Abstract
Studies using genetic mouse models that have defective autophagy have led to the conclusion that macroautophagy/autophagy serves as a tumor suppressor. One of such models is the liver-specific Atg5 or Atg7 knockout mice, and these knockout mice develop spontaneous liver tumors. It has been generally agreed that p62-mediated Nrf2 activation plays a critical role in promoting autophagy deficiency-induced liver injury and liver tumorigenesis. The mechanisms of how persistent Nrf2 activation induces liver injury and tumorigenesis are incompletely known. We discuss the recent progress on the new roles of HMGB1 and Yap in regulating liver injury and tumorigenesis in mice with liver-specific autophagy deficiency.
Key words: Hepatomegaly, HMGB1, mTOR, Nrf2, p62, Proteotoxicity, Yap
INTRODUCTION
Liver cancer is the third leading cause of cancer-related deaths worldwide and is one of the major cancers that accounts for more than 600,000 deaths per year1. The mortality rate of hepatocellular carcinoma (HCC) in the US increased approximately 40% between 2000 and 2016 according to a more recent study2. Many environmental factors such as chronic alcohol consumption, hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, and obesity-related nonalcoholic fatty liver diseases (NAFLD) are risk factors for HCC. The development of liver tumors involves a chronic multihit process, including hepatocyte death, inflammatory response, fibrosis, cirrhosis, and cycles of cell death and compensatory hepatocyte proliferation, which creates an environment that is permissive to genetic changes leading to neoplastic transformation3–5. Autophagy is a lysosomal degradation pathway that degrades cellular proteins, lipid, and organelles to sustain metabolism and homeostasis in the liver6,7. Interestingly, the above environmental risk factors leading to chronic liver pathogenesis are often associated with impaired hepatic autophagy6–9. Indeed, others and we have previously demonstrated that liver-specific Atg7 (L-Atg7) or L-Atg5 knockout (KO) mice have increased liver injury, severe hepatomegaly, inflammation, fibrosis and developed spontaneous liver tumors10–14.
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a transcription factor that regulates the expression of genes for cellular antioxidants, drug metabolism enzymes, and transporters to protect against oxidative and electrophilic stresses15,16. Under normal conditions, Keap1 (Kelch-like ECH-associated protein 1) binds to Nrf2 to negatively regulate Nrf2 stability by promoting its ubiquitination and degradation by the ubiquitin proteasome system. Nrf2 can be activated via the canonical and noncanonical pathways. For the canonical Nrf2 activation, in response to oxidative and electrophilic stress, certain key cysteine residues of Keap1 is modified/oxidized that disrupts the Keap1-Nrf2 interaction resulting in the increased Nrf2 stability and nuclear translocation. For the noncanonical Nrf2 activation, p62/SQSTM1 (hereafter referred to as p62) directly interacts with the Kelch-repeat domain of Keap1 via its Keap1-interacting region (KIR) and thus competes with Keap1 for the binding with Nrf217–19. p62 is an autophagy substrate protein as it directly interacts with the autophagosome membrane protein microtubule light chain 3 protein (LC3), and increased autophagy generally leads to increased p62 degradation. Therefore autophagy-deficient livers often have accumulation of p62 that causes the dissociation of Nrf2 from Keap1, which allows Nrf2 translocation to the nucleus for the noncanonical Nrf2 activation20,21.
Interestingly, deletion of p62 in L-Atg7 KO mice inhibited liver injury, hepatomegaly, and liver tumorigenesis in L-Atg7 KO mice10,20. Because p62 can activate Nrf2 by sequestrating Keap1, the improvement on the liver pathology by the deletion of p62 is likely associated with the impaired Nrf2 activation in hepatic autophagy-deficient mice. Indeed, others and we have demonstrated that deletion of Nrf2 in L-Atg7 or L-Atg5 mice almost completely abolishes hepatomegaly, liver injury, and liver tumorigenesis. These observations indicate that persistent activation of Nrf2 due to the accumulation of p62 is likely the major player for the liver injury and tumorigenesis in autophagy-deficient livers12,19. However, it is unknown whether other molecules or signaling pathways would also contribute to liver injury and tumorigenesis in autophagy-deficient livers in addition to Nrf2 activation. Recently, two studies have independently identified the HMGB1 (high-mobility group box 1) and the Hippo–Yap (yes-associated protein) signaling pathways as new important players that contribute to the pathogenesis of autophagy-deficient livers13,14.
HMGB1 PROMOTES HEPATIC DUCTULAR REACTION AND TUMORIGENESIS
HMGB1 is a nonhistone DNA binding protein that can regulate DNA replication, DNA repair, and transcription22. While it is mainly a nuclear protein, a small portion of HMGB1 can also be found in the cytoplasm. More importantly, HMGB1 can be released into the extracellular space either passively during cell death or actively following cytokine stimulation. Released HMGB1 acts as the prototypic damage-associated molecular pattern (DAMP) molecule to activate immune cells by binding with specific cell-surface receptors including TLR4 (toll-like receptor 4), TLR2, and RAGE (receptor for advanced glycation endproducts)22,23. Therefore, HMGB1 is considered as a proinflammatory cytokine and is associated with liver injury, sepsis, inflammation, and fibrosis24,25. Because L-Atg7 and L-Atg5 KO mice have liver injury, inflammation, and fibrosis, it is natural to hypothesize that HMGB1 could be involved in the pathogenesis of autophagy-deficient livers. To test this hypothesis, Khambu et al.13 performed a series of elegant systemic experiments to investigate the role of HMGB1 in liver injury, inflammation, ductular reaction, fibrosis, and tumorigenesis in L-Atg7 and L-Atg5 KO mice. Khambu et al.13 first found that the hepatic levels of HMGB1 were much lower in L-Atg7 KO mice than in matched WT mice. Decreased HMGB1 was not due to the decreased transcription of HMGB1 since no mRNA changes were found in L-Atg7 KO mice. Interestingly, ELISA and mass spectrometry analysis revealed increased serum levels of HMGB1 in L-Atg7 KO mice, suggesting an increased release of hepatic HMGB1 into the extracellular space and blood. Results from the immunostaining of HMGB1 in liver tissues indicated that HMGB1 is mainly released from the hepatocytes but not from the nonparenchymal cells. In addition to L-Atg7 KO mice, release of HMGB1 was also observed in L-Atg5 or L-VPS34 KO mice and even from Atg7-deficeint renal proximal tubules, suggesting that release of HMGB1 is a general event in autophagy-deficient cells regardless of tissue specificity.
To further determine the functional contribution of HMGB1 in the pathogenesis of autophagy-deficient liver, Khambu et al.13 deleted HMGB1 in L-Atg7 KO mice. Surprisingly, deletion of HMGB1 did not improve liver injury (based on the serum aminotransferase activities), hepatomegaly, inflammation, and fibrosis, which are typical pathological changes of autophagy deficiency in mouse livers. Deletion of HMGB1 also did not attenuate the accumulation of hepatic p62 and activation of Nrf2 in autophagy-deficient livers. As discussed above, the p62–Nrf2 axis has been previously shown to play critical roles in autophagy deficiency-induced hepatomegaly, liver injury, inflammation, and fibrosis10,12,19, which perhaps explains why deletion of HMGB1 did not improve these phenotypes in L-Atg7 KO mice.
Ductular reaction is originally defined as a reaction of ductular phenotypes, possibly but not necessarily of ductular origin, which is generally characterized as the proliferation of reactive bile ducts in response to liver injuries26,27. While it is commonly observed in biliary disorders, ductular reaction is now found to be associated with various liver diseases including alcoholic and nonalcoholic hepatitis, chronic viral hepatitis, and HCC. Ductular reaction is also closely associated with liver fibrosis, and the extent of ductular reaction is correlated with patient mortality. It is generally thought that ductular reaction plays a critical role in liver regeneration, and it involves transdifferentiation of liver cells from cholangiocytes, hepatic progenitor cells, and/or hepatocytes to repair damages on cholangiocytes or hepatocytes. The signaling pathways that are involved in ductular reaction-associated transdifferentiation include Notch, Hippo/Yap, Wnt/β-catemin, HGF/c-MET, and TWEAK/Fn14 pathways27. Ductular reaction has been observed in the autophagy-deficient mouse livers12. Interestingly, Khambu et al.13 found that deletion of HMGB1 markedly blunted increased ductular reaction in L-Atg7 KO mice. Deletion of the HMGB1 receptor RAGE did not affect the release of HMGB1 but significantly blunted ductular reaction in L-Atg7 KO mice, suggesting that HMGB1-mediated ductular reaction requires RAGE signaling. Hedgehog signaling was elevated in Mdr2-deficient mouse livers that have increased ductular reaction28. However, the Hedgehog signaling was only mildly increased in L-Atg7 KO mice, and deletion of HMGB1 did not affect the increased Hedgehog signaling in L-Atg7 KO mice. When isolated cellular fractions enriched with ductular cells and hepatic progenitor cells were further treated with recombinant HMGB1, increased levels of phosphorylated ERK1/2 (extracellular signal-regulated kinases 1/2) were observed, which could be blocked by a RAGE antibody. In a separate study29, recombinant HMGB1 promoted the proliferation of bipotential murine oval liver (BMOL) cells, which are well-characterized bipotential liver progenitor cells. Interestingly, increased ERK activation (but not Yap and Notch) was also found in recombinant HMGB1-treated BMOL cells, which could be blocked by inhibiting RAGE. These results suggest that ERK activation may play a critical role in HMGB1-mediated ductular reaction in autophagy-deficient livers.
In early studies, L-Atg7 and L-Atg5 KO mice were generated by crossing Atg7 or Atg5 floxed mice with albumin Cre mice, and these knockouts have increased cell death and liver injury11,21. To determine whether the release of HMGB1 from autophagy-deficient livers was due to cell death, Khambu et al.13 generated an inducible mouse model by crossing Atg7 floxed mice with Alb-Cre ERT2 mice. This inducible mouse model can delete Atg7 in adult mice and can also avoid the possible deletion of Atg7 in cholangiocytes. Using this inducible mouse model, Khambu et al. found that significant HMGB1 release started by day 5 and day 7 after tamoxifen injection, which was coincidental with the deletion of hepatic Atg7. However, obvious liver injury only developed at day 15 and day 20 based on the serum values of alanine aminotransferase after the tamoxifen injection, suggesting that HMGB1 release from Atg7-deficient hepatocytes may occur independent of cell death. Further studies revealed that release of HMGB1 from Atg7-deficient livers required Nrf2, since codeletion of Nrf2 and Atg7 suppressed HMGB1 release. Inflammasome has been shown to mediate HMGB1 secretion in monocytes and macrophages30. Caspase 1 is a key component of inflammasome; Khambu et al. also found increased caspase 1 activation and cleavage of gasdermin D in liver-specific Atg7 KO mice. Cleaved gasdermin D can translocate to the plasma membrane to form pores for either the release of cytokines such as IL-1β or trigger cell death called pyroptosis31,32. Deletion of caspase 1 in L-Atg7 KO mice significantly suppressed HMGB1 release and ductular reaction but not the liver injury and hepatomegaly, similar to the mice with the codeletion of HMGB1 and Atg7. Interestingly, deletion of Nrf2 in L-Atg7 KO mice inhibited caspase 1/11 activation and gasdermin D cleavage, suggesting that Nrf2 activation promotes the inflammasome activation and subsequent gasdermin D cleavage and HMGB1 release in L-Atg7 KO mice. However, whether Nrf2 activation can directly activate caspase 1/11 or indirectly due to the increased cell death and inflammation in autophagy-deficient livers remains to be investigated in the future.
As discussed above, ductular reaction has been linked to cell proliferation, fibrosis, and tumorigenesis in the liver27. Indeed, Khambu et al.13 found that deletion of HMGB1 decreased the number of tumors in L-Atg7 KO mice, suggesting that the release of HMGB1 and ductular reaction may promote liver tumorigenesis in autophagy-deficient livers. Importantly, in a recent separate study, Hernandez et al.29 also reported that HMGB1 promoted ductular reaction and liver tumorigenesis in three other different models [Tak1 liver-specific KO, Mdr2 KO, and 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) diet], although autophagy and Nrf2 activation was not determined in this study. Interestingly, Hernandez et al.29 also found that HMGB1 did not play a role in liver inflammation and fibrosis in these models. Taken together, the studies from Khambu et al. indicate that HMGB1 release due to the Nrf2-mediated inflammasome activation in autophagy-deficient livers promotes ductular reaction and liver tumorigenesis but is dispensable for liver injury, hepatomegaly, inflammation, and fibrosis.
HIPPO–YAP PATHWAY IN AUTOPHAGY-DEFICIENT LIVER
The Hippo pathway was originally discovered from genetic screening in Drosophila for regulators of growth, which was later found that the Hippo pathway is highly conserved from Drosophila to mammals33–35. Yap is the primary target of Hippo signaling, which acts as a transcriptional coactivator and binds to the TEAD family of transcription factors for regulating the transcription of a set of genes for cell proliferation, antiapoptosis, and “stemness”36,37. Yap is mainly regulated at the posttranslational level via Hippo signaling-mediated phosphorylation and sequestration in the cytoplasm. Hippo pathway mutants or liver-specific deletion of Hippo components (e.g., Mst1/2, Nf2) or overexpression of Yap leads to liver overgrowth phenotype and development of liver cancer35,38,39. Yap is highly expressed in biliary cells, and increased Yap activity in the liver promotes ductular reaction40. Therefore, many of the phenotypes from the Yap-activating livers including hepatomegaly, ductular reaction, and liver tumorigenesis were similar to the liver pathologies of autophagy-deficient livers. In a recent study, Lee at al.14 systematically investigated the role of Yap in the pathogenesis of L-Atg7 KO mice. By performing immunostaining for Yap, Lee et al. found that both cytoplasmic and nuclear Yap increased in L-Atg7 KO mouse livers and in primary cultured hepatocytes isolated from Atg7 KO mice. Moreover, gene set enrichment analysis of L-Atg7 KO livers also revealed enrichment signature of Yap target genes, and increased expression of Yap target genes was further confirmed by qRT-PCR. These results support the notion that Yap is accumulated and activated in L-Atg7 KO mouse livers. To test whether autophagy could directly degrade Yap to cause the accumulation of Yap in L-Atg7 KO mice, Lee et al. inhibited autophagy either pharmacologically (using leupeptin and NH4Cl) or genetically knockdown Atg7 (using shRNA) in AML12 cells, and both conditions led to the increased levels of Yap protein. Moreover, Yap protein also colocalized with Lysotracker-positive lysosomes and GFP–LC3-positive autophagosomes in cultured THLE5B human hepatocytes. These observations suggest that Yap could be degraded by autophagy, and livers with impaired autophagy may lead to the accumulation of Yap. To further determine the role of Yap in the pathogenesis of autophagy-deficient livers, Lee et al. generated tamoxifen inducible L-Yap/Atg7 double knockout (DKO) mice. Unlike the HMGB1/Atg7 DKO mice reported by Khambu et al.13, Yap/Ag7 DKO mice have decreased hepatocyte size, hepatomegaly, portal and lobular inflammation, ductular reaction, progenitor cell expansion, and fibrosis compared with L-Atg7 KO mice. Subsequently, Yap/Atg7 DKO mice also had decreased tumor size and numbers compared with L-Atg7 KO mice, although tumors still developed in the Yap/Atg7 DKO mice, which are similar to the HMGB1/Atg7 DKO mice. Interestingly, p62-Nrf2 signaling pathway was still activated in Yap/Atg7 DKO mice, suggesting that Yap may act in a parallel pathway that contributes to the hepatomegaly, liver injury, and tumorigenesis independent of Nrf2 activation in L- Atg7 KO mice.
SUMMARY AND FUTURE PERSPECTIVES
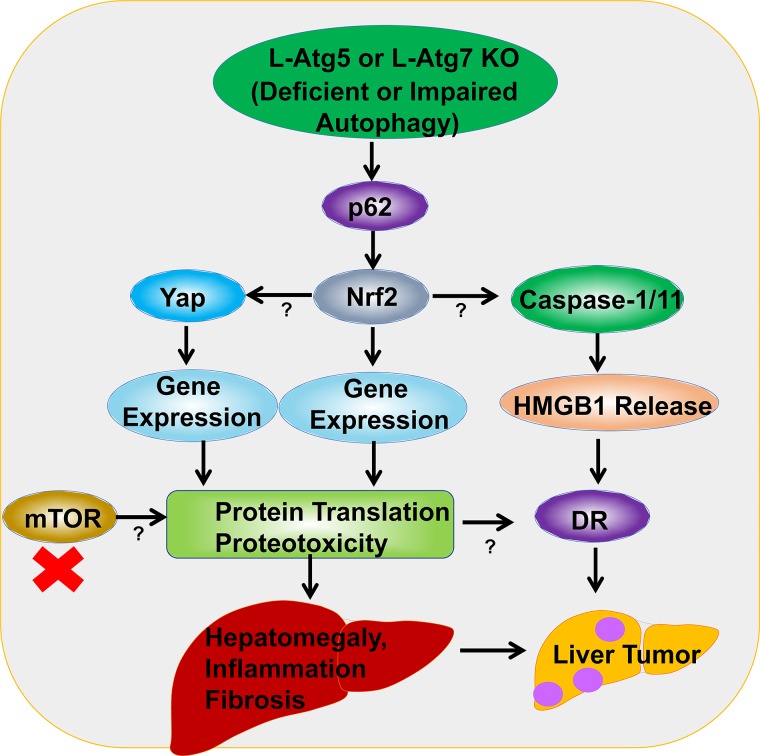
In summary, autophagy-deficient livers have accumulated p62, increased Nrf2 and Yap activation, as well as elevated release of hepatic HMGB1, which are responsible for hepatomegaly, inflammation, ductular reaction, fibrosis, and liver tumorigenesis. However, it appears that p62, Nrf2, Yap, and HMGB1 may play specific distinctive roles and contribute to the different pathologies in the autophagy-deficient livers. HMGB1 seems to act downstream of Nrf2 and contributes to the ductular reaction and tumor progression but does not affect hepatomegaly, inflammation, and fibrosis. In contrast, both Nrf2 and Yap contribute to all the phases of liver pathogenesis including hepatomegaly, inflammation, ductular reaction, fibrosis, and tumorigenesis in autophagy-deficient livers. It should be noted that deletion of Nrf2 completely abolishes liver tumorigenesis in L-Atg5 KO and L-Atg7 KO mice, but deletion with p62, HMGB1, or Yap only decreases the number of tumors in L-Atg5 KO and L-Atg7 KO mice. These observations suggest that Nrf2 activation plays a central and predominate role in contributing to the pathogenesis of autophagy-deficient livers. While deletion of p62 inhibits the persistent Nrf2 activation, liver injury, hepatomegaly, and liver tumorigenesis in L-Atg7 KO mice, the p62/Atg7 DKO mice still have intact Nrf2 pathway that may be accountable for the occurrence of tumors in these DKO mice, although the number of tumors are decreased markedly. Similarly, deletion of either HMGB1 or Yap also has no or mild effects on Nrf2 activation in L-Atg7 KO mice, which may explain why deletion of HMGB1 and Yap cannot completely eliminate the tumorigenesis in L-Atg7 KO mice. Why would deletion of p62-Nrf2 or Yap improve the hepatomegaly and liver injury in autophagy-deficient livers but not HMGB1? It is likely that both Nrf2 (transcription factor) and Yap (transcription coactivator) increase a large set of gene expression and in turn increase the robust newly synthesized proteins. Proteins generally can be timely and efficiently cleared by autophagy, but instead these proteins are accumulated in autophagy-deficient livers resulting in the increased hepatic proteotoxicity and hepatocyte cell death. Therefore, blocking protein synthesis such as the inhibition of mTOR may be beneficial for the improvement of the pathogenesis in autophagy-impaired livers. Since impaired hepatic autophagy has been associated with many chronic liver diseases including alcoholic and nonalcoholic fatty liver disease and viral hepatitis, targeting p62-Nrf2, HMGB1 and Yap may be a promising avenue for preventing the pathogenesis of these chronic liver diseases. The molecular events and signaling pathways that may contribute to the pathogenesis of autophagy-deficient or autophagy-impaired livers are summarized in Figure 1.
Figure 1.
A proposed model of the molecular events in autophagy deficiency-induced liver pathogenesis. Liver-specific Atg5 (L-Atg5) or L-Atg7 knockout (KO) mice have increased accumulation of hepatic p62 resulting in persistent activation of nuclear factor (erythroid-derived 2)-like 2 (Nrf2). Nrf2 activation activates caspase 1/11 and inflammasome that promotes the release of hepatic high-mobility group box 1 (HMGB1). Nrf2 activation also promotes yes-associated protein (Yap) accumulation and activation in autophagy-deficient livers. However, the exact mechanisms of how Nrf2 activates caspase 1/11 and Yap are unclear. Release of HMGB1 promotes ductular reaction (DR) and liver tumorigenesis without affecting hepatomegaly, liver injury, inflammation, and fibrosis. Nrf2 and Yap upregulate the expression of a large set of their target genes and subsequent protein translation resulting in the accumulation of hepatic proteins and proteotoxicity in autophagy-deficient livers. Accumulation of hepatic proteins and increased proteotoxicity then promote hepatomegaly, inflammation, DR, fibrosis, and tumorigenesis. Inhibition of mTOR may suppress overall protein translation and protein input, which may in turn inhibit proteotoxicity and subsequent liver injury in autophagy-deficient livers.
ACKNOWLEDGMENT
Grant support: R01 AA020518, R01 DK102142, U01 AA024733, and P20GM103549 and P30GM118247.
REFERENCES
- 1. Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2. Xu J. Trends in liver cancer mortality among adults aged 25 and over in the United States, 2000–2016. NCHS Data Brief 2018;1–8. [PubMed] [Google Scholar]
- 3. Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology 2012;56:769–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–56. [DOI] [PubMed] [Google Scholar]
- 5. Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–8. [PMC free article] [PubMed] [Google Scholar]
- 6. Czaja MJ, Ding WX, Donohue TM Jr., et al. Functions of autophagy in normal and diseased liver. Autophagy 2013;9:1131–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology 2008;47:1773–85. [DOI] [PubMed] [Google Scholar]
- 8. Ueno T, Komatsu M. Autophagy in the liver: Functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–84. [DOI] [PubMed] [Google Scholar]
- 9. Ding WX, Manley S, Ni HM. The emerging role of autophagy in alcoholic liver disease. Exp Biol Med. (Maywood) 2011;236:546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ni HM, Woolbright BL, Williams J, et al. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khambu B, Huda N, Chen XY, et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J Clin Invest. 2018;128:2419–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee YA, Noon LA, Akat KM, et al. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat Commun. 2018;9(1):4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011;16:123–40. [DOI] [PubMed] [Google Scholar]
- 16. Lau A, Villeneuve NF, Sun Z, et al. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain A, Lamark T, Sjottem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lau A, Wang XJ, Zhao F, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–23. [DOI] [PubMed] [Google Scholar]
- 20. Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007;131:1149–63. [DOI] [PubMed] [Google Scholar]
- 21. Ni HM, Boggess N, McGill MR, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sims GP, Rowe DC, Rietdijk ST, et al. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–88. [DOI] [PubMed] [Google Scholar]
- 23. Martinotti S, Patrone M, Ranzato E. Emerging roles for HMGB1 protein in immunity, inflammation, and cancer. Immunotargets Ther. 2015;4:101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arriazu E, Ge X, Leung TM, et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut 2017;66:1123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999;285:248–51. [DOI] [PubMed] [Google Scholar]
- 26. Roskams TA, Theise ND, Balabaud C, et al. Nomenclature of the finer branches of the biliary tree: Canals, ductules, and ductular reactions in human livers. Hepatology 2004;39:1739–45. [DOI] [PubMed] [Google Scholar]
- 27. Sato K, Marzioni M, Meng F, et al. Ductular reaction in liver diseases: Pathological mechanisms and translational significances. Hepatology 2019;69(1):420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pusterla T, Nemeth J, Stein I, et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology 2013;58:363–73. [DOI] [PubMed] [Google Scholar]
- 29. Hernandez C, Huebener P, Pradere JP, et al. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J Clin Invest. 2018;128:2436–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamkanfi M, Sarkar A, Vande Walle L, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185:4385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015;526:660–5. [DOI] [PubMed] [Google Scholar]
- 32. Kayagaki N, Stowe IB, Lee BL, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015;526:666–71. [DOI] [PubMed] [Google Scholar]
- 33. Wu S, Huang J, Dong J, et al. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003;114:445–56. [DOI] [PubMed] [Google Scholar]
- 34. Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010;19:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol. 2015;63:1491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao B, Ye X, Yu J, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao B, Kim J, Ye X, et al. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–98. [DOI] [PubMed] [Google Scholar]
- 38. Lu L, Li Y, Kim SM, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA 2010;107:1437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benhamouche S, Curto M, Saotome I, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yimlamai D, Christodoulou C, Galli GG, et al. Hippo pathway activity influences liver cell fate. Cell 2014;157:1324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]