ABSTRACT
Tissue homeostasis is maintained by differentiated progeny of residential stem cells. Both extrinsic signals and intrinsic factors play critical roles in the proliferation and differentiation of adult intestinal stem cells (ISCs). However, how extrinsic signals are transduced into ISCs still remains unclear. Here, we find that heparan sulfate (HS), a class of glycosaminoglycan (GAG) chains, negatively regulates progenitor proliferation and differentiation to maintain midgut homeostasis under physiological conditions. Interestingly, HS depletion in progenitors results in inactivation of Decapentaplegic (Dpp) signaling. Dpp signal inactivation in progenitors resembles HS-deficient intestines. Ectopic Dpp signaling completely rescued the defects caused by HS depletion. Taken together, these data demonstrate that HS is required for Dpp signaling to maintain midgut homeostasis. Our results provide insight into the regulatory mechanisms of how extrinsic signals are transduced into stem cells to regulate their proliferation and differentiation.
KEY WORDS: Intestinal stem cell, Heparan sulfate, Drosophila, Tissue homeostasis, Dpp signaling
Summary: Heparan sulfate chains in Drosophila adult intestinal stem cells negatively regulate stem cell proliferation and differentiation to control tissue homeostasis under physiological conditions.
INTRODUCTION
Adult stem cells are responsible for tissue homeostasis in the tissues in which they reside; frequently lost cells are constantly replenished by the progeny of stem cells. The proliferation and differentiation of adult stem cells must be tightly balanced. Disruption of this balance will lead to either excessive stem cells or stem cell depletion, eventually resulting in various diseases, such as cancer (Lin, 2008; Morrison and Spradling, 2008; Radtke and Clevers, 2005; Xie and Spradling, 1998). Therefore, understanding of the underlying mechanisms controlling adult stem cell proliferation and differentiation will provide insight into the potential development of therapeutic applications for human diseases.
The posterior midgut of the adult Drosophila intestine is an excellent system to study how stem cell proliferation and differentiation are regulated. Mammalian and Drosophila intestines show marked similarities in terms of development, cellular make-up and genetic control (Casali and Batlle, 2009; Edgar, 2012; Stainier, 2005; Wang and Hou, 2010). Adult intestinal stem cells (ISCs) are interspersed along the base membrane of the Drosophila adult midgut (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Initial studies proposed that ISCs constantly undergo asymmetric divisions and produce non-dividing enteroblasts (EBs) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). The ligand of the Notch pathway, Delta (Dl), is specifically expressed in ISCs, while Notch receptor is expressed in both ISCs and EBs. ISCs signal via Dl to activate Notch signaling in EBs (Ohlstein and Spradling, 2007). EBs terminally differentiate into either an absorptive enterocyte (EC) or a secretory enteroendocrine cell (ee) depending on their signaling environments (Beebe et al., 2010; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2007; Perdigoto et al., 2011; Yeung et al., 2011). Recent studies demonstrate that in response to differentiation and subsequent loss of a neighboring ISC (or vice versa), a significant proportion of ISCs divide symmetrically (de Navascués et al., 2012; Goulas et al., 2012; O'Brien et al., 2011). Moreover, ee cells may not be generated from EBs, but directly from ISCs or ee progenitor cells (EEPs) (Biteau and Jasper, 2014; Chen et al., 2018; Zeng et al., 2015). Interestingly, unlike in other systems in which differentiated cells can de-differentiate into stem cells, we found that no regeneration of new ISCs could be observed after all the progenitors were ablated in the intestines, indicating that fully differentiated cells are likely unable to de-differentiate into ISCs when all the progenitors are depleted (Brawley and Matunis, 2004; Lu and Li, 2015; Raff, 2003).
Numerous studies have shown that ISC proliferation and differentiation under physiological conditions and during tissue regeneration are regulated by many signaling pathways and intrinsic factors, including the Notch, Wingless (Wg), Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT), Epidermal Growth Factor Receptor (EGFR), Hippo (Hpo), Insulin, Hedgehog (Hh) and Bone Morphogenetic Protein (BMP) signaling pathways (Amcheslavsky et al., 2009; Biteau and Jasper, 2011; Buchon et al., 2009; Chakrabarti et al., 2016; Chen et al., 2016; Choi et al., 2011; Cordero et al., 2012; Guo and Ohlstein, 2015; Han et al., 2015; Jiang et al., 2011, 2009; Jin et al., 2017; Karpowicz et al., 2010; Lee et al., 2009; Li et al., 2013a,b, 2014; Lin and Xi, 2008; Lin et al., 2008; Martorell et al., 2014; Ohlstein and Spradling, 2006, 2007; Rahman et al., 2017; Ren et al., 2010, 2015; Schell et al., 2017; Shaw et al., 2010; Singh et al., 2016; Staley and Irvine, 2010; Tian and Jiang, 2014; Tian et al., 2015, 2017; Xu et al., 2011; Zhai et al., 2017; Zhou et al., 2015). However, it remains unclear how extrinsic signals are transduced into ISCs to regulate their proliferation and differentiation under physiological conditions.
Heparan sulfate chains are attached to the core protein of heperan sulfate proteoglycans (HSPGs), macromolecules presented on the cell surface and in the extracellular matrix (ECM). There are three evolutionarily conserved families of HSPGs: Glypicans and Syndecans are two major cell surface HSPGs, while Perlecans are secreted HSPGs that are mainly distributed in the ECM (Esko and Lindahl, 2001; Esko and Selleck, 2002; Lin, 2004). HS chain biosynthesis is initiated in the Golgi apparatus at the GAG attachment site(s) of the core protein. HS is synthesized by a series of conserved HS biosynthetic and modifying enzymes, including Gal transferases, the exostosin (EXT) proteins [Tout-velu (Ttv), Sister of ttv (Sotv), Brother of ttv (Botv)], Sulfateless (Sfl) and Sugarless (Sgl) (Esko and Lindahl, 2001; Esko and Selleck, 2002; Lin, 2004). Previous studies demonstrate that HSPGs are required for the distribution of several well-known morphogens, including Wg, Hh, Upd and Dpp (Belenkaya et al., 2004; Bellaiche et al., 1998; Binari et al., 1997; Bornemann et al., 2004; Dani et al., 2012; Filmus et al., 2008; Fujise et al., 2003; Jackson et al., 1997; Kamimura et al., 2006; Levings and Nakato, 2017; Lin and Perrimon, 1999, 2000, 2002; Liu et al., 2010; Mii et al., 2017; Takei et al., 2004; Yan and Lin, 2009; Yu et al., 2017; Zhang et al., 2013). Although HS plays important roles in the functions of HSPGs, its role(s) in regulating ISC proliferation and differentiation under physiological conditions remains elusive.
In this study, we provide evidence that HS in progenitors restricts ISC proliferation and differentiation under normal homeostasis. Importantly, we demonstrate that HS is required for Dpp signal activation. Thus, our data uncover a mechanism of HS to maintain midgut homeostasis.
RESULTS
Loss of HS in progenitors leads to disruption of midgut homeostasis
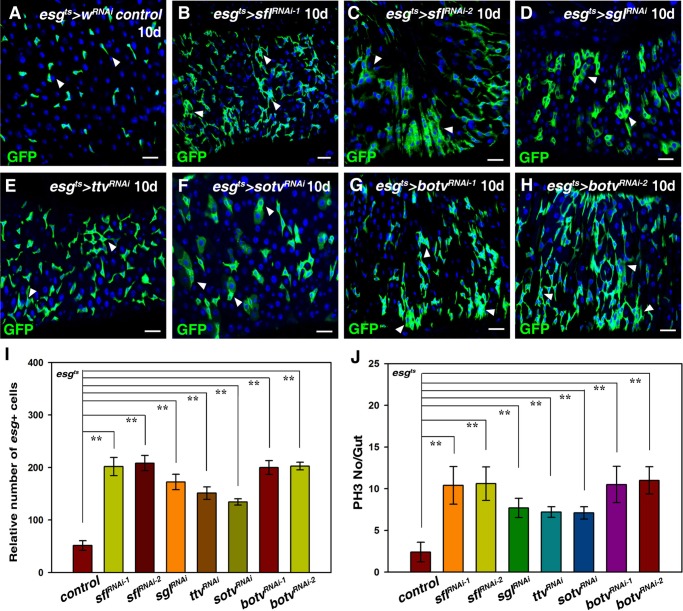
In order to identify intrinsic factors regulating the proliferation and differentiation of ISCs, we carried out a genome-wide RNAi screen using the esgGal4, UAS-GFP, tubGal80ts (esgts) driver in the posterior midgut. esgGal4 is expressed in progenitors (ISCs and EBs) in the midgut. 11,316 RNAi lines from Vienna Drosophila RNAi Center (VDRC), Fly stocks of National Institute of Genetics (NIG-FLY), and the Transgenic RNAi Project (TRiP) at Harvard Medical School/Tsinghua University were screened (manuscript in preparation). Many factors affecting ISC maintenance, viability and proliferation/differentiation were identified from this screen. Among these factors, HS biosynthetic enzymes (including Sfl, Sgl and the EXT proteins) were identified as candidates. Only single or paired esg+ cells are observed in control flies (Fig. 1A). However, the number of esg+ cells was significantly increased when sfl was depleted in progenitors. esg+ cells formed clusters and GFP was expressed in polyploid cells, indicative of midgut homeostasis loss (Fig. 1B,C,I). sfl encodes the only Drosophila HS N-deacetylase/N-sulfotransferase, which catalyzes the first step of HS modification (Esko and Lindahl, 2001; Lin, 2004). The RNAi off-target effect could be excluded as induction of two independent RNAi constructs against sfl produced a similar phenotype (Fig. 1B,C,I) and the knockdown efficacy of these RNAi lines was confirmed by qRT-PCR (Fig. S1). When sgl and EXT genes were knocked down, midgut homeostasis was also lost (Fig. 1D,I). Consistent with the increase of progenitors, we observed a significant increase of the number of cells undergoing mitosis in these intestines (Fig. 1J; Fig. S2). Meanwhile, many large esg+ cells expressed mature EC-marker PDM1, indicative of intestinal homeostasis loss (Fig. S3). Previous studies had demonstrated that mutations in these HS biosynthetic genes led to striking reductions in HS levels (Bornemann et al., 2004; Han et al., 2004; Takei et al., 2004; Toyoda et al., 2000a,b). Consistently, we found HS was effectively abrogated in tubts>botvRNAi intestines (Fig. S4). These data demonstrate that HS in progenitors restricts ISC proliferation, thereby maintaining midgut homeostasis. As EXT3/Botv participates in the earliest steps of HS biosynthesis, we mainly focused on botv for further analysis.
Fig. 1.
HS in progenitors restricts ISC proliferation. (A) esg+ cells (green) in control flies at 29°C for 10 days (white arrowheads). (B,C) The number of esg+ cells (green) is dramatically increased in esgts>sflRNAi flies at 29°C for 10 days (white arrowheads). (D) The number of esg+ cells (green) is dramatically increased in esgts>sglRNAi flies at 29°C for 10 days (white arrowheads). (E) The number of esg+ cells (green) is dramatically increased in esgts>ttvRNAi flies at 29°C for 10 days (white arrowheads). (F) The number of esg+ cells (green) is dramatically increased in esgts>sotvRNAi flies at 29°C for 10 days (white arrowheads). (G,H) The number of esg+ cells (green) is dramatically increased in esgts>botvRNAi flies at 29°C for 10 days (white arrowheads). (I) Quantification of the relative number of esg+ cells in the different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. (J) Quantification of the number of pH3 per gut in the different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. In all panels except graphs, GFP is in green and blue indicates DAPI staining for DNA. Scale bars: 20 μm.
HS in progenitors negatively regulates ISC proliferation and differentiation
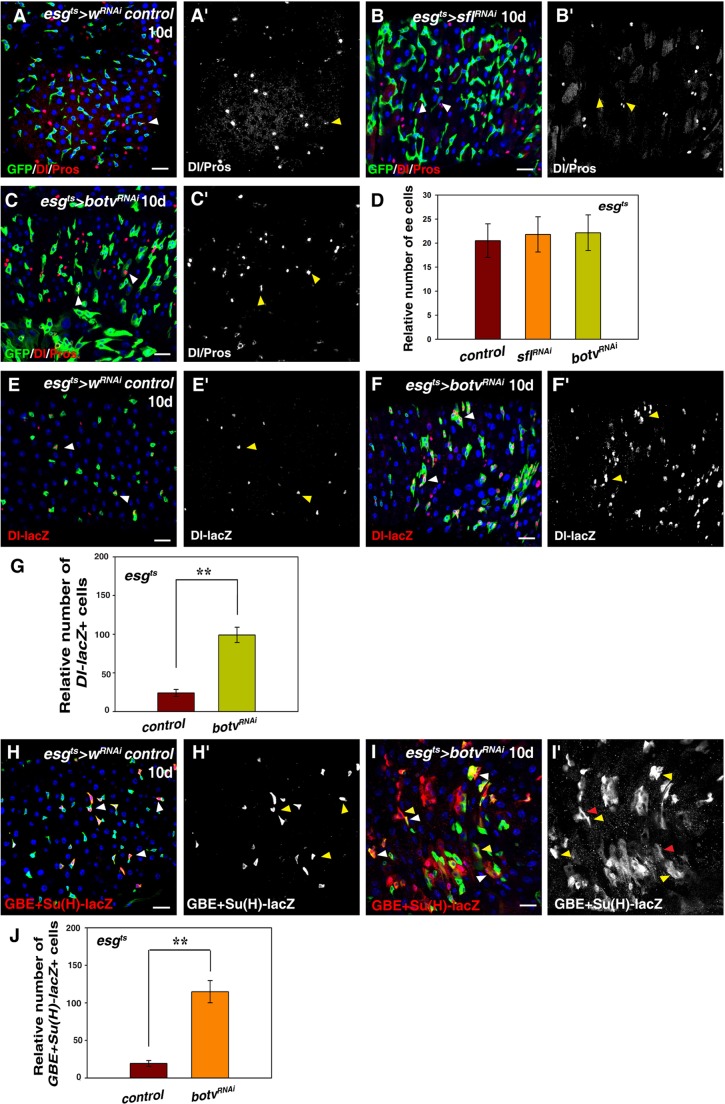
We examined the identity of these esg+ cells in the absence of HS. We found that the number of ISCs (by Dl and Dl-lacZ) in esgts>sflRNAi and esgts>botvRNAi intestines was significantly increased compared to those in the control flies (Fig. 2A–C,E–F). The number of EBs [by GBE+Su(H)-lacZ] was also significantly increased in esgts>botvRNAi intestines compared to those in the control flies (Fig. 2H–J). However, no obvious change in the number of ee cells (by Pros) was observed in these intestines (Fig. 2A–D). We found that the size of the large GFP+ cells (premature/mature ECs) was smaller than that of fully differentiated ECs, indicating that HS may also regulate EC maturation (Fig. 2H,I). We also observed a significant increase of ISCs when HS synthesis was disrupted in ISCs using ISC-specific driver Dlts (data not shown). Taken together, these data demonstrate that HS in progenitors negatively regulates ISC proliferation and differentiation under physiological conditions.
Fig. 2.
HS in progenitors negatively regulates ISC proliferation and differentiation. (A) Dl (ISC marker) and Pros (ee marker) (red) in control intestines (white arrowhead). Split channel for Dl and Pros (A′, in grayscale) in control intestines (yellow arrowhead). (B) Dl and Pros (red) in esgts>sflRNAi intestines (white arrowheads). Split channel for Dl and Pros (B′, in grayscale) in esgts>sflRNAi intestines (yellow arrowheads). (C) Dl and Pros (red) in esgts>botvRNAi intestines (white arrowheads). Split channel for Dl and Pros (C′, in grayscale) in esgts>botvRNAi intestines (yellow arrowheads). (D) Quantification of the relative number of ee cells in intestines with indicated phenotypes. n=10–15 intestines. mean±s.d. is shown. No obvious change in the number of ee cells is observed. (E) Dl-lacZ (red) in control intestines (white arrowheads). Split channel for Dl-lacZ (E′, in grayscale) in control intestines (yellow arrowheads). (F) Dl-lacZ (red) in esgts>botvRNAi intestines (white arrowheads). Split channel for Dl-lacZ (F′, in grayscale) in esgts>botvRNAi intestines (yellow arrowheads). (G) Quantification of the relative number of Dl-lacZ+ cells in control and esgts>botvRNAi intestines. n=10–15 intestines. mean±s.d. is shown. **P<0.01. (H) EBs [by GBE+Su(H)-lacZ in red] in control intestines (white arrowheads). Split channel for GBE+Su(H)-lacZ (H′, in grayscale) in control intestines (yellow arrowheads). (I) The number of EBs [by GBE+Su(H)-lacZ in red] is dramatically increased in esgts>botvRNAi intestines (white and yellow arrowheads). Split channel for GBE+Su(H)-lacZ (I′, in grayscale) in esgts>botvRNAi intestines (yellow and red arrowheads). Note that the size of the larger GBE+Su(H)-lacZ+ cells is smaller compared to the size of the neighboring wild-type EC cells [polyploid GBE+Su(H)-lacZ− cells], indicating that HS chains also affect EC maturation (yellow arrowheads in I). (J) Quantification of the relative number of GBE+Su(H)-lacZ+ cells in control and esgts>botvRNAi intestines. n=10–15 intestines. mean±s.d. is shown. **P<0.01. GFP in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm.
Consistent with increased ISC proliferation, we found the number of 10xSTATGFP+ cells and the intensity of 10xSTATGFP signal in esgts>botvRNAi intestines were both significantly increased compared to those in the control flies (Fig. S5A–D). However, the expression of the JAK/STAT pathway cytokines (Upd1-3) was only mildly increased in esgts>botvRNAi intestines compared to those in control intestines as determined by qRT-PCR (Fig. S6A). We examined whether the accumulation of esg+ cells observed in esgts>botvRNAi intestines resulted from the expression of cytokines in progenitors. We found that neither individual nor simultaneous depletion of cytokines could suppress the accumulation of esg+ cells observed in those intestines (Fig. S6B–L). These data indicate that cytokines are unlikely produced in these HS-deficient progenitors.
HSPGs (except Perlecan) may play redundant roles in progenitor proliferation
HSPGs comprise a core protein to which HS chains are attached. Our data demonstrate that HS in progenitors inhibits ISC proliferation. We explored which HSPG(s) are required for ISC proliferation. A previous study indicated that loss of the core protein of Perlecan (Per) caused detachment of ISC from basement membrane, resulting in loss of ISC proliferation (You et al., 2014). Dally and Dally-like (Dlp) are two major Glypicans in Drosophila (Esko and Lindahl, 2001; Esko and Selleck, 2002; Lin, 2004). We examined whether the other HSPGs, except Per, are required for ISC proliferation. We first examined the expression pattern of Dally and Dlp in the intestines. We found that Dally was mainly expressed in ECs (Fig. S7A,B), while Dlp was mainly expressed in the visceral muscles (VMs), and at low levels in the midgut epithelium (Fig. S7C,D). Effective RNAi constructs against dally and dlp were utilized to explore whether these HSPGs are required for ISC proliferation (Zhang et al., 2013). We found that neither individual nor combinational knockdown of dally and dlp affected ISC proliferation significantly (Fig. S7E–H). We also found that neither individual nor combinational knockdown of the other HSPGs, sdc and cow, as significantly affected ISC proliferation as those observed in HS-deficient progenitors (data not shown). These data indicate that these HSPGs (except Per) may play redundant roles in ISC proliferation. Therefore, we focused on HS chains, but not individual HSPGs, for further investigation.
HS is required for Dpp signal activation in progenitors
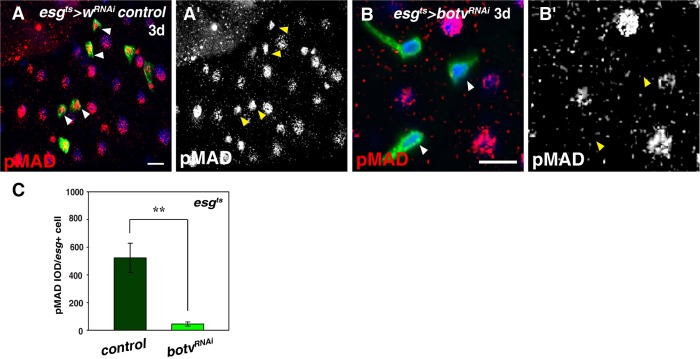
The above mentioned experiments demonstrate that HS in progenitors is required for midgut homeostasis under normal conditions. We explored the underlying mechanism of how HS controls midgut homeostasis. HS is required for the activation of many signaling pathways, including Wg, JAK/STAT, Notch and BMP (Belenkaya et al., 2004; Bellaiche et al., 1998; Binari et al., 1997; Bornemann et al., 2004; Dani et al., 2012; Filmus et al., 2008; Fujise et al., 2003; Jackson et al., 1997; Kamimura et al., 2006; Levings and Nakato, 2017; Lin and Perrimon, 1999, 2000, 2002; Liu et al., 2010; Mii et al., 2017; Takei et al., 2004; Yan and Lin, 2009; Yu et al., 2017; Zhang et al., 2013). Interestingly, the majority of these signaling pathways positively regulate ISC proliferation and differentiation, while Dpp signaling negatively regulates ISC proliferation and differentiation (Guo et al., 2013; Jiang and Edgar, 2009; Jiang et al., 2011, 2009; Li et al., 2013a,b, 2014; Lin et al., 2008; Tian and Jiang, 2014; Tian et al., 2015; Zhou et al., 2015). Notch signaling is not blocked upon loss of HS as the number of GBE+Su(H)-lacZ+ cells was dramatically increased, indicating that Notch signaling is not affected by HS depletion (Fig. 2H–J). Therefore, we speculated that HS may be required for Dpp signal activation in progenitors. To confirm our hypothesis, we examined Dpp signal activation in the absence of HS. Dpp signaling (by pMAD) is mainly activated in ECs, but also in progenitors in control flies (Fig. 3A) (Li et al., 2013b; Tian and Jiang, 2014; Zhou et al., 2015). When HS was depleted in progenitors, pMAD signal in progenitors was abolished, supporting the notion that HS is required for Dpp signal activation in progenitors (Fig. 3A–C).
Fig. 3.
Loss of HS leads to Dpp signal inactivation. (A) Dpp signaling (by pMAD in red) in control intestines. Dpp signaling is highly activated in ECs and progenitors (green) (white arrowheads). Split channel for pMAD (A′, in grayscale) (yellow arrowheads). (B) Dpp signaling (by pMAD in red) in progenitors is eliminated in esgts>botvRNAi intestines (white arrowheads). Split channel for pMAD (B′, in grayscale) (yellow arrowheads). (C) Quantification of by pMAD signal after knockdown of botv in progenitors. IOD was used. n≥4. mean±s.d. is shown. **P<0.01. In all panels except graphs, GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm.
Dpp signaling negatively regulates ISC proliferation and midgut homeostasis under physiological conditions
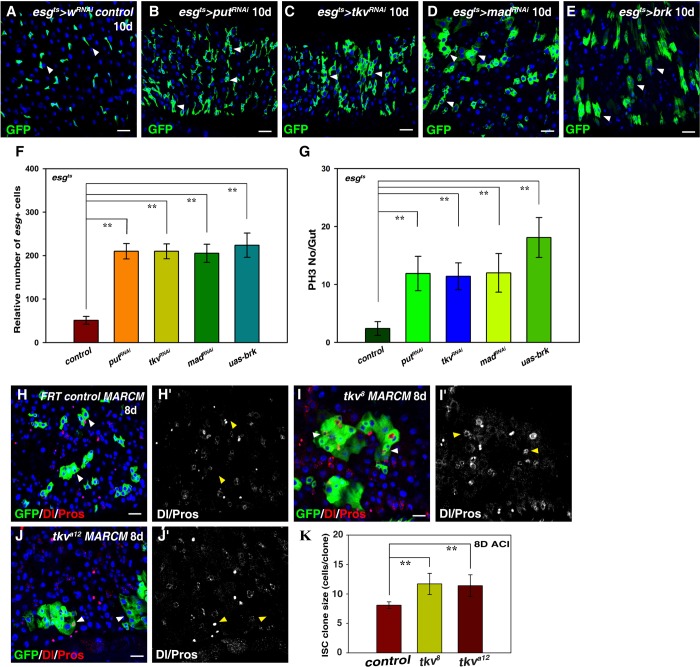
We then explored whether the defects observed in HS-depletion intestines are direct consequences of Dpp signal inactivation. We first depleted the expression of several key components of the Dpp signaling pathway, including the type II receptor Punt (Put), the type I receptor Thickveins (Tkv) and Mother against Dpp (Mad), in the progenitors using functional RNAi constructs by the esgts driver (Li et al., 2013b; Xu et al., 2018). We found that the number of esg+ cells was dramatically increased in these intestines and many polyploid cells expressed GFP, indicative of midgut homeostasis loss (Fig. 4A–D,F). The observed phenotypes are almost identical to those of HS depletion. We further ectopically expressed brinker (brk) in progenitors to block Dpp signaling (Jaźwińska et al., 1999). Similarly, the number of esg+ cells was dramatically increased in esgts>brk intestines, and esg+ cells formed clusters with many polyploid cells expressing GFP (Fig. 4E,F). Consistently, we observed a significant increase of the number of pH3+ cells in these intestines (Fig. 4F; Fig. S8). These data show that Dpp signaling negatively regulates ISC proliferation and midgut homeostasis under physiological conditions.
Fig. 4.
Dpp signaling in ISCs restricts ISC proliferation. (A) esg+ cells (green) in control flies at 29°C for 10 days (white arrowheads). (B) The number of esg+ cells (green) is dramatically increased in esgts>putRNAi flies at 29°C for 10 days (white arrowheads). (C) The number of esg+ cells (green) is dramatically increased in esgts>tkvRNAi flies at 29°C for 10 days (white arrowheads). (D) The number of esg+ cells (green) is dramatically increased in esgts>madRNAi flies at 29°C for 10 days (white arrowheads). (E) The number of esg+ cells (green) is dramatically increased in esgts>brk flies at 29°C for 10 days (white arrowheads). (F) Quantification of the relative number of esg+ cells in different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. (G) Quantification of the number of pH3/gut in different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. (H) ISC MARCM clones (green) in FRT control (8 days at 25°C, 8D ACI) (white arrowheads). Dl and Pros in red. Split channel for Dl and Pros (H′, in grayscale) (yellow arrowheads). (I,J) The size of tkv8 (I) and tkva12 (J) ISC MARCM clones (green) is significantly increased (8D ACI) (white arrowheads). Dl and Pros in red. Split channel for Dl and Pros (I′,J′, in grayscale) (yellow arrowheads). (K) Quantification of ISC clone size in control and tkv mutants (8D ACI). Note that the size of tkv mutant ECs is smaller than neighboring wild-type ECs. mean±s.d. is shown. n=10. **P<0.01. In all panels except graphs, GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm. Note that images for the control flies in A are reproduced from Fig. 1.
We examined the identity of the esg+ cells upon Dpp signaling inactivation in progenitors. We found that the number of ISCs (by Dl and Dl-lacZ) in esgts>tkvRNAi intestines was significantly increased compared to those in the control flies (Fig. S9A,B, and data not shown). No obvious change in the number of ee cells was observed in these intestines (Fig. S9A,B). We found the number of EBs [by GBE+Su(H)-lacZ] was also significantly increased in esgts>putRNAi and esgts>tkvRNAi intestines compared to those in the control flies (Fig. S9C–F). Consistently, we found that the size of the polyploid GFP+ cells was smaller than fully differentiated ECs, indicating that Dpp signaling may also regulate EC maturation (Fig. S9C–E). Furthermore, we generated tkv ISC clones using the MARCM technique (Lee and Luo, 2001). We utilized two amorphic tkv mutants, tkv8 and tkva12. The ISC clone size of both tkv mutants was significantly increased compared with control clones (Fig. 4H–K). Together, these data demonstrate that Dpp signaling in progenitors negatively regulates ISC proliferation and differentiation to maintain midgut homeostasis under normal conditions.
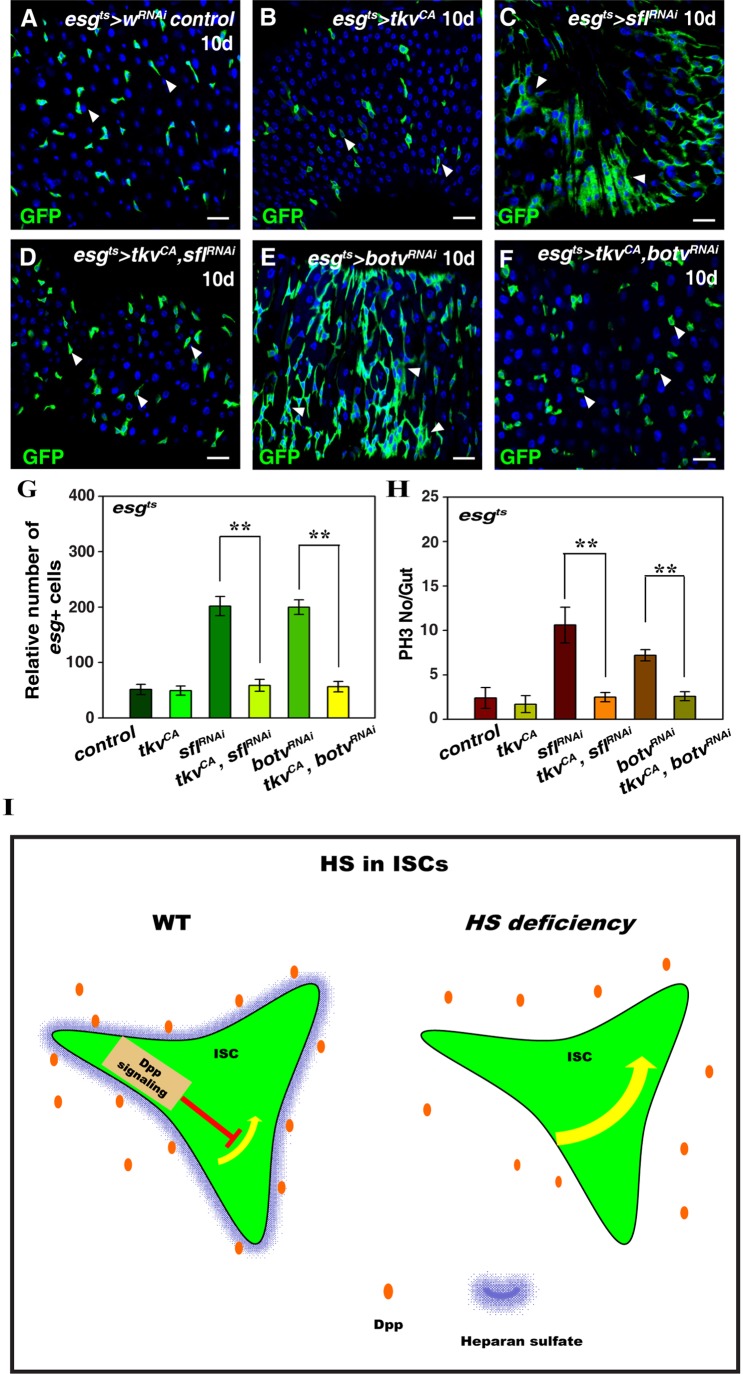
To further confirm that Dpp signaling inactivation is the direct consequence of HS disruption, we performed rescue experiments. Our rationale is: if Dpp signal inactivation is the direct consequence of HS disruption, then restoring Dpp signaling in HS-depleted intestines will completely rescue the defects observed in HS-deficient intestines. Interestingly, co-expression of a constitutively active form of tkv (tkvCA) with either sflRNAi or botvRNAi completely rescued the defects observed in esgts>sflRNAi and esgts>botvRNAi intestines respectively (Fig. 5A–G). Moreover, the increased number of ISCs undergoing mitosis was also completely rescued in these intestines (Fig. 5H; Fig. S10). These data indicate that Dpp signal inactivation is very likely the direct consequence of HS depletion. Taken together, these data show that HS is required for Dpp signaling in progenitors to maintain midgut homeostasis under physiological conditions (Fig. 5I).
Fig. 5.
Ectopic activation of Dpp signaling completely rescued defects observed in the absence of HS. (A) esg+ cells (green) in control flies at 29°C for 10 days (white arrowheads). (B) Progenitors (ISC+EBs, green) in esgts>tkvCA intestines (white arrowheads). (C) The number of esg+ cells (green) is dramatically increased in esgts>sflRNAi flies at 29°C for 10 days (white arrowheads). (D) Expression of tkvCA could completely rescue increased ISC proliferation observed in esgts>sflRNAi intestines (white arrowheads). (E) The number of esg+ cells (green) is dramatically increased in esgts>botvRNAi flies at 29°C for 10 days (white arrowheads). (F) Expression of tkvCA could completely rescue increased ISC proliferation observed in esgts>botvRNAi intestines (white arrowheads). (G) Quantification of the relative number of esg+ cells in different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. (H) Quantification of the number of pH3 per gut in different genotypes indicated. mean±s.d. is shown. n=10–15 intestines. **P<0.01. (I) Model of HS function in progenitor cells. In all panels except graphs, GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm. Note that images for the control flies in A,C and E are reproduced from Fig. 1.
DISCUSSION
Residential stem cells must respond to extrinsic signals to properly adjust their proliferation and differentiation rate to maintain tissue homeostasis under normal conditions. However, how extrinsic signals are transduced into ISCs is poorly understood. Here we reveal that HS is required for Dpp signaling in progenitors to maintain midgut homeostasis under normal conditions.
HSPGs are involved in multiple biological processes, including cell proliferation/differentiation, cell adhesion, extracellular matrix assembly and growth factor diffusion/storage (Esko and Selleck, 2002; Gomes et al., 2013; Iozzo, 1998; Kamimura et al., 2011; Lin, 2004; Lin and Perrimon, 2000; Bernfield et al., 1999; Sarrazin et al., 2011; Yan et al., 2010). The highly diversified functions of HSPGs are mediated by the varied nature of the core proteins and specific HS modifications (Esko and Lindahl, 2001; Esko and Selleck, 2002; Lin, 2004). Although HS plays important roles in the diversified functions of HSPGs, the role of HS in ISC proliferation and differentiation under normal homeostasis has not been systemically explored (Guo et al., 2014; Takemura and Nakato, 2017; You et al., 2014). Our data demonstrate that HS in progenitors negatively regulates ISC proliferation and differentiation to maintain midgut homeostasis under normal conditions (Fig. 1). Consistent with our results, a previous study revealed that heparan sulfate 3-O sulfotransferases (Hs3sts) negatively regulate ISC proliferation (Guo et al., 2014). The extracellular endosulfatases, Sulfs (Sulf1 in Drosophila), specifically remove 6-O sulfate groups (at the 6-O position of glucosamine residues) from highly sulfated regions of HS. Interestingly, a recent study found that ISC proliferation was increased in the absence of sulf1 (Takemura and Nakato, 2017). These data indicate that the levels of HS need to be properly controlled for adequate ISC proliferation.
How does HS restrict ISC proliferation? Our data support the notion that under normal conditions, HS is required for Dpp signal activation, which in turn negatively regulates ISC proliferation and differentiation to maintain midgut homeostasis based on the following observations: (1) Dpp signaling was greatly diminished in the absence of HS in ISCs (Figs 1 and 2); (2) Dpp signaling inactivation in progenitors led to increased ISC proliferation and midgut homeostasis loss under normal conditions (Fig. 3); and (3) most importantly, restoring Dpp signaling in the absence of HS in progenitors completely rescued increased ISC proliferation and tissue homeostasis loss (Fig. 5). Although we cannot exclude the possibility that HS may be required for activating other signaling pathways in progenitors, our data favor the notion that HS mainly activates Dpp signaling in progenitors, regardless of the sources of Dpp ligand. Previous studies showed that injury-induced BMP signaling negatively regulates midgut homeostasis; our results indicate that under physiological conditions, Dpp signaling also negatively regulates ISC proliferation and differentiation to maintain tissue homeostasis (Guo et al., 2013; Tian et al., 2017; Zhou et al., 2015). Consistent with previous studies, our results show that Dpp signaling negatively regulates ISC proliferation and differentiation (Guo et al., 2013; Zhou et al., 2015). However, Guo et al.’s, Zhou et al's and our data are contradictory to Tian et al's findings (Tian et al., 2017). We speculate that the paradox may be resulted from the differences in genetic backgrounds, the drivers used and the experimental conditions.
Interestingly, we found that co-expression of dpp could completely rescue the defects observed in esgts>botvRNAi intestines, while co-expression of gbbGFP could only partially rescue the defects observed in esgts>botvRNAi intestines, indicating that Dpp may be more potent than Gbb (Fig. S11). Our finding that HS is required for Dpp signal activation is not unique for ISCs. Previous studies showed that both sfl and dally are required for Dpp signal activation to control germline stem cell (GSC) maintenance in ovary (Guo and Wang, 2009; Hayashi et al., 2009). Therefore, regardless of the sources of Dpp molecules, we reveal that HS is required for Dpp signal activation to maintain midgut homeostasis under normal conditions.
MATERIALS AND METHODS
Fly lines and cultures
Flies were maintained on standard media at 25°C. Crosses were raised at 18°C in humidity controlled incubators, or as otherwise noted. Flies hatched in 18°C incubators (2–3 days old) were picked and transferred to 29°C incubator, unless otherwise specified. Flies were transferred to new vials with fresh food every day, and dissected at time points specified in the text. In all experiments, only the female posterior midgut was analyzed. Information for alleles and transgenes used in this study can be found either in FlyBase or as noted: esgGal4, UAS-GFP, tubGal80ts (esgts, gift from N. Perrimon, Harvard University), esgGal4, UAS-RFP, tubGal80ts, sflRNAi (BL34601, BL50538), tubGal80ts, tubGal4 (tubts), sglRNAi (BL65348), ttvRNAi (BL51480), sotvRNAi (BL52883), botvRNAi (GD2083, BL61257), dallyRNAi (BL33952), dlpRNAi (BL34089, BL34091), Upd1RNAi (BL33680, BL28722), Upd2RNAi (BL33949, BL33988), Upd3RNAi (BL32859, BL28575), GBE+Su(H)-lacZ (gift from S. Bray, University of Cambridge) (Furriols and Bray, 2001), 10XSTATGFP (gift from G. Baeg, National University of Singapore) (Bach et al., 2007), esg-lacZB7–2-22, tubGal4ts, Dl05151 (Dl-lacZ), tkvRNAi (VDRC3059, NIG 14026R-1, and HMS02185), putRNAi (GL00069, HMS01944 and NIG 7904R-2), madRNAi (GL01527 and GLV21013), w(white)RNAi (BL33623) and/or Gal4RNAi (HMS504, from TRiP at Harvard Medical School) were used as control, FRT40A-tkv8, FRT40A-tkva12, UAS-brk, UAS-tkvQ253D (tkvCA), dallyP1 (Nakato et al., 1995), dally06464 (dallyP2, BL11685). hsFlp, ActGal4, UAS-GFP; FRT40A-tubGal80 (for MARCM clonal analysis), UAS-dpp, UAS-gbb-GFP (BL63507 and BL63508).
RNAi knockdown and overexpression experiments
To address gene function in ISCs, esgGal4, UAS-GFP, tubGal80ts (esgts) was used, and crosses (unless stated otherwise) were maintained at 18°C to bypass potential requirements during early developmental stages. 2–3 days old progeny with the desired genotypes were collected after eclosion and maintained at 29°C to inactivate Gal80ts before dissection and immunostaining. The flies were transferred to new vials with fresh food every day. Both UAS-dsRNA and UAS-shRNA transgene stocks were used in this study. If possible, several dsRNA or shRNA lines were tested for each gene (the lines listed above showed similar phenotypes), and one or two RNAi lines were used for detailed study. To detect JAK/STAT signaling, esgGal4, UAS-RFP; 10XSTATGFP, tubGal80ts driver was used. The time points that the flies are analyzed/dissected were indicated in the text.
Immunostainings and fluorescence microscopy
For standard immunostaining, intestines were dissected in 1× PBS (10 mM NaH2PO4/Na2HPO4, 175 mM NaCl, pH 7.4), and fixed in 4% paraformaldehyde for 25 min at room temperature. Samples were rinsed, washed with 1× PBT (0.1% Triton X-100 in 1× PBS) and blocked in 5% horse serum in 1× PBT for 45 min. Embryos were fixed and stained following standard protocol. Primary antibodies were added to the samples and incubated at 4°C overnight. The following primary antibodies were used: mouse mAb anti-Dl [C594.9B, 1:50, developed by S. Artavanis-Tsakonas, Developmental Studies Hybridoma Bank (DSHB)], mouse mAb anti-Prospero (MR1A, 1:100, developed by C.Q. Doe, DSHB), mouse mAb anti-Dlp (13G8, 1:100, developed by P. A. Beachy, DSHB), rabbit mAb anti-pMAD3 (Epitomics, 1:200), rabbit anti-β-glactosidase (Cappel, 1:5000), mouse anti-β-glactosidase (Cell Signaling, 1:1000), rabbit anti-PDM1 (gift from Xaohang Yang, Zhejiang University, 1:1000), rabbit anti-pH3 (pSer10, Millipore, 1:2000, USA) and mouse anti-HS (clone F58-10E4 and F69-3G10, 1:100, Amsbio). For 3G10 staining, fixed intestines were pretreated by heparanase III (2 U/ml, Sigma-Aldrich) at 37°C for 2 h to expose the neo-epitope site. The primary antibodies were detected by fluorescent-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories. Secondary antibodies were incubated for 2 h at room temperature. DAPI (Sigma-Aldrich, 0.1 μg/ml) was added after secondary antibody staining. The samples were mounted in mounting medium (70% glycerol containing 2.5% DABCO). All images were captured by a Zeiss LSM780 inverted confocal microscope, and were processed in Adobe Photoshop and Illustrator.
MARCM ISC clone analysis
The clonal analyses were achieved using the MARCM system. The ISC clones were induced by heat shocking 3–5 day-old adult flies at 37°C for 60 min. The flies were maintained at 25°C incubator and transferred to new vials with fresh food every day. The sizes of the marked clones were assayed at 8 days after clone induction (8D ACI, clones from at least 10 midguts for each genotype were assayed).
Signal quantification
Image-Pro-Plus 6.0 software was used for pMAD signal quantification. Two parameters, integrated optical density (IOD) and area, were used in the analysis. For pMAD signal quantification in ISCs, a pixel filter was set to ensure that the area of interest did not include objects larger than 20 pixels, excluding esg− cells (which are ECs and ee cells). IOD value per ISC was used. At least four different images were analyzed for each sample.
qRT-PCR
RNA was extracted from 30 flies or guts using TRIzol (Invitrogen). RNA was cleaned using RNAeasy (QIAGEN), and complementary DNA (cDNA) was synthesized using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed using the iScript one-step RT-PCR SYBR green kit (Bio-Rad). Data were acquired using an iQ5 System (Bio-Rad). qRT-PCR was performed in duplicate on each of three independent biological replicates. All results are presented as mean±s.d. of the biological replicates. The ribosomal gene RpL11 was used as the normalization control. Primers used for qRT-PCR can be found in Table S1.
Data analysis
pH3 numbers were scored manually under Zeiss Imager Z2/LSM780 microscope for indicated genotypes. To determine the relative number of esg+ cells per confocal image (including esg>GFP+, esg-lacZ+ and 10xSTATGFP+), confocal images of 40× lens/1.0 zoom from a defined posterior midgut region (R4-R5 regions) of different genotypes indicated were acquired. The relative number of esg+ and GBE+Su(H)-lacZ+ cells was determined using Image-Pro Plus software from each confocal image. The number of intestines scored is indicated in the text. Fluorescence intensity of 10xSTATGFP and HS was measured using Image Pro Plus 6.0 (measure/count function). The data are presented as the means±standard deviation and two-tailed Student's t-tests were performed for statistical comparisons. PEMS 3.1 software was used for s.d. analyses and Sigma Plot software for graph generation. *P<0.05; **P<0.01. The graphs were further modified using Adobe Photoshop and Illustrator.
Supplementary Material
Acknowledgements
We are grateful to Norbert Perrimon, Sarah Bray, Xinhua Lin, Steven Hou, Gyeong-Hun Baeg, Rongwen Xi, Xiaohang Yang and Yu Cai for generous gifts of reagents; the Bloomington Stock Center, VDRC, NIG-FLY Center, TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and Tsinghua Fly Center (THFC) for fly stocks; and DSHB for antibodies. We apologize to the colleagues whose work could not be cited because of space restrictions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Z.L.; Methodology: R.K., L.S.; Formal analysis: H.M., Huiqing Zhao, F.L., R.K., Z.L.; Investigation: H.M., Huiqing Zhao, F.L., Hang Zhao, R.K., L.S., M.W.; Data curation: Z.L.; Writing - original draft: Z.L.; Writing - review & editing: Z.L.; Supervision: Z.L.; Project administration: Z.L.; Funding acquisition: Z.L.
Funding
This work is supported by grants from the National Natural Science Foundation of China (nos. 31471384, 31972893 and 31271582), and the Beijing Natural Science Foundation (nos. KZ201910028040 and 5162004).
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.047126.supplemental
References
- Amcheslavsky A., Jiang J. and Ip Y. T. (2009). Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49-61. 10.1016/j.stem.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach E. A., Ekas L. A., Ayala-Camargo A., Flaherty M. S., Lee H., Perrimon N. and Baeg G.-H. (2007). GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323-331. 10.1016/j.modgep.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Beebe K., Lee W.-C. and Micchelli C. A. (2010). JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 338, 28-37. 10.1016/j.ydbio.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Belenkaya T. Y., Han C., Yan D., Opoka R. J., Khodoun M., Liu H. and Lin X. (2004). Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell 119, 231-244. 10.1016/j.cell.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Bellaiche Y., The I. and Perrimon N. (1998). Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85 10.1038/27932 [DOI] [PubMed] [Google Scholar]
- Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J. and Zako M. (1999). Functions of cell surface Heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729-777. 10.1146/annurev.biochem.68.1.729 [DOI] [PubMed] [Google Scholar]
- Binari R. C., Staveley . E., Johnson W. A., Godavarti R., Sasisekharan R. and Manoukian A. S. (1997). Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development 124, 2623-2632. [DOI] [PubMed] [Google Scholar]
- Biteau B. and Jasper H. (2011). EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045-1055. 10.1242/dev.056671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B. and Jasper H. (2014). Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 7, 1867-1875. 10.1016/j.celrep.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann D. J., Duncan J. E., Staatz W., Selleck S. and Warrior R. (2004). Abrogation of heparan sulfate synthesis in Drosophila disrupts the Wingless, Hedgehog and Decapentaplegic signaling pathways. Development 131, 1927-1938. 10.1242/dev.01061 [DOI] [PubMed] [Google Scholar]
- Brawley C. and Matunis E. (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331-1334. 10.1126/science.1097676 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Chakrabarti S. and Lemaitre B. (2009). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333-2344. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A. and Batlle E. (2009). Intestinal stem cells in mammals and Drosophila. Cell Stem Cell 4, 124-127. 10.1016/j.stem.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Dudzic J. P., Li X., Collas E. J., Boquete J.-P. and Lemaitre B. (2016). Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 12, e1006089 10.1371/journal.pgen.1006089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu N., Huang H., Cai T. and Xi R. (2016). A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. eLife 5, e14330 10.7554/eLife.14330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu N., Wang C., Huang P., Huang H., Jin Z., Yu Z., Cai T., Jiao R. and Xi R. (2018). Transient Scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat. Cell Biol. 20, 152-161. 10.1038/s41556-017-0020-0 [DOI] [PubMed] [Google Scholar]
- Choi N. H., Lucchetta E. and Ohlstein B. (2011). Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl. Acad. Sci. USA 108, 18702-18707. 10.1073/pnas.1109348108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J. B., Stefanatos R. K., Scopelliti A., Vidal M. and Sansom O. J. (2012). Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 31, 3901-3917. 10.1038/emboj.2012.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N., Nahm M., Lee S. and Broadie K. (2012). A targeted glycan-related gene screen reveals heparan sulfate proteoglycan sulfation regulates WNT and BMP trans-synaptic signaling. PLoS Genet. 8, e1003031 10.1371/journal.pgen.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Navascués J., Perdigoto C. N., Bian Y., Schneider M. H., Bardin A. J., Martínez-Arias A. and Simons B. D. (2012). Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 31, 2473-2485. 10.1038/emboj.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A. (2012). Intestinal stem cells: no longer immortal but ever so clever. EMBO J. 31, 2441-2443. 10.1038/emboj.2012.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko J. D. and Lindahl U. (2001). Molecular diversity of heparan sulfate. J. Clin. Invest. 108, 169-173. 10.1172/JCI200113530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko J. D. and Selleck S. B. (2002). Order out of chaos: assembly of ligand binding sites in Heparan sulfate. Annu. Rev. Biochem. 71, 435-471. 10.1146/annurev.biochem.71.110601.135458 [DOI] [PubMed] [Google Scholar]
- Filmus J., Capurro M. and Rast J. (2008). Glypicans. Genome Biol. 9, 224 10.1186/gb-2008-9-5-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise M., Takeo S., Kamimura K., Matsuo T., Aigaki T., Izumi S. and Nakato H. (2003). Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development 130, 1515-1522. 10.1242/dev.00379 [DOI] [PubMed] [Google Scholar]
- Furriols M. and Bray S. (2001). A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11, 60-64. 10.1016/S0960-9822(00)00044-0 [DOI] [PubMed] [Google Scholar]
- Gomes A. M., Stelling M. P. and Pavão M. S. G. (2013). Heparan sulfate and heparanase as modulators of breast cancer progression. BioMed Res. Int. 2013, 11 10.1155/2013/852093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas S., Conder R. and Knoblich J. A. (2012). The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529-540. 10.1016/j.stem.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Ohlstein B. (2015). Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science 350 10.1126/science.aab0988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Wang Z. (2009). The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development 136, 3627-3635. 10.1242/dev.036939 [DOI] [PubMed] [Google Scholar]
- Guo Z., Driver I. and Ohlstein B. (2013). Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell Biol. 201, 945-961. 10.1083/jcb.201302049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Li Z. and Lin X. (2014). Hs3st-A and Hs3st-B regulate intestinal homeostasis in Drosophila adult midgut. Cell. Signal. 26, 2317-2325. 10.1016/j.cellsig.2014.07.015 [DOI] [PubMed] [Google Scholar]
- Han C., Belenkaya T. Y., Khodoun M., Tauchi M., Lin X. and Lin X. (2004). Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development 131, 1563-1575. 10.1242/dev.01051 [DOI] [PubMed] [Google Scholar]
- Han H., Pan C., Liu C., Lv X., Yang X., Xiong Y., Lu Y., Wu W., Han J., Zhou Z. et al. (2015). Gut–neuron interaction via Hh signaling regulates intestinal progenitor cell differentiation in Drosophila. Cell Discov. 1, 15006 10.1038/celldisc.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Kobayashi S. and Nakato H. (2009). Drosophila glypicans regulate the germline stem cell niche. J. Cell Biol. 187, 473-480. 10.1083/jcb.200904118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo R. V. (1998). Matrix proteoglycans: from molecular design to cellular function. Annu. Rev. Biochem. 67, 609-652. 10.1146/annurev.biochem.67.1.609 [DOI] [PubMed] [Google Scholar]
- Jackson S. M., Nakato H., Sugiura M., Jannuzi A., Oakes R., Kaluza V., Golden C. and Selleck S. B. (1997). dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development 124, 4113-4120. [DOI] [PubMed] [Google Scholar]
- Jaźwińska A., Kirov N., Wieschaus E., Roth S. and Rushlow C. (1999). The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96, 563-573. 10.1016/S0092-8674(00)80660-1 [DOI] [PubMed] [Google Scholar]
- Jiang H. and Edgar B. A. (2009). EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 136, 483-493. 10.1242/dev.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G. and Edgar B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343-1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M. O., Bravo M.-J., Blumhagen R. Z. and Edgar B. A. (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84-95. 10.1016/j.stem.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Patel P. H., Kohlmaier A., Pavlovic B., Zhang C. and Edgar B. A. (2017). Intestinal stem cell pool regulation in Drosophila. Stem Cell Rep. 8, 1479-1487. 10.1016/j.stemcr.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K., Koyama T., Habuchi H., Ueda R., Masu M., Kimata K. and Nakato H. (2006). Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J. Cell Biol. 174, 773-778. 10.1083/jcb.200603129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K., Maeda N. and Nakato H. (2011). In vivo manipulation of heparan sulfate structure and its effect on Drosophila development. Glycobiology 21, 607-618. 10.1093/glycob/cwq202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Perez J. and Perrimon N. (2010). The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135-4145. 10.1242/dev.060483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. and Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251-254. 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Lee W.-C., Beebe K., Sudmeier L. and Micchelli C. A. (2009). Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development 136, 2255-2264. 10.1242/dev.035196 [DOI] [PubMed] [Google Scholar]
- Levings D. C. and Nakato H. (2017). Loss of heparan sulfate in the niche leads to tumor-like germ cell growth in the Drosophila testis. Glycobiology 28, 32-41. 10.1093/glycob/cwx090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Qi Y. and Jasper H. (2013a). Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell Rep. 4, 10-18. 10.1016/j.celrep.2013.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Han L., Shi L. and Lin X. (2013b). Trachea-derived Dpp controls adult midgut homeostasis in Drosophila. Dev. Cell 24, 133-143. 10.1016/j.devcel.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Li Z., Guo Y., Han L., Zhang Y., Shi L., Huang X. and Lin X. (2014). Debra-mediated Ci degradation controls tissue homeostasis in Drosophila adult midgut. Stem Cell Rep. 2, 135-144. 10.1016/j.stemcr.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. (2004). Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131, 6009-6021. 10.1242/dev.01522 [DOI] [PubMed] [Google Scholar]
- Lin H. (2008). Cell biology of stem cells: an enigma of asymmetry and self-renewal. J. Cell Biol. 180, 257-260. 10.1083/jcb.200712159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. and Perrimon N. (1999). Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature 400, 281 10.1038/22343 [DOI] [PubMed] [Google Scholar]
- Lin X. and Perrimon N. (2000). Role of heparan sulfate proteoglycans in cell-cell signaling in Drosophila. Matrix Biol. 19, 303-307. 10.1016/S0945-053X(00)00073-1 [DOI] [PubMed] [Google Scholar]
- Lin X. and Perrimon N. (2002). Developmental roles of heparan sulfate proteoglycans in Drosophila. Glycoconj. J. 19, 363-368. 10.1023/A:1025329323438 [DOI] [PubMed] [Google Scholar]
- Lin G. and Xi R. (2008). Intestinal stem cell, muscular niche and Wingless signaling. Fly (Austin) 2, 310-312. 10.4161/fly.7428 [DOI] [PubMed] [Google Scholar]
- Lin G., Xu N. and Xi R. (2008). Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119-1123. 10.1038/nature07329 [DOI] [PubMed] [Google Scholar]
- Liu M., Lim T. M. and Cai Y. (2010). The Drosophila female germline stem cell lineage acts to spatially restrict DPP function within the Niche. Sci. Signal. 3, ra57 10.1126/scisignal.2000740 [DOI] [PubMed] [Google Scholar]
- Lu Y. and Li Z. (2015). No intestinal stem cell regeneration after complete progenitor ablation in Drosophila adult midgut. J. Genet. Genomics 42, 83-86. 10.1016/j.jgg.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Martorell Ò., Barriga F. M., Merlos-Suárez A., Stephan-Otto Attolini C., Casanova J., Batlle E., Sancho E. and Casali A. (2014). Iro/IRX transcription factors negatively regulate Dpp/TGF-β pathway activity during intestinal tumorigenesis. EMBO Rep. 15, 1210-1218. 10.15252/embr.201438622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A. and Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475-479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Mii Y., Yamamoto T., Takada R., Mizumoto S., Matsuyama M., Yamada S., Takada S. and Taira M. (2017). Roles of two types of heparan sulfate clusters in Wnt distribution and signaling in Xenopus. Nat. Commun. 8, 1973 10.1038/s41467-017-02076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. J. and Spradling A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598-611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakato H., Futch T. A. and Selleck S. B. (1995). The division abnormally delayed, dally, gene: a putative integral membrane proteoglycan required for cell division patterning during postembryonic development of the nervous system in Drosophila. Development 121, 3687-3702. [DOI] [PubMed] [Google Scholar]
- O'Brien, Lucy E., Soliman S. S., Li X. and Bilder D. (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603-614. 10.1016/j.cell.2011.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B. and Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Ohlstein B. and Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science 315, 988-992. 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Perdigoto C. N., Schweisguth F. and Bardin A. J. (2011). Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138, 4585-4595. 10.1242/dev.065292 [DOI] [PubMed] [Google Scholar]
- Radtke F. and Clevers H. (2005). Self-renewal and cancer of the gut: two sides of a coin. Science 307, 1904-1909. 10.1126/science.1104815 [DOI] [PubMed] [Google Scholar]
- Raff M. (2003). Adult stem cell plasticity: fact or artifact? Annu. Rev. Cell Dev. Biol. 19, 1-22. 10.1146/annurev.cellbio.19.111301.143037 [DOI] [PubMed] [Google Scholar]
- Rahman M. M., Franch-Marro X., Maestro J. L., Martin D. and Casali A. (2017). Local Juvenile Hormone activity regulates gut homeostasis and tumor growth in adult Drosophila. Sci. Rep. 7, 11677 10.1038/s41598-017-11199-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Wang B., Yue T., Yun E.-Y., Ip Y. T. and Jiang J. (2010). Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA 107, 21064-21069. 10.1073/pnas.1012759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Zhang Y., Li M., Wu L., Wang G., Baeg G.-H., You J., Li Z. and Lin X. (2015). Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet. 11, e1005180 10.1371/journal.pgen.1005180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W. C. and Esko J. D. (2011). Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell J. C., Wisidagama D. R., Bensard C., Zhao H., Wei P., Tanner J., Flores A., Mohlman J., Sorensen L. K., Earl C. S. et al. (2017). Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat. Cell Biol. 19, 1027 10.1038/ncb3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R. L., Kohlmaier A., Polesello C., Veelken C., Edgar B. A. and Tapon N. (2010). The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147-4158. 10.1242/dev.052506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. R., Zeng X., Zhao J., Liu Y., Hou G., Liu H. and Hou S. X. (2016). The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 538, 109 10.1038/nature19788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D. Y. R. (2005). No organ left behind: tales of gut development and evolution. Science 307, 1902-1904. 10.1126/science.1108709 [DOI] [PubMed] [Google Scholar]
- Staley B. K. and Irvine K. D. (2010). Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580-1587. 10.1016/j.cub.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y., Ozawa Y., Sato M., Watanabe A. and Tabata T. (2004). Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development 131, 73-82. 10.1242/dev.00913 [DOI] [PubMed] [Google Scholar]
- Takemura M. and Nakato H. (2017). Drosophila Sulf1 is required for the termination of intestinal stem cell division during regeneration. J. Cell Sci. 130, 332-343. 10.1242/jcs.195305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A. and Jiang J. (2014). Intestinal epithelium-derived BMP controls stem cell self-renewal in Drosophila adult midgut. eLife 3, e01857 10.7554/eLife.01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A., Shi Q., Jiang A., Li S., Wang B. and Jiang J. (2015). Injury-stimulated Hedgehog signaling promotes regenerative proliferation of Drosophila intestinal stem cells. J. Cell Biol. 208, 807-819. 10.1083/jcb.201409025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A., Wang B. and Jiang J. (2017). Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc. Natl. Acad. Sci. USA 114, E2699-E2708. 10.1073/pnas.1617790114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Kinoshita-Toyoda A., Fox B. and Selleck S. B. (2000a). Structural analysis of Glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu: Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J. Biol. Chem. 275, 21856-21861. 10.1074/jbc.M003540200 [DOI] [PubMed] [Google Scholar]
- Toyoda H., Kinoshita-Toyoda A. and Selleck S. B. (2000b). Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects Heparan sulfate in vivo. J. Biol. Chem. 275, 2269-2275. 10.1074/jbc.275.4.2269 [DOI] [PubMed] [Google Scholar]
- Wang P. and Hou S. X. (2010). Regulation of intestinal stem cells in mammals and Drosophila. J. Cell. Physiol. 222, 33-37. 10.1002/jcp.21928 [DOI] [PubMed] [Google Scholar]
- Xie T. and Spradling A. C. (1998). Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251-260. 10.1016/S0092-8674(00)81424-5 [DOI] [PubMed] [Google Scholar]
- Xu N., Wang S. Q., Tan D., Gao Y., Lin G. and Xi R. (2011). EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev. Biol. 354, 31-43. 10.1016/j.ydbio.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Xu R., Li J., Zhao H., Kong R., Wei M., Shi L., Bai G. and Li Z. (2018). Self-restrained regulation of stem cell niche activity by niche components in the Drosophila testis. Dev. Biol. 439, 42-51. 10.1016/j.ydbio.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Yan D. and Lin X. (2009). Shaping morphogen gradients by proteoglycans. Cold Spring Harb. Perspect. Biol. 1, a002493 10.1101/cshperspect.a002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wu Y., Yang Y., Belenkaya T. Y., Tang X. and Lin X. (2010). The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development 137, 2033-2044. 10.1242/dev.045740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T. M., Chia L. A., Kosinski C. M. and Kuo C. J. (2011). Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell. Mol. Life Sci. 68, 2513-2523. 10.1007/s00018-011-0687-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Zhang Y., Li Z., Lou Z., Jin L. and Lin X. (2014). Drosophila perlecan regulates intestinal stem cell activity via cell-matrix attachment. Stem Cell Rep. 2, 761-769. 10.1016/j.stemcr.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Griffiths L. R. and Haupt L. M. (2017). Exploiting Heparan sulfate proteoglycans in human neurogenesis—controlling lineage specification and fate. Front. Integr. Neurosci. 11 10.3389/fnint.2017.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Han L., Singh S. R., Liu H., Neumüller R. A., Yan D., Hu Y., Liu Y., Liu W., Lin X. et al. (2015). Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 10, 1226-1238. 10.1016/j.celrep.2015.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Boquete J.-P. and Lemaitre B. (2017). A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila. PLoS Genet. 13, e1006854 10.1371/journal.pgen.1006854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., You J., Ren W. and Lin X. (2013). Drosophila glypicans Dally and Dally-like are essential regulators for JAK/STAT signaling and Unpaired distribution in eye development. Dev. Biol. 375, 23-32. 10.1016/j.ydbio.2012.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Florescu S., Boettcher A.-L., Luo L., Dutta D., Kerr G., Cai Y., Edgar B. A. and Boutros M. (2015). Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev. Biol. 399, 189-203. 10.1016/j.ydbio.2014.12.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.