Abstract
Background
46,XX male syndrome is a rare disorder that usually causes infertility. This study was established to identify the genetic causes of this condition in a series of 46,XX males through the combined application of cytogenetic and molecular genetic techniques.
Case presentation
We identified eight azoospermic 46,XX males who underwent infertility-related consultations at our center. They all presented normal male phenotypes. In seven of the eight 46,XX males (87.5%), translocation of the SRY gene to the terminal short arm of the X chromosome was clearly involved in their condition, which illustrated that this translocation is the main mechanism of 46,XX sex reversal, in line with previous reports. However, one patient presented a homozygous DAX1 mutation (c.498G > A, p.R166R), which was not previously reported in SRY-negative XX males.
Conclusions
We proposed that this synonymous DAX1 mutation in case 8 might not be associated with the activation of the male sex-determining pathway, and the male phenotype in this case might be regulated by some unidentified genetic or environmental factors. Hence, the detection of genetic variations associated with sex reversal in critical sex-determining genes should be recommended for SRY-negative XX males. Only after comprehensive cytogenetic and molecular genetic analyses can genetic counseling be offered to 46,XX males.
Keywords: 46,XX male; Sex reversal; SRY; DAX1 mutation
Background
46,XX male syndrome, also called de la Chapelle syndrome, is a genetic disorder encountered infrequently in a clinical context [1]. This syndrome is found in approximately 1 in 20,000–25,000 males, who exhibit a male phenotype but a 46,XX karyotype [2]. These patients have three main phenotypic manifestations: (1) classic XX males with infertility presenting normal male internal and external genitalia; (2) XX males with ambiguous genitalia presenting apparently external genital ambiguities at birth, such as hypospadias, micropenis, or hyperclitoridy; and (3) XX true hermaphrodites presenting internal or external genital ambiguities at birth [3–5].
Molecular findings on the presence of the sex-determining region Y (SRY) gene can be used to divide XX males into SRY-positive and -negative groups [6]. About 90% of XX males have the SRY gene, which plays a critical role in encoding the testis-determining factor (TDF) [7]. These SRY-positive XX individuals present normal genitalia and a male phenotype at birth [4]. However, in the SRY-negative 46,XX males, external genital ambiguities of different degrees are presented [5].
According to a literature review, several genes such as SRY, SOX9, DAX1, and WNT4 are associated with sex reversal. Herein, we describe eight azoospermic 46,XX males presenting a normal male phenotype and masculinization. We also perform a literature review to investigate the correlation between DAX1 mutation and XX males, aiming to explain the genetic cause of SRY-negative XX males.
Case presentation
Participants and clinical data
From 2015 to 2017, eight Chinese males underwent consultations for infertility at the Center for Reproductive Medicine and Center for Prenatal Diagnosis of the First Hospital of Jilin University because of no pregnancy resulting from regular unprotected coitus. The results of physical and routine clinical examinations were listed in Table 1. All patients were finally diagnosed with azoospermia based on routine semen examination [8]. The Ethics Committee of the First Hospital of Jilin University approved our study protocol (No. 2016–422) and all patients provided written informed consent to participate in this study.
Table 1.
Summary of the physical and clinical examinations of our patients
| Case | Age | Height (cm) | Weight (kg) | Testicle volume (mL) | Serum levels | Male signs | Diagnosis | ART follow-up | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | Right | FSH | LH | E2 | PRL | T | |||||||
| 1 | 23 | 165 | 49 | 2 | 2 | N.A. | N.A. | N.A. | N.A. | N.A. | Sparse beard | Azoospermia | AID |
| 2 | 36 | 171 | 57 | 2 | 2 | 45 | 29.6 | 19.27 | 196 | 3.9 | Normal | Azoospermia | AID |
| 3 | 28 | 166 | 60 | 2 | 2 | 18.8 | 14.1 | 33.36 | 269 | 4.7 | Normal | Azoospermia | AID |
| 4 | 32 | 169 | 50 | 1 | 1 | 45.4 | 28.5 | 31.8 | 219 | 5.4 | Sparse beard | Azoospermia | N.A. |
| 5 | 26 | 175 | 80 | 3 | 5 | 21.2 | 14.3 | 22.63 | 147 | 6.5 | Normal | Azoospermia | N.A. |
| 6 | 30 | 170 | 60 | 4 | 4 | 62.83 | 27.12 | 17.9 | 316.6 | 9.19 | Normal | Azoospermia | N.A. |
| 7 | 23 | 165 | 56 | 3 | 3 | 40.05 | 22.11 | 13.6 | 316.6 | 2.22 | Normal | Azoospermia | N.A. |
| 8 | 27 | 168 | 51 | 10 | 10 | 6.6 | 9.6 | 33.14 | 485 | 3.6 | Normal | Azoospermia | N.A. |
The reference values were obtained from electrochemiluminescence immunoassays (ECLIA) using Roche Elecsys1010 (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. N.A. Not available, ART Assisted reproductive technology, AID Artificial insemination by donor sperm
FSH (follicle-stimulating hormone): 1.5–12.4 mIU/ml; LH(luteinizing hormone): 1.7–8.5 mIU/ml; E2(estradiol): 28–248 pg/ml; PRL(prolactin): 86–258 uIU/ml; T(testosterone): 9.9–27.8 nmol/l
Methods
Karyotype analysis
Conventional G-banding techniques were applied on the cultured peripheral blood cells for chromosomal karyotyping. We described all of the chromosomal karyotypes according to the ISCN 2016 nomenclature [9].
AZF microdeletion analysis
Microdeletions in the AZF region were detected using polymerase chain reaction (PCR), as previously described in accordance with the recommendations of the European Academy of Andrology and the European Molecular Genetics Quality Network. Specific sequence-tagged sites (STSs) were mapped in the AZF region, including SY84 and SY86 for AZFa; SY27, SY134, and SY143 for AZFb; SY157, SY254, and SY255 for AZFc; and SY152 for AZFd [10].
Fluorescence in situ hybridization analysis (FISH)
FISH specific for the Y chromosome was performed on metaphase slides for the patients to further confirm the existence of SRY through the standard operating protocol (Cytocell Technologies, Cambridge, UK). The detecting probes were as follows: red-labeled sex-determining region Y (SRY) probe with two nonoverlapping probes, green-labeled probe for a heterochromatic region (DYZ1) in Yq12, and blue-labeled probe for the X centromere (DXZ1).
Sanger sequencing
Some genes had been shown to be critically involved in sex differentiation, such as SOX9, DAX1, and WNT4 [11, 12]. Sequencing was performed to detect mutations in these genes on an ABI 3730xl DNA analyzer (Applied Biosystems) by BGI (Beijing, China) for the SRY-negative patients [13].
Results
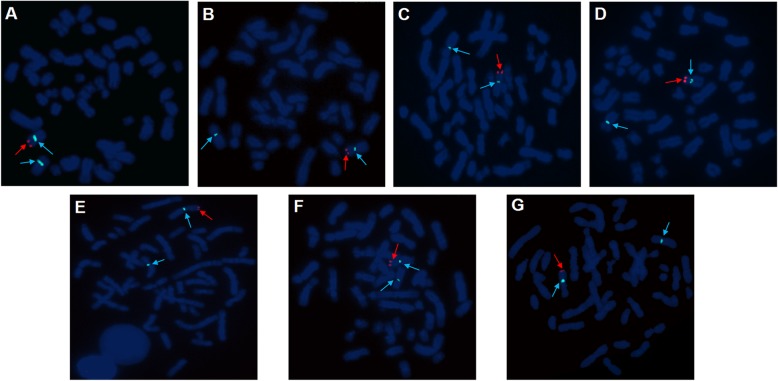
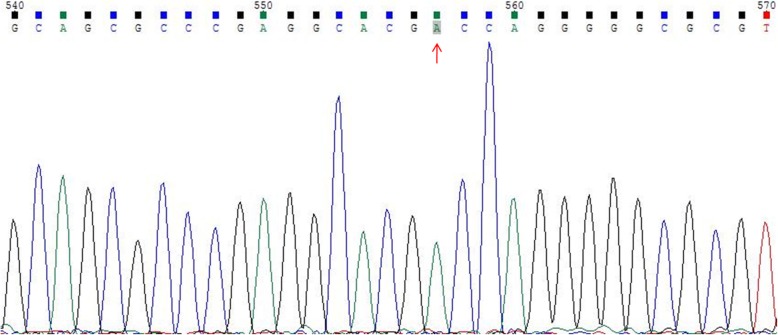
The results of cytogenetic G-banding and AZF microdeletion analyses were listed in Table 2. All of the 46,XX males reported here exhibited AZFa+b + c microdeletion. FISH confirmed the presence of a translocated SRY region located on the distal tip of the short arm of the X chromosome in seven patients (cases 1–7, Fig. 1). For case 8, the PCR analysis demonstrated the absence of the SRY gene; as such, Sanger sequencing was performed on three key genes (SOX9, WNT4, and DAX1) associated with sex reversal. No mutations were discovered in the coding regions of SOX9 and WNT4. However, a homozygous variant in exon 1 of DAX1 (c.498G > A, p.R166R) was detected (Fig. 2), which was not previously reported in SRY-negative 46,XX males. Only cases 1 to 3 chose artificial insemination with donor sperm, according to the assisted reproductive technology follow-up outcomes.
Table 2.
Sequence-tagged site deletions and chromosomal analysis
| Case | AZFa | AZFb | AZFc | Karyotype analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SY86 | SY84 | SY127 | SY134 | SY143 | SY152 | SY254 | SY255 | SY157 | ||
| 1 | – | – | – | – | – | – | – | – | – | 46,XX |
| 2 | – | – | – | – | – | – | – | – | – | 46,XX |
| 3 | – | – | – | – | – | – | – | – | – | 46,XX |
| 4 | – | – | – | – | – | – | – | – | – | 46,XX |
| 5 | – | – | – | – | – | – | – | – | – | 46,XX |
| 6 | – | – | – | – | – | N.A. | – | – | N.A. | 46,XX |
| 7 | – | – | – | – | – | – | – | – | N.A. | 46,XX |
| 8 | – | – | – | – | – | – | – | – | – | 46,XX |
AZF Azoospermic factor, STS Sequence-tagged site; −, deletion of specific STS, N.A. Not available
Fig. 1.
FISH demonstrated that the SRY region was located on the distal tip of the short arm of the chromosome X. a:case 1. b:case 2. c: case 3. d: case 4. e: case 5. f: case 6. g: case 7. Red arrows indicated SRY signal (red), and blue arrows indicated X centromere (blue)
Fig. 2.
Sanger sequencing analysis of DAX1 gene for case 8: a synonymous mutation of DAX1 (c.498G > A, p.R166R)
Discussion and conclusions
In this paper, we reported eight azoospermic cases of 46,XX males with normal male genitalia and no apparent abnormalities. All subjects presented 46,XX karyotypes and AZFa+b + c microdeletion. Among them, seven cases were SRY-positive (87.5%), with the SRY gene being translocated to the short end of the X chromosome, while the eighth case was SRY-negative (12.5%) with a synonymous mutation of DAX1 (c.498G > A, p.R166R).
The first case of 46,XX male syndrome was reported in 1964 [1]. The clinical manifestations are mainly characterized by a normal phenotype in newborns, but delayed puberty, gynecomastia, or infertility in adolescents. In addition, hypospadias, cryptorchidism, and severely ambiguous genitalia can also be observed [14]. The majority of XX males were found to be SRY-positive, presenting sterility with normal male genitalia and SRY translocation to the X chromosome or autosomes. However, reports on SRY-negative XX males have been limited, with these cases usually presenting infertility with immature/ambiguous to normal genitalia, incomplete testicular development or ovotestis, and varying degrees of masculinization [15].
The mechanism of 46,XX male syndrome in SRY-positive cases could be summarized as involving cross-over errors in the pseudoautosomal regions of the sex chromosome during paternal meiosis [16]. However, the mechanism in SRY-negative 46,XX males has remained unclear. Several possibilities were proposed to explain these cases: mosaicism for SRY in hidden gonads, the inhibition of male pathways resulting from mutations of autosomal or X-linked genes, and mutations of other sex-determining genes downstream of SRY [14, 15].
The sex of individuals is known to be determined by the SRY gene in most mammals, but the existence of SRY-negative males demonstrates the involvement of other genes in determining maleness in the absence of SRY. Mutations of some critical genes, such as SOX9, DAX1, and WNT4, have been proven to be associated with sex reversal [11, 12]. DAX1 (dosage-sensitive sex reversal adrenal hypoplasia congenital critical region of the X chromosome gene 1), also called NR0B1, is located on chromosome Xp21.3-p21.2 (ΟMIM#300473). It contains two exons and one intron, which encode an orphan nuclear receptor. DAX1 is widely expressed in the adrenals, hypothalamus, pituitary, and testis, playing critical roles in testicular and ovarian development. Mutations of DAX-1 are usually associated with primary adrenal insufficiency or congenital adrenal hypoplasia (CAH) and hypogonadotropic hypogonadism (MIM #300200) [17, 18]. DAX1 was initially recognized as a dosage-sensitive ovarian-determining gene. An increased number of copies of DAX1 could lead to high expression of its encoded proteins, which would result in sex reversal [11]. Further research revealed that DAX1 was actually necessary for testis differentiation and spermatogenesis [19, 20]. Zenteno et al. [21] assumed that SRY-negative XX males with normal genitalia were homozygous for deletions or loss-of-function mutations in dosage-sensitive sex reversal. In addition, Domenice et al. [12] proposed that the loss of function of the DAX1 gene might prevent its repressive effect on masculinizing genes and thus determine testicular development in XX individuals, which probably explained the presence of 46,XX sex reversal. In addition, Dangle et al. [22] reported a 46,XX SRY-negative case with a heterozygous deletion encompassing DAX1.
In case 8 presented here, Sanger sequencing of the coding regions of the DAX1 gene showed a synonymous mutation of this gene (c.498G > A, p.R166R), which was also described in other reports. For example, Mou et al. [23] reported a series of patients with secretory azoospermia and fertility who presented synonymous mutation (c.498 G > A) in DAX1. In addition, Achermann et al. [24] described the same DAX1 mutation in patients with hypogonadotropic hypogonadism or pubertal delay. Moreover, patients with CAH could also present DAX1 mutation (c.498 G > A), in those with disease onset in either infancy or adulthood [25, 26]. Xu et al. [27] reported a 3-year-old boy who was diagnosed with X-linked CAH, with three novel mutations detected in DAX1: a missense mutation (c.376G > A, p.Val126Met), a synonymous mutation (c.498G > A, p.Arg166Arg), and a nonsense mutation (c.1225C > T, p.Gln409X). Currently, the mechanisms triggering testis development in SRY-negative 46,XX males remain unknown. Overexpression of the DAX1 gene could cause female-to-male sex reversal [28], but this was not analyzed in case 8, as we failed to investigate probably hidden gonadal mosaicism for SRY or mutations in autosomes, or other functional mutations of unknown sex-determining genes (e.g., SF1, RSPO1, SOX3, SOX10, ROCK1, and DMRT) [14]. Considering the pathogenicity of the polymorphism as recorded in the ClinVar database, the potential risk of CAH in case 8 should be considered, besides the sex reversal. Despite the DAX1 mutation detected in case 8 not previously being reported in SRY-negative 46,XX males, the potential association between DAX1 mutation and SRY-negative 46,XX males still requires further investigation. We also speculated that other unidentified genetic or environmental factors might play critical roles in regulating sex determination and gonad sex differentiation.
The SOX9 and WNT4 genes were also sequenced in case 8. The SRY-box 9 (SOX9) gene, located in 22q13, is a widely expressed transcription factor involved in male sex determination. Normal expression of SOX9 was found to be associated with testicular differentiation. However, its overexpression or duplication might lead to 46,XX male sex reversal and testicular differentiation in the absence of SRY [29, 30]. With regard to WNT4, located in 1p36.12, it has also been shown to play a critical role in the development of the reproductive system as a candidate ovary-determining gene or antitestis gene [31]. Moreover, it has been reported that loss-of-function mutation in WNT4 could result in partial XX male sex reversal [32]. Given their lack of clear causative mutations in this study, SOX9 and WNT4 might not be critical factors in SRY-negative XX males.
In other words, irrespective of the status as SRY-positive or -negative, 46,XX males would always present infertility due to the absence of the AZFa, AZFb, and AZFc regions located on chromosome Yq11, which are involved in regulating normal spermatogenesis [14].
In the present study, eight cases of 46,XX male syndrome were identified based on cytogenetic and molecular genetic analyses. One SRY-negative XX male carried a homozygous p.R166R synonymous mutation in DAX1, while the other seven SRY-positive 46,XX individuals had SRY translocated to the terminal of the X chromosome. Our findings enrich the understanding of the genotype–phenotype correlation in 46,XX males, especially in patients with SRY-negative female-to-male sex reversal. The combined application of chromosomal analysis, AZF microdeletion evaluation, SRY detection, and sequencing of key sex-determining genes should be recommended for these patients.
Acknowledgements
We express our sincere gratitude to all the staff of the Genetics Laboratory and Andrology Laboratory for their excellent work. We thank Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Abbreviations
- AZF
Azoospermia factor
- FISH
Fluorescent in situ hybridization
- ISCN 2016
International System for Human Cytogenetic Nomenclature 2016
- SRY
Sex-determining region Y
- STS
Specific sequence-tagged sites
Authors’ contributions
FY wrote the first draft of the manuscript. HZ and QX collected the data of all the patients. YJ and LL participated in analysis and interpretation of data. RL and RW reviewed the manuscript and were involved in its critical revision before submission. All authors read and approved the final manuscript.
Funding
This work was supported by The National Key Research and Development Program of China (2016YFC1000601).
Availability of data and materials
The data and material used or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This report was approved by the Ethics Committee of the First Hospital of Jilin University (No.2016–422). The patients provided written informed consent for participating in this study.
Consent for publication
Written informed consents were obtained from the patients for publication of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Delachapelle A, Hortling H, Niemi M, et al. XX sex chromosomes in a human male. First case. Acta Med Scand. 1964;175:25–28. doi: 10.1111/j.0954-6820.1964.tb04630.x. [DOI] [PubMed] [Google Scholar]
- 2.de la Chapelle A. The etiology of maleness in XX men. Hum Genet. 1981;58(1):105–116. doi: 10.1007/BF00284157. [DOI] [PubMed] [Google Scholar]
- 3.Abbas NE, Toublanc JE, Boucekkine C, et al. A possible common origin of “Y-negative” human XX males and XX true hermaphrodites. Hum Genet. 1990;84(4):356–360. doi: 10.1007/BF00196234. [DOI] [PubMed] [Google Scholar]
- 4.McElreavey K, Rappaport R, Vilain E, et al. A minority of 46, XX true hermaphrodites are positive for the Y-DNA sequence including SRY. HumGenet. 1992;90(1–2):121–125. doi: 10.1007/BF00210754. [DOI] [PubMed] [Google Scholar]
- 5.Boucekkine C, Toublanc JE, Abbas N, et al. Clinical and anatomical spectrum in XX sex reversed patients. Relationship to the presence of Y specific DNA-sequences. Clin Endocrinol. 1994;40(6):733–742. doi: 10.1111/j.1365-2265.1994.tb02506.x. [DOI] [PubMed] [Google Scholar]
- 6.Abusheikha N, Lass A, Brinsden P. XX males without SRY gene and with infertility. Hum Reprod. 2001;16(4):717–718. doi: 10.1093/humrep/16.4.717. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson-Smith MA, Cooke A, Affara NA, et al. Genotype-phenotype correlations in XX males and their bearing on current theories of sex determination. Hum Genet. 1990;84(2):198–202. doi: 10.1007/BF00208942. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . WHO laboratory manual for the examination and processing of human semen. 5. Geneva: World Health Organization; 2010. [Google Scholar]
- 9.McGowan-Jordan J, Simons A, Schmid M, editors. An international system for human cytogenomic nomenclature. S. Karger, Basel. 2016. [Google Scholar]
- 10.Zhang YS, Dai RL, Wang RX, et al. Analysis of Y chromosome microdeletion in 1738 infertile men from northeastern China. Urology. 2013;82(3):584–588. doi: 10.1016/j.urology.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Ding XP, Wei X, et al. Investigation of mutations in the SRY, SOX9, and DAX1 genes in sex reversal patients from the Sichuan region of China. Genet Mol Res. 2014;13(1):1518–1526. doi: 10.4238/2014.March.12.4. [DOI] [PubMed] [Google Scholar]
- 12.Domenice S, Corrêa RV, Costa EM, et al. Mutations in the SRY, DAX1, SF1 and WNT4 genes in Brazilian sex-reversed patients. Braz J Med Biol Res. 2004;37(1):145–150. doi: 10.1590/S0100-879X2004000100020. [DOI] [PubMed] [Google Scholar]
- 13.Baetens D, Stoop H, Peelman F, et al. NR5A1 is a novel disease gene for 46,XX testicular and ovotesticular disorders of sex development. Genet Med. 2017;19(4):367–376. doi: 10.1038/gim.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li TF, Wu QY, Zhang C, Li WW, et al. 46,XX testicular disorder of sexual development with SRY-negative caused by some unidentified mechanisms: a case report and review of the literature. BMC Urol. 2014;14:104. doi: 10.1186/1471-2490-14-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajender S, Rajani V, Gupta NJ, et al. SRY-negative 46,XX male with normal genitals, complete masculinization and infertility. Mol Hum Reprod. 2006;12(5):341–346. doi: 10.1093/molehr/gal030. [DOI] [PubMed] [Google Scholar]
- 16.Alves C, Braid Z, Coeli FB, et al. 46,XX male-testicular disorder of sexual differentiation (DSD): hormonal, molecular and cytogenetic studies. Arq Bras Endocrinol Metabol. 2010;54(8):685–689. doi: 10.1590/S0004-27302010000800004. [DOI] [PubMed] [Google Scholar]
- 17.Seminara SB, Achermann JC, Genel M, et al. X-linked adrenal hypoplasia congenita: a mutation in DAX1 expands the phenotypic spectrum in males and females. J Clin Endocrinol Metab. 1999;84(12):4501–4509. doi: 10.1210/jcem.84.12.6172. [DOI] [PubMed] [Google Scholar]
- 18.El-Khairi R, Martinez-Aguayo A, Ferraz-de-Souza B, et al. Role of DAX-1 (NR0B1) and steroidogenic factor-1 (NR5A1) in human adrenal function. Endocr Dev. 2011;20:38–46. doi: 10.1159/000321213. [DOI] [PubMed] [Google Scholar]
- 19.Meeks JJ, Weiss J, Jameson JL. Dax1 is required for testis determination. Nat Genet. 2003;34(1):32–33. doi: 10.1038/ng1141. [DOI] [PubMed] [Google Scholar]
- 20.Yu RN, Ito M, Saunders TL, et al. Role of Ahch in gonadal development and gametogenesis. Nat Genet. 1998;20(4):353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- 21.Zenteno JC, López M, Vera C, et al. Two SRY-negative XX male brothers without genital ambiguity. Hum Genet. 1997;100(5–6):606–610. doi: 10.1007/s004390050561. [DOI] [PubMed] [Google Scholar]
- 22.Dangle P, Touzon MS, Reyes-Múgica M, et al. Female-to-male sex reversal associated with unique Xp21.2 deletion disrupting genomic regulatory architecture of the dosage-sensitive sex reversal region. J Med Genet. 2017;54(10):705–709. doi: 10.1136/jmedgenet-2016-104128. [DOI] [PubMed] [Google Scholar]
- 23.Mou L, Xie N, Yang L, et al. A novel mutation of DAX-1 associated with secretory Azoospermia. PLoS One. 2015;10(7):e0133997. doi: 10.1371/journal.pone.0133997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achermann JC, Gu WX, Kotlar TJ, et al. Mutational analysis of DAX1 in patients with hypogonadotropic hypogonadism or pubertal delay. J Clin Endocrinol Metab. 1999;84(12):4497–4500. doi: 10.1210/jcem.84.12.6269. [DOI] [PubMed] [Google Scholar]
- 25.Reutens AT, Achermann JC, Ito M, et al. Clinical and functional effects of mutations in the DAX-1 gene in patients with adrenal hypoplasia congenita. J Clin Endocrinol Metab. 1999;84(2):504–511. doi: 10.1210/jcem.84.2.5468. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler B, George PM, Mackenzie K, et al. Three cases of congenital adrenal hypoplasia with novel mutations in the (NROB1) DAX-1 gene. Ann Clin Biochem. 2008;45(Pt 6):606–609. doi: 10.1258/acb.2008.008038. [DOI] [PubMed] [Google Scholar]
- 27.Xu XQ, Feng YY, Yuan WX, et al. Novel mutations in DAX1 of X-linked adrenal hypoplasia congenita over several generations in one family. Endocr Pract. 2013;19(4):e105–e111. doi: 10.4158/EP12368.CR. [DOI] [PubMed] [Google Scholar]
- 28.Sukumaran A, Desmangles JC, Gartner LA, et al. Duplication of dosage sensitive sex reversal area in a 46, XY patient with normal sex determining region of Y causing complete sex reversal. J Pediatr Endocrinol Metab. 2013;26(7–8):775–779. doi: 10.1515/jpem-2012-0354. [DOI] [PubMed] [Google Scholar]
- 29.Vetro A, Ciccone R, Giorda R, et al. XX males SRY negative: a confirmed cause of infertility. J Med Genet. 2011;48(10):710–712. doi: 10.1136/jmedgenet-2011-100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia XY, Zhang C, Li TF, et al. A duplication upstream of SOX9 was not positively correlated with the SRY-negative 46,XX testicular disorder of sex development: A case report and literature review. Mol Med Rep. 2015;12(4):5659–5664. doi: 10.3892/mmr.2015.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima Y, Hayashi Y, Mizuno K, et al. Up-regulation of SOX9 in human sex-determining region on the Y chromosome (SRY)-negative XX males. Clin Endocrinol. 2008;68(5):791–799. doi: 10.1111/j.1365-2265.2007.03101.x. [DOI] [PubMed] [Google Scholar]
- 32.Mandel H, Shemer R, Borochowitz ZU, et al. SERKAL syndrome: an autosomal-recessive disorder caused by a loss-of-function mutation in WNT4. Am J Hum Genet. 2008;82(1):39–47. doi: 10.1016/j.ajhg.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and material used or analysed during the current study are available from the corresponding author on reasonable request.