Abstract
Calcium-dependent activator protein for secretion 1 (CAPS1) has been reported to promote metastasis in colorectal cancer (CRC), however, the underlying mechanisms have not yet been elucidated. The present study revealed that exosomes derived from CAPS1-overexpressing CRC cells could enhance the migration of normal colonic epithelial FHC cells. GW4869, an inhibitor of exosomes, could attenuate the migration of FHC cells. Furthermore, liquid chromatography-mass spectrometry (LC-MS) and bioinformatics analysis demonstrated that overexpression of CAPS1 could alter the expression pattern of exosomal proteins involved in cell migration. Bone morphogenetic protein 4, which may serve vital roles in the process of CAPS1-induced cell migration, was downregulated in the exosomes. In summary, the present results demonstrated that CAPS1 promotes cell migration by regulating exosomes. Inhibiting the secretion of exosomes may be helpful for the treatment of patients with metastatic CRC.
Keywords: CAPS1, exosome, colorectal cancer, liquid chromatography-mass spectrometry, proteomics, BMP4
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancer types, and it accounts for 10% of all cancer cases, with an occurrence of >1.8 million cases every year worldwide (1). Treatment strategies involving surgery, chemotherapy and radiotherapy have increased the overall survival rates for patients with early stage CRC (2,3). However, ~60% of patients with CRC develop metastasis, which is the major cause of mortality in patients with CRC (4,5). Thus, it is important to elucidate the mechanisms underlying the metastasis of CRC, and to investigate new CRC treatments based on the discoveries.
The activation of pro-oncogenes KRAS, c-Src and c-Myc, and inactivation of tumor suppressor genes, such as the loss of adenomatous polyposis coli (APC) and tumor suppressor p53 (TP53), all contribute to CRC tumorigenesis and development (2). Exosomes, as mediators of intercellular communication, have been reported to be involved in CRC progression (6,7). Exosomes are extracellular vesicles (EVs) with a diameter of 30–180 nm, secreted by various cells (8–10). A recent study demonstrated that exosomes derived from the HCT-8 CRC cell line contributed to the EMT process via miR-210 (6). Exosomes derived from CRC cells (HT29, SW480 and SW620) were revealed to increase the migration and invasion capacities of colonic mesenchymal cells (7,11). In addition, drug resistance during tumor treatment could also be attributed to exosomes. miR-145 and miR-34a released via exosomes are hypothesized to enhance the 5-fluorouracil resistance in DLD-1 cells (12). Epidermal growth factor receptor has been identified to be expressed on the surface of exosomes and was able to bind to Cetuximab, which reduced its therapeutic efficacy in Caco2 and HCT-116 cells (13).
Exosomes form as intraluminal vesicles (ILVs) by budding into early endosomes and multi-vesicular bodies (MVBs). Subsequently, MVBs are released upon fusion with the plasma membrane (PM) (14,15). The SNARE complex may serve a role in the fusion of MVBs with the PM. However, the underlying mechanism remains to be fully elucidated (16). On the other hand, the intracellular adaptor protein syntenin, endosomal sorting complex required for transport (ESCRT), heparanase and tetraspanins demonstrate key roles in the loading of exosomes (7). In addition, sumoylation is considered to be a mechanism involved in the loading of miRNAs into exosomes (17).
Ca2+-dependent activator protein for secretion 1 (CAPS1) is characterized as a 145-kDa brain protein containing a central pleckstrin homology domain, a Munc-13 homology (MH) domain, a C-terminal domain, a C-terminal membrane-association domain and a C2 domain (18). As a special regulator of the Ca2+-dependent large dense-core vesicle fusion process, CAPS1 promotes the assembly of the SNARE complex via the MH domain, which regulates vesicle exocytosis (19,20). Our previous study demonstrated that CAPS1 was upregulated in CRC, and that increased CAPS1 expression was associated with frequent metastasis and poor prognosis in patients with CRC (18). Furthermore, CAPS1 was revealed to promote CRC cell migration and invasion in vitro and facilitate CRC liver metastasis in vivo (18). However, whether exosomes are required for CAPS1-induced CRC metastasis requires further investigation.
BMP4 is a member of bone morphogenetic proteins (BMPs), which are multi-functional cytokines belonging to the transforming growth factor-β (TGF-β) family (21). Previous studies suggest that BMP4 is closely associated with tumorigenesis (22,23). In CRC, BMP4 was revealed to be frequently upregulated due to aberrant activation of Wnt-β-catenin signaling, and promoted cell migration and invasion (22,24). Knockdown of BMP4 inhibited tumor formation of CRC cells in vivo through apoptosis induction (22). The expression of BMP4 was significantly increased in hepatocellular carcinoma (HCC) tissues (25). Increased BMP4 was correlated with high metastasis of HCC cells (25). BMP4 facilitated HCC cell invasion and metastasis though ID2-mediated EMT and promoted HCC cell proliferation via autophagy activation (23,25). In breast cancer, BMP4 promoted cell migration and invasion possibly via induction of MMP-1 and CXCR4 expression (26). The function of BMP4 appears to be divergent but with clear evidence supporting tumor suppressing functions in lung squamous cell carcinoma (SQC). For example, decreased BMP4 induced by SOX2 enhanced lung SQC cell growth (27). This finding indicated a tissue context specific role of BMP4.
The present study revealed that CAPS1 promoted FHC cell migration by altering the protein expression profile of exosomes derived from CRC cells. GW4869, an exosome inhibitor, inhibited CAPS1-induced cell migration.
Materials and methods
Cell culture and conditioned medium preparation
The cell lines, HT29, SW480, FHC and 293T, used in the present study were purchased from The American Type Culture Collection. The cells were cultured in complete DMEM containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 mg/ml penicillin, and 10 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.), at 37°C and 5% CO2. Conditioned medium (CM) was harvested at 48 h from confluent cultures with exosome-depleted medium and centrifuged at 1,400 × g for 2 min at 4°C to remove cellular debris. To inhibit exosome secretion, cells were treated with 10 µM GW4869 (MedChemExpress) before collecting the CM.
Exosome isolation and characterization
Exosomes were isolated from HT29/SW480 CM by serial centrifugation. The medium was subjected to ultracentrifugation at 100,000 × g for 6 h at 4°C and washed with PBS (100,000 × g for 20 min) (7,28). Subsequently, the exosomes were re-suspended in PBS. The presence of exosomes was confirmed by particle size with a Nanoparticle Tracking Analysis (NTA) system (NTA 3.2 Dev Build 3.2.16, Malvern Panalytical Ltd., Malvern, UK), and the expression of exosome-specific markers such as tumor susceptibility gene 101 protein (TSG101) and CD81 was evaluated by western blot analysis.
Electron microscopy
For electron microscopy, exosomes were fixed with 2% paraformaldehyde and loaded on carbon-coated copper grids. The grids were placed on 2% gelatin for 20 min at 37°C and washed with 0.15 M glycine in PBS. Subsequently, the sections were blocked with 1% cold water fish-skin gelatin (11,29,30). The grids were viewed under a Philips CM120 transmission electron microscope (Philips Research).
Exosome uptake assay
The exosomes were fluorescently labeled using an ExoGlow-Protein EV Labeling kit (System Biosciences), according to the manufacturer's instructions. Approximately 100–500 µg of labeled exosomes were added to 1×105 293T/FHC cells. The red fluorescent signal was observed at 24 and 48 h.
Cell migration assay
Transwell migration assays were performed in 24-well chambers (pore size, 0.8 µm; EMD Milipore). FHC cells were treated with HT29 cell-derived exosomes, HT29/SW480-CM or HT29/SW480-CM+GW4869 for 24 h. FHC cells were then re-suspended in FBS-free DMEM and added to the upper chamber. DMEM containing 10% FBS was added to the lower chamber. The cells were incubated for 5 days, and subsequently, the cells on the undersurface of the membrane were fixed by 4% paraformaldehyde for 30 min at room temperature and stained with 0.1% crystal violet overnight at room temperature. The cells that had migrated through the filter were counted in four randomly selected regions per filter.
Western blot analysis
The cells or exosomes were homogenized in RIPA lysis buffer supplemented with protease inhibitors (Roche Diagnostics). The concentration of proteins was measured using the BCA Protein assay kit (Beyotime Institute of Biotechnology). Western blot analysis was performed as previously described (18). A total of 40 µg proteins per lane were loaded. The cell or exosome lysates were immunoblotted with antibodies against CAPS1 (cat. no. ab32011; Abcam), β-actin (cat. no. ab8227), CD63 (cat. no. ab59479), CD81 (cat. no. ab35026), TSG101 (cat. no. ab133586; all from Abcam) or BMP4 (cat. no. A0425; ABclonal).
Quantitative proteomics and bioinformatics analyses of exosomal proteins
Proteins were extracted from the isolated exosomes, digested with trypsin, and analyzed by LC-MS. The mass spectra were searched against the Swiss-Prot Human Proteome database (https://www.uniprot.org/). Gene Ontology (GO) (http://www.omicsbean.com:88/) and Ingenuity Pathway Analysis (IPA) analysis software were used to analyze differentially expressed proteins.
Statistical analysis
The data are presented as the means ± standard deviation. GraphPad Prism 7 software (GraphPad Prism, Inc.) was used for statistical analysis. Student's t-test was used for comparisons between groups. P<0.05 (two-sided) was considered to indicate a statistically significant difference.
Results
Characterization of exosomes isolated from CAPS1-overexpressing CRC cells
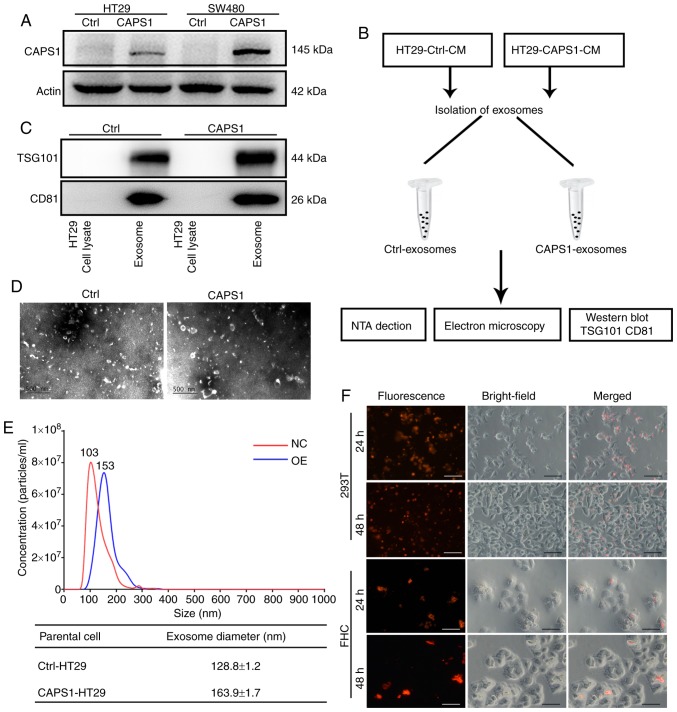
Our previous study indicated that CAPS1 was upregulated in CRC, and that increased CAPS1 facilitated CRC metastasis (18). The previous study generated stable CAPS1-overexpressing HT29 and SW480 cell lines (CAPS1-HT29 and CAPS1-SW480) by lentiviral infection, as well as corresponding control cell lines (18). The protein level of CAPS1 was detected in CAPS1-HT29 and CAPS1-SW480 cells in the present study (Fig. 1A). In order to confirm the role of exosomes in the process of CAPS1-induced CRC metastasis, exosomes were isolated from the CM of CAPS1-overexpressing HT29 cells (CAPS1-HT29-CM) and control cells (Fig. 1B). The harvested exosomes were detected by the presence of exosome-specific markers, TSG101 and CD81, using western blot analysis (Fig. 1C). The morphology of the exosomes was characterized by transmission electron microscopy, which revealed no noticeable differences (Fig. 1D). The particles of exosomes were measured by NTA system, revealing a diameter range of 30–180 nm. In comparison, exosomes derived from CAPS1-overexpressing cells exhibited a larger mean diameter. (Fig. 1E). To investigate whether HT29-derived exosomes (HT29-exo) could be internalized by target cells, the exosomes were pre-labeled fluorescently and co-cultured with 293T and FHC cells. Red fluorescence was observed within target cells at 24 and 48 h, indicating that exosomes were endocytosed successfully by target cells (Fig. 1F).
Figure 1.
Characterization of exosomes isolated from CAPS1-overexpressing CRC cells. (A) The protein level of CAPS1was assessed in CAPS1-overexpressing CRC cells (HT29 and SW480) and normalized to the level of β-actin. (B) The workflow of exosome isolation and characterization. (C) The exosome markers TSG101 and CD81 were identified within the exosome fraction but not in the HT29 total cell fraction. (D) Electron micrograph of exosomes isolated from CAPS1-HT29 and control cells, revealing their typical morphology and size. Scale bar, 500 nm. (E) The diameters of the exosomes isolated from CAPS1-HT29 and control cells were measured by the NTA system. (F) Exosomes isolated from HT29 cells were fluorescently pre-labeled and incubated with 293T and FHC cells. Images were captured at 24 and 48 h after the addition of exosomes. Scale bar, 50 µm. CAPS1, calcium-dependent activator protein for secretion 1; CRC, colorectal cancer; TSG101, tumor susceptibility gene 101 protein; NTA, Nanoparticle Tracking Analysis.
Exosomes derived from CAPS1-overexpressing CRC cells enhance FHC migration
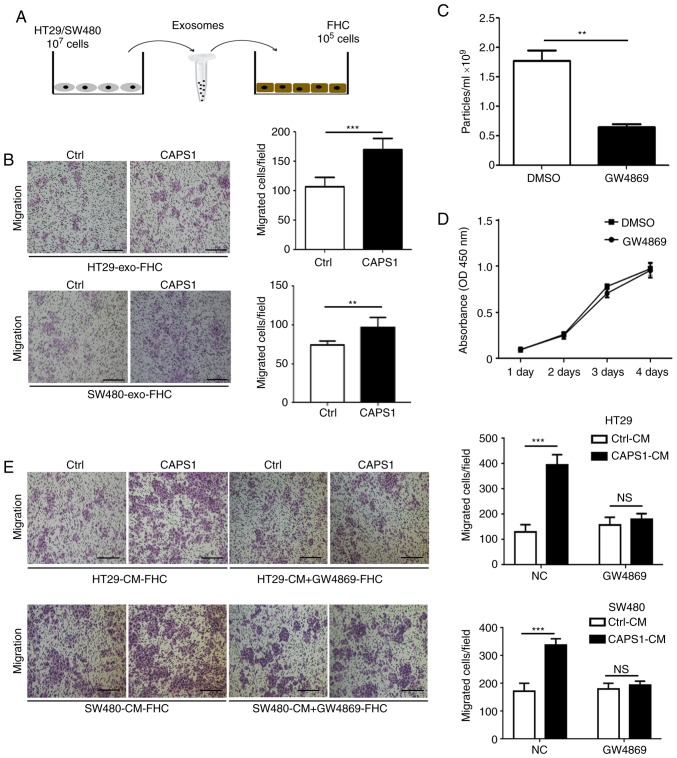
To investigate whether CAPS1 promotes CRC metastasis via exosomes. CAPS1-HT29-derived exosomes were co-cultured with FHC cells for 24 h (Fig. 2A). The FHC cell line is from normal fetal colonic mucosa, which exhibits a tumorigenic phenotype and has a metastatic potential in vivo (31,32). A Transwell migration assay was performed to detect the migration of FHC cells, which demonstrated that exosomes derived from CAPS1-overexpressing CRC cells (HT29 and SW480) significantly enhanced the migration of FHC cells (HT29, P<0.001 SW480, P=0.0067; Fig. 2B).
Figure 2.
Exosomes from CAPS1-overexpressing CRC cells enhance FHC migration. (A) The workflow of FHC cells treated with HT29/SW480-derived exosomes. (B) Exosomes derived from CAPS1-HT29/SW480 promoted FHC migration as determined by a Transwell migration assay. (C) GW4869 inhibited the secretion of exosomes in HT29 cells. (D) The viability of HT29 cells pretreated with GW4869 was not affected in the CCK-8 assay. (E) GW4869 attenuated the migration of FHC cells. Scale bar, 50 µm (left panel). The number of cells that migrated to the underside of the membrane was counted in four randomly selected regions per filter (right panel). Results of the Transwell migration assay are presented as the means ± standard deviation (n=3). Magnification, ×100. **P<0.01 and ***P<0.001. CAPS1, calcium-dependent activator protein for secretion 1; CRC, colorectal cancer.
To further investigate whether CRC cell-derived exosomes participate in FHC cell migration, GW4869, an inhibitor of exosomes, was used to inhibit exosome secretion in HT29 cells (P=0.0038; Fig. 2C). It was determined that GW4869 had no effect on HT29 cell viability (P=0.4652; Fig. 2D). FHC cells were cultured with CM derived from CAPS1-overexpressing CRC cells (HT29 and SW480), as well as control cells, for 24 h. CM derived from CAPS1-overexpressing cells significantly increased FHC migration in the Transwell migration assay (HT29, P<0.001; SW480, P<0.001), an effect that was attenuated following GW4869-treatment (HT29, P=0.2275; SW480, P=0.227; Fig. 2E), which indicated that CAPS1 promoted FHC cell migration via exosomes.
Proteomic profiling of exosomes derived from CAPS1-overexpressing CRC cells
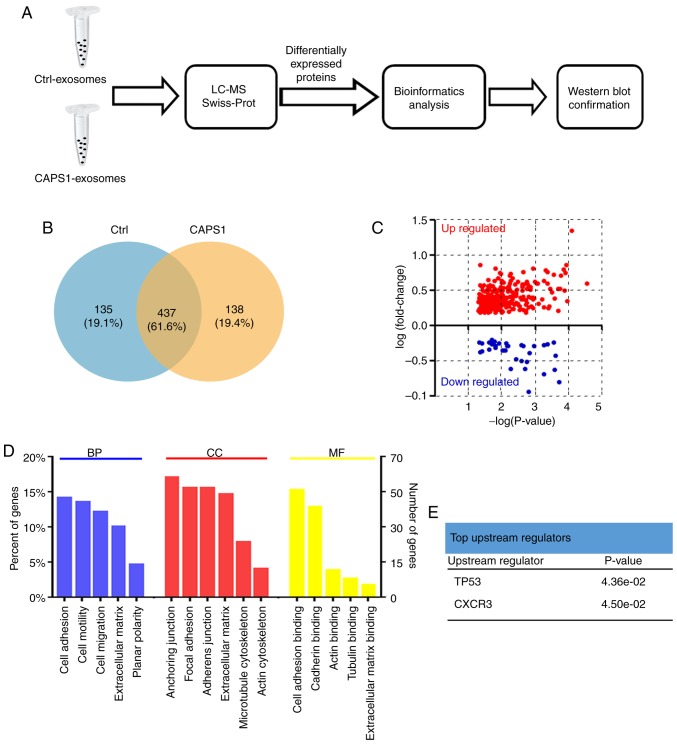
The present study further characterized the changes in the exosome protein profile induced by CAPS1 overexpression (Fig. 3A). Proteins were extracted from exosomes derived from CAPS1-HT29 and control cells. LC-MS was applied to assess the proteomics of exosomes, and the raw LC-MS data files were searched against the Swiss-Prot Human Proteome database. A total of 1,024 proteins were identified and quantified (Table SI) (As aforementioned a total of 1035 proteins was identified, but only 1,024 proteins could be quantified since the intensity value of the remaining 11 proteins was extremely low and had to be ruled out for further analysis). CAPS1-HT29 and control cells had in total 437 exosomal proteins in common, while 138 exosomal proteins from CAPS1-HT29 differed from 135 proteins from control exosomes (Fig. 3B; Table SII). Further analysis revealed that 297 proteins were upregulated and 45 proteins were downregulated, with the cut-off threshold set at fold-change=1.5 and P<0.05 (Fig. 3C; Table SIII).
Figure 3.
LC-MS and bioinformatics analysis of the differentially expressed exosomal proteins. (A) The workflow of LC-MS and bioinformatics analysis of differentially expressed exosomal proteins. (B) Venn diagram revealing the differentially expressed proteins identified in CAPS1-HT29 and control cell-derived exosomes. (C) The exosomal protein profile was altered by CAPS1-overexpression. In total, 297 proteins were significantly upregulated and 45 proteins were significantly downregulated. (D) GO analysis of differentially expressed exosomal proteins. (E) The upstream regulators of differentially expressed exosomal proteins in the IPA analysis. LC-MS, liquid chromatography-mass spectrometry; CAPS1, calcium-dependent activator protein for secretion 1; GO, Gene Ontology; IPA, Ingenuity Pathway Analysis.
GO enrichment analysis and the UniProt-GOA database were utilized to further classify the differentially expressed proteins (Fig. 3D). The enriched terms were cell adhesion, anchoring junction and cell adhesion binding for biological processes, cell components and molecular functions, respectively. Using IPA based on the Ingenuity Knowledge Base, p53 and CXCR3 were identified as the top upstream regulators (Fig. 3E).
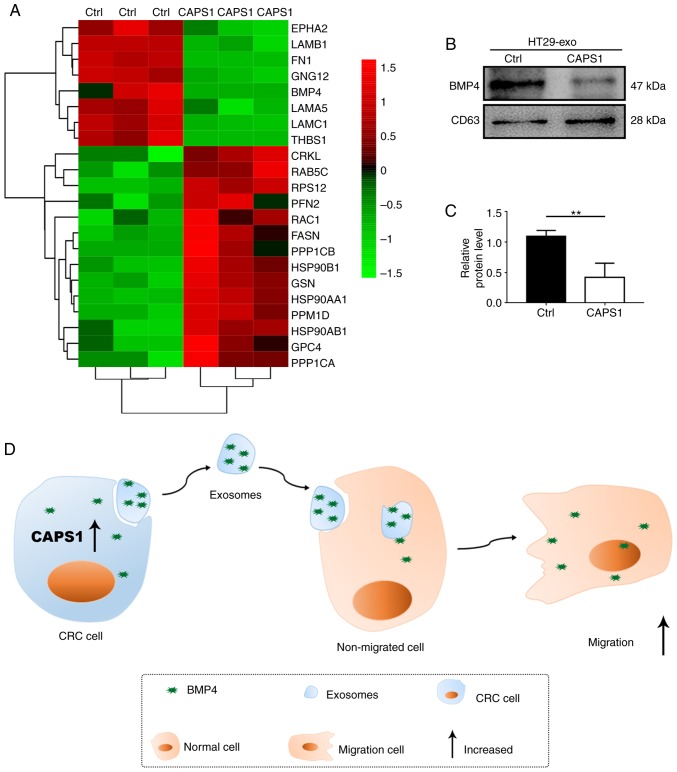
According to the bioinformatics analysis, the candidate proteins that may participate in cell migration were identified (Fig. 4A). Previously, it has been reported that BMP4 is involved in metastasis in breast cancer, prostate cancer, HCC and CRC (24–26,33). Thus, BMP4 may serve vital roles in the process of CAPS1-induced cell migration. To investigate the variation of BMP4 in the exosomes derived from CAPS1-overexpressing CRC cells, western blot analysis was performed. It was revealed that BMP4 was downregulated ~0.5-fold compared with Ctrl, which was consistent with the LC-MS results (Fig. 4B and C). These data indicated that exosomes are required for CAPS1-mediated cell migration (Fig. 4D).
Figure 4.
Candidate exosomal proteins that contribute to FHC cell migration. (A) Representative heatmap of the differentially expressed genes. The genes and samples are presented in the rows and columns, respectively. Red and green indicates upregulated and downregulated genes, respectively. The color scale bar indicates the log10 ratio of the intensities. (B) BMP4 protein level was assessed in exosomes derived from CAPS1-HT29 and control cells, and normalized to the level of CD63. (C) Quantification of the western blots was performed using ImageJ software and presented as the mean ± standard deviation (n=3). **P<0.01. (D) A proposed working model for exosomes derived from CAPS1-overexpressing CRC cells in the promotion of cell migration. CAPS1, calcium-dependent activator protein for secretion 1; BMP4, bone morphogenetic protein 4.
Discussion
The present study demonstrated that exosomes derived from CAPS1-overexpressing CRC cells can enhance cell migration. Overexpression of CAPS1 led to alterations in the expression pattern of exosomal proteins, including the downregulation of BMP4, which was further confirmed.
CAPS1 is the vertebrate homologue of the Caenorhabditis elegans UNC-31 protein, which is involved in vesicle secretion (34). Previously, the essential role of CAPS1 in the development of cancer was investigated. Our previous study revealed that CAPS1 promoted CRC metastasis via the PI3K/Akt/GSK3β/Snail signaling pathway-mediated EMT process (18). In addition, it was demonstrated that CAPS1 serves as a biomarker of HCC, and the expression of CAPS1 is decreased in patients with aggressive HCC (35). Further investigation revealed that CAPS1 likely inhibits HCC development via alteration of the exocytosis-associated tumor microenvironment (36). In addition, decreased CAPS1 expression was revealed to be correlated with poor prognosis of patients with central nervous system primitive neuro-ectodermal tumors (37).
Metastasis is the predominant pathological reason for the mortality of patients with CRC. Notably, increasing evidence suggests that exosomes serve as key mediators in tumor metastasis (11,38). It was reported that exosomes from highly liver metastatic CRC cell lines could significantly enhance the migration ability of Caco-2 cells, a CRC cell line with poor liver metastatic potential (11). Similarly, the present study revealed that exosomes derived from CAPS1-overexpressing CRC cells facilitated FHC migration. The function of exosomes in tumors predominantly depends on their communication between cells. For example, exosomes can transfer oncogenes, such as lncRNA H19 and miR-193a, to CRC, which promotes cancer progression (39,40). Exosomes isolated from the serum of CRC patients were revealed to transfer malignant traits and confer the same phenotype of the primary tumor cells to the target cells (41). Exosomes, as a communication media for genetic exchange between cells, likely contribute to the process of metastasis mediated by CAPS1 in CRC.
CAPS1 is multi-domain protein. Among these domains, MH domain drives trans-SNARE complex formation and mediates membrane fusion through syntaxin interactions (42,43). Notably, SNARE complex protein YKT6 has been identified to potentially be involved in the fusion of MVBs with the PM during the biogenesis of exosomes (16). Whether CAPS1 regulates the secretion and loading of exosomes via the SNARE complex in CRC requires further investigation.
Bioinformatics strategies have frequently been used to evaluate the function of exosomes (35,44). In the present bioinformatics analysis, the differentially expressed exosomal proteins involved in MAPK, Wnt, TGF-β, PI3K-AKT, or RAS signaling, which were frequently dysregulated in CRC and were associated with CRC metastasis (45–50), were all considered as candidate proteins involved in cell migration induced by CAPS1 (Fig. 4A). In view of the role of BMP4 in metastasis, the present study examined the expression level of BMP4 in the exosomes, and the downregulation of BMP4 was further confirmed by western blot analysis (Fig. 4B and C). However, the role of BMP4 in the process of CAPS1-induced cell migration should be further investigated. Furthermore, p53 and CXCR3 were identified as upstream regulators. p53 is activated by stress stimuli, and in turn, it governs a complex anti-proliferative transcriptional program to serve a tumor suppressive role (51,52). However, TP53 is highly mutated and loses its function in CRC (53). CXCR3 is a G protein-coupled receptor that binds to chemokines to mediate leukocyte trafficking, integrin activation, cytoskeletal changes and chemotactic migration (54). A previous study reported that CXCR3 was upregulated in CRC and facilitated tumor metastasis via lymph nodes (55). Fibronectin 1 (FN1), which was downregulated in the present LC-MS analysis, is a target molecule of CXCR3. Furthermore, FN1 has been revealed to regulate numerous molecules that are involved in cytoskeletal organization and integrin signaling during tumor progression and metastasis (56). Thrombospondin-1 (THBS-1), a target gene of p53, is an endogenous inhibitor of angiogenesis and was also downregulated in the current LC-MS analysis (57). BMP4, FN1 and THBS-1 transferred by exosomes may serve an essential role in the process of CAPS1-induced CRC cell migration.
CAPS1 had an effect on the diameter of the exosomes according to the analysis of mean diameter by NTA detection. The exosomes derived from CAPS1-HT29 cells exhibited a higher mean diameter. Similarly, the majority of CAPS1-HT29-derived exosomes presented with a diameter of 153 nm, whereas those from control cells exhibited a diameter of 103 nm (Fig. 1E).
In summary, the present study demonstrated that CAPS1 promoted cell migration by remodeling the protein profile of exosomes. Furthermore, several candidate proteins were identified that may participate in the process of CRC metastasis mediated by CAPS1. However, the RNA profile of CAPS1-CRC-derived exosomes, and the proteomics and transcriptomic profiles of target cells require further analysis. The present data may improve understanding of a new target for the treatment of patients with CRC metastasis via the inhibition of exosome secretion.
Supplementary Material
Acknowledgements
We thank Dr Xiaoxiao Wang and Dr Yaohua Li from Key Laboratory of Glycoconjugate Research (Ministry of Public Health), Department of Biochemistry and Molecular Biology, Shanghai Medical college, Fudan University for technical assistance.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81772615, 81672334, 81573423, 81770137 and 81802302) and the Shanghai Science and Technology Commission (grant no. 15410710100).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
BW, DS, LD and SC designed and conceived this project. BW, DS and LM developed methodology; BW, DS and YD performed experiments and generated data; SC, SZ analyzed and interpreted data; SC, BW wrote the manuscript. All authors contributed to and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
There authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Mundade R, Imperiale TF, Prabhu L, Loehrer PJ, Lu T. Genetic pathways, prevention, and treatment of sporadic colorectal cancer. Oncoscience. 2014;1:400–406. doi: 10.18632/oncoscience.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody H. Colorectal cancer. Nature. 2015;521:S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 4.Xu F, Tang B, Jin T, Dai C. Current status of surgical treatment of colorectal liver metastases. World J Clin Cases. 2018;6:716–734. doi: 10.12998/wjcc.v6.i14.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 6.Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Ther. 2016;17:1062–1069. doi: 10.1080/15384047.2016.1219815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-López L, Blancas I, Garrido JM, Mut-Salud N, Moya-Jódar M, Osuna A, Rodríguez-Serrano F. The role of exosomes on colorectal cancer: A review. J Gastroenterol Hepatol. 2018;33:792–799. doi: 10.1111/jgh.14049. [DOI] [PubMed] [Google Scholar]
- 8.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR. Mass-spectrometry-based molecular characterization of extracellular vesicles: Lipidomics and proteomics. J Proteome Res. 2015;14:2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 9.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: A common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 10.Schorey JS, Bhatnagar S. Exosome function: From tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Ding X, Nan L, Wang Y, Wang J, Yan Z, Zhang W, Sun J, Zhu W, Ni B, et al. Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol Rep. 2015;33:2445–2453. doi: 10.3892/or.2015.3843. [DOI] [PubMed] [Google Scholar]
- 12.Akao Y, Khoo F, Kumazaki M, Shinohara H, Miki K, Yamada N. Extracellular disposal of tumor-suppressor miRs-145 and −34a via microvesicles and 5-FU resistance of human colon cancer cells. Int J Mol Sci. 2014;15:1392–1401. doi: 10.3390/ijms15011392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragusa M, Statello L, Maugeri M, Barbagallo C, Passanisi R, Alhamdani MS, Li Destri G, Cappellani A, Barbagallo D, Scalia M, et al. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: Biomolecular, genetic and translational implications. Oncoscience. 2014;1:132–157. doi: 10.18632/oncoscience.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 16.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 17.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao GX, Xu YY, Weng SQ, Zhang S, Chen Y, Shen XZ, Dong L, Chen S. CAPS1 promotes colorectal cancer metastasis via Snail mediated epithelial mesenchymal transformation. Oncogene. 2019;38:4574–4589. doi: 10.1038/s41388-019-0740-7. [DOI] [PubMed] [Google Scholar]
- 19.Speidel D, Varoqueaux F, Enk C, Nojiri M, Grishanin RN, Martin TF, Hofmann K, Brose N, Reim K. A family of Ca2+-dependent activator proteins for secretion: Comparative analysis of structure, expression, localization, and function. J Biol Chem. 2003;278:52802–52809. doi: 10.1074/jbc.M304727200. [DOI] [PubMed] [Google Scholar]
- 20.Grishanin RN, Kowalchyk JA, Klenchin VA, Ann K, Earles CA, Chapman ER, Gerona RR, Martin TF. CAPS acts at a prefusion step in dense-core vesicle exocytosis as a PIP2 binding protein. Neuron. 2004;43:551–562. doi: 10.1016/j.neuron.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 21.Ehata S, Yokoyama Y, Takahashi K, Miyazono K. Bi-directional roles of bone morphogenetic proteins in cancer: Another molecular Jekyll and Hyde? Pathol Int. 2013;63:287–296. doi: 10.1111/pin.12067. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama Y, Watanabe T, Tamura Y, Hashizume Y, Miyazono K, Ehata S. Autocrine BMP-4 signaling is a therapeutic target in colorectal cancer. Cancer Res. 2017;77:4026–4038. doi: 10.1158/0008-5472.CAN-17-0112. [DOI] [PubMed] [Google Scholar]
- 23.Deng G, Zeng S, Qu Y, Luo Q, Guo C, Yin L, Han Y, Li Y, Cai C, Fu Y, Shen H. BMP4 promotes hepatocellular carcinoma proliferation by autophagy activation through JNK1-mediated Bcl-2 phosphorylation. J Exp Clin Canc Res. 2018;37:156. doi: 10.1186/s13046-018-0828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Liu H, Zhang L, Liu X, Zhang C, Wang Y, He Q, Zhang Y, Li Y, Chen Q, et al. DJ-1 promotes colorectal cancer progression through activating PLAGL2/Wnt/BMP4 axis. Cell Death Dis. 2018;9:865. doi: 10.1038/s41419-018-0883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng S, Zhang Y, Ma J, Deng G, Qu Y, Guo C, Han Y, Yin L, Cai C, Li Y, et al. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial-mesenchymal transition via upregulating ID2. Cancer Lett. 2017;390:67–76. doi: 10.1016/j.canlet.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 26.Guo D, Huang J, Gong J. Bone morphogenetic protein 4 (BMP4) is required for migration and invasion of breast cancer. Mol Cell Biochem. 2012;363:179–190. doi: 10.1007/s11010-011-1170-1. [DOI] [PubMed] [Google Scholar]
- 27.Fang WT, Fan CC, Li SM, Jang TH, Lin HP, Shih NY, Chen CH, Wang TY, Huang SF, Lee AY, et al. Downregulation of a putative tumor suppressor BMP4 by SOX2 promotes growth of lung squamous cell carcinoma. Int J Cancer. 2014;135:809–819. doi: 10.1002/ijc.28734. [DOI] [PubMed] [Google Scholar]
- 28.Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 29.Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44:1741–1748. doi: 10.1159/000485780. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. 2014;13:256. doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucek K, Gajdusková P, Brázdová M, Hýzd'alová M, Kocí L, Vydra D, Trojanec R, Pernicová Z, Lentvorská L, Hajdúch M, et al. Fetal colon cell line FHC exhibits tumorigenic phenotype, complex karyotype, and TP53 gene mutation. Cancer Genet Cytogenet. 2010;197:107–116. doi: 10.1016/j.cancergencyto.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqui KM, Chopra DP. Primary and long term epithelial cell cultures from human fetal normal colonic mucosa. In Vitro. 1984;20:859–868. doi: 10.1007/BF02619632. [DOI] [PubMed] [Google Scholar]
- 33.Lin SC, Lee YC, Yu G, Cheng CJ, Zhou X, Chu K, Murshed M, Le NT, Baseler L, Abe JI, et al. Endothelial-to-osteoblast conversion generates osteoblastic metastasis of prostate cancer. Dev Cell. 2017;41:467–480.e3. doi: 10.1016/j.devcel.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ann K, Kowalchyk JA, Loyet KM, Martin TF. Novel Ca2+-binding protein (CAPS) related to UNC-31 required for Ca2+-activated exocytosis. J Biol Chem. 1997;272:19637–19640. doi: 10.1074/jbc.272.32.19637. [DOI] [PubMed] [Google Scholar]
- 35.Liu T, Xue R, Huang X, Zhang D, Dong L, Wu H, Shen X. Proteomic profiling of hepatitis B virus-related hepatocellular carcinoma with magnetic bead-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Acta Biochim Biophys Sin (Shanghai) 2011;43:542–550. doi: 10.1093/abbs/gmr044. [DOI] [PubMed] [Google Scholar]
- 36.Xue R, Tang W, Dong P, Weng S, Ma L, Chen S, Liu T, Shen X, Huang X, Zhang S, et al. CAPS1 negatively regulates hepatocellular carcinoma development through alteration of exocytosis-associated tumor microenvironment. Int J Mol Sci. 2016;17(pii):E1626. doi: 10.3390/ijms17101626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller S, Rogers HA, Lyon P, Rand V, Adamowicz-Brice M, Clifford SC, Hayden JT, Dyer S, Pfister S, Korshunov A, et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol. 2011;13:866–879. doi: 10.1093/neuonc/nor070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459–1475. doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 39.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdouh M, Hamam D, Gao Z, Arena V, Arena M, Arena GO. Exosomes isolated from cancer patients' sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J Exp Clin Canc Res. 2017;36:113. doi: 10.1186/s13046-017-0587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James DJ, Kowalchyk J, Daily N, Petrie M, Martin TF. CAPS drives trans-SNARE complex formation and membrane fusion through syntaxin interactions. Proc Natl Acad Sci USA. 2009;106:17308–17313. doi: 10.1073/pnas.0900755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch H, Hofmann K, Brose N. Definition of Munc13-homology-domains and characterization of a novel ubiquitously expressed Munc13 isoform. Biochem J. 2000;349:247–253. doi: 10.1042/0264-6021:3490247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 45.Fang YJ, Lu ZH, Wang GQ, Pan ZZ, Zhou ZW, Yun JP, Zhang MF, Wan DS. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24:875–884. doi: 10.1007/s00384-009-0725-z. [DOI] [PubMed] [Google Scholar]
- 46.Urosevic J, Nebreda AR, Gomis RR. MAPK signaling control of colon cancer metastasis. Cell Cycle. 2014;13:2641–2642. doi: 10.4161/15384101.2014.946374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y, Liu XQ, Rajput A, Geng L, Ongchin M, Zeng Q, Taylor GS, Wang J. Phosphatase PRL-3 is a direct regulatory target of TGFbeta in colon cancer metastasis. Cancer Res. 2011;71:234–244. doi: 10.1158/0008-5472.CAN-10-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pancione M, Forte N, Fucci A, Sabatino L, Febbraro A, Di Blasi A, Daniele B, Parente D, Colantuoni V. Prognostic role of beta-catenin and p53 expression in the metastatic progression of sporadic colorectal cancer. Hum Pathol. 2010;41:867–876. doi: 10.1016/j.humpath.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 49.Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, Wu L, Zhang T, Sun X, Zhong L, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Canc Res. 2016;35:148. doi: 10.1186/s13046-016-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahrami A, Hassanian SM, ShahidSales S, Farjami Z, Hasanzadeh M, Anvari K, Aledavood A, Maftouh M, Ferns GA, Khazaei M, Avan A. Targeting RAS signaling pathway as a potential therapeutic target in the treatment of colorectal cancer. J Cell Physiol. 2018;233:2058–2066. doi: 10.1002/jcp.25890. [DOI] [PubMed] [Google Scholar]
- 51.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mills KD. Tumor suppression: Putting p53 in context. Cell Cycle. 2013;12:3461–3462. doi: 10.4161/cc.26806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 54.Ma B, Khazali A, Wells A. CXCR3 in carcinoma progression. Histol Histopathol. 2015;30:781–792. doi: 10.14670/HH-11-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawada K, Hosogi H, Sonoshita M, Sakashita H, Manabe T, Shimahara Y, Sakai Y, Takabayashi A, Oshima M, Taketo MM. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene. 2007;26:4679–4688. doi: 10.1038/sj.onc.1210267. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y, Wu B, Ge W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. 2017;140:900–913. doi: 10.1002/ijc.30496. [DOI] [PubMed] [Google Scholar]
- 57.Jia L, Waxman DJ. Thrombospondin-1 and pigment epithelium-derived factor enhance responsiveness of KM12 colon tumor to metronomic cyclophosphamide but have disparate effects on tumor metastasis. Cancer Lett. 2013;330:241–249. doi: 10.1016/j.canlet.2012.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.