, 2019; Pardoll, 2012). Yet, while these ‘immune checkpoint inhibitors’ have been demonstrated to successfully produce anti-tumor immunity for several different types of cancer, only the minority of patients show responses resulting in long-term survival (Fares et al., 2019; Schadendorf et al., 2015; Sharma et al., 2017). As a consequence, current research is focused on understanding the biology that enables immune evasion in these non-responsive tumors (Spranger and Gajewski, 2018). A series of recent studies have conclusively demonstrated that tumor-derived EVs substantially contribute to immunosuppressive phenotypes in a variety of cancer types owing to the fact they carry the immune checkpoint receptor ligand PD-L1 (Chen et al., 2018; Poggio et al., 2019; Ricklefs et al., 2018; Theodoraki et al., 2018). Ricklefs et al. (2018) demonstrated in vitro that co-incubation of EVs derived from glioblastoma stem cell-like cells (GSCs) inhibits activation of CD8+ T cells presented with antigen by DCs and further, inhibition of T cell activation by tumor-derived EVs could be rescued by co-culturing with anti-PD-L1 antibody blockade.
In order to assess the significance of exosomal PD-L1 to cancer progression in vivo, Poggio et al. (2019) turned to a genetic model and generated Rab27a or nSMase2 (also known as Smpd3) CRISPR/Cas9 knockouts of TRAMP-C2, a syngeneic model of prostate cancer that is resistant to anti-PD-L1 blockade (Poggio et al., 2019). When tested in fully immunocompetent mouse models, Rab27a-KO and nSMase2-KO cells failed to form tumors in a similar fashion to Pd-l1-KO cells (Poggio et al., 2019). These findings indicate that the exosome-mediated inhibition of tumor growth occurs in a process dependent on immune cell interactions. Further, injection of exogenous wild-type exosomes – but not exosomes from PD-L1-KO cells – into syngeneic mice rescues the immunosuppressive phenotype and enables tumor growth of Rab27a-KO tumors (Poggio et al., 2019).
These studies establish a role for tumor-derived EVs in inducing systemic immunosuppression by conveying immune checkpoint signals to distant immune cells. In terms of clinical applications, exosomal PD-L1 could also potentially be utilized in the future as a diagnostic marker to predict tumor responses (Chen et al., 2018; Theodoraki et al., 2018). Future studies should also investigate whether there is a difference in the biological activity of currently utilized antibody-based therapies in inhibiting target receptors on the whole cell versus those on EVs. A variety of factors such as binding affinity, target antigen density, flow rate and buffer conditions can impact the binding of monoclonal antibodies (Rhoden et al., 2016; Tabrizi et al., 2010; Wemhoff et al., 1992), and it is currently unknown whether the collective actions of these factors differentially impact the activity of commercial therapeutic antibodies against stationary tumor cells and circulating EVs.
Weapons for tumor counter-attack
Tumor cells appear to not simply endure being continuously targeted for destruction by immunocytes; rather, they actively secrete substances to eliminate such threatening cell types as a means of immune suppression. This is sometimes referred to as the ‘counter-attack’ hypothesis. While contested (Restifo, 2001), this concept has been used to explain the finding that some tumor types express Fas ligand (FASLG), which induces apoptosis in contacted cells that express its receptor, FasR. Interestingly, some have suggested that this mechanism is also utilized in normal physiology by immune-privileged tissues, such as retina and brain (O'Connell et al., 1996).
Early studies investigating the interactions of tumor-derived EVs with lymphocytes found that EVs harbor Fas ligand as cargo and can stimulate apoptosis of T cells (Andreola et al., 2002). These findings are supported by reports that EVs derived from a diverse set of cancer cell types stimulate apoptosis in CD8+ T cells, in a process that is dependent on the activity of Fas ligand, or another apoptosis-inducing ligand, TNF-related apoptosis-inducing ligand (TRAIL, also known as TNFSF10), within its cargo (Bergmann et al., 2009; Huber et al., 2005; Kim et al., 2005; Martínez-Lorenzo et al., 2004; Taylor et al., 2003; Wieckowski et al., 2009). Bergmann et al. (2009) showed that EVs isolated from the sera of patients with head and neck squamous cell carcinoma (HNSCC) had significantly higher expression of Fas ligand compared to EVs in sera from healthy controls and were capable of inducing cell death in CD8+ T cells.
Tumors that show a high density of infiltrated CD8+ T lymphocytes generally correlate with a better prognosis and therapeutic responses for a variety of cancers (Gibney et al., 2016; Katsuya et al., 2017; Sato et al., 2005; Xu et al., 2019). However, there are many tumors that lack T-cell infiltration, often referred to as ‘cold tumors’, and this represents a major barrier for their targeting by immunotherapy (Bonaventura et al., 2019). Therefore, current research efforts are directed at developing strategies for improving T-cell trafficking and infiltration (Bonaventura et al., 2019). A number of studies have also argued that Fas-mediated apoptosis of T cells is one barrier to immune infiltration in some tumor cases (Ichinose et al., 2001; Motz et al., 2014; Zhu et al., 2019).
Concealing tumor cell presence with decoys
Similar in some ways to the counter-attack mechanism, some groups have suggested that EVs act as cellular ‘decoys’ that enable a tumor cell to avoid detection by a patrolling immunocyte. MICA and MICB are two of the many ligands for the NKG2D (also known as KLRK1) activating receptor expressed on the surface of natural killer (NK) cells and CD8+ T cells. While expressed at low levels in normal cell types, NKG2D ligands are upregulated in response to cell stress, marking the cell for destruction by the innate immune response (Lanier, 2015). In the case of cancer, genotoxic stress likewise can induce the expression of NKG2D ligands, making tumor cells vulnerable to targeting by immune cells (Gasser et al., 2005). To circumvent this issue, tumor cells release NKG2D ligands such as MICA from their surface, either by proteolytic cleavage into a soluble isoform or by shedding EVs (Ashiru et al., 2010).
Several studies have shown that incubation of NK cells with tumor exosomes results in downregulation of NKG2D receptor on NK cells and CD8+ T cells, and thus impairs their cytotoxicity against MICA-positive target cells (Ashiru et al., 2010; Clayton et al., 2008; Lundholm et al., 2014). NKG2D downregulation by EVs has been found to occur in a dose-dependent manner (Lundholm et al., 2014). In addition, these authors also provide in vivo evidence from patients with castration-resistant prostate cancer that circulating NK cells and CD8+ T cells display a lower surface expression of NKG2D than healthy controls (Lundholm et al., 2014). However, a soluble isoform of the NKG2D ligand MULT1 (also known as Ulbp1) was shown to activate NK cells and inhibit tumor growth when injected with B16 cells into mice (Deng et al., 2015). A recent study demonstrated that treatment of A375 melanoma cells with an α3-domain-specific antibody that inhibits the proteolytic release of MICA and/or MICB from the plasma membrane significantly inhibited tumor growth when applied to fully immunocompetent mouse models (Ferrari de Andrade et al., 2018). Thus the proteolytic shedding of MICA or MICB from the cell surface – and potentially also the surfaces of EVs themselves – may actually be the dominant biological process in desensitizing NK cells.
It should also be mentioned that many actively utilized antibodies in cancer therapies target tumor antigens and are capable of provoking anti-tumor immune responses by antibody-dependent cell-mediated cytotoxicity (ADCC) (Natsume et al., 2009). In this mechanism, antibodies bound to tumor cells can activate cells of the innate immune response (Wang et al., 2015). Previously, studies pointed out that the serum of some patients with cancer could inhibit NK cell activation and ADCC (Matsuzaki et al., 1985). Later, it was determined that tumor-derived exosomes sequester tumor-reactive antibodies and consequently reduce ADCC activity against tumor cells (Aung et al., 2011; Battke et al., 2011). This has been demonstrated to occur for multiple commonly used therapeutics, such as rituximab, which targets CD20 on B-cell lymphoma cells, and trastuzumab, which targets HER2 on breast cancer cells (Aung et al., 2011; Battke et al., 2011).
Controlling cellular phenotypes
Tumor EVs have potent effects on altering the behavior of recipient cell types, typically in a fashion that supports disease progression. For instance, cancer cells will phenocopy the behavior of more aggressive subpopulations within the tumor upon receiving microvesicles originating from these groups of cells (Zomer et al., 2015). Tumor-derived exosomes also interact with recipient cell types at distant organ sites, thereby creating a pre-metastatic niche (Costa-Silva et al., 2015).
Tumor EVs induce highly differential behavioral effects based on the particular recipient immunocyte. As discussed above, tumor EVs inhibit proliferation and induced apoptosis in CD8+ T cells; however, surprisingly, opposite effects were observed when CD4+ T cells were tested (Wieckowski et al., 2009). Instead, EV-treated CD4+ T cells biased their maturation towards CD25high/FOXP3+ T-regulatory cells (Tregs), which are known to maintain self-tolerance and suppress immune responses (Szajnik et al., 2010; Wieckowski et al., 2009). Further, tumor EVs appear to even promote the proliferation of Treg cells and enhance their immunosuppressive activity (Szajnik et al., 2010). Tregs were the cell type most sensitive to exposure to exosomes, which resulted in gene expression changes (Muller et al., 2016). In addition to all of these findings, it was reported that T cells largely do not take up tumor-derived exosomes when compared to other tested immune cell types, indicating that the effects mediated by tumor-derived EVs are restricted to surface interactions (Muller et al., 2017, 2016). However, the full mechanism of what specific surface interactions with tumor EVs mediate Treg stimulatory activity has not yet been elucidated.
Tumor-associated macrophages (TAMs) are another highly abundant blood cell found within tumor microenvironments. Mature macrophages have been conventionally categorized as being either a ‘classically activated’ (M1) phenotype (often considered pro-inflammatory and cytotoxic) or an ‘alternatively activated’ (M2) phenotype (considered anti-inflammatory and immunosuppressive) (Ostuni et al., 2015). In the past, it has been suggested that TAMs are produced by circulating monocytes that have undergone maturation towards an immunosuppressive M2 phenotype, although it is now known that the true TAM phenotype is not well-captured by this categorization (Franklin et al., 2014; Sica et al., 2006). Numerous studies have shown that tumor-derived EVs bias monocyte polarization towards an immunosuppressive TAM phenotype (Gabrusiewicz et al., 2018; Ham et al., 2018; Hsu et al., 2018; Wang et al., 2018a,b; Ying et al., 2016). Accordingly, EV-treated monocytes display increased markers that are associated with M2 macrophages, such as CD163 and CD206, and increased expression of immunosuppressive molecules, such as secretion of IL-10 and production of PD-L1 (Gabrusiewicz et al., 2018; Hsu et al., 2018). In addition, treatment with an N-SMase inhibitor to disrupt exosome biogenesis altered macrophage polarization in co-culture experiments with gastric cancer cells, and in tumor-bearing mice (Wang et al., 2018a). With regard to the underlying mechanism, these studies consistently implicated particular miRNA species in the reprogramming of macrophages (Chen et al., 2018; Cooks et al., 2018; Hsu et al., 2018).
While there remain many unanswered questions of how tumor-derived EVs enable cellular reprogramming, this coercive strategy of immune suppression may be particularly effective for tumor cells, given the potential for amplification of suppressive signaling by newly recruited immune cells. For instance, it was recently shown that Tregs themselves shed EVs that also modify the behavior of the recipient immune cells, such as promoting immune tolerance in dendritic cells (Tung et al., 2018).
Perspectives
As discussed here, EVs contain a diverse set of selectively recruited cargo, including proteins and nucleic acids, which enable their function as entities of intercellular communication. Tumor-derived EVs significantly contribute to the progression of cancer by multiple means, including modulating immune responses. To date, several mechanisms have been described for how tumor-derived EVs support immunosuppression in cancer including: (i) direct suppression of immune cytotoxicity by mediating immune checkpoint signaling or dampening stimuli of the innate immune response, (ii) eliminating threatening cell types through tumor counter-attack, and (iii) coercing other resident cell types into supporting immunosuppressive functions (see Fig. 3).
EV-mediated immune suppression is an interesting example of how cancers subvert normal biological function. In the context of infection and cell injury, EVs function to alert the immune system by distributing foreign antigens or danger-associated molecular patterns, thereby helping to mount an effective response. The remnants of this physiological response can also be observed in cancer, for instance, the preference of solid tumor-derived EVs to travel towards draining lymph nodes, or neoantigen priming of dendritic cells by tumor-derived EVs. Yet in cancer, communication to immune cells through tumor-derived EVs may actually exacerbate the progression of the disease.
As some of these studies have shown, in some cases, the effects of tumor-derived EVs appear to be mediated by particular units of cargo contained by them, such as immune checkpoint ligands. However, as is the case for much of EV biology, the mechanisms underlying EV-mediated effects on recipient immune cells are not fully understood. The difficulty of this task is based on multiple factors including: the heterogeneity of cargo contained in EVs, the variation in the cell types interacting with them, and the mixed populations pervading most EV samples. As a consequence of commonly utilized isolation procedures, the majority of those studies mentioned here likely involved the use of samples containing mixed populations of EVs. In addition, there is a disproportionate focus on exosomes, leaving the functional roles of less commonly studied large EVs like microvesicles an open question. Indeed, the study of microvesicles carries with it its own unique set of challenges (see Box 1); however, determining the physiological roles of these types of underappreciated EVs will be necessary to develop our understanding of EV biology. Interestingly, we are now beginning to understand that the molecular cargo may differ between these types of EVs; further, EVs present within the tumor microenvironment contain cargo species that typically are only found within the context of disease or injury. Notably, this is the case with DNA and unique noncoding RNAs, which may facilitate DAMP-recognition-mediated signaling in recipient cell types. Tumor-derived EVs carrying these elements could facilitate pro-inflammatory signaling within the tumor microenvironment, perhaps through such a mechanism (Matei et al., 2017; Nabet et al., 2017).
Box 1. Challenges and outstanding questions in tumor microvesicle (TMV) research.
Challenges
A lack of defined biomarkers
Heterogeneous in size, resulting in overlap with both other small EVs (i.e. exosomes and ARMMs) as well as other large EVs (i.e. apoptotic bodies and oncosomes)
Rudimentary understanding of dedicated machinery for TMV biogenesis, resulting in confounded results with genetic manipulation in functional studies
Confusion over the distinction between exosomes, microvesicles and other EV species
Outstanding questions
What are the mechanisms and regulators of TMV biogenesis?
Are there distinct functional roles between exosomes and TMVs in intercellular communication?
Do TMVs possess DNA? What is the physiological purpose of the reported genetic exchange by EVs?
What mechanisms direct recipient cell uptake of EVs?
Is EV-destined cargo distributed equally in the cell or sorted into distinct subpopulations?
How does EV quantity and contents collectively result in a change in recipient cell behavior?
Given the importance of tumor-derived EVs in supporting immunosuppression as discussed here, there has been speculation as to whether inhibiting exosome or microvesicle biogenesis may prove an effective therapeutic strategy. However, given the thus far undetermined role of EV-mediated communication during normal human physiological processes and the potential functional redundancy of EVs, this seems less probable. More likely, future treatments might involve bioengineered EVs, with purposefully packaged cargo aimed at eliciting anti-tumor immune responses and blocking disease progression. In this regard, exosomes isolated from non-metastatic melanoma cultures could effectively inhibit lung metastases of metastatic melanoma by expanding patrolling monocyte populations that promote cancer cell clearance at the pre-metastatic niche (Plebanek, et al., 2017). Lingering but essential questions include: how are individual cargo species packaged into EVs? What are the molecular regulators of EV uptake? And, collectively, how does EV cargo translate into recipient cell function? While an exacting task, fully understanding the cell biology of EVs will ultimately reveal new ways in which they can be leveraged in clinical settings, including stimulating anti-tumor immune responses in cancer.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
We acknowledge support from the National Institutes of Health, the Catherine Peachey Fund and 100 Voices of Hope, for EV-related research in the D'Souza-Schorey lab. Deposited in PMC for release after 12 months.
References
- Abels E. R. and Breakefield X. O. (2016). Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 36, 301-312. 10.1007/s10571-016-0366-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers J. C., Ramakrishnan V., Kim R., Phillips S., Kaimal V., Mao Y., Hua W., Yang I., Fu C.-C., Nolan J. et al. (2015). miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J. Neurooncol. 123, 205-216. 10.1007/s11060-015-1784-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A. and Rak J (2008). Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 10, 619-624. 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K., Meehan B., Kerbel R. S., Allison A. C. and Rak J (2009). Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. USA 106, 3794-3799. 10.1073/pnas.0804543106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreola G., Rivoltini L., Castelli C., Huber V., Perego P., Deho P., Squarcina P., Accornero P., Lozupone F., Lugini L. et al. (2002). Induction of lymphocyte apoptosis by tumor cell secretion of fasl-bearing microvesicles. J. Exp. Med. 195, 1303-1316. 10.1084/jem.20011624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonyak M. A., Li B., Boroughs L. K., Johnson J. L., Druso J. E., Bryant K. L., Holowka D. A. and Cerione R. A. (2011). Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA 108, 4852-4857. 10.1073/pnas.1017667108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., Mitchell P. S., Bennett C. F., Pogosova-Agadjanyan E. L., Stirewalt D. L. et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 108, 5003-5008. 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiru O., Boutet P., Fernández-Messina L., Agüera-González S., Skepper J. N., Valés- Gómez M. and Reyburn H. T. (2010). Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 70, 481-489. 10.1158/0008-5472.CAN-09-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung T., Chapuy B., Vogel D., Wenzel D., Oppermann M., Lahmann M., Weinhage T., Menck K., Hupfeld T., Koch R. et al. (2011). Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc. Natl. Acad. Sci. USA 108, 15336-15341. 10.1073/pnas.1102855108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L., Lessard R., Dai L., Cho Y.-J., Pomeroy S. L., Breakefield X. O. and Skog J (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2, 180 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti F., Napoletano C., Rahimi Koshkaki H., Belleudi F., Zizzari I. G., Ruscito I., Palchetti S., Bellati F., Benedetti Panici P., Torrisi M. R. et al. (2017). Tumor-derived microvesicles modulate antigen cross- processing via reactive oxygen species-mediated alkalinization of phagosomal compartment in dendritic cells. Front. Immunol. 8, 1179 10.3389/fimmu.2017.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battke C., Ruiss R., Welsch U., Wimberger P., Lang S., Jochum S. and Zeidler R (2011). Tumour exosomes inhibit binding of tumour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol. Immunother. 60, 639-648. 10.1007/s00262-011-0979-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C., Strauss L., Wieckowski E., Czystowska M., Albers A., Wang Y., Zeidler R., Lang S. and Whiteside T. L. (2009). Tumor-derived microvesicles in sera of patients with head and neck cancer and their role in tumor progression. Head Neck 31, 371-380. 10.1002/hed.20968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsmedh A., Szeles A., Henriksson M., Bratt A., Folkman M. J., Spetz A.-L. and Holmgren L (2001). Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA 98, 6407-6411. 10.1073/pnas.101129998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C., Sausen M., Leary R. J., Kinde I., Wang Y., Agrawal N., Bartlett B. R., Wang H., Luber B., Alani R. M. et al. (2014). Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolukbasi M. F., Mizrak A., Ozdener G. B., Madlener S., Ströbel T., Erkan E. P., Fan J.-B., Breakefield X. O. and Saydam O (2012). miR-1289 and “Zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 1, e10 10.1038/mtna.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura P., Shekarian T., Alcazer V., Valladeau-Guilemond J., Valsesia-Wittmann S., Amigorena S., Caux C. and Depil S. (2019). Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol. 10, 168 10.3389/fimmu.2019.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekman M. L., Maas S. L. N., Abels E. R., Mempel T. R., Krichevsky A. M. and Breakefield X. O. (2018). Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 14, 482-495. 10.1038/s41582-018-0025-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu N., Wu H., Sun B., Zhang G., Zhan S., Zhang R. and Zhou L (2011). Exosome-loaded dendritic cells elicit tumor-specific CD8+ cytotoxic T cells in patients with glioma. J. Neurooncol. 104, 659-667. 10.1007/s11060-011-0537-1 [DOI] [PubMed] [Google Scholar]
- Chen G., Huang A. C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H. et al. (2018). Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382 10.1038/s41586-018-0392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet J. R., Kang Q., Ruf I. K., Briggs H. A., Vojtech L. N., Hughes S. M., Cheng H. H., Arroyo J. D., Meredith E. K., Gallichotte E. N. et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 111, 14888-14893. 10.1073/pnas.1408301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou N.-T., Kageyama R. and Ansel K. M. (2018). Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 25, 3356-3370.e4. 10.1016/j.celrep.2018.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J. W., Sedgwick A., Rosse C., Muralidharan-Chari V., Raposo G., Method M., Chavrier P. and D'Souza-Schorey C (2015). Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 6, 6919 10.1038/ncomms7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J. W., Tricarico C. J., Marous D. R. and D'Souza-Schorey C. (2019a). Coordinated regulation of intracellular fascin distribution governs tumor microvesicle release and invasive cell capacity. Mol. Cell Biol. 39, e00264-18 10.1128/MCB.00264-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J. W., Zhang Y., Sheehan C. and D'Souza-Schorey C. (2019b). An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 21, 856-866. 10.1038/s41556-019-0345-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A., Mitchell J. P., Court J., Linnane S., Mason M. D. and Tabi Z (2008). Human tumor- derived exosomes down-modulate NKG2D expression. J. Immunol. 180, 7249-7258. 10.4049/jimmunol.180.11.7249 [DOI] [PubMed] [Google Scholar]
- Colombo M., Raposo G. and Théry C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255-289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- del Conde I., Shrimpton C. N., Thiagarajan P. and López J. A. (2005). Tissue-factor–bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 106, 1604-1611. 10.1182/blood-2004-03-1095 [DOI] [PubMed] [Google Scholar]
- Cooks T., Pateras I. S., Jenkins L. M., Patel K. M., Robles A. I., Morris J., Forshew T., Appella E., Gorgoulis V. G. and Harris C. C. (2018). Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 9, 771 10.1038/s41467-018-03224-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N. M., Ocean A. J., Singh S., Zhang H., Thakur B. K., Becker A., Hoshino A., Mark M. T., Molina H. et al. (2015). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816-826. 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Gowen B. G., Zhang L., Wang L., Lau S., Iannello A., Xu J., Rovis T. L., Xiong N. and Raulet D. H. (2015). A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science 348, 136-139. 10.1126/science.1258867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J. M., Vanpouille-Box C., Spada S., Rudqvist N.-P., Chapman J. R., Ueberheide B. M., Pilones K. A., Sarfraz Y., Formenti S. C. and Demaria S (2018). Exosomes shuttle TREX1-sensitive IFN-stimulatory dsDNA from irradiated cancer cells to DCs. Cancer Immunol. Res. 6, 910-920. 10.1158/2326-6066.CIR-17-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi M., De Archangelis C., Battisti F., Rahimi Koshkaki H., Belleudi F., Zizzari I. G., Ruscito I., Albano C., Di Filippo A., Torrisi M. R. et al. (2018). Tumor-derived microvesicles enhance cross- processing ability of clinical grade dendritic cells. Front. Immunol. 9, 2481 10.3389/fimmu.2018.02481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C. and Schorey J. S. (2018). Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 62, 125-133. 10.1042/EBC20170078 [DOI] [PubMed] [Google Scholar]
- Fares C. M., Van Allen E. M., Drake C. G., Allison J. P. and Hu-Lieskovan S. (2019). Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am. Soc. Clin. Oncol. Educ. Book 39, 147-164. 10.1200/EDBK_240837 [DOI] [PubMed] [Google Scholar]
- Ferrari de Andrade L., Tay R. E., Pan D., Luoma A. M., Ito Y., Badrinath S., Tsoucas D., Franz B., May K. F. et al. (2018). Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell–driven tumor immunity. Science 359, 1537-1542. 10.1126/science.aao0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S., Cornils K., Speiseder T., Badbaran A., Reimer R., Indenbirken D., Grundhoff A., Brunswig-Spickenheier B., Alawi M. and Lange C (2016). Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS ONE 11, e0163665 10.1371/journal.pone.0163665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskaa T., Knutsen E., Nikolaisen M. A., Jørgensen T. E., Johansen S. D., Perander M. and Seternes O. M. (2016). Distinct small RNA signatures in extracellular vesicles derived from breast cancer cell lines. PLoS ONE 11, e0161824 10.1371/journal.pone.0161824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flörcken A., Kopp J., van Lessen A., Movassaghi K., Takvorian A., Jöhrens K., Möbs M., Schönemann C., Sawitzki B., Egerer K. et al. (2013). Allogeneic partially HLA-matched dendritic cells pulsed with autologous tumor cell lysate as a vaccine in metastatic renal cell cancer: a clinical phase I/II study. Hum. Vaccin. Immunother. 9, 1217-1227. 10.4161/hv.24149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel E. B. and Audhya A (2018). ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 74, 4-10. 10.1016/j.semcdb.2017.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R. A., Liao W., Sarkar A., Kim M. V., Bivona M. R., Liu K., Pamer E. G. and Li M. O. (2014). The cellular and molecular origin of tumor-associated macrophages. Science 344, 921-925. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz J. M. and Lenardo M. J. (2019). Development of immune checkpoint therapy for cancer. J. Exp. Med. 216, 1244-1254. 10.1084/jem.20182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. T., Sun Z., Zhuang F. and Robb G. B. (2015). Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLOS ONE 10, e0126049 10.1371/journal.pone.0126049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrusiewicz K., Li X., Wei J., Hashimoto Y., Marisetty A. L., Ott M., Wang F., Hawke D., Yu J., Healy L. M. et al. (2018). Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. OncoImmunology 7, e1412909 10.1080/2162402X.2017.1412909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Olmo D. C., Domínguez C., García-Arranz M., Anker P., Stroun M., García-Verdugo J. M. and García-Olmo D (2010). Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 70, 560-567. 10.1158/0008-5472.CAN-09-3513 [DOI] [PubMed] [Google Scholar]
- García-Romero N., Carrión-Navarro J., Esteban-Rubio S., Lázaro-Ibáñez E., Peris-Celda M., Alonso M. M., Guzmán-De-Villoria J., Fernández-Carballal C., de Mendivil A. O. et al. (2016). DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 8, 1416-1428. 10.18632/oncotarget.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S., Orsulic S., Brown E. J. and Raulet D. H. (2005). The DNA damage pathway regulates innate immune system ligands for the NKG2D receptor. Nature 436, 1186-1190. 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney G. T., Weiner L. M. and Atkins M. B. (2016). Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 17, e542-e551. 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestra A., La Placa M. D., Saladino F., Cassarà D., Nagase H. and Vittorelli M. L. (1998). The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 18, 3433-3437. [PubMed] [Google Scholar]
- Giusti I., D'Ascenzo S., Millimaggi D., Taraboletti G., Carta G., Franceschini N., Pavan A. and Dolo V (2008). Cathepsin B mediates the pH-dependent proinvasive activity of tumor- shed microvesicles. Neoplasia N. Y. N 10, 481-488. 10.1593/neo.08178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D. W., Xu R., Gopal S. K., Rai A. and Simpson R. J. (2017). Proteomic insights into extracellular vesicle biology-defining exosomes and shed microvesicles. Expert Rev. Proteomics 14, 69-95. 10.1080/14789450.2017.1260450 [DOI] [PubMed] [Google Scholar]
- Gu X., Erb U., Büchler M. W. and Zöller M (2015). Improved vaccine efficacy of tumor exosome compared to tumor lysate loaded dendritic cells in mice. Int. J. Cancer 136, E74-E84. 10.1002/ijc.29100 [DOI] [PubMed] [Google Scholar]
- Haderk F., Schulz R., Iskar M., Cid L. L., Worst T., Willmund K. V., Schulz A., Warnken U., Seiler J., Benner A. et al. (2017). Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2, eaah5509 10.1126/sciimmunol.aah5509 [DOI] [PubMed] [Google Scholar]
- Hallal S., Mallawaaratchy D. M., Wei H., Ebrahimkhani S., Stringer B. W., Day B. W., Boyd A. W., Guillemin G. J., Buckland M. E. and Kaufman K. L. (2019). Extracellular vesicles released by glioblastoma cells stimulate normal astrocytes to acquire a tumor-supportive phenotype via p53 and MYC signaling pathways. Mol. Neurobiol. 56, 4566-4581. 10.1007/s12035-018-1385-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham S., Lima L. G., Chai E. P. Z., Muller A., Lobb R. J., Krumeich S., Wen S. W., Wiegmans A. P. and Möller A. (2018). Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front. Immunol. 9, 871 10.3389/fimmu.2018.00871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-L., Hung J.-Y., Chang W.-A., Jian S.-F., Lin Y.-S., Pan Y.-C., Wu C.-Y. and Kuo P.-L. (2018). Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol. Ther. 26, 568-581. 10.1016/j.ymthe.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber V., Fais S., Iero M., Lugini L., Canese P., Squarcina P., Zaccheddu A., Colone M., Arancia G., Gentile M. et al. (2005). Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology 128, 1796-1804. 10.1053/j.gastro.2005.03.045 [DOI] [PubMed] [Google Scholar]
- Ichinose M., Masuoka J., Shiraishi T., Mineta T. and Tabuchi K (2001). Fas ligand expression and depletion of T-cell infiltration in astrocytic tumors. Brain Tumor Pathol. 18, 37-42. 10.1007/BF02478923 [DOI] [PubMed] [Google Scholar]
- Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T. D., Tridandapani S. et al. (2013). Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984-995. 10.1182/blood-2011-08-374793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen D. K., Fenix A. M., Franklin J. L., Higginbotham J. N., Zhang Q., Zimmerman L. J., Liebler D. C., Ping J., Liu Q., Evans R. et al. (2019). Reassessment of exosome composition. Cell 177, 428-445.e18. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C., Melo S. A., Protopopov A., Tang J., Seth S., Koch M., Zhang J., Weitz J., Chin L., Futreal A et al. et al. (2014). Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 289, 3869-3875. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. and LeBleu V. S. (2016). Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harbor Symp. Quant. Biol. 81, 275-280. 10.1101/sqb.2016.81.030932 [DOI] [PubMed] [Google Scholar]
- Kanada M., Bachmann M. H., Hardy J. W., Frimannson D. O., Bronsart L., Wang A., Sylvester M. D., Schmidt T. L., Kaspar R. L., Butte M. J. et al. (2015). Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 112, E1433-E1442. 10.1073/pnas.1418401112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuya Y., Horinouchi H., Goto Y., Kanda S., Fujiwara Y., Nokihara H., Yamamoto N., Watanabe S., Motoi N. and Ohe Y (2017). Prognostic value of CD8+ tumor-infiltrating lymphocyte density and PD-L1 expression in tumor cells in thymic epithelial tumors. J. Clin. Oncol. 35, 21-21 10.1016/j.lungcan.2016.05.007 [DOI] [Google Scholar]
- Kim J. W., Wieckowski E., Taylor D. D., Reichert T. E., Watkins S. and Whiteside T. L. (2005). Fas ligand–positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 11, 1010-1020. [PubMed] [Google Scholar]
- Kreger B. T., Dougherty A. L., Greene K. S., Cerione R. A. and Antonyak M. A. (2016). Microvesicle cargo and function changes upon induction of cellular transformation. J. Biol. Chem. 291, 19774-19785. 10.1074/jbc.M116.725705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge A., Arif S. and Moulin V. J. (2018). Microvesicles: intercellular messengers in cutaneous wound healing. J. Cell. Physiol. 233, 5550-5563. 10.1002/jcp.26426 [DOI] [PubMed] [Google Scholar]
- Lanier L. L. (2015). NKG2D receptor and its ligands in host defense. Cancer Immunol Res 3, 575-582. 10.1158/2326-6066.CIR-15-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Sanz-Garcia A., Visakorpi T., Escobedo-Lucea C., Siljander P., Ayuso-Sacido Á. and Yliperttula M (2014). Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate 74, 1379-1390. 10.1002/pros.22853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Chennakrishnaiah S., Audemard E., Montermini L., Meehan B. and Rak J (2014). Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem. Biophys. Res. Commun. 451, 295-301. 10.1016/j.bbrc.2014.07.109 [DOI] [PubMed] [Google Scholar]
- Lee T. H., Chennakrishnaiah S., Meehan B., Montermini L., Garnier D., D'Asti E., Hou W., Magnus N., Gayden T., Jabado N. et al. (2016). Barriers to horizontal cell transformation by extracellular vesicles containing oncogenic H-ras. Oncotarget 7, 51991-52002. 10.18632/oncotarget.10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Li C., Zhang Y., Zhang D., Otterbein L. E. and Jin Y. (2019). Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 216, 2202-2220. 10.1084/jem.20182313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. C. Y., Eaton S. A., Young P. E., Lee M., Shuttleworth R., Humphreys D. T., Grau G. E., Combes V., Bebawy M., Gong J. et al. (2013). Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 10, 1333-1344. 10.4161/rna.25281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yan X., Pan Y., Wang Y., Wang N., Li L., Liu Y., Chen X., Zhang C.-Y., Gu H. et al. (2015). MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol. Cancer 14, 58 10.1186/s12943-015-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Chen L., Peng Y., Yu S., Liu J., Wu L., Zhang L., Wu Q., Chang X., Yu X et al. et al. (2017). Dendritic cells loaded with tumor derived exosomes for cancer immunotherapy. Oncotarget 9, 2887-2894. 10.18632/oncotarget.20812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente A., Skotland T., Sylvänne T., Kauhanen D., Róg T., Orłowski A., Vattulainen I., Ekroos K. and Sandvig K (2013). Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 1831, 1302-1309. 10.1016/j.bbalip.2013.04.011 [DOI] [PubMed] [Google Scholar]
- Lundholm M., Schröder M., Nagaeva O., Baranov V., Widmark A., Mincheva-Nilsson L. and Wikström P (2014). Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: mechanism of immune evasion. PLoS ONE 9, e108925 10.1371/journal.pone.0108925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K. J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D. J., Olova N., Sutcliffe H., Rainger J. K., Leitch A. et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461-465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Lorenzo M. J., Anel A., Alava M. A., Piñeiro A., Naval J., Lasierra P. and Larrad L (2004). The human melanoma cell line MelJuSo secretes bioactive FasL and APO2L/TRAIL on the surface of microvesicles. Possible contribution to tumor counterattack. Exp. Cell Res. 295, 315-329. 10.1016/j.yexcr.2003.12.024 [DOI] [PubMed] [Google Scholar]
- Matei I., Kim H. S. and Lyden D (2017). Unshielding exosomal RNA unleashes tumor growth and metastasis. Cell 170, 223-225. 10.1016/j.cell.2017.06.047 [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Kagimoto T., Oda T., Kawano F. and Takatsuki K (1985). Natural killer activity and antibody-dependent cell-mediated cytotoxicity in multiple myeloma. Jpn. J. Clin. Oncol. 15, 611-617. 10.1093/oxfordjournals.jjco.a039094 [DOI] [PubMed] [Google Scholar]
- McKenzie A. J., Hoshino D., Hong N. H., Cha D. J., Franklin J. L., Coffey R. J., Patton J. G. and Weaver A. M. (2016). KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15, 978-987. 10.1016/j.celrep.2016.03.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A., Sugimoto H., O'Connell J. T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L. T., Melo C. A. et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707-721. 10.1016/j.ccell.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D., Gubin M. M., Schreiber R. D. and Smyth M. J. (2014). New insights into cancer immunoediting and its three component phases – elimination, equilibrium and escape. Curr. Opin. Immunol. 27, 16-25. 10.1016/j.coi.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz G. T., Santoro S. P., Wang L.-P., Garrabrant T., Lastra R. R., Hagemann I. S., Lal P., Feldman M. D., Benencia F. and Coukos G (2014). Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 20, 607-615. 10.1038/nm.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L., Mitsuhashi M., Simms P., Gooding W. E. and Whiteside T. L. (2016). Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 6, 20254 10.1038/srep20254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L., Simms P., Hong C.-S., Nishimura M. I., Jackson E. K., Watkins S. C. and Whiteside T. L. (2017). Human tumor-derived exosomes (TEX) regulate Treg functions via cell surface signaling rather than uptake mechanisms. Oncoimmunology 6, e1261243 10.1080/2162402X.2016.1261243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V., Clancy J., Plou C., Romao M., Chavrier P., Raposo G. and D'Souza- Schorey C. (2009). ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 19, 1875-1885. 10.1016/j.cub.2009.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B. Y., Qiu Y., Shabason J. E., Wu T. J., Yoon T., Kim B. C., Benci J. L., DeMichele A. M., Tchou J., Marcotrigiano J. et al. (2017). Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170, 352-366.e13. 10.1016/j.cell.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume A., Niwa R. and Satoh M (2009). Improving effector functions of antibodies for cancer treatment: enhancing ADCC and CDC. Drug Des Devel Ther 3, 7-16. 10.2147/DDDT.S4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell J., O'Sullivan G. C., Collins J. K. and Shanahan F (1996). The Fas counterattack: Fas- mediated T cell killing by colon cancer cells expressing Fas ligand. J. Exp. Med. 184, 1075-1082. 10.1084/jem.184.3.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R., Kratochvill F., Murray P. J. and Natoli G (2015). Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 36, 229-239. 10.1016/j.it.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Oushy S., Hellwinkel J. E., Wang M., Nguyen G. J., Gunaydin D., Harland T. A., Anchordoquy T. J. and Graner M. W. (2018). Glioblastoma multiforme-derived extracellular vesicles drive normal astrocytes towards a tumour-enhancing phenotype. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160477 10.1098/rstb.2016.0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka A. K., Ueno H., Connolly J., Kerneis-Norvell F., Blanck J.-P., Johnston D. A., Fay J. and Banchereau J (2006). Dendritic cells loaded with killed allogeneic melanoma cells can induce objective clinical responses and MART-1 specific CD8+ T-cell immunity. J. Immunother. 29, 545-557. 10.1097/01.cji.0000211309.90621.8b [DOI] [PubMed] [Google Scholar]
- Pardoll D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252-264. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebanek M. P., Angeloni N. L., Vinokour E., Li J., Henkin A., Martinez-Marin D., Filleur S., Bhowmick R., Henkin J., Miller S. D. et al. (2017). Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 8, 1319 10.1038/s41467-017-01433-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggio M., Hu T., Pai C.-C., Chu B., Belair C. D., Chang A., Montabana E., Lang U. E., Fu Q., Fong L et al. et al. (2019). Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177, 414-427.e13. 10.1016/j.cell.2019.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F., Garris C., Lai C. P., Newton A., Pfirschke C., Engblom C., Alvarez D., Sprachman M., Evavold C., Magnuson A. et al. (2016). SCS macrophages suppress melanoma by restricting tumor-derived vesicle–B cell interactions. Science 352, 242-246. 10.1126/science.aaf1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak J. and Guha A (2012). Extracellular vesicles–vehicles that spread cancer genes. BioEssays 34, 489-497. 10.1002/bies.201100169 [DOI] [PubMed] [Google Scholar]
- Raposo G. and Stoorvogel W (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373-383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J. and Geuze H. J. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161-1172. 10.1084/jem.183.3.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Sobreiro M., Chen J.-F., Novitskaya T., You S., Morley S., Steadman K., Gill N. K., Eskaros A., Rotinen M., Chu C.-Y. et al. (2018). Emerin deregulation links nuclear shape instability to metastatic potential. Cancer Res. 78, 6086-6097. 10.1158/0008-5472.CAN-18-0608 [DOI] [PubMed] [Google Scholar]
- Restifo N. P. (2001). Countering the ‘counterattack’ hypothesis. Nat. Med. 7, 259 10.1038/85357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoden J. J., Dyas G. L. and Wroblewski V. J. (2016). A modeling and experimental investigation of the effects of antigen density, binding affinity, and antigen expression ratio on bispecific antibody binding to cell surface targets. J. Biol. Chem. 291, 11337-11347. 10.1074/jbc.M116.714287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs F. L., Alayo Q., Krenzlin H., Mahmoud A. B., Speranza M. C., Nakashima H., Hayes J. L., Lee K., Balaj L., Passaro C. et al. (2018). Immune evasion mediated by PD-L1 on glioblastoma- derived extracellular vesicles. Sci. Adv. 4, eaar2766 10.1126/sciadv.aar2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. D. and Morelli A. E. (2014). Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195-208. 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P. A. and Furuta K (2015). The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203-216. 10.1038/nri3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rughetti A., Rahimi H., Belleudi F., Napoletano C., Battisti F., Zizzari I. G., Antonilli M., Bellati F., Wandall H. H., Benedetti Panici P. et al. (2014). Microvesicle cargo of tumor-associated MUC1 to dendritic cells allows cross- presentation and specific carbohydrate processing. Cancer Immunol. Res. 2, 177-186. 10.1158/2326-6066.CIR-13-0112-T [DOI] [PubMed] [Google Scholar]
- Sabado R. L., Balan S. and Bhardwaj N (2017). Dendritic cell-based immunotherapy. Cell Res. 27, 74-95. 10.1038/cr.2016.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A. and Tripodi M (2016). The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep 17, 799-808. 10.1016/j.celrep.2016.09.031 [DOI] [PubMed] [Google Scholar]
- Sato E., Olson S. H., Ahn J., Bundy B., Nishikawa H., Qian F., Jungbluth A. A., Frosina D., Gnjatic S., Ambrosone C. et al. (2005). Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 102, 18538-18543. 10.1073/pnas.0509182102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadendorf D., Hodi F. S., Robert C., Weber J. S., Margolin K., Hamid O., Patt D., Chen T.-T., Berman D. M. and Wolchok J. D. (2015). Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol. 33, 1889-1894. 10.1200/JCO.2014.56.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher T. N. and Schreiber R. D. (2015). Neoantigens in cancer immunotherapy. Science 348, 69-74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- Sedgwick A. E. and D'Souza-Schorey C (2018). The biology of extracellular microvesicles. Traffic 19, 319-327. 10.1111/tra.12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick A. E., Clancy J. W., Olivia Balmert M. and D'Souza-Schorey C (2015). Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 5, 14748 10.1038/srep14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P., Seo A. Y., Pasolli H. A., Song Y. E., Johnson M. C. and Lippincott-Schwartz J (2019). A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 21, 452 10.1038/s41556-019-0300-y [DOI] [PubMed] [Google Scholar]
- Sharma P., Hu-Lieskovan S., Wargo J. A. and Ribas A (2017). Primary, adaptive and acquired resistance to cancer immunotherapy. Cell 168, 707-723. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M. J., Temoche-Diaz M. M., Karfilis K. V., Ri S. and Schekman R. (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction [WWW Document]. eLife 5, e19276 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M. J., Yao J., Qin Y., Nottingham R. M., Temoche-Diaz M. M., Schekman R. and Lambowitz A. M. (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 114, E8987-E8995. 10.1073/pnas.1712108114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M. J., Temoche-Diaz M. M. and Schekman R (2018). Extracellular vesicles and cancer: caveat lector. Annu. Rev. Cancer Biol. 2, 395-411. 10.1146/annurev-cancerbio-030617-050519 [DOI] [Google Scholar]
- Sica A., Schioppa T., Mantovani A. and Allavena P (2006). Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Potential targets of anti- cancer therapy. Eur. J. Cancer, Cancer and inflammation 42, 717-727. 10.1016/j.ejca.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Sork H., Corso G., Krjutskov K., Johansson H. J., Nordin J. Z., Wiklander O. P. B., Lee Y. X. F., Westholm J. O., Lehtiö J., Wood M. J. A. et al. (2018). Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 8, 10813 10.1038/s41598-018-28485-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S. and Gajewski T. F. (2018). Mechanisms of tumor cell–intrinsic immune evasion. Annual Review of Cancer Biology 2, 213-228. 10.1146/annurev-cancerbio-030617-050606 [DOI] [Google Scholar]
- Steffen A., Dez G. L., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T. and Chavrier P (2008). MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18, 926-931. 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Szajnik M., Czystowska M., Szczepanski M. J., Mandapathil M. and Whiteside T. L. (2010). Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 5, e11469 10.1371/journal.pone.0011469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi M., Bornstein G. G. and Suria H (2010). Biodistribution mechanisms of therapeutic monoclonal antibodies in health and disease. AAPS J. 12, 33-43. 10.1208/s12248-009-9157-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Okada R., Nagao K., Kawamata Y., Hanyu A., Yoshimoto S., Takasugi M., Watanabe S., Kanemaki M. T., Obuse C et al. et al. (2017). Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8, 15287 10.1038/ncomms15287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboletti G., D'Ascenzo S., Borsotti P., Giavazzi R., Pavan A. and Dolo V (2002). Shedding of the matrix metalloproteinases MMP-2, MMP-9, and MT1-MMP as membrane vesicle- associated components by endothelial cells. Am. J. Pathol. 160, 673-680. 10.1016/S0002-9440(10)64887-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. D., Gerçel-Taylor Ç., Lyons K. S., Stanson J. and Whiteside T. L. (2003). T-cell apoptosis and suppression of T-cell receptor/CD3-ζ by fas ligand-containing membrane vesicles shed from ovarian tumors. Clin. Cancer Res. 9, 5113-5119. [PubMed] [Google Scholar]
- Thakur B. K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J. et al. (2014). Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766-769. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki M.-N., Yerneni S. S., Hoffmann T. K., Gooding W. E. and Whiteside T. L. (2018). Clinical significance of PD-L1+ exosomes in plasma of head and neck cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 24, 896-905. 10.1158/1078-0432.CCR-17-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torralba D., Baixauli F., Villarroya-Beltri C., Fernández-Delgado I., Latorre-Pellicer A., Acín-Pérez R., Martín-Cófreces N. B., Jaso-Tamame Á. L., Iborra S., Jorge I. et al. (2018). Priming of dendritic cells by DNA- containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 9, 2658 10.1038/s41467-018-05077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricarico C., Clancy J. and D'Souza-Schorey C (2017). Biology and biogenesis of shed microvesicles. Small GTPases 8, 220-232. 10.1080/21541248.2016.1215283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung S. L., Boardman D. A., Sen M., Letizia M., Peng Q., Cianci N., Dioni L., Carlin L. M., Lechler R., Bollati V. et al. (2018). Regulatory T cell-derived extracellular vesicles modify dendritic cell function. Sci. Rep. 8, 6065 10.1038/s41598-018-24531-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Drapkina O. and Tonevitsky A. (2019). Transcriptome of extracellular vesicles: state-of-the-art. Front. Immunol. 10, 202 10.3389/fimmu.2019.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T., Spinelli C., Minciacchi V. R., Balaj L., Zandian M., Conley A., Zijlstra A., Freeman M. R., Demichelis F., De S. et al. (2018). Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 7, 1505403 10.1080/20013078.2018.1505403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T., Chin A., Mariscal J., Bannykh S., Engman D. M. and Vizio D. D. (2019). Protein composition reflects extracellular vesicle heterogeneity. Proteomics 19, 1800167 10.1002/pmic.201800167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J. and Lötvall J. O. (2007). Exosome- mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654-659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- van Niel G., D'Angelo G. and Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213-228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Vanpouille-Box C., Demaria S., Formenti S. C. and Galluzzi L (2018). Cytosolic DNA sensing in organismal tumor control. Cancer Cell 34, 361-378. 10.1016/j.ccell.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D. J., Pascual-Montano A., Mittelbrunn M. and Sánchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Erbe A. K., Hank J. A., Morris Z. S. and Sondel P. M. (2015). NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front. Immunol. 6, 368 10.3389/fimmu.2015.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li B., Wei Y., Zhao Y., Wang L., Zhang P., Yang J., He W., Chen H., Jiao Z. et al. (2018a). Tumor-derived exosomes induce PD1+ macrophage population in human gastric cancer that promotes disease progression. Oncogenesis 7, 41 10.1038/s41389-018-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L. and Qiu Z (2018b). Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 78, 4586-4598. 10.1158/0008-5472.CAN-17-3841 [DOI] [PubMed] [Google Scholar]
- Wei Z., Batagov A. O., Schinelli S., Wang J., Wang Y., El Fatimy R., Rabinovsky R., Balaj L., Chen C. C., Hochberg F. et al. (2017). Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8, 1145 10.1038/s41467-017-01196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Malcor J.-D. M. and Harper M. T. (2018). Lipid rafts are essential for release of phosphatidylserine-exposing extracellular vesicles from platelets. Sci. Rep. 8, 9987 10.1038/s41598-018-28363-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemhoff G. A., Rabbany S. Y., Kusterbeck A. W., Ogert R. A., Bredehorst R. and Ligler F. S. (1992). Kinetics of antibody binding at solid-liquid interfaces in flow. J. Immunol. Methods 156, 223-230. 10.1016/0022-1759(92)90029-S [DOI] [PubMed] [Google Scholar]
- Wieckowski E. U., Visus C., Szajnik M., Szczepanski M. J., Storkus W. J. and Whiteside T. L. (2009). Tumor-derived microvesicles promote regulatory t cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 183, 3720-3730. 10.4049/jimmunol.0900970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers J., Lozier A., Raposo G., Regnault A., Théry C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T. et al. (2001). Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 7, 297-303. 10.1038/85438 [DOI] [PubMed] [Google Scholar]
- Wortzel I., Dror S., Kenific C. M. and Lyden D (2019). Exosome-mediated metastasis: communication from a distance. Dev. Cell 49, 347-360. 10.1016/j.devcel.2019.04.011 [DOI] [PubMed] [Google Scholar]
- Xu X., Tan Y., Qian Y., Xue W., Wang Y., Du J., Jin L. and Ding W. (2019). Clinicopathologic and prognostic significance of tumor-infiltrating CD8+ T cells in patients with hepatocellular carcinoma. Medicine (Baltim.) 98, e13923 10.1097/MD.0000000000013923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying X., Wu Q., Wu X., Zhu Q., Wang X., Jiang L., Chen X. and Wang X (2016). Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor- associated macrophages. Oncotarget 7, 43076-43087. 10.18632/oncotarget.9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Freitas D., Kim H. S., Fabijanic K., Li Z., Chen H., Mark M. T., Molina H., Martin A. B., Bojmar L. et al. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric-flow field-flow fractionation. Nat. Cell Biol. 20, 332-343. 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Regnault A., Lozier A., Wolfers J., Flament C., Tenza D., Ricciardi-Castagnoli P., Raposo G. and Amigorena S (1998). Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 4, 594-600. 10.1038/nm0598-594 [DOI] [PubMed] [Google Scholar]
- Zhu J., Petit P.-F. and Van den Eynde B. J. (2019). Apoptosis of tumor-infiltrating T lymphocytes: a new immune checkpoint mechanism. Cancer Immunol. Immunother. 68, 835-847. 10.1007/s00262-018-2269-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A., Maynard C., Verweij F. J., Kamermans A., Schäfer R., Beerling E., Schiffelers R. M., de Wit E., Berenguer J., Ellenbroek S. I. J. et al. (2015). In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046-1057. 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]