Abstract
Platinum-containing doublet chemotherapy is the cornerstone of lung cancer treatment; however, cisplatin resistance is a major obstacle in the treatment of lung cancer. However, the mechanism underlying this resistance has not been fully elucidated. Previous studies have shown that serum apurinic/apyrimidinic endonuclease 1 (APE1) levels in patients with NSCLC are inversely associated with progression-free survival after platinum-containing doublet chemotherapy, and can serve as a biomarker for predicting disease prognosis and treatment efficacy. The present study was designed to investigate the role played by APE1 in the resistance of lung cancer to cisplatin. The levels of mitochondrial apurinic endonuclease 1 (m-APE1) and total APE1 (t-APE1) protein in a cisplatin-resistant A549 cell line (A549/DDP) and cisplatin-sensitive A549 cells were analyzed by western blotting. Mitochondrial membrane potential was detected by using the JC-1 staining method. The cisplatin-resistance of APE1-overexpressing A549 cells and APE1-silenced A549/DDP cells was assessed by cell apoptosis and colony formation assays. The results revealed that cisplatin-resistant A549 cells contained high levels of APE1, and exhibited elevated levels of autophagy. The levels of m-APE1 and t-APE1 protein were increased in the A549/DDP cells when compared with these levels in the A549 cells. Overexpression of APE1 and Mia40 enhanced the cisplatin resistance and autophagy of the A549 cells. APE1 knockdown restored the cisplatin sensitivity and reduced the levels of LC3II and Parkin in the A549/DDP cells, but promoted the release of cytochrome c. Furthermore, Parkin silencing or treatment with 3-methyladenine (3-MA, an autophagy inhibitor) promoted the apoptosis of APE1-overexpressing A549 cells, indicating that Parkin-mediated mitophagy plays an important role in the APE1-induced cisplatin resistance of A549 cells. In conclusion, APE1 promotes the cisplatin resistance of lung cancer cells by inducing Parkin-mediated mitophagy.
Keywords: lung cancer, APE1, cisplatin resistance, mitophagy, autophagy
Introduction
Lung cancer is one of the most common causes of cancer-related morbidity and mortality, and approximately 1.5 million new cases of lung cancer are diagnosed annually worldwide. Furthermore, lung cancer accounts for 13% of all newly diagnosed cancers worldwide (1,2). Approximately 85% of lung tumors are non-small cell lung cancers (NSCLCs), and adenocarcinoma is the most prevalent cancer subtype (3). The high incidences of lung cancer recurrence and metastasis are the two main causes of the failure to successfully treat NSCLC. Standard therapies for patients with locally advanced or metastatic lung cancer consist of radiotherapy and chemotherapy. However, most patients fail to achieve long-term survival with such treatments as they become resistant to chemotherapy (4). Chemotherapy resistance plays an important role in the recurrence and metastasis of NSCLC.
Cisplatin, approved by the FDA in 1978, is one of most effective chemotherapy drugs commonly used in the treatment of various types of solid tumors, including testicular, head and neck, ovarian, esophageal, cervical, and non-small cell lung cancer tumors (5,6). Cisplatin binds to DNA in cellular nuclei and mitochondria to form cisplatin-DNA adducts, which exert cytotoxic effects by blocking the replication and transcription of DNA in cancer cells (7). As cisplatin-based chemotherapy is commonly used to treat patients with advanced NSCLC, cisplatin resistance leads to the failure of this type of chemotherapy (8). Therefore, it is critical that we gain a better understanding of the mechanisms of acquired cisplatin resistance, and develop safe methods for reversing drug resistance. Apurinic/apyrimidinic endonuclease 1 (APE1) is a key, multi-functional DNA repair enzyme that plays important roles in repairing DNA damage, controlling protein reduction/oxidation, and modulating the activity of transcription factors (9,10). The mitochondrial intermembrane space assembly (MIA) pathway and Mia40 protein play pivotal roles in trafficking APE1 into mitochondria via the formation of disulfide bonds (11). APE1 provides most of the total AP endonuclease activity found in human cells, and is essential for proper functioning of the base excision repair (BER) pathway. APE1 also helps to regulate gene transcription, and participates in many important pathways, (e.g. the NF-κB, AP-1, Myb, HIF-1α and p53 pathways) via redox-dependent mechanisms (10,12). Importantly, APE1 accumulates in the cell nuclei and cytoplasm of various tumors, including NSCLC tumors. Abnormal APE1 expression in cancer patients is associated with a poor prognosis, tumor cell invasion, metastasis and angiogenesis, as well as resistance to radiotherapy and chemotherapy (13–15). Previous research has shown that serum APE1 levels in patients with NSCLC are inversely associated with progression-free survival after platinum-containing doublet chemotherapy, and can serve as a biomarker for predicting disease prognosis and treatment efficacy (16). However, the role played by APE1 in the resistance of lung cancer to cisplatin remains unclear.
To explore the role of APE1 in cisplatin resistance, the levels of APE1 and Mia40 expression were assessed in a cisplatin-resistant A549 cell line. Moreover, the cellular behaviors of cisplatin-resistant A549 cells were examined after altering their levels of APE1 and Mia40 expression, and a subsequent Parkin-mediated autophagy analysis was also performed. We believe that our results provide new insights into the cisplatin resistance of NSCLC cells, and will facilitate the development of new clinical therapies.
Materials and methods
Cell culture and establishment of a cisplatin-resistant subline
A549 lung cancer cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone; GE Healthcare), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere containing 5% CO2. A cisplatin-resistant A549 cell line (A549/DDP) was derived from A549 cells by incubating A549 cells with progressively increasing concentrations of cisplatin. After each treatment, the surviving cells were re-expanded and conventionally propagated for 4 generations in cisplatin-free medium. The relative levels of cisplatin resistance were determined using the clonogenic assay.
CCK-8 assay
Viability of the treated cells was determined by using a CCK-8 assay kit (Dojindo, Japan) according to the manufacturer's instructions. Briefly, 5,000 cells in 5 or 100 µl of medium were seeded into each well of a 96-well culture plate. Next, 10 µl of CCK-8 reagent was added to each well and incubated for 2 h at 37°C. After incubation, the absorbance of each well at 450 nm was measured with a microplate reader (Thermo Fisher, Finland).
Colony formation assay
Cells were seeded into the wells of a 12-well plate (1×103 cells per well) and cultured for 10–14 days. After culture, the cells were fixed with 20% methanol and stained with 0.1% crystal violet. Representative microscopic fields of the stained cells were photographed, and the numbers of colonies were counted.
Quantitative real-time polymerase chain reaction (qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from cells according to the manufacturer's instructions; after which, a reverse transcription reagent kit (Takara, Tokyo, Japan) was used to reverse transcribe the total RNA into cDNA. The cDNA was detected according to the protocol used for SYBR Green Master Mix (Thermo Fisher Scientific, Inc.). qPCR analysis for relative expression level of mRNA was determined according to the procedures as follows: 95°C for 2 min, followed by 40 cycles of 94°C for 20 sec, 58°C for 20 sec, 72°C for 20 sec, and finally extension at 72°C for 4 min. The Agilent Stratagene Mx3000P Sequence Detection system (Agilent Technologies) was used to perform qPCR analysis. The primers used for qPCR were synthesized by Invitrogen/Thermo Fisher Scientific, Inc. and their sequences were as follows: APE1 forward, 5′-CCAGCCCTGTATGAGGACC-3′ and reverse, 5′-GGAGCTGACCAGTATTGATGAGA-3′; Mia40 forward, 5′-ATGACCCCAACGATCCATACG-3′ and reverse, 5′-GGGGATAGAGGTCTGGGTATTT-3′; GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′. GAPDH was used as an internal reference gene. The relative levels of gene expression were calculated using the 2−∆∆Cq method (17).
Cell apoptosis assay
The A549 cells (5×105) were seeded in 6-well plates (Corning Inc.) and cultured for 24 h, then the A549 cells were transfected as described below. Forty-eight hours later, the treated cells were collected, washed, fixed, and permeabilized; after which, they were stained using Annexin V-FTIC/PI (KeyGen) according to the manufacturer's instructions. After 15 min of staining, the number of apoptotic cells was determined by flow cytometry (BD Biosciences).
Mitochondrial membrane potential
The mitochondrial membrane potential was detected by using the JC-1 staining method. Briefly, the A549 cells (5×105) were seeded in 6-well plates (Corning Inc.) and cultured for 24 h. Then the A549 cells were transfected as described below. Forty-eight hours later, the cells were harvested, washed with PBS, and then incubated for 30 min with 5 µM JC-1 (Cell Signaling Technology). After incubation, the cells were analyzed by flow cytometry (BD Biosciences).
Isolation of subcellular fractionation
The cytosolic fractions from the cultured A549 cells were isolated using a biochemical fractionation method using Mitochondria Isolation Kit for Cultured Cells (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Briefly, the cells after the indicated treatment were lysed in lysis buffer A [20 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 2 mmol/l EDTA, and 1 % Triton X-100 with protease and phosphatase inhibitors] for 20 min at 4°C, and mitochondria were extracted in a Dounce homogenizer in mitochondrial buffer. The supernatant was further centrifuged to pellet the mitochondria, and the resulting supernatant was stored as the cytosolic fraction. Subcellular fractionation and western blot analysis were used to detect the cytochrome c content in the cytosol and mitochondria.
Western blot analysis
Cells (2×106) were collected and lysed in RIPA buffer. A BCA Protein Assay Kit (Pierce Biotechnology; Thermo Fisher Scientific, Inc.) was used to measure the protein concentrations in the lysates. Next, 50 µg samples of total protein were separated by 12% SDS-PAGE, and the separated protein bands were transferred onto PVDF membranes (EMD Millipore). The membranes were first incubated with primary antibodies against APE1 (Abcam, cat. no. ab137708; dilution 1:1,000), Mia40 (Abcam; cat. no. ab87033, dilution 1:1,000), GAPDH (Abcam; ab8245, dilution 1:5,000), COX4 (Abcam; cat. no. ab33985, dilution 1:1,000), LC3 (Abcam; cat. no. ab48394, dilution 1:1,000), cytochrome c (Abcam; cat. no. ab133504, dilution 1:1,000), and Parkin (Abcam; cat. no. ab77924, dilution 1:1,000), followed by incubation with an HRP-conjugated goat anti-rabbit antibody (Wuhan Boster Biological Technology, Ltd.; cat. no. BA1054, dilution 1:20,000) or HRP-conjugated goat anti-mouse antibody (Wuhan Boster Biological Technology, Ltd.; cat. no. BA1051, dilution 1:20,000). Immunostaining of the protein bands was detected by enhanced chemiluminescence (ECL) reaction (Kibbutz Beit Haemek), and staining intensity was analyzed with an Alpha Innotech Flour Chem-FC2 imaging system (Alpha Innotech).
Cell transfection
To force overexpression of APE1 and Mia40 in cells, pcDNA 3.1 vectors (Genechem) containing APE1 or Mia40 plasmids were co-transfected into the A549 cells using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). An empty vector served as a negative control. Specific small interfering RNAs (siRNAs) purchased from RiboBio, Inc. were used to knock down APE1 and Parkin expression in the cells. The siRNAs used were si-APE1 (5′-UACUCCAGUCGUACCAGACCUdTdT-3′), si-Parkin (5′-AUUUCUUGACCUUUUCUCCACdTdT-3′), and a scrambled control siRNA (5′-CCAUGAGGUCAUGGUCUGdTdT-3′). Lipofectamine 2000 transfection reagent was used for all siRNA transfections. After 48 h of transfection with plasmid or siRNA, the transfected A549 cells were used for subsequent experiments.
Immunofluorescence and confocal microscopy
Briefly, treated cells were fixed in 4% paraformaldehyde and then permeabilized with 0.5% Triton X-100 for 15 min. Next, the cells were washed with PBS, blocked with 5% BSA in PBS, and then incubated with the primary antibody (anti-APE1, dilution 1:500), overnight at 4°C, followed by incubation with a secondary antibody that was conjugated with Alexa Fluorescence 568 (Invitrogen, Thermo Fisher Scientific, Inc.; cat. no. A-11011; dilution 1:1,000.). DAPI was used for nuclear staining (Sigma-Aldrich, Merck KGaA; cat. no. D9542, dilution 1:5,000). The stained cells were visualized by confocal fluorescence microscopy.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.) and SPSS version 16.0 software (SPSS, Inc.). Results are presented as the mean ± SEM of values obtained from at least three independent experiments. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Dunnett's test. A P-value <0.05 was considered statistically significant.
Results
Cisplatin-resistant A549 cells exhibit high levels of APE1 and autophagy
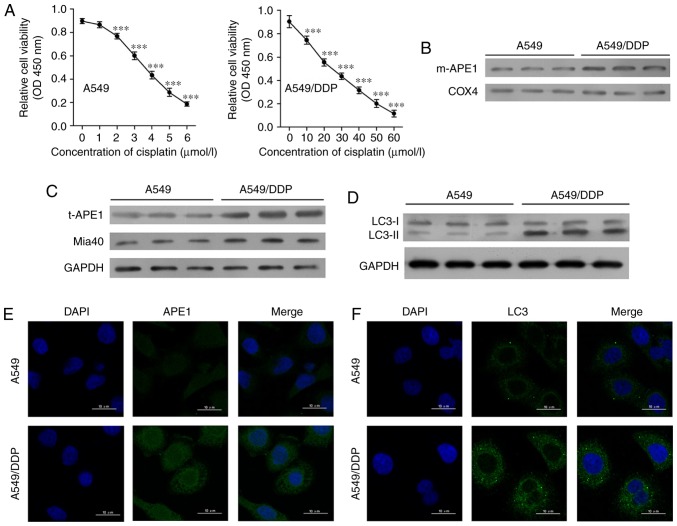
First, we established a cisplatin-resistant A549 cell line (A549/DDP) by incubating A549 cells with progressively higher concentrations of cisplatin. The A549 cells were assessed for viability after they had been treated with 0, 1, 2, 3, 4, 5 and 6 µmol/l cisplatin, respectively (Fig. 1A). A549/DDP cells were assessed for viability after treatment with 0, 10, 20, 30, 40, 50, and 60 µmol/l cisplatin, respectively. The A549/DDP cell line was significantly more resistant to cisplatin, as the IC50 values for the A549 and A549/DDP cells were 4.10 and 25.91 µmol/l, respectively. Next, we examined the levels of mitochondrial APE1 (m-APE1) and total APE1 (t-APE1) in both cell lines. We found that both the m-APE1 and t-APE1 protein levels were increased in the A549/DDP cells when compared with those in the A549 cells (Fig. 1B and C). Immunofluorescence assays confirmed the increased t-APE1 protein levels in A549/DDP cells (Fig. 1E), suggesting that APE1 might be transported into the mitochondria. As the MIA pathway is responsible for APE1 trafficking into the mitochondria, and Mia40 is directly involved in APE1's mitochondrial translocation, we analyzed the levels of Mia40 protein. We found that the levels of Mia40 protein were also increased in the A549/DDP cells when compared with those in the A549 cells (Fig. 1C). Furthermore, our findings suggest that the increased m-APE1 protein levels in A549/DDP cells are mediated by Mia40.
Figure 1.
Cisplatin-resistant A549 cells exhibit high levels of APE1 and autophagy. (A) A549 cells were treated with 0, 1, 2, 3, 4, 5 and 6 µmol/l cisplatin for 24 h, and the A549/DDP cells were treated with 0, 10, 20, 30, 40, 50 and 60 µmol/l cisplatin for 24 h. Cell viability was assessed by the CCK-8 assay. (B) Western blot analysis was performed to analyze the levels of mitochondrial APE1 (m-APE1) protein in A549/DDP and A549 cells. COX4 was used as loading control for the mitochondrial APE1 protein. (C) Western blot analysis of the total APE1 (t-APE1) and Mia40 protein levels in A549/DDP and A549 cells. GAPDH was used as a loading control. (D) Western blot analysis of the LC3 protein levels in A549/DDP and A549 cells. (E and F) Immunofluorescence assays were performed to assess the total APE1 and total LC3 levels in A549 and A549/DDP cells. Data represent results obtained from three independent experiments (mean ± SEM of triplicate samples). ***P<0.001, vs. 0 µmol/l cisplatin-treated A549 or A549/DDP cells. APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
Accumulating evidence suggests that the autophagy process in cancer cells is involved in their resistance to chemotherapy (18–20). As shown in Fig. 1D and F, the levels of LC3II were increased in the A549/DDP cells. This finding suggests that cisplatin-resistant A549 cells have higher levels of autophagy.
APE1 and Mia40 overexpression enhances the cisplatin resistance of A549 cells
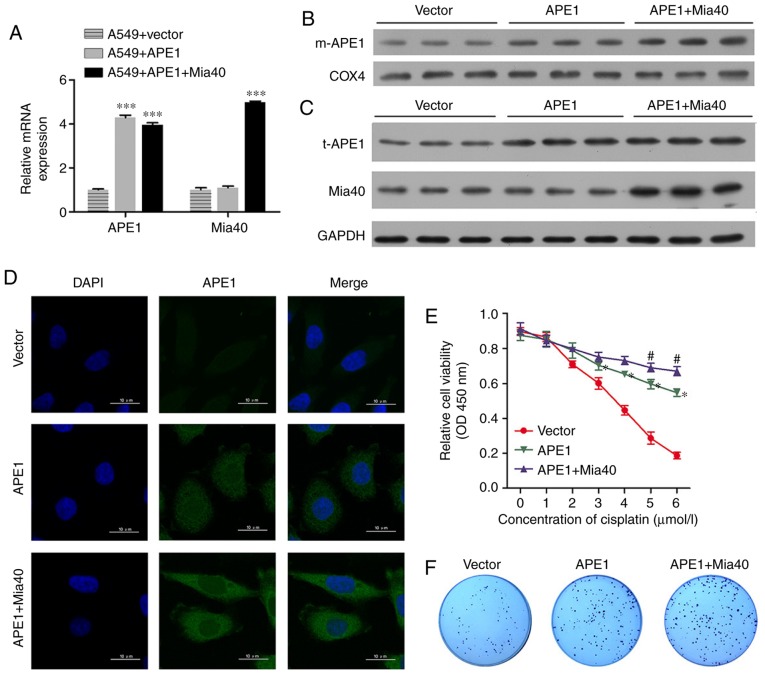
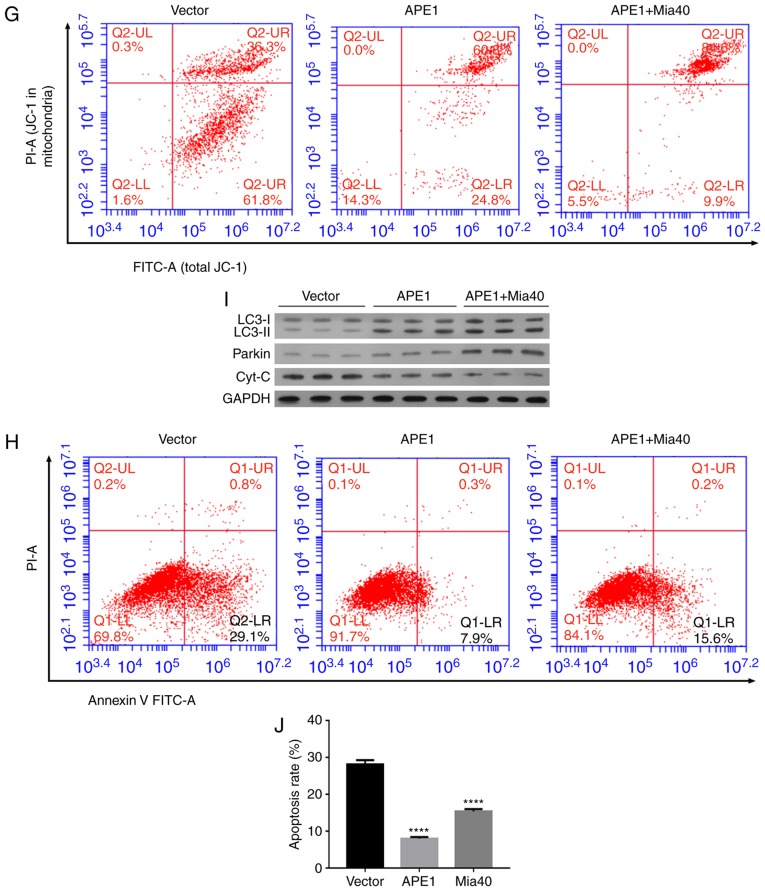
To investigate the roles of APE1 and Mia40 in A549 cells, we performed transfection studies that forced the overexpression of APE1 and Mia40 in A549 cells. The transfection efficiencies are shown in Fig. 2A and Fig. S1A. Interestingly, we found that overexpression of Mia40 increased the levels of mitochondrial APE1 (m-APE1) protein in the A549 cells (Fig. 2B). Moreover, the levels of t-APE1 and Mia40 protein in the A549 cells were increased after the cells were transfected with APE1 and Mia40 plasmids, respectively (Fig. 2C). These results indicate that Mia40 promotes the translocation of APE1 into mitochondria. Immunofluorescence assays confirmed the increase in APE1 expression (Fig. 2D). We also assessed the viability of A549 cells overexpressing APE1 and Mia40, and found that overexpression of both APE1 and Mia40 enhanced the cisplatin resistance of the A549 cells (Fig. 2E). Colony-forming assays showed that overexpression of both APE1 and Mia40 enhanced the colony formation efficiency of the A549 cells (Fig. 2F). In addition, APE1 and Mia40 overexpression decreased the apoptosis rate of the A549 cells after treatment with cisplatin (Fig. 2H and J). As APE1 and Mia40 interact with each other via disulfide bond formation, and Mia40 levels directly affect the translocation of APE1 into mitochondria, where it helps to maintain the integrity of mitochondrial DNA (11), it is possible that the overexpression of Mia40 enhances cisplatin resistance by promoting the translocation of APE1 into mitochondria. We next used flow cytometry to assess the mitochondrial membrane potential of transfected A549 cells. As shown in Fig. 2G, the overexpression of both APE1 and Mia40 enhanced the mitochondrial membrane potential. Furthermore, overexpression of APE1 and Mia40 also promoted autophagy in A549 cells by upregulating LC3-II expression, and decreasing the levels of cytochrome c protein in the cytosolic fraction (Fig. 2I). It is possible that APE1 induced Parkin-mediated mitophagy, because as shown in Fig. 2I, overexpression of APE1 increased Parkin expression, and overexpression of Mia40 further increased the levels of Parkin protein in the A549 cells.
Figure 2.
APE1 and Mia40 overexpression increases the cisplatin resistance of A549 cells. A549 cells were transfected with vectors containing an APE1 overexpression plasmid and Mia40 overexpression plasmid as indicated. (A) Transfection efficiency was assessed by qPCR. (B) Western blot analysis of the mitochondrial APE1 (m-APE1) protein levels in A549 cells after transfection. COX4 was used as loading control for mitochondrial APE1. (C) Western blot analysis was used to analyze the total APE1 (t-APE1) and Mia40 protein levels in A549 cells after transfection. GAPDH was used as a loading control. (D) Immunofluorescence assays were performed to assess the total APE1 levels in A549 cells after transfection. (E) The transfected A549 cells were treated with 0, 1, 2, 3, 4, 5, and 6 µmol/l cisplatin for 24 h, and their viability was assessed by the CCK-8 assay. (F) Colony-formation assays were performed to analyze the colony formation efficiency of the transfected A549 cells. *P<0.05, ***P<0.001, vs. the vector group. #P<0.05, vs. the APE1 group. APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3. (G) Flow cytometry was used to analyze the cellular distribution of JC-1 after transfection. (H) Transfected A549 cells were treated with cisplatin, and their rate of apoptosis was analyzed by flow cytometry. (I) The transfected A549 cells were subjected to fractionation to obtain the cytosolic fraction. Western blot analysis was performed to analyze the levels of LC3, total cytochrome c, and Parkin in the transfected A549 cells. (J) The cell apoptosis rate is shown in a histogram. GAPDH was used as loading control. Data represent results obtained from three independent experiments (mean ± SEM of triplicate samples). ****P<0.0001 vs. the vector group. APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
APE1 knockdown promotes the cisplatin sensitivity of A549/DDP cells
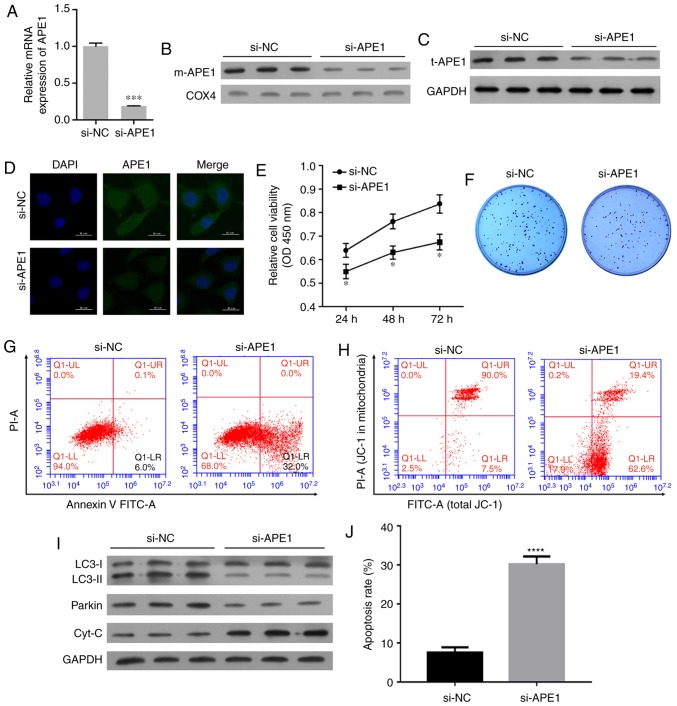
To further investigate the role of APE1 in A549/DDP cells, we performed transfections to knock down APE1 expression those cells. The transfection efficiency is shown in Fig. 3A. Both the m-APE1 and total APE1 (t-APE1) protein levels in A549/CDD cells were significantly decreased by APE1 knockdown (Fig. 3B and C). The decrease in t-APE1 protein was confirmed by an immunofluorescence assay (Fig. 3D). APE1 knockdown increased the cisplatin sensitivity of A549/DDP cells (Fig. 3E). In addition, APE1 knockdown reduced the colony formation efficiency and promoted the apoptosis of A549/DDP cells (Fig. 3F, G and J). The mitochondrial membrane potential of A549/DDP cells was significantly downregulated after APE1 knockdown (Fig. 3H). With regards to cell mitophagy, APE1 knockdown reduced the levels of LC3-II and Parkin in the A549/DDP cells; however, it significantly upregulated the levels of cytochrome c protein in the cytosolic fraction (Fig. 3I). Collectively, APE1 was shown to play an important role in the cisplatin resistance of A549 cells, and regulate Parkin-mediated mitophagy.
Figure 3.
APE1 knockdown increases the cisplatin sensitivity of A549/DDP cells. A549/DDP cells were transfected with small interfering RNA (si-APE1 or si-NC). (A) Transfection efficiency was assessed by qPCR. (B) The levels of mitochondrial APE1 (m-APE1) in the transfected A549/DDP cells were analyzed by western blotting. COX4 was used as loading control for mitochondrial APE1. (C) The total APE1 (t-APE1) protein levels in transfected A549/DDP cells were analyzed by western blotting. GAPDH was used as loading control. (D) Immunofluorescence assays were performed to assess the total APE1 levels in A549/DDP cells after transfection. (E) Transfected A549/DDP cells were treated with 15 µmol/l cisplatin for 24 h, and their viability was assessed by the CCK-8 assay. (F) Colony formation assays were performed to analyze the colony formation efficiency of the transfected A549/DDP cells. (G) The transfected A549/DDP cells were treated with cisplatin, and their rate of apoptosis was analyzed by flow cytometry. (H) Flow cytometry was used to analyze the distribution of JC-1 after transfection. (I) The transfected A549/DDP cells were subjected to fractionation to obtain the cytosolic fraction. Western blot analysis was performed to analyze the levels of LC3, total cytochrome c, and Parkin. (J) The cell apoptosis rate is shown in a histogram. GAPDH was used as loading control. Data represent the results obtained from three independent experiments (mean ± SEM of triplicate samples). *P<0.05, ***P<0.001 and ****P<0.0001 vs. the si-NC group. APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
Parkin-mediated mitophagy plays an important role in the APE1-induced cisplatin resistance of A549 cells
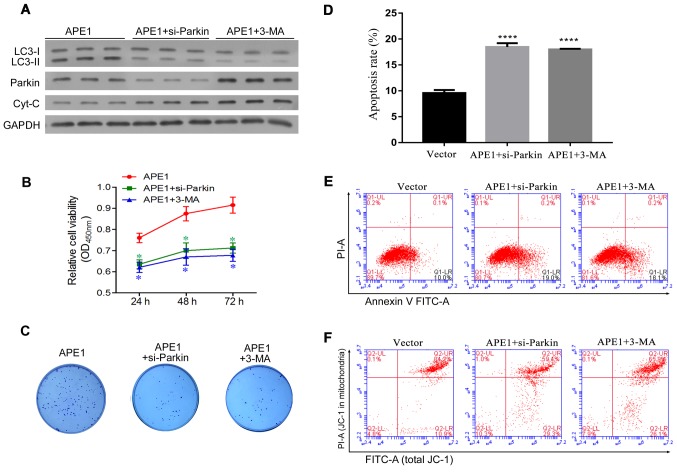
Given that APE1 overexpression and knockdown were shown to regulate Parkin-mediated mitophagy, we next investigated whether Parkin-mediated mitophagy is involved in cisplatin resistance. Here, the study was divided into three groups as follows: a) APE1 overexpression group; b) APE1 overexpression+Parkin siRNA group; c) APE1 overexpression+3-MA group. 3-MA (5 mM) (Sigma-Aldrich; Merck KGaA, M9298) was used as an inhibitor of autophagy according to the product introduction. In A549 cells, Parkin was significantly knocked down by si-Parkin transfection (Fig. S1B). As shown in Fig. 4A, Parkin knockdown and 3-MA treatment significantly decreased the levels of autophagy in the APE1-overexpressing A549 cells, and also increased the levels of cytochrome c protein in these cells (Fig. 4A). With regards to cisplatin resistance, both Parkin knockdown and 3-MA treatment significantly increased the cisplatin sensitivity of the APE1-overexpressing A549 cells (Fig. 4B). Moreover, Parkin knockdown and 3-MA treatment reduced the colony formation efficiency and significantly promoted the apoptosis of the APE1-overexpressing A549 cells (Fig. 4C-E). We also assessed the mitochondrial membrane potential of these cells, and found that their mitochondrial membrane potential was reduced by Parkin knockdown and 3-MA treatment (Fig. 4F). In brief, Parkin-mediated mitophagy was shown to play an important role in the APE1-induced cisplatin resistance of A549 cells.
Figure 4.
Parkin-mediated mitophagy plays an important role in the APE1-induced cisplatin resistance of A549 cells. A549 cells were first transfected with an APE1 overexpression plasmid. Next, the APE1-overexpressing A549 cells were transfected with small interfering RNA (si-Parkin or si-NC) or treated with 3-MA. (A) A549 cells after the relevant transfections (APE1, si-Parkin or si-NC) and their indicated treatment were subjected to fractionation to obtain the cytosolic fraction. Western blot analysis was performed to analyze the levels of LC3, total cytochrome c, and Parkin in the cytosolic fraction. GAPDH was used as loading control. (B) Following the indicated transfection and treatment, the A549 cells were co-cultured with 3 µmol/l cisplatin for 24 h, and their viability was assessed by the CCK-8 assay. (C) Colony formation assays were performed to analyze the colony formation efficiency of the transfected A549 cells after they had received their indicated treatment. (D) The cell apoptosis rates are shown in a histogram. (E) The apoptosis rates of the transfected A549 cells treated or not with 3-MA were analyzed by flow cytometry. (F) The distribution of JC-1 in the transfected A549 cells was analyzed by flow cytometry. *P<0.05 and ****P<0.0001 vs. the APE1 group. 3-MA, 3-methyladenine (an autophagy inhibitor); APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
Discussion
Autophagy is a selective autophagic process that provides metabolites needed for biosynthesis and energy production in response to stressful conditions (21). Autophagy induced by metabolic and therapeutic stress can play a dual role (22). For its pro-death role, excessive autophagy in cancer cells induces ‘autophagic cell death’ or ‘type II programmed cell death.’ For its pro-survival role, autophagy eliminates damaged organelles and recycles cellular degradation products (23,24).
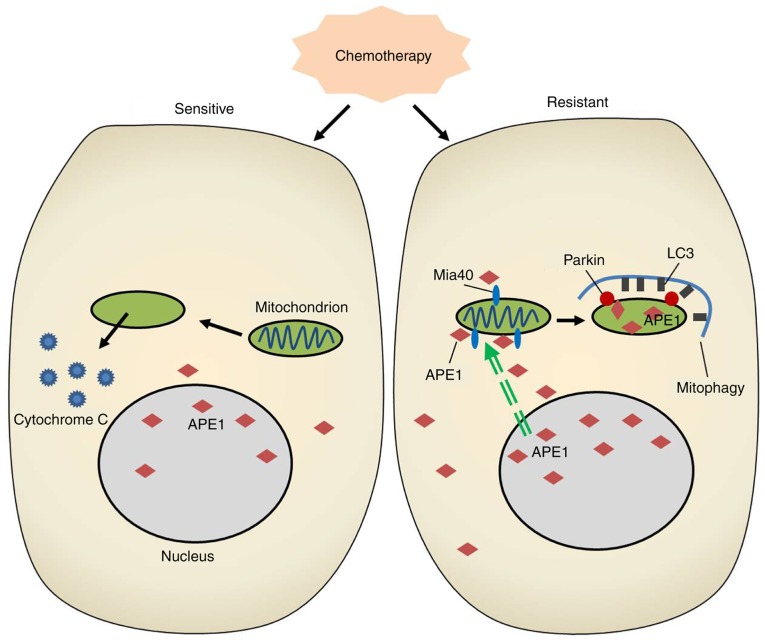
Mitochondrial-specific autophagy (mitophagy) is an important mitochondrial quality control system that selectively degrades excess or damaged mitochondria via an autophagic process. Similarly, mitophagy plays a dual role in cancer cell drug resistance that depends on the specific micro-environmental condition and cell type (25). Mitophagy can promote the survival of various types of cancer cells and tumors exposed to cytotoxic stress by degrading damaged mitochondria and reducing the levels of reactive oxygen species in mitochondria (26,27). Paradoxically, excessively damaged mitochondria may induce cell metabolic disorders and force cancer cells to undergo ‘autophagic cell death.’ While chemotherapeutic agents such as cisplatin induce oxidative stress and mitochondrial dysfunction in cancer cells, mitophagy selectively degrades excessive or damaged mitochondria via autophagy (28). Therefore, the progress of mitophagy helps to mediate drug resistance in cancer cells. In our study, the cisplatin-resistant A549 cell line displayed an increased level of mitophagy when compared with the classical A549 cell line, and inhibition of autophagy by 3-MA treatment or Parkin knockdown reduced the cisplatin resistance of A549 cells. Interestingly, we found accumulations of APE1 throughout cisplatin-resistant A549 cells, and in the mitochondria of those cells. In addition, our findings demonstrated for the first time that APE1 can induce Parkin-mediated mitophagy in A549 cells (Fig. 5).
Figure 5.
A schematic diagram showing how APE1 functions in the signaling circuit involved in lung cancer cisplatin resistance. APE1, apurinic/apyrimidinic endonuclease 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
Parkin, an E3 ubiquitin ligase, selectively degrades damaged mitochondria via mitophagy in mammalian cells (29,30). For Parkin-mediated mitophagy, Parkin requires PINK1, a Ser/Thr kinase, to assist in sensing a mitochondria's functional status (31–33). The efficiency of mitophagy depends on the levels of ubiquitin and deubiquitinases present in mammalian cells (33–35). Defects of Parkin are often associated with various pathologies, including cancer. A previous study showed that Parkin deficiency promoted tumorigenesis in a mouse model. However, Parkin is often inactivated in many tumors, and its role remains unclear. Parkin was reported to promote resistance to apoptosis independent of mitophagy (36). However, Villa et al (25) reported that E3 ubiquitin ligase (ARIH1/HHARI) protects against chemotherapy-induced cell death by Parkin-mediated mitophagy (25). Our study showed that the levels of Parkin in cisplatin-resistant A549 cells were increased, and that Parkin knockdown increased the cisplatin sensitivity of APE1-overexpressing A549 cells. Thus, Parkin-mediated mitophagy in A549 cells promoted cisplatin resistance of these cells. Our results are in accordance with the conclusion previously reached by Villa et al (25).
As we found that Parkin-mediated mitophagy in A549 cells promotes the cisplatin resistance of these cells, strategies for inhibiting mitophagy should receive increased attention as methods for overcoming cisplatin resistance and improving the efficacy of cancer therapy. However, the functional outcomes produced by mitophagy-induced cell death or survival can depend on the type of cancer treatment administered, and the results achieved can be confusing. In breast cancer, inhibition of mitophagy by liensinine markedly promoted the sensitivity of the cancer cells to classical chemotherapeutic agents, including doxorubicin, paclitaxel, vincristine, and cisplatin (37). Moreover, treatment with liensinine significantly inhibited late-stage mitophagy and reduced the viability of breast cancer cells, but increased their rate of apoptosis. An in vivo assay showed that inhibition of mitophagy inhibited tumor growth in the MDA-MB-231 ×enograft model (37). Our study demonstrated that inhibition of mitophagy by Parkin knockdown and 3-MA treatment significantly enhanced the cisplatin sensitivity of tumor cells. Our findings provide a potential therapeutic strategy for treating lung cancer.
Our study has some limitations that should be mentioned. First, due to limited research funds, only one cell line was used in the study, and this may weaken the reliability of our findings. Thus, it is important to verify our findings in other NSCLC cell lines before reaching a final conclusion. Second, although we had intended to clarify the molecular mechanism of APE1 in a cisplatin-resistant cell model, some differences exist between the results shown by in vitro and in vivo assays. Hence, some further in vivo assays should be conducted to confirm our findings. Despite these weaknesses, our study provides novel information concerning cisplatin resistance in NSCLC.
In conclusion, our study revealed a novel mechanism by which APE1 promotes cisplatin resistance in lung cancer cells. In the cisplatin-resistant cell line, APE1 was highly expressed and was translocated into the mitochondria via its interaction with Mia40. This mitochondrial translocation of APE1 increased the mitochondrial membrane potential, decreased the levels of cytochrome c protein, and induced Parkin-mediated mitophagy, resulting in the cisplatin resistance of lung cancer cells.
Supplementary Material
Acknowledgements
L.A.S. edited and revised the manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (no. 81501993).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZL and NL conceived and designed the research methods, drafted and revised the manuscript. ZL, YW, LW and YD performed experiments. ZL, JZ and FC analyzed the data and prepared the figures. WX and JH assisted with the analysis of the data and interpreted the results of the experiments. All authors approved the final version of the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ren J, Nie Y, Lv M, Shen S, Tang R, Xu Y, Hou Y, Zhao S, Wang T. Estrogen upregulates MICA/B expression in human non-small cell lung cancer through the regulation of ADAM17. Cell Mol Immunol. 2015;12:768–776. doi: 10.1038/cmi.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao L, Zhang H, Zhang B, Zhang L, Wang C. Prognostic value of combination of preoperative platelet count and mean platelet volume in patients with resectable non-small cell lung cancer. Oncotarget. 2017;8:15632–15641. doi: 10.18632/oncotarget.14921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proto C, Imbimbo M, Gallucci R, Brissa A, Signorelli D, Vitali M, Macerelli M, Corrao G, Ganzinelli M, Greco FG, et al. Epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of central nervous system metastases from non-small cell lung cancer: The present and the future. Transl Lung Cancer Res. 2016;5:563–578. doi: 10.21037/tlcr.2016.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 6.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nature Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 8.Gridelli C, Aapro M, Ardizzoni A, Balducci L, De Marinis F, Kelly K, Le Chevalier T, Manegold C, Perrone F, Rosell R, et al. Treatment of advanced non-small-cell lung cancer in the elderly: Results of an international expert panel. J Clin Oncol. 2005;23:3125–3137. doi: 10.1200/JCO.2005.00.224. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Wilson DM., III Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Signal. 2014;20:678–707. doi: 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barchiesi A, Wasilewski M, Chacinska A, Tell G, Vascotto C. Mitochondrial translocation of APE1 relies on the MIA pathway. Nucleic Acids Res. 2015;43:5451–5464. doi: 10.1093/nar/gkv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen KY, Chen YM, Huang MS, Hu CY, et al. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:223–229. doi: 10.1158/1055-9965.EPI-08-0749. [DOI] [PubMed] [Google Scholar]
- 13.Woo J, Park H, Sung SH, Moon BI, Suh H, Lim W. Prognostic value of human apurinic/apyrimidinic endonuclease 1 (APE1) expression in breast cancer. PLoS One. 2014;9:e99528. doi: 10.1371/journal.pone.0099528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren T, Qing Y, Dai N, Li M, Qian C, Yang Y, Cheng Y, Li Z, Zhang S, Zhong Z, Wang D. Apurinic/apyrimidinic endonuclease 1 induced upregulation of fibroblast growth factor 2 and its receptor 3 induces angiogenesis in human osteosarcoma cells. Cancer Sci. 2014;105:186–194. doi: 10.1111/cas.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, He L, Dai N, Guan W, Shan J, Yang X, Zhong Z, Qing Y, Jin F, Chen C. Serum APE1 as a predictive marker for platinum-based chemotherapy of non-small cell lung cancer patients. Oncotarget. 2016;7:77482–77494. doi: 10.18632/oncotarget.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018;285:1751–1766. doi: 10.1111/febs.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T, Liu H, Ming L. Multiple roles of autophagy in the sorafenib resistance of hepatocellular carcinoma. Cell Physiol Biochem. 2017;44:716–727. doi: 10.1159/000485285. [DOI] [PubMed] [Google Scholar]
- 20.Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, Hung SC, Hsiao M, Yao CJ, Shieh MJ. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy. 2014;10:1179–1192. doi: 10.4161/auto.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: Always two sides to a problem. J Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarauta V, Jaime P, Gonzalo O, de Miguel D, Ramírez-Labrada A, Martínez-Lostao L, Anel A, Pardo J, Marzo I, Naval J. Inhibition of autophagy with chloroquine potentiates carfilzomib-induced apoptosis in myeloma cells in vitro and in vivo. Cancer Lett. 2016;382:1–10. doi: 10.1016/j.canlet.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Gewirtz DA. The four faces of autophagy: Implications for cancer therapy. Cancer Res. 2014;74:647–651. doi: 10.1158/0008-5472.CAN-13-2966. [DOI] [PubMed] [Google Scholar]
- 25.Villa E, Proics E, Rubio-Patino C, Obba S, Zunino B, Bossowski JP, Rozier RM, Chiche J, Mondragón L, Riley JS, et al. Parkin-Independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20:2846–2859. doi: 10.1016/j.celrep.2017.08.087. [DOI] [PubMed] [Google Scholar]
- 26.Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, Sachleben JR, Asara JM, Locasale JW, Karczmar GS, Macleod KF. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16:1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan C, Luo L, Guo CY, Goto S, Urata Y, Shao JH, Li TS. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Yan C, Li TS. Dual role of mitophagy in cancer drug resistance. Anticancer Res. 2018;38:617–621. doi: 10.21873/anticanres.12266. [DOI] [PubMed] [Google Scholar]
- 29.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 30.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 31.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 32.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits parkin to damaged mitochondria and activates latent parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll RG, Hollville E, Martin SJ. Parkin sensitizes toward apoptosis induced by mitochondrial depolarization through promoting degradation of Mcl-1. Cell Rep. 2014;9:1538–1553. doi: 10.1016/j.celrep.2014.10.046. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL, Huang C, Li P, Gao N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy. 2015;11:1259–1279. doi: 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.