Abstract
NLRP (NACHT, LRR and PYD domain-containing proteins) family plays pivotal roles in mammalian reproduction. Mutation of NLRP7 is often associated with human recurrent hydatidiform moles. Few studies regarding the functions of NLRP7 have been performed in other mammalian species rather than humans. In the current study, for the first time, the function of NLRP7 has been explored in ovine ovary. NLRP7 protein was mainly located in ovarian follicles and in in vitro pre-implantation embryos. To identify its origin, 763 bp partial CDS of NLRP7 deriving from sheep cumulus oocyte complexes (COCs) was cloned, it showed a great homology with Homo sapiens. The high levels of mRNA and protein of NLRP7 were steadily expressed in oocytes, parthenogenetic embryos or IVF embryos. NLRP7 knockdown by the combination of siRNA and shRNA jeopardized both the parthenogenetic and IVF embryo development. These results strongly suggest that NLRP7 plays an important role in ovine reproduction. The potential mechanisms of NLRP7 will be fully investigated in the future.
Introduction
NLRs (nucleotide-binding and oligomerization domain (NOD)-like receptors), also called CATERPILLER (caspase recruitment domain, transcription enhancer, purine binding, pyrin, lots of leucine repeats), serve as the intracellular guards to coordinate the innate immunity and inflammatory responses after perception of adverse signals within the cell (Barbé et al. 2014, Meunier & Broz 2017). Recently, NLRs have also emerged as the key regulators of folliculogenesis and early embryonic development in mammals. A subset of phylogenetically related NLRs represents a new category of maternal genes which are highly expressed in oocytes and pre-implantation embryos. Mutations of these genes might lead to hereditary reproductive defects and imprinting diseases (Van Gorp et al. 2014). Four subfamilies of NLRs are classified based on different N-terminal effector domain: NLRA, NLRB, NLRC and NLRP. For example, the NLRP family contains a pyrin domain (PYD) (Ting et al. 2008). Fourteen members of NLRP family are found in Macaca mulatta and Homo sapiens and 20 members are present in Mus musculus (Zhang et al. 2008, McDaniel & Wu 2009). In the process of evolution, NLRP1, 4, 9 have duplicated, while NLRP7, 8, 11 and 13 have been lost. Interestingly, in humans, nine NLRP proteins (NLRP2, 4, 5, 7, 8, 9, 11, 13 and 14) are reproduction related and play the crucial roles in the reproductive system (Tian et al. 2009).
NLRP7, also known as NALP7 (neuronal apoptosis LRR pyrin domain protein 7), NOD12 (nucleotide-binding oligomerization domain protein 12), PYPAF3 (PYRIN-containing APAF1-like protein 3), CLR19.4, PAN7 and HYDM, belongs to the NLRP family and locates into human 19q13.4. It plays multiple roles including immunity and reproduction (Van Gorp et al. 2014). The structure of NLRP7 has a central large NACHT (NAIP, CIITA, HET-E, TP1) domain with a nuclear localization signal, an N-terminal PYD (pyrin) domain involving protein–protein interactions and downstream signal pathways and an LRR (leucine-rich repeats) domain that varies in length depending on splicing isoforms (Slim & Wallace 2013, Reddy et al. 2016). NLRP7 has no ortholog in the rodents, but has a paralog, NLRP2. NLRP7 probably emerged from NLRP2 by gene duplication during evolution (Duéñez-Guzmán & Haig 2014). In the mouse oocytes, NLRP2 knockdown leads to embryonic arrest between two- and eight-cell stages (Peng et al. 2012).
Previously, NLRP7 was described as an inhibitor of the inflammasome signal pathway, its overexpression in HEK-293T cells impaired caspase-1-mediated IL-1β production (Kinoshita et al. 2005). In contrast, recent evidence suggested that NLRP7 induced an inflammasome formation in response to microbial acylated lipopeptides and promoted inflammatory cytokines production (Khare et al. 2012, Radian et al. 2013, 2015, Zhou et al. 2016). In peripheral blood mononuclear cells, NLRP7 localizes to the microtubule-organizing center, the Golgi apparatus and associates with microtubules. This suggests that it may coordinate cytokines secretion and transportation (Messaed et al. 2011). In addition, NLRP7 is referred to as a maternal-effect gene, whose mutations commonly result in recurrent hydatidiform moles (RHMs), a gestational trophoblastic disease characterized by a mass exhibiting trophoblastic hyperplasia and swelling of chorionic villi as well as impaired embryonic development (Murdoch et al. 2006, Sebire et al. 2013, Nguyen et al. 2014, Carey et al. 2015, Ito et al. 2016, Sills et al. 2017, Kalogiannidis et al. 2018). The homozygous or compound heterozygous NLRP7 missense and non-sense mutations in male do not jeopardize their normal reproductive outcomes and this indicates that NLRP7 may specifically regulate female reproduction (Qian et al. 2007, Wang et al. 2009). Thus, excepting for an inflammatory response, NLRP7 may have another important role related to female reproduction, that is, it is present in the oocyte paralleling other maternal-effect genes to regulate female reproductive activities. For example, ovum donation has rescued defects in patients with recessive mutations in NLRP7 (Fisher et al. 2011, Nguyen et al. 2014, Akoury et al. 2015). It is still controversial whether mutations in NLRP7 also contribute to the etiology of other forms of molar pregnancies and reproductive wastage syndromes (Slim et al. 2011, Andreasen et al. 2012, Brown et al. 2013, Manokhina et al. 2013, Slim & Wallace 2013, Mahadevan et al. 2014). As yet, the exact mechanisms of NLRP7 in imprinting defects on abnormal pregnancies are still in debate (Sanchez-Delgado et al. 2015, Singer et al. 2015, Soellner et al. 2017, Reynaud et al. 2018) and are complicated by the variety of disease phenotypes identified.
Majority of the studies on NLRP7 were focused on its effects on human recurrent hydatidiform moles and a few on other animals. Thus, NLRP7 was referred to as a primate-specific NLR (Van Gorp et al. 2014). Until now, there was no report on the expression, localization, and function of NLRP7 in the ovine species. Therefore, the main purpose of this study was to elucidate the potential roles of NLRP7 in the non-primates, that is, sheep.
Materials and methods
Chemicals
Unless otherwise stated, reagents were purchased from Sigma Chemical Co..
Animal studies and ethics statement
All experimental procedures concerning the handling of sheep strictly followed protocols approved by the Animal Welfare Committee of China Agricultural University (Permit Number: SYXK2015002), and this study was carried out in strict accordance with the guidelines and regulations established by this committee.
Sheep tissues collection
The tissues used in this study were taken from slaughterhouse in the Dachang Hui Autonomous County in Langfang City, Hebei Province, China. Heart, liver, spleen, kidney, stomach (rumen), uterus, oviduct, testis (head of epididymis), ovary, placenta (from ewes with 3 months of pregnancy), hypothalamus, pituitary, pineal gland and corpus luteum were randomly sampled from three individual adult (4–6 years old) small-tailed Han sheep (Ovis aries). Tissues were quickly frozen in liquid nitrogen and stored at −80°C until RNA extraction.
RT-PCR and quantitative real-time PCR
Total RNA was isolated from adult ovine tissues using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol and the total RNA concentration was quantified using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was conducted using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara) with Oligo dT Primers (50 µM) and Random 6 mers (50 µM).
The primer sequences used for RT-PCR and quantitative real-time PCR were listed in Table 1. cDNA samples were amplified by RT-PCR with Ex Taq DNA polymerase (Takara). The cycling parameters were an initial denaturation of 2 min, 95°C, followed by 30 cycles of 94°C for 30 s, 60°C for 35 s, 72°C for 40 s and a final extension at 72°C for 10 min. The amplified products were evaluated by a 1.5% agarose gel containing ethidium bromide. After purification, the PCR products were sequenced and sequence identities were confirmed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The mRNA abundance was quantified on the LightCycler 480 II instrument (Roche) using LightCycler® 480 SYBR Green I Master (Roche). The thermal program included a 10-min incubation at 95°C to activate FastStart DNA polymerase, followed by 35 cycles of 95°C for 10 s, 60°C for 15 s and 72°C for the appropriate extension time with single fluorescence acquisition.
Table 1.
Primer sets for relative quantification of NLRP7/β-actin/GAPDH genes.
| Gene | Accession number | Forward primer | Reverse primer | Tm (°C) | Length (bp) |
|---|---|---|---|---|---|
| NLRP7 | XM_004015893.4 | 5′-TGCTTACCGGGACTTCTGTC-3′ | 5′-CCACTGCCAAGTGGTGTCA-3′ | 60 | 181 (spanning the third intron) |
| β-actin | NM_001009784.3 | 5′-CTCTTCCAGCCTTCCTTCCT-3′ | 5′-GGGCAGTGATCTCTTTCTGC-3′ | 58 | 178 (spanning the fourth intron) |
| GAPDH | XM_027961471.1 | 5′-CTGGCCAAGGTCATCCAT-3′ | 5′-ACAGTCTTCTGGGTGGCAGT-3′ | 60 | 86 (spanning the seventh intron) |
Comparisons among several tissues should be performed with at least two endogenous references (MiQE guidelines: http://www.rdml.org/miqe.php). The relative expression was normalized by the corresponding geometric average of the two housekeeping genes (reference genes): β-actin and GAPDH. All samples were performed on three independent occasions. Quantification of gene expression was calculated by Microsoft Excel using the standards as described above or according to the 2^−△△CT method (Schmittgen & Livak 2008).
Quantification of NLRP7 mRNA in oocytes and pre-implantation embryos
The oocytes and embryo samples used in the experiment included germinal vesicle (GV), 8 h, 16 h and metaphase II (MII) stage oocytes and parthenogenetic or in vitro-fertilized 2-cell, 4-cell, 8-cell, 16-cell, morula and blastocyst-stage embryos (n = 4 pools of 20 embryos). Procedures used for RNA isolation, cDNA synthesis, and quantitative real-time PCR analysis of mRNA abundance during in vitro oocyte maturation and pre-implantation embryonic development were conducted as described previously (Bettegowda et al. 2006, Tejomurtula et al. 2009
Western blotting
Western blot was performed as described previously (Pisani et al. 2010). In brief, proteins were extracted from ovine tissues following homogenization in RIPA lysis buffer and the tissue lysates were centrifuged at 12,000 × g for 15 min at 4°C. Protein concentration was measured using a bicinchoninic acid protein assay kit (Pierce). Equal amounts of protein were loaded per lane in SDS-PAGE and transferred onto a PVDF membrane (Immobilon; Millipore). The membrane was incubated with a blocking solution containing 5% skim milk in TTBS (10 mM Tris–HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween 20), followed by incubation with the rabbit anti-NLRP7 antibody (N3C2, GeneTex, USA) at 1:1000 overnight at 4°C. After several rinses, membrane was incubated with goat-anti-rabbit IgG antibody (1:3000 dilution) labeled with horseradish peroxidase in blocking solution for 40 min at 37°C. Finally, the proteins were visualized with an enhanced chemiluminescence detection system according to the manufacturer’s instructions (GE Healthcare). α-actin (Bioss, 1:3000, Beijing, China) was used as a loading control.
Immunohistochemistry
Ovaries were fixed in 4% paraformaldehyde overnight, dehydrated through gradient alcohol solutions and embedded in paraffin wax. Seven-micrometer sections were deparaffinized in xylene, rehydrated through gradient alcohol solutions and heated in 10 mM sodium citrate buffer (pH 6.0) with microwaves (10 min at high power) and cooled to room temperature for antigen retrieval. Immunostaining was performed using PV-9001IHC kits (ZSGB-Bio, China). In brief, sections were incubated with 1% BSA for 30 min at room temperature to block nonspecific binding, followed by incubation with the NLRP7 polyclonal antibody (GeneTex, N3C2, USA) (1:50 dilution of rabbit anti-NLRP7 antibody) in blocking solution for overnight at 4°C and with goat anti-rabbit secondary antibody (1:1500 dilution) for 30 min at room temperature. After several washes, the diaminobenzidine substrate solution was applied to the sections. The resultant sections were counterstained lightly with hematoxylin. Negative controls were performed via processing sections as above without the primary antibody. Previously published criteria were used to classify ovine follicles into specific developmental stages including the number of layers and appearance of granulosa cells (Fortune et al. 2003).
Immunofluorescence
All steps were performed at room temperature unless mentioned. The collected oocytes and embryos were fixed with 4% paraformaldehyde for at least 30 min, permeated for 20 min with 0.5% Triton X-100 in PBS, and all samples were incubated in the blocking solution (2% BSA, 2% skimmed milk powder, 0.15 mmol/L glycine and 0.05% Tween-20 in PBS) for 1 h, and then they were incubated with anti-NLRP7 (GeneTex, N3C2) antibody with a dilution of 1:200 for overnight at 4°C. After three times of washing with PBS containing 0.1% Tween-20 and 0.01% Triton X-100 for 5 min, the samples were incubated with FITC conjugated goat anti-rabbit second antibody (Abcam, dilution 1:500) for 2 h at 37°C. After three times of washing with PBS containing 0.1% Tween-20 and 0.01% Triton X-100 for 5 min, the DNA was stained with DAPI (C1005, Beyotime Institute of Biotechnology). Finally, the oocytes or embryos were mounted on glass slides and were observed with a fluorescent microscope.
Partial CDS cloning and sequence analysis
A local Chinese breed, namely small-tailed Han sheep (genesis and species) was used in the present study. Ovaries were collected and total RNA was extracted from COCs using an RNA isolation kit (Invitrogen) according to the manufacturer’s instruction. Approximately 5 μg of total RNA were used as template for the synthesis of the first strand of cDNA by M-MLV reverse transcriptase (Promega). Based on the conserved regions of Homo sapiens (NM_001127255.1), Bos taurus (XM_002695413.4), Capra hircus (XM_018063100.1) and Ovis aries (XM_004015893.3) NLRP7 mRNA sequences, a pair of primers was designed as the following: 5′- TAGTGCGATTGGGGTCTTG-3′ (forward) and 5′- CGCTATCTGGGATTGTTCTC-3′ (reverse). PCR was performed using 25 μL of PrimeSTAR GXL DNA polymerase in a final volume of 50 μL, containing 4 μL of dNTP mixture (2.5 mM each), 2 μL of primers (20 pmol/L), 2 μL of cDNA template (50 ng) and 5 μL of sterile water in 10 μL of 5× PrimeSTAR GXL Buffer, as instructed by the manufacturer (Takara). Amplification was performed in a DNA Thermal cycler (Mini Cycler Elite Thermal Cycler, MJ Research, Waltham, USA) under the following procedures: 35 cycles of denaturation at 98°C for 5 min, annealing at 60°C for 15 s, and extension at 68°Cfor 20 s. The final extension reaction was 5 min at 68°C. The PCR products were analyzed by electrophoresis on a 1% agarose gel, followed by purification and recovery using an agarose gel DNA purification and recovery kit (TaKaRa). The recovered fragments were cloned into the pMD18T vector (TaKaRa) and finally transformed into competent cells of Escherichia coli strain DH5α. White colonies were checked and the positive colonies were then sequenced (Sangon Biological Engineering and Technology and Service, Shanghai, China). Nucleotide sequences were analyzed using DNAMAN software. Multiple alignments of the NLRP7 proteins were performed using BLAST (www.ncbi.nlm.nih.gov/BLAST) and ClustalX (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) program. Phylogenetic analyses were conducted using neighbor-joining (NJ) methods using MEGA 7 (Kumar et al. 2016).
Oocyte collection and in vitro maturation (IVM)
Ovine ovaries were collected from a commercial abattoir and transported to the laboratory in a thermos flask containing sterile saline solution supplemented with penicillin (100 IU/mL) and streptomycin (100 IU/mL) at 26–29°C within 2.5 h after slaughter. After three washes in fresh saline, COCs were recovered from the ovary with a blade in a culture dish of 90 mm diameter. COCs with a homogenous, evenly granulated ooplasm surrounded by at least three layers of compact cumulus cells were collected into Hepes-buffered tissue culture medium-199 (HTCM-199) supplemented with 0.1% polyvinyl alcohol (PVA), 0.04% of sodium bicarbonate, 25 IU/mL heparin, 0.065 g/L penicillin as well as 0.05 g/L streptomycin under a stereomicroscope for experiments. The tissue culture medium-199 (TCM-199) supplemented with sodium pyruvate (2.5 mM), l-glutamine (1.0 mM), penicillin (100 IU/mL), streptomycin (100 IU/mL), 10% fetal bovine serum (v/v, Gibco) and l-cysteine (0.1 mM) was used as the basal culture medium. Maturation medium was modified from basal culture medium by adding 100 ng/mL epidermal growth factor (EGF), 10 µg/mL follicle-stimulating hormone (FSH), 10 µg/mL luteinizing hormone (LH), and 1 µg/mL estradiol-17β. COCs were incubated for 24 h at 38.5°C in a humidified atmosphere under 5% CO2 in groups of 100 containing 800 µL maturation medium in four-well Petri dishes (Nunclon; Nalge Nunc, Roskilde, Denmark) covered with mineral oil.
Parthenogenetic activation (PA) of oocytes
COCs were denuded with 0.1% hyaluronidase by pipetting and washed in maturation medium after 24 h of IVM. Parthenogenetic activation (PA) of ovine oocytes was performed as described previously (Quan et al. 2017), with some modification. Briefly, oocytes with a polar body were exposed for 5 min in 5 μM ionomycin in TCM 199 supplemented with 10% FBS followed by 4 h in 2 mM 6-dimethylaminopurine (6-DMAP) in TCM 199 supplemented with 10% FBS. The presumptive zygotes were then washed in modified synthetic oviduct fluid (mSOF) and transferred separately into pre-equilibrated 50 µL IVC droplets (10–15 zygotes/droplet) containing mSOF without serum and glucose for 2 days. Then, they were further treated with 10% fetal serum (10%) and glucose (1.5 mM) for another 5 days (sequential mSOF).
In vitro fertilization (IVF)
IVM oocytes were fertilized in the synthetic oviductal fluid (SOF) medium with 2% ESS (estrous sheep serum) for 23 h at 38.5°C in an atmosphere of 5% CO2, 5% O2 and 90% N2 in four-well Petri dishes (Nunclon). Frozen-thawed sperm was selected by the swim-up method and was pelleted by centrifugation at 650 g for 10 min. The concentration of sperm in the resulting pellet was adjusted to 1×106 spermatozoa/mL. Thereafter, the presumptive zygotes were transferred in SOFaa medium with 0.3% bovine serum albumin (BSA) and cultured for 8 days (Bebbere et al. 2008). The first cleavage was observed between 24 and 26 h after the start of fertilization.
RNA interference (RNAi) experiments
Three candidate shRNAs, NLRP7-2504shRNA, NLRP7-3122shRNA and NLRP7-3728shRNA, specially targeting ovine NLRP7 gene (gene ID: 101102259) as well as a scrambled shRNA were designed by BLOCK-iT™ RNAi Designer (http://rnaidesigner.lifetechnologies.com/rnaiexpress). The sequences for them were listed in Table 2. These sequences were synthesized by Shanghai Sangon Biological Engineering Technology & Service Co. Ltd. (China) and were cloned into pZDonor expressing vector (Syngentech, China), which containing U6 promoter, shRNA, and termination sequences. The publicly available siRNA design algorithm (siRNA target finder, Ambion) was used to design siRNA species targeting the ORF of NLRP7 mRNA in the Ovis aries. The candidate siRNA and shRNA species were interrogated by using the BLAST (Basic Local Alignment Search Tool) program to rule out homology to any other known genes (especially NLRP2) in the ovine EST and genomic database. Three distinct NLRP7 siRNA species and one non-sense siRNA were synthesized by Shanghai GenePharma Co. Ltd. (China) and dissolved in nuclease free water. The sequences for them were listed in Table 3.
Table 2.
NLRP7 shRNA sequence.
| shRNAs | Targets | shRNA sequence | TargetGene |
|---|---|---|---|
| pHS-ASR-LJ031 | NLRP-7 Ovis aries 2504 | ACCGCAGGTAGGAAAGAAGATATTCGAAAATATCTTCTTTCCTAC | NLRP-7 |
| pHS-ASR-LJ032 | NLRP-7 Ovis aries 3122 | ACCGCTGTCTTCTGCTTTGATTGTCGAAACAATCAAAGCAGAAG | NLRP-7 |
| pHS-ASR-LJ033 | NLRP-7 Ovis aries 3728 | ACCGCACTGAGATATTTCTCTTACCGAAGTAAGAGAAATATCTCA | NLRP-7 |
| pHS-ASR-LJ006 | Control | ACCGTAATTGTCAAATCAGAGTGCTTCAAGAGAAAGCACTCTGAT | Control |
Table 3.
NLRP7 siRNA sequence.
| Targets | Sense (5′-3′) | Sense (5′-3′) |
|---|---|---|
| LOC101102259-Ovis-1464 | GGAAGUUUACUGAAGAGAATT | UUCUCUUCAGUAAACUUCCTT |
| LOC101102259-Ovis-2614 | GCUCUGUGCUCAGUUCAAATT | UUUGAACUGAGCACAGAGCTT |
| LOC101102259-Ovis-3602 | GGACAAAUCUACCGUGGAATT | UUCCACGGUAGAUUUGUCCTT |
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Injection pipettes were made from sterile borosilicate glass with filament using a PN-30 micropipette puller (Narishige, Co., Tokyo, Japan), and angled at MF-900 with a microforge (Narishige, Co., Tokyo, Japan). Oocyte microinjections were performed on an Olympus IX71 inverted microscope (Olympus Co.) equipped with Transferman NK2 micromanipulators and CellTram Air/Oil pressure control systems (Eppendorf Co., Hamburg, Germany). The microinjection of sheep oocytes (n = 10–30 per treatment) with siRNAs and shRNA-plasmids was based on former studies (Paradis et al. 2005), with some modifications. Metaphase II stage oocytes were pretreated for 15 min in HB-TCM (Hepes buffered) supplemented with 7.5 mg/mL cytochalasin B before microinjection in droplets of prewarmed cycloheximide-supplemented HB-TCM under mineral oil. Three of the NLRP7 siRNA duplexes as well as shRNAs plasmids were combined at a final concentration of 20 mM and 1 µg/µL, respectively, which was denoted as NLRP7-siRNA+shRNA group. Control groups consisted of oocytes injected with scrambled siRNA (nonspecific) mixed with nonsense shRNA plasmid (Nc-siRNA+shRNA group) and non-injected oocytes (non-injected group). Approximately 10 pl of mixture were injected into the cytoplasm of each oocyte using a microinjection pipette and a pneumatic PicoPump (World Precision Instruments, Stevenage, UK). After injection, the oocytes were maintained in HB-TCM medium at 38.5°C with a humidified atmosphere of 5% CO2 for 30 min followed by parthenogenetic activation or in vitro fertilization. Experiment was repeated four times.
After microinjection, parthenogenetic or IVF embryos at the 2-, 8-cell stage (n = 4 pools of 20 embryos per treatment) from NLRP7-siRNA + shRNA-injected group and control groups were collected to perform quantitative real-time RT-PCR for identification of NLRP7 mRNA knockdown efficiency. The development of the uninjected or injected embryos (with NLRP7 siRNA + shRNA or negative control siRNA + shRNA) was evaluated by recording the population of embryos 7 days after PA or IVF.
Statistical analysis
All statistical analyses were performed using the IBM SPSS 22.0 software package (IBM corp. NY, USA, 2013). One-way ANOVA was used followed by a multiple pairwise comparison (Duncan’s test) to determine significant differences between the comparative groups. The results are expressed as the means ± s.e.m., and P values <0.05 were considered statistically significant. In addition, t-test was also applied to pairwise comparisons.
Results
Tissue distribution and intraovarian localization of ovine NLRP7
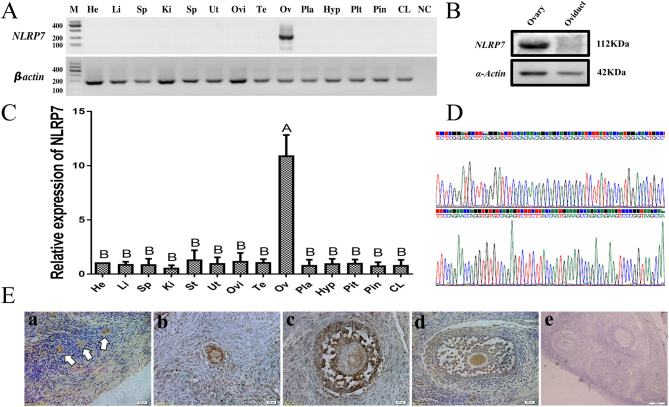
To determine the tissue distribution of NLRP7, NLRP7 mRNA expression in 14 different sheep tissues was analyzed using reverse transcription polymerase chain reaction (RT-PCR) with NLRP7-specific primers. As shown in Fig. 1A, NLRP7 transcripts were strongly expressed in sheep ovary, with no detectable expression in the other tissues (including the heart, liver, spleen, kidney, stomach, uterus, oviduct, testis, placenta, hypothalamus, pituitary, pineal gland and corpus luteum). Similarly, quantitative real-time PCR analysis with two reference genes (β-actin and GAPDH) demonstrated that NLRP7 mRNA was also mainly expressed in the ovary, with a neglected expression in other tissues (P < 0.01)(Fig. 1C). Western blot of NLRP7 protein in ovary and randomly picked control tissue (oviduct) shows the same results as RT-PCR (Fig. 1B). To validate the specificity of NLRP7 primer, RT-PCR products were sequenced (Fig. 1D) and the sequences correspond to the NCBI database XM_004015893 as expected.
Figure 1.
Tissue distribution and intraovarian localization of ovine NLRP7.(A) RT-PCR analysis of NLRP7 mRNA transcripts in multiple ovine tissues. Samples of heart (He), liver (Li), spleen (Sp), kidney (Ki), stomach (St), uterus (Ut), oviduct (Ovi), testis (Te), ovary (Ov), placenta (Pla), hypothalamus (Hyp), pituitary (Pit), pineal gland (Pin) and corpus luteum (CL) were subjected to RT-PCR. M, Marker; NC, no PCR substrate as negative control. β-actin was used as a loading control. (B) Western blot of extracts obtained from sheep ovary and a control tissue (oviduct, randomly selected). Molecular masses (kDa) are indicated on the right. α-actin was used as an internal control.(C) Distribution of NLRP7 mRNA in different ovine tissues demonstrated by quantitative real-time RT-PCR using β-actin and GAPDH as the reference genes; Experiments were repeated three times (n = 3) independently. Results have been normalized to the abundance in the heart (calibrator sample, randomly selected) and expressed as the mean ± s.e.m. Bars with different capital superscripts are significantly different (P < 0.01). (D) Sequencing results of NLRP7 RT-PCR products.(E) Immunohistochemical localization of NLRP7 protein in the ovary sections. primordial follicles (a), primary follicle (b) and antral follicles (c, d,), e, negative control. Arrows are (a) indicators of primordial follicle. Scale bar = 200 µm.
To further validate the expression of NLRP7 in the ovary, immunohistochemistry staining was performed. Immunohistochemical localization of NLRP7 within adult ovary sections revealed that the protein was present in follicles regardless of their developmental stages (including primordial, primary and antral follicles) or the inner cell types (including oocytes, granulosa and theca cells) (Fig. 1E). It seems that the positive signal of NLRP7 in oocyte is weaker than that in granulosa/theca cells before the early antral stage (Fig. 1B, C and E). As the cavitation increases, the signal of NLRP7 in the oocytes becomes comparable to granulosa/theca cells (Fig. 1E-d). The ovarian stromal cells also showed a weak positive signal compared to the strong expression in follicles. Immunoreactivity was not detected in control tissue sections incubated in the absence of NLRP7 antibody.
Expression pattern of NLRP7 protein in ovine COC, oocyte and parthenogenetic embryos
Immunofluorescence staining confirmed that NLRP7 proteins were expressed in ovine COC, denuded oocyte (Fig. 2A) as well as in parthenogenetic embryos (two-cell, four-cell, eight-cell, morula and blastocyst) (Fig. 2B). In detail, NLRP7 was predominant at the outer cortical region in growing oocytes. After cleavage, NLRP7 distributed to the cytoplasm and was excluded from the cell-to-cell contact region until the morula stage. Furthermore, a clear nuclear signal is only evident starting from the blastocyst stage.
Figure 2.
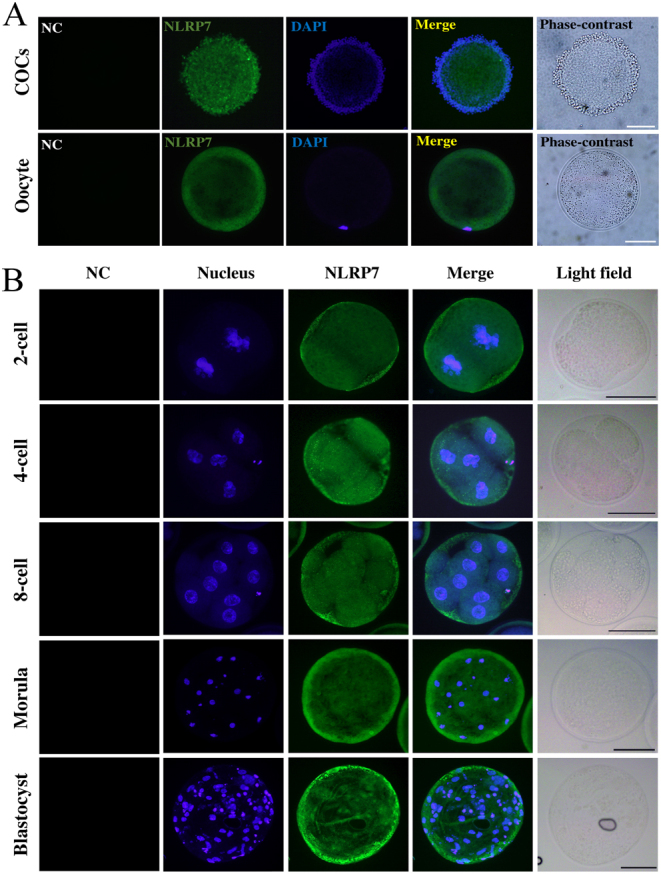
Expression pattern of NLRP7 protein in ovine COC, oocyte and parthenogenetic embryos. (A) Immunofluorescence with NLRP7 antibody (N3C2, Gene Tex) in COCs (cumulus oocyte complexes) and denuded oocyte.(B) Immunofluorescence of NLRP7 protein in parthenogenetic embryos. Scale bar = 50 µm. Omission of NLRP7 primary antibody shows no signal as negative control (NC). Each sample was counterstained with DAPI to visualize nucleus (blue).
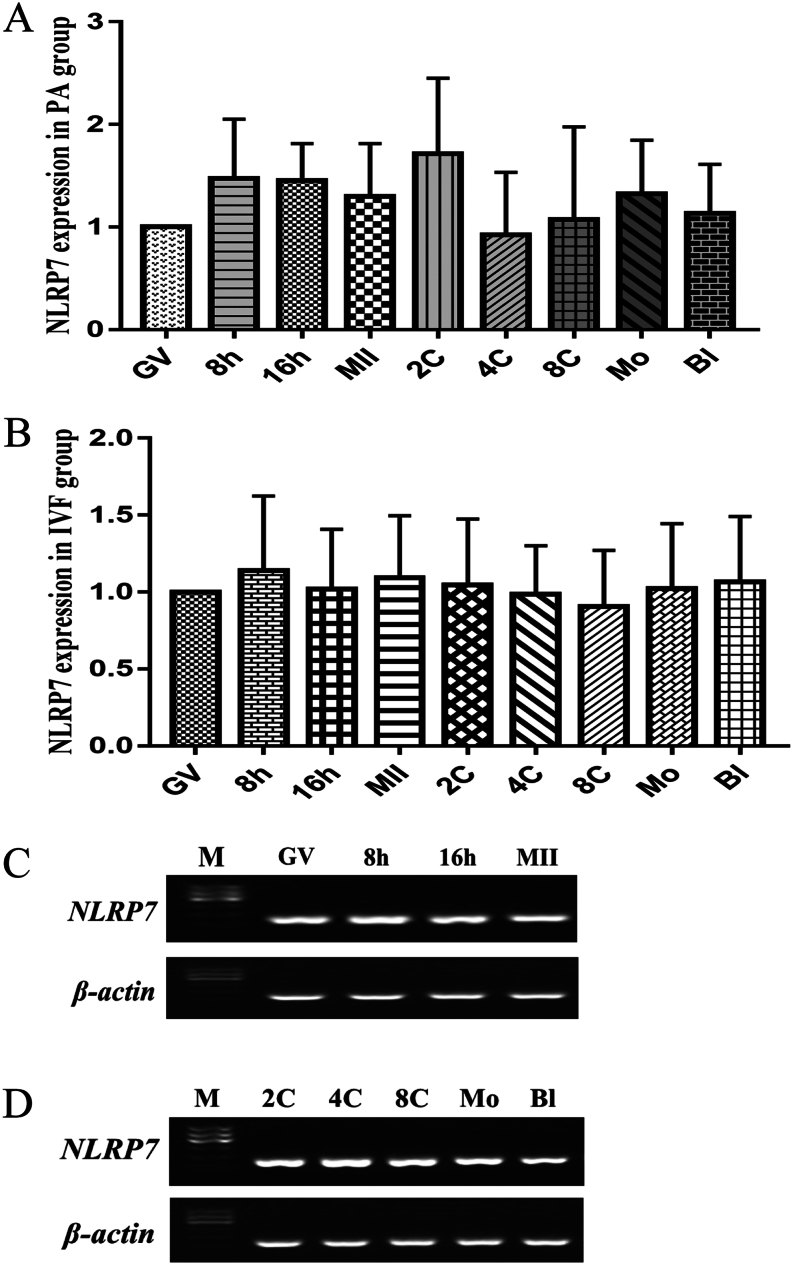
Expression pattern of NLRP7 mRNA in IVM oocytes and PA/IVF embryos
For a better understanding of NLRP7 mRNA abundance in oocyte maturation and in vitro pre-implantation embryos, its expression in in vitro maturation (IVM), parthenogenetic activation (PA) and in vitro fertilization (IVF) was evaluated by either real-time RT-PCR or RT-PCR. As shown in Fig. 3A, B, and C, the transcripts of NLRP7 were abundant at GV (germinal vesicle), 8 h, 16 h and MII (Metaphase II) during oocyte maturation. Moreover, NLRP7 mRNA also steadily expressed from two-cell to the blastocyst stage not only in PA but also in IVF embryos (Fig. 3A, B, and D). There were no significant differences at the stages between oocyte maturation and in vitro pre-implantation embryos development regarding the abundance of NLRP7 mRNA expression (P > 0.05).
Figure 3.
Expression pattern of NLRP7 mRNA in IVM oocytes and PA/IVF embryos.(A) Relative abundance of NLRP7 transcripts in samples collected from GV (germinal vesicle), 8 h, 16 h, MII (Metaphase II) stage oocytes and PA (parthenogenetic activation) 2-cell (2C), 4-cell (4C), 8-cell (8C), morula (Mo) and blastocyst (Bl) stage embryos (n = 4 pools of 20 each). (B) Relative abundance of NLRP7 transcripts in samples collected from GV (germinal vesicle), 8 h, 16 h, MII (Metaphase II) stage oocytes and IVF 2-cell (2C), 4-cell (4C), 8-cell (8C), morula (Mo) and blastocyst (Bl) stage embryos (n = 4 pools of 20 each). (C) RT-PCR of NLRP7 in IVM oocytes.(D) RT-PCR of NLRP7 in PA embryos. M, Marker. β-actin was used as an internal control.
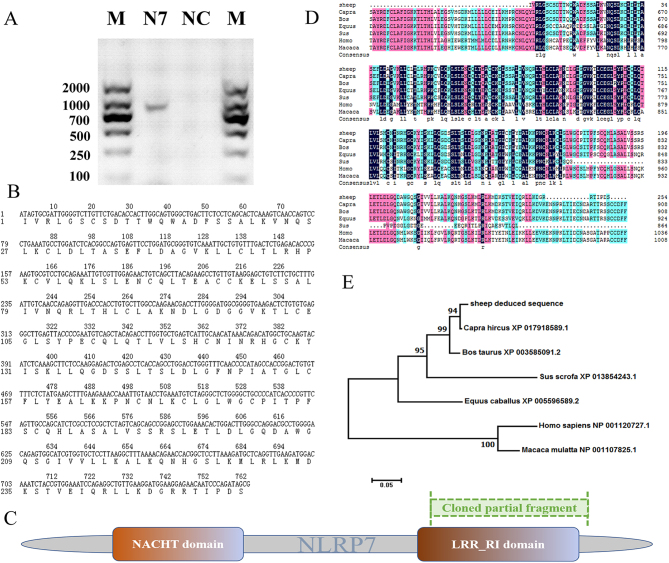
Cloning and characterization of ovine NLRP7 partial CDS
We have tried hard to clone the complete CDS of NLRP7 (XM_004015893.3) gene by using different primers and programs, but failed to get the aiming amplicon. However, the partial coding sequence (CDS) was subsequently cloned using reverse transcript cDNA derived from ovine COCs. A 763-bp length sequence of sheep NLRP7 was obtained (Fig. 4A), which containing 254 putative amino acid residues (Fig. 4B) and was submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/; accession numbers: MF197687). The NLRP7 protein consists of two major domains: NACHT and LRR_RI. Figure 4C shows the scheme of the cloned fragment (marked in green) relative to the entire length of NLRP7 protein (marked in gray). The sequence obtained in sheep was analyzed with Blast and ClustalX program, which confirmed homology with the orthologous gene of public databases (Table 4 and Fig. 4D). The deduced protein sequence of NLRP7 shared 98% identity with Ovis aries (XP_004015942.2), 97% with Capra hircus (XP_017918589.1), 94% with Bos taurus (XP_002695459.2), 56% with Homo sapiens (NP_001120727.1), 57% with Macaca mulatta (NP_001107825.1), 82% with Sus scrofa (XP_020950638.1) and 76% with Equus caballus (XP_005596589.2) sequences. To determine the phylogenetic relationship of NLRP7 between sheep and other known or predicted sequences from other mammals, a phylogenetic tree was constructed using the deduced ovine NLRP7 protein sequence with MEGA 7 program. The analysis revealed that the highest homology was between sheep and goat and the lowest was between sheep and primates (Fig. 4E). In general, the amino acid sequence of NLRP7 protein is relatively well conserved among the mammals during evolution.
Figure 4.
Cloning and characterization of ovine NLRP7 partial CDS.(A) Reverse transcription-polymerase chain reaction (RT-PCR) amplification of the ovine NLRP7 partial CDS. Lane N7, NLRP7 amplicon; NC, negative control; M, 2000 bp DNA marker. The amplified ovine CDS is 763 bp.(B) Nucleotide and amino acid sequences of cloned NLRP7 partial fragment of sheep. The deduced amino acid sequence (254 residues) is under the nucleotide sequence and numbered on the left. These sequence data have been submitted to the GenBank under an accession number MF197687.(C) Scheme of the cloned fragment (marked in green) compared to the full length of NLRP7 (marked in gray). NACHT and LRR_RI are two major domains of NLRP7. (D) Multiple alignments of NLRP7 amino acid sequence in different species including Capra hircus, Bos Taurus, Homo sapiens, Macaca mulatta, Sus scrofa, Equus caballus. The multiple alignments were produced using ClustalX, in which black color indicates positions that have a single, fully conserved residue (100% similarity) as opposed to pink (>75%), aqua (>50%), respectively. Dashes indicate gaps. (E) Phylogenetic tree of mammalian NLRP7 homologs was conducted in MEGA7 using the Neighbor-Joining method, with the sum of branch length = 0.86954469. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches.
Table 4.
Multi-alignment of NLRP7 with putative protein between different species.
| Organism | Protein | Max score | Total score | Query cover | E value | Ident |
|---|---|---|---|---|---|---|
| Ovis aries | XP_004015942.2 | 494 | 764 | 98% | 1e-174 | 98% |
| Capra hircus | XP_017918589.1 | 491 | 730 | 98% | 9e-174 | 97% |
| Bos taurus | XP_002695459.2 | 473 | 624 | 98% | 1e-166 | 94% |
| Homo sapiens | NP_001120727.1 | 269 | 608 | 99% | 1e-87 | 56% |
| Macaca mulatta | NP_001107825.1 | 275 | 549 | 99% | 4e-90 | 57% |
| Sus scrofa | XP_020950638.1 | 403 | 601 | 99% | 1e-139 | 82% |
| Equus caballus | XP_005596589.2 | 372 | 480 | 98% | 2e-127 | 76% |
Effect of NLRP7 knockdown on in vitro pre-implantation embryo development
Considering the continuous expression of NLRP7 in oocytes and embryos, we hypothesized that NLRP7 is required for normal embryo development in ovine. To investigate the function of NLRP7 in pre-implantation embryo development in vitro, RNAi to reduce NLRP7 expression in PA and IVF embryos was carried out. Three NLRP7 siRNA and shRNA species targeting different regions of the NLRP7 gene were mixed into a cocktail to achieve potent and stable gene knockdown. Metaphase II oocytes were subjected to microinjection followed by PA or IVF. Microinjection of a cocktail of NLRP7-siRNA + shRNA reduced NLRP7 mRNA levels by 51.6% (PA embryo)/54.9% (IVF embryo) at two-cell stage (P < 0.01) and 68.1% (PA embryo)/74.8% (IVF embryo) at eight-cell stage compared to non-injected control (P < 0.01)(Fig. 5A and B). To identify whether knockdown of NLRP7 in ovine embryos has any effect on embryo development, PA/IVF embryos population on day 7 for NLRP7-siRNA+shRNA-injected vs non-injected and NC-siRNA + shRNA-injected group were determined. It turned out that the NLRP7-siRNA + shRNA injection jeopardized the normal development both in PA and IVF embryos (Fig. 5C, D, and E) compared to control group. These results clearly indicate that NLRP7 is associated with ovine normal pre-implantation embryo development in vitro.
Figure 5.
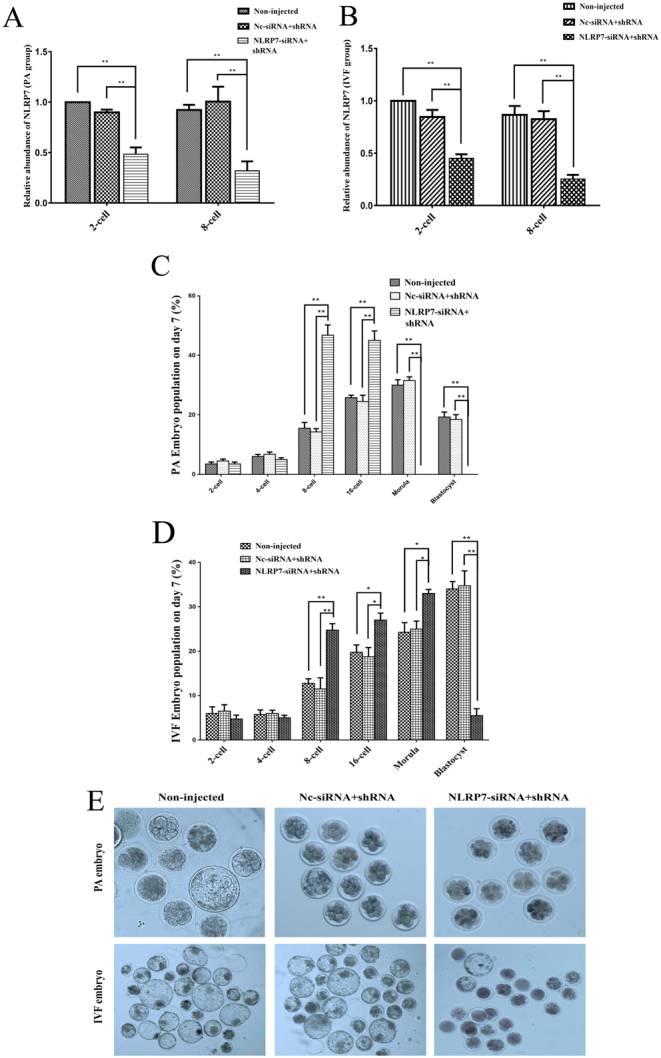
Effect of NLRP7 knockdown on in vitro pre-implantation embryo development. Metaphase II oocytes were subjected to Non-injected control, NC-siRNA+shRNA injection or NLRP7-siRNA+shRNA injection (n = 10–30 embryos per treatment) following by PA or IVF. Microinjected embryos were cultured in vitro for 7 days. (A and B) Validation of NLRP7 knockdown efficacy by quantitative real-time PCR in samples of 2-cell, 8-cell embryos in PA and IVF group respectively. Results were normalized to the 2-cell stage Non-injected control group and expressed as the mean ± s.e.m. Experiments were repeated four times (n = 4), **P < 0.01. (C and D) Percentage (mean ± s.e.m.) of PA and IVF embryos in three groups at different stages after culture for 7 days. Results were normalized to the 2-cell stage non-injected control group, **P < 0.01, *P < 0.05. (E) Morphology of PA and IVF embryos after 7 days of culture. The original magnification was 100×.
Discussion
Previous studies reported that NLRP7 was expressed in human immune cells lines of B, T, and monocytic cells (Messaed et al. 2011), as well as in variety of human tissues, including lung, spleen, thymus, and reproductive organs (testis and ovaries) (Kinoshita et al. 2005, Duéñez-Guzmán & Haig 2014). Furthermore, NLRP7 expression was also detected in the human testicular seminomas cells and endometrial cancer tissues (Okada et al. 2004, Ohno et al. 2008). Current study indicated that NLRP7 is mainly expressed in sheep ovaries and its expression pattern differed from that in human (Kinoshita et al. 2005). The expression pattern of NLRP7 in sheep was similar to its paralog of Nlrp2 in the mouse which was only detected in ovaries, but not in other tested tissues, even the testes (Peng et al. 2012). NLRP7 expression was not restricted in the oocytes at various follicular stages but also in granulosa cells and theca cells. This expression pattern is also consistent with the observation on Nlrp2 in mouse ovaries (Peng et al. 2012). Interestingly, immunohistochemistry staining showed that NLRP7 signal in oocyte was weaker than that in granulosa/theca cells before the early antral stage. This indicated that it might play some roles also in the autocrine/paracrine systems. The expression of NLRP7 in ovine ovary indicates that its potential roles in the ovary function. Immunofluorescence analysis demonstrated a cytoplasm localization of NLRP7 protein close to the subcortex of oocytes, implying that this protein might be involved in cell cytoskeleton maintenance or signal transduction. Intracellular localization of NLRP7 protein in sheep oocytes was similar to the Nlrp2 protein in the mouse (Peng et al. 2012), as well as NLRP7 in the human (Akoury et al. 2015). In fact, among the human NLRP families, in addition to NLRP5, both NLRP2 and NLRP7 were considered as the subcortical maternal complex (SCMC) members (Mahadevan et al. 2017). Our study reveals that in pre-morula embryos, NLRP7 distributed into the cytoplasm and little signal was detected in the cell-to-cell contact region, which was very similar to the human NLRP7. In humans, at blastocyst stage, NLRP7 translocates from the nucleus to cytoplasm (Akoury et al. 2015). However, this appears not the case in sheep since there was a clear nuclear signal of NLRP7 protein in the blastocyst stage. This difference is presumably species specific. As a matter of fact, NACHT domain of NLRP7 possesses a nuclear localization signal (Slim & Wallace 2013, Reddy et al. 2016); thus, it is understandable that NLRP7 is located in the nucleus. Furthermore, the results also indicated that ovine NLRP7 blastocyst localization resembles KHDC3L in human (Akoury et al. 2015) and Dnmt1o in mice (Howell et al. 2001).
It was reported that the mouse Nlrp2 mRNA accumulates during oogenesis and degrades (undetectable) after the two-cell stage in embryos, but the proteins persisted through to the blastocyst stage (Peng et al. 2012). However, in human, NLRP4, 5, 8, 9, 11, 12, 13 and 14 were highly expressed in oocytes and then gradually decreased in embryos with a very low level in day 5 (D5) embryos, while NLRP2 and NLRP7 progressively decreased from oocytes to day 3 (D3) embryos, and then showed a sharp increase in D5 (Zhang et al. 2008). Our study showed that NLRP7 mRNAs were abundant in oocyte maturation and PA or IVF embryos at different developmental stages, with no significant fluctuation, which was different from that observed in human pre-implantation embryos (Zhang et al. 2008).
In the current study, a 763 bp fragment (GenBank accession number: MF197687) of the NLRP7 CDS from ovine COCs was cloned, which shared 99% identity (data not shown) to the predicted Ovis aries NLRP7 mRNA in the NCBI database, with accession number XM_004015893. The deduced 254 amino acids peptide covered most of the LRR_RI domain (leucine-rich repeats, ribonuclease inhibitor-like), which was implicated in mediating autoregulation and protein–protein interaction as well as being typical of the NLRP family (Wagner et al. 2009). In accordance, the homology of the ovine NLRP7 protein with other mammalian species ranges from 97% identity in a 98% query cover with Capra hircus, to the 56 of 99% query cover with Homo sapiens, implying that the NLRP7 gene is evolutionarily conserved among different species. In fact, many NLRP members possess conserved domains (Ye & Ting 2008). Moreover, the phylogenetic relationship of ovine NLRP7 showed more affinity in ruminants than in primates, which is congruent with mammalian lineages topologies (Hedges et al. 2002, Reyes et al. 2004). On the other hand, to further study the structure and characteristics regarding ovine NLRP7 gene in the ovaries, full length coding sequence including 3′- and 5′- ends remained to be investigated.
To study the function of NLRP7 in ovine pre-implantation embryo development, RNAi was performed in Metaphase II stage oocytes followed by parthenogenetic activation or IVF. RNAi is a sequence-specific mRNA degradation induced by double-stranded RNA (dsRNA), which is usually induced either by delivering siRNA or shRNA into cells (Rao et al. 2009). RNAi became a favorable tool to study gene function also in mammalian oocytes and embryos. As a matter of fact, mouse oocytes were the first mammalian cell type where RNAi was conducted (Svoboda et al. 2000, Wianny & Zernicka-Goetz 2000). siRNA exhibits its effect immediately with transient gene silencing which is suitable for mouse embryos knockdown in which their developmental time was short. In contrast, shRNA is continuously synthesized by the host cell with much more durable effect, but it requires more time to transcribe from the vectors. In this study, the combination of siRNA and shRNA method was used to fulfill high potent and sustainable NLRP7 gene knockdown. Our data demonstrated that NLRP7 suppression in ovine embryos resulted in developmental impairment, indicating that NLRP7 is required for normal embryo development in vitro. The results were similar to the report by Peng et al. (2012). Nlrp2 knockdown in mouse leads to blockage at the two-cell stage both in fertilized and parthenogenetic embryos. Furthermore, others reported Nlrp2-deficient oocytes of mature adult mice showing defective parthenogenetic development (Kuchmiy et al. 2016). The similar phenomenon was also observed in Nlrp4e, Nlrp5, Nlrp14-deficient mice (Tong et al. 2000, Hamatani et al. 2004, Chang et al. 2013), as well as in rhesus macaque monkey (Wu et al. 2009) and sow (Peng et al. 2015).
The RNAi outcomes indicate that NLRP7 mRNA knockdown does not preclude embryo development up to the 8–16-cells stage, while further development is arrested. These results can be interpreted in at least two ways: (1) NLRP7 mRNA degradation is not sufficient to significantly decrease the cellular content of NLRP7 protein for several days, and thus, the effects of RNAi occur only when the endogenous protein is also degraded and cannot be replaced. (2) NLRP7 functions in the late stage of pre-implantation embryo development (morula and blastocyst). This interpretation seems compatible with the translocation of NLRP7 to a nuclear localization only at the blastocyst stage. Which is the case requires further investigations.
It was reported that maternal heterozygous NLRP7 variant caused recurrent reproductive failure and imprinting disturbances in the offspring (Soellner et al. 2017)). Actually, NLRP7 mutation impairs the normal protein biosynthesis, which is similar to its knockdown effect. In other ways, the NLRP7 knockdown may disturb the natural DNA methylation status (Mahadevan et al. 2014) or influence the regular protein–protein interactions between the SCMC members (Mahadevan et al. 2017) in oocytes or embryos. Besides, previous studies have shown that NLRP7 is important for the formation of inflammasome to protect against an immediate danger in the processing and maturation of pro-inflammatory cytokines, the interleukin, IL-1β and IL-18 (Davis et al. 2011). Accordingly, knockdown of NLRP7 probably weakened the embryo immunity. Certainly, to further investigate the NLRP7 function in ovine, the over-expression, knock-out or other targeted inhibition experiments should be tested.
In conclusion, we identified that NLRP7 was present in the ovine and its high expression was only detected in the ovary. The localization and expression patterns of NLRP7 suggest its unique role in the female reproduction. Indeed, NLRP7 knockdown is unfavorable for the pre-implantation embryo development in vitro. The functions of NLRP7 in animal reproduction, particularly in husbandry important animals, deserve further exploration. The identification of NLRP7 downstream signaling pathway to elucidate its molecular mechanisms is necessary and it is our future goal.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research was supported by funds from the National Science and Technology Major Project of China (2018ZX0800801B, 2016ZX08008003), National Natural Science Foundation of China (31830091), and the Beijing Dairy Industry Innovation Team (BAIC06-2017).
Acknowledgements
The authors thank Prof. Wei Cui for helpful discussions.
References
- Akoury E, Gupta N, Bagga R, Brown S, Déry C, Kabra M, Srinivasan R, Slim R. 2015. Live births in women with recurrent hydatidiform mole and two NLRP7 mutations. Reproductive Biomedicine Online 120–124. ( 10.1016/j.rbmo.2015.03.011) [DOI] [PubMed] [Google Scholar]
- Akoury E, Zhang L, Ao A, Slim R. 2015. NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Human Reproduction 159–169. ( 10.1093/humrep/deu291) [DOI] [PubMed] [Google Scholar]
- Andreasen L, Bolund L, Niemann I, Hansen ES, Sunde L. 2012. Mosaic moles and non-familial biparental moles are not caused by mutations in NLRP7, NLRP2 or C6orf221. Molecular Human Reproduction 593–598. ( 10.1093/molehr/gas036) [DOI] [PubMed] [Google Scholar]
- Barbé F, Douglas T, Saleh M. 2014. Advances in Nod-like receptors (NLR) biology. Cytokine & Growth Factor Reviews 681–697. ( 10.1016/j.cytogfr.2014.07.001) [DOI] [PubMed] [Google Scholar]
- Bebbere D, Bogliolo L, Ariu F, Fois S, Leoni GG, Tore S, Ledda S, Berlinguer F, Naitana S, Ledda S. 2008. Expression pattern of zygote arrest 1 (ZAR1), maternal antigen that embryo requires (MATER), growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15) genes in ovine oocytes and in vitro-produced preimplantation embryos. Reproduction, Fertility, and Development 908–915. ( 10.1071/RD08095) [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Patel OV, Ireland JJ, Smith GW. 2006. Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, β-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, β-actin, and histone H2A during bovine oocyte maturation and early embryogenesis in vitro. Molecular Reproduction & Development 267–278. ( 10.1002/mrd.20333) H2A. [DOI] [PubMed] [Google Scholar]
- Brown L, Mount S, Reddy R, Slim R, Wong C, Jobanputra V, Clifford P, Merrill L, Brown S. 2013. Recurrent pregnancy loss in a woman with NLRP7 mutation: not all molar pregnancies can be easily classified as either “partial” or “complete” hydatidiform moles. International Journal of Gynecological Pathology 399–405. ( 10.1097/PGP.0b013e31826cbf6a) [DOI] [PubMed] [Google Scholar]
- Carey L, Nash BM, Wright DC. 2015. Molecular genetic studies of complete hydatidiform moles. Translational Pediatrics 181–188. ( 10.3978/j.issn.2224-4336.2015.04.02) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BH, Liu X, Liu J, Quan FS, Guo ZK, Zhang Y. 2013. Developmental expression and possible functional roles of mouse Nlrp4e in preimplantation embryos. In Vitro Cellular & Developmental Biology. Animal 548–553. ( 10.1007/s11626-013-9638-9) [DOI] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual Review of Immunology 707–735. ( 10.1146/annurev-immunol-031210-101405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duéñez-Guzmán EA, Haig D. 2014. The evolution of reproduction-related NLRP genes. Journal of Molecular Evolution 194–201. ( 10.1007/s00239-014-9614-3) [DOI] [PubMed] [Google Scholar]
- Fisher RA, Lavery SA, Carby A, Abu-Hayyeh S, Swingler R, Sebire NJ, Seckl MJ. 2011. What a difference an egg makes. Lancet 1974. ( 10.1016/S0140-6736(11)61751-0) [DOI] [PubMed] [Google Scholar]
- Fortune JE. 2003. The early stages of follicular development: activation of primordial follicles and growth of preantral follicles. Animal Reproduction Science 135–163. ( 10.1016/S0378-4320(03)00088-5) [DOI] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. 2004. Age-associated alteration of gene expression patterns in mouse oocytes. Human Molecular Genetics 2263–2278. ( 10.1093/hmg/ddh241) [DOI] [PubMed] [Google Scholar]
- Hedges SB. 2002. The origin and evolution of model organisms. Nature Reviews. Genetics 838–849. ( 10.1038/nrg929) [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 829–838. ( 10.1016/s0092-8674(01)00280-x) [DOI] [PubMed] [Google Scholar]
- Ito Y, Maehara K, Kaneki E, Matsuoka K, Sugahara N, Miyata T, Kamura H, Yamaguchi Y, Kono A, Nakabayashi K. et al. 2016. Novel nonsense mutation in the NLRP7 gene associated with recurrent hydatidiform mole. Gynecologic and Obstetric Investigation 353–358. ( 10.1159/000441780) [DOI] [PubMed] [Google Scholar]
- Kalogiannidis I, Kalinderi K, Kalinderis M, Miliaras D, Tarlatzis B, Athanasiadis A. 2018. Recurrent complete hydatidiform mole: where we are, is there a safe gestational horizon? Opinion and mini-review. Journal of Assisted Reproduction & Genetics 967–973. ( 10.1007/s10815-018-1202-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C. 2012. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 464–476. ( 10.1016/j.immuni.2012.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. 2005. PYPAF3, a pyrin-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1β secretion. Journal of Biological Chemistry 21720–21725. ( 10.1074/jbc.M410057200) [DOI] [PubMed] [Google Scholar]
- Kuchmiy AA, D’Hont J, Hochepied T, Lamkanfi M. 2016. NLRP2 controls age-associated maternal fertility. Journal of Experimental Medicine 2851–2860. ( 10.1084/jem.20160900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 1870–1874. ( 10.1093/molbev/msw054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S, Sathappan V, Utama B, Lorenzo I, Kaskar K, Van den Veyver IB. 2017. Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Scientific Reports 44667. ( 10.1038/srep44667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S, Wen S, Wan YW, Peng HH, Otta S, Liu Z, Iacovino M, Mahen EM, Kyba M, Sadikovic B. et al. 2014. NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Human Molecular Genetics 706–716. ( 10.1093/hmg/ddt457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokhina I, Hanna CW, Stephenson MD, McFadden DE, Robinson WP. 2013. Maternal NLRP7 and C6orf221 variants are not a common risk factor for androgenetic moles, triploidy and recurrent miscarriage. Molecular Human Reproduction 539–544. ( 10.1093/molehr/gat019) [DOI] [PubMed] [Google Scholar]
- McDaniel P, Wu X. 2009. Identification of oocyte-selective NLRP genes in rhesus macaque monkeys (Macaca mulatta). Molecular Reproduction & Development 151–159. ( 10.1002/mrd.20937) [DOI] [PubMed] [Google Scholar]
- Messaed C, Akoury E, Djuric U, Zeng J, Saleh M, Gilbert L, Seoud M, Qureshi S, Slim R. 2011. NLRP7, a nucleotide oligomerization domain-like receptor protein, is required for normal cytokine secretion and co-localizes with Golgi and the microtubule-organizing center. Journal of Biological Chemistry 43313–43323. ( 10.1074/jbc.M111.306191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Broz P. 2017. Evolutionary convergence and divergence in NLR function and structure. Trends in Immunology 744–757. ( 10.1016/j.it.2017.04.005) [DOI] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B. et al. 2006. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nature Genetics 300–302. ( 10.1038/ng1740) [DOI] [PubMed] [Google Scholar]
- Nguyen NMP, Zhang L, Reddy R, Déry C, Arseneau J, Cheung A, Surti U, Hoffner L, Seoud M, Zaatari G. et al. 2014. Comprehensive genotype–phenotype correlations between NLRP7 mutations and the balance between embryonic tissue differentiation and trophoblastic proliferation. Journal of Medical Genetics 51 623–634. ( 10.1136/jmedgenet-2014-102546) 25097207. [DOI] [PubMed] [Google Scholar]
- Ohno S, Kinoshita T, Ohno Y, Minamoto T, Suzuki N, Inoue M, Suda T. 2008. Expression of NLRP7 (PYPAF3, NALP7) protein in endometrial cancer tissues. Anticancer Research 2493–2497. [PubMed] [Google Scholar]
- Okada K, Hirota E, Mizutani Y, Fujioka T, Shuin T, Miki T, Nakamura Y, Katagiri T. 2004. Oncogenic role of NALP7 in testicular seminomas. Cancer Science 949–954. ( 10.1111/j.1349-7006.2004.tb03182.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis F, Vigneault C, Robert C, Sirard MA. 2005. RNA interference as a tool to study gene function in bovine oocytes. Molecular Reproduction & Development 111–121. ( 10.1002/mrd.20193) [DOI] [PubMed] [Google Scholar]
- Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, Wang Y, Liu J, Zhang B, Quan F. et al. 2012. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLOS ONE e30344. ( 10.1371/journal.pone.0030344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Liu F, Li W, Zhang W. 2015. Knockdown of NLRP5 arrests early embryogenesis in sows. Animal Reproduction Science 151–156. ( 10.1016/j.anireprosci.2015.11.004) [DOI] [PubMed] [Google Scholar]
- Pisani LF, Ramelli P, Lazzari B, Braglia S, Ceciliani F, Mariani P. 2010. Characterization of maternal antigen that embryos require (MATER/NLRP5) gene and protein in pig somatic tissues and germcells. Journal of Reproduction & Development 41–48. ( 10.1262/jrd.09-098a) [DOI] [PubMed] [Google Scholar]
- Qian J, Deveault C, Bagga R, Xie X, Slim R. 2007. Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Human Mutation 741. ( 10.1002/humu.9498) [DOI] [PubMed] [Google Scholar]
- Quan GB, Ma Y, Ni YN, Wu GQ, Lv CR, Hong QH. 2017. Effects of synthetic polymers on in vitro maturation of sheep oocytes and subsequent developmental capacity after parthenogenetic activation or fertilization. Small Ruminant Research 153–157. ( 10.1016/j.smallrumres.2017.06.008) [DOI] [Google Scholar]
- Radian AD, de Almeida L, Dorfleutner A, Stehlik C. 2013. NLRP7 and related inflammasome activating pattern recognition receptors and their function in host defense and disease. Microbes and Infection 630–639. ( 10.1016/j.micinf.2013.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radian AD, Khare S, Chu LH, Dorfleutner A, Stehlik C. 2015. ATP binding by NLRP7 is required for inflammasome activation in response to bacterial lipopeptides. Molecular Immunology 294–302. ( 10.1016/j.molimm.2015.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DD, Vorhies JS, Senzer N, Nemunaitis J. 2009. siRNA vs. shRNA: similarities and differences. Advanced Drug Delivery Reviews 746–759. ( 10.1016/j.addr.2009.04.004) [DOI] [PubMed] [Google Scholar]
- Reddy R, Nguyen NMP, Sarrabay G, Rezaei M, Rivas MCG, Kavasoglu A, Berkil H, Elshafey A, Nunez KP, Dreyfus H. et al. 2016. The genomic architecture of NLRP7 is Alu rich and predisposes to disease-associated large deletions. European Journal of Human Genetics 1445–1452. ( 10.1038/ejhg.2016.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Catzeflis F, Nevo E, Pesole G, Saccone C. 2004. Congruent mammalian trees from mitochondrial and nuclear genes using Bayesian methods. Molecular Biology and Evolution 397–403. ( 10.1093/molbev/msh033) [DOI] [PubMed] [Google Scholar]
- Reynaud D, Abi Nahed R, Traboulsi W, Aboussaouira T, Benharouga M, Alfaidy N. 2018. Characterization of the role of NLRP7 protein in normal and tumor pregnancies: case of choriocarcinoma. Annales d'Endocrinologie 317. ( 10.1016/j.ando.2018.06.367) [DOI] [Google Scholar]
- Sanchez-Delgado M, Martin-Trujillo A, Tayama C, Vidal E, Esteller M, Iglesias-Platas I, Deo N, Barney O, Maclean K, Hata K. et al. 2015. Absence of maternal methylation in biparental hydatidiform moles from women with NLRP7 maternal-effect mutations reveals widespread placenta-specific imprinting. PLOS Genetics e1005644. ( 10.1371/journal.pgen.1005644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nature Protocols 1101–1108. ( 10.1038/nprot.2008.73) [DOI] [PubMed] [Google Scholar]
- Sebire NJ, Savage PM, Seckl MJ, Fisher RA. 2013. Histopathological features of biparental complete hydatidiform moles in women with NLRP7 mutations. Placenta 50–56. ( 10.1016/j.placenta.2012.11.005) [DOI] [PubMed] [Google Scholar]
- Sills ES, Obregon-Tito AJ, Gao H, McWilliams TK, Gordon AT, Adams CA, Slim R. 2017. Pathogenic variant in NLRP7 (19q13. 42) associated with recurrent gestational trophoblastic disease: data from early embryo development observed during in vitro fertilization. Clinical and Experimental Reproductive Medicine 40–46. ( 10.5653/cerm.2017.44.1.40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer H, Biswas A, Nuesgen N, Oldenburg J, El-Maarri O. 2015. NLRP7, Involved in hydatidiform molar pregnancy (HYDM1), interacts with the transcriptional repressor ZBTB16. PLOS ONE e0130416. ( 10.1371/journal.pone.0130416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slim R, Ao A, Surti U, Zhang L, Hoffner L, Arseneau J, Cheung A, Chebaro W, Wischmeijer A. 2011. Recurrent triploid and dispermic conceptions in patients with NLRP7 mutations. Placenta 409–412. ( 10.1016/j.placenta.2011.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slim R, Wallace EP. 2013. NLRP7 and the genetics of hydatidiform moles: recent advances and new challenges. Frontiers in Immunology 242. ( 10.3389/fimmu.2013.00242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soellner L, Begemann M, Degenhardt F, Geipel A, Eggermann T, Mangold E. 2017. Maternal heterozygous NLRP7 variant results in recurrent reproductive failure and imprinting disturbances in the offspring. European Journal of Human Genetics 924–929. ( 10.1038/ejhg.2017.94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 4147–4156. [DOI] [PubMed] [Google Scholar]
- Tejomurtula J, Lee KB, Tripurani SK, Smith GW, Yao J. 2009. Role of importin alpha8, a new member of the importin alpha family of nuclear transport proteins, in early embryonic development in cattle. Biology of Reproduction 333–342. ( 10.1095/biolreprod.109.077396) [DOI] [PubMed] [Google Scholar]
- Tian X, Pascal G, Monget P. 2009. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evolutionary Biology 202. ( 10.1186/1471-2148-9-202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JPY, Willingham SB, Bergstralh DT. 2008. NLRs at the intersection of cell death and immunity. Nature Reviews. Immunology 372–379. ( 10.1038/nri2296) [DOI] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. 2000. Mater, a maternal effect gene required for early embryonic development in mice. Nature Genetics 267–268. ( 10.1038/81547) [DOI] [PubMed] [Google Scholar]
- Van Gorp H, Kuchmiy A, Van Hauwermeiren F, Lamkanfi M. 2014. NOD‐like receptors interfacing the immune and reproductive systems. FEBS Journal 4568–4582. ( 10.1111/febs.13014) [DOI] [PubMed] [Google Scholar]
- Wagner RN, Proell M, Kufer TA, Schwarzenbacher R. 2009. Evaluation of Nod-like receptor (NLR) effector domain interactions. PLOS ONE e4931. ( 10.1371/journal.pone.0004931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CM, Dixon PH, Decordova S, Hodges MD, Sebire NJ, Ozalp S, Fallahian M, Sensi A, Ashrafi F, Repiska V. et al. 2009. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. Journal of Medical Genetics 569–575. ( 10.1136/jmg.2008.064196) [DOI] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biology 70–75. ( 10.1038/35000016) [DOI] [PubMed] [Google Scholar]
- Wu X. 2009. Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta). Human Reproduction 415–424. ( 10.1093/humrep/den403) [DOI] [PubMed] [Google Scholar]
- Ye Z, Ting JP. 2008. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Current Opinion in Immunology 3–9. ( 10.1016/j.coi.2008.01.003) [DOI] [PubMed] [Google Scholar]
- Zhang P, Dixon M, Zucchelli M, Hambiliki F, Levkov L, Hovatta O, Kere J. 2008. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLOS ONE e2755. ( 10.1371/journal.pone.0002755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shah SZA, Yang L, Zhang Z, Zhou X, Zhao D. 2016. Virulent Mycobacterium bovis Beijing strain activates the NLRP7 inflammasome in THP-1 macrophages. PLOS ONE e0152853. ( 10.1371/journal.pone.0152853) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a