Abstract
Typically, N-glycosylation studies done on cultured cells require up to millions of cells followed by lengthy preparation to release, isolate, and profile N-glycans. To overcome these limitations, we report a rapid array-based workflow for profiling N-glycan signatures from cells, adapted from imaging mass spectrometry (IMS) used for on-tissue N-glycan profiling. Using this approach, N-glycan profiles from a low-density array of 8 cell chambers could be reported within four hours of completing cell culture. Approaches are demonstrated that account for background N-glycans due to serum media. Normalization procedures are shown. The method is robust and reproducible, requiring as few as 3,000 cells per replicate with 3–20% coefficient of variation to capture label-free profiles of N-glycans. Quantification by stable isotopic labeling of N-glycans in cell culture is demonstrated and adds no additional time to preparation. Utility of the method is demonstrated by measurement of N-glycan turnover rates due to induction of oxidative stress in human primary aortic endothelial cells. The developed method and ancillary tools serve as a foundational launching point for rapid profiling of N-glycans ranging from high-density arrays down to single cells in culture.
Keywords: N-glycan, N-glycosylation, single cell, imaging mass spectrometry, array, stable isotope, label-free
Introduction
N-glycosylation is a complex carbohydrate post-translational modification that modulates protein function, alters cell signaling, influences cell-cell and cell-extracellular matrix activity1–3. The process of N-glycosylation is a dynamic process involving metabolic, transcriptional and translational events, allowing for exquisite control of biological status3. More than half of the human proteome may be N-glycosylated based on the presence of the N-glycan consensus sequence N-X-S/T≠P4; the consensus site may or may not be occupied dependent on the biology4–6. The presence or absence of specific N-glycoforms results in profound effects on cellular processes of invasion, metastasis, immune modulation, and differentiation7–9.
Within the past decade, imaging mass spectrometry (IMS) has been developed as a technique to understand N-glycan regulation in tissue10, 11. These studies have demonstrated that N-glycoforms may be used for prognostic and diagnostic markers of disease changing with cell status, disease progression, and useful for prediction of survival status10–18. However, once a distinct N-glycan has been identified as a biomarker or is modulated by specific biology or drug, there are limited approaches to understanding the cellular mechanisms of a target N-glycan. A main approach to N-glycan profiling of cells utilizes liquid chromatography coupled to mass spectrometry (LC-MS)19–21. A problem is that even for a simple profiling experiment, sample processing for LC-MS can be lengthy, requiring significant hands-on time and may require large numbers of cells. Alternatively, matrix-assisted laser desorption/ionization (MALDI) mass spectrometry approaches to profiling N-glycans mix the released N-glycans with matrix in solution, then spot onto a target for MALDI MS analysis. However, this approach still requires significant time for isolation of N-glycans and results in large variation in signal due to heterogeneous matrix-analyte crystallization. In cases where the research necessitates a survey of multiple cell experiments, the resources and time required can become restrictive. Efficiently and reproducibly defining the complexity of N-glycan synthesis, regulation, and function in cultured cells remains an ongoing analytical challenge in glycomics.
Here, we report a simple array-based method for N-glycan profiling of cultured cells that is adapted from imaging mass spectrometry workflows. Cells are cultured on a low density chamber slide array with 8 wells, followed by simple steps of fixation and delipidation. Deglycosylation is done by spraying PNGase F onto the cells, followed by N-glycan profiling using a MALDI imaging mass spectrometry platform. The method is robust and reproducible, requiring as few as 3,000 human cells per replicate to capture quantitative profiles of N-glycans. Supporting approaches are demonstrated that account for serum media contributions and allow normalization based on protein binding stains. We further show that the method is useful for stable isotopic labeling of N-glycans in cell culture. Utility of the method is demonstrated by measurement of N-glycan turnover due to oxidative stress in primary human aortic endothelial cells. It is anticipated that this method serves as a foundational launching point for high density array-based N-glycan profiling of cultured cells, useful for studies in basic, translational and pharmaceutical investigations on mechanisms of N-glycan function and regulation in cultured cells.
Methods
Cell culture.
All cells were cultivated on sterile 8-well Lab-Tek ® II Chamber Slide System (Electron microscopy, Hatfield, PA) according to manufacturer’s protocols (Supplemental Data 1). Cell types were human aortic endothelial cells (HAEC) derived from a 40-year-old male, HepG2/C3A cells, a clonal derivative of HepG2 cells derived from HepG2 hepatoblastoma cells22, primary prostate adenocarcinoma (PPC1), Polynuclear Giant Cancer Cells (PGCCs) derived from PPC123, and the mouse 4T1 model stage IV human breast cancer cell line. For each cell type, cells were plated onto 8-well array, and allowed to adhere overnight at minimum prior to analysis. For 15N labeling of HAEC N-glycans, L-glutamine (Amide-15N, 98%+, Cambridge Isotope Laboratories, Inc) was substituted for 14N glutamine in complete media. For 15N labeling under oxidative stress, cells were plated into cell chambers, allowed to adhere overnight, followed by cultivating for up to one week in 15N media spiked with 50 μM H2O2 or in 15N media untreated. Media was changed daily.
Imaging Mass Spectrometry Sample Preparation of Cultured Cells
Cell media was removed from the array, cells were washed three times in cold PBS and fixed in neutral buffered formalin for 20 minutes. Cell chambers were removed and arrays incubated in Carnoy’s Solution (10% glacial acetic acid, 30% chloroform, 60% 200 proof ethanol) for three minutes without mixing or agitating the array. Arrays were removed from the solution, placed flat on the benchtop and allowed to air dry, typically taking 3 minutes to dry in a standard laboratory benchtop hood. Carnoy’s Solution wash was repeated a total of three times, air-drying in between each wash. Arrays were either stored in a desiccator, freezer, or immediately deglycosylated. Microscopy imaging was done using an EVOS imaging system (Fisher Scientific).
Deglycosylation was done by spraying each array with PNGase F Prime (Bulldog Bio) using an M3 TM-Sprayer (HTX Technologies) with 10 passes at 25 μL/min, 1200 mm/s, 45°C, 3 mm spacing between passes with 10 psi nitrogen gas and incubated for 2 hours at 37°C and ≥80% relative humidity. MALDI matrix α-cyano-4-hydroxycinnamic acid (CHCA, Sigma; 50% acetonitrile/0.1% trifluoroacetic acid) was sprayed onto the cells with an M3 TM-Sprayer (HTX Technologies) using 10 passes at 70 μL/min, 1300 mm/s, 79°C, 2.5 mm spacing between passes and 10 psi nitrogen gas. Two passes of ammonium phosphate monobasic (5 mM) were applied to limit matrix cluster formation that suppressed signal. This was done at 70 μL/min, 1300 mm/s, 60°C, 3 mm spacing between passes and 10 psi nitrogen gas.
Normalization was done post-acquisition using an approach developed in the current study towards protein quantification on a solid surface. Briefly, MALDI matrix was removed after data acquisition by incubating 1 minute in 100 % ethanol, repeating once. The arrays were washed four times with PBS, gently tapping excess liquid off between each wash. A rapid dip (<1 second) in HPLC-grade H2O was required remove salts, followed by air-drying. Arrays were placed in a microscope slide mailer filled 80% with Simply Blue Stain (Thermo Fisher Scientific) and incubated for 1 hour. Arrays were rinsed twice with PBS and stored in PBS until imaging. Brightfield slide scanning was used to capture stained images of cells using a Nanozoomer slide scanner at 20X (Hamamatsu). Quantitative densitometry was done using an Odyssey Imaging system (Li-Cor Biosciences) to scan slides at 700 nm with an 84 μm scan line resolution. Images were analyzed using Image Studio (Li-Cor Biosciences) or ImageJ software24.
Mass Spectrometry Profiling
N-glycans were profiled as image data of array chambers using a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (FT-ICR; 7 Tesla solariX™, Bruker Scientific, LLC) equipped with a MALDI source. Transients of 512 kiloword were acquired in broadband positive ion mode over m/z 500–5,000, with a calculated on-tissue mass resolution at full width half maximum of 81,000 at m/z 1400. Data for each pixel was collected with a total of 300 laser shots rastered over a 300 μm diameter area. Mass accuracy during acquisition was maintained at 10 ppm by lockmass on primary N-glycan peak m/z 1663.5814 (composition Hex5HexNAc4 + 1Na).
Data Analysis
Image data was uploaded into SCiLS 2019c Pro (Bruker Scientific, LLC). Recalibrated peaks (DataAnalysis 5.0, Bruker Scientific, LLC) were matched within ± 3 ppm to an in-house database of N-glycoforms and exported as peak areas. Extracted peak areas were visualized after natural log transformation with MultiExperiment Viewer (http://www.tm4.org/).25 Serum media profiles were subtracted from cell profiles using mMass26 or DataAnalysis 5.0 (Bruker Scientific, LLC). Serum media profiles were also segmented out by hierarchical clustering based on spatial localization to the media blank well using SCiLS 2019c Pro (Bruker Scientific, LLC). Extracted data as peak areas or intensities were further analyzed using GraphPad 8.0, comparing significance in expression levels by Mann-Whitney exact p-values corrected for multiple comparisons using the method of Benjamini, and Hochberg27.
Results
A rapid array-based workflow for N-glycan profiling from cultured cells.
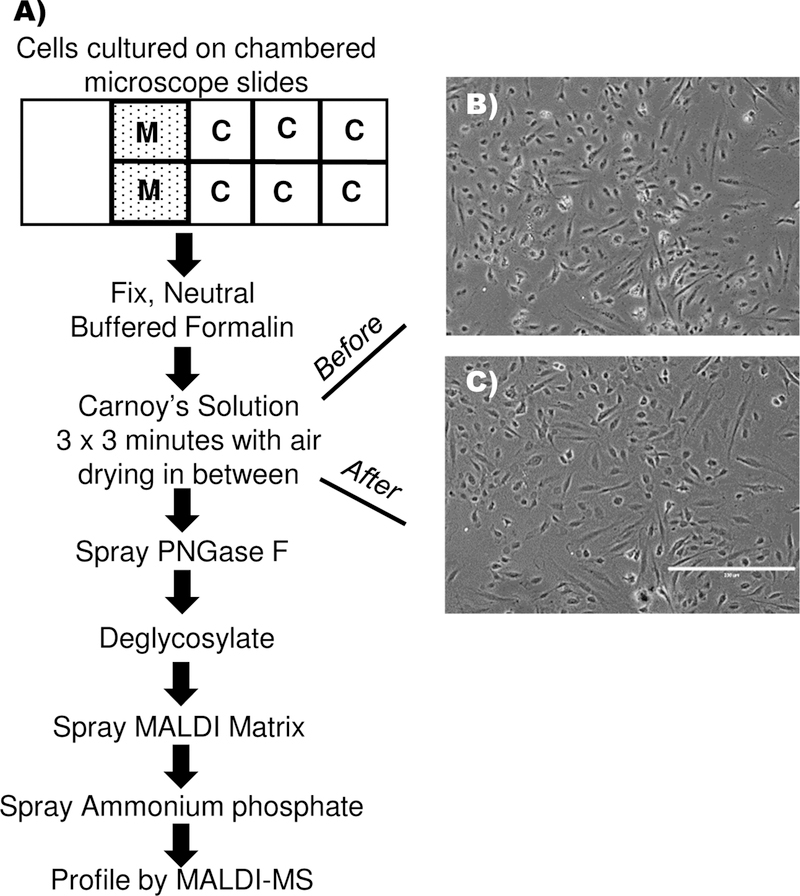
A method was developed for array-based N-glycan profiling of cultured cells, adapted from tools and protocols used for N-glycan profiling on tissue sections. Human aortic endothelial cells (HAEC) were used for initial method development, grown at 5,000 cells in monolayer to 45–60% cell confluency on chambered slides (Fig. 1A). Cells were fixed in place on the 8-well array to minimize loss of cells during subsequent preparation steps. Delipidation by static incubation of the slide in Carnoy’s solution was a critical factor to detecting N-glycan signal. Cell morphology remained intact after dilipidation (Fig. 1B, C). Enzyme and matrix application used an automated sprayer. A total of 0.0069 mg/cm2 PNGAse F sprayed onto cells produced optimal N-glycan signal, a decrease of 33% compared to the 0.0104 mg/cm2 required for tissue spraying10, 28. A 20% decrease of MALDI matrix from 2.585 mg/cm2, to 1.508 mg/cm2 MALDI matrix produced optimal N-glycan signal from cells compared to on-tissue application for N-glycans at 2.585 mg/cm2. Sensitivity of N-glycan detection was increased by applying a buffer solution of ammonium phosphate monobasic (AP) onto the matrix-coated slide to reduce matrix-cluster formation. Dipping the array into the AP solution resulted in the hydrophilic N-glycan signal observed outside of chambered wells, thus the automated sprayer was also used to apply AP onto the slides. N-glycans could be analyzed immediately after spraying AP or the prepared array could be stored for up to 7 days in a vacuum desiccator. Overall, the general workflow took approximately 5 hours for four arrays containing 8 chambers each, effectively 32 experiments per 4 arrays, including one hour of MS analysis. Not counting automated spraying time, incubation and MS analysis, hands-on time was approximately 1.5 hours. It is feasible to accomplish N-glycan analysis of 88 cell chambers (11 arrays/8 chambered wells each) in one day with overnight MS analysis.
Figure 1.
Workflow for N-glycan profiling from a low-density array of cultured cells. A) Cells are cultured on chambered microscope slides. Media blanks are necessary to account for media contributions. Neutral buffered formalin is used to fix cells to the slide. Carnoy’s solution delipidates the cells and is critical for sensitive detection of N-glycans. Air drying ensures that cells remain adhered to the slide. Both PNGase F and matrix application are applied at less volume per area than for tissue sections. Rapid acquisition of N-glycan profiles is done by MALDI MS. B) Cell morphology prior to incubation in Carnoy’s solution; C) Cell morphology remains intact after incubation Carnoy’s solution, shown prior to spraying PNGase F onto the cells.
Media N-glycan measurements are reproducible and allow determination of media contribution to cell N-glycan profiles.
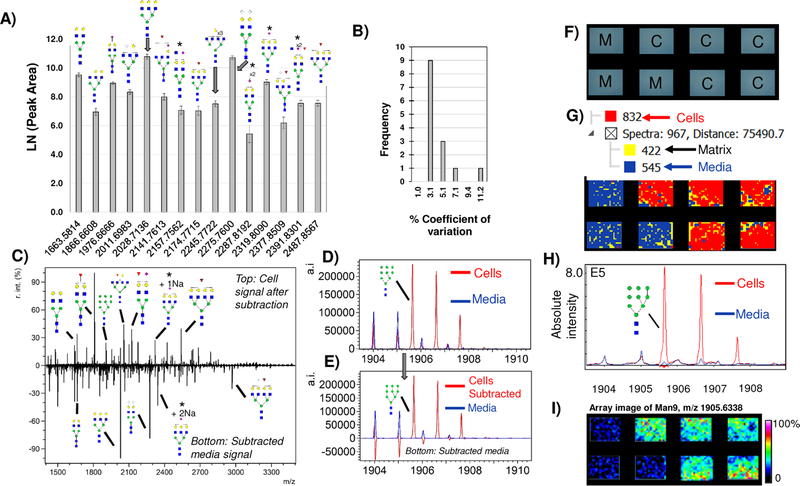
Cell culture typically uses animal sera, e.g., fetal bovine serum (FBS), which contains proteins that are also N-glycosylated. All analyses on cells cultured with serum included a well that was a serum media blank to aid in determining serum contributions. Testing of serum media blanks produced reproducible N-glycan signal, which could be used to account for N-glycan contributions from serum media (Fig. 2A). Interestingly, the contribution of serum media was not removed during washing using aqueous or organic based solutions; this may be dependent on the soda lime (CaHNaO2) surface chemistry of the array. Serum media measurements were highly reproducible (≤3% CV, Fig. 2B) regardless of different amounts of media placed in each chamber, suggesting that serum media glycoproteins were binding to the surface of the slide. Two ways to differentiate serum N-glycans from cellular N-glycans were assessed. First, since serum media measurements were highly reproducible, media contributions could be accounted for by subtracting of the media as a background signal (Fig 2C, D). A second approach for serum media contribution leveraged the image aspect of the data. Hierarchical spectral clustering was used to calculate peaks localizing only to serum media blanks (Fig 2E-H). Peaks detected only in serum media blanks formed unique spectral signatures that could be subtracted from cells upon exporting peak area or intensity. The use of high mannose Man5–9 N-glycans was helpful in assessing subtraction or data integrity of the hierarchical clustering. Overall, serum N-glycans were detected as a reproducible and constant background from sera containing media and this is what allows a serum media background to be accounted for during N-glycan profiling of cells.
Figure 2.
Media contribution of N-glycans. A) Major peaks with putative N-glycoform structures detected in HAEC media blanks, illustrating the need to account for background N-glycans due to media when measuring cellular profiles of N-glycans. B) Media measurements are reproducible with 3.1% coefficient of variation. This allows media to be accounted for as a background measurement in cells grown with serum. C) Example background subtraction with major peaks annotated by putative N-glycoform structure. Top half of the spectrum is cells only; bottom half is media only. Subtracted media spectrum contains peaks that are NeuGc only (Hex4HexNAc5NeuGc1 + 1Na and Hex5HexNAc4NeuGc2 + 1Na). D-E) Example evaluation of media subtraction. (D) shows original overlap in spectra from media (blue) and cells (Red). E) Subtraction of media from cells retains Man9 intensity values while minimizing media contributions (bottom half of spectrum). F-I) A second approach to accounting for media contributions uses the image data to determine media contribution based on spatially oriented hierarchical clustering, shown on 4T1 mouse breast tumor cells. E) Schematic of array layout, M= media blank, C= cells. F) Hierarchical clustering of image spectra. Spectra are clustered based on localization to chamber and intensity. Each hierarchical cluster region is colorized for visualization showing cells as red, media as blue and MALDI matrix as yellow. G) Man9 signal from hierarchical cluster correlating to cells only (red) overlaid with media hierarchical cluster (blue). H) Man9 viewed as a heatmap image array. * = isobaric masses from N-glycan configurations that could contain either Neu5Ac and/or NeuGc. r.int. - relative intensity; a.i. – absolute intensity.
Normalization by staining of cellular proteins.
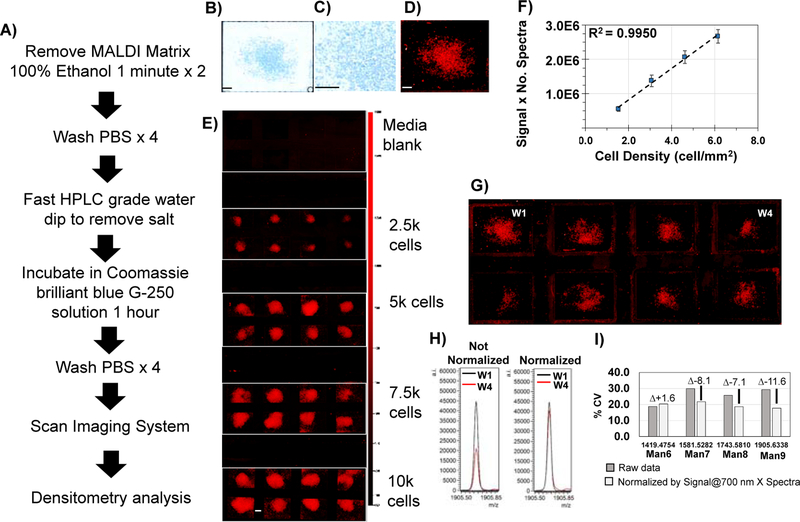
Protein regulation may occur independent of N-glycan regulation and thus N-glycomic studies normalize to protein expression. We investigated approaches for normalization (Figure 3) that incorporated protein-binding stains. Coomassie G-250 stain (abbreviated as Coomassie), a stain known for decades as quantitative with protein expression and used for proteomic studies29, 30, was determined to be a quantitative and reproducible normalization approach. Cells in the array were stained with Coomassie after MS data acquisition. Amount of protein stain could be detected using an infrared (IR) imaging system or brightfield microscopy (Fig. 3B-D). IR signal due to Coomassie protein binding was linear with cell densities of 1.5 – 6.1 cells/mm2 (R2=0.9950) (Fig. 3E, F). Normalization was assessed using IR data that showed normal variation due to cell growth conditions varying by 2,500 to 5,000 cells (Fig. 3G). A normalization factor of IR signal multiplied by number of spectral counts per well was found to reduce variability up to 12% CV based on evaluation of high mannose N-glycans (Fig. 3E). We observed that regardless of normalization factor used, some data demonstrated a very small increase in variation (see Man6, Fig. 3G). This was determined to be due to variability at lower peak areas correlating with lower cell density and could not be excluded as outlier data. To summarize, array detection of Coomassie protein binding staining applied post-acquisition provides an accessible normalization factor from the same cells that are profiled for N-glycan signal that is quantitative down to cell densities of 1.5 cell/mm2 and may be used to minimize variability.
Figure 3.
Normalization of data from array-based N-glycan profiling of cells. Normalization strategy using a protein-binding stain, Coomassie brilliant blue staining G-250. This workflow can be done after mass spectrometry profiling. A) Workflow for the method. B) Coomassie blue staining visualized by brightfield. One cell chamber is shown. C) 20X magnification demonstrates staining of individual cells. D) Infrared imaging of Coomassie blue stain at 700 nm. Same cell chamber is shown. Example scan of stain applied to differing cell amounts detected by Odyssey Imaging System, scanning at 700 nm with 84 μm line width. E) Infrared signal detects down to 2,500 cells. F) Calibration curve based on infrared signal from the Coomassie stain detected at 700 nm is linear with cell numbers. G) Infrared detection of cells from normal cell plating at 5,000 cells. Adherent cells varied from ~2,500 to 5,000. H) Use of infrared data as a normalization factor demonstrated on Man9 from well #1 and well #4. I) Overall changes in %CV due to normalization by cell number and protein signal. Normalization using Signal @ 700 nm multiplied by number of spectra collected per chamber minimizes overall variation in measurement. Signal = Intensity –(Background x Area) where Intensity background = ()/m, Ii= signal intensity of ith pixel within region, Ij = signal intensity of jth pixel within region, area = n = number pixels within region, m = number of pixels in the background area. Bar - 500 μm.
N-glycan profiling of different cultured cell types.
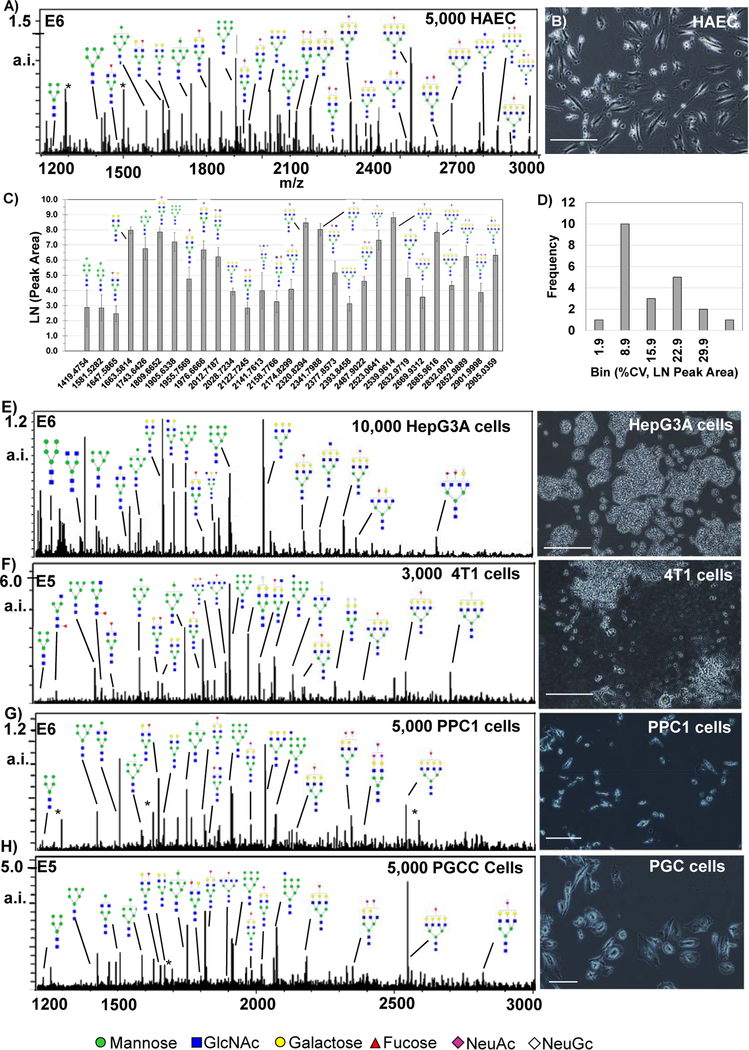
N-glycan profiling tested across cell types grown as 8-chamber arrays demonstrated unique and complex N-glycan profiles per cell type (Fig. 4). Initial N-glycan profiling of 5,000 HAEC at approximately 45% confluency illustrated abundant signal from branched N-glycans (3.08 cells/ mm2) (Fig. 4A, B). N-glycan profiles were reproducible, the majority of which were ≤10% CV (Figure 4B,D).Tests measuring N-glycan signal with increasing HAEC cell numbers demonstrated that numbers of cells beyond 10,000 in the 0.7 × 0.7 cm2 chambers resulted in apparent suppression of N-glycan signal (Supplemental Figure 1). N-glycan profiling of other cell types included human and mouse cells grown with serum-containing media and one cell line grown at endpoint in serum-free media (HepC3A) (Fig. 5E–G). Significantly, N-glycan profiles from different cell types were collected at their normal confluency required for biological studies. Plated cell counts ranged from 3,000–10,000 cells per well. A total of 70 N-glycoforms were detected in common after serum media subtraction from cell types including the mannose series Man5-Man9, bi-tri- and tetra-antennary, with variations on fucose and sialic acid residues (Supplemental Table 1). Overall, the approach allowed rapid detection and measurement of complex N-glycan profiles across species, cell types, and culture conditions without change to normal conditions required for cell culture.
Figure 4.
N-glycan profiles from cells in culture. Major N-glycan peaks are annotated by putative structure. Cells were grown at normal confluency levels prior to N-glycoform profiling experiments and intensity levels vary per cell type. A) Human aortic endothelial cells (HAEC) showing N-glycan profiles by peak intensity. B) Photomicrograph of HAEC showing cell confluency at ~65%. C) Label free quantification of HAEC by peak area, n=8. D) Reproducibility of HAEC was mostly <10% CV. E-F, major N-glycoforms from different cell lines with examples of cell morphology to the right of N-glycan profiles. E) HepC3A cells grown in animal free serum. F) mouse 4T1 animal stage IV human breast cancer. G) PPC-1 cells demonstrating signal detection from small parental cells with low cell density. H) PGCC derived from PPC1 cells by radiation stress. * = matrix peak. a.i. – absolute intensity.
Figure 5.
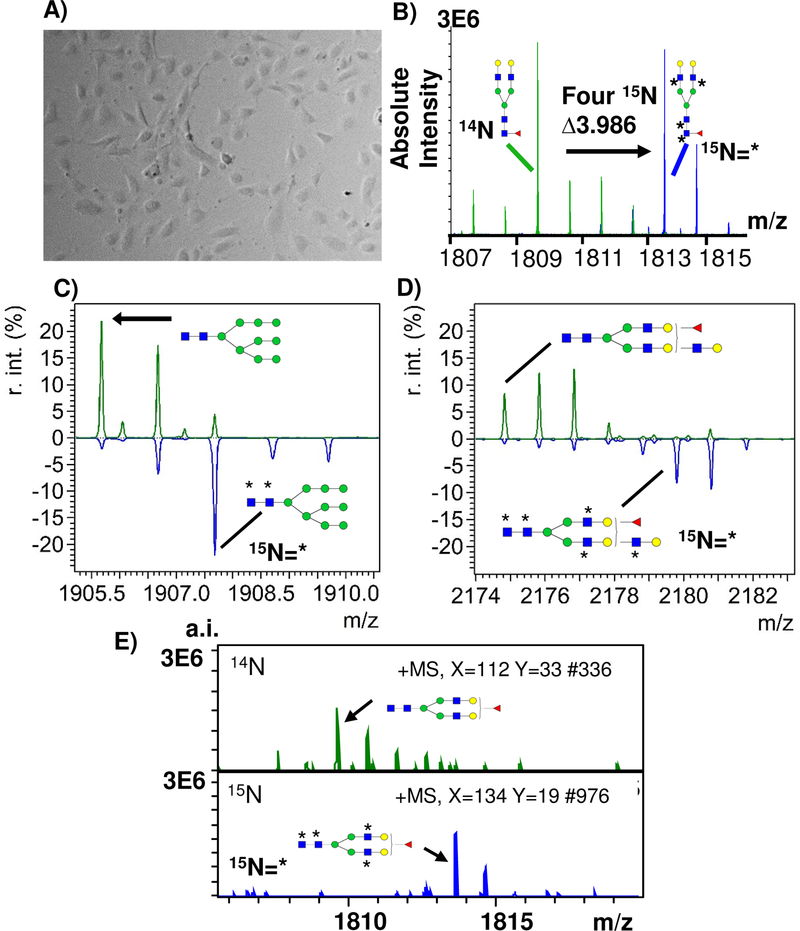
Detection of stable isotopic labeling in cell culture (SILAC) using Isotopic Detection of Aminosugars With Glutamine (IDAWG) labeling. A) Representative image of human aortic endothelial cells plated at 5,000 cells and cultured for 96 hours with 15N glutamine. 15N incorporates into GlcNac, GalNAc, and sialic acids. B) 15N incorporated into 4 GlcNAc residues of Hex5dhex1HexNac4 bi-antennary N-glycan resulting in a mass shift of 3.986 Da. C) 15N incorporated into 2 GlcNAc residues of Man9, resulting in a 1.9941 Da shift; D) 15N is incorporated into 5 GlcNAc residues of a Hex6dHexHexNAc5 tri-antennary N-glycan resulting in a 4.9852 Da shift. * indicates 15N incorporation. F) Example single spectra from HAEC 14N compared to single spectra of HAEC with 15N labeling. r.int. - relative intensity; a.i. – absolute intensity.
Stable isotopic labeling of N-glycans by Isotopic Detection of Aminosugars With Glutamine (IDAWG).
As further proof of concept for detection of cellular patterns of N-glycosylation, the stable isotopic labeling by amino acids in cell culture (SILAC) was tested by incorporating 15N into N-glycan structures using the Isotopic Detection of Aminosugars With Glutamine (IDAWG) labeling approach31, 32. In the IDAWG approach, the 15N stable isotope is localized to the amide in the essential nutrient glutamine. The use of 15N-amide glutamine in cell media thus incorporates a 15N label into all N-acetylglucosamine, N-acetylgalactosamine, and N-acetyl-neuraminic acids. IDAWG applied for one week to 10,000 HAEC in cell culture with a confluency of around 55% demonstrated >95% incorporation of the 15N label into the N-glycan m/z 1809.6393 (composition Hex5dHexHexNAc4 + 1Na) (Fig. 5A-D). The IDAWG tag was detected well above noise in single pixels, suggesting that it may be possible to measure N-glycan turnover of single cell types (Fig. 5E). Importantly, the use of the IDAWG tag did not add to the sample preparation time required for mass spectrometry. A generalized conclusion was that stable isotopic labeling could be easily detected by mass spectrometry profiling of cells down to single spectra without altering normal cell culture conditions.
Measurement of N-glycan turnover rates due to oxidative stress using IDAWG labeling.
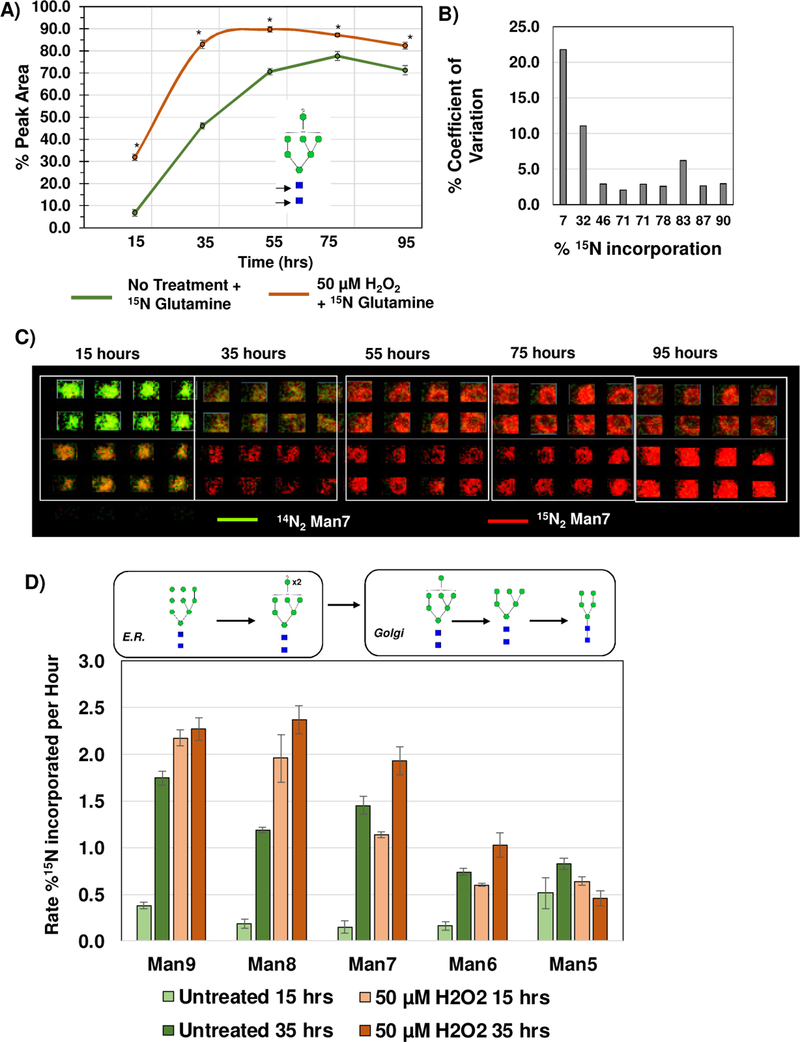
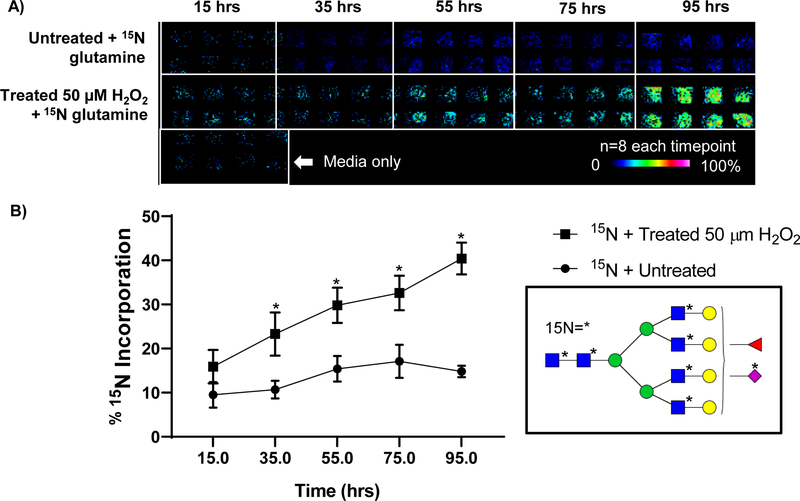
One advantage of the developed approach was that cellular incorporation of the IDAWG label was measured independently, contrasting with conventional SILAC LC-MS methods that typically mix equal ratios of labeled cells. We hypothesized that the independent collection of stable isotopic label incorporation would facilitate array-based measurement of N-glycan turnover rates since the rate of 15N incorporation to a single N-glycan corresponds to synthesis of a new 15N labeled N-glycan. As proof of principle, IDAWG was used to measure the N-glycan turnover due to oxidative stress conditions applied to HAEC in culture. Oxidative stress (OS) was induced over a time course using 50 μM hydrogen peroxide in cell culture media compared to an untreated control group that was also 15N labeled (Fig. 6). Higher levels of hydrogen peroxide (≥75 μm) produced significant levels of cell death. Initial low levels of 15N incorporation (7% 15N) showed up to 20% CV, which dropped to around 10% when measuring incorporation of 32% or more 15N label (Fig. 6A, B). This was consistent with label free measurements where low abundant peaks showed higher variability. Array-based data of the time course illustrates the dramatic differences in 15N incorporation due to oxidative stress conditions (Fig. 6C). High mannose N-glycans showed four times faster turnover rates compared to untreated cells at 15 hours of treatment and complete turnover (e.g., incorporation of ≥90% of the 15N label) by 4 days of oxidative stress. Interestingly, Man9-Man7 N-glycans were detected with significantly higher rates of 15N incorporation than untreated HAEC. At the same time, Man6 and Man5 demonstrated equalization of mannose trimming between untreated and the oxidative stress condition. We suggest that these differences in rates may reflect a higher unfolded protein response due to OS treatment, with proteins that are incorrectly folded being shunted off for degradation. Indeed, transcription levels of CHOP increased in the OS treated cells compared to the untreated cells, along with increases in TGFβ1 and COL1A1 that suggested processes of differentiation were occurring (Supplemental Figure 2). Additionally, we found that array-based N-glycan profiling could detect the emergence of new N-glycan synthesis due to OS (Fig. 7). A peak consistent with seven 15N labels on m/z 2830.999 (abbreviated m/z 2830) tetra-antennary, fucosylated and sialylated N-glycan increased 1.7-fold in expression level (p-value 3.0E-3) by 35 hours of OS treatment compared to the untreated cells (Fig. 7A, B). Over the time course of 95 hours, expression levels of m/z 2830 remained constant at a 10% incorporation of 15N7 (Fig. 7B) while OS treated HAEC demonstrated a 2.7-fold increase in the same N-glycan (p-value 2.0E-4). Additional investigation reported 14 N-glycoforms significantly increased by OS treatment at 35 hours (Supplemental Table 2). A point should be made that the preparation of 11 arrays with 8 wells each took around 6 hours and MS profiling was done overnight. In conclusion, IDAWG labeling combined with the array-based approach allows rapid measurement of N-glycan turnover rates during biological studies.
Figure 6.
Stable isotopic labeling in cell culture (SILAC) used to measure N-glycan turnover due to oxidative stress. Oxidative stress conditions were 50 μM H2O2 over a time course of 95 hours. Rate is reported as percent incorporated between 0–15 hours and 15–35 hours. A) Example, Man7 15N label (Glutamine with 15N at the amide residue) incorporation has the highest turnover rate between 15 and 30 hours of oxidative stress. B) Comparison of 15N label and % coefficient of variation (% CV), n=8 per timepoint. Low abundant levels of incorporation showed higher variability. C) Visualization of Man7 15N exchange in array format, colorized by green as the 14N red and red as the 15N. D) Evaluation of N-glycan turnover in Man9-Man5 as measured by % incorporation of 15N under oxidative stress conditions. Man9–7 levels are significantly higher in oxidative stress conditions, but normalize to levels of untreated cells by Man5. This data suggests that 50 μM H2O2 may increase initial rates of N-glycosylation of proteins that is normalized by protein degradation mechanisms. This data further demonstrates high reproducibility of individual N-glycan measurement under differing biological conditions.
Figure 7.
Oxidative stress applied to primary human aortic endothelial cells may induce significant N-glycan variation. Oxidative stress conditions are 50 μM H2O2 applied with 15N isotopic labeling using Glutamine with 15N at the amide residue. A) Example turnover rate of a tetra-antennary, fucose and NeuAc containing N-glycan at m/z 2830.9991 (abbreviated m/z 2830) with seven 15N labels. Data visualized in array format demonstrates clear emergences of the 15N labeled structure due OS-mediated turnover. The media only blank is critical for accurate interpretation of data. B) Rate of 15N incorporation for m/z 2830 increases steadily under oxidative stress conditions while remaining constant in untreated conditions. * on N-glycan structure indicates 15N label.
Discussion
Analysis of changes in N-glycans in tissues associated with disease progression is an evolving area10–18. However, tools for investigating cellular mechanisms of a particular N-glycan are limited, requiring a significant amount of processing time and large numbers of cells. In the current study, we establish foundational rapid array-based workflows for N-glycan profiling of cultured cells. The approach was created by adapting tools used by imaging mass spectrometry workflows to use in MS scanning of cultured cells grown in arrays. A total of four laboratories provided cells in amounts from 2,500 to 10,000 for testing throughout the study, demonstrating robustness in the approach. We found that most laboratories used a common source for low-density arrays of 8 cell chambers; this lower density array was used for method development. We expect that the approach will be readily adaptable to higher well density arrays with even lower cell numbers. Preparation techniques of delipidation (Fig. 1B,C) and deglycosylation (Fig. 4B) maintained cell morphology, supporting that high spatial resolution, high speed scanning imaging type instruments (≤1 μm)33, 34 could be used to increase both spatial resolution and acquisition speed of single cell N-glycan profiles. The approach also stands to be highly automatable as the major portions of the workflow, spraying of enzyme, spraying of matrix, incubation for deglycosylation, and MS profiling are already automated and do not require significant hands-on interaction.
Several challenges were addressed during development of the method. First, the single cell layers presented a low amount of biological material that required unique approaches to increase sensitivity. For N-glycan profiling of cells, less enzyme (−33%) and less MALDI matrix (−20%) were used compared to what is used for tissue imaging of N-glycans10, 11. Second, for low abundant analytes matrix-cluster formation plays a known role in limiting analyte detection due to ion suppression35, 36. Ammonium phosphate monobasic (AP) has been used to reduce matrix cluster formation and improve detection of very low molecular amounts of peptides 36–38. Here, AP was applied as a spray to limited overwhelming matrix cluster formation without delocalization of N-glycans from chambered wells. Another challenge was determination of a way to differentiate serum media N-glycans from cellular N-glycans. We found that the constant serum media background could be subtracted using current software programs. Imaging mass spectrometry software could be used to determine serum media contributions by statistically clustering acquired spectra as derived from media or from cells. Imaging data was also useful as through array visualization, it allowed rapid assessment of whether or not a peak was also present in the media blank. A conclusion was that a serum media blank must be part of the array-based N-glycan profiling in order to account for serum media contributions. During the course of investigating ways to handle serum media contributions, we also found that serum media N-glycan profiles were highly reproducible. We therefore suggest that the adapted IMS approach may be useful as a quality control check on N-glycans derived from sera used in cell culture 39. The use of other types of solid surfaces compatible with cell growing, mass spectrometry high vacuum and without leachable chemicals could be options to limit media binding to the surface and minimize serum media signal. Finally, the last challenge was that even growth of the same cell type in different chambers could result in variability that affected analytical analyses. Here, we developed an approach to normalization that accounted for that variability. Normalization utilized data derived from a common protein binding dye, Coomassie blue G-250, which has been used for decades to quantify proteins for proteomic studies. The developed ancillary test for normalization was quantitative down to a cell density of 1.5 cells/mm2 and could be performed after the array had been scanned by mass spectrometry. Normalization reduced variability over 10% for measurements on cell densities ranging from 1.5–6.1 cells/mm2. In total, the approach allowed reproducible profiling of 70 N-glycoforms from our in-house database of 154 N-glycoforms across cell types derived from primary human sources or mouse models. Although we did not attempt to define N-glycan structure, it is anticipated that tandem mass spectrometry experiments could be done directly off the array as has been done in multiple tissue imaging experiments11, 40. We expect that high-speed scanning instruments capable of tandem mass spectrometry could be used to monitor for specific N-glycoforms in the arrays41. Furthermore, the use of post-ionization separation by ion mobility is expected to increase both the number and sensitivity of the analysis by defining N-glycoform variation42, 43.
The ability to rapidly quantify N-glycan profiles from cells on arrays is a significant advancement. The developed approach allows quantitation by both label-free and stable isotopic labeling of cells in culture. This will facilitate detailed studies on specific N-glycans involved in cell signaling mechanisms. We also demonstrated that combining the new method with IDAWG allowed measurements of N-glycan turnover rates. The ability to rapidly detect turnover rate of a single N-glycan due to cell manipulation is a powerful advancement to understanding regulation of biomaker N-glycans as it allows direct assessment of changes in levels of N-glycans that influence cell signaling. Here, primary human aortic endothelial cells derived from a 40 year old male were highly sensitive to oxidative stress, altering N-glycan turnover within 15 hours of exposure. Induction of significantly different N-glycans was observed over four days of oxidative stress exposure. We anticipate that the ability to easily profile small cell populations on high density arrays will be useful for drug screening. Such an approach could be used at a patient specific level to determine if a patients N-glycan biomarker is responsive to drug treatment.
Conclusions
To summarize, we have established foundational workflows for rapid array-based acquisition of complex N-glycan profiles from cultured cells. Cells are maintained in the same wells used during culture and do not lose morphology during preparation. This strategy so far has been useful on all tested cultured cell types with minimal to no variation in mass spectrometry sample preparation. Workflows are already partially automated and we expect that the same method will be applicable to fully automated high-speed acquisition of N-glycan profiles in high-density arrays. Quantification can be done as label-free approaches and with SILAC approaches, demonstrated using IDAWG labeling. We anticipate that the combination of cell profiling and metabolic labeling will be useful to many applications investigating target N-glycans, e.g., drug screening, congenital defects of glycosylation in cells, cell differentiation, and cell-cell recognition.
Supplementary Material
Supplemental Data 1, Cell Culture Methods
Supplemental Figure 1, Increased cell counts result in ion suppression
Supplemental Figure 2, PCR measurement of markers eNOS, COL1A1, TGFβ due to oxidative stress in human aortic endothelial cells.
Supplemental Table 1, Common N-glycans profiled in cell types.
Supplemental Table 2, N-glycan turnover increases due to oxidative stress
Acknowledgements
Funding for the experiments was made possible by the U01CA242096 (NIH/NCI) to RRD, ASM and PMA. PMA and CLC supported by P20GM103542 (NIH/NIGMS), HL007260 (NHLBI), and in part by pilot research funding, Hollings Cancer Center’s Cancer Center Support Grant P30 CA138313 at the Medical University of South Carolina. ESY’s work in this publication was supported by the South Carolina Clinical & Translational Research (SCTR) Institute, with an academic home at the Medical University of South Carolina CTSA, NIH/NCATS Grant Number UL1 TR001450, in the form of a pilot award to ESY as well as funding from Medical University of South Carolina and NIH. This project was also supported by a grant from the NIH/NCI P01 CA203628. Additional support was provided by the South Carolina Centers of Economic Excellence SmartState program to RRD and ASM. Harmin Herrara is thanked for assistance with cell culture of specific cells from ASM. Jeremy Wolff and Shannon Cornett (Bruker Scientific) are thanked for the kind use of their instrument in completing the experiments.
Abbreviations:
- MALDI IMS
matrix-assisted laser desorption / ionization imaging mass spectrometry
- SILAC
stable isotopic labeling by amino acids in culture
References
- 1.Stanley P; Schachter H; Taniguchi N, N-Glycans Varki, A.; Cummings RD; Esko JD; Freeze HH; Stanley P; Bertozzi CR; Hart GW; Etzler ME, Eds. Essentials of Glycobiology. 2nd edition Cold Spring Harbor Laboratory Press: Cold Spring Harbor (NY), 2009. Chapter 8. [Google Scholar]
- 2.Varki A, Biological roles of glycans. Glycobiology 2017, 27, (1), 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moremen KW; Tiemeyer M; Nairn AV, Vertebrate protein glycosylation: diversity, synthesis and function. Nature Reviews Molecular Cell Biology 2012, 13, (7), 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall RD In The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins, Biochem Soc Sym, 1974, 17–26. [PubMed] [Google Scholar]
- 5.Apweiler R; Hermjakob H; Sharon N, On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta (BBA)-General Subjects 1999, 1473, (1), 4–8. [DOI] [PubMed] [Google Scholar]
- 6.An HJ; Froehlich JW; Lebrilla CB, Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Current Opinion in Chemical Biology 2009, 13, (4), 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinho SS; Reis CA, Glycosylation in cancer: mechanisms and clinical implications. Nature Reviews Cancer 2015, 15, (9), 540–555. [DOI] [PubMed] [Google Scholar]
- 8.Marth JD; Grewal PK, Mammalian glycosylation in immunity. Nature Reviews Immunology 2008, 8, (11), 874–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger RP; Dookwah M; Steet R; Dalton S, Glycosylation and stem cells: Regulatory roles and application of iPSCs in the study of glycosylation‐related disorders. Bioessays 2016, 38, (12), 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers TW; Jones EE; Betesh LR; Romano PR; Gao P; Copland JA; Mehta AS; Drake RR, Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Analytical Chemistry 2013, 85, (20), 9799–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR, MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 2014, 9, (9), e106255, pp1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powers TW; Holst S; Wuhrer M; Mehta AS; Drake RR, Two-dimensional N-glycan distribution mapping of hepatocellular carcinoma tissues by MALDI-imaging mass spectrometry. Biomolecules 2015, 5, (4), 2554–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake RR; Powers TW; Jones EE; Bruner E; Mehta AS; Angel PM, MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Advances in Cancer Research 2017, 134, 85–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson OJR; Briggs MT; Condina MR; Winderbaum LJ; Pelzing M; McColl SR; Everest-Dass AV; Packer NH; Hoffmann P, MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney. Analytical and Bioanalytical Chemistry 2015, 407, (8), 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott DA; Casadonte R; Cardinali B; Spruill L; Mehta AS; Carli F; Simone N; Kriegsmann M; Mastro LD; Kriegsmann J, Increases in Tumor N‐glycan Polylactosamines Associated with Advanced HER2 Positive and Triple Negative Breast Cancer Tissues. PROTEOMICS–Clinical Applications 2018, e1800014, 1800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briggs MT; Kuliwaba JS; Muratovic D; Everest‐Dass AV; Packer NH; Findlay DM; Hoffmann P, MALDI mass spectrometry imaging of N‐glycans on tibial cartilage and subchondral bone proteins in knee osteoarthritis. Proteomics 2016, 16, (11–12), 1736–1741. PMID: . [DOI] [PubMed] [Google Scholar]
- 17.de Haan N; Reiding KR; Haberger M; Reusch D; Falck D; Wuhrer M, Linkage-specific sialic acid derivatization for MALDI-TOF-MS profiling of IgG glycopeptides. Analytical Chemistry 2015, 87, (16), 8284–8291.PMID: . [DOI] [PubMed] [Google Scholar]
- 18.West CA; Wang M; Herrera H; Liang H; Black A; Angel PM; Drake RR; Mehta AS, N-linked glycan branching and fucosylation are increased directly in hepatocellular carcinoma tissue as determined through in situ glycan imaging. Journal of Proteome Research 2018, 17(10), 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aich U; Lakbub J; Liu A, State‐of‐the‐art technologies for rapid and high‐throughput sample preparation and analysis of N‐glycans from antibodies. Electrophoresis 2016, 37, (11), 1468–1488. [DOI] [PubMed] [Google Scholar]
- 20.Shajahan A; Heiss C; Ishihara M; Azadi P, Glycomic and glycoproteomic analysis of glycoproteins—a tutorial. Analytical and Bioanalytical Chemistry 2017, 409, (19), 4483–4505. PMCID: PMC5498624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domann PJ; Pardos‐Pardos AC; Fernandes DL; Spencer DIR; Radcliffe CM; Royle L; Dwek RA; Rudd PM, Separation‐based glycoprofiling approaches using fluorescent labels. Proteomics 2007, 7, (S1), 70–76. [DOI] [PubMed] [Google Scholar]
- 22.Sells MA; Chen ML; Acs G, Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proceedings of the National Academy of Sciences 1987, 84, (4), 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White-Gilbertson S; Lu P; Norris JS; Voelkel-Johnson C, Genetic and pharmacological inhibition of acid ceramidase prevents asymmetric cell division by neosis. Journal of Lipid Research 2019, 60(7), 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins TJ, ImageJ for microscopy. Biotechniques 2007, 43, (1 Suppl), 25–30. PMID: . [DOI] [PubMed] [Google Scholar]
- 25.Saeed AI; Bhagabati NK; Braisted JC; Liang W; Sharov V; Howe EA; Li J; Thiagarajan M; White JA; Quackenbush J, TM4 Microarray Software Suite. Methods in Enzymology 2006, 411, 134–193. [DOI] [PubMed] [Google Scholar]
- 26.Strohalm M; Kavan D; Novak P; Volny M; Havlicek V, mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Analytical Chemistry 2010, 82, (11), 4648–4651. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y; Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995, 57, (1), 289–300. [Google Scholar]
- 28.Drake RR; Powers TW; Norris‐Caneda K; Mehta AS; Angel PM, In Situ Imaging of N‐Glycans by MALDI Imaging Mass Spectrometry of Fresh or Formalin‐Fixed Paraffin‐Embedded Tissue. Current Protocols in Protein Science 2018, 94, (1), e68. [DOI] [PubMed] [Google Scholar]
- 29.Burgess-Cassler A; Johansen JJ; Santek DA; Ide JR; Kendrick NC, Computerized quantitative analysis of coomassie-blue-stained serum proteins separated by two-dimensional electrophoresis. Clinical Chemistry 1989, 35, (12), 2297–2304. [PubMed] [Google Scholar]
- 30.Butt RH; Coorssen JR, Coomassie blue as a near-infrared fluorescent stain: A systematic comparison with Sypro Ruby for in-gel protein detection. Molecular & Cellular Proteomics 2013, 12, (12), 3834–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlando R; Lim J-M; Atwood Iii JA; Angel PM; Fang M; Aoki K; Alvarez-Manilla G; Moremen KW; York WS; Tiemeyer M, IDAWG: Metabolic incorporation of stable isotope labels for quantitative glycomics of cultured cells. Journal of Proteome Research 2009, 8, (8), 3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang M; Lim JM; Wells L, Quantitative Glycomics of Cultured Cells Using Isotopic Detection of Aminosugars with Glutamine (IDAWG). Current Protocols in Chemical Biology 2010, 2, (2), 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanni EJ; Rubakhin SS; Sweedler JV, Mass spectrometry imaging and profiling of single cells. Journal of Proteomics 2012, 75, (16), 5036–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zavalin A; Yang J; Hayden K; Vestal M; Caprioli RM, Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Analytical and Bioanalytical Chemistry 2015, 407, (8), 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller BO; Li L, Detection of 25,000 molecules of substance P by MALDI-TOF mass spectrometry and investigations into the fundamental limits of detection in MALDI. Journal of the American Society for Mass Spectrometry 2001, 12, (9), 1055–1063. [Google Scholar]
- 36.Zhu X; Papayannopoulos IA, Improvement in the detection of low concentration protein digests on a MALDI TOF/TOF workstation by reducing α-cyano-4-hydroxycinnamic acid adduct ions. Journal of Biomolecular Techniques 2003, 14, (4), 298–307. [PMC free article] [PubMed] [Google Scholar]
- 37.Smirnov IP; Zhu X; Taylor T; Huang Y; Ross P; Papayanopoulos IA; Martin SA; Pappin DJ, Suppression of α-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry. Analytical Chemistry 2004, 76, (10), 2958–2965. [DOI] [PubMed] [Google Scholar]
- 38.Asara JM; Allison J, Enhanced detection of phosphopeptides in matrix-assisted laser desorption/ionization mass spectrometry using ammonium salts. Journal of the American Society for Mass Spectrometry 1999, 10, (1), 35–44. [DOI] [PubMed] [Google Scholar]
- 39.McGillicuddy N; Floris P; Albrecht S; Bones J, Examining the sources of variability in cell culture media used for biopharmaceutical production. Biotechnology letters 2018, 40, (1), 5–21. [DOI] [PubMed] [Google Scholar]
- 40.Angel PM; Spraggins JM; Scott Baldwin H; Caprioli R, Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry. Analytical Chemistry 2012, 84, (3), 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prentice BM; Chumbley CW; Caprioli RM, High‐speed MALDI MS/MS imaging mass spectrometry using continuous raster sampling. Journal of Mass Spectrometry 2015, 50, (4), 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fenn LS; McLean JA, Structural separations by ion mobility-MS for glycomics and glycoproteomics In Mass Spectrometry of Glycoproteins, Springer: 2013; pp 171–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenn LS; McLean JA, Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Molecular BioSystems 2009, 5, (11), 1298–1302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data 1, Cell Culture Methods
Supplemental Figure 1, Increased cell counts result in ion suppression
Supplemental Figure 2, PCR measurement of markers eNOS, COL1A1, TGFβ due to oxidative stress in human aortic endothelial cells.
Supplemental Table 1, Common N-glycans profiled in cell types.
Supplemental Table 2, N-glycan turnover increases due to oxidative stress