Abstract
Adipocyte infiltration in pancreatic cancer (PC) has been demonstrated to be independently associated with PC risk and an active contributor to tumor progression. However, to date, little is known about these unique pancreatic tumor-surrounding adipocytes, or their response to cancer cells. The present study utilized an in vitro indirect coculture model in which the phenotypic changes of adipocytes following exposure to PC cells were directly observed. RNA-sequencing was performed on 3T3-L1 adipocytes cultured with or without Panc-1 cancer cells, and significant changes were identified at the transcriptional level. In terms of delipidation and the impaired function of glucose and lipid metabolism, coculture with tumor cells resulted in an altered metabolic phenotype in mature adipocytes. In co-cultured adipocytes, the appearance of fibroblast-like cells was observed, and the mesenchymal cell differentiation pathway was enriched following the integrated analysis into the transcriptome. In addition, reverse transcription-quantitative PCR analyses of co-cultured adipocytes revealed a loss in gene expression of mature adipocyte markers, and a gain in gene expression of fibroblast-specific markers. It was also confirmed that newly generated cancer-associated adipocytes could facilitate the invasive capacities of the tumor, and may contribute to PC stromal remodeling. The present study supports a novel concept that reprogramming of stromal adipocytes orchestrated by PC cells may generate cancer-associated adipocytes with activated phenotypes, which may ultimately drive pancreatic tumor progression.
Keywords: pancreatic cancer, adipocyte, tumor microenvironment, delipidation, dedifferentiation, transcriptome
Introduction
Pancreatic cancer (PC) is the fourth leading cause of cancer-associated mortality worldwide (1), and the majority of patients succumb within 6 months of the initial diagnosis (2). Poor prognosis is closely associated with the unsatisfactory results of currently available treatments, which can be largely ascribed to the unique pancreatic tumor microenvironment (TME) (3). Various stromal components act in concert with tumor cells, allowing a dynamic response to changing environments during tumor development and following exposure to chemotherapeutic drugs (4). PC cells usually promote a supportive TME by activating a fibro-inflammatory reaction (3). Conversely, normal host cells can be educated and activated by PC cells to promote tumor progression through the secretion of growth factors and chemokines. The majority of studies have highlighted the involvement of fibroblasts and inflammatory cells (5–8). However, to the best of our knowledge, little attention has been paid to the function of mature adipocytes in the pancreatic TME. Normal adipocytes have primarily been recognized as an energy reservoir to store surplus fuel, but now, there is solid evidence to suggest that they are vital endocrine cells with the ability to produce multiple hormones and chemokines involved in inflammatory and immune responses (9), indicating that stromal adipocytes may be excellent candidates to impact pancreatic tumor behavior and the TME.
Tumor-surrounding host cells can be educated to undergo morphological, functional and epigenetic changes and form unique tumor-associated cell types (10,11). Both in vitro and in vivo studies in breast cancer have confirmed the identity of distinct cancer-associated adipocytes, which are characterized by decreased lipid droplets and a fibroblast-like shape (12–14). In metastatic ovarian cancer, omental adipocytes at the invasive front can also undergo a lipolytic process (15). Furthermore, it has been demonstrated that cancer-associated adipocytes acquire the ability to produce numerous proinflammatory factors (12,15,16). However, little is known about the potentially altered phenotypes of PC-surrounding adipocytes. It is noteworthy that intrapancreatic fatty infiltration (pancreatic steatosis) has been demonstrated to be independently associated with the increased risk of PC precursor lesions and pancreatic carcinoma from clinical investigations (17,18). In addition, various studies have confirmed that cancer-associated adipocytes are positively involved in PC progression (19–22). Therefore, in order to further reveal the mechanisms of adipocytes in PC progression, research focusing on pancreatic cancer-associated adipocytes is required. The present study demonstrated that pancreatic cancer-associated adipocytes exhibit profound phenotypic alterations in human cancer and in vitro. The transcriptome profiling of these modified adipocytes is also outlined, which provides the basic information required for further studies.
Materials and methods
Pathological examination of adipocytes in PC specimens
The human tissue samples used in the present study were obtained from consecutive archival paraffin blocks of patients who were diagnosed with pancreatic ductal adenocarcinoma and underwent surgery in Huadong Hospital (Shanghai, China) between November 2018 and January 2019. The demographic and pathological features of the patients are listed in Table I. Written informed consent for the use of tissues in scientific research was obtained from all patients. The collection of the pathological data from patients with PC was approved by the Institutional Research Ethics Committee of Huadong Hospital, Fudan University (approval no. 2018K098). Immunohistochemical staining of paraffin-embedded tissues with rabbit anti-FABP4 polyclonal antibody (pAb; cat. no. 12802-1-AP; ProteinTech Group, Inc.) was performed in order to observe the characteristics of the adipocytes. For each slide, representative images of adipocytes surrounding the normal tissue and adipocytes in the vicinity of the PC cells were obtained. Adipocyte cell sizes were assessed using ImageJ software (National Institutes of Health) (23).
Table I.
The demographic and pathological features of the patients.
| No. | Sex | Age | Location | Tumor sizea | Tumor differentiation | TNM stageb |
|---|---|---|---|---|---|---|
| 1 | Male | 48 | Head | 4.6 cm | Moderate | IIB |
| 2 | Male | 66 | Head | 6.8 cm | Poor | III |
| 3 | Female | 76 | Head | 4.9 cm | Poor | III |
| 4 | Male | 67 | Head | 4.1 cm | Well | IIA |
| 5 | Male | 62 | Tail | 4.3 cm | Moderate | IIB |
| 6 | Female | 61 | Body | 5.7 cm | Poor | IIA |
| 7 | Male | 69 | Tail | 3.3 cm | Moderate | IB |
| 8 | Female | 79 | Head | 2.4 cm | Well | IB |
| 9 | Female | 64 | Head | 3.6 cm | Poor | IIA |
| 10 | Female | 66 | Body | 5.1 cm | Poor | IIB |
Maximum tumor dimension
8th edition of AJCC TNM staging for PC. AJCC, American Joint Committee on Cancer; PC, pancreatic cancer.
Cell culture and coculture
The human PC cell lines Panc-1 and Mia PaCa2 were purchased from the Cell Bank of the Type Culture Collection of Chinese Academy of Sciences and cultured according to standard protocols. In brief, Panc-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.), containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Mia PaCa2 cells were cultured in DMEM, supplemented with 10% FBS and 2.5% horse serum. Murine 3T3-L1 preadipocytes were purchased from the American Type Culture Collection and cultured in DMEM supplemented with 10% newborn calf serum. All cells were cultured at 37°C (5% CO2) in a humidified incubator. Two days after reaching 100% confluence (designated as Day 0), the 3T3-L1 preadipocytes were then induced to differentiate in DMEM containing 10% FBS, 0.5 mM 3-isobutyl-1-methylxanthine (M), 1 µM dexamethasone (D) and 1 µg/ml insulin (I). After 48 h (Day 2), these cells were sustained in DMEM supplemented with 10% FBS and 1 µg/ml insulin for another 2 days. Next, 3T3-L1 mature adipocytes were cultured in DMEM supplemented with 10% FBS. After induction (at Day 8), mature adipocytes were cocultured with PC cells using a Transwell indirect coculture system (0.4 µm pore size; Corning), and mature adipocytes cultured alone at the same time-points were used as controls.
RNA sequencing (RNA-seq) of 3T3-L1 cells
Total RNA was extracted from 3T3-L1 mature adipocytes using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA-seq libraries were prepared according to the TruSeq™ RNA Sample Preparation kit from Illumina. Briefly, 5 µg total RNA was isolated according to the polyA mRNA selection method by oligo(dT) beads and then fragmented. Double-stranded cDNA was synthesized using a SuperScript Double-stranded cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) with random primers (Illumina). Libraries were size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose, and the cDNA library was amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. Quantified by TBS380, amplified cDNA fragments were sequenced with the Illumina HiSeq X10 platform (2×150 bp read length). The expression level of each gene was calculated using the fragments per kilobase of transcript per million (FPKM) base pairs sequenced method (24). In order to identify the differentially expressed genes (DEGs) between 2 groups (3 biological replicates per group), Cuffdiff was used (25). Genes with a fold change ≥2 and a false discovery rate (FDR) <0.05 were identified as significant DEGs. In order to comprehend the functions of these DEGs, Gene Ontology (GO) functional enrichment and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using Goatools and KOBAS, respectively (26). Heat-map representations of the expression of selected genes expression were generated using the ‘pheatmap’ R package.
Oil Red O staining
Following three washes with PBS, 3T3-L1 adipocytes were fixed with 3.7% formaldehyde for 20 min. Stock Oil Red O solution (0.5% in isopropanol) was diluted with distilled water (3:2). Fixed cells were subsequently stained with Oil Red O solution at room temperature for 1 h (27). The cells were then washed with water and visualized by light microscopy.
Reverse transcriptase-quantitative PCR (RT-qPCR)
Following total RNA extraction, PrimeScript RT Master Mix (Takara Bio) was used for first-strand cDNA synthesis with random primers. The expression of the considered genes was determined via RT-qPCR, which was carried out with SYBR-Green PCR Master Mixture reagent (Applied Biosystems; Thermo Fisher Scientific, Inc.) using an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling conditions were as follows: 10 min denaturation at 95°C, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Relative RNA quantity was calculated using the 2−ΔΔCq method with 18S rRNA as an endogenous control (28). All reactions were performed in triplicate. The primer sequences used in the present study are listed in Table II.
Table II.
Primers used in the present study.
| Gene name | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) |
|---|---|---|
| 18S | CGCCGCTAGAGGTGAAATTCT | CATTCTTGGCAAATGCTTTCG |
| C/EBPα | CAAGAACAGCAACGAGTACCG | GTCACTGGTCAACTCCAGCAC |
| PPAR-γ | CCGAAGAACCATCCGATTGA | TTTGTGGATCCGGCAGTTAAG |
| FABP4 | TTCGATGAAATCACCGCAGA | GGTCGACTTTCCATCCCACTT |
| HSL | GGCTTACTGGGCACAGATACCT | CTGAAGGCTCTGAGTTGCTCAA |
| ATGL | CTGAGAATCACCATTCCCACATC | CACAGCATGTAAGGGGGAGA |
| FASN | AGAGATCCCGAGACGCTTCT | GCTTGGTCCTTTGAAGTCGAAGA |
| CIDEA | GCCTGCAGGAACTTATCAGC | AGAACTCCTCTGTGTCCACCA |
| Glut4 | ACACTGGTCCTAGCTGTATTC | CCAGCCACGTTGCATTGTA |
| IRS1 | CCAGCCTGGCTATTTAGCTG | CCCAACTCAACTCCACCACT |
| APN | TGGAATGACAGGAGCTGAAGG | TATAAGCGGCTTCTCCAGGCT |
| RESISTIN | TCGTGGGACATTCGTGAAGA | GGGCTGCTGTCCAGTCTATCC |
| LEPTIN | GGGCTTCACCCCATTCTGA | TGGCTATCTGCAGCACATTTTG |
| α-SMA | GGAGAAGCCCAGCCAGTCGC | AGCCGGCCTTACAGAGCCCA |
| FSP-1 | GCTGCCCAGATAAGGAACCC | TGCGAAGAAGCCAGAGTAAGG |
| MMP-9 | TGAGTCCGGCAGACAATCCT | CGCCCTGGATCTCAGCAATA |
| PAI-1 | TCTCCAATTACTGGGTGAGTCAGA | GCAGCCGGAAATGACACAT |
| MMP11 | GCCCTCATGTCCCCTTTCTAC | CCTTCGGTCATCTGGGCTAA |
Western blot analysis
Total lysates were extracted using lysis buffer containing 50 mM Tris-HCl (pH 6.8), 2% SDS, protease inhibitor mixture (all from Roche Diagnostics), 100 mmol/l NaF, and 1 mmol/l PMSF. The protein concentration was determined by the bicinchoninic acid assay. Equal amounts (20 µg) of protein were separated via 12% SDS-PAGE, transferred to polyvinylidene fluoride membranes (EMD Millipore), blocked with 5% skim milk at room temperature for 1 h and immunoblotted with the indicated primary antibodies (1:1,000) at 4°C overnight. Rabbit anti-PPAR-γ pAb (cat. no. 16643-1-AP), rabbit anti-FABP4 pAb (cat. no. 12802-1-AP), rabbit anti-HSL pAb (LIPE; cat. no. 17333-1-AP), rabbit anti-FASN pAb (cat. no. 10624-2-AP) and rabbit anti-α-tubulin pAb (cat. no. 11224-1-AP) were purchased from ProteinTech Group, Inc. Rabbit anti-phospho-Akt monoclonal antibody (mAb; cat. no. 4060) and rabbit anti-Akt mAb (cat. no. 4691) were purchased from Cell Signaling Technology, Inc. The following day, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG antibodies (1:5,000; cat. no. SA00001-2; ProteinTech Group, Inc.) at 37°C for 1 h. Blots were then visualized with enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Inc.) using the ImageQuant LAS 4000 System (GE Healthcare). Semi-quantification analysis was conducted by Quantity One software, version 4.6 (Bio-Rad Laboratories).
Transwell cell migration assay
PC cells (Panc-1 and Mia PaCa2) alone and cocultured with mature adipocytes were assessed for their ability to migrate through an 8 µm pore membrane (24-well insert; Corning). After 5 days of coculture with adipocytes, tumor cells were trypsinized and seeded (5×104) into the upper chamber in serum-free DMEM. The lower chamber was filled with medium containing 10% FBS. After incubation for 36 h, cells on the bottom of the insert were fixed with 100% methanol and stained in 0.1% crystal violet at room temperature for 30 min. The number of migrating cells was counted separately in three randomly selected fields using a light microscope (IX71; Olympus).
Statistical analysis
Statistical analysis of all data was completed using GraphPad Prism software (version 5.0; GraphPad Software, Inc.). All experiments were performed in triplicate. The results are expressed as the mean ± standard deviation. Unpaired two-tailed Student's t-test was performed to determine significance of differences between two groups. P<0.05 was considered to indicate a statistically significant difference. P-values are presented in the figures (*P<0.05; **P<0.01; ***P<0.001).
Results
Pancreatic cancer cells stimulate delipidation in mature adipocytes
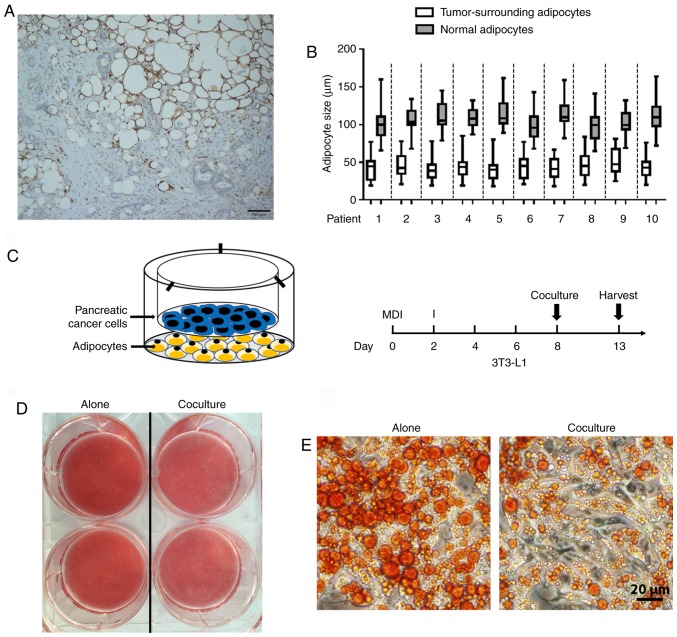
A series of studies have demonstrated crosstalk between cancer cells and adipocytes, but these have largely focused on the postive effect of adiopocytes on tumor progression (29). In order to determine the direct impact of PC cells on mature adipocytes, the presence of adipocytes in human pancreatic tumors was first investigated. To address this issue, immunohistochemical staining of FABP4 (highly expressed in mature adipocytes) was performed in tumor samples. The presence of FABP4-stained adipocytes at the invasive front of pancreatic tumors was observed (Fig. 1A). Furthermore, tumor-neighboring adipocytes were smaller in size compared with those normal adipocytes further away (Fig. 1B). In order to assess the morphological changes of PC-related adipocytes, an adipocyte-PC cell indirect coculture system was established (Fig. 1C, left panel). First, 3T3-L1 cells were induced to differentiate into mature adipocytes, and human PC cells were then cultured on top (Fig. 1C, right panel). As indicated by Oil Red O staining, following coculture with Panc-1 for 5 days, mature adipocytes exhibited a decrease in lipid content (Fig. 1D) with a marked decrease in the size of lipid droplets (Fig. 1E) compared with those cultured alone, which is consistent with the clinical features of small adipocytes adjacent to the tumor tissues.
Figure 1.
PC cells stimulate delipidation in mature adipocytes. (A) FABP4-stained adipocytes at the invasive front of the tumor exhibited smaller lipid droplets compared with normal adipocytes. (B) The cell diameters of tumor-surrounding adipocytes and normal adipocytes are presented. For each patient, fifty adipocytes were randomly selected in tumor-surrounding adipose tissue or normal adipose tissue, respectively, and the maximum diameter of each adipocyte was calculated. Data are presented in the box plots as the mean ± standard deviation. (C) Left, schematic of the indirect coculture system. Right, experimental timeline of the coculture approach. After 8 days of differentiation, mature 3T3-L1 adipocytes were cultured with or without pancreatic cancer cells for another 5 days and harvested. (D) Mature adipocytes cocultured with or without Panc-1 tumor cells were stained with red oil. (E) A decrease in the size of lipid droplets in adipocytes following coculture with Panc-1 for 5 days was observed. PC, pancreatic cancer.
Altered gene expression profiling is identified in pancreatic cancer-associated adipocytes
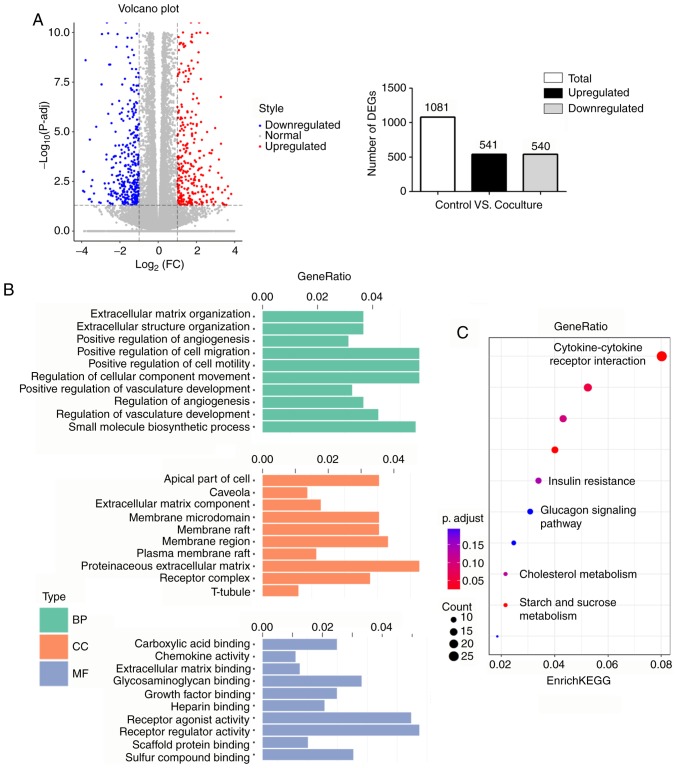
Given the great phenotypic changes of cancer-associated adipocytes in other types of tumor, such as breast and prostate cancer (13,16), we reasoned that PC-associated adipocytes may change their gene expression patterns. RNA-sequencing was performed on 3T3-L1 cells cultured with or without PC cells, and identified 1,081 genes that were differentially expressed in cancer-associated adipocytes, including 541 upregulated and 540 downregulated genes (Fig. 2A). Next, all DEGs were characterized through GO term mapping according to their major GO categories [i.e., the GO terms ‘biological processes’ (BP), ‘cellular components’ (CC), and ‘molecular functions’ (MF)] (Fig. 2B). The top BP GO annotation was ‘positive regulation of cell migration/motility/cellular component movement’. Within the CC of ontology, the most abundant term was ‘proteinaceous extracellular matrix’. Furthermore, ‘receptor regulator activity’ was primarily enriched in the MF group. DEGs were also mapped to the KEGG pathway database (Fig. 2C). Numerous metabolic pathways, such as ‘insulin resistance’, ‘glucagon signaling pathway’ and ‘cholesterol metabolism’, were enriched, indicating that diverse metabolic changes were occurring in the cancer-associated adipocytes. It was also revealed that ‘cytokine-cytokine receptor interaction’ was the most significantly enriched KEGG pathway.
Figure 2.
Altered gene expression profiling is identified in mature adipocytes following coculture with Panc-1 cancer cells. (A) Left, volcano plot of DEGs. Upregulated genes in coculture adipocytes are colored in red, and downregulated genes in blue. Right, schematic representation of the DEGs. Of 1,081 DEGs, 541 were upregulated, while 540 were downregulated in the coculture group compared with the control group. (B) Histogram of GO annotations. The three major categories, BP, CC and MF, were analyzed. (C) A scatter diagram of enriched KEGG pathways. The level of KEGG enrichment was measured using GeneRatio. The color and size of the dots represent the range of P-adjusted values and gene counts, respectively. DEGs, differentially expressed genes; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Pancreatic cancer induces dysregulated metabolism in adipocytes
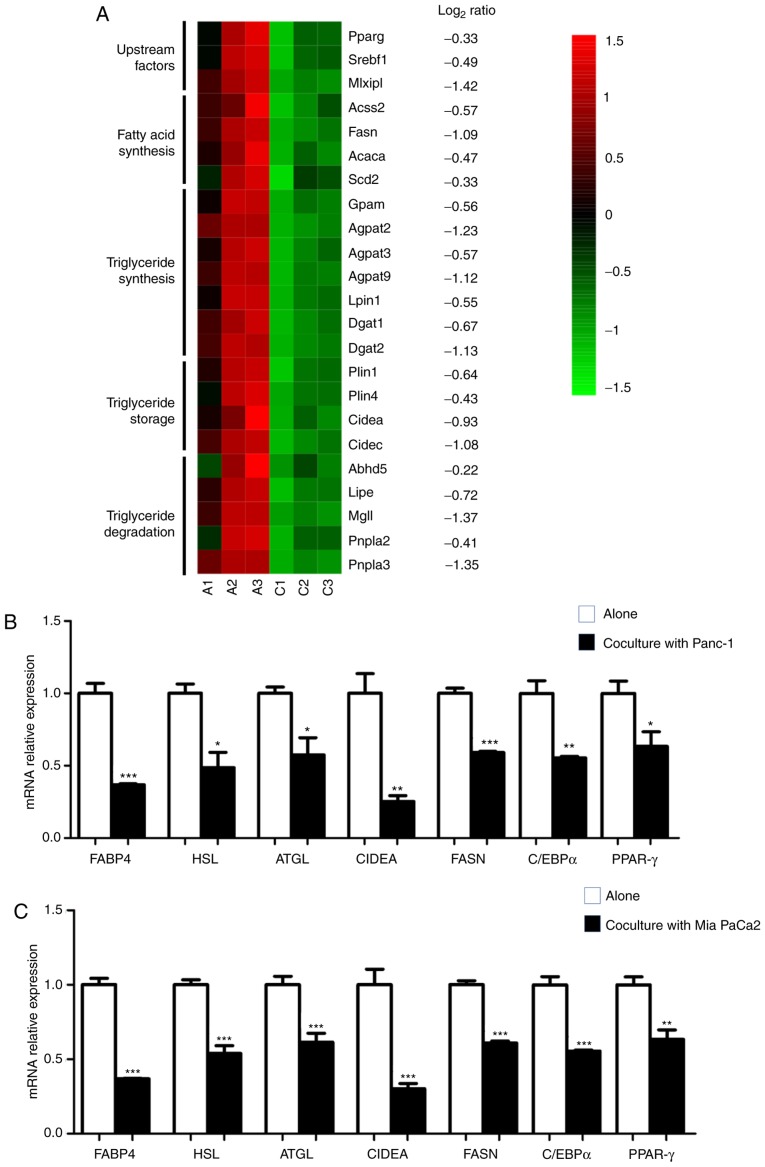
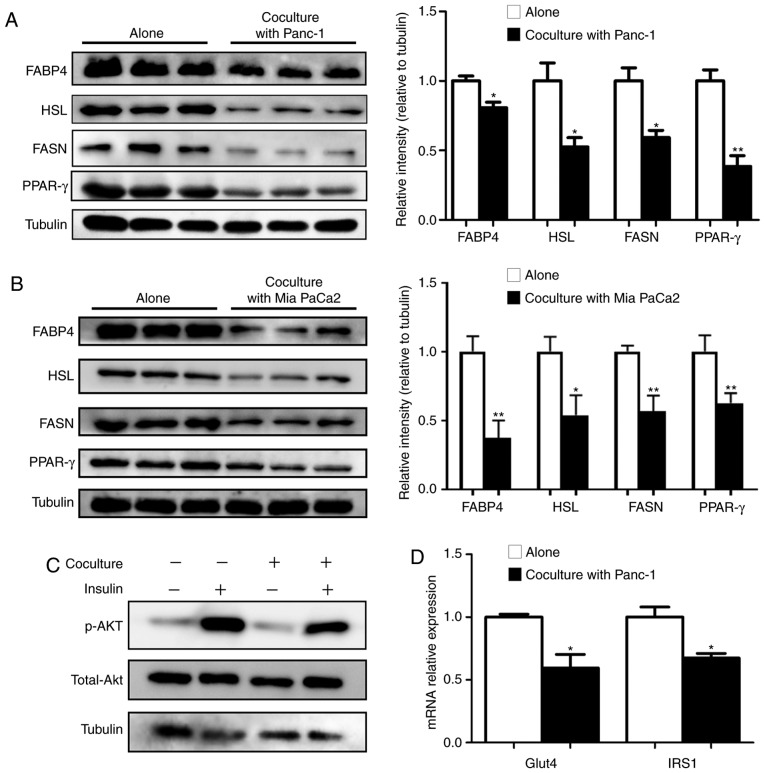
The aforementioned data revealed that upon coculture with PC cells, a marked decrease in lipid accumulation and a profound impact on gene expression networks associated with metabolism were exhibited in adipocytes. Furthermore, a broad spectrum of genes involved in fatty acid and triglyceride synthesis for lipogenesis, and their upstream regulators, as well as triglyceride storage and degradation, were significantly decreased (Fig. 3A), revealing the dysregulated lipid homeostasis in cocultured adipocytes. RT-qPCR analysis of adipocytes cocultured with Panc-1 (Fig. 3B) or with another PC cell line Mia PaCa2 (Fig. 3C) was performed, which confirmed the substantial inhibition of lipid metabolism-associated genes, including FABP4, HSL, ATGL, CIDEA and FASN, and of two key adipogenic upstream factors, PPAR-γ and C/EBPα. In line with the RT-qPCR data, immunoblotting analysis also confirmed decreased levels of FABP4, HSL, FASN and PPAR-γ in cocultured adipocytes (Fig. 4A and B). The present study also investigated the effect of coculture on adipocyte glucose metabolism and revealed that there was a marked decrease in the insulin-induced phosphorylation of Akt (Fig. 4C), as well as the transcriptional expression of Glut4 and IRS1 (Fig. 4D).
Figure 3.
PC induces dysregulated metabolism in adipocytes. (A) Heatmap of RNA-sequencing expression of genes associated with lipid metabolism signature between adipocytes grown alone (A1-A3) and cocultured with Panc-1 (C1-C3). (B and C) Reverse transcription-quantitative PCR of lipid metabolism-associated genes in adipocytes grown alone, and cocultured in the presence of (B) Panc-1 or (C) Mia PaCa2. Data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001. PC, pancreatic cancer.
Figure 4.
Impaired lipid metabolism function in cocultured adipocytes is accompanied by insulin resistance. Immunoblots and quantification of key proteins associated with lipid metabolism in adipocytes cultured alone, and cocultured with (A) Panc-1 or with (B) Mia PaCa2. (C) The mature adipocytes were cultivated with/without Panc-1 for 5 days before incubation with 100 nM insulin for 30 min. The expression of total and p-Akt was analyzed via western blotting. (D) Reverse transcription-quantitative PCR analysis of Glut4 and IRS-1 mRNA expression in mature adipocytes grown with/without Panc-1 for 5 days. Data are presented as the mean ± standard deviation. *P<0.05; **P<0.01. p-Akt, phosphorylated Akt.
Pancreatic cancer-associated adipocytes undergo dedifferentiation
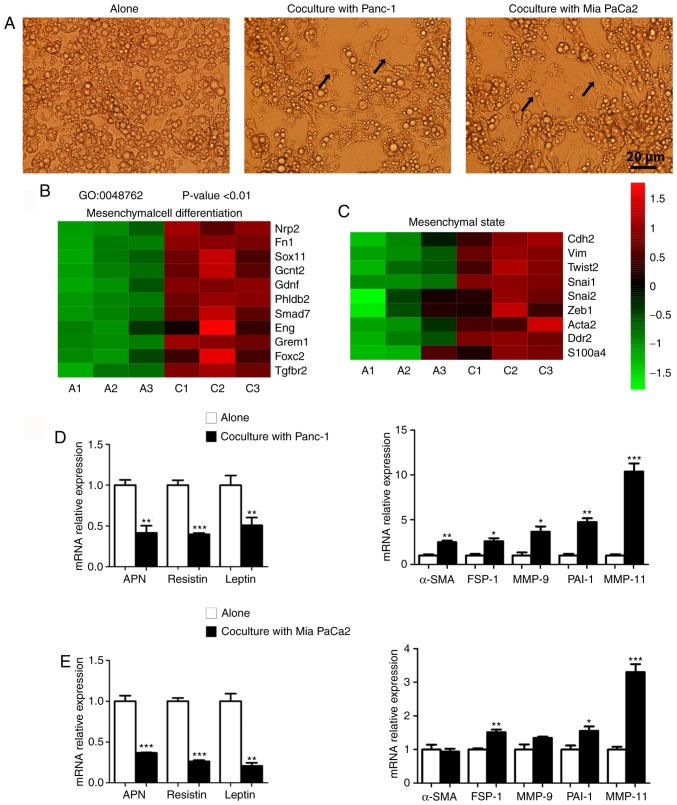
Adipocytes are commonly regarded as terminally differentiated cells, but increasing evidence suggests they may have greater plasticity under certain circumstances (12,30). In the present study, the ‘fat cell differentiation’ pathway was highly enriched in cocultured adipocytes. A total of 25 DEGs were involved in this process (Table III) among which several mature adipocyte markers, such as leptin and resistin, were significantly downregulated. It was also observed that, following coculture with pancreatic cancer cells for 5 days, numerous adipocytes exhibited a stretched, fibroblastic shape (Fig. 5A). Further analysis using the GO BP demonstrated that the mesenchymal cell differentiation process was activated in cocultured adipocytes (Fig. 5B). There was also a marked increase in the expression of mesenchymal state-associated genes (31,32), indicating a shift towards more mesenchymal phenotypes in these cocultured adipocytes (Fig. 5C). The RT-qPCR results demonstrated that adipocytes cocultured with Panc-1 exhibited a loss in expression of mature adipocyte markers, including adiponectin (APN), leptin and resistin, and a gain in expression of fibroblast markers, including α-SMA, FSP-1, MMP-9, PAI-1 and MMP-11 (Fig. 5D). Similar results were obtained by coculturing adipocytes with Mia PaCa2 (Fig. 5E).
Table III.
DEGs involved in the fat differentiation pathway in coculture adipocytes.
| Gene ID | Description | log2 FC | Regulation |
|---|---|---|---|
| RNASEL | Ribonuclease L | 1.034174 | Up |
| ENPP1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 1.108244 | Up |
| MMP11 | Matrix metallopeptidase 11 | 2.89554 | Up |
| VSTM2A | V-set and transmembrane domain containing 2A | 1.242257 | Up |
| ARL4A | ADP-ribosylation factor-like 4A | −1.43843 | Down |
| ERO1L | ERO1-like | 1.709092 | Up |
| KLF5 | Kruppel-like factor 5 | 1.345372 | Up |
| CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | 1.382325 | Up |
| ADGRF5 | Adhesion G protein-coupled receptor F5 | 1.84409 | Up |
| ZBTB7C | Zinc finger and BTB domain containing 7C | 1.041932 | Up |
| CCDC85B | Coiled-coil domain containing 85B | 1.669037 | Up |
| SCD1 | Stearoyl-Coenzyme A desaturase-1 | −1.01976 | Down |
| ADIG | Adipogenin | −1.21847 | Down |
| NOCT | Nocturnin | −1.14398 | Down |
| KLF4 | Kruppel-like factor 4 | 1.189328 | Up |
| TRPV4 | Transient receptor potential cation channel subfamily V, member 4 | 1.235702 | Up |
| TMEM120A | Transmembrane protein 120A | −1.1772 | Down |
| MEDAG | Mesenteric estrogen dependent adipogenesis | 1.111338 | Up |
| LEP | Leptin | −1.84249 | Down |
| CREB5 | cAMP responsive element binding protein 5 | 1.663459 | Up |
| CLIP3 | CAP-GLY domain containing linker protein 3 | 1.930421 | Up |
| AAMDC | Adipogenesis associated Mth938 domain containing | −1.15184 | Down |
| RETN | Resistin | −1.56133 | Down |
| ADRB3 | Adrenergic receptor, β3 | −1.69788 | Down |
| ANGPTL8 | Angiopoietin-like 8 | −1.03671 | Down |
DEGs, differentially expressed genes.
Figure 5.
PC-associated adipocytes undergo a dedifferentiation process. (A) The morphological changes of mature adipocytes towards fibroblast-like cells were observed following coculture with Panc-1 (middle) or Mia PaCa2 cells (right). Arrows indicate typical fibroblast-like cells. (B) Heatmap of enriched GO: Mesenchymal cell differentiation signature in adipocytes grown with Panc-1 vs. cultured alone. (C) Heatmap of RNA-sequencing expression of genes associated with the mesenchymal state in adipocytes grown alone and cocultured with Panc-1. (D and E) Reverse transcription-quantitative PCR analysis of mature adipocyte markers (APN, Resistin and Leptin) and fibroblast-specific markers (α-SMA, FSP-1, MMP-9, PAI-1 and MMP-11) in adipocytes cultured alone, and cocultured with (D) Panc-1 or with (E) Mia PaCa2. Data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001. PC, pancreatic cancer; GO, Gene Ontology.
Cancer-associated adipocytes increase PC cell aggressiveness
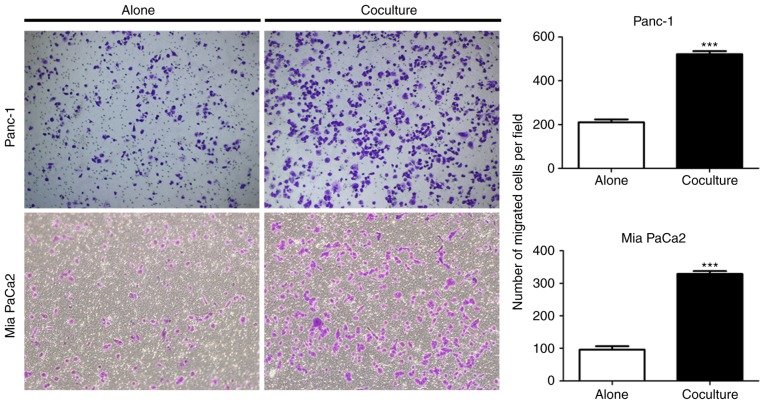
Previous research has demonstrated that adipocytes activated by tumor cells exerted an active role in tumor progression and TME remodeling (33). In the present study, bioinformatics analysis of the whole transcriptome of adipocytes revealed several enriched pathways associated with the remodeling of the pancreatic cancer stroma (Table IV), including the cytokine-mediated signaling pathway, angiogenesis and extracellular matrix organization. In order to directly assess the role of cancer-associated adipocytes in PC progression, the present study performed Transwell assays to assess the ability of the PC cells to traverse the membrane. PC cells were cocultured with or without adipocytes for 5 days, and it was revealed that both adipocyte-exposed Panc-1 and Mia PaCa2 tumor cells had a significantly increased ability to fully migrate through a Transwell membrane (Fig. 6). Overall, these results indicated that adipocytes can increase PC cell aggressiveness.
Table IV.
Summary of enriched pathways potentially related to the remodeling of the pancreatic tumor microenvironment in cocultured adipocytes.
| Category | Gene count | Genes | P-value |
|---|---|---|---|
| Cytokine-mediated signaling pathway | 28 | ACKR3/GPR35/GREM2/IRAK3/CCL11/CCL8/CCL9/CCL6/SPHK1SOCS3/LRTM1/OSMR/PRLR/IL2RB/PDGFB/LRRC15/TREM2/RTN4RL2/ECM1/CXCL9/CXCL10/CXCL13/EPO/LEP/IRF7/ACSL1/CX3CL1/CASP4 | 2.65 e-6 |
| Angiogenesis | 37 | NRP2/FN1/IHH/ACKR3/CHIL1/ITGB2/ADORA2B/CCL11/SPHK1/AHR/PRL2C2/PLAU/KLF5/RUNX1/DLL1/FGF1/ENG/GREM1/ANGPT4/ECM1/CYR61/KLF4/NOS3/CXCL10/ANXA3/HSPB1/SERPINE1/EPO/LEP/AQP1/ADM/HMOX1/CX3CL1/FOXC2/THY1/CAMP/TGFBR2 | 7.28 e-8 |
| Extracellular matrix organization | 27 | FN1/IHH/LAMC2/MMP11/ECM2/ERO1L/EGFLAM/FBLN1/MYH11 PHLDB2/HAS1/LOX/ADAMTSL2/ENG/OLFML2A/WT1/GREM1/FERMT1/NPNT/CYR61/EMILIN1/ELN/FSCN1/BCL3/APLP1/FOXC2/MMP13 | 4.73 e-10 |
Figure 6.
Coculture of pancreatic cancer cells with adipocytes increases their aggressiveness. Migration of Panc-1 and Mia PaCa2 cancer cells grown alone or cocultured with adipocytes. Representative images (left) and quantification (right) of three independent experiments are presented. Data are presented as the mean ± standard deviation. ***P<0.001.
Discussion
Research focusing on the occult PC stroma has resulted in an increased level of knowledge and understanding; however, the current understanding of cancer-associated adipocytes remains unclear compared with that of other cell types, forming the impetus for the research presented. It was consistently observed that adipocytes adjacent to human pancreatic tumors had smaller lipid droplet sizes. In the in vitro coculture system of the present study, adipocytes demonstrated a status of delipidation and impaired lipid homeostasis, indicating that PC cells are able to blunt energy-consuming activities, including adipogenesis in cocultured adipocytes. The impact of the tumor shutting down adipocyte metabolism should serve to save nutrients (such as glucose) that would be accessible for the tumor. Regarding the mechanisms whereby PC cells decreased energy utilization in adipocytes, it was observed that dysregulated lipid metabolism was accompanied by insulin resistance, demonstrated by a decrease in insulin-stimulated phosphorylation of Akt, as well as decreased Glut4 and IRS1 mRNA expression in cocultured adipocytes. Previous studies have revealed that Glut4-mediated adipose tissue glucose influx could activate de novo lipogenesis, and the latter favored enhanced systemic insulin sensitivity (34,35). This is of clinical relevance since it has been recognized that PC is a powerful diabetogenic state and new-onset diabetes in patients with PC is likely to be induced by the secreted products of cancer cells (36). The metabolic reprogramming of adipocytes orchestrated by cancer cells would help PC deal with the high energy demand during tumor progression through the control of non-cancer cell insulin sensitivity. Furthermore, it is well recognized that adipocyte-derived lipids are potent energy sources that fulfill the energetic needs of tumor cells (15,37,38). In this regard, the phenomenon of delipidation could also result from the abundant release of free fatty acids in cocultured adipocytes. In fact, the increased accumulation of lipid in PC cells during coculturing with adipocytes was observed in the present study (data not shown). Collectively, these results indicated that multiple modes of metabolic crosstalk occur between stromal adipocytes and PC cells.
Another key concern arising from the present study is the process of dedifferentiation in adipocytes induced by PC cells. A number of studies have suggested that adipocytes retain their capacity to return to a precursor state. Cell lineage tracing studies in mouse models have revealed that adipocytes could undergo reprogramming into fibroblasts within fibrotic dermis (39), or into preadipocytes during lactation (40). In agreement with previously reported results demonstrating such adipocyte plasticity during adipocyte-cancer crosstalk (13,41–43), the present study revealed that cocultured adipocytes underwent dedifferentiation, forming the mesenchymal cell type and giving rise to fibroblast-like cells. It will be of great interest to determine the underlying mechanisms of this process. In this regard, the factors secreted by tumor cells are likely to be the key regulators. Both Wnt3a released by breast cancer cells (13) and Wnt5a secreted from melanoma cells (42) have been reported as triggering factors of adipocyte dedifferentiation in the tumor microenvironment. In addition, the aforementioned role of adipocyte metabolic alteration on the dedifferentiation process cannot be excluded. It is not currently known whether metabolic changes have a critical impact on the adipocyte differentiation and vice versa, therefore further research is required.
Notably, for the adipocytes exposed to PC cells, in addition to the impairment of metabolism and concomitant dedifferentiation process, it is of great interest to determine whether they have acquired pluripotency, and can reprogram their genomes to form new subsets. The data from the present study demonstrated that adipocytes cocultured with cancer cells maintain a mesenchymal state, which may facilitate the entrance into a stem-cell state (44). In addition, bioinformatics analysis of the transcriptome of adipocytes revealed, following coculture, a marked change in gene expression networks associated with the modulation of matrix remodeling. These adipocytes could obtain new competencies that contribute to the remodeling of the tumor stroma. In breast cancer, cancer-associated adipocytes revert to a fibroblastic cell population and take part in the desmoplastic reaction (13). PC is a highly desmoplastic malignancy characterized by excessive deposition of the extracellular matrix (ECM) (45). Obesity is positively correlated with increased ECM deposition in pancreatic TME, and adipocytes in the fatty PC stroma can activate pancreatic stellate cells, indirectly leading to excessive desmoplasia (19). Collectively, these results highlight the role of cancer-associated adipocytes in the PC desmoplastic stroma. The present study also characterized the cytokine-mediated signaling and the positive regulation of angiogenesis in PC-induced adipocytes. The inappropriate release of proinflammatory factors by adipose tissue has been extensively recognized as the primary cause of obesity-associated disorders, including cancer (46,47). In adipose tissue, the presence of pancreatic tumor cells may result in a marked increased inflammatory response. In addition, adipose tissue is one of the most vascularized tissues in the body with the highest capacity for angiogenesis (48), and highly vascularized adipose tissue may provide a predetermined environment to foster tumors. Thus, it is reasonable to speculate that upon interplay with adipose tissue, tumors may mobilize the angiogenetic process of adipocytes for their own sake.
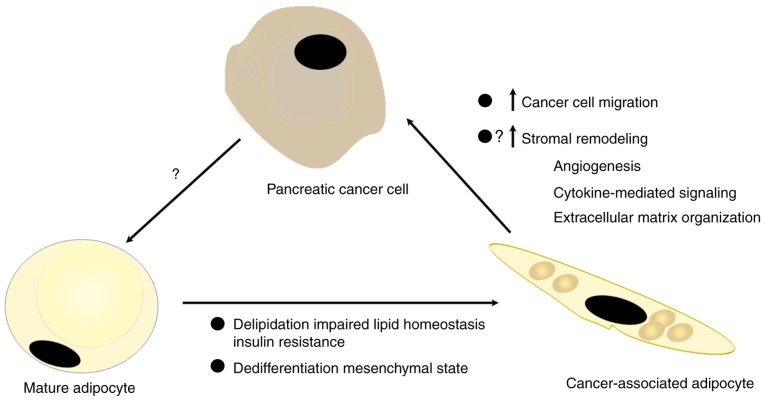
In summary, the present study revealed the distinct phenotypes of pancreatic cancer-associated adipocytes. The model utilized, as presented in Fig. 7, proposes systemic crosstalk between PC and adipocytes, providing critical insights towards the understanding of the contribution of adipocytes to the formation of a unique pancreatic tumor stroma, which ultimately leads to increased tumor malignancy.
Figure 7.
Schematic representation of the crosstalk between pancreatic cancer cells and mature adipocytes. Cancer cells promote delipidation and dedifferentiation in adipocytes, and then newly generated cancer-associated adipocytes could facilitate the tumor invasive capacities and stromal remodeling.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The sequencing data have been deposited in the Gene Expression Omnibus with the assigned accession number, GSE123939. The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZC, YL, WW and CJ participated in the conception and design of the present study. ZC, YL, CX performed the statistical analysis and were involved in the preparation of the figures. HW, JL and PH participated in the analysis of the figures and data. ZC, YL, WW and CJ prepared and revised the manuscript. HH reviewed the results and revised the manuscript. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Institutional Research Ethics Committee of Huadong Hospital, Fudan University (approval no. 2018K098). Written informed consent for the use of tissues in scientific research was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62:317–326. doi: 10.1136/gutjnl-2012-303588. [DOI] [PubMed] [Google Scholar]
- 3.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, Holland EC, Sutton JC, Joyce JA. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352:6288. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol. 2017;10:76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, Ungefroren H, Vogel I, Kruger U, Becker T, Ebsen M, Rocken C, et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int J Cancer. 2014;135:843–861. doi: 10.1002/ijc.28736. [DOI] [PubMed] [Google Scholar]
- 7.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, Logsdon CD. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Re. 2008;68:918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T, Ouellette MM, Hollingsworth MA. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng Y, Scherer PE. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 2010;1212:E1–E19. doi: 10.1111/j.1749-6632.2010.05875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Q, Zhou D, Rucki AA, Williams J, Zhou J, Mo G, Murphy A, Fujiwara K, Kleponis J, Salman B, et al. Cancer-associated fibroblasts in pancreatic cancer are reprogrammed by tumor-induced alterations in genomic DNA methylation. Cancer Res. 2016;76:5395–5404. doi: 10.1158/0008-5472.CAN-15-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144:1210–1219. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirat B, Bochet L, Dabek M, Daviaud D, Dauvillier S, Majed B, Wang YY, Meulle A, Salles B, Le Gonidec S, et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011;71:2455–2465. doi: 10.1158/0008-5472.CAN-10-3323. [DOI] [PubMed] [Google Scholar]
- 13.Bochet L, Lehuede C, Dauvillier S, Wang YY, Dirat B, Laurent V, Dray C, Guiet R, Maridonneau-Parini I, Le Gonidec S, et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013;73:5657–5668. doi: 10.1158/0008-5472.CAN-13-0530. [DOI] [PubMed] [Google Scholar]
- 14.Wang YY, Attane C, Milhas D, Dirat B, Dauvillier S, Guerard A, Gilhodes J, Lazar I, Alet N, Laurent V, et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2:e87489. doi: 10.1172/jci.insight.87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent V, Guerard A, Mazerolles C, Le Gonidec S, Toulet A, Nieto L, Zaidi F, Majed B, Garandeau D, Socrier Y, et al. Periprostatic adipocytes act as a driving force for prostate cancer progression in obesity. Nat Commun. 2016;7:10230. doi: 10.1038/ncomms10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori M, Takahashi M, Hiraoka N, Yamaji T, Mutoh M, Ishigamori R, Furuta K, Okusaka T, Shimada K, Kosuge T, et al. Association of pancreatic fatty infiltration with pancreatic ductal adenocarcinoma. Clin Transl Gastroenterol. 2014;5:e53. doi: 10.1038/ctg.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebours V, Gaujoux S, d'Assignies G, Sauvanet A, Ruszniewski P, Levy P, Paradis V, Bedossa P, Couvelard A. Obesity and fatty pancreatic infiltration are risk factors for pancreatic precancerous lesions (PanIN) Clin Cancer Res. 2015;21:3522–3528. doi: 10.1158/1078-0432.CCR-14-2385. [DOI] [PubMed] [Google Scholar]
- 19.Incio J, Liu H, Suboj P, Chin SM, Chen IX, Pinter M, Ng MR, Nia HT, Grahovac J, Kao S, et al. Obesity-Induced inflammation and desmoplasia promote pancreatic cancer progression and resistance to chemotherapy. Cancer Discov. 2016;6:852–869. doi: 10.1158/2159-8290.CD-15-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura T, Ohuchida K, Sada M, Abe T, Endo S, Koikawa K, Iwamoto C, Miura D, Mizuuchi Y, Moriyama T, et al. Extra-pancreatic invasion induces lipolytic and fibrotic changes in the adipose microenvironment, with released fatty acids enhancing the invasiveness of pancreatic cancer cells. Oncotarget. 2017;8:18280–18295. doi: 10.18632/oncotarget.15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler KM, Considine RV, True E, Swartz-Basile DA, Pitt HA, Zyromski NJ. Adipocytes enhance murine pancreatic cancer growth via a hepatocyte growth factor (HGF)-mediated mechanism. Int J Surg. 2016;28:179–184. doi: 10.1016/j.ijsu.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbone C, Piro G, Gaianigo N, Ligorio F, Santoro R, Merz V, Simionato F, Zecchetto C, Falco G, Conti G, et al. Adipocytes sustain pancreatic cancer progression through a non-canonical WNT paracrine network inducing ROR2 nuclear shuttling. Int J Obes (Lond) 2018;42:334–343. doi: 10.1038/ijo.2017.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 27.Huang H, Song TJ, Li X, Hu L, He Q, Liu M, Lane MD, Tang QQ. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proce Natl Acad Sci USA. 2009;106:12670–12675. doi: 10.1073/pnas.0906266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Park J, Morley TS, Kim M, Clegg DJ, Scherer PE. Obesity and cancer-mechanisms underlying tumour progression and recurrence. Nat Rev Endocrinol. 2014;10:455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corsa CAS, MacDougald OA. Cyclical dedifferentiation and redifferentiation of mammary adipocytes. Cell Metab. 2018;28:187–189. doi: 10.1016/j.cmet.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Lengyel E, Makowski L, DiGiovanni J, Kolonin MG. Cancer as a matter of fat: The crosstalk between adipose tissue and tumors. Trends Cancer. 2018;4:374–384. doi: 10.1016/j.trecan.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MA, Peroni OD, Villoria J, Schon MR, Abumrad NA, Bluher M, Klein S, Kahn BB. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sah RP, Nagpal SJ, Mukhopadhyay D, Chari ST. New insights into pancreatic cancer-induced paraneoplastic diabetes. Nat Rev Gastroenterol Hepatol. 2013;10:423–433. doi: 10.1038/nrgastro.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoy AJ, Balaban S, Saunders DN. Adipocyte-tumor cell metabolic crosstalk in breast cancer. Trends Mol Med. 2017;23:381–392. doi: 10.1016/j.molmed.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Miranda F, Mannion D, Liu S, Zheng Y, Mangala LS, Redondo C, Herrero-Gonzalez S, Xu R, Taylor C, Chedom DF, et al. Salt-inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell. 2016;30:273–289. doi: 10.1016/j.ccell.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Marangoni RG, Korman BD, Wei J, Wood TA, Graham LV, Whitfield ML, Scherer PE, Tourtellotte WG, Varga J. Myofibroblasts in murine cutaneous fibrosis originate from adiponectin-positive intradermal progenitors. Arthritis Rheumatol. 2015;67:1062–1073. doi: 10.1002/art.38990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang QA, Song A, Chen W, Schwalie PC, Zhang F, Vishvanath L, Jiang L, Ye R, Shao M, Tao C, et al. Reversible de-differentiation of mature white adipocytes into preadipocyte-like precursors during lactation. Cell Metab. 2018;28:282–288. doi: 10.1016/j.cmet.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoico E, Darra E, Rizzatti V, Budui S, Franceschetti G, Mazzali G, Rossi AP, Fantin F, Menegazzi M, Cinti S, Zamboni M. Adipocytes WNT5a mediated dedifferentiation: A possible target in pancreatic cancer microenvironment. Oncotarget. 2016;7:20223–20235. doi: 10.18632/oncotarget.7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoico E, Darra E, Rizzatti V, Tebon M, Franceschetti G, Mazzali G, Rossi AP, Fantin F, Zamboni M. Role of adipose tissue in melanoma cancer microenvironment and progression. Int J Obes (Lond) 2018;42:344–352. doi: 10.1038/ijo.2017.218. [DOI] [PubMed] [Google Scholar]
- 43.Chirumbolo S, Bjorklund G. Can wnt5a and wnt non-canonical pathways really mediate adipocyte de-differentiation in a tumour microenvironment? Eur J Cancer. 2016;64:96–100. doi: 10.1016/j.ejca.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 44.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whatcott CJ, Diep CH, Jiang P, Watanabe A, LoBello J, Sima C, Hostetter G, Shepard HM, Von Hoff DD, Han H. Desmoplasia in primary tumors and metastatic lesions of pancreatic cancer. Clin Cancer Res. 2015;21:3561–3568. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bluher M. Adipose tissue dysfunction in obesity. Exp Clin Endocrinol Diabetes. 2009;117:241–250. doi: 10.1055/s-0029-1192044. [DOI] [PubMed] [Google Scholar]
- 47.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer. 2011;11:886–895. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 48.Lemoine AY, Ledoux S, Larger E. Adipose tissue angiogenesis in obesity. Thromb Haemost. 2013;110:661–668. doi: 10.1160/TH13-01-0073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data have been deposited in the Gene Expression Omnibus with the assigned accession number, GSE123939. The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.