Abstract
MicroRNAs (miRNAs) have been shown to play a crucial role in the progression of human cancers, including urothelial carcinoma (UC), the sixth-most common cancer in the world. Among them, miR-34a has been implicated in the regulation of cancer stem cells (CSCs); however, its role in UC has yet to be fully elucidated. In this study, bioinformatics and experimental analysis confirmed that miR-34a targets CD44 (a CSC surface marker) and c-Myc (a well-known cell cycle regulator) in UC. We found that, surprisingly, most UC cell lines and patient samples did express miR-34a, although epigenetic silencing by promoter hypermethylation of miR-34a expression was observed only in UMUC3 cells, and a subset of patient samples. Importantly, overexpression of c-Myc, a frequently amplified oncogene in UC, was shown to upregulate CD44 expression through a competing endogenous RNA (ceRNA) mechanism, such that overexpression of the c-Myc 3′UTR upregulated CD44, and vice versa. Importantly, we observed a positive correlation between the expression of c-Myc and CD44 in clinical samples obtained from UC patients. Moreover, overexpression of a dominant-negative p53 mutant downregulated miR-34a, but upregulated c-Myc and CD44, in UC cell lines. Functionally, the ectopic expression of miR-34a was shown to significantly suppress CD44 expression, and subsequently, suppression of cell growth and invasion capability, while also reducing chemoresistance. In conclusion, it appears that aberrant promoter methylation, and c-Myc-mediated ceRNA mechanisms, may attenuate the function of miR-34a, in UC. The tumor suppressive role of miR-34a in controlling CSC phenotypes in UC deserves further investigation.
Keywords: miR-34a, CD44, c-Myc, ceRNA, urothelial carcinoma
1. Introduction
Urothelial carcinoma (UC) is the sixth most common cancer in the world, with a particularly high incidence in southwestern Taiwan. The majority of UCs are found in the urinary bladder, while tumors from the upper urinary tract (ureter or renal pelvis, known as UTUC) account for 5–10% of all UCs worldwide [1], but 20% in Taiwan [2]. The two principle forms of UC are generally differentiated according to the depth of muscle invasion: non-muscle invasive and muscle invasive cancer. One of the major challenges in treating UC is dealing with the high recurrence rate of superficial cancers, and more than 40% of all UC patients will recur within 5 years, even after surgical treatment [3]. This may be attributed to the presence of drug-resistant cancer stem cells (CSCs), which are characterized by the presence of CD44 [4,5].
CD44 is an integral cell membrane glycoprotein identified as a key biomarker for the isolation and characterization of CSCs [4,6,7]. CD44, a receptor for the extracellular matrix polysaccharide hyaluronate, is involved in several physiological processes—including intercellular adhesion, cell-matrix adhesion, and cell migration—as well as tumor cell invasion and metastasis [8]. Therefore, elucidating the molecular mechanism(s) underlying the formation of CSCs in UC could lead to the development of novel therapeutic methods to deal with this deadly disease.
Epigenetic modification is considered a hallmark of cancer, due to its roles in cellular carcinogenesis and tumor progression [9]. One such modification involves the regulation of microRNAs (miRNAs or miRs), resulting in the generation of small (20–23 nucleotides), non-coding, single-stranded RNAs, which induce the degradation or translational inhibition of homologous target mRNAs by binding their 3′-untranslated region (UTR) [10]. Researchers have also recently identified another mechanism of miRNA regulation, namely, competing endogenous RNAs (ceRNAs), in which an mRNA sharing a common miRNA response element (MRE) with another mRNA, regulate one another by competing for the same miRNA [11]. In this ceRNA mechanism, the 3′UTR of the mRNA could act in trans to modulate gene expression, epigenetically.
Numerous studies have reported that miRNAs can regulate cancer progression, particularly the processes of growth, invasion, and metastasis [12]. For example, miR-34a, which resides on chromosome 1q36.22 and belongs to the miR-34a family, has been shown to target the cell cycle regulator, c-Myc, in several human cancers [13,14]. In addition, miR-34a has also been found to participate in controlling cancer stemness, by targeting CD44 [15,16,17,18].
In this context, miR-34a would be considered a tumor suppressor miR in human cancer. Moreover, the promoter region of miR-34a contains a CpG island commonly associated with promoter hypermethylation in human cancer [19]. It has also been suggested that miR-34a negatively associates with urothelial carcinoma (UC); however, the relationship between the expression of miR-34a and its target genes, in UC, has yet to be elucidated. Researchers also have yet to determine the role of epigenetic modification in the expression of miR-34a. Consequently, our objective in this study was to determine the role of epigenetic modification in the expression of miR-34a, and its target genes, in UC. Unexpectedly, we observed that miR-34a is epigenetically controlled by a ceRNA mechanism through the 3′UTR of c-Myc, leading to upregulation of CD44 and cancer progression.
2. Results
2.1. miR-34a Is Epigenetically Silenced in the UMUC3 Cell Line
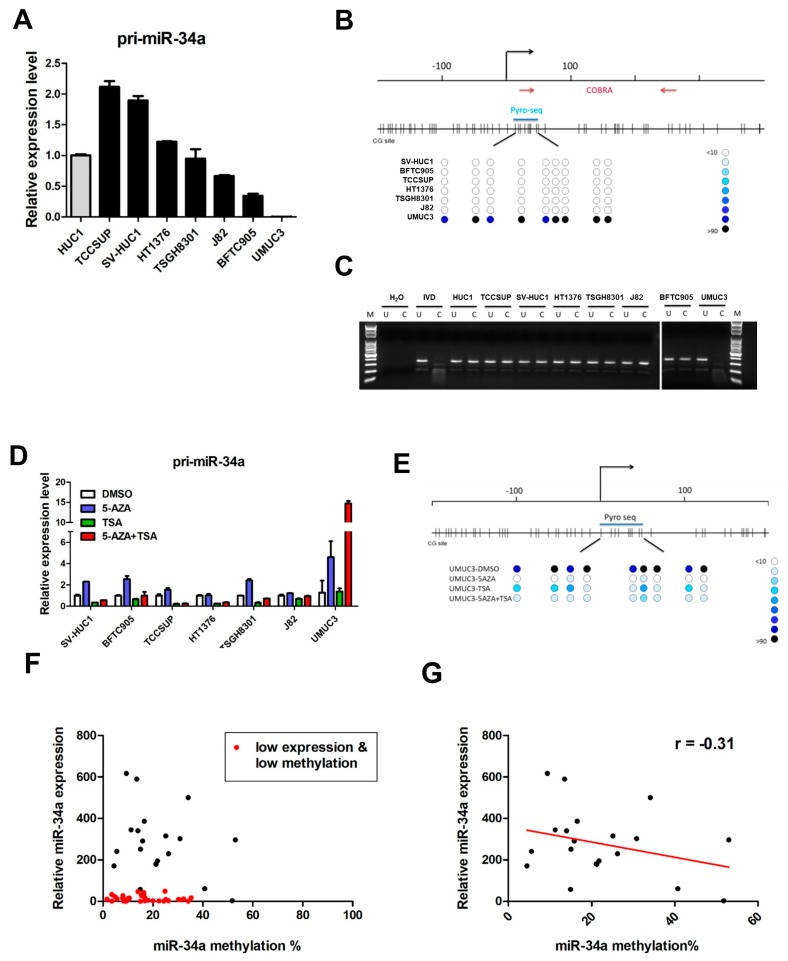
We first examined expression of the immature miR-34a transcript, pri-miR-34a, in a panel of normal bladder urothelial cells (HUCs), and seven human UC cell lines. Surprisingly, compared to primary and immortalized (SV-HUC1) HUC cells, there was a dramatic downregulation of miR-34a only in UMUC3 cells, while miR-34a expression levels varied in the other cell lines (Figure 1A).
Figure 1.
miR-34a is epigenetically silenced by promoter DNA methylation in UMUC3 cell lines, and a subset of urothelial carcinoma (UC) patient samples. (A) Expression of pri-miR-34a in normal HUC1 bladder cells and various UC cell lines, relative to GAPDH, as an internal control. Methylation analysis of the miR-34a promoter in various UC cell lines, using (B) bisulphite pyrosequencing and (C) COBRA assay. The upper panel in (B) presents the genomic structure of the miR-34a promoter, with the corresponding locations of all CpG sites from −200 to +400. The lower panel in (B) illustrates DNA methylation at each CpG site (circle) of various cells, where the intensity of the blue color indicates the degree of methylation. The CpG sites interrogated by bisulphite prosequencing are also indicated. In the COBRA assays in (C), bisulphite-modified DNA was amplified via PCR and digested using BstUI. U, undigested control; C, digested using BstUI; M, DNA ladder marker; IVD, in vitro methylated DNA. (D) Expression of pri-miR-34a in UC cell lines, following epigenetic treatment. UC cell lines treated with 5-aza-2’-deoxycytidine (5aza) and/or trichostatin A (TSA) were examined for pri-miR-34a expression, using qRT-PCR. Error bars represent standard deviations calculated from duplicates. (E) Bisulphite pyrosequencing analysis of the miR-34a promoter in UMUC3 cells treated with various epigenetic drugs, as in (D). (F) Scatter plot showing expression and promoter methylation of miR-34a in 55 urothelial carcinoma patient samples, showing a distinct group of patient samples (red dots) with both low expression and low promoter methylation of miR-34a. Analysis of the other patient samples (methylated) showed an inverse correlation (r = −0.31) between expression and methylation of miR-34a, as shown in (G).
Previous studies have demonstrated that dysregulation of miRNA, through promoter CpG island methylation, is an important mechanism in carcinogenesis [20]. Thus, we sought to identify an epigenetic mechanism downregulating miR-34a, in UMUC3 cells. Bisulphite pyrosequencing (Figure 1B), and combined bisulfite restriction analysis (COBRA, Figure 1C) assays were used to determine the methylation status of the CpG island in which miR-34a resides. In agreement with the RT-PCR results (Figure 1A), high miR-34a methylation levels were observed only in UMUC3 cells. In determining whether DNA methylation was responsible for epigenetic silencing of miR-34a, we found that epigenetic treatment with a DNA methyltransferase inhibitor, 5′aza-2′-deoxycytidine (5-AZA), and/or histone deacetylase (HDAC) inhibitor (trichostatin A, TSA), led to robust re-expression of miR-34a, in UMUC3 cells only (Figure 1D), due to demethylation of the miR-34a promoter region (Figure 1E). These results suggest that miR-34a is epigenetically silenced in UMUC3 cells.
Further clinical studies, using 55 UC patient samples, found the miR-34a promoter to be devoid of DNA hypermethylation, with only few samples having methylation levels > 50% (Figure 1F). Interestingly, one group of samples (n = 35) had low miR-34a expression, regardless of promoter methylation level (Figure 1F, red dot). After exclusion of those samples, the remaining samples (n = 20) showed an inverse correlation between miR-34a expression and promoter methylation (Figure 1G, r = −0.31). Together, these studies of UC cell lines and patient samples showed that even lowly expressing miR-34a samples were devoid of high levels of DNA methylation. Thus, an alternative mechanism may be involved in the suppression of miR-34a function in UC.
2.2. c-Myc Acts as a ceRNA of CD44 in Urothelial Carcinoma
One recent study revealed that mRNAs compete for binding to a shared set of miRNAs, thereby modulating miRNA-based regulation by acting as competing endogenous RNAs (ceRNAs) [21]. We therefore hypothesized that a ceRNA mechanism may be responsible for regulating miR-34a in UC. We then conducted a search of the intersection of three databases (TargetScan, miRanda, and PicTar) to identify any miR-34a targets overexpressed in UC. Interestingly, numerous studies have reported that c-Myc, a known miR-34a target [22], is amplified and upregulated in UC [23,24]. To determine whether miR-34a targets the c-Myc 3′UTR, in UC, we overexpressed miR-34a in UMUC3 cells with low miR-34a expression (Figure 2A). As expected, transfection of miR-34a reduced mRNA and protein levels of c-Myc in UMUC3 cells (Figure 2B). Studies have also shown that the stem cell marker, CD44 is a miR-34a target in UC [17]. Indeed, overexpression of miR-34a was shown to reduce both mRNA and protein of CD44 in UMUC3 cells (Figure 2C). Taken together, these results suggest that miR-34a targets c-Myc and CD44 in UC.
Figure 2.
miR-34a targets both c-Myc and CD44, in UMUC3 urothelial cancer cells. (A) Expression of mature miR-34a in UMUC3 cells overexpressing pri-miR-34a or an empty vector (NC). Overexpression of miR-34a led to the downregulation of (B) c-Myc and (C) CD44. Upper panel, RT-PCR; lower panel, western blot analysis. GAPDH was used as a loading control for western blot analysis. (*** p < 0.001).
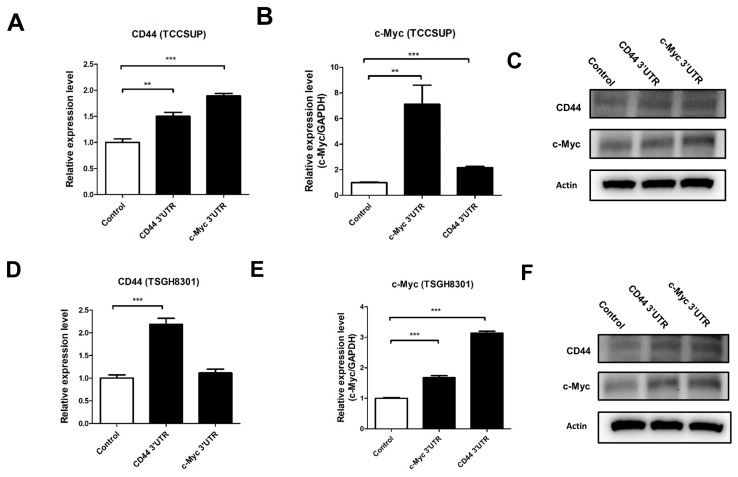
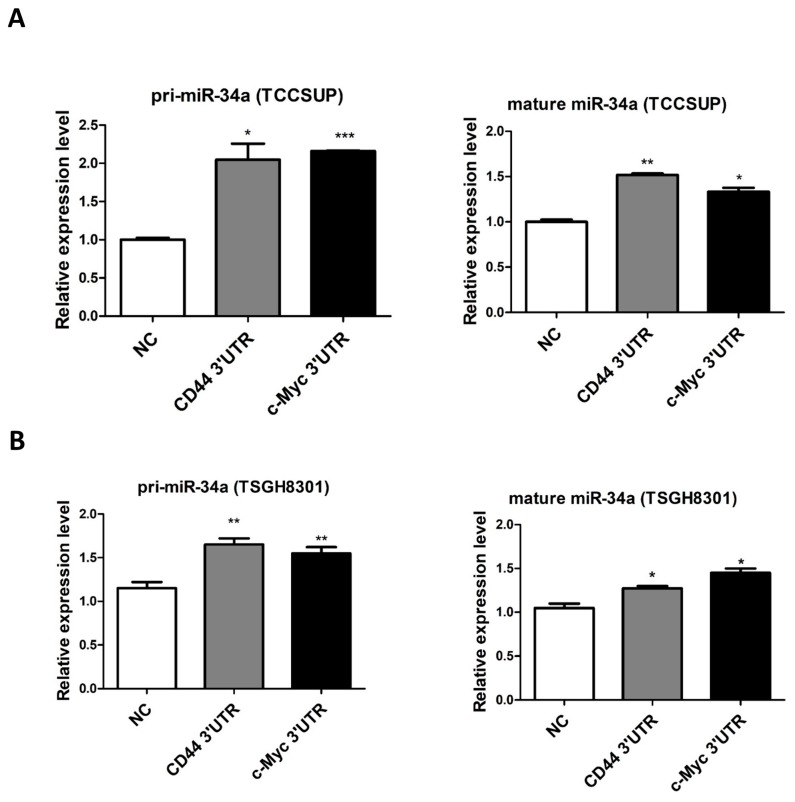
The above results indicate that the ectopic expression of miR-34a repressed both c-Myc and CD44 in UC. Therefore, it was reasonable to assume a significantly positive correlation between CD44 and c-Myc (both targets of miR-34a and thus ceRNAs), in UC. To elucidate this ceRNA mechanism, we exogenously overexpressed the c-Myc and CD44 3′-UTRs, in TCCSUP and TSGH8301 UC cells having high endogenous miR-34a expression. Interestingly, overexpression of these 3′-UTRs significantly upregulated the expression of both CD44 and c-Myc mRNA and protein, in TCCSUP (Figure 3A–C) and TSGH8301 (Figure 3D,E) cells. Notably, these events were not due to a decrease, but slight increase, in miR-34a expression, both in pre-miR-34a and fully mature miR-34a (Figure 4A,B). Taken together, these results demonstrate that overexpression of c-Myc 3′UTR leads to increased activation of CD44, and vice versa.
Figure 3.
c-Myc acts as a ceRNA of CD44 in urothelial carcinoma cells. TCCSUP (A–C) or TSGH8301 (D–F) UC cells overexpressing the c-Myc 3′UTR or CD44 3′UTR. Expression of CD44 and c-Myc, as evaluated using RT-PCR (mRNA, A,B,D,E), and eestern blot analysis (protein, C,F). GAPDH was used as loading control for western blot analysis. (** p < 0.01, *** p < 0.001).
Figure 4.
Overexpression of c-Myc or CD44 3′-UTRs do not suppress miR-34a expression in urothelial carcinoma cells. Pri-miR-34a and mature miR-34a were both detected in (A) TCCSUP and (B) TSGH8301 urothelial carcinoma cells overexpressing c-Myc or CD44 3′-UTRs. (* p < 0.05; ** p < 0.01; *** p < 0.001).
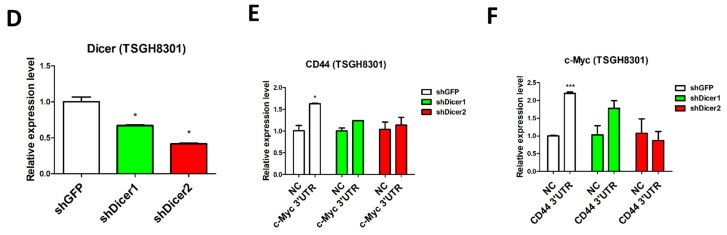
2.3. Dicer Knockdown Abolishes ceRNA Effect in UC
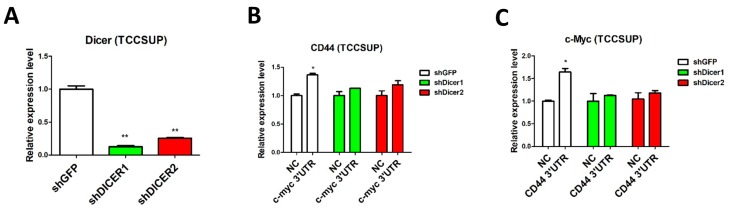
To determine whether c-Myc acts as a ceRNA of CD44 through miR-34a, we disrupted miRNA biogenesis via knockdown of the double-strand RNA endoribonuclease, Dicer [25], in TCCSUP and TSGH8301 cells. RT-PCR confirmed significant suppression of Dicer mRNA expression in both cell types transiently infected with anti-Dicer shRNAs (Figure 5A,D). Compared to the negative-control (shGFP), overexpression of the c-Myc 3′UTR did not increase CD44 levels in Dicer-depleted TCCSUP (Figure 5B) or TSGH8301 (Figure 5E) cells. Reciprocally, overexpression of the CD44 3’UTR did not increase c-Myc levels in either Dicer-knocked down cell line (Figure 5C,F). It is also noteworthy that TSGH8301 cells (with lower Dicer knockdown efficiency), had increased c-Myc and CD44 expression levels, following overexpression of either 3′UTR, in poorly knocked down Dicer cells (Figure 5D–F). Taken together, these results confirm that c-Myc and CD44 act as ceRNAs, through miR-34a, in UC.
Figure 5.
Dicer knockdown diminishes the ceRNA effects of c-Myc and CD44 in urothelial carcinoma cells. Dicer was depleted by shRNA in TCCSUP and TSGH8301 urothelial carcinoma cells. Relative expression of Dicer was assessed in (A) TCCSUP and (D) TSGH8301 cells using RT-PCR. Expression of CD44 (B,E) and c-Myc (C,F) in the control and Dicer-depleted cells was also assessed using RT-PCR. (* p < 0.05; ** p < 0.01, *** p < 0.001).
2.4. Expression of c-Myc Correlates with Expression of CD44, in Samples from UC patients and Cell Lines
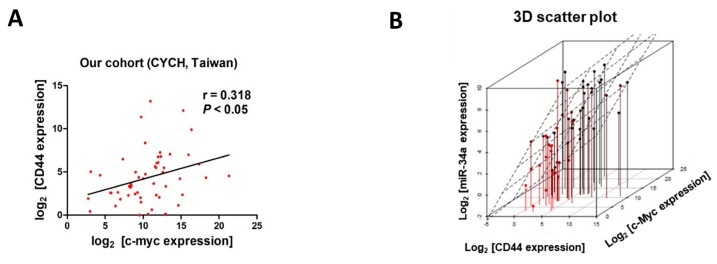
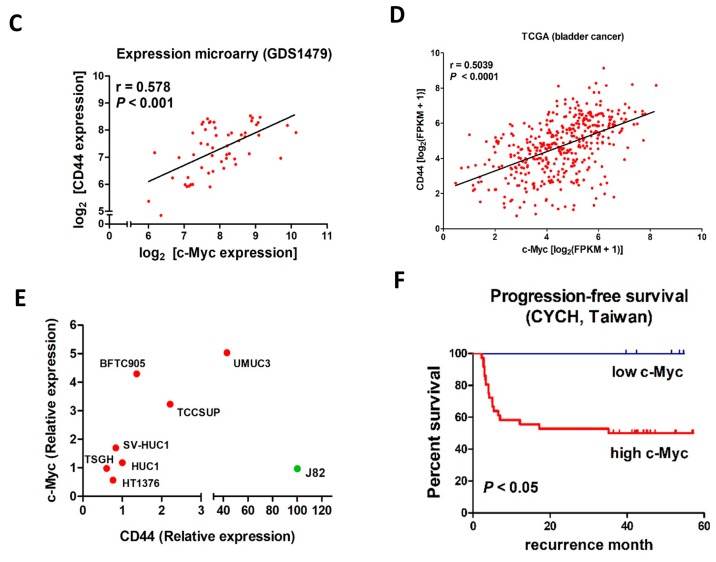
We also examined the ceRNA mechanism, in vivo, by assessing the expression of c-Myc and CD44 in samples obtained from 55 UC patients. Consistent with our cell line study, the expression of c-Myc in the clinical samples positively correlated with the expression of CD44 (R2 = 0.1015, p < 0.05, Figure 6A). 3D scatter plot analysis of these samples also revealed a positive correlation between the expression of miR-34a and the expression of c-Myc and CD44 (Figure 6B), although no correlation between expression of these genes with any of the clinical-pathological parameters was found (Table 1). Similar findings were obtained from a gene expression array (GDS1479, Figure 6C) and RNA-Seq dataset (from TCGA, Figure 6D). We also investigated the expression of c-Myc and CD44 in HUC1, normal urothelial cells and various UC cell lines (Figure 6E). Except for the J82 cell line, which has surprisingly low levels of c-Myc expression (Figure 6E, green dot), all cells demonstrated a positive correlation between c-Myc and CD44 expression, in UC cells (r = 0.672). In light of the strong association between c-Myc and the aggressiveness of cancer and eventual prognosis [26], we also examined the correlation between the expression of c-Myc and tumor recurrence in this sample cohort. Kaplan–Meier survival analysis revealed an association between higher c-Myc expression levels and poor recurrence-free survival (p < 0.05, Figure 6F).
Figure 6.
Positive correlation between c-Myc and CD44 expression in samples from urothelial carcinoma patients. (A) Scatter plot analysis revealed a positive association between the expression of c-Myc and CD44 (r = 0.318, p < 0.05) in samples obtained from 55 UC cancer patients at Chia-Yi Christian Hospital (CYCH), Taiwan. (B) 3D scatter plot showing positive correlations between c-Myc, CD44, and miR-34a in this cohort of UC patient samples. A positive correlation between the expression of c-Myc and CD44 was also observed in samples obtained from two publicly available databases (C) GEO GDS1479 (60 samples, expression microarray, r = 0.578, p < 0.05) and (D) TCGA (411 samples, RNA-Seq, r = 0.5039, p < 0.001). (E) Scatter plot showing the expression CD44 in UC cell lines. Except for J82 cells (green dot), which showed an exceptionally low expression of c-Myc, all other cells showed a positive correlation between expression of these genes (r = 0.672). (F) Kaplan–Meier survival analysis indicating that higher c-Myc expression associates with poor recurrence-free survival (p < 0.05).
Table 1.
Correlation between the expression of miR-34a, c-Myc, CD44, and clinical-pathological data in 55 UC samples.
| miR-34a | c-Myc | CD44 | |
|---|---|---|---|
| Gender | |||
| Male | 130.2 ± 173.6 1 | 8.4 ± 4.1 | 457.3 ± 1.6 |
| Female | 31.1 ± 67.5 | 6.1 ± 1.8 | 23.8 ± 42.6 |
| Location | |||
| Bladder | 106.9 ± 157.8 | 1.7 ± 5.5 | 512.5 ± 1.7 |
| Upper Tract | 105.1 ± 169.1 | 1.6 ± 6.2 | 33.8 ± 44.2 |
| Histological Grade | |||
| Low Grade | 143.7 ± 198.1 | 2.3 ± 7.6 | 108.1 ± 270.1 |
| High Grade | 100.2 ± 149.8 | 1.7 ± 5.5 | 413.5 ± 1.6 |
| Pathological Stage | |||
| Stage T1 | 90.9 ± 149.2 | 1.7 ± 5.9 | 68.9 ± 179.1 |
| ≥Stage T2 | 132.2 ± 173.5 | 1.2 ± 5.3 | 680.5 ± 2.1 |
| Relapse | |||
| Yes | 108.6 ± 153.1 | 1.4 ± 3.4 | 566.1 ± 1.8 |
| No | 102.7 ± 174.4 | 1.5 ± 5.8 | 17.9 ± 22.9 |
1 Relative expression value, Mean ± SD.
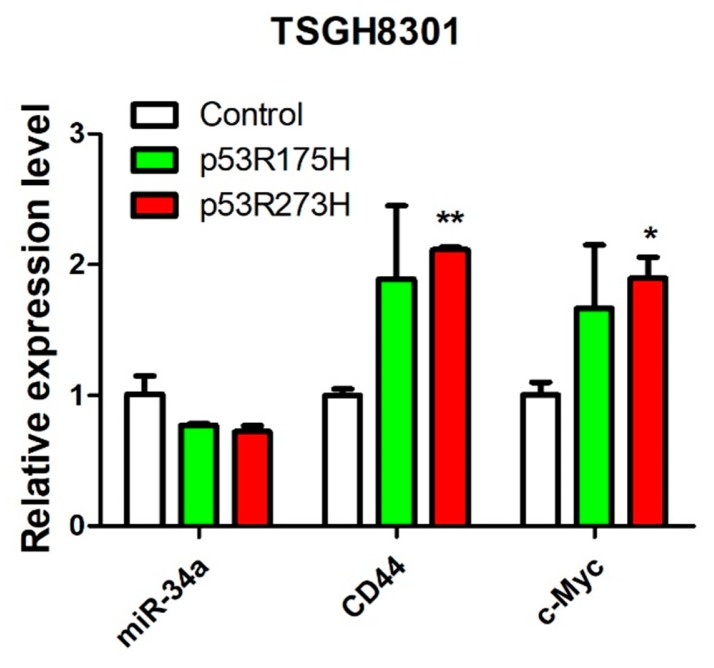
2.5. Dominant-Negative p53 Mutation Downregulates miR-34a in UC
Our previous experiment, using clinical samples, found a group of UC patient samples with low miR-34a expression, but also low miR-34a promoter methylation. This occurrence suggested an alternative mechanism(s) of controlling miR-34a expression in UC. In that regard, the miR-34a promoter was found to possess a perfect p53-binding site [27] and be directly regulated by the tumor suppressor p53 [28]. To determine whether p53 could regulate miR-34a expression, and the subsequent expression of c-Myc and CD44, we stably transfected p53 mutants (p53R175H and p53R273H) into TSGH8301 UC cells, with endogenous wild-type p53 [29], to create a dominant-negative effect. Compared to the control, overexpression of the p53 mutant downregulated miR-34a expression, while upregulating CD44 and c-Myc (Figure 7). These results confirmed that p53 regulates miR-34a expression directly, and subsequently, miR-34a’s capability of downregulating CD44 and c-Myc expression, in UC cells.
Figure 7.
p53 controls miR-34a expression in TSGH8301 urothelial carcinoma cells. Co-expression of p53 mutants p53R175H or p53R273H, with wild-type p53, in TSGH8301 cells, downregulated miR-34a and upregulated CD44 and c-Myc, as determined by RT-PCR. * p < 0.05; ** p < 0.01.
2.6. miR-34a Suppresses Tumorigenicity and Drug Resistance in UC
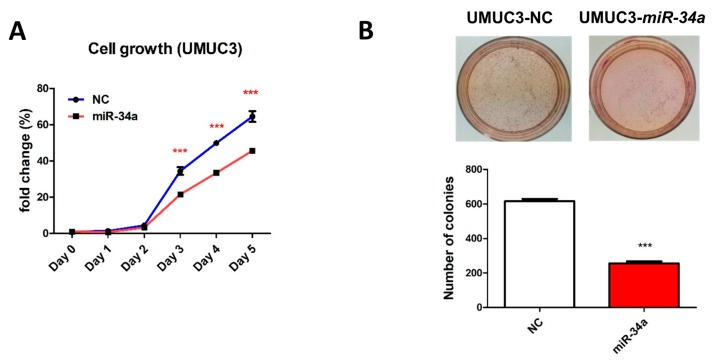
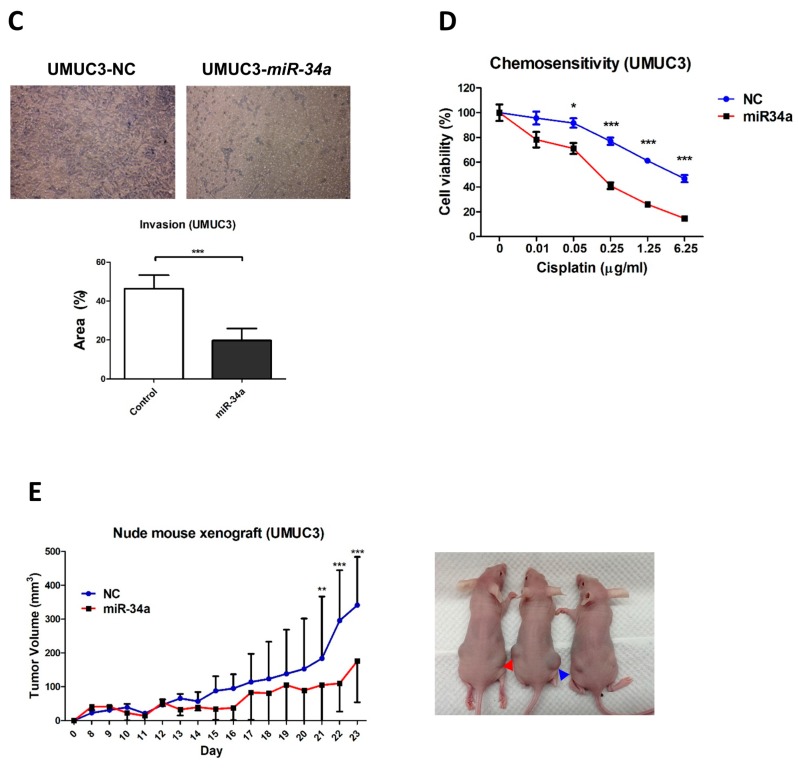
miR-34a has been reported to be a key regulator of tumor suppression, and its downregulation has been linked to the occurrence and development of several cancers [30]. Here, we sought to elucidate the functional role of miR-34a, in UC, by stably overexpressing miR-34a in UMUC3 cells. Such overexpression significantly inhibited cell growth (Figure 8A), and anchorage-independent growth (Figure 8B). It also significantly inhibited the invasive ability of UMUC3 cells (Figure 8C) and resensitized them to the effects of cisplatin (Figure 8D). Finally, a nude mouse model demonstrated that miR-34a overexpression inhibited the growth of UMUC3 cell xenografts in vivo (Figure 8E). Overall, these findings revealed that miR-34a overexpression inhibited both the growth and invasive ability of cancer cells, while also reducing chemoresistance in UC cells.
Figure 8.
Overexpression of miR-34a suppresses tumorigenicity and drug resistance in UMUC3 UC cells. (A) Cell proliferation of UMUC3 cells stably expressing miR-34a or an empty vector was measured using cell counting kits (CCKs), on the designated days. Anchorage-independent growth in UMUC3 cells overexpressing miR-34a was determined using soft-agar (B) and cell invasion (magnification 100×) (C) assays. Quantitative analysis of the number of colonies in the soft agar and invasion assays indicated below. (D) Chemosensitivity to cisplatin in UMUC3 cells overexpressing miR-34a, as determined using a CCK. (E) Nude mouse xenograft model showing tumor sizes of UMUC3 cells overexpressing miR-34a (red line) or a control plasmid (blue line). Representative examples of nude mice bearing tumors are also presented (left: miR-34a, red arrow head; right: control, blue arrow head). * p < 0.05; ** p < 0.01, *** p < 0.001.
3. Discussion
miR-34a plays an important role in cancer progression and drug resistance. Previous studies have reported miR-34a downregulation in many cancer types [31], including urothelial cancer (UC) [32]. Epigenetic silencing of miR-34a has also been addressed in several previous studies of human cancer [19,33,34]; however, the mechanism underlying miR-34a downregulation, in UC, has yet to be elucidated. In this study, when evaluating miR-34a expression in seven UC cell lines, we were surprised to observe epigenetic silencing of miR-34a, via promoter hypermethylation, only in UMUC3 UC cells. Clinical analyses also found that most UC patient samples were devoid of promoter miR-34a hypermethylation. For UC cells (BFTC905 and J82) and patient samples showing low expression, but also low miR-34a promoter methylation, the possibility of other epigenetic mechanisms (e.g., histone modifications), in regulating miR-34a, cannot be excluded. Since miR-34a is expressed in most UC cell lines and patient samples, alternative mechanisms likely exist for controlling miR-34a function in UC.
A new type of miRNA regulation, referred to as a competitive endogenous RNA (ceRNA) mechanism, involves crosstalk between miRNA targets and the regulation of gene expression, through competition in binding to shared miRNAs has recently been described [35]. Several research groups, including ours, have reported this phenomenon in human cancers [36,37,38,39]. In particular, we recently demonstrated that estrogen receptor-driven upregulation of E2F6 can ‘sponge’ miR-193a, to facilitate c-Kit overexpression, in ovarian cancer [36]. In the current study, we demonstrated that overexpression of the c-Myc 3′UTR could upregulate CD44 through miR-34a. Importantly, we also observed a positive correlation between the expression of c-Myc and CD44 in samples from UC patients and cell lines. Taken together, our results indicate that c-Myc may act as a ceRNA of CD44, in UC.
Several miR-34a targets have been shown to control the function of miR-34a, through ceRNA mechanisms. For example, one oncogenic, long noncoding (lnc)RNA (lncRNA-SNHG7) has been shown to sponge miR-34a, upregulating the expression of the oncogene GALNT7 in colorectal cancer [40]. In another study, lnc015192 was shown to sponge miR-34a to upregulate Adam12 expression in breast cancer [41]. However, the current study was the first to demonstrate that c-Myc mRNA can act as a ceRNA, to upregulate CD44 expression, by targeting miR-34a, in UC. Our results indicate that c-Myc participates in cancer stemness, not only via protein regulation [42,43,44], but also via mRNA. Note that the genomic amplification of chromosome 8q (where c-Myc resides), commonly associated with UC, provides an additional driving force for the overexpression of CD44 [23,24]. This may also explain the significantly shorter recurrence-free survival of patients presenting high c-Myc expression.
As in previous studies, we observed that miR-34a overexpression in UC cells inhibited tumor invasion and reduced drug resistance, probably through the inhibition of CD44 [15,16,17]. In the future, it may be possible to implement epigenetic interventions to restore miR-34a expression, and inhibit cancer stemness, in a subset of UC patients with epigenetic silencing of this micro-RNA [16].
On the other hand, the tumor suppressor gene p53, the most frequently mutated tumor suppressor in human cancer, including urothelial carcinoma [45,46], also directly upregulates miR-34a [47,48,49], thereby explaining the fact that p53 signaling can control miR-34a expression, in UC samples, without miR-34a promoter methylation. In this regard, overexpression of a dominant-negative p53 mutant resulted in miR-34a downregulation, and the subsequent upregulation of CD44 and c-Myc, in UC cells with wild-type p53.
It is also noteworthy to point out that the ceRNA effect of c-Myc and CD44 overexpression, by the c-Myc 3′UTR, in UC cell lines (Figure 4A,B), was not due to a decrease—but surprisingly, a slight increase—of mR-34a expression in those cells. This phenomenon may be due to the presence of a feedback loop that increases miR-34a expression upon the increase of c-Myc/CD44 [50]. However, further experiments need to be performed to validate such feedback loop.
4. Materials and Methods
4.1. Cell Cultures and Patient Samples
Human UC cell lines (SV-HUC, BFTC905, HT1376, TSGH8301, TCCSUP, J82, and UMUC3) were used in this study. SVHUC, BFTC905, HT1376, and TSGH8301 cell lines were maintained in RPMI 1640 medium (Gibco, Grand Island, NY), TCCSUP was maintained in DMEM medium, and J82 and UMUC3 were maintained in MEM medium (Gibco). Cultured cells were supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin, in a humidified atmosphere of 5% CO2, at 37 ℃. Fifty-five urothelial cancer samples from bladders or upper urinary tracts (ureter or renal pelvis) were procured from Chia-Yi Christian Hospital, Chia Yi, Taiwan. The clinical-pathological data of all samples is summarized in Table 2. All tissue samples were acquired from either transurethral resection (TURBT) or radical surgery. Patients were then followed up by either cystoscopy or radiographic detection (CT or MRI) for recurrence. All patients provided informed consent. This human sample study was approved by the Institutional Review Board of Chia-Yi Christian hospital, Taiwan.
Table 2.
Summary of clinical-pathological data of urothelial carcinoma samples.
| Clinical Parameter | Patient Samples (n = 55) |
|---|---|
| Age * | 70.94 ± 11.8 4 |
| Median | 74 |
| Range | 40–90 |
| Gender * | |
| Male | 41 |
| Female | 13 |
| Location 1,* | |
| Bladder | 36 |
| Upper Tract | 18 |
| Histological Grade 2 | |
| Low Grade | 12 |
| High Grade | 43 |
| Pathological Stage | |
| Stage T1 | 30 |
| ≥ Stage T2 | 25 |
| Relapse * | |
| Yes | 33 |
| No | 21 |
| Treatment | |
| TURBT | 34 |
| Non-TURBT 3 | 21 |
1 Upper urinary tract UC from ureter or renal pelvis; 2 Grading, low grade: G1, high grade: ≥G2; 3 Radial cystectomy (n = 2) or nephroureterectomy with bladder cuff excision (n = 19); 4 Mean ± SD. * 1 case was missing a value.
4.2. Plasmid Constructs and shRNA Knockdown
The miR-34a full-length DNA sequence was amplified via PCR with specific primers (Table 3) inserted into pSilencer4.1 as miRNA expression vectors. The CD44 3′UTR and c-Myc 3′UTR regions (each of which contains two miR-34a binding sites) were ligated into EGFP-C1. All shRNA plasmids in the pLKO.1 plasmid were purchased from the National RNAi Core Facility at Academia Sinica, Taiwan (rnai.genmed.sinica.edu.tw) and the clone IDs were Dicer-1 TRCN0000290426 and Dicer-2 TRCN0000290486).
Table 3.
Primers used in this study
| Primer Name | Primer Sequence (5′ to 3′) | Annealing TEMP (°C) | Product Size (bp) |
|---|---|---|---|
| COBRA | |||
| miR-34a-BS-F | TTTTTTTTTTTAGGTGGAGGAG | 60 | 270 |
| miR-34a-BS-R | ATACAAACTTCCAAACCTCTCC | ||
| Bisulphite Pyrosequeincing | |||
| miR-34a-Pyro-F | TAGGTGGGGGTTAGGTAG | 64 | 87 |
| miR-34a-Pyro-R 1 | agctggacatcacctcccacaacgCAAACTCCCACCCCTCCC | ||
| miR-34a-SEQ | GGTAGGGAGTATGAAG | ||
| RT-PCR | |||
| pri-miR-34a RT-F | TGTGATTAACCCCGTCTTGCA | 60 | 101 |
| pri-miR-34a RT-R | GCAGATTCTTGAGCCAGATTGC | ||
| CD44-RT-F | TCCCAGTATGACACATATTGC | 60 | 129 |
| CD44-RT-R | CACCTTCTTCGACTGTTGAC | ||
| c-Myc-RT-F | CTCGGATTCTCTGCTCTCTCCTCG | 60 | 177 |
| c-Myc-RT-R | TCTGACCTTTTGCCAGGAGCCT | ||
| Dicer-RT-F | TTAACCTTTTGGTGTTTGATGAGTGT | 60 | 98 |
| Dicer-RT-R | GCGAGGACATGATGGACAATT | ||
| GAPDH RT-F | CCCCTTCATTGACCTCAACTACAT | 60 | 135 |
| GAPDH RT-R | CGCTCCTGGAAGATGGTGA | ||
| Plasmid construction | |||
| miR-34a construct | |||
| miR-34a-BamHI F | CGCGGATCCGTAGAGATGGAGTCTTGCTAGTTGC | 64 | 514 |
| miR-34a-HindIII R | CCCAAGCTTTTCTCCCTACGTGCAAACTTCT | ||
| 3′UTR construct | |||
| CD44-XhoI F | CCTCGAGTGATCGTTCCAGTTCCCACTTG | 66 | 263 |
| CD44-PstI R | CCCCTGCAGGGGGGTCTGTTGAAGAT | ||
| c-Myc-XhoI F | CCTCGAGCTTGAGACTGAAAGATTTAGCCAT | 66 | 369 |
| c-Myc-PstI R | CCCCTGCAGTAAGATTTGGCTCAATGATATATTTG | ||
1 Primer sequence of the 5′tailed universal primer (UNIVR) is shown as lower case.
4.3. RNA Expression
All RNA samples were extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. The expression of CD44 and c-Myc, in UC cell lines, was analyzed by RT-PCR, using the StepOne Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with specific primers (Table 3). Relative gene expression levels were determined by comparing the threshold cycle (i.e., Ct value) of the test gene, normalized to the Ct of GAPDH, in each sample.
4.4. DNA Methylation Analysis
All DNA was extracted using Genomic DNA Mini Kits (Geneaid, Taipei, Taiwan) according to the manufacturer’s instructions, and underwent bisulfite modification, using EZ DNA Methylation-Gold™ kits (Zymo Research, Orange, CA, USA), according to the manufacturer’s protocol. Combined Bisulfite Restriction Analysis (COBRA) was used to determine miR-34a promoter methylation status, through digestion using BstUI (New England BioLabs, Beverly, MA, USA). Digested products were separated on 1.5% agarose gels for visualization. Pyrosequencing was performed using the PyroMark Q24 (Qiagen, GmbH, Hilden, Germany), with Pyro Gold Reagents (Qiagen), according to the manufacturer’s instructions. The methylation status of nine CpG sites, proximal to miR-34a, was measured. The methylation percentage of each cytosine was determined by the fluorescence intensity of a cytosine, divided by the sum of fluorescence intensity of cytosines and thymines, at each CpG site (multiplied by 100%). Primers for methylation analysis are shown in Table 3.
4.5. Cell Invasion Assay
2 × 104 cells were seeded in 1% FBS medium in the upper chamber on 2 Millicell™ matrigel-coated (BD Bioscience, NJ, USA) cell culture inserts (Millipore, Billerica, MA). After 48 h, the cells were fixed using methanol and stained using Giemsa reagent (Sigma, St. Louis, MO, USA).
4.6. Cell Viability Assay
Cells were seeded in 96-well plates for 4 d without or with cisplatin in various concentrations. Cell growth was determined using a cell counting kit (CCK-8, Sigma), according to the manufacturer’s instructions. Cell absorbances were then determined at 450 nm, using a microplate reader.
4.7. Soft Agar Assay
For soft agar assays, a 3-mL base layer of agar (0.5% agar in culture medium) was allowed to solidify in a six-well flat-bottomed plate, before the addition of 2.0-mL cell suspensions (10,000 cells) in 0.3% agar in culture medium. The cell-containing layer was then solidified at room temperature for 20 min. Colonies were allowed to grow for 14–21 days at 37 °C with 5% CO2 before imaging. Plates were then stained with 1 mg/mL iodonitrotetrazolium chloride (Sigma) overnight at 37 °C.
4.8. Western Blotting
All samples were lysed using PRO-PREP Protein Extraction Solution (iNtRON Biotechnology, Korea), and then immunoblotted, using monoclonal primary antibodies (CD44, GAPDH, and Dicer), purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and Myc (Abcam, Cambridge, UK). Horseradish peroxidase-conjugated secondary antibodies, including mouse IgG and rabbit IgG antibodies (Abcam), were also used. Specific signals were visualized using a chemiluminescence (ECL) detection kit (Millipore) and the BioSpectrum® 2D Imaging System (BioSpectrum 800, UVP, Upland, CA, USA).
4.9. Bioinformatics Analysis
The output results of multiple prediction programs were integrated using TargetScan (http://www.targetscan.org/), PicTar (http://pictar.org/), and miRanda (http://www.microrna.org/microrna/), to precisely identify miRNAs targeting c-Myc and CD44. Moreover, results of other microRNAs found in these three databases were retrieved and compared.
4.10. In Vivo Tumorigenicity Assay
Eight-week-old, athymic nude mice (BALB/cByJNarl) were obtained from the Taiwan National Laboratory Animal Center or BioLASCO Taiwan Co., Ltd., Taipei, Taiwan. All mice were kept under specific pathogen-free conditions, using a laminar airflow rack, with free access to sterilized food and autoclaved water. All animal experiments were approved by the Animal Experimentation Ethics Committee of National Chung Cheng University (Taiwan). 1 × 106 cells, in a 1:1 mixture of 0.1 mL medium and Matrigel (BD Biosciences), were injected subcutaneously into each flank of each mouse (day 0). Tumor size was measured daily, using calipers, in length (L) and width (W). Tumor volumes were calculated using the formula (L × W2/2). At the end of the experiments, all mice were sacrificed by cervical dislocation, as previously approved.
4.11. Statistical Calculation
All statistical analysis was performed using GraphPad (San Diego, CA, USA) Prism 5 software (version 5.01). Non-parametric variables were assessed using the Mann–Whitney test, when appropriate. Recurrence-free survival was assessed using Kaplan-Meier analysis with log-rank test. A p value of less than 0.05 was considered significant.
5. Conclusions
In conclusion, miR-34a was epigenetically silenced only in a subset of UC cell lines and patient samples. Moreover, overexpression of c-Myc was shown to upregulate CD44, via a miR-34a-mediated ceRNA mechanism, in UC cell lines and clinical samples. The overexpression of miR-34a was also shown to suppress tumor growth and invasive capability, while reducing drug resistance. Thus, amplification of c-Myc is an important mechanism in controlling cancer stemness, through ceRNA mechanisms, in UC. These novel modes of endogenous competition could explain the regulation of any number of genes, both in healthy and diseased tissues.
Acknowledgments
The authors would like to thank Po-Yen Hsu for assistance.
Author Contributions
P.-C.C., W.-Y.H., W.-H.H., R.-I.L., Y.-C.J., and C.-H.S. performed the experiments. P.-C.C., Y.-C.J., and C.-H.S. collected patient samples. H.-Y.L., J.M.J.L. and Y.-M.C. performed data analysis, while C.-H.S. and M.W.Y.C. designed the experiments. C.-C.Y., W.-Y.H., and M.W.Y.C. wrote the manuscript.
Funding
This research was funded by the Ministry of Science and Technology, Taiwan (MOST 104-2320-B-194-001-MY3, 106-2923-B-194-001-MY3, 108-2314-B-194-003-MY2), Ditmanson Medical Foundation Chiayi Christian Hospital, Taiwan (grant no. R107-001 and R101-9) and the Center for Innovative Research on Aging Society (CIRAS) from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by Ministry of Education (MOE) in Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Green D.A., Rink M., Xylinas E., Matin S.F., Stenzl A., Roupret M., Karakiewicz P.I., Scherr D.S., Shariat S.F. Urothelial carcinoma of the bladder and the upper tract: Disparate twins. J. Urol. 2013;189:1214–1221. doi: 10.1016/j.juro.2012.05.079. [DOI] [PubMed] [Google Scholar]
- 2.Tan L.B., Chang L.L., Cheng K.I., Huang C.H., Kwan A.L. Transitional cell carcinomas of the renal pelvis and the ureter: Comparative demographic characteristics, pathological grade and stage and 5-year survival in a Taiwanese population. BJU Int. 2009;103:312–316. doi: 10.1111/j.1464-410X.2008.07985.x. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen D.P., Thalmann G.N. Contemporary update on neoadjuvant therapy for bladder cancer. Nat. Rev. Urol. 2017;14:348–358. doi: 10.1038/nrurol.2017.30. [DOI] [PubMed] [Google Scholar]
- 4.Chan K.S., Espinosa I., Chao M., Wong D., Ailles L., Diehn M., Gill H., Presti J., Jr., Chang H.Y., van de Rijn M., et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. USA. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C.T., Lin W.Y., Chang Y.H., Chen W.C., Chen M.F. Impact of CD44 expression on radiation response for bladder cancer. J. Cancer. 2017;8:1137–1144. doi: 10.7150/jca.18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H., Tian Y., Yuan X., Wu H., Liu Q., Pestell R.G., Wu K. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther. 2015;8:3783–3792. doi: 10.2147/ott.s95470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Horst G., Bos L., van der Pluijm G. Epithelial plasticity, cancer stem cells, and the tumor-supportive stroma in bladder carcinoma. Mol. Cancer Res. 2012;10:995–1009. doi: 10.1158/1541-7786.MCR-12-0274. [DOI] [PubMed] [Google Scholar]
- 8.Ponta H., Sherman L., Herrlich P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 9.Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 10.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranwal S., Alahari S.K. miRNA control of tumor cell invasion and metastasis. Int. J. Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C., Lu P., Sun G., Yang L., Wang Z., Wang Z. miR-34a sensitizes lung cancer cells to cisplatin via p53/miR-34a/MYCN axis. Biochem. Biophys. Res. Commun. 2017;482:22–27. doi: 10.1016/j.bbrc.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Christoffersen N.R., Shalgi R., Frankel L.B., Leucci E., Lees M., Klausen M., Pilpel Y., Nielsen F.C., Oren M., Lund A.H. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 15.Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., Patrawala L., Yan H., Jeter C., Honorio S., et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Yu G., Shi R., Lang B., Chen X., Xia D., Xiao H., Guo X., Guan W., Ye Z., et al. Cisplatin-induced epigenetic activation of miR-34a sensitizes bladder cancer cells to chemotherapy. Mol. Cancer. 2014;13:8. doi: 10.1186/1476-4598-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G., Yao W., Xiao W., Li H., Xu H., Lang B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting CD44. J. Exp. Clin. Cancer Res. 2014;33:779. doi: 10.1186/s13046-014-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G., Li H., Wang J., Gumireddy K., Li A., Yao W., Tang K., Xiao W., Hu J., Xiao H., et al. miRNA-34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J. Urol. 2014;192:1229–1237. doi: 10.1016/j.juro.2014.05.094. [DOI] [PubMed] [Google Scholar]
- 19.Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki H., Maruyama R., Yamamoto E., Kai M. DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarringhalam K., Tay Y., Kulkarni P., Bester A.C., Pandolfi P.P., Kulkarni R.V. Identification of competing endogenous RNAs of the tumor suppressor gene PTEN: A probabilistic approach. Sci. Rep. 2017;7:7755. doi: 10.1038/s41598-017-08209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamura S., Saini S., Majid S., Hirata H., Ueno K., Deng G., Dahiya R. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS ONE. 2012;7:e29722. doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahdy E., Pan Y., Wang N., Malmstrom P.U., Ekman P., Bergerheim U. Chromosome 8 numerical aberration and C-MYC copy number gain in bladder cancer are linked to stage and grade. Anticancer Res. 2001;21:3167–3173. [PubMed] [Google Scholar]
- 24.Zaharieva B., Simon R., Ruiz C., Oeggerli M., Mihatsch M.J., Gasser T., Sauter G., Toncheva D. High-throughput tissue microarray analysis of CMYC amplificationin urinary bladder cancer. Int. J. Cancer. 2005;117:952–956. doi: 10.1002/ijc.21253. [DOI] [PubMed] [Google Scholar]
- 25.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Navarro F., Lieberman J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS ONE. 2015;10:e0132767. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang F.L., Ling Y.F., Lai M.D. Exogenous mutant p53 DNA enhanced cisplatin-induced apoptosis in TSGH-8301 human bladder cancer cells. Anticancer Res. 2000;20:329–336. [PubMed] [Google Scholar]
- 30.Chen A.H., Qin Y.E., Tang W.F., Tao J., Song H.M., Zuo M. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 2017;17:63. doi: 10.1186/s12935-017-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slabakova E., Culig Z., Remsik J., Soucek K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi: 10.1038/cddis.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Li T., Han G., Li Y., Shi L.H., Li H. Expression and role of miR-34a in bladder cancer. Indian J. Biochem. Biophys. 2013;50:87–92. [PubMed] [Google Scholar]
- 33.Kwon H., Song K., Han C., Zhang J., Lu L., Chen W., Wu T. Epigenetic Silencing of miRNA-34a in Human Cholangiocarcinoma via EZH2 and DNA Methylation: Impact on Regulation of Notch Pathway. Am. J. Pathol. 2017;187:2288–2299. doi: 10.1016/j.ajpath.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siemens H., Neumann J., Jackstadt R., Mansmann U., Horst D., Kirchner T., Hermeking H. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 2013;19:710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- 35.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng F.H.C., Lin H.Y., Hwang T.W., Chen Y.C., Huang R.L., Chang C.B., Yang W., Lin R.I., Lin C.W., Chen G.C.W., et al. E2F6 functions as a competing endogenous RNA, and transcriptional repressor, to promote ovarian cancer stemness. Cancer Sci. 2018 doi: 10.1111/cas.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsushima K., Natsume A., Ohka F., Shinjo K., Hatanaka A., Ichimura N., Sato S., Takahashi S., Kimura H., Totoki Y., et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang R., Xing L., Zheng X., Sun Y., Wang X., Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Yang F., Shen Y., Zhang W., Jin J., Huang D., Fang H., Ji W., Shi Y., Tang L., Chen W., et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018;25:2209–2220. doi: 10.1038/s41418-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Zeng C., Hu J., Pan Y., Shan Y., Liu B., Jia L. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J. Hematol. Oncol. 2018;11:89. doi: 10.1186/s13045-018-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X., Xie X., Liu P., Yang L., Chen B., Song C., Tang H., Xie X. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating miR-34a. Oncogene. 2018;37:6316–6326. doi: 10.1038/s41388-018-0410-1. [DOI] [PubMed] [Google Scholar]
- 42.Zviran A., Mor N., Rais Y., Gingold H., Peles S., Chomsky E., Viukov S., Buenrostro J.D., Scognamiglio R., Weinberger L., et al. Deterministic Somatic Cell Reprogramming Involves Continuous Transcriptional Changes Governed by Myc and Epigenetic-Driven Modules. Cell Stem Cell. 2019;24:328–341. doi: 10.1016/j.stem.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 2018;37:173. doi: 10.1186/s13046-018-0835-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Schaijik B., Davis P.F., Wickremesekera A.C., Tan S.T., Itinteang T. Subcellular localisation of the stem cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: A review. J. Clin. Pathol. 2018;71:88–91. doi: 10.1136/jclinpath-2017-204815. [DOI] [PubMed] [Google Scholar]
- 45.Muller P.A., Vousden K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Vida A., Lerner S.P., Bellmunt J. The Cancer Genome Atlas Project in Bladder Cancer. Cancer Treat. Res. 2018;175:259–271. doi: 10.1007/978-3-319-93339-9_12. [DOI] [PubMed] [Google Scholar]
- 47.Rokavec M., Li H., Jiang L., Hermeking H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014;6:214–230. doi: 10.1093/jmcb/mju003. [DOI] [PubMed] [Google Scholar]
- 48.Menges C.W., Kadariya Y., Altomare D., Talarchek J., Neumann-Domer E., Wu Y., Xiao G.H., Shapiro I.M., Kolev V.N., Pachter J.A., et al. Tumor suppressor alterations cooperate to drive aggressive mesotheliomas with enriched cancer stem cells via a p53-miR-34a-c-Met axis. Cancer Res. 2014;74:1261–1271. doi: 10.1158/0008-5472.CAN-13-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siemens H., Jackstadt R., Kaller M., Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai X., Wolkenhauer O., Vera J. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016;44:6019–6035. doi: 10.1093/nar/gkw550. [DOI] [PMC free article] [PubMed] [Google Scholar]