ABSTRACT
PUF RNA-binding proteins have diverse roles in animal development, with a broadly conserved role in stem cells. Two paradigmatic PUF proteins, FBF-1 and FBF-2, promote both self-renewal and differentiation in the C. elegans germline. The LST-1 protein is a pivotal regulator of self-renewal and is oncogenic when mis-expressed. Here, we demonstrate that LST-1 self-renewal activity resides within a predicted disordered region that harbors two KXXL motifs. We find that the KXXL motifs mediate the binding of LST-1 to FBF, and that point mutations of these motifs abrogate LST-1 self-renewal activity. The LST-1-FBF partnership is therefore crucial to stem cell maintenance and is a key element in the FBF regulatory network. A distinct region within LST-1 determines its spatial expression and size of the GSC pool. Most importantly, the molecular understanding of how an IDR-rich protein works in an essential partnership with a conserved stem cell regulator and RNA-binding protein suggests broad new avenues for combinatorial control.
KEY WORDS: Intrinsically disordered region, PUF RNA-binding protein, PUF partnership, Stem cell self-renewal, Zinc finger, Stem cell pool
Summary: Two short motifs in a disordered C. elegans protein are central to its self-renewal activity and to its partnership with a conserved PUF RNA-binding protein.
INTRODUCTION
A central paradigm in stem cell biology is that niche signaling regulates key target genes to promote self-renewal. Examples of niches and niche signaling pathways abound (e.g. Lander et al., 2012), but direct targets of niche signaling – the key genes activated in stem cells to drive self-renewal – have been elusive, with a handful of exceptions. One such is the Myc gene, a target of Notch, BMP and Wnt signaling in mammalian stem cells (Moore and Lemischka, 2006). Another is the lst-1 (lateral signaling target 1) gene, a target of GLP-1/Notch signaling in nematode germline stem cells (GSCs) (Fig. 1A) (Kershner et al., 2014; Lee et al., 2016). The LST-1 protein stands out as being pivotal for self-renewal and acts redundantly with another target of niche signaling, SYGL-1 (Kershner et al., 2014). LST-1 protein is normally restricted to the GSC pool region but drives a germline tumor when ubiquitously expressed (Shin et al., 2017). Although the biological significance of LST-1 is unambiguous, the challenge now is to understand, in molecular terms, how it executes its key role in stem cell self-renewal and how it is regulated.
Fig. 1.
LST-1 isoforms and their role in germline stem cells. (A) The lst-1 gene plays a central role in germline stem cell self-renewal. (B) Hypothesis that LST-1 functions post-transcriptionally via interaction with FBF. (C) Hypothesis that LST-1 loss (downward black arrow) defines the extent of the GSC pool. (D) The lst-1 locus encodes two transcripts, lst-1L(long) and lst-1S(short), which make two proteins, LST-1L and LST-1S. For locus and transcripts: exons (boxes); introns (lines); open reading frame (yellow); untranslated regions (white); start codons (black arrows); zinc finger (ZnF) (orange). For proteins: intrinsically disordered regions (IDRs) (dark blue) predicted using DISOPRED3 (Buchan and Jones, 2019; Jones and Cozzetto, 2015). (E-H) Left, alleles used to study LST-1L in GSC self-renewal. Conventions as in D; epitope tags (magenta); frameshift mutation (asterisk, ATG to ATΔ). Middle, predicted proteins. Right, GSC maintenance was scored positive (+) if the vast majority (>90%) produced many progeny and negative (−) if all lacked GSCs. See Fig. S1B for more information. (I) Western blot from whole worm lysate probed with anti-V5 antibody to detect epitope-tagged LST-1. LST-1L and LST-1S are labeled with black arrowheads. Actin is the loading control. The ratio between LST-1L and actin was calculated using Fiji/ImageJ. Control is wild-type N2. Uncropped blots are available in Fig. S1C. (J,K) Representative single confocal z-slices from the middle plane of the distal region of an extruded gonad, stained using anti-FLAG antibody to detect epitope-tagged LST-1 protein. Dotted line delineates gonad boundary, asterisk marks the distal end. Left, FLAG staining alone (magenta); right, merge of FLAG (magenta) and DAPI (cyan). Inset shows perinuclear staining. Scale bar in inset: 2 µm. (J) N-terminal tag detects LST-1L. (K) C-terminal tag detects both isoforms. (E-K) Alleles: lst-1(wildtype(wt))V5 is q1004; lst-1(frameshift(fs))V5 is q1198; lst-1(L)FLAG is q926; and lst-1(L/S)FLAG is q895 (see Table S1 for strain names).
In addition to LST-1, PUF RNA-binding proteins are central to self-renewal of germline stem cells (Crittenden et al., 2002; Forbes and Lehmann, 1998; Lin and Spradling, 1997; Wickens et al., 2002). This role is best understood in Caenorhabditis elegans, where two PUF proteins, FBF-1 and FBF-2 (collectively FBF), control self-renewal and differentiation by regulating hundreds of target RNAs (Kershner et al., 2013; Porter et al., 2019; Prasad et al., 2016). LST-1 was recently proposed to work as an FBF partner to repress differentiation-promoting RNAs and thus to promote self-renewal (Fig. 1B) (Shin et al., 2017). This idea was based on several lines of evidence. LST-1 protein harbors a predicted Nanos-like zinc finger (Kershner et al., 2014) and is cytoplasmic and granular (Shin et al., 2017), features that are consistent with a role in RNA regulation; LST-1 interacts in yeast with both FBF-1 and FBF-2 (Shin et al., 2017); LST-1 cannot form tumors in the absence of FBF, revealing dependence on FBF for its self-renewal activity (Shin et al., 2017); and LST-1 contributes to repression of gld-1, an established FBF target mRNA (Brenner and Schedl, 2016; Shin et al., 2017). Although this model is attractive, it has not yet been tested in vivo in nematodes, nor is it known whether LST-1 must bind FBF directly to exert its self-renewal activity.
The restriction of LST-1 expression to the GSC pool region suggested that its expression might help determine the size of the GSC pool (Fig. 1C) (Shin et al., 2017). Consistent with that idea, ubiquitous expression of full-length LST-1 throughout the germline drove formation of a tumor. One established lst-1 regulator is Notch signaling, which activates lst-1 transcription within the niche (Kershner et al., 2014; Lee et al., 2016). However, prior to this work, little was known about how LST-1 protein is spatially restricted or whether that restriction was biologically significant.
The LST-1 amino acid sequence provides few clues to the molecular basis of its self-renewal activity or its regulation (Kershner et al., 2014). Here, we identify one region sufficient for stem cell self-renewal and another region required for spatial regulation. Within the self-renewal region, we find two short sequence motifs that mediate direct binding to FBF and that are essential for LST-1 self-renewal activity. The regulatory region includes a Nanos-like zinc finger and loss of this zinc finger expands LST-1 distribution and generates a larger than normal GSC pool. Thus, LST-1 works within the stem cell regulatory network as a key FBF partner, and its spatial regulation helps determine the size of the GSC pool.
RESULTS
LST-1L isoform is critical for self-renewal
The lst-1 locus encodes two transcripts, which generate longer LST-1L and shorter LST-1S proteins (Fig. 1D) (Kershner et al., 2014). The LST-1L and LST-1S amino acid sequences overlap extensively and harbor multiple predicted intrinsically disordered regions [IDRs; regions with a high proportion of polar and charged amino acids and a low proportion of nonpolar amino acids (Dyson, 2016)] plus a CCHC Nanos-like zinc finger (Fig. 1D, Fig. S1A) (Kershner et al., 2014). To differentiate between the roles of LST-1L and LST-1S, we compared self-renewal activities of wild-type LST-1, lst-1(wt) (Fig. 1E), with a mutant LST-1, lst-1(frameshift) that harbors a single base pair deletion in the lst-1L start codon, and only makes LST-1S (Fig. 1F). For protein visualization, both lst-1(wt) and lst-1(fs) carried a V5 epitope tag at the shared C terminus (Fig. 1E,F), which we indicate henceforth in superscript.
We assayed self-renewal activities in lst-1(wt)V5 and lst-1(fs)V5 animals in the absence of SYGL-1, where GSC maintenance relies on LST-1 alone (Kershner et al., 2014). Virtually all lst-1(wt)V5 animals make a healthy fertile germline in the absence of SYGL-1, while strong loss-of-function lst-1 mutants are all sterile with no GSCs without SYGL-1 (Fig. S1B), as shown previously (Kershner et al., 2014; Shin et al., 2017). The lst-1(fs)V5 were also all sterile with no GSCs in the absence of SYGL-1 (Fig. 1E,F, Fig. S1B). Western blot demonstrated that LST-1L was nearly eliminated in lst-1(fs)V5, while LST-1S remained (Fig. 1I, Fig. S1C). Immunostaining showed that LST-1V5 proteins were similarly restricted in lst-1(wt)V5 and lst-1(fs)V5 germlines (Fig. S1D-G). We conclude that LST-1L is necessary for GSC self-renewal activity, and that LST-1S not sufficient despite expression in GSCs.
Consistent with its role in GSC self-renewal, we expected LST-1L to be expressed in GSCs. To visualize this isoform specifically, we introduced a FLAG epitope tag at the unique LST-1L N-terminus to create lst-1(L)FLAG. FLAG was used because attempts to insert an N-terminal V5 failed (Fig. 1G). As a control, we made C-terminal lst-1(L/S)FLAG to visualize LST-1L and LST-1S collectively (Fig. 1H). Both N-terminal and C-terminal LST-1FLAG variants maintained GSCs in the absence of SYGL-1 and therefore were functional (Fig. 1G,H, Fig. S1B). Upon immunostaining, both LST-1(L)FLAG and LST-1(L/S)FLAG were expressed in the GSC region with similar subcellular localization (Fig. 1J,K). We conclude that the LST-1L isoform is present in GSCs and that it harbors self-renewal activity.
N-terminal LST-1 fragment is sufficient for stem cell maintenance
To delineate the region within LST-1 required for self-renewal activity, we generated a series of lst-1 variant alleles (Fig. 2). Each was introduced into the lst-1(wt)V5 locus (Fig. 2A). Because all lst-1 variants from this point carry V5, we henceforth omit the V5 superscript in allele designations. As a control, we created an lst-1(ø) protein null mutant by deleting the entire open reading frame at the endogenous locus (Fig. 2B). As expected (Kershner et al., 2014; Shin et al., 2017), all lst-1(ø) homozygotes maintained GSCs in the presence of SYGL-1 due to redundancy, but none maintained GSCs in the absence of SYGL-1 (Fig. 2B, Fig. S2A).
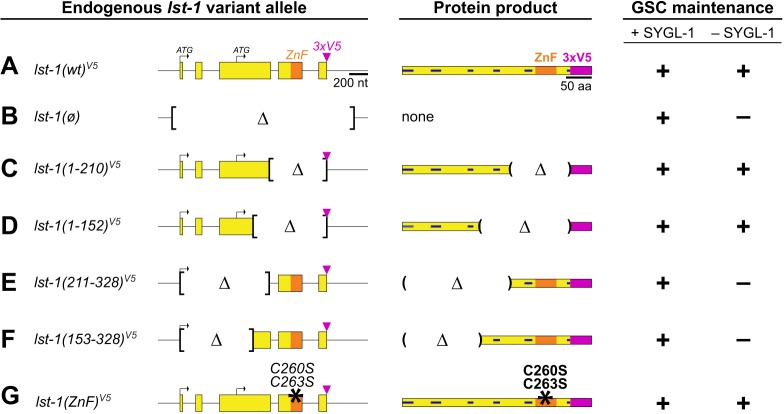
Fig. 2.
Identification of LST-1 region required for GSC maintenance. (A-G) Left, alleles. Only lst-1L is depicted for simplicity. All variants were created at the endogenous locus in lst-1(wild-type)V5, an otherwise wild-type allele carrying 3× V5 at its C terminus (Fig. 1E). Internal deletion boundaries (brackets). Middle, protein products. For amino acid changes in each variant, see Fig. S1A. Right, GSC maintenance was scored positive (+) if the vast majority (>90%) produced many progeny and negative (−) if all lacked GSCs. For result details, see Fig. S2A. Alleles: lst-1(wild-type)V5 is q1004; lst-1(ø) is q869 and removes entire open reading frame plus 139 bp upstream of start codon and 228 bp downstream of stop codon; lst-1(1-210)V5 is q1115; lst-1(1-152)V5 is q1060; lst-1(211-328)V5 is q1044; lst-1(153-328)V5 is q1119; and lst-1(ZnF)V5 is q1032 and harbors missense mutations in two amino acids that are crucial for zinc finger architecture (black asterisks) (Hashimoto et al., 2010; Weidmann et al., 2016).
To test lst-1 variants for function in vivo, each was assayed for GSC maintenance with and without SYGL-1. Two variants harboring N-terminal regions, lst-1(1-210) and lst-1(1-152), maintained GSCs in the absence of SYGL-1 (Fig. 2C,D, Fig. S2A). Complementary variants harboring C-terminal regions, lst-1(211-328) and lst-1(153-328), did not maintain GSCs (Fig. 2E,F, Fig. S2A) (see Fig. 4 and Fig. S5 for confirmation of germline expression). Analogous transgenic experiments performed prior to the CRISPR/Cas9 revolution produced similar results (Fig. S2B-E). Although both sets of truncation experiments showed that the zinc finger was dispensable for self-renewal, we explored this domain specifically by mutating two cysteine residues required for its architecture (Hashimoto et al., 2010; Weidmann et al., 2016). The lst-1(C260S C263S) missense mutant, dubbed lst-1(ZnF), maintained GSCs in the absence of SYGL-1, confirming that the zinc finger is not crucial for GSC maintenance (Fig. 2G). We conclude that LST-1 self-renewal activity resides in the N-terminal half of the protein.
Fig. 4.
Biological readout and protein expression of LST-1 variants. (A) Schematic of progenitor zone (PZ). Somatic niche (gray); germline stem cells in naïve state (yellow); GSC daughters primed for differentiation (graded yellow to green); early meiotic prophase (crescents). Double-headed arrow indicates PZ size, measured by conventional methods (see Materials and methods) in germ cell diameters (gcd) from the distal end. (B) Progenitor zone size in lst-1 variants measured in average gcd from the distal end. Individual data points are plotted as pink circles; middle line, median; whiskers, minimum and maximum values; boxes, 25-75% quantile. n≥9 animals for each sample. Asterisks indicate statistically significant differences (one-way ANOVA with Tukey's post-hoc test) compared with lst-1(wt)V5: ****P<0.0001, ***P<0.001; ns, not significant (P>0.05). (C-G) LST-1 variant protein expression in the distal gonad. Representative confocal z-projections of extruded gonads stained using anti-V5 antibodies to detect LST-1 variants (magenta) and with DAPI (cyan). Left, anti-V5 antibody immunostaining; right, merge of anti-V5 antibody and DAPI staining. White arrowhead marks the proximal boundary of the progenitor zone. (H) LST-1 quantitation as a function of distance from the distal end measured using Fiji/ImageJ (see Materials and methods for details). Each line represents mean values of at least three independent replicates, each with at least five gonads; shading shows the s.e.m. Total germlines were at least 23 per genotype. After background from the lst-1(ø) negative control was subtracted for each replicate, LST-1(wt)V5 was set to 1.0 at its peak and variants were normalized to this value. The x-axis shows distance from distal end in μm at the bottom and in germ cell diameters (gcd) at the top. (B-H) Alleles are indicated as follows: lst-1(wt)V5 is q1004; lst-1(1-210)V5 is q1115; lst-1(1-152)V5 is q1060; lst-1(211-328)V5 is q1044; lst-1(153-328)V5 is q1119; lst-1(ZnF)V5 is q1032; lst-1(A)V5 is q1124; lst-1(B)V5 is q1086; lst-1(AB)V5 is q1125; and lst-1(ø) is q869.
Interestingly, although both lst-1(1-210) and lst-1(1-152) maintain GSCs, these two variants differ with respect to the hermaphrodite sperm/oocyte switch: in the absence of SYGL-1, all lst-1(1-210) animals made the switch and were self-fertile, whereas no lst-1(1-152) animals made the switch and had a sterile Mog (Masculinization of Germline) phenotype (Fig. S2A). A role for LST-1 in the sperm/oocyte decision was known: indeed, even with SYGL-1 present, a few lst-1(ø) homozygotes (≤5%) failed to make the switch and had a Mog phenotype (Fig. S2A), as previously shown for two other lst-1 strong loss-of-function alleles (Kershner et al., 2014; Shin et al., 2017). A low penetrance Mog defect can be associated with an increased brood size (Lamont et al., 2004), but this was not the case for lst-1 strong loss-of-function alleles (Shin et al., 2017). This LST-1-mediated effect on germline sex determination likely reflects the common molecular basis for regulation of germline self-renewal and germline sex determination as both rely on FBF RNA regulation (Crittenden et al., 2002; Zhang et al., 1997). Yet for the purposes of this study, we focus on LST-1 regulation of GSC maintenance via its N-terminal half.
Two FBF-binding motifs are essential for LST-1 self-renewal activity
LST-1 has been proposed to function with FBF for self-renewal (Fig. 1B, see Introduction). We therefore investigated the N-terminal half of LST-1 for FBF interaction. Two other FBF partners, CPB-1 and GLD-3, possess a consensus KTXL motif that is crucial for FBF binding, where X is any amino acid (Fig. 3A) (Menichelli et al., 2013; Wu et al., 2013). We scanned LST-1(1-152) for a KTXL FBF-binding motif but found none. Instead, we found two similar sequences: KQLL (amino acids 32-35) and KLHL (amino acids 80-83), which we dub A and B potential motifs, respectively (Fig. 3B, Fig. S1A). Intriguingly, A and B reside in the region unique to LST-1L.
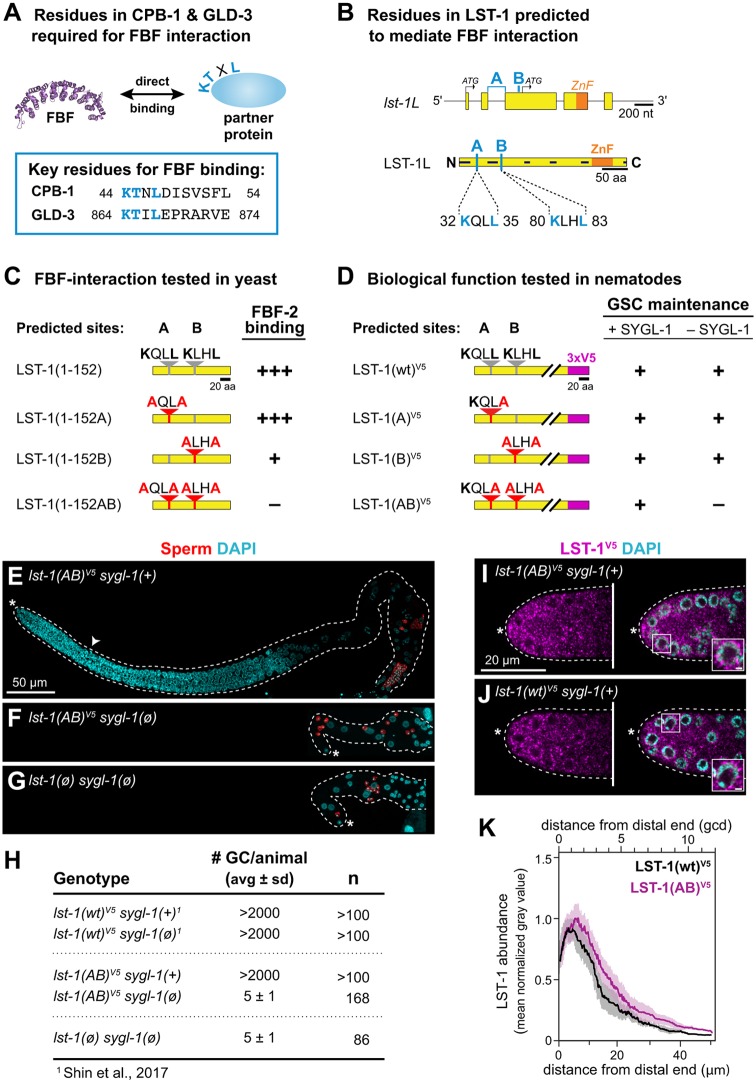
Fig. 3.
FBF-binding regions in LST-1 and their role in stem cell regulation. (A) KTXL motif identified in vitro for two FBF partner proteins: CPB-1 and GLD-3 (Menichelli et al., 2013; Wu et al., 2013). PDB ID for FBF is 3K5Q (Wang et al., 2009). (B) Two predicted FBF interaction motifs, A and B (light blue), in the LST-1L-specific sequence. Top: nucleotides encoding the A motif span an intron. Bottom: positions of A and B in LST-1L (see also Fig. S1A). (C) Yeast interaction between LST-1 and FBF-2. Red letters indicate missense mutations. Binding: strong (+++), weak (+) or none (−) (see Fig. S3 for data). (D) GSC maintenance scored in LST-1 A and B motif variant nematodes. For result details, see Fig. S4B. (E-G) Representative z-projected confocal images of extruded gonads immunostained using an anti-SP56 sperm antibody (red) (Ward et al., 1986) and DAPI (cyan). (E) lst-1(AB)V5 in a strain harboring wild-type SYGL-1. The germline and progenitor zone (white arrowhead, boundary) are both of normal size. (F) lst-1(AB)V5 in a strain lacking SYGL-1. The germline is tiny and has only a few sperm (red). Other DAPI-stained nuclei belong to the somatic gonad. (G) lst-1(ø) sygl-1(ø) mutants have tiny germlines and only a few sperm. (H) Quantitation and comparison of the number of germ cells (# GC) per animal. Total number of GCs in lst-1(AB)V5 sygl-1(ø) is indistinguishable from lst-1(ø) sygl-1(ø). (I,J) Representative single confocal z-slice from the middle plane of the distal region of an extruded gonad, stained with anti-V5 antibodies to detect LST-1 (magenta) and DAPI (cyan). Inset shows perinuclear staining (scale bar: 2 µm). (I) LST-1(AB)V5 protein is restricted to distal germline, with distribution similar to wild type (compare with I). (J) LST-1(wt)V5 protein is restricted to distal germline, as previously reported (Shin et al., 2017). (K) Quantitation of LST-1(wt)V5 and LST-1(AB)V5 protein as a function of distance from the distal end (by Fiji/ImageJ; see Materials and methods for details). Lines represent mean value of three independent replicates, each with at least seven gonads; shading shows s.e.m. Sample sizes: LST-1(wt)V5, 25 germlines; LST-1(AB)V5, 36 germlines. After background from wild-type N2 control was subtracted for each replicate, LST-1(wt)V5 was set to 1.0 at its peak and LST-1(AB)V5 was normalized to this value. The x-axis shows distance from distal end in μm at the bottom and germ cell diameters (gcd) at the top. (D-K) Alleles: lst-1(wt)V5 is q1004; lst-1(A)V5 is q1124; lst-1(B)V5 is q1086; lst-1(AB)V5 is q1125; lst-1(ø) is q869; and sygl-1(ø) is q828.
We first tested the A and B motifs for FBF binding in yeast (Fig. S3A). Because full-length, wild-type LST-1 bound to FBF-1 and FBF-2 similarly in a yeast two-hybrid assay (Shin et al., 2017), we focused on FBF-2 here. We first found that full-length LST-1(wt) and LST-1(1-152) interacted similarly with FBF-2 in yeast (Fig. S3B) and therefore used LST-1(1-152) for subsequent assays. To test the A and B sites, we made site-specific mutants, changing their first and fourth amino acids to alanine (Fig. 3C, Fig. S3B). Mutation of site A had no appreciable effect on the yeast interaction, mutation of site B reduced the interaction, and mutation of A and B abolished the interaction (Fig. 3C, Fig. S3B) both in yeast growth assays (Fig. S3C) and β-gal assays (Fig. S3D). We conclude that the LST-1 self-renewal fragment has two FBF interaction motifs, and that, at least in yeast, the B motif is quantitatively more important than the A motif.
To probe the significance of LST-1 A and B sites for self-renewal activity in nematodes, we engineered point mutations at the lst-1 endogenous locus, both individually and together (Fig. 3D). The nematode B motif mutation was identical to that made in yeast (K80A L83A), but the nucleotide sequence encoding the A motif straddles an intron (Fig. 3B, top), making simultaneous mutation of both K32 and L35 challenging. Because the leucine at the fourth position stood out as crucial for CPB-1 and GLD-3 interactions with FBF (Menichelli et al., 2013; Wu et al., 2013), we chose to disrupt L35 alone to test the importance of the A motif in nematodes. We introduced mutations into lst-1(wt)V5 (Fig. 3D) and made three alleles, lst-1(A)[L35A], lst-1(B)[K80A L83A] and lst-1(AB)[L35A K80A L83A]. All were fertile and healthy when SYGL-1 was present [Fig. 3D,E,H for lst-1(AB); Fig. S4B]. When SYGL-1 was removed, lst-1(A) and lst-1(B) mutants maintained GSCs and were fertile, but lst-1(AB) did not maintain GSCs and was sterile (Fig. 3D, Fig. S4B). Indeed, the lst-1(AB) sygl-1(ø) germline was indistinguishable from that of lst-1(ø) sygl-1(ø) (Fig. 3F-H); it was tiny and differentiated to sperm (Fig. 3D,F,H, Fig. S4B). Importantly, LST-1(AB) protein was present with an abundance and distribution similar to LST-1(wt) (Fig. 3I-K). We conclude that LST-1 depends on its two KXXL FBF-binding motifs for self-renewal activity.
The C-terminal region controls spatial restriction
We next examined the lst-1 variants for more subtle effects on GSC maintenance. Specifically, we assayed size of the progenitor zone (PZ) in all variants (Fig. 4A), because PZ size is a rough measure of the switch from stem cell state to differentiation (Crittenden et al., 2006). All PZ measurements were carried out in the presence of wild-type SYGL-1 to ensure healthy germline size and organization. Most variants had a PZ size similar to wild type, but lst-1(1-210) and lst-1(ZnF) PZs were larger than normal (Fig. 4B).
We reasoned that longer PZs in lst-1(1-210) and lst-1(ZnF) might reflect aberrant LST-1 expression. To test this idea, we immunostained for LST-1, again with wild-type SYGL-1 present to ensure robust germline size and organization. LST-1(wt) was restricted to the same distal germline region reported previously (Shin et al., 2017) (Fig. 4C). By contrast, LST-1(1-210) and LST-1(ZnF) were more abundant and their expression extended more proximally than wild type (Fig. 4D,F). LST-1(211-328) was of lower abundance (Fig. 4E). lst-1(ø) provided a negative control (Fig. 4G). To quantify protein abundance as a function of position within the distal gonad, we generated z-projections (n≥5) from at least three independent experiments. We then used Fiji/ImageJ to score LST-1 abundance along the gonadal axis (see Materials and methods for details). This quantitation confirmed that LST-1(1-210) and LST-1(ZnF) proteins were both more abundant and expanded proximally compared with LST-1(wt) (Fig. 4H). This altered expression pattern offers a likely explanation for the enlarged PZ in the two mutants.
The increased abundance of LST-1(1-210) and LST-1(ZnF) proteins could be due to increased lst-1 mRNA stability, to increased translation or to changes in LST-1 protein turnover. To query the abundance of lst-1 mRNAs, we performed single-molecule fluorescence in situ hybridization (smFISH) with probes designed against sequences that were identical in the three variants tested, but absent in the control (Fig. 5A). The lst-1 RNAs in lst-1(wt), lst-1(1-210) and lst-1(ZnF) were all restricted to the distal-most four or five rows of germ cells and absent in the lst-1(ø) control (Fig. 5B-E). Therefore, expansion of LST-1 protein was not due to expansion of lst-1 RNA. Curiously, quantitation revealed that lst-1(1-210) and lst-1(ZnF) RNAs were modestly more abundant (∼30% higher) than lst-1(wt) RNA (Fig. 5F,G), suggesting that the LST-1 zinc finger has an autoregulatory effect on lst-1 mRNA abundance. Together, these analyses indicate that the LST-1 C-terminal region mediates two distinct regulatory activities: the zinc finger downregulates lst-1 RNA abundance and a broader region, yet to be defined but including the zinc finger, downregulates LST-1 protein abundance and controls its extent, likely through an effect on protein turnover (see Discussion).
Fig. 5.
lst-1 variant RNA expression. (A) lst-1 smFISH probe (red) anneals to sequences present in all strains, except negative control lst-1(ø). (B-E) Representative z-projected confocal images of lst-1 RNA distribution. Extruded gonads were stained for lst-1 RNA (red) using smFISH and DAPI (cyan). Left, lst-1 RNA; right, merge of lst-1 RNA and DAPI. (F) Quantitation of lst-1 RNA as a function of distance from distal end (see Materials and methods for details). Each line represents mean values of three independent replicates, with a total of 30 gonads per variant; shading shows the s.e.m. After background subtraction using lst-1(ø) for each replicate, lst-1(wt)V5 was set to 1.0 at its peak and variants were normalized to this value. The x-axis shows distance from distal end in μm at the bottom and in germ cell diameters (gcd) at the top. (G) Comparison of mean lst-1 RNA values from Fig. 4F (lines) and mean LST-1 protein values from Fig. 3F (shading) in the distal germline. (A-G) Alleles indicated are as follows: lst-1(wt)V5 is q1004; lst-1(1-210)V5 is q1115; lst-1(ZnF)V5 is lst-1(q1032); and lst-1(ø) is q869.
Spatial extent of LST-1 determines GSC pool size
Finally, we asked whether the increased PZ size in lst-1(1-210) and lst-1(ZnF) mutants reflects a shift in the regulatory network from self-renewal to differentiation. To this end, we conducted emb-30 assays (Fig. 6A) (Cinquin et al., 2010). Briefly, this assay blocks the cell cycle and stops migration through the progenitor zone so that germ cells reveal their naïve or differentiated state in situ. This is the only functional assay available for GSC pool size, and provides a rough size estimate. We focused on lst-1(wt) and lst-1(1-210) for this experiment, and tested both CRISPR-induced lst-1 mutants (Fig. 2A,C) as well as single-copy transgenic variants inserted at a MosSCI site (Fig. S2B,C). While the LST-1 fragments assayed were identical, two differences existed between these experiments, for historical reasons. First, the endogenous alleles were assayed in a sygl-1(ø) background so that GSCs were dependent on LST-1 alone, while transgenes were assayed in an lst-1(ø) sygl-1(+) background so that all LST-1 protein came from the transgenic allele. Second, the endogenous alleles were tagged with V5 while transgenes were tagged with HA. Remarkably, these two experiments gave virtually the same result: lst-1(wt) possessed an average of ∼40 germ cells in its GSC pool, whereas lst-1(1-210) had an average of ∼55-75 (Fig. 6B). Thus, lst-1(1-210) increases GSC pool size, suggesting that LST-1 extent modulates size of the GSC pool.
Fig. 6.
Effect of LST-1 distribution on GSC pool size. (A) Schematic of emb-30 assay to estimate GSC pool size. This assay resolves two pools in the PZ, one inferred to be the GSC pool (Cinquin et al., 2010). Left: at permissive temperature, the emb-30(ts) PZ appears normal with M-phase germ cells (pink) scattered throughout and GLD-1 levels (green) gradually increasing as germ cells move proximally towards meiotic entry. Middle: after shifting emb-30(ts) from permissive to restrictive temperature, PZ germ cells stop dividing, stop moving proximally and arrest in either of two states. Distal PZ cells arrest in mitotic M-phase and express the PH3 M-phase marker, but not the GLD-1 differentiation marker; proximal PZ cells enter meiotic prophase and express abundant GLD-1 but no PH3. Right: inference of emb-30 results for wild-type PZ. (B) Quantitation of GSC pool sizes for CRISPR/Cas9-induced endogenous LST-1 variants (left) or LST-1 transgenes (right). After emb-30 temperature shift, we visualized the distal pool by morphology of DAPI-stained nuclei (see Materials and methods). Individual data points are plotted as pink or blue circles; boxes indicate 25-75% quantile; middle line indicates the median; whiskers indicate minimum and maximum values. P-values were calculated using a two-tailed t-test, assuming equal variance and comparing each LST-1(1-210) variant to its respective LST-1(wt) control. ***P<0.001. Endogenous variants were scored in a sygl-1(ø) background; genotypes and sample sizes: wtV5 is lst-1(q1004) sygl-1(q828); emb-30(tn377ts) (n=25) and 1-210V5 is lst-1(q1115) sygl-1(q828); emb-30(tn377ts) (n=30). Transgenic variants were scored in sygl-1(+) with genotypes and sample sizes as follows: wtHA is lst-1(ok814); qSi22; emb-30(tn377ts) (n=7) and 1-210HA is lst-1(ok814); qSi300; emb-30(tn377ts) (n=8). (C) Schematic of transgene driving ubiquitous germline expression of LST-1(1-210). (D) Representative z-projected confocal image of a germline tumor driven by a single-copy transgene as in C and lacking endogenous LST-1. Anti-PH3 antibodies (magenta); DAPI (cyan). (E-G) Schematics illustrating the effects of LST-1 spatial extent on GSC pool size. Top: lst-1 RNA (black) and LST-1 protein (magenta), with line thickness corresponding to quantity and line length to extent of expression. Bottom: effects on GSC pool size. Black arrow, switch from self-renewal to differentiation. (E) Wild type: lst-1 RNA and LST-1 protein are both restricted to distal germline and GSC pool size is similarly restricted. (F) LST-1(1-210): lst-1(1-210) RNA is slightly more abundant than in wild type, but still restricted to distal germline (from Fig. 4). By contrast, LST-1(1-210) protein is more abundant than wild type and expands proximally (from Fig. 3). The GSC pool expands correspondingly (from Fig. 5). (G) Ubiquitous LST-1(1-210): major expansion of LST-1 leads to germline tumor formation (from Fig. 5). The switch to differentiation fails to occur (red X).
To further interrogate the potency of the LST-1(1-210) protein for GSC maintenance, we assayed its effect when ubiquitously expressed. This assay was essentially the same as that carried out earlier with full-length LST-1(wt) protein, which makes a massive germline tumor when placed under control of a ubiquitous germline promoter, mex-5 and the tbb-2 3′UTR (Shin et al., 2017). To ask whether LST-1(1-210) is similarly oncogenic, we made an analogous transgene, placing LST-1(1-210) under the same regulatory elements (Fig. 6C). The strain was created and maintained with lst-1(RNAi) to prevent expression of the potentially oncogenic LST-1. Upon removal from lst-1(RNAi), ubiquitous LST-1(1-210) drove formation of germline tumors, as evidenced by M-phase nuclear morphology and PH3-marked cells (Fig. 6D). We conclude that ubiquitous LST-1(1-210) mimics LST-1(wt) in its oncogenicity. This result dovetails with the more moderate effect of LST-1(1-210) expansion on GSC pool size and suggests that LST-1 downregulation facilitates the molecular switch from stem cell state to differentiation (Fig. 6E-G).
DISCUSSION
C. elegans LST-1 provides a key link between niche signaling and an RNA regulatory network driving stem cell self-renewal (Kershner et al., 2014; Shin et al., 2017). Here, we report the molecular basis of the LST-1–FBF partnership, the significance of the partnership for self-renewal activity and the significance of LST-1 spatial restriction to GSC pool size.
Dual FBF-binding motifs may provide plasticity to the RNA regulatory network
The LST-1 self-renewal region harbors two short KXXL motifs that mediate FBF binding (Fig. 7A). Each motif has biological activity in nematodes: LST-1 self-renewal activity remains intact when either is mutated, but when both are mutated, self-renewal activity is lost. The discovery of two motifs was unexpected, because other FBF partners GLD-3 and CPB-1 possess only a single KXXL motif (Campbell et al., 2012; Menichelli et al., 2013; Wu et al., 2013). Yet the two LST-1 motifs are conserved throughout the Caenorhabditis (Fig. S4A), suggesting biological significance.
Fig. 7.
LST-1 molecular function: summary and speculation. (A) LST-1 protein possesses one region responsible for self-renewal and another that restricts its spatial expression. The LST-1 self-renewal region is composed largely of intrinsically disordered regions (IDRs) and harbors two KXXL motifs that mediate interaction with FBF. The spatial regulatory region possesses a Nanos-related zinc finger. (B-D) Possible modes of LST-1-FBF complex formation. The FBF RNA-binding domain (RBD) contacts its target RNA directly via the FBF-binding element (FBE). The A and B KXXL motifs are functionally redundant for FBF binding and for self-renewal activity, which means that complex formation can rely on the A motif only (B) or on the B motif only (C). Formation of these A-specific and B-specific complexes may be subject to differential regulation or offer distinct platforms for recruitment of effectors. LST-1-FBF complexes may also form via interaction with both A and B motifs (D), which may provide additional possibilities for regulation and function. (E-G) Proposals for function of the LST-1–FBF complex, depicted here in a mode that relies on B motif binding. (E) LST-1-FBF may enhance or inhibit recruitment of an effector. (F) Model for yeast Puf3p, the disordered N-terminal tail of which acts in cis as an integral part of Puf3 to recruit the CCR4-Not complex and promote deadenylase activity [modified from Webster et al. (2019)]. (G) LST-1 IDRs, or small linear motifs not yet identified, may function in trans with FBF to recruit an effector.
The existence of two KXXL motifs may afford plasticity to the LST-1–FBF complex and its role in the FBF regulatory network. We do not yet know where within FBF these motifs bind, but clues exist. The motifs in GLD-3 and CPB-1 interact in vitro at the loop between PUF repeats 7 and 8, dubbed the R7/8 loop (Menichelli et al., 2013; Wu et al., 2013). By analogy, the dual LST-1 motifs may each be able to bind the same loop. Indeed, a small purified fragment harboring one LST-1 motif, called in this work the B site, binds to the R7/8 loop in the crystal structure of an FBF-2/LST-1B-site/RNA complex (Qiu et al., 2019). However, the LST-1 A and B motifs are unlikely to bind the same site simultaneously, raising the possibility that these dual motifs provide other opportunities. For example, some LST-1-FBF complexes may rely on the A site binding to the R7/8 loop (Fig. 7B), while others rely on B site binding to the same loop (Fig. 7C). This scenario introduces the possibility of considerable plasticity in the regulation and molecular configuration of the two complexes. Alternatively, the dual motifs may bind at distinct sites in FBF (Fig. 7D). More radically, they may link two FBF proteins together, with LST-1 binding at each of their R7/8 loops. These various scenarios have important implications for configuration, stability and regulation of this critical LST-1–FBF partnership and thus for FBF combinatorial control of RNAs and stem cell self-renewal.
LST-1 self-renewal activity: a series of IDRs that work with FBF
The LST-1 self-renewal region possesses two FBF-binding motifs plus predicted intrinsically disordered regions (IDRs) (Fig. 7A). The LST-1(1-152) variant contains three IDRs and LST-1(1-210) has four (Fig. 7A). Therefore, the LST-1 self-renewal region brings a series of IDRs to FBF (Fig. 7E). IDRs are commonly found as integral parts of RNA-binding proteins (Calabretta and Richard, 2015) and have been implicated in diverse steps of RNA regulation: splicing (Chen and Moore, 2014), decapping (Jonas and Izaurralde, 2013), deadenylation (Webster et al., 2019) and RNP granule formation (Mittag and Parker, 2018; Uversky, 2017). However, IDRs in RNA-binding proteins act within the same polypeptide and hence in cis with the RNA-binding domain. The LST-1 IDRs, by contrast, are not in the same polypeptide as FBF and hence work in trans.
The LST-1 trans activity could act via a range of mechanisms (Fig. 7E). However, we suggest that LST-1 IDRs, or perhaps not yet identified small linear motifs interspersed within or among the IDRs, stabilize the formation of a complex that represses target mRNAs. Our thinking is guided by three previous studies. First, FBF-binding elements regulate poly(A) tail length (Ahringer and Kimble, 1991). Second, FBF, like other PUF proteins, interacts in vitro with the CCR4-Not deadenylase complex (Suh et al., 2009), suggesting that FBF represses RNAs, at least in part, via deadenylation. Third, LST-1 promotes destabilization of an FBF target mRNA in vivo (Shin et al., 2017), suggesting that it works with FBF to promote deadenylation. Indeed, yeast Puf3 (yPuf3), a PUF RNA-binding protein from S. cerevisiae, possesses an N-terminal tail composed largely of IDRs and critical for interactions with the CCR4-Not complex. Remarkably, the longer the yPuf3 tail, and hence the more IDRs, the greater the deadenylase activity in vitro (Webster et al., 2019) (Fig. 7F). We suggest that trans-acting LST-1 protein may work similarly to stabilize the interaction with an effector protein or complex (Fig. 7G). The CCR4-Not complex is a strong candidate because of its in vitro interaction with FBF (Suh et al., 2009). Other possibilities exist and they are not mutually exclusive. For example, LST-1 may prevent recruitment of a positive-acting regulatory factor, such as the GLD-2/GLD-3 poly(A) polymerase (Wang et al., 2002). Regardless, the discovery of an IDR-rich fragment that works in trans with an RNA-binding protein suggests new avenues for combinatorial control.
LST-1 downregulation and the molecular switch from stem cell state to differentiation
Normally, lst-1 mRNA and LST-1 protein are restricted to a germline region corresponding roughly to the GSC pool, but ubiquitous LST-1 expression drives formation of a germline tumor (Kershner et al., 2014; Shin et al., 2017; this work). Therefore, spatial distribution of LST-1 must be highly regulated. Here, we report one key aspect of that spatial regulation. Two LST-1 variants, LST-1(1-210) and LST-1(ZnF), both lacking a functional zinc finger, are more abundant and expand more proximally than normal. Moreover, both variants increase the size of the progenitor zone (Fig. 4B) and LST-1(1-210) increases the size of the GSC pool (Fig. 6B,D). Our interpretation is that downregulation of LST-1 protein is essential for proper cell fate determination and the switch between self-renewal and differentiation (Fig. 6E).
The primary mechanism of LST-1 downregulation is likely regulated protein instability. RNAs encoding LST-1(1-210) and LST-1(ZnF) were restricted spatially as in wild type, but the proteins were dramatically expanded (Fig. 5G). Germ cells move proximally at a rate of about 0.5 to 1 cell per hour in the progenitor zone (Rosu and Cohen-Fix, 2017), which provides a useful space-time axis. Wild-type lst-1 RNA and LST-1 protein disappear at about the same position along this axis (Shin et al., 2017; this work), suggesting a tight and precise regulation with RNA and protein turned over about the same time. By contrast, LST-1(1-210) and LST-1(ZnF) proteins persist for 10-20 cells rows further proximally than their RNA, and thus protein turnover is delayed up to 20 h. This change in protein turnover was particularly noticeable with LST-1(1-210). Therefore, loss of the zinc finger is likely to affect LST-1 stability, but loss of the C-terminal third is more dramatic. Earlier studies found that decreased proteasome activity leads to increased germline proliferation (Gupta et al., 2015; Macdonald et al., 2008; Mohammad et al., 2018). However, identification of the crucial E3 ligase or ligases for LST-1 protein turnover remains a challenge for the future.
Regulation of protein stability as a determining factor in the fate switch from stem cell to differentiation is likely a broadly used mechanism (Werner et al., 2017). Although few cases are thoroughly understood, examples exist in flies and human cells in addition to nematodes. In flies, cyclin A protein is downregulated by the Bam-dependent deubiquitinase complex to promote differentiation (Ji et al., 2017); in human embryonic stem cells, Nanog is downregulated by ERK MAP kinase to promote differentiation (Kim et al., 2014). As more examples are uncovered, the regulation of protein stability may emerge as a ubiquitous mechanism for triggering fate switches.
LST-1 and its role in FBF combinatorial control of stem cell regulation
We have found that LST-1 partnership with the FBF RNA-binding protein is pivotal to GSC self-renewal. This remarkable protein therefore provides an important new window into FBF combinatorial control of stem cell regulation. Indeed, two FBF partners, LST-1 and SYGL-1, drive GSC self-renewal (Shin et al., 2017; this work). Each partner is sufficient, and at least one must be present to maintain stem cells (Kershner et al., 2014). Intriguingly, both full-length SYGL-1 protein and the LST-1(1-210) self-renewal fragment consist largely of IDRs and are of comparable size. The SYGL-1 protein possesses one KXXL motif, although its significance has not yet been tested. Nonetheless, we suggest that LST-1 and SYGL-1 are both trans-acting FBF partners that bring IDRs to their respective complexes. Analogous short, trans-acting RNA regulators may be more common than appreciated.
The existence of two IDR-rich FBF partners might simply reflect redundancy but also might have a more interesting role and expand the FBF repertoire for combinatorial control. Their functional redundancy is well established (Kershner et al., 2014), but in favor of individual roles, the LST-1 and SYGL-1 amino acid sequences bear no similarity to each other, and some genetic interactions differ between the two (Brenner and Schedl, 2016; Shin et al., 2017). Moreover, LST-1 localizes to perinuclear granules while SYGL-1 localizes to smaller cytoplasmic puncta, and spatial regulation of LST-1 is tighter than SYGL-1 (Shin et al., 2017). We therefore suggest that additional layers of regulation remain to be discovered. Accordingly, we note that stem cell maintenance must proceed under widely divergent physiological and environmental circumstances. Study of the factors that orchestrate responses to those circumstances, likely including LST-1 and SYGL-1, provide a tantalizing entrée to the complexities of regulation in metazoans.
MATERIALS AND METHODS
Nematode strains and maintenance
C. elegans were maintained at 20°C on Nematode Growth Medium (NGM) plates spotted with E. coli OP50, following established protocols (Brenner, 1974), except that strains containing emb-30(tn377ts) were maintained at 15°C and the strain containing the qSi291 tumor transgene was maintained on lst-1(RNAi) plates (see germline tumor assays section). Wild-type was N2 Bristol strain. See Table S1 for list of strains used in this study. We also used the balancer LGI; LGIII hT2[qIs48] (Siegfried and Kimble, 2002).
CRISPR/Cas9 genome editing to generate lst-1 alleles
See Table S2 for list of CRISPR-induced alleles, and Tables S4 and S5 for additional details about their generation. We used two CRISPR/Cas9 editing methods to create alleles at the endogenous lst-1 locus. Three alleles, lst-1(q867), lst-1(q869) and lst-1(q926), were generated using a DNA-based CRISPR/Cas9 approach with a co-conversion strategy (Arribere et al., 2014; Dickinson et al., 2013). Briefly, the following components were microinjected into wild-type germlines: an lst-1 sgRNA plasmid (25 ng/µl), a repair oligo designed to incorporate the desired lst-1 mutations (500 nM) and a plasmid encoding Cas9 (pDD162, 50 ng/μl) (Dickinson et al., 2013) along with a dpy-10 sgRNA (pJA58, 10 ng/µl) and repair oligo targeting the dpy-10 locus (AF-ZF-827, 500 nM) (Arribere et al., 2014). Progeny of injected hermaphrodites were visually screened for co-injection marker editing and subsequently screened by PCR and Sanger sequencing for editing at the lst-1 locus.
Other alleles, lst-1(q895), lst-1(q1032), lst-1(q1044), lst-1(q1060), lst-1(q1086), lst-1(q1115), lst-1(q1119), lst-1(q1124), lst-1(q1125) and lst-1(q1198), were generated using RNA-protein complex CRISPR/Cas9 editing with a co-conversion strategy (Arribere et al., 2014; Paix et al., 2015). The following were microinjected into wild-type N2 (for q895), JK6154 (for q1125), JK5596 (for q1198) or JK5929 [lst-1(q1004), which we call lst-1(wt)V5 for simplicity] (all other alleles): lst-1 crRNAs (10 µM), dpy-10 or unc-58 co-CRISPR crRNAs (4 µM) and tracrRNA (13.6 µM) (all Alt-R from Integrated DNA Technologies); repair oligos encoding the desired lst-1 mutation (4 µM) and targeting the respective co-CRISPR locus (1.34 µM); and recombinant Cas9 protein (24.5 µM). Progeny of injected hermaphrodites were first visually screened for co-injection marker editing and next screened by PCR and Sanger sequencing for editing at the lst-1 locus. All CRISPR/Cas9-generated alleles were outcrossed with wild type at least twice prior to experimentation.
MosSCI to generate lst-1 transgenes
See Table S3 for list of transgenes generated for this study, and Table S5 for details about plasmids. The Mos1-mediated single-copy insertion (MosSCI) method was used to generate all transgenes (Frøkjær-Jensen et al., 2012, 2008, 2014). Briefly, repair plasmids containing the gene of interest flanked by sequence targeting the ttTi5605 insertion site were cloned using the Gibson assembly method (Gibson et al., 2009). The repair plasmids were microinjected at 50 ng/μl together with Mos1 transposase and co-injection marker plasmids into JK4950. At least three successful insertions were isolated and analyzed in our experiments, and we report one representative line in this work. During strain generation and maintenance, lst-1(qSi291) [Pmex-5::lst-1(1-210)::GGSGG linker::3xFLAG::tbb-2 3′ UTR, unc-119(+)] and related strains were grown on lst-1(RNAi) feeding bacteria to prevent germline tumorigenesis (see RNA interference).
RNA interference
RNA interference (RNAi) was performed by feeding as described previously (Timmons and Fire, 1998). We used sygl-1 or lst-1 clones from the Ahringer RNAi library (Fraser et al., 2000) and L4440 plasmid lacking a gene of interest insertion (‘empty’ RNAi) when an experimental control was required. HT115(DE3) bacteria cultures harboring the RNAi vectors were grown at 37°C in 2xYT media containing 25 μg/μl carbenicillin and 50 μg/μl tetracycline overnight, and were then concentrated and seeded onto NGM plates containing 1 mM IPTG. Bacteria were induced overnight at room temperature before plating worms.
DAPI staining
To visualize nuclear morphology, we stained extruded gonads with DAPI (4′,6-diamidino-2-phenylindole) as described previously (Crittenden et al., 2017), with minor modifications. Briefly, we dissected animals in PBStw [PBS+0.1% (v/v) Tween-20] with 0.25 mM levamisole to extrude gonads, then fixed at room temperature for at least 15 min in ∼2% (w/v) paraformaldehyde diluted in PBStw. Samples were incubated overnight in −20°C methanol, washed with PBStw, then incubated with 0.5 ng/μl DAPI in PBStw to stain DNA. We mounted in either Vectashield (Vector Laboratories) or ProLong Gold (Thermo Fisher Scientific).
GSC maintenance and masculinization assays
For Figs 1E,F, 2, 3D, Figs S1B, S2A, S2E, S4B, mid-L4 hermaphrodites were placed on NGM plates at 20°C. After 3-4 days, their F1 progeny were assayed for embryo production, which requires a functional germline. All fertile animals made many embryos and young larvae and were scored positive for GSC maintenance. Sterile animals were analyzed further with DAPI staining and compound microscopy. Two types of steriles were found: Mog (for Masculinization of Germline) and Glp (Germline proliferation defective). Mog germlines had a roughly normal size, harbored mitotically dividing GSCs in the distal germline, but made only sperm (no oocytes); Mogs were scored positive for GSC maintenance. Glp steriles had a very small germline made of only a few sperm. In Glp animals, we counted sperm number after DAPI staining and divided by four to estimate germ cell number. We removed sygl-1 in some cases by feeding RNAi and in others by crossing into a sygl-1 loss-of-function or null mutant. For RNAi, strains were plated onto sygl-1(RNAi) plates as mid-L4 hermaphrodites at 20°C and their F1 progeny were scored for GSC maintenance as described. In the case of Glp germlines, we quantitated the number of germ cells by DAPI staining, counting the number of mature sperm, and dividing by four (since one germ cell differentiates into four sperm).
Progenitor zone size
Progenitor zone (PZ) size was assessed in nematodes staged to 24 h past mid-L4 at 20°C. Extruded gonads were DAPI stained and imaged with a confocal microscope (see Microscopy). We examined nuclear morphology to determine PZ size, according to convention (Crittenden et al., 2006; Seidel and Kimble, 2015). Briefly, when germ cells exit the PZ and begin meiotic prophase, their nuclear morphology takes on a distinctive crescent shape (see Fig. 4A). We selected a central focal plane in the distal gonad and then counted the number of cells along each edge of the tissue until we reached the distal-most cell with crescent morphology. We counted manually using the FIJI/ImageJ multi-point tool, calling each DAPI-stained nucleus a unique cell row. We averaged the two values from the two edges of the gonad together to determine PZ size.
Immunostaining
We performed immunostaining of extruded gonads as described previously (Crittenden et al., 2017) with minor modifications. All strains (except the strain containing the qSi291 tumor transgene, see Germline Tumor Assays section) were grown at 20°C and staged to 24 h past mid-L4 stage, then dissected in PBStw with 0.25 mM levamisole to extrude gonads. Tissue was fixed in 2.5% (w/v) paraformaldehyde diluted in PBStw for 10 min, then permeabilized with PBStw+0.2% (v/v) Triton-X for 10-15 min. Samples were blocked for at least 1 h and not more than 4 h in 0.5% (w/v) bovine serum albumin diluted in PBStw, except α-FLAG which was blocked in 30% (v/v) goat serum diluted in PBStw. Next, samples were incubated overnight at 4°C with primary antibodies diluted in blocking solution as follows: mouse anti-FLAG (M2, 1:1000, F1804-1MG, Millipore Sigma), rabbit anti-GLD-1 (1:100, a gift from E. Goodwin, University of Wisconsin, Madison, USA), mouse anti-phospho-histone H3 (Ser10) (6G3, 1:200, 9706L, Cell Signaling Technology), mouse anti-V5 (SV5-Pk1, 1:1000, MCA1360, Bio-Rad) and mouse anti-SP56 (1:200, a gift from Susan Strome, University of California, Santa Cruz, CA, USA). Secondary antibodies were diluted in blocking solution and incubated with samples for at least 1 h and not more than 4 h as follows: Alexa 488 donkey anti-mouse (1:1000, A21202, Thermo Fisher Scientific), Alexa 647 goat anti-rabbit (1:1000, A21245, Thermo Fisher Scientific). To visualize DNA, DAPI was included at a final concentration of 0.5-1 ng/μl during a final PBStw wash performed after secondary antibody incubation. Samples were mounted in ProLong Gold (Thermo Fisher Scientific) and allowed to cure overnight before imaging. All steps were performed at room temperature unless otherwise indicated.
smFISH
Single molecule fluorescence in situ hybridization (smFISH) (Raj et al., 2008; Voronina et al., 2012) was performed as described previously (Lee et al., 2016). Custom Stellaris FISH probes were designed using the Stellaris Probe Designer Tool (Biosearch Technologies). The lst-1 probe set contains 40 probes targeting the 5′UTR of lst-1L, the coding sequence for amino acids 1-210 and the 3′UTR. Probes were labeled with CAL Fluor Red 610 and used at a final concentration of 0.25 μM. Probe sequences are provided in Table S6.
emb-30 assay
The assay was performed as previously described (Cinquin et al., 2010; Shin et al., 2017) with minor modifications. Briefly, strains were maintained in a programmable incubator at 15°C until 36 h beyond L4, then transitioned to 25°C for an additional 12 h. Gonads were extruded, fixed and stained using anti-PH3 and -GLD-1 antibodies, and DAPI (see Immunostaining). We imaged gonads by confocal microscopy (see Microscopy). To analyze the images, we used the DAPI channel to determine the ‘M-phase boundary’ between presence and absence of arrested M-phase cells. In cases where a single M-phase cell was found more than three cell rows proximal to all other M-phase cells, that cell was disregarded for determining the boundary. We counted all cells distal to the M-phase boundary, including arrested M-phase cells and cells likely still arrested but not with a typical M-phase morphology, using the multipoint tool in Fiji/ImageJ (Schindelin et al., 2012). Germlines with excessively fragmented distal nuclei were excluded from the counts as cell numbers could not be determined (20-60% per experiment).
Germline tumor assays
To induce ubiquitous expression of LST-1(1-210) using the qSi291 tumor transgene, L4 P0 animals were transferred from lst-1 RNAi bacteria to OP50-seeded NGM plates. Experiments were performed at 15°C to maximize tumor penetrance (Shin et al., 2017). After removal from RNAi, subsequent generations were assayed under a dissection microscope and showed increasing tumor penetrance (n>100 for all): in F1, we observed no animals with germline tumors; in F2, ∼60% had tumors; in F3, ∼90% had tumors. For Fig. 6D, we dissected and stained F3 generation L4-staged animals.
Microscopy
All gonad images were taken using a laser scanning Leica TCS SP8 confocal microscope fitted with Photomultiplier (PMT) and Hybrid (HyD) detectors, and run with LAS software version 3.3.1 or X. A 63×/1.40 CS2 HC Plan Apochromat oil immersion objective was used. All images were taken using the standard scanner with 400-700 Hz scanning speed and 100-300% zoom. To prepare figures, Adobe Photoshop was used to equivalently and linearly adjust contrast among samples to be compared.
Fluorescence quantitation
Immunostaining quantitation in Figs 3K and 4H was performed using Fiji/ImageJ (Schindelin et al., 2012) with images taken under identical conditions across all samples. In Fig. 3K, we performed three independent experiments consisting of at least seven gonads per genotype for a total of at least 21 gonads per genotype. In Fig. 4H, we performed at least three independent experiments consisting of at least five gonads per genotype, with a total of at least 23 gonads per genotype. To collect intensity data from our images, we adapted a workflow from the literature (Brenner and Schedl, 2016). First, the sum of all z-slices for each gonad was projected onto a single plane. A freehand line, 50 pixels wide and at least 80 μm long that bisected the gonad, was drawn manually starting from the distal tip of the tissue. Next, the pixel intensity data for the V5 channel along the line was obtained using the Plot Profile feature. We averaged the raw pixel intensity at every x value to generate an average protein expression plot for each genotype in a given experiment. Next, to adjust for non-specific background staining, we subtracted the average intensity of the respective negative control from the average expression curves at each x value. We then normalized each average expression curve using the maximum and minimum values of the respective average wild-type [lst-1(wtV5)] plot. Finally, to generate the plots shown, the adjusted (background subtracted and normalized) protein expression plots for each genotype were averaged among at least three experiments. Standard error at each x value was calculated among the three independent replicates for each genotype. The number of germ cell diameters (gcd) along the x-axis were calculated using a conversion factor of 4.4 gcd/µm (Lee et al., 2016).
smFISH quantitation in Fig. 5F was performed similarly to the immunostaining quantitation described above, with minor modifications. After z-projection, an average gonad-specific background level was also collected and subtracted from the raw values. This was carried out by using the rectangle tool to create a 2 µm square box that was manually placed on the image where no transcripts could be seen by eye. This was repeated for three separate locations along the gonad: one distally within 50 µm of the distal tip, one centrally between 50-100 µm from the distal tip and one proximally between 100-150 µm from the distal tip. For each location, the measure feature was used to collect the average pixel intensity within the 2×2 µm box. The values obtained for each location were then averaged together to yield the final background value for the individual gonad. This gonad-specific background value was subtracted from the raw values of the respective gonad and we proceeded with quantitation as described above (i.e. plot profile, averaging, background subtraction and normalization). We performed three independent experiments consisting of at least nine gonads per genotype for a total of 30 gonads per genotype analyzed. To compare between datasets that were collected using different zoom factors, we condensed each average RNA expression plot by calculating a rolling average of either four or five x- and y-values. After adjustment, the respective x-values across all data sets were essentially equal and differed by no more than 0.02 µm. For smFISH, the genotype of the negative control was lst-1(q869), which harbors a deletion in the lst-1 locus spanning from 139 bp upstream of the start codon to 228 bp downstream of the stop codon. Of note, five of the 40 smFISH probes used were predicted to anneal in the lst-1(ø) negative control.
Yeast two-hybrid
Modified yeast two-hybrid assays were performed as described previously (Bartel and Fields, 1997). Briefly, LST-1 variants were amplified from cDNA and cloned into the Gal4 activation domain plasmid pACT2 using the Gibson assembly method (Gibson, 2009). We also used plasmid pJK2017, which is FBF-2 cDNA (amino acids 121-632) fused to the LexA-binding domain in the pBTM116 backbone (Shin et al., 2017). Activation and binding domain plasmid pairs were co-transformed into L40-ura3 strain [MATa, ura3-52, leu2-3,112, his3Δ200, trp1Δ1, ade2, LYS2::(LexA-op)4-HIS3, ura3::(LexA-op)8-LacZ] using the LiOAc method (Gietz and Schiestl, 2007). His3 reporter activity was assayed on synthetic defined medium –Leu–Trp–His plates supplemented with varying concentrations of 3-amino-1,2,4-triazole (3-AT) (Millipore Sigma) and compared with –Leu–Trp plates as controls. We measured LacZ reporter activity using the Beta-Glo Assay System following the commercially available protocols and the yeast literature (Promega) (Hook et al., 2005) and luminescence was quantitated using a Biotek Synergy H4 Hybrid plate reader with Gen5 software. A complete list of plasmids used in yeast two-hybrid assays is available in Table S5.
Western blots
For Fig. 1I, samples were prepared by boiling ∼50 unstaged adult worms in sample buffer [60 mM Tris (pH 6.8), 25% glycerol, 2% SDS, 0.1% bromophenol blue with 700 mM β-mercaptoethanol]. For Fig. S3E, we grew yeast transformants in –Leu–Trp liquid media and prepared samples by boiling yeast in sample buffer. Subsequent analysis was conducted on a 12% SDS-PAGE gel and blots were probed with either mouse anti-V5 (SV5-Pk1, 1:1000, MCA1360, Bio-Rad), mouse anti-HA (HA.11, 1:1000, MMS-101P, Covance) or mouse anti-actin (C4, 1:40,000, MAB1501, Millipore Sigma) followed by donkey anti-mouse horseradish peroxidase (1:10,000, 715-035-150, Jackson ImmunoResearch). Immunoblots were developed using SuperSignal West Pico/Femto Sensitivity substrate (Thermo Fisher Scientific) and developed using a Konica Minolta SRX-101A medical film processor. For final figure preparations, contrast of the blot was linearly adjusted in Adobe Photoshop. For Fig. 1I, Fiji/ImageJ was used for quantitation.
Statistics
Where appropriate, statistical analyses are described in figure legends. Homogeneity of variance was established using Levine's test. One-way ANOVA and Tukey's post-hoc tests were performed to calculate statistical significance for multiple samples. A two-tailed t-test assuming equal variance was performed when comparing two samples. All statistical tests were performed in R and the P-value cut-off was 0.05.
Supplementary Material
Acknowledgements
We thank Jonathan Doenier, Michael Green, Sarah Jayawardene, Peggy Kroll-Conner, Kimberley Law, Alex Murphy and Brandon Taylor for help generating strains central to this work, as well as Jadwiga Forster, Kyle Krueger and Charlotte Kanzler for technical assistance. We also thank members of the Kimble and Wickens laboratories for insightful discussions, and Sarah Crittenden and Brian Carrick for comments on our manuscript. We are grateful to Laura Vanderploeg and Anne Helsley-Marchbanks for assistance with figures and manuscript preparation. We thank Susan Strome (University of California, Santa Cruz, CA, USA) for SP56 antibodies. Some strains used in the study were provided by the Caenorhabditis Genetics Center, supported by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.A.H., A.S.F., A.M.K., H.S., J.K.; Methodology: K.A.H., A.M.K., H.S., J.K.; Validation: K.A.H., A.L.E., A.S.F.; Formal analysis: A.S.F., H.S.; Investigation: K.A.H., A.L.E., A.S.F.; Resources: K.A.H., A.L.E., A.S.F., A.M.K., H.S.; Writing - original draft: K.A.H., J.K.; Writing - review & editing: K.A.H., A.L.E., A.S.F., A.M.K., H.S., M.W., J.K.; Visualization: K.A.H., A.L.E., A.S.F.; Supervision: J.K.; Funding acquisition: M.W., J.K.
Funding
This work was supported by the National Institutes of Health (GM050942 to M.W.). J.K. is an Investigator with the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.181644.supplemental
References
- Ahringer J. and Kimble J. (1991). Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature 349, 346-348. 10.1038/349346a0 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S. and Fire A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P. L. and Fields S. (eds) (1997). The Yeast Two-Hybrid System. New York: Oxford University Press. [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J. L. and Schedl T. (2016). Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics 202, 1085-1103. 10.1534/genetics.115.185678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D. W. A. and Jones D. T. (2019). The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402-W407. 10.1093/nar/gkz297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta S. and Richard S. (2015). Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 40, 662-672. 10.1016/j.tibs.2015.08.012 [DOI] [PubMed] [Google Scholar]
- Campbell Z. T., Menichelli E., Friend K., Wu J., Kimble J., Williamson J. R. and Wickens M. (2012). Identification of a conserved interface between PUF and CPEB proteins. J. Biol. Chem. 287, 18854-18862. 10.1074/jbc.M112.352815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. and Moore M. J. (2014). The spliceosome: disorder and dynamics defined. Curr. Opin. Struct. Biol. 24, 141-149. 10.1016/j.sbi.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O., Crittenden S. L., Morgan D. E. and Kimble J. (2010). Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. USA 107, 2048-2053. 10.1073/pnas.0912704107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., Petcherski A. G., Moulder G., Barstead R., Wickens M. and Kimble J. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660-663. 10.1038/nature754 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T. and Kimble J. (2006). Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17, 3051-3061. 10.1091/mbc.e06-03-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Seidel H. S. and Kimble J. (2017). Analysis of the C. elegans germline stem cell pool. Methods Mol. Biol. 1463, 1-33. 10.1007/978-1-4939-4017-2_1 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J. and Goldstein B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J. (2016). Making sense of intrinsically disordered proteins. Biophys. J. 110, 1013-1016. 10.1016/j.bpj.2016.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A. and Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679-690. [DOI] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M. and Ahringer J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325-330. 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S.-P., Grunnet M. and Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M. and Jorgensen E. M. (2012). Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117-118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., LaBella M., Pozniakovsky A., Moerman D. G. and Jorgensen E. M. (2014). Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11, 529-534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G. (2009). Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 37, 6984-6990. 10.1093/nar/gkp687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. III and Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gietz R. D. and Schiestl R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31-34. 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- Gupta P., Leahul L., Wang X., Wang C., Bakos B., Jasper K. and Hansen D. (2015). Proteasome regulation of the chromodomain protein MRG-1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development 142, 291-302. 10.1242/dev.115147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Hara K., Hishiki A., Kawaguchi S., Shichijo N., Nakamura K., Unzai S., Tamaru Y., Shimizu T. and Sato M. (2010). Crystal structure of zinc-finger domain of Nanos and its functional implications. EMBO Rep. 11, 848-853. 10.1038/embor.2010.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook B., Bernstein D., Zhang B. and Wickens M. (2005). RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11, 227-233. 10.1261/rna.7202705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S., Li C., Hu L., Liu K., Mei J., Luo Y., Tao Y., Xia Z., Sun Q. and Chen D. (2017). Bam-dependent deubiquitinase complex can disrupt germ-line stem cell maintenance by targeting cyclin A. Proc. Natl. Acad. Sci. USA 114, 6316-6321. 10.1073/pnas.1619188114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S. and Izaurralde E. (2013). The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27, 2628-2641. 10.1101/gad.227843.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. and Cozzetto D. (2015). DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857-863. 10.1093/bioinformatics/btu744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A., Crittenden S. L., Friend K., Sorensen E. B., Porter D. F. and Kimble J. (2013). Germline stem cells and their regulation in the nematode Caenorhabditis elegans. Adv. Exp. Med. Biol. 786, 29-46. 10.1007/978-94-007-6621-1_3 [DOI] [PubMed] [Google Scholar]
- Kershner A. M., Shin H., Hansen T. J. and Kimble J. (2014). Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 111, 3739-3744. 10.1073/pnas.1401861111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Kim M. O., Cho Y.-Y., Yao K., Kim D. J., Jeong C.-H., Yu D. H., Bae K. B., Cho E. J., Jung S. K. et al. (2014). ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Res. 13, 1-11. 10.1016/j.scr.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Lamont L. B., Crittenden S. L., Bernstein D., Wickens M. and Kimble J. (2004). FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7, 697-707. 10.1016/j.devcel.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Lander A. D., Kimble J., Clevers H., Fuchs E., Montarras D., Buckingham M., Calof A. L., Trumpp A. and Oskarsson T. (2012). What does the concept of the stem cell niche really mean today? BMC Biol. 10, 19 10.1186/1741-7007-10-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Sorensen E. B., Lynch T. R. and Kimble J. (2016). C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. eLife 5, e18370 10.7554/eLife.18370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. and Spradling A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463-2476. [DOI] [PubMed] [Google Scholar]
- Macdonald L. D., Knox A. and Hansen D. (2008). Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180, 905-920. 10.1534/genetics.108.091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menichelli E., Wu J., Campbell Z. T., Wickens M. and Williamson J. R. (2013). Biochemical characterization of the Caenorhabditis elegans FBF•CPB-1 translational regulation complex identifies conserved protein interaction hotspots. J. Mol. Biol. 425, 725-737. 10.1016/j.jmb.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T. and Parker R. (2018). Multiple modes of protein-protein interactions promote RNP granule assembly. J. Mol. Biol. 430, 4636-4649. 10.1016/j.jmb.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Vanden Broek K., Wang C., Daryabeigi A., Jantsch V., Hansen D. and Schedl T. (2018). Initiation of meiotic development is controlled by three post-transcriptional pathways in Caenorhabditis elegans. Genetics 209, 1197-1224. 10.1534/genetics.118.300985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. A. and Lemischka I. R. (2006). Stem cells and their niches. Science 311, 1880-1885. 10.1126/science.1110542 [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D. and Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47-54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. F., Prasad A., Carrick B. H., Kroll-Connor P., Wickens M. and Kimble J. (2019). Toward identifying subnetworks from FBF binding landscapes in Caenorhabditis spermatogenic or oogenic germlines. G3 (Bethesda) 9, 153-165. 10.1534/g3.118.200300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Porter D. F., Kroll-Conner P. L., Mohanty I., Ryan A. R., Crittenden S. L., Wickens M. and Kimble J. (2016). The PUF binding landscape in metazoan germ cells. RNA 22, 1026-1043. 10.1261/rna.055871.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C., Bhat V. D., Rajeev S., Zhang C., Lasley A. E., Wine R. N., Campbell Z. T. and Tanaka Hall T. M. T. (2019). A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity. eLife 8, e48968 10.7554/eLife.48968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A. and Tyagi S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877-879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S. and Cohen-Fix O. (2017). Live-imaging analysis of germ cell proliferation in the C. elegans adult supports a stochastic model for stem cell proliferation. Dev. Biol. 423, 93-100. 10.1016/j.ydbio.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S. and Kimble J. (2015). Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. Elife 4, e10832 10.7554/eLife.10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Haupt K. A., Kershner A. M., Kroll-Conner P., Wickens M. and Kimble J. (2017). SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. PLoS Genet. 13, e1007121 10.1371/journal.pgen.1007121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K. and Kimble J. (2002). POP-1 controls axis formation during early gonadogenesis in C. elegans. Development 129, 443-453. [DOI] [PubMed] [Google Scholar]
- Suh N., Crittenden S. L., Goldstrohm A. C., Hook B., Thompson B., Wickens M. and Kimble J. (2009). FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181, 1249-1260. 10.1534/genetics.108.099440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L. and Fire A. (1998). Specific interference by ingested dsRNA. Nature 395, 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Uversky V. N. (2017). Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18-30. 10.1016/j.sbi.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Voronina E., Paix A. and Seydoux G. (2012). The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139, 3732-3740. 10.1242/dev.083980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M. and Kimble J. (2002). A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312-316. 10.1038/nature01039 [DOI] [PubMed] [Google Scholar]
- Wang Y., Opperman L., Wickens M. and Hall T. M. T. (2009). Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. USA 106, 20186-20191. 10.1073/pnas.0812076106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Roberts T. M., Strome S., Pavalko F. M. and Hogan E. (1986). Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J. Cell Biol. 102, 1778-1786. 10.1083/jcb.102.5.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M. W., Stowell J. A. W. and Passmore L. A. (2019). RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not. eLife 8, e40670 10.7554/eLife.40670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann C. A., Qiu C., Arvola R. M., Lou T.-F., Killingsworth J., Campbell Z. T., Tanaka Hall T. M. and Goldstrohm A. C. (2016). Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. eLife 5, e17096 10.7554/eLife.17096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Manford A. G. and Rape M. (2017). Ubiquitin-dependent regulation of stem cell biology. Trends Cell Biol. 27, 568-579. 10.1016/j.tcb.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D. S., Kimble J. and Parker R. (2002). A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18, 150-157. 10.1016/S0168-9525(01)02616-6 [DOI] [PubMed] [Google Scholar]
- Wu J., Campbell Z. T., Menichelli E., Wickens M. and Williamson J. R. (2013). A protein•protein interaction platform involved in recruitment of GLD-3 to the FBF•fem-3 mRNA complex. J. Mol. Biol. 425, 738-754. 10.1016/j.jmb.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J. and Wickens M. P. (1997). A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390, 477-484. 10.1038/37297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.