Abstract
In the present study, to evaluate the role of Ku80 in thyroid carcinoma (TC), 86 thyroid tissue samples from patients with a spectrum of thyroid disorders were examined for protein levels of Ku80, nuclear factor-κB (NF-κB) and RET/TC by immunohistochemistry. Furthermore, in TC cells, Ku80 mRNA was detected by reverse transcription-quantitative PCR analysis and silenced using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technique to assess its role. An antibody array was used to identify Ku80-related regulatory genes. The protein levels of Ku80 in the TC tissues were significantly higher than those in non-neoplastic adjacent tissue samples (P<0.01). The activation of NF-kB and expression of RET/TC in the TC group were significantly increased (P<0.05) and were correlated with the protein expression of Ku80 (P<0.05). In papillary TC cells, the mRNA levels of Ku80 were high; Ku80 knockdown resulted in reductions in proliferation, invasion and colony formation, increased apoptosis, and reduced levels of proteins involved in MAPK signaling, cell proliferation and apoptosis. The high expression of Ku80 in TC was found to be associated with the expression of RET/TC and activation of NF-κB, and Ku80 knockdown decreased the malignancy of TC cells.
Keywords: thyroid carcinoma, Ku80, immunohistochemical, CRISPR/Cas9
Introduction
According to reports, ~1–2% of all of cancer cases occur in the thyroid (1). Papillary thyroid carcinoma (PTC) is the most common malignancy of the thyroid gland. The incidence of PTC in 30–50-year-old women is twice that of age-matched men (2). The prevalence of PTC has been increasing at an alarming rate of ~4% per year in the United States (3). Although patients with PTC have a good prognosis, half of patients are <40 years of age and more likely to develop lymph node metastasis; patients may experience recurrence or even succumb to disease. Therefore, examining the molecular mechanism underlying the occurrence and development of TC is of high significance.
Ku is a DNA-binding protein that is important in double-stranded DNA break repair via the non-homologous end-joining pathway (4,5). It is also essential for V(D)J recombination (5). The abnormal expression of Ku has been reported in a variety of tumors. Ku gene mutation may result in altered DNA repair mechanisms and genomic instability. The abnormal expression of Ku is considered to be closely related to the occurrence and development of malignant tumors. However, related reports are not concordant and some are contradictory. For example, the expression of Ku70/Ku80 has been shown to be decreased in various types of malignant tumor, such as colon adenocarcinoma, malignant melanoma, lung cancer and cervical cancer (6), suggesting that the downregulation of Ku is correlated with the occurrence and development of these tumors. By contrast, the levels of Ku in ovarian cancer, bladder cancer, malignant lymphoma, esophageal cancer, breast cancer, non-melanomatous skin cancer, oral cancer and other tumor tissues have been shown to be elevated (6), revealing Ku as an oncogenic protein that can lead to tumorigenesis and development through transcriptional regulation, the inhibition of apoptosis and the induction of cell proliferation. The role of Ku in the occurrence and development of TC, and the underlying mechanism remain to be fully elucidated.
Previous studies have shown that the protein expression of Ku is altered in human tumor tissues, with Ku presumably associated with tumorigenesis; however, the specific role of Ku in tumor development and the underlying mechanisms remain undefined. In the present study, the expression of Ku80 and its associated molecules were assessed in TC and paracancerous tissues. Subsequently, Ku80 was knocked down to examine its effects on cell proliferation, apoptosis, invasion and other associated molecular mechanisms, which may provide a potential a therapeutic target for the early prevention and treatment of TC.
Materials and methods
Cell culture
The K1 human PTC cell line (cat. no. 92030501) was purchased from the European Collection of Authenticated Cell Cultures and the B-CPAP TC cell line (cat. no. SCSP-543) was purchased from the Library of Chinese Academy of Sciences (Shanghai, China). The two cell lines were authenticated using short tandem repeat analysis, as described in the 2012 ANSI Standard (ASN-0002) by the ATCC Standards Development Organization and by Capes-Davis et al (7). The K1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum/Ham's F12/MCDB105 (2:1:1; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA); the B-CPAP cells were maintained in RPMI 1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS, Ausbian) in a humidified incubator with 5% CO2 at 37°C.
Clinical tissue sample collection
TC and normal adjacent paracancerous samples from 20 patients, Hashimoto's thyroiditis (HT) specimens from 20 patients, HT with TC samples from 15 patients, and normal thyroid specimens from 18 patients were evaluated for the protein expression levels of Ku80, RET/TC and NF-κB. These samples, derived from the depository of the Department of Pathology were histopathologically and clinically diagnosed at Xi'an Central Hospital (Shanxi, China) between January 2010 and December 2013, following the provision of written informed consent from the patients or their guardians. The Institutional Review Board of Xi'an Central Hospital approved the use of specimens in this study.
Knockdown of Ku80 using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology
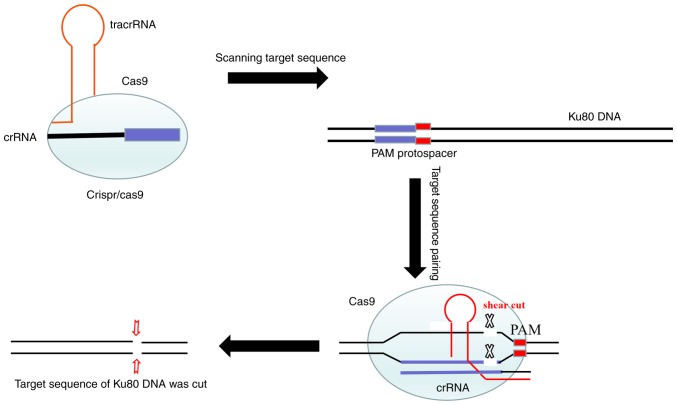
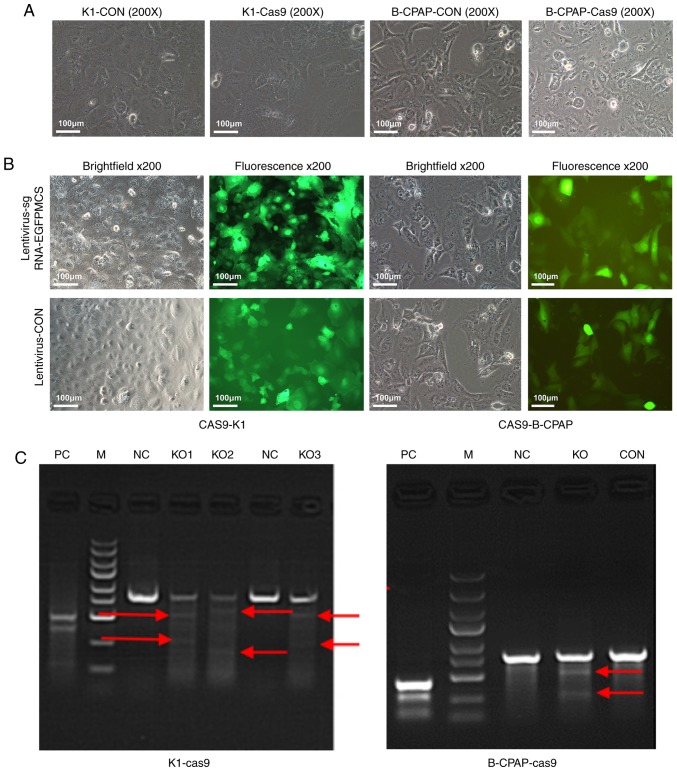
CRISPR/Cas9 technology consists of the Cas9-puromycin (puro) lentivirus and single guide RNA (sgRNA)-EGFPMCS lentivirus (Fig. 7). The Cas9-puro lentivirus carries the puromycin resistance gene, and enhanced green fluorescent protein (EGFP) is present for labeling the sgRNA-EGFPMCS lentivirus (8). Three sgRNAs targeting the Ku80 gene were designed. The vectors corresponding to these lentiviruses were constructed by Shanghai GenePharma Co., Ltd. The sgRNA sequences were 5′-GTCATAAGCATATCGAACTA-3′, 5′-TCATATCAAGCATAACTATG-3′ and 5′-GGTTCAGAGAAGATTCTTCA-3′. Initially, the K1 and B-CPAP cells were seeded in 6-well plates and infected with the Cas9-puro lentivirus. In order to obtain cells stably expressing Cas9, at 72 h post-infection, 2.0 µg/ml of puromycin was used for 48 h to kill uninfected cells, and K1-Cas9 and B-CPAP-Cas9 were generated (Fig. 1A). Subsequently, the log-phase K1-Cas9 and B-CPAP-Cas9 cells were infected with the three sgRNA-EGFPMCS lentiviral vectors, respectively. After 72 h, EGFP was observed under an inverted fluorescence microscope (Fig. 1B). Reverse transcription-PCR was used to detect the expression levels of LV-Cas9-puro in the K1-cas9 and B-CPAP-Cas9 cells. A cruiser enzyme digestion assay was performed for Cas9-sgRNA activity assessment (Fig. 1C), as described below.
Figure 7.
Schematic diagram of CRISPR/Cas9. The CRISPR/Cas9 dual-vector lentivirus knocked out the target sequence of the Ku80 gene. CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated protein 9; tracrRNA, trans-activating RNA; crRNA, CRISPR-derived RNA; PAM, proto-spacer adjacent motifs.
Figure 1.
Microscopy and PCR results of K1 and B-CPAP cells infected with Cas9. (A) K1 and B-CPAP cells were grown and infected with the lentivirus Cas9-puro or empty virus for 16 h and subjected to bright field microscopy. (B) Cas9-K1 and Cas9-B-CPAP cells were infected with lentivirus-sgRNA-EGFPMCS. Samples were assessed by bright field or fluorescence microscopy (magnification, ×200). (C) Cruiser enzyme digestion assay for Cas9-sgRNA activity in the K1-cas9 and B-CPAP-Cas9-stably expressing strains. K1 and B-CPAP cells were successfully infected with the Cas9-sgRNA lentivirus. (KO1-3, three potential targets for Ku80 of K1 cells; KO, one potential target for the Ku80 of B-CPAP cells). Red arrows represent potential target bands of target genes. M, DL2502 ladder marker (small to large 100, 250, 500, 750, 1,000, 1,500, 2,000, 3,000 and 5,000 bp); KO, knockout; NC, negative control; CON, blank control; PC, positive reference; sgRNA, single guide RNA; EGFP, enhanced green fluorescent protein.
Cruiser enzyme digestion assay
The cruiser enzyme digestion assay was performed using a knockout and mutation detection kit (Shanghai Jisheng Medical Technology Co., Ltd. cat. no. MB001-100420rnx). In brief, the steps were as follows: i) Cells were collected for detection, and the sample genome was extracted using a genomic DNA extraction kit; ii) primers were designed for PCR amplification (primer design included cutting target sites and the recommended length of products was 350–500 bp); hybridized DNA products were obtained by PCR, according to the manufacturer's protocol; iii) enzyme digestion and screening of positive clones was performed; and iv) positive PCR products were detected.
Total RNA was extracted from cells using TRIzol (Shanghai Pufei Biotech Co.; cat. no. 3101-100). The reference gene was GAPDH, and the primers were designed and synthesized by Shanghai GenePharma Co., Ltd. GenePharma (Shanghai, China). The detection primer sequences are listed in Table I. The qPCR conditions were as follows: 95°C for 2 min, then 35 cycles of 95°C for 20 sec and 72°C for 30 sec, followed by a last cycle at 95°C for 5 min for denaturation of the primers. Analysis of the products was conducted by 2% agarose gel electrophoresis. The amplified fragment size of the positive reference was 493 bp, and the digested fragment size was 122 BP and 371 bp, respectively. If the expected size of cleavage bands appeared in the experimental group, this was considered a positive clone screened by restriction enzyme digestion. The experiment was performed in duplicate in at least two independent repeats.
Table I.
Primers used for the Cruiser enzyme digestion assay.
| Target point | Upstream sequence (5′-3′) | Downstream sequence (5′-3′) | Amplified fragment size (bp) | Enzyme digestion fragment 1 | Enzyme digestion fragment 2 |
|---|---|---|---|---|---|
| KO1 (K1 cell) | AGGTGATGGGTGACTTCAGAGG | CCGAAAGTGTGGTTCACAGACC | 801 | 528 | 273 |
| KO2 (K1 cell) | AGGTGATGGGTGACTTCAGAGG | CCGAAAGTGTGGTTCACAGACC | 801 | 597 | 204 |
| KO3 (K1 cell) | CACTGTACTACTCCAAGAGCAG | TACTTCCTTGGTCTCCATGTCC | 801 | 541 | 260 |
| KO (B-CPAP cell) | AGGTGATGGGTGACTTCAGAGG | CCGAAAGTGTGGTTCACAGACC | 801 | 528 | 273 |
Immunohistochemistry (IHC)
IHC was conducted on 4-mm FFPE histologic sections using mouse monoclonal anti-Ku80 (1:300, cat. no. 2753s; Cell Signaling Technology, Inc., Danvers, MA, USA), mouse monoclonal anti-RET/TC (1:250, cat. no. Sc-365943; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse monoclonal anti-NF-κB P65 (1:500, cat. no. Sc-8008; Santa Cruz Biotechnology, Inc.). The HT, HT + TC, TC and normal paracancerous tissues were routinely dewaxed and hydrated, and antigen retrieval was performed according to the requirements of the manufacturers of primary antibodies. Subsequently, 1% hydrogen peroxide was added for 20 min to quench intrinsic peroxidase activity, and goat serum (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) was used for 30 min to block nonspecific antibody binding. The slides were then incubated with primary antibodies at 37°C for 90 min. Following washing with PBS three times, the slides were incubated for 30 min with secondary antibodies at room temperature (MaxVision TM HRP-Polymer anti-mouse IHC kit; cat. no. KIT-5002; Fuzhou Maixin Biotech Co., Ltd.). Following three additional washes, DAB solution was added to the slides, which were then observed under a microscope. Ku80 was observed in the nucleus. Immunofluorescence double-labeling and laser confocal microscopy were used to localize NF-κB and RET/TC, determining whether they were co-expressed at the cellular level. Two independent observers assessed and scored the extent of immunostaining. The ratio of stained cells and the degree of staining were used as evaluation criteria. For each case, at least 1,000 tumor cells were analyzed, and the percentage of tumor cells with positively stained nuclei was recorded. For each sample, the ratios of cells expressing Ku80, RET/TC and NF-κB P65 varied between 0 and 100%, and the intensity of nuclear staining varied from weak to strong. A score was given based on the percentage of positive cells: <5% cells, 1 point; 6–35% of cells, 2 points; 36–70% of cells, 3 points; >70% of cells, 4 points. Another score was assigned according to the intensity of staining: Negative staining, 1 point; weak staining (light yellow), 2 points; moderate staining (yellow brown), 3 points; and strong staining (brown), 4 points. The final score was calculated as the product of the above two scores. A final score ≥4 indicated high protein expression in the tumor; otherwise, protein expression in the tumor was considered to be low.
Cell proliferation, colony formation, cell invasion and cell apoptosis assays
An MTT assay was used to evaluate cell proliferation. The cells were seeded at 2×103 cells per well in 96-well plates. At the end of each treatment, the cells were treated with 20 µl sterile MTT (5 mg/ml, Sigma; Merck KGaA, Darmstadt, Germany) for 4 h at 37°C. The medium was then removed and 100 µl of dimethyl sulfoxide (Sigma; Merck KGaA) was added. The optical density (OD) at 450 nm was measured on a microplate reader (Tecan Infinite, Tecan Group, Ltd., Groedig, Austria) according to the manufacturer's instructions. Data are expressed as a percentage of the control cells. The experiments were performed in triplicate. For the colony formation assay, 500 cells were seeded in each well of 6-well plates and incubated for 14 days. The colonies were fixed with 4% paraformaldehyde (Sinopharm Chemical Reagent Co. Ltd., Shanghai, China), stained with 500 µl Giemsa (Shanghai Dingguo Biotechnology Co. Ltd., Shanghai, China) for 10 min and counted. An invasion assay was performed with Boyden chambers (Corning, Inc., NY, USA) according to the manufacturer's protocol. To assess the invasive ability of the cells, filters were precoated with Matrigel prior to cell seeding into the chambers (5×104/well). The culture system consisted of a 24-well plate with 500 and 750 µl per well in the inner and outer chambers, respectively. The cells were divided into three groups and assessed in triplicate. Following incubation at 37°C for 16 h, images of the invaded cells were captured and counted under an inverted microscope. To assess apoptosis, logarithmic growth-phase cells were harvested and washed twice with PBS. Sequential staining was then performed with 10 µl of Annexin V-APC with an Annexin V-APC staining kit purchased from eBioscience; Thermo Fisher Scientific, Inc. (cat. no. 88-8007). Finally, apoptosis was detected by flow cytometry according to the manufacturer's instructions (Beckman Coulter, Inc.).
RT-qPCR analysis
To assess gene expression levels, total RNA was isolated from the cultured cells using TRIzol (Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA with oligo (dT) primers and the M-MLV reverse transcriptase kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions. qPCR analysis was performed on a LightCycler 480 II Real-time PCR instrument (Roche Diagnostics, Basel, Switzerland) with SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan). Relative mRNA expression was determined using the human GAPDH gene as an endogenous reference standard. The qPCR conditions were as follows: An initial cycle at 95°C for 5 min; 35 cycles of 95°C for 50 sec and 55°C for 1 min; final extension at 72°C for 1 min. The specificity of the PCR products was confirmed by melting-curve analysis. The mRNA levels of Ku80 were assessed using the 2−ΔΔCq method (9). The primer sequences used in this experiment are shown in Table II. To assess gene expression levels of RET/TC, total RNA was isolated from clinical tissue samples of the HT, HT + TC, TC and normal thyroid tissue specimens with a total RNA isolation kit (AP-MNMS-RNA; Axygen, Union City, CA, USA) as described by the manufacturer.
Table II.
Primers used in the reverse transcription-quantitative PCR analysis.
| Gene | Upstream sequence (5′-3′) | Downstream sequence (5′-3′) | Amplified fragment size (bp) |
|---|---|---|---|
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA | 121 |
| XRCC5 | GCACTGACAATCCCCTTTCTG | TGTTGAGCTTCAGCTTTAACCTG | 97 |
Automatic western blot quantitative analysis system (WES)
A total of 168 h after infection of the K1 cells with the Cas9-sgRNA lentivirus, total protein was extracted using a lysis buffer (Beyotime Institute of Biotechnology) and quantitated using a BCA protein assay kit (Beyotime Institute of Biotechnology). Equal quantities of protein (20 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The membranes were first blocked with 5% skim milk in TBS at room temperature for 1 h, then incubated with rabbit anti-human Ku80 antibody (1:300; cat. no. 2753S; Cell Signaling Technology, Inc.) at 4°C overnight. The membranes were then washed with TBS/Tween 20 (TBS-T) and incubated with secondary antibodies (rabbit IgG from the Plonox kit; 1:10,000; cat. no. ab205718; Abcam) at room temperature for 2 h. Following incubation, the membrane was washed with TBS-T three times (10 min each). The target band was 83 kDa, and data were analyzed using Compass Software for Simple Western (version 4; ProteinSimple).
Cancer phenotype, PathScan stress and apoptosis signaling antibody array
The PathScan Antibody Array kit (Cell Signaling Technology, Inc.) was used to assess changes in key molecules of the signaling pathways in K1 cells co-infected with Cas9 and the sgRNA lentivirus, according to the manufacturer's protocol. The experiment was performed with two chips, including the cancer phenotype and the stress and apoptotic pathways. The samples were divided into negative control (NC) and experimental knockout (KO) groups, and data were evaluated by chemiluminescence imaging and gamma analysis. Each experiment was repeated three times.
Statistical analysis
Data are presented as the mean ± standard deviation and were analyzed using SPSS 22.0 statistical software (IBM Corp., Armonk, NY, USA). Comparisons between groups for statistical significance were made using Student's t-test. (two-tailed, paired) P<0.05 was considered to indicate a statistically significant difference.
Results
High expression of Ku80 in TC tissues and cells is associated with RET/TC rearrangement and the activation of NF-κB
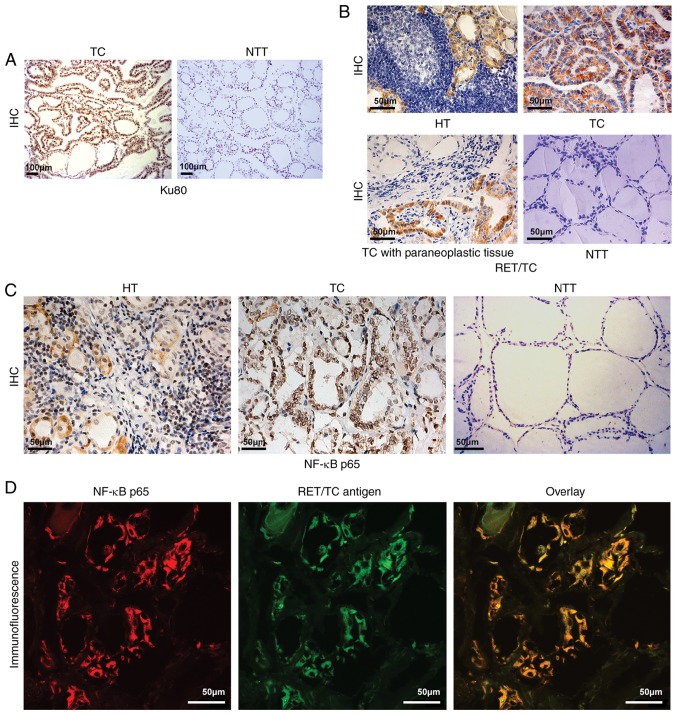
Studies have found that Ku80 gene polymorphism is associated with susceptibility to PTC (10). The fusion oncogene RET/TC, expressed in most TC cells, activates nuclear transcription factor NF-κB, which regulates Ku70 and Ku80 (11,12). Therefore, the present study examined the expression of Ku80 in TC and paracancerous tissue samples and found significantly higher levels of Ku80 in the tumor specimens (16/20, 80%) compared with that in the normal samples (4/20, 20%) (Fig. 2A). RT-qPCR analysis showed that the levels of RET/TC were higher in HT (7/20, 35%) and TC (12/20, 60%) tissues compared with those in paracancerous normal samples, in which no expression of RET/TC was detected (P<0.01). This indicated that RET/TC rearrangement may occur from the early lesion of HT to TC. This finding was also confirmed by IHC in the above tissues. The expression of RET/TC in HT (6/20, 30%) tissues was lower than that in TC (13/20, 65%) tissues (P<0.01). This further suggested that the expression of RET/PTC in HT may occur from the early lesion of HT, increasing in TC (Table III, Fig. 2B). IHC was also used to detect the expression of NF-κB. Compared with that in the normal thyroid tissue, the expression of NF-κB in the diseased thyroid tissue was elevated, with nuclear translocation of NF-κB present in the HT (18/20, 90%) and TC (19/20, 95%) tissues with the expression of RET (P<0.01; Fig. 2C), indicating that NF-κB was in the active state. These findings suggested that the protein expression of RET may transform HT into TC by activating NF-κB. To confirm this result, the co-expression of NF-κB and RET/TC was detected by immunofluorescence double labeling and laser confocal microscopy. The results showed that NF-κB and RET/TC were co-expressed in the HT and TC thyroid follicular epithelial cells, confirming that the protein expression of RET may transform HT into TC by activating NF-κB. Representative photomicrographs are shown in Fig. 2D and the results are summarized in Table IV.
Figure 2.
Ku80, RET/TC and NF-κB p65 expression in NTT, TC, HT and paracancerous tissues. (A) IHC staining for Ku80 assessment in TC and adjacent tissues (magnification, ×100). Ku80 was expressed in TC tissues (strong positive staining of nuclei) but not in adjacent tissues (no positive staining of nuclei or cytoplasm). (B) IHC detection of RET/TC protein expression in the diseased thyroid tissue (magnification, ×40). HT exhibited weak positive staining of the cytoplasm; TC exhibited strong positive staining of the cytoplasm; TC with paraneoplastic (slightly weakly positive staining of cytoplasm; NTT exhibited negative staining of the cytoplasm and nuclei. (C) Assessment of NF-κB p65 protein expression in the diseased thyroid tissue by IHC (magnification, ×40). HT exhibited weak positive cytoplasmic and nuclear staining; TC exhibited strong positive cytoplasmic and nuclear staining; NTT exhibited almost negative staining of the cytoplasm or nuclei. (D) Immunofluorescence double-labeling and laser confocal microscopy for RET/TC antigen and NF-κB detection (magnification, ×600). NF-κB expression is shown as red fluorescence, RET/TC antigen expression is shown as green fluorescence. Laser confocal microscopy results show NF-κB and RET/TC were co-expressed in the cytoplasm and/or nuclei of thyroid follicular epithelial cells. TC, thyroid carcinoma; HT, Hashimoto's thyroiditis; NTT, normal thyroid tissue; IHC, immunohistochemistry; NF-κB, nuclear factor-κB.
Table III.
Positive rate of RET/PTC (%) in diseased and normal thyroid tissue.
| P-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Factor | A | B | C | D | A vs. D | B vs. D | C vs. D | B vs. C | A vs. C vs. (B+C) |
| Disease | HT | HT + PTC | PTC | NTT | |||||
| N | 20 | 15 | 20 | 18 | |||||
| PCR | 7 (35%) | ND | 12 (60%) | 0 (0) | 0.009a | ND | 0.0001a | ND | 0.113 |
| IHC | 6 (30%) | 9 (60%) | 13 (65%) | 0 (0) | 0.021a | 0.0001a | 0.0001a | 0.762 | 0.019a |
P<0.05 (χ2 test or Fisher's exact test). HT, Hashimoto's thyroiditis; PTC, thyroid papillary carcinoma; NTT, normal thyroid; N, number of cases; IHC, immunohistochemistry; ND, not determined.
Table IV.
Immunohistochemical data for NF-κB and its co-expression with the RET/TC antigen.
| Positive cases/total cases (%) | |||
|---|---|---|---|
| Group | RET antigen | NF-κB | RET + NF-κB |
| HT | 6/20 (30)a | 18/20 (90)a | 6/20 (30) |
| HT + TC | 9/15 (60)a | 14/15 (93.3)a | 9/15 (60) |
| TC | 13/20 (65)a | 19/20 (95)a | 13/20 (65) |
| NTT | 0/18 (0) | 5/18 (27.8) | 0/18 (0) |
P<0.05, compared with the NTT group. HT, Hashimoto's thyroiditis; TC, thyroid carcinoma; NTT, normal thyroid; NF-κB, nuclear factor-κB.
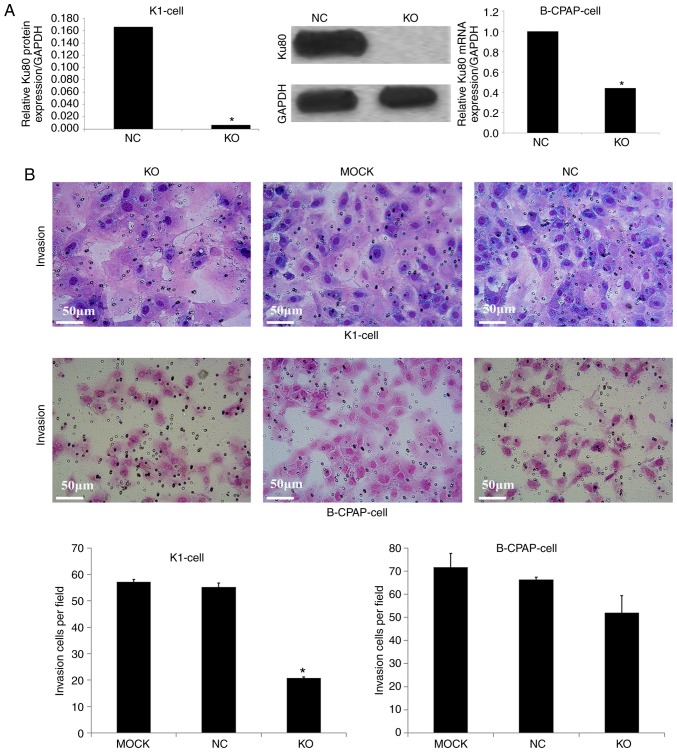
Knockdown of Ku80 suppresses TC cell proliferation, invasion and colony formation
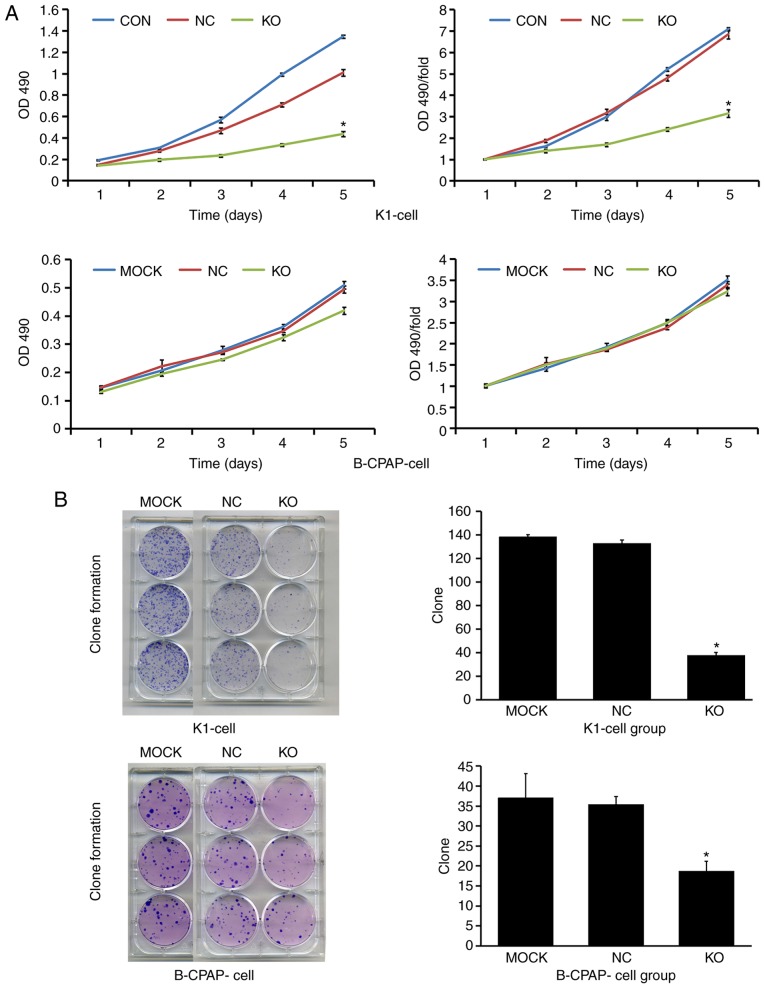
To assess the role of Ku80 in TC cell proliferation, invasion and colony formation, the CRISPR/Cas9 technique was used to knock down Ku80 in K1 and B-CPAP cells. A total of 168 h after infection, the knockdown efficacy of Ku80 was determined by western blotting and RT-qPCR analysis, respectively. As shown in Fig. 3A, compared with the NC group, the Ku80 sgRNA-infected K1 cells exhibited a 99.3% reduction in protein levels of Ku80, indicating a high knockdown efficacy. In addition, RT-qPCR analysis showed that the mRNA levels of Ku80 were reduced by 56% in the Ku80 sgRNA-infected B-CPAP cells. To further evaluate the effect of Ku80 on proliferation, the proliferative ability of sgRNA-infected K1 and sgRNA-infected B-CPAP cells were then assessed using an MTT assay, which revealed that the proliferation rates of the K1 cells were significantly decreased on days 4 and 5 following Ku80 knockdown compared with those of the control cells (P<0.05). The proliferative ability of the B-CPAP cells on the fifth day did not differ significantly from that on the first day (Fig. 4A). These results were confirmed by colony formation and invasion assays; in K1 cells, the numbers of invasive cells and cell clones formed were markedly decreased following Ku80 knockdown compared with numbers in the control group. The invasive ability of the sgRNA-B-CPAP cells was not significantly decreased (P>0.05), however, the clone formation ability was significantly decreased (P<0.05) (Figs. 3B and 4B). The proliferation and invasiveness of the sgRNA-B-CPAP cells were not decreased compared with those in the control group, possibly due to the rate of Ku80 knockdown being lower than that of the K1 cells. Collectively, these findings suggested that Ku80 was important in the proliferation, invasion and colony formation of TC cells in vitro.
Figure 3.
Effect of Ku80 knockdown on TC cell invasion. (A) Expression levels of Ku80 in Cas9-sgRNA lentivirus-infected B-CPAP and Cas9-sgRNA lentivirus-infected K1 cells. Peak area analysis of Ku80 protein in K1-Cas9 cells; protein expression of Ku80 in K1-Cas9 cells, detected by western blotting; Detection of Ku80 mRNA in B-CPAP-Cas9 cells following Ku80 knockdown by quantitative PCR. (B) Effect of Ku80 knockdown on K1 and B-CPAP cell invasion. K1 and B-CPAP cells were grown and infected with the Cas9-sgRNA lentivirus for 16 h and subjected to an invasion assay. Transwell photomicrographs of K1-Cas9 and B-CPAP-Cas9 cells in various experimental groups are shown. The graphs show the summarized data of the invasion assay of K1 and B-CPAP cells, respectively (*P<0.05 vs. NC cells). KO, experimental group; NC, negative control group; MOCK, blank control group.
Figure 4.
Effect of Ku80 knockdown on TC cell proliferation and clone formation. (A) Effect of Ku80 knockdown on K1 and B-CPAP cell proliferation. K1 and B-CPAP cells were grown and infected with the Cas9-sgRNA or control lentivirus, and subjected to an MTT assay. Proliferation and fold curves of K1 and B-CPAP cells in various experimental groups are shown. (B) Effect of Ku80 knockdown on K1 and B-CPAP cell clonal formation ability. Digital photographs of colonies formed in various experimental groups of K1-Cas9 and B-CPAP-Cas9 cells are shown with quantitation of the digital photographs on the right (*P<0.05 vs. NC cells). KO, experimental group; NC, negative control group; MOCK, blank control group; OD, optical density.
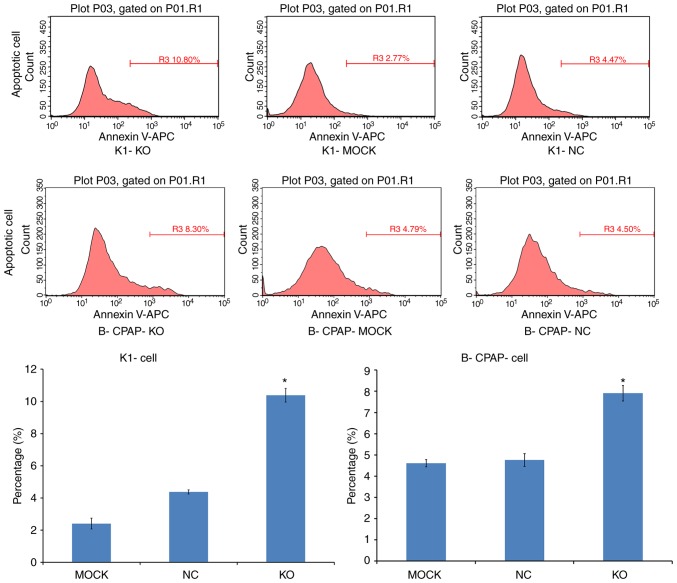
Knockdown of Ku80 promotes TC cell apoptosis
Apoptosis was assessed by Annexin V-APC single staining. Flow cytometric analysis demonstrated that, compared with the control, the apoptotic rates of the sgRNA-K1 cells were 4.38±0.12 and 10.38±0.42% in the NC and KO cells, respectively. The percentages of apoptotic sgRNA-B-CPAP cells were 5.01±0.24 and 7.82±0.22% in the NC and KO groups, respectively. Both infected cell lines exhibited significantly increased apoptotic rates (P<0.05; Fig. 5), indicating that Ku80 suppressed TC cell apoptosis.
Figure 5.
Effect of Ku80 knockdown on K1 and B-CPAP cell apoptosis. Peak figures of K1-Cas9 and B-CPAP-Cas9 cells in various experimental groups are shown above, with the respective quantitation shown in graphs below (*P<0.05 vs. NC cells). KO, experimental group; NC, negative control group; MOCK, blank control group.
Identification of genes mediating the activity of Ku80 in TC cells
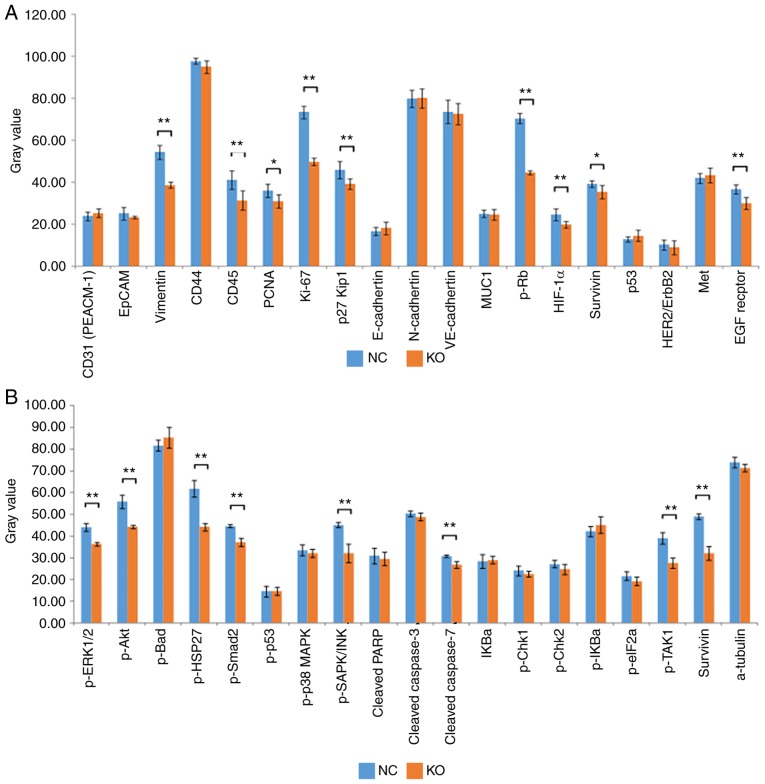
To detect and compare key signaling molecules in K1 cells, PathScan stress and apoptosis signaling antibody array analysis was performed for K1 cells infected with CRISPR/Cas9 or control lentiviruses. Chemiluminescence and gray scale data indicated that, compared with the NC group, the KO group had significantly reduced protein expression levels of vimentin, CD45, proliferating cell nuclear antigen (PCNA), Ki-67, phosphorylated (p-)retinoblastoma (Rb), hypoxia-inducible factor 1α (HIF-1α), survivin and EGF receptor in cancer phenotype pathways (Fig. 6A, P<0.05). Similarly, the protein expression levels of phosphorylated (p-)ERK1/2, p-Akt, p-HSP27, p-Smad2, p-SAPK/JNK, p-TGF-β-activated kinase 1 (TAK1) and survivin (involved in apoptotic signaling pathways) were also downregulated in the KO group (Fig. 6B, P<0.05). These data further indicated that Ku80 altered apoptosis by regulating multiple genes in the MAPK pathway.
Figure 6.
Identification of Ku80 knockdown-related gene expression. (A) Statistical analysis of gray values revealed that the expression levels of vimentin, CD45, PCNA, Ki-67, p-Rb, HIF-1α, survivin and EGF receptor in cancer phenotype pathways were significantly decreased in the KO group compared with those in the control group (*P<0.05, **P<0.01). (B) Statistical analysis of gray values revealed that the expression levels of p-ERK1/2, p-Akt, p-HSP27, p-Smad2, p-SAPK/JNK, p-TAK1 and survivin involved in the molecular profile of stress and apoptotic signaling pathways were significantly decreased in the KO group compared with those in the control group (**P<0.01). KO, experimental group; NC, negative control group; p-, phosphorylated.
Discussion
TC is the most common type of endocrine malignancy and represents the leading cause of endocrine organ cancer-related mortality. As with other types of cancer, early diagnosis of TC is important for improving patient survival rate and treatment outcomes (13). Although TC has a good prognosis, there is a risk of recurrence which represents a threat to human life. In recent years, the incidence of PTC has been steadily increasing worldwide (14). The etiology of PTC is most closely associated with exposure to radiation (15). Little progress has been made in mechanistic research. The present study demonstrated that the occurrence of TC is closely related to the Ku80 protein. It was found that silencing Ku80 significantly inhibited tumor cell proliferation, invasion and colony formation, and induced apoptosis. In addition, statistical analysis of gray scale values showed that the protein levels of vimentin, CD45, PCNA, Ki-67, p-Rb, HIF-1α, EGF receptor, p-ERK1/2, p-Akt, p-HSP27, p-Smad2, p-SAPK, JNK, p-TAK1 and survivin were significantly lower following Ku80 knockdown compared with those in the control (P<0.05). These findings reveal potential novel targets for TC treatment.
The Ku protein is the main repair protein for DNA double-strand breaks (DSBs). It is composed of Ku80 and Ku70 monomers and serves an important role in DNA DSB injury repair (16). In addition to repairing DNA DSBs, Ku has other important biological functions, including involvement in cell cycle regulation, transcriptional regulation, immunoglobulin-encoding gene V(D)J chain rearrangement and maintenance of telomere structural stability (17). Studies have shown that the protein expression of Ku70/Ku80 is increased in various tumors, including ovarian cancer, bladder cancer, colon cancer, cervical cancer, malignant lymphoma, esophageal cancer, lung cancer, breast cancer, non-melanomatous skin cancer and cancer of the oral cavity (6), revealing Ku as an oncogenic protein. The increased activity of Ku inhibits apoptosis and promotes cell proliferation through transcriptional regulation, leading to tumorigenesis and cancer development. However, the mechanisms underlying the effects of Ku in TC have rarely been reported. In order to clarify the association between Ku and TC, the present study analyzed the level of Ku in human TC tissue specimens and TC cells. As shown above, compared with the control groups, tumor samples and cells showed significantly increased expression of Ku80, corroborating previous reports. To further determine the specific role of Ku80, a series of experiments were performed following Ku80 silencing. Ku80 knockdown resulted in reduced proliferation, invasion and colony formation in tumor cells, whereas apoptosis was induced. Although the proliferative activity and invasion rate of B-CPAP cells in the MTT and invasion assays, respectively, did not differ significantly to those in the respective control groups, the overall trends of cell proliferation and invasion were downward. This suggests that the expression of Ku80, encoded by the XRCC5 gene, is extremely high in tumor cells, and the Ku80-knockdown efficiency in B-CPAP cells was not high enough, causing residual Ku80 to exert effects. In addition, following Ku80 knockdown, the protein expression levels of vimentin, CD45, PCNA, Ki-67, p-Rb, HIF-1α, EGF receptor, p-ERK1/2, p-Akt, p-HSP27, p-Smad2, p-SAPK/JNK, p-TAK1, survivin and others, which promote cell proliferation, metastasis, invasion and angiogenesis, were significantly reduced. This further demonstrates that Ku80 serves a critical role in the development of TC.
Cancer occurrence is a complex process. The mutation or ectopicity of related genes in a pathway can lead to the proliferation or apoptosis of tumor cells. Common mutations include those in BRAF, RET, RAS, PTEN and RET/PTC (18). The RET proto-oncogene, located on the long arm of chromosome 10 (10q11.2), encodes a 150-kD receptor tyrosine kinase that was discovered in 1985 in experiments involving lymphoma DNA extracts; transfection into NIH3T3 mouse fibroblast cells results in malignant transformation (19,20). Subsequent analysis of human tumors has revealed that the RET/TC fusion protein is a characteristic alteration found in ~20% of patients with PTC (21). Oncogenes of the RET/PTC family, particularly RET/PTC1 and RET/PTC3, serve key roles in the development of several PTCs, particularly pediatric PTC and those resulting from exposure to ionizing radiation (22). The low RET/PTC oncogene expression observed in certain benign thyroid diseases suggests that the local cellular microenvironment may be important in regulating RET/PTC oncogenic activity. Based on previous reports, the present study assessed the gene expression levels of RET/TC remodeling in TC, normal and HT tissues by RT-qPCR and IHC analyses. Compared with levels in the normal control group, the HT and TC samples had significantly higher expression levels of RET/TC. However, its expression was lower in HT samples than that in TC samples, suggesting that the protein expression of RET in HT may be an early precursor of TC, and may be involved in the subsequent development of TC.
NF-κB consists of heterodimers of various members of the Rel family, including p50, c-Rel, v-rel, p52, p-65 (RelA) and Rel B (23). It is constitutively activated in several types of cancer (24,25). As NF-κB regulates numerous genes associated with cell transformation, survival, proliferation, invasion, metastasis and inflammation, the constitutive activation of NF-κB in cancer cells is key in various aspects of tumor progression (26). Numerous genes necessary for tumor growth have NF-κB-binding sites and are targeted by this transcription factor (27). The present study found that, compared with levels in normal thyroid tissues, the expression levels of NF-κB in the diseased thyroid tissues were elevated, with RET protein expression observed in HT and TC tissues. In addition, NF-κB was found to undergo nuclear transcription and activation in TC cells, suggesting that the protein expression of RET may cause HT cancer to develop into TC by activating NF-κB.
In conclusion, the present study demonstrated that the expression of Ku80 was significantly increased in TC, which exhibited high expression levels of RET/TC and NF-KB. In addition, the expression of Ku80 was shown to be positively correlated with the levels of RET/TC and NF-KB. Furthermore, Ku80 knockdown significantly reduced proliferation, invasion and tumor formation in TC cells, and markedly increased the apoptotic rate. It was also shown that Ku80 is involved in the pathogenesis of TC. Finally, Ku80 knockdown significantly downregulated MAPK signaling pathway-related molecules in addition to proliferation and metastasis-associated proteins, including EGFR, p-ERK1/2, p-SAPK/JNK, p-Akt, p-TAK1, PCNA, Ki-67, p-Rb, HIF-1a, survivin, vimentin, CD45, p-HSP27 and p-Smad2. These findings suggest that the increased expression of Ku80 is closely related to the occurrence and development of TC. Upregulating Ku80 modulates the transcription of a variety of genes which promote cell proliferation, invasion and tumor formation, and inhibit apoptosis via the MAPK pathway. The present study provides a theoretical basis for TC targeted therapy. However, the work was limited by the absence of mechanistic investigation; screening for possible molecules was only performed through gene chip technology and the results were not confirmed. Further investigation is required to determine which molecules regulate the function of Ku80.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- IHC
immunohistochemistry
- TC
thyroid carcinoma
- NF-κB
nuclear factor-κB
- NJEJ
non-homologous end-joining
- EGFP
enhanced green fluorescent protein
- OD
optical density
- TBS-T
TBS-Tween 20
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81372857), the Science and Technology Research and Development Program of Shaanxi Province (grant no. 2008K09-09), the Xi'an Science and Technology Program [grant no. SF09027(9)] and the Xi'an Science and Technology Program [grant no. 2016047SF/YX039(3)].
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YF and JL performed the experiments, participated in data collection and drafted the manuscript. WW, HF, YD and NL performed the statistical analysis and participated in study design. YZ, JY and JW helped draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committees of Xi'an Central Hospital (LP2018-08-27).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu S, Semenciw R, Ugnat AM, Mao Y. Increasing thyroid cancer incidence in Canada, 1970–1996: time trends and age-period-cohort effects. Br J Cancer. 2001;85:1335–1339. doi: 10.1054/bjoc.2001.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Homayouni M, Mohammad Arabzadeh SA, Nili F, Razi F, Amoli MM. Evaluation of the presence of Epstein-Barr virus (EBV) in Iranian patients with thyroid papillary carcinoma. Pathol Res Pract. 2017;213:854–856. doi: 10.1016/j.prp.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM, Schlumberger M. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American thyroid association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Bertinato J, Tomlinson JJ, Schild-Poulter C, Hache RJ. Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene. 2003;302:53–64. doi: 10.1016/S0378111902010892. [DOI] [PubMed] [Google Scholar]
- 6.Gullo C, Au M, Feng G, Teoh G. The biology of Ku and its potential oncogenic role in cancer. Biochim Biophys Acta. 2006;1765:223–234. doi: 10.1016/j.bbcan.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Capes-Davis A, Reid YA, Kline MC, Storts DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A, et al. Match criteria for human cell line authentication: Where do we draw the line? Int J Cancer. 2013;132:2510–2519. doi: 10.1002/ijc.27931. [DOI] [PubMed] [Google Scholar]
- 8.Cui H, Lan X, Lu S, Zhang F, Zhang W. Bioinformatic prediction and functional characterization of human KIAA0100 gene. J Pharm Anal. 2017;7:10–18. doi: 10.1016/j.jpha.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Gomes BC, Silva SN, Azevedo AP, Manita I, Gil OM, Ferreira TC, Limbert E, Rueff J, Gaspar JF. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol Rep. 2010;24:1079–1085. [PubMed] [Google Scholar]
- 11.Neely RJ, Brose MS, Gray CM, McCorkell KA, Leibowitz JM, Ma C, Rothstein JL, May MJ. The RET/PTC3 oncogene activates classical NF-κB by stabilizing NIK. Oncogene. 2011;30:87–96. doi: 10.1038/onc.2010.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim JW, Kim H, Kim KH. Expression of Ku70 and Ku80 mediated by NF-kappa B and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J Biol Chem. 2002;277:46093–46100. doi: 10.1074/jbc.M206603200. [DOI] [PubMed] [Google Scholar]
- 13.Erdogdu IH, Yumrutas O, Ozgur Cevik M, Bozgeyik I, Erdogdu M, Inan HM, Bagis H. Differential expression of PIWIL2 in papillary thyroid cancers. Gene. 2018;649:8–13. doi: 10.1016/j.gene.2018.01.066. [DOI] [PubMed] [Google Scholar]
- 14.Stewart BW, Kleihues P, editors. IARC Press; Lyon: 2003. World Cancer Report. [Google Scholar]
- 15.Williams D. Radiation carcinogenesis: Lessons from Chernobyl. Oncogene. 2008;27(Suppl 2):S9–S18. doi: 10.1038/onc.2009.349. [DOI] [PubMed] [Google Scholar]
- 16.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuteja R, Tuteja N. Ku autoantigen: A multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol. 2000;35:1–33. doi: 10.1080/10409230091169177. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Zhang B, Zhao Y, Chen P, Ji M, Hou P, Shi B. Association of BRAFV600E mutation with clinicopathological features of papillary thyroid carcinoma: A study on a Chinese population. Int J Clin Exp Pathol. 2014;7:6922–6928. [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizaka Y, Itoh F, Tahira T, Ikeda I, Sugimura T, Tucker J, Fertitta A, Carrano AV, Nagao M. Human ret proto-oncogene mapped to chromosome 10q11.2. Oncogene. 1989;4:1519–1521. [PubMed] [Google Scholar]
- 20.Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 21.Nikiforov YE. RET/PTC rearrangement in thyroid tumors. Endocr Pathol. 2002;13:3–16. doi: 10.1385/EP:13:1:03. [DOI] [PubMed] [Google Scholar]
- 22.Prescott JD, Zeiger MA. The RET oncogene in papillary thyroid carcinoma. Cancer. 2015;121:2137–2146. doi: 10.1002/cncr.29044. [DOI] [PubMed] [Google Scholar]
- 23.Thanos D, Maniatis T. NF-kappa B: A lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 24.Amit S, Ben-Neriah Y. NF-kappaB activation in cancer: A challenge for ubiquitination- and proteasome-based therapeutic approach. Semin Cancer Biol. 2003;13:15–28. doi: 10.1016/S1044-579X(02)00096-2. [DOI] [PubMed] [Google Scholar]
- 25.Aggarwal BB. Nuclear factor-kappaB: The enemy within. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799:775–787. doi: 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uwagawa T, Yanaga K. Effect of NF-κB inhibition on chemoresistance in biliary-pancreatic cancer. Surg Today. 2015;45:1481–1488. doi: 10.1007/s00595-015-1129-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.