Abstract
Multiplex assays of variant effect (MAVEs), such as deep mutational scans and massively parallel reporter assays, test thousands of sequence variants in a single experiment. Despite the importance of MAVE data for basic and clinical research, there is no standard resource for their discovery and distribution. Here, we present MaveDB (https://www.mavedb.org), a public repository for large-scale measurements of sequence variant impact, designed for interoperability with applications to interpret these datasets. We also describe the first such application, MaveVis, which retrieves, visualizes, and contextualizes variant effect maps. Together, the database and applications will empower the community to mine these powerful datasets.
Keywords: Deep mutational scanning, Massively parallel reporter assays, Large-scale mutagenesis, MAVE, Multiplexed assay of variant effect, Genome interpretation, Personalized medicine
Background
Experimentally interrogating the effects of genetic variation has helped reveal the mechanisms by which genes function and facilitate an understanding of the clinical consequences of human genetic variation. Multiplex assays of variant effect (MAVEs) leverage high-throughput DNA sequencing to greatly increase the scale at which variants can be experimentally investigated [1–3]. A MAVE yields a set of scores that describe the functional effect of thousands to tens of thousands of variants of a coding sequence, promoter, enhancer, or another genetic element relative to a reference sequence. MAVEs are being adopted rapidly for both basic research and clinical applications [4]. As a consequence, the total number of variants with functional data generated by MAVEs was predicted to surpass 200,000 by the end of 2018 [3], meaning there are now more MAVE-annotated variants in the literature than classified missense variants available in ClinVar [5].
These large-scale variant effect maps are yielding insights into protein function, structure, and evolution [6–10]; exploring gene regulation and promoter function [11–13]; improving computational variant effect prediction [14, 15]; and guiding variant interpretation in the clinic [16–21]. However, the impact of variant effect maps has been limited by shortcomings in data availability, dissemination, and discoverability. Nearly half of the publications describing large-scale variant effect mapping do not provide variant effect scores for all variants that were assayed (Additional file 1: Figure S1) [6–8, 11, 13, 16–169]. When variant effect scores are provided, they are typically accessible only as a supplementary table or via a bespoke web interface [13, 18–20, 70, 170] leading to a proliferation of inconsistent formats. Some publications, instead of including variant effect scores, deposit the associated high-throughput DNA sequencing data in the Sequence Read Archive or Gene Expression Omnibus [171, 172]. This raw data can be used to reconstruct variant effect scores, but accurately replicating the original analysis can be non-trivial. While databases of variant effect information exist, they are typically designed for a specific application [173–175] or a specific group of target genes [176–178]. Larger and more general databases can sometimes contain variant effect data [179, 180], but these platforms were not developed with large-scale variant effect maps in mind, so valuable context for the variant effect scores and associated metadata may be lost. Furthermore, most existing resources lack support for noncoding targets entirely.
To overcome these challenges and facilitate future advances, we are establishing an open-source platform for MAVE resources. The foundation is MaveDB, a central repository that allows researchers to store and publish processed MAVE datasets, associated metadata, and linked raw data using a machine-readable, standardized, and searchable format. An easy-to-use web interface maximizes the impact and usefulness of researchers’ work by making the data readily accessible to the whole community, whether for clinical applications, meta-analysis, or reanalysis as computational techniques are refined.
This platform is designed to allow additional applications to communicate directly with MaveDB. The first of potentially many such applications, MaveVis, visualizes and provides context to protein variant effect maps by generating heatmaps and automatically integrating them with secondary structure, surface accessibility, interaction interfaces, and conservation data.
Construction and content
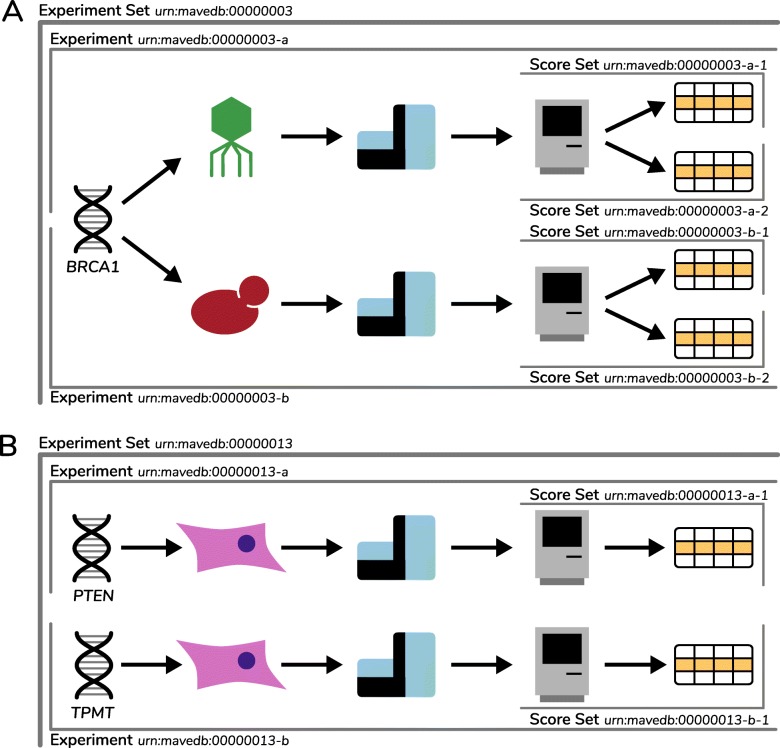
To capture the structure of real-world study designs, MaveDB is organized hierarchically into score sets, experiments, and experiment sets (Fig. 1). Score sets, the most basic unit of organization, contain the variant effect scores and additional metadata such as target sequence information and detailed methods. Each variant effect score is a numeric value. Optional data columns containing values related to each variant effect score such as variant counts and measures of uncertainty can also be included and named by the user.
Fig. 1.
Representative structures of MaveDB entries. Each panel depicts a single experiment set and its associated accession numbers. a A single target sequence analyzed using two different MAVEs. For example, MAVEs in phage display and yeast two-hybrid formats were performed on BRCA1 (experiment accessions ending in “-a” and “-b”) [16]. Each of these yielded two score sets, one for amino acid variants and one for nucleotide variants (score set accessions ending in “-1” and “-2”). b Two distinct target sequences analyzed using the same MAVE. For example, the VAMP-seq assay was performed on the PTEN and TPMT target sequences (experiment accessions ending in “-a” and “-b”) [18], yielding an amino acid score set for each target (score set accessions ending in “-1”)
Most experimental designs in MaveDB involve multiple score sets. For example, protein MAVEs commonly have one score set for nucleotide variants and another for amino acid variants [23]. Experiments with tiled designs [54, 92] can have score sets for each tile, and experiments with multiple distinct reference sequences [147] can have score sets for each reference sequence [26, 147]. In addition, we envision that reanalysis and renormalization of the existing datasets using updated methods will be commonplace [14, 15, 181, 182]. By grouping all analyses of a single raw dataset under one experiment, MaveDB ensures that the number of assays performed on each target sequence can be tracked accurately.
Each experiment describes one or more analyses arising from a single MAVE, including any technical and biological replicates. In addition to the links to score sets, experiments contain metadata including methodological details, links to raw data, and associated publications (Table 1), but no variant score information. Experiment sets contain one or more related experiments, for example, multiple MAVEs performed on the same target sequence under different conditions or multiple experiments from the same publication (Fig. 1).
Table 1.
MaveDB metadata fields
| Field name | Experiment | Score set | Type | Searchable | Link |
|---|---|---|---|---|---|
| Keyword | ✓ | ✓ | String | ✓ | |
| Abstract | ✓ | ✓ | Markdown | ✓ | |
| Method | ✓ | ✓ | Markdown | ✓ | |
| Short description | ✓ | ✓ | String | ✓ | |
| Title | ✓ | ✓ | String | ✓ | |
| PubMed ID | ✓ | ✓ | Accession | ✓ | ✓ |
| DOI | ✓ | ✓ | Accession | ✓ | ✓ |
| SRA accession | ✓ | Accession | ✓ | ✓ | |
| RefSeq accession | ✓ | Accession | ✓ | ✓ | |
| Ensembl accession | ✓ | Accession | ✓ | ✓ | |
| UniProt accession | ✓ | Accession | ✓ | ✓ | |
| Created by | ✓ | ✓ | Contributor | ✓ | ✓ |
| Last modified by | ✓ | ✓ | Contributor | ✓ | ✓ |
| Creation date | ✓ | ✓ | DateTime | ✓ | |
| Modification date | ✓ | ✓ | DateTime | ✓ | |
| Publication date | ✓ | ✓ | DateTime | ✓ | |
| License | ✓ | License | ✓ | ✓ | |
| Has replacement | ✓ | Boolean | ✓ |
MaveDB currently contains over one million variant effect scores across 45 unique targets. We welcome both new and previously described datasets from the community and have implemented a conversion tool, mavedb-convert, for datasets generated by Enrich [183], Enrich2 [182], and EMPIRIC [184] (see the “Availability of data and materials” section).
MaveDB is implemented in Python using the Django Python Web framework [185, 186]. The relational database backend is PostgreSQL [187]. The full database schema is shown in Additional file 1: Figure S2. Asynchronous tasks such as handling file uploads and sending emails are managed using RabbitMQ and Celery [188, 189]. Variant score and count data are stored using PostgreSQL JSONField objects, which offer additional flexibility for storing arbitrarily named data columns compared to a more traditional relational database design. Database accession numbers for publicly accessible entries are assigned in Universal Resource Name (URN) format [190].
Differences between each variant sequence and the target sequence are described using HGVS format [191]. MaveDB supports DNA and protein variant strings that describe substitutions or small indels in most sequence contexts, including splice site variants and multi-mutants.
Contributors are authenticated using their ORCID ID via the OAuth2 protocol [192, 193]. Consequently, an individual must have an ORCID ID to be named as a contributor to a MaveDB dataset. Users do not need to log in to browse or download publicly available data. MaveDB allows users to provide a private contact email address if they want to be contacted by administrators or receive alerts, but all other details are pulled from their public ORCID record.
Abstract and methods sections support Markdown [194] blocks for formatted text with support for mathematical notation. Markdown blocks are rendered to HTML using Pandoc [195].
MaveVis is implemented using R [196] and Docker [197]. Surface accessibility and interface burial are calculated using FreeSasa [198]. Secondary structure is calculated using DSSP [199]. Conservation tracks are calculated using the AMAS algorithm [200], based on multiple alignments computed using ClustalOmega [201] for the appropriate UniRef90 set of orthologous proteins with at least 90% sequence identity from UniProtKB [180].
Utility and discussion
Accessing datasets
MaveDB can be accessed through a standard web browser that allows users to explore by keyword, target gene, or organism. Alternatively, the advanced search function allows users to query all metadata fields, including the full text of methods and abstracts. Complete sets of variant effect scores and related values can be downloaded from any score set page in a comma-separated value format. These files can be parsed easily in most scientific programming environments or imported into spreadsheet applications.
The advanced search function is also accessible programmatically through the Representational State Transfer-Application Programming Interface (REST API). The API returns structured data, including full score sets and metadata, in JSON format, suitable for deserialization by most programming languages. Users of the R programming environment [196] can access MaveDB’s REST API using the rapimave library, which also includes a suite of exploration, searching, parsing, and filtering functions (see the “Availability of data and materials” section).
Creating new entries
Typically, a user starts by logging in using their ORCID ID and creating an experiment. The experiment can be added to an existing experiment set if desired; otherwise, a new one will be created automatically. The user provides a description of the assay used to generate the raw data, adds links to the raw data if available, and can then add contributors. After the experiment is created, the user creates one or more associated score sets. Here, the user enters the required information about the target such as its name and sequence and also describes the methods used to calculate the variant effect scores from the raw data. Variant effect scores and optional count files are then uploaded via the web interface and validated by the server.
Publishing datasets
When first created, score sets, experiments, and experiment sets are private and have temporary accession numbers. Private entries are only viewable by their contributors, and all values may be modified. Private entries can be accessed through the API by providing a contributor’s private access token generated on the contributor’s user profile page.
Completed private score sets can be published, making the score set publicly viewable. Publication creates a stable accession number and freezes the target sequence and variant effect score data, ensuring that all subsequent analyses based on the data are recomputable. Associated experiment and experiment sets are also published automatically if they are still private. Users may continue to edit some metadata such as the methods and descriptions after publication.
Published scores cannot be changed, but in case a correction is necessary, MaveDB allows score sets to be deprecated when creating a replacement. Users browsing MaveDB will only see the most recent version, but deprecated score sets will remain available by accession number to ensure that previous analyses are reproducible.
Contributor permissions
MaveDB supports three contributor roles: administrator, editor, and viewer. Administrators can add or remove contributors, modify entries, and publish score sets. Editors can modify entries but cannot affect the contributor list or make entries public. Viewers can see their private entries in the database but cannot change them.
All three roles appear in the contributor list with no visual distinction between them, and administrators can continue to change the contributor list for each score set or experiment after publication. Since score sets and experiments have independent contributor lists, MaveDB maintains clear attribution when datasets are reanalyzed.
Data licensing
Administrators may select one of several Creative Commons licenses for each score set [202–204], and additional licensing options may be added in response to user requests. The license information is included as score set metadata and as part of the header of each downloaded file. Administrators can relicense after publication, although users who download under a more permissive license would not be subject to a more restrictive license.
Visualizing variant effect maps
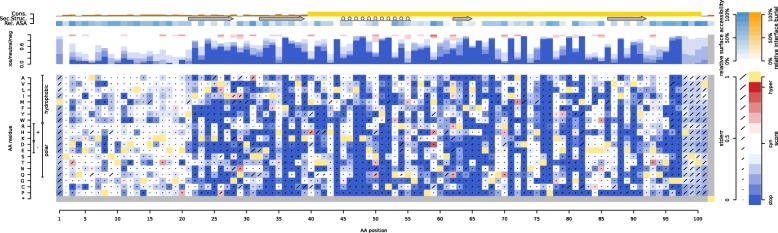
The MaveVis application allows users to quickly visualize score sets retrieved directly from MaveDB. One example of MaveVis output for a variant effect map of the protein SUMO1 [146] is shown in Fig. 2. Score sets are rendered as heatmaps with additional tracks representing integrated structural and conservation information from PDB [205] and UniprotKB [180]. The heatmap shows all possible amino acid changes at each protein sequence position, with colors reflecting the variant effect scores. The color scale is automatically calibrated based on the scores of reference and null alleles in the dataset or set manually by the user. Error bars are drawn directly on the heatmap fields to represent the measurement error provided in the score set, if present. Additional tracks show burial in protein interaction interfaces, residue-specific solvent accessibility, protein secondary structure, and sequence conservation.
Fig. 2.
Heatmap for the SUMO1 MAVE dataset rendered by MaveVis. The x-axis iterates over amino acid positions in the protein, while the y-axis lists all possible amino acid changes organized by their physicochemical properties. The heatmap color reflects the variant effect score, with blue being as deleterious as a full deletion (labeled “stop” in the color bar), white being equivalent to the reference allele (labeled “syn” in the color bar), and red representing a stronger phenotype than the reference residue at that position (labeled “hyper” in the color bar). Yellow cells indicate the reference amino acid at each position. Error bars represent standard error of the mean. The stacked bars above the heatmap represent the relative frequencies for each phenotype bin of the corresponding color at each position. Additional tracks show data integrated from other databases: orange heatmap tracks represent burial in protein interaction interfaces, the steel blue heatmap track represents solvent accessibility, the arrows and spirals correspond to secondary structure, and the yellow bar chart at the top shows sequence conservation
MaveVis is hosted at http://varianteffect.org, a portal for applications built on MaveDB. Users can follow the MaveVis link on each MaveDB score set page or navigate directly to http://vis.varianteffect.org and search for datasets. Once a score set is selected, the corresponding UniProt accession from MaveDB is suggested when available. MaveVis automatically presents potentially relevant PDB structures for the selected protein that overlap with the score set target sequence, allowing users to select which structures to include in the visualization. The resulting plot can be downloaded in PNG, PDF, or SVG format.
In addition to the web interface, MaveVis also exists as an R package for local use (see the “Availability of data and materials” section). The R package provides direct access to both the visualization and underlying data integration functions, making it easy to automatically compile structural and conservation feature tables for individual proteins.
The MaveVis server automatically synchronizes with MaveDB at regular intervals via its API, caching any new score sets, automatically obtaining relevant PDB and UniProt data, and pre-calculating partial results for a more responsive user experience. MaveVis also exposes its own API, allowing it to be used within more complex workflows.
To facilitate communication between MaveVis and MaveDB, we developed an R package, hgvsParseR, to parse or assemble HGVS [191] strings that describe alleles (see the “Availability of data and materials” section). In addition to its utility for visualizing variant effect maps, we expect that this package will be generally useful for working with data from ClinVar [5], gnomAD [206], and other important sequence variation resources.
Conclusions
MaveDB is the foundation of an open-source platform for the collection, distribution, and analysis of variant effect maps. Designed to be flexible and extensible, the MaveDB repository can accommodate data from diverse target sequences and experimental methods as the field evolves. Using MaveDB to combine variant effect data with external contextual information, MaveVis is the first application built on this resource. We envision developing additional applications such as tertiary structure analysis, automatic imputation of missing values in variant effect maps [207], and a broadly applicable dashboard to assist dataset interpretation.
MaveDB, MaveVis, and Enrich2 simplify, standardize, and democratize MAVE data analysis. These tools are the beginnings of a community-driven, open-source platform that allows researchers to explore these comprehensive datasets. The impact of each dataset will continue to increase as the number of assayed variants grows, contributing to a more complete understanding of genetic variation and sequence function.
Supplementary information
Additional file 1: Figure S1. Data unavailability is a persistent feature of the MAVE literature. We compiled a list of 159 publications that contained at least one new deep mutational scanning or massively parallel reporter assay dataset and manually inspected the publication’s supplementary data and methods to determine whether counts or scores for the assayed variants were present. Refer to https://github.com/VariantEffect/MaveReferences for the full table. This figure was generated from release v0.1.1. Of the 159 total publications, 91 (57%) provide scores or counts. Figure S2. UML (Unified Markup Language) diagram of the complete MaveDB schema in PDF format. The diagram was generated using the Django Extensions package and visualized using Graphviz.
Acknowledgements
We would like to acknowledge Bernie Pope, Peter Georgeson, Nick Moore, Matthew Wakefield, and Dan Bolon for their helpful advice and guidance. We would also like to acknowledge the University of Washington Genome Sciences Information Technology Services team for their outstanding service and support of MaveDB during the development process.
Review history
Review history is available as Additional file 2.
Peer review information
Anahita Bishop was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Authors’ contributions
AFR, DMF, and FPR conceived the project. DE and AFR built the database and its web interface. JW built the MaveVis application. DE and JW built the APIs. All authors wrote the manuscript and approved the final version.
Funding
This work was supported by the National Institutes of Health (NIH; R01GM109110 and RM1HG010461 to DMF) and the Brotman Baty Institute for Precision Medicine. DMF was a CIFAR Azrieli Global Scholar. ATP was supported by the Lorenzo and Pamela Galli Charitable Trust and by an Australian National Health and Medical Research Council (NHMRC) Program Grant (1054618) and NHMRC Senior Research Fellowship (1116955). The research benefitted by support from the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support. FPR and JW gratefully acknowledge the funding by the One Brave Idea Initiative, the National Human Genome Research Institute of the NIH Center of Excellence in Genomic Science Initiative (HG004233), the Canadian Excellence Research Chairs Program, and a Canadian Institutes of Health Research Foundation Grant.
Availability of data and materials
MaveDB is hosted at https://www.mavedb.org/.
MaveVis is hosted at http://vis.varianteffect.org/.
Source code for all websites, tools, and packages is available on GitHub at https://github.com/VariantEffect/.
• MaveDB: GitHub [208] and Zenodo [209]
• MaveVis: GitHub [210] and Zenodo [211]
• mavedb-convert: GitHub [212] and Zenodo [213]
• rapimave: GitHub [214] and Zenodo [215]
• hgvsParseR: GitHub [216] and Zenodo [217]
A pre-compiled docker image for MaveVis is also available on DockerHub at https://hub.docker.com/r/jweile/mavevis/.
The full list of MAVE studies is available at GitHub [218] and Zenodo [219].
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniel Esposito and Jochen Weile contributed equally to this work.
Contributor Information
Frederick P. Roth, Email: fritz.roth@utoronto.ca
Douglas M. Fowler, Email: dfowler@uw.edu
Alan F. Rubin, Email: alan.rubin@wehi.edu.au
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13059-019-1845-6.
References
- 1.Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasperini M, Starita L, Shendure J. The power of multiplexed functional analysis of genetic variants. Nat Protoc. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weile Jochen, Roth Frederick P. Multiplexed assays of variant effects contribute to a growing genotype–phenotype atlas. Human Genetics. 2018;137(9):665–678. doi: 10.1007/s00439-018-1916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, et al. Variant interpretation: functional assays to the rescue. Am J Hum Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Lee JM, Huddleston J, Doud MB, Hooper KA, Wu NC, Bedford T, et al. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc Natl Acad Sci. 2018;201806133. [DOI] [PMC free article] [PubMed]
- 8.Cantor AJ, Shah NH, Kuriyan J. Deep mutational analysis reveals functional trade-offs in the sequences of EGFR autophosphorylation sites. Proc Natl Acad Sci. 2018;115:E7303–E7312. doi: 10.1073/pnas.1803598115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmiedel JM, Lehner B. Determining protein structures using deep mutagenesis. Nat Genet. 2019;51:1177–1186. doi: 10.1038/s41588-019-0431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rollins NJ, Brock KP, Poelwijk FJ, Stiffler MA, Gauthier NP, Sander C, et al. Inferring protein 3D structure from deep mutation scans. Nat Genet. 2019;51:1170–1176. doi: 10.1038/s41588-019-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Ahituv N. Gene regulatory elements, major drivers of human disease. Annu Rev Genomics Hum Genet. 2017;18:45–63. doi: 10.1146/annurev-genom-091416-035537. [DOI] [PubMed] [Google Scholar]
- 13.Kircher M, Xiong C, Martin B, Schubach M, Inoue F, Bell RJA, et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nat Commun. 2019;10:3583. doi: 10.1038/s41467-019-11526-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray VE, Hause RJ, Fowler DM. Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions. Genetics. 2017;207:53–61. doi: 10.1534/genetics.117.300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray VE, Hause RJ, Luebeck J, Shendure J, Fowler DM. Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 2018;6:116–124. doi: 10.1016/j.cels.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starita LM, Young DL, Islam M, Kitzman JO, Gullingsrud J, Hause RJ, et al. Massively parallel functional analysis of BRCA1 RING domain variants. Genetics. 2015;200:413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majithia AR, Tsuda B, Agostini M, Gnanapradeepan K, Rice R, Peloso G, et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay Gregory M., Daza Riza M., Martin Beth, Zhang Melissa D., Leith Anh P., Gasperini Molly, Janizek Joseph D., Huang Xingfan, Starita Lea M., Shendure Jay. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562(7726):217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starita Lea M., Islam Muhtadi M., Banerjee Tapahsama, Adamovich Aleksandra I., Gullingsrud Justin, Fields Stanley, Shendure Jay, Parvin Jeffrey D. A Multiplex Homology-Directed DNA Repair Assay Reveals the Impact of More Than 1,000 BRCA1 Missense Substitution Variants on Protein Function. The American Journal of Human Genetics. 2018;103(4):498–508. doi: 10.1016/j.ajhg.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mighell TL, Evans-Dutson S, O’Roak BJ. A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotype relationships. Am J Hum Genet. 2018;102:943–955. doi: 10.1016/j.ajhg.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patwardhan RP, Lee C, Litvin O, Young DL, Pe’er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst A, Gfeller D, Kan Z, Seshagiri S, Kim PM, Bader GD, et al. Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol Biosyst. 2010;6:1782–1790. doi: 10.1039/c0mb00061b. [DOI] [PubMed] [Google Scholar]
- 25.Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc Natl Acad Sci. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Torkamani A, Jones TM, Ruiz DI, Pons J, Lerner RA. Phenotype-information-phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems. Proc Natl Acad Sci. 2011;108:13456–13461. doi: 10.1073/pnas.1111218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adkar BV, Tripathi A, Sahoo A, Bajaj K, Goswami D, Chakrabarti P, et al. Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure. 2012;20:371–381. doi: 10.1016/j.str.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlinkmann KM, Honegger A, Türeci E, Robison KE, Lipovšek D, Plückthun A. Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc Natl Acad Sci. 2012;109:9810–9815. doi: 10.1073/pnas.1202107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traxlmayr MW, Hasenhindl C, Hackl M, Stadlmayr G, Rybka JD, Borth N, et al. Construction of a stability landscape of the CH3 domain of human IgG1 by combining directed evolution with high throughput sequencing. J Mol Biol. 2012;423:397–412. doi: 10.1016/j.jmb.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z, Huang W, Bakkalbasi E, Brown NG, Adamski CJ, Rice K, et al. Deep sequencing of systematic combinatorial libraries reveals β-lactamase sequence constraints at high resolution. J Mol Biol. 2012;424:150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujino Y, Fujita R, Wada K, Fujishige K, Kanamori T, Hunt L, et al. Robust in vitro affinity maturation strategy based on interface-focused high-throughput mutational scanning. Biochem Biophys Res Commun. 2012;428:395–400. doi: 10.1016/j.bbrc.2012.10.066. [DOI] [PubMed] [Google Scholar]
- 37.Wu NC, Young AP, Dandekar S, Wijersuriya H, Al-Mawsawi LQ, Wu T-T, et al. Systematic identification of H274Y compensatory mutations in influenza A virus neuraminidase by high-throughput screening. J Virol. 2013;87:1193–1199. doi: 10.1128/JVI.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liachko I, Youngblood RA, Keich U, Dunham MJ. High-resolution mapping, characterization, and optimization of autonomously replicating sequences in yeast. Genome Res. 2013;23:698–704. doi: 10.1101/gr.144659.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DNA. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starita LM, Pruneda JN, Lo RS, Fowler DM, Kim HJ, Hiatt JB, et al. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci. 2013;110:E1263–E1272. doi: 10.1073/pnas.1303309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gold MG, Fowler DM, Means CK, Pawson CT, Stephany JJ, Langeberg LK, et al. Engineering A-kinase anchoring protein (AKAP)-selective regulatory subunits of protein kinase A (PKA) through structure-based phage selection. J Biol Chem. 2013;288:17111–17121. doi: 10.1074/jbc.M112.447326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsyth CM, Juan V, Akamatsu Y, DuBridge RB, Doan M, Ivanov AV, et al. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. mAbs. 2013;5:523–532. doi: 10.4161/mabs.24979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DNA. Latent effects of Hsp90 mutants revealed at reduced expression levels. PLOS Genet. 2013;9:e1003600. doi: 10.1371/journal.pgen.1003600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Procko E, Hedman R, Hamilton K, Seetharaman J, Fleishman SJ, Su M, et al. Computational design of a protein-based enzyme inhibitor. J Mol Biol. 2013;425:3563–3575. doi: 10.1016/j.jmb.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacquier H, Birgy A, Nagard HL, Mechulam Y, Schmitt E, Glodt J, et al. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc Natl Acad Sci. 2013;110:13067–13072. doi: 10.1073/pnas.1215206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim I, Miller CR, Young DL, Fields S. High-throughput analysis of in vivo protein stability. Mol Cell Proteomics. 2013;12:3370–3378. doi: 10.1074/mcp.O113.031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mogno I, Kwasnieski JC, Cohen BA. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res. 2013;23:1908–1915. doi: 10.1101/gr.157891.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melamed D, Young DL, Gamble CE, Miller CR, Fields S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA. 2013;19:1537–1551. doi: 10.1261/rna.040709.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagenaar TR, Ma L, Roscoe B, Park SM, Bolon DN, Green MR. Resistance to vemurafenib resulting from a novel mutation in the BRAFV600E kinase domain. Pigment Cell Melanoma Res. 2014;27:124–133. doi: 10.1111/pcmr.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rockah-Shmuel L, Tóth-Petróczy Á, Sela A, Wurtzel O, Sorek R, Tawfik DS. Correlated occurrence and bypass of frame-shifting insertion-deletions (InDels) to give functional proteins. PLoS Genet. 2013;9:e1003882. doi: 10.1371/journal.pgen.1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hietpas Ryan T., Bank Claudia, Jensen Jeffrey D., Bolon Daniel N. A. SHIFTING FITNESS LANDSCAPES IN RESPONSE TO ALTERED ENVIRONMENTS. Evolution. 2013;67(12):3512–3522. doi: 10.1111/evo.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strauch E-M, Fleishman SJ, Baker D. Computational design of a pH-sensitive IgG binding protein. Proc Natl Acad Sci. 2014;111:675–680. doi: 10.1073/pnas.1313605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liachko I, Youngblood RA, Tsui K, Bubb KL, Queitsch C, Raghuraman MK, et al. GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris. PLOS Genet. 2014;10:e1004169. doi: 10.1371/journal.pgen.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. Massively parallel functional annotation of 3′ untranslated regions. Nat Biotechnol. 2014;32:387–391. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oikonomou P, Goodarzi H, Tavazoie S. Systematic identification of regulatory elements in conserved 3′ UTRs of human transcripts. Cell Rep. 2014;7:281–292. doi: 10.1016/j.celrep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi H, Olson CA, Wu NC, Ke R, Loverdo C, Chu V, et al. A quantitative high-resolution genetic profile rapidly identifies sequence determinants of hepatitis C viral fitness and drug sensitivity. PLOS Pathog. 2014;10:e1004064. doi: 10.1371/journal.ppat.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 2014;31:1956–1978. doi: 10.1093/molbev/msu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roscoe BP, Bolon DNA. Systematic exploration of ubiquitin sequence, E1 activation efficiency, and experimental fitness in yeast. J Mol Biol. 2014;426:2854–2870. doi: 10.1016/j.jmb.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014;42:e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife. 2014;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharon E, van Dijk D, Kalma Y, Keren L, Manor O, Yakhini Z, et al. Probing the effect of promoters on noise in gene expression using thousands of designed sequences. Genome Res. 2014;24:1698–1706. doi: 10.1101/gr.168773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gajula KS, Huwe PJ, Mo CY, Crawford DJ, Stivers JT, Radhakrishnan R, et al. High-throughput mutagenesis reveals functional determinants for DNA targeting by activation-induced deaminase. Nucleic Acids Res. 2014;42:9964–9975. doi: 10.1093/nar/gku689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guy MP, Young DL, Payea MJ, Zhang X, Kon Y, Dean KM, et al. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014;28:1721–1732. doi: 10.1101/gad.245936.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noderer WL, Flockhart RJ, Bhaduri A, AJD d A, Zhang J, Khavari PA, et al. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol. 2014;10:748. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reich L“L”, Dutta S, Keating AE. SORTCERY—a high–throughput method to affinity rank peptide ligands. J Mol Biol. 2015;427:2135–2150. doi: 10.1016/j.jmb.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bank C. Hietpas RT, Jensen JD, Bolon DNA. A systematic survey of an intragenic epistatic landscape. Mol Biol Evol. 2015;32:229–238. doi: 10.1093/molbev/msu301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doolan KM, Colby DW. Conformation-dependent epitopes recognized by prion protein antibodies probed using mutational scanning and deep sequencing. J Mol Biol. 2015;427:328–340. doi: 10.1016/j.jmb.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 2014;24:2643–2651. doi: 10.1016/j.cub.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al-Mawsawi LQ, Wu NC, Olson CA, Shi VC, Qi H, Zheng X, et al. High-throughput profiling of point mutations across the HIV-1 genome. Retrovirology. 2014;11:124. doi: 10.1186/s12977-014-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Massively parallel single-amino-acid mutagenesis. Nat Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Podgornaia AI, Laub MT. Pervasive degeneracy and epistasis in a protein-protein interface. Science. 2015;347:673–677. doi: 10.1126/science.1257360. [DOI] [PubMed] [Google Scholar]
- 79.Shalem O, Sharon E, Lubliner S, Regev I, Lotan-Pompan M, Yakhini Z, et al. Systematic dissection of the sequence determinants of gene 3′ end mediated expression control. PLOS Genet. 2015;11:e1005147. doi: 10.1371/journal.pgen.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romero PA, Tran TM, Abate AR. Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc Natl Acad Sci. 2015;112:7159–7164. doi: 10.1073/pnas.1422285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu NC, Olson CA, Du Y, Le S, Tran K, Remenyi R, et al. Functional constraint profiling of a viral protein reveals discordance of evolutionary conservation and functionality. PLOS Genet. 2015;11:e1005310. doi: 10.1371/journal.pgen.1005310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kretz CA, Dai M, Soylemez O, Yee A, Desch KC, Siemieniak D, et al. Massively parallel enzyme kinetics reveals the substrate recognition landscape of the metalloprotease ADAMTS13. Proc Natl Acad Sci. 2015;112:9328–9333. doi: 10.1073/pnas.1511328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doud MB, Ashenberg O, Bloom JD. Site-specific amino acid preferences are mostly conserved in two closely related protein homologs. Mol Biol Evol. 2015;32:2944–2960. doi: 10.1093/molbev/msv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rockah-Shmuel L, Tóth-Petróczy Á, Tawfik DS. Systematic mapping of protein mutational space by prolonged drift reveals the deleterious effects of seemingly neutral mutations. PLOS Comput Biol. 2015;11:e1004421. doi: 10.1371/journal.pcbi.1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kowalsky CA, Faber MS, Nath A, Dann HE, Kelly VW, Liu L, et al. Rapid fine conformational epitope mapping using comprehensive mutagenesis and deep sequencing. J Biol Chem. 2015;290:26457–26470. doi: 10.1074/jbc.M115.676635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klesmith JR, Bacik J-P, Michalczyk R, Whitehead TA. Comprehensive sequence-flux mapping of a levoglucosan utilization pathway in E. coli. ACS Synth Biol. 2015;4:1235–1243. doi: 10.1021/acssynbio.5b00131. [DOI] [PubMed] [Google Scholar]
- 87.Aakre CD, Herrou J, Phung TN, Perchuk BS, Crosson S, Laub MT. Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell. 2015;163:594–606. doi: 10.1016/j.cell.2015.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 2015;163:698–711. doi: 10.1016/j.cell.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 89.Shen SQ, Myers CA, Hughes AEO, Byrne LC, Flannery JG, Corbo JC. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016;26:238–255. doi: 10.1101/gr.193789.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vvedenskaya IO, Zhang Y, Goldman SR, Valenti A, Visone V, Taylor DM, et al. Massively systematic transcript end readout, “MASTER”: transcription start site selection, transcriptional slippage, and transcript yields. Mol Cell. 2015;60:953–965. doi: 10.1016/j.molcel.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin Z, Rienzi SCD, Janzon A, Werner JJ, Angenent LT, Dangl JL, et al. Novel rhizosphere soil alleles for the enzyme 1-aminocyclopropane-1-carboxylate deaminase queried for function with an in vivo competition assay. Appl Environ Microbiol. 2016;82:1050–1059. doi: 10.1128/AEM.03074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang L, Liu P, Bank C. Renzette N, Prachanronarong K, Yilmaz LS, et al. A balance between inhibitor binding and substrate processing confers influenza drug resistance. J Mol Biol. 2016;428:538–553. doi: 10.1016/j.jmb.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 93.Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, et al. Engineering an allosteric transcription factor to respond to new ligands. Nat Methods. 2016;13:177–183. doi: 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sahoo A, Khare S, Devanarayanan S, Jain PC, Varadarajan R. Residue proximity information and protein model discrimination using saturation-suppressor mutagenesis. eLife. 2015;4:e09532. doi: 10.7554/eLife.09532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu NC, Du Y, Le S, Young AP, Zhang T-H, Wang Y, et al. Coupling high-throughput genetics with phylogenetic information reveals an epistatic interaction on the influenza A virus M segment. BMC Genomics. 2016;17:46. doi: 10.1186/s12864-015-2358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foight GW, Keating AE. Comparison of the peptide binding preferences of three closely related TRAF paralogs: TRAF2, TRAF3, and TRAF5. Protein Sci. 2016;25:1273–1289. doi: 10.1002/pro.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elazar A, Weinstein J, Biran I, Fridman Y, Bibi E, Fleishman SJ. Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. eLife. 2016;5:e12125. doi: 10.7554/eLife.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoggard T, Liachko I, Burt C, Meikle T, Jiang K, Craciun G, et al. High throughput analyses of budding yeast ARSs reveal new DNA elements capable of conferring centromere-independent plasmid propagation. G3. 2016;6:993–1012. doi: 10.1534/g3.116.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rich MS, Payen C, Rubin AF, Ong GT, Sanchez MR, Yachie N, et al. Comprehensive analysis of the SUL1 promoter of Saccharomyces cerevisiae. Genetics. 2016;203:191–202. doi: 10.1534/genetics.116.188037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Winkelman JT, Vvedenskaya IO, Zhang Y, Bird JG, Taylor DM, et al. Multiplexed protein-DNA cross-linking: scrunching in transcription start site selection. Science. 2016;351:1090–1093. doi: 10.1126/science.aad6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mishra P, Flynn JM, Starr TN, Bolon DNA. Systematic mutant analyses elucidate general and client-specific aspects of Hsp90 function. Cell Rep. 2016;15:588–598. doi: 10.1016/j.celrep.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Puchta O, Cseke B, Czaja H, Tollervey D, Sanguinetti G, Kudla G. Network of epistatic interactions within a yeast snoRNA. Science. 2016;352:840–844. doi: 10.1126/science.aaf0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li C, Qian W, Maclean CJ, Zhang J. The fitness landscape of a tRNA gene. Science. 2016;352:837–840. doi: 10.1126/science.aae0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mavor D, Barlow K, Thompson S, Barad BA, Bonny AR, Cario CL, et al. Determination of ubiquitin fitness landscapes under different chemical stresses in a classroom setting. eLife. 2016;5:e15802. doi: 10.7554/eLife.15802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Julien P, Miñana B, Baeza-Centurion P, Valcárcel J, Lehner B. The complete local genotype–phenotype landscape for the alternative splicing of a human exon. Nat Commun. 2016;7:11558. doi: 10.1038/ncomms11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vvedenskaya IO, Vahedian-Movahed H, Zhang Y, Taylor DM, Ebright RH, Nickels BE. Interactions between RNA polymerase and the core recognition element are a determinant of transcription start site selection. Proc Natl Acad Sci. 2016;113:E2899–E2905. doi: 10.1073/pnas.1603271113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steinberg B, Ostermeier M. Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway. J Mol Biol. 2016;428:2730–2743. doi: 10.1016/j.jmb.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 109.Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, et al. Local fitness landscape of the green fluorescent protein. Nature. 2016;533:397–401. doi: 10.1038/nature17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses. 2016;8:155. doi: 10.3390/v8060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016;166:679–690. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu NC, Dai L, Olson CA, Lloyd-Smith JO, Sun R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife. 2016;5:e16965. doi: 10.7554/eLife.16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tripathi A, Gupta K, Khare S, Jain PC, Patel S, Kumar P, et al. Molecular determinants of mutant phenotypes, inferred from saturation mutagenesis data. Mol Biol Evol. 2016;33:2960–2975. doi: 10.1093/molbev/msw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harris DT, Wang N, Riley TP, Anderson SD, Singh NK, Procko E, et al. Deep mutational scans as a guide to engineering high affinity T cell receptor interactions with peptide-bound major histocompatibility complex. J Biol Chem. 2016;291:24566–24578. doi: 10.1074/jbc.M116.748681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kowalsky CA, Whitehead TA. Determination of binding affinity upon mutation for type I dockerin–cohesin complexes from Clostridium thermocellum and Clostridium cellulolyticum using deep sequencing. Proteins Struct Funct Bioinforma. 2016;84:1914–1928. doi: 10.1002/prot.25175. [DOI] [PubMed] [Google Scholar]
- 118.Ernst J, Melnikov A, Zhang X, Wang L, Rogov P, Mikkelsen TS, et al. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat Biotechnol. 2016;34:1180–1190. doi: 10.1038/nbt.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brenan L, Andreev A, Cohen O, Pantel S, Kamburov A, Cacchiarelli D, et al. Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 2016;17:1171–1183. doi: 10.1016/j.celrep.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qiu C, Erinne OC, Dave JM, Cui P, Jin H, Muthukrishnan N, et al. High-resolution phenotypic landscape of the RNA polymerase II trigger loop. PLOS Genet. 2016;12:e1006321. doi: 10.1371/journal.pgen.1006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haddox HK, Dingens AS, Bloom JD. Experimental estimation of the effects of all amino-acid mutations to HIV’s envelope protein on viral replication in cell culture. PLOS Pathog. 2016;12:e1006114. doi: 10.1371/journal.ppat.1006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Rosmalen M, Janssen BMG, Hendrikse NM, van der Linden AJ, Pieters PA, Wanders D, et al. Affinity maturation of a cyclic peptide handle for therapeutic antibodies using deep mutational scanning. J Biol Chem. 2017;292:1477–1489. doi: 10.1074/jbc.M116.764225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernandes JD, Faust TB, Strauli NB, Smith C, Crosby DC, Nakamura RL, et al. Functional segregation of overlapping genes in HIV. Cell. 2016;167:1762–1773. doi: 10.1016/j.cell.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keskin A, Akdoğan E, Dunn CD. Evidence for amino acid snorkeling from a high-resolution, in vivo analysis of Fis1 tail-anchor insertion at the mitochondrial outer membrane. Genetics. 2017;205:691–705. doi: 10.1534/genetics.116.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kelsic ED, Chung H, Cohen N, Park J, Wang HH, Kishony R. RNA structural determinants of optimal codons revealed by MAGE-Seq. Cell Syst. 2016;3:563–571. doi: 10.1016/j.cels.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yartseva V, Takacs CM, Vejnar CE, Lee MT, Giraldez AJ. RESA identifies mRNA-regulatory sequences at high resolution. Nat Methods. 2017;14:201–207. doi: 10.1038/nmeth.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koenig P, Lee CV, Walters BT, Janakiraman V, Stinson J, Patapoff TW, et al. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc Natl Acad Sci. 2017;114:E486–E495. doi: 10.1073/pnas.1613231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Klesmith JR, Bacik J-P, Wrenbeck EE, Michalczyk R, Whitehead TA. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. Proc Natl Acad Sci. 2017;114:2265–2270. doi: 10.1073/pnas.1614437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45:e16. doi: 10.1093/nar/gkw942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Levo M, Avnit-Sagi T, Lotan-Pompan M, Kalma Y, Weinberger A, Yakhini Z, et al. Systematic investigation of transcription factor activity in the context of chromatin using massively parallel binding and expression assays. Mol Cell. 2017;65:604–617. doi: 10.1016/j.molcel.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 132.Chan YH, Venev SV, Zeldovich KB, Matthews CR. Correlation of fitness landscapes from three orthologous TIM barrels originates from sequence and structure constraints. Nat Commun. 2017;8:14614. doi: 10.1038/ncomms14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matreyek KA, Stephany JJ, Fowler DM. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res. 2017;45:e102. doi: 10.1093/nar/gkx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ashenberg O, Padmakumar J, Doud MB, Bloom JD. Deep mutational scanning identifies sites in influenza nucleoprotein that affect viral inhibition by MxA. PLoS Pathog. 2017;13:e1006288. doi: 10.1371/journal.ppat.1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wrenbeck EE, Azouz LR, Whitehead TA. Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded. Nat Commun. 2017;8:15695. doi: 10.1038/ncomms15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu NC, Xie J, Zheng T, Nycholat CM, Grande G, Paulson JC, et al. Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe. 2017;21:742–753. doi: 10.1016/j.chom.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bhagavatula G, Rich MS, Young DL, Marin M, Fields S. A massively parallel fluorescence assay to characterize the effects of synonymous mutations on TP53 expression. Mol Cancer Res. 2017;15:1301–1307. doi: 10.1158/1541-7786.MCR-17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bandaru P, Shah NH, Bhattacharyya M, Barton JP, Kondo Y, Cofsky JC, et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 2017;6:e27810. doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Starr TN, Picton LK, Thornton JW. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. doi: 10.1038/nature23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Woodsmith J, Apelt L, Casado-Medrano V, Özkan Z, Timmermann B, Stelzl U. Protein interaction perturbation profiling at amino-acid resolution. Nat Methods. 2017;14:1213–1221. doi: 10.1038/nmeth.4464. [DOI] [PubMed] [Google Scholar]
- 141.Ma L, Boucher JI, Paulsen J, Matuszewski S, Eide CA, Ou J, et al. CRISPR-Cas9–mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc Natl Acad Sci. 2017;114:11751–11756. doi: 10.1073/pnas.1708268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cuperus JT, Groves B, Kuchina A, Rosenberg AB, Jojic N, Fields S, et al. Deep learning of the regulatory grammar of yeast 5′ untranslated regions from 500,000 random sequences. Genome Res. 2017;27:2015–2024. doi: 10.1101/gr.224964.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Spencer JM, Zhang X. Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci Rep. 2017;7:16836. doi: 10.1038/s41598-017-17081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rabani M, Pieper L, Chew G-L, Schier AF. A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Mol Cell. 2017;68:1083–1094. doi: 10.1016/j.molcel.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ke S, Anquetil V, Zamalloa JR, Maity A, Yang A, Arias MA, et al. Saturation mutagenesis reveals manifold determinants of exon definition. Genome Res. 2018;28:11–24. doi: 10.1101/gr.219683.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weile J, Sun S, Cote AG, Knapp J, Verby M, Mellor JC, et al. A framework for exhaustively mapping functional missense variants. Mol Syst Biol. 2017;13:957. doi: 10.15252/msb.20177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Plesa C, Sidore AM, Lubock NB, Zhang D, Kosuri S. Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Science. 2018;359:343–347. doi: 10.1126/science.aao5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vainberg Slutskin I, Weingarten-Gabbay S, Nir R, Weinberger A, Segal E. Unraveling the determinants of microRNA mediated regulation using a massively parallel reporter assay. Nat Commun. 2018;9:529. doi: 10.1038/s41467-018-02980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Staller MV, Holehouse AS, Swain-Lenz D, Das RK, Pappu RV, Cohen BA. A high-throughput mutational scan of an intrinsically disordered acidic transcriptional activation domain. Cell Syst. 2018;6:444–455. doi: 10.1016/j.cels.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Diss G, Lehner B. The genetic landscape of a physical interaction. eLife. 2018;7:e32472. [DOI] [PMC free article] [PubMed]
- 151.Hartman EC, Jakobson CM, Favor AH, Lobba MJ, Álvarez-Benedicto E, Francis MB, et al. Quantitative characterization of all single amino acid variants of a viral capsid-based drug delivery vehicle. Nat Commun. 2018;9:1385. doi: 10.1038/s41467-018-03783-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mason DM, Weber CR, Parola C, Meng SM, Greiff V, Kelton WJ, et al. High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis. Nucleic Acids Res. 2018;46:7436–7449. doi: 10.1093/nar/gky550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kotler E, Shani O, Goldfeld G, Lotan-Pompan M, Tarcic O, Gershoni A, et al. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol Cell. 2018;71:178–190. doi: 10.1016/j.molcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 155.Salinas VH, Ranganathan R. Coevolution-based inference of amino acid interactions underlying protein function. eLife. 2018;7:e34300. doi: 10.7554/eLife.34300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mavor D, Barlow KA, Asarnow D, Birman Y, Britain D, Chen W, et al. Extending chemical perturbations of the ubiquitin fitness landscape in a classroom setting reveals new constraints on sequence tolerance. Biol Open. 2018;7:bio036103. doi: 10.1242/bio.036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dorrity MW, Cuperus JT, Carlisle JA, Fields S, Queitsch C. Preferences in a trait decision determined by transcription factor variants. Proc Natl Acad Sci. 2018;115:E7997–E8006. doi: 10.1073/pnas.1805882115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Castaldi PJ, Guo F, Qiao D, Du F, Naing ZZC, Li Y, et al. Identification of functional variants in the FAM13A chronic obstructive pulmonary disease genome-wide association study locus by massively parallel reporter assays. Am J Respir Crit Care Med. 2018;199:52–61. doi: 10.1164/rccm.201802-0337OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wong MS, Kinney JB, Krainer AR. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol Cell. 2018;71:1012–1026. doi: 10.1016/j.molcel.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Phillips AM, Doud MB, Gonzalez LO, Butty VL, Lin Y-S, Bloom JD, et al. Enhanced ER proteostasis and temperature differentially impact the mutational tolerance of influenza hemagglutinin. eLife. 2018;7:e38795. [DOI] [PMC free article] [PubMed]
- 161.Blaesi EJ, Palowitch GM, Hu K, Kim AJ, Rose HR, Alapati R, et al. Metal-free class Ie ribonucleotide reductase from pathogens initiates catalysis with a tyrosine-derived dihydroxyphenylalanine radical. Proc Natl Acad Sci. 2018;115:10022–10027. doi: 10.1073/pnas.1811993115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Giacomelli Andrew O., Yang Xiaoping, Lintner Robert E., McFarland James M., Duby Marc, Kim Jaegil, Howard Thomas P., Takeda David Y., Ly Seav Huong, Kim Eejung, Gannon Hugh S., Hurhula Brian, Sharpe Ted, Goodale Amy, Fritchman Briana, Steelman Scott, Vazquez Francisca, Tsherniak Aviad, Aguirre Andrew J., Doench John G., Piccioni Federica, Roberts Charles W. M., Meyerson Matthew, Getz Gad, Johannessen Cory M., Root David E., Hahn William C. Mutational processes shape the landscape of TP53 mutations in human cancer. Nature Genetics. 2018;50(10):1381–1387. doi: 10.1038/s41588-018-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Naftaly S, Cohen I, Shahar A, Hockla A, Radisky ES, Papo N. Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries. Nat Commun. 2018;9:3935. doi: 10.1038/s41467-018-06403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bassalo MC, Garst AD, Choudhury A, Grau WC, Oh EJ, Spindler E, et al. Deep scanning lysine metabolism in Escherichia coli. Mol Syst Biol. 2018;14:e8371. doi: 10.15252/msb.20188371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cheung R, Insigne KD, Yao D, Burghard CP, Wang J, Hsiao Y-HE, et al. A multiplexed assay for exon recognition reveals that an unappreciated fraction of rare genetic variants cause large-effect splicing disruptions. Mol Cell. 2019;73:183–194. doi: 10.1016/j.molcel.2018.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Michaels YS, Barnkob MB, Barbosa H, Baeumler TA, Thompson MK, Andre V, et al. Precise tuning of gene expression levels in mammalian cells. Nat Commun. 2019;10:818. doi: 10.1038/s41467-019-08777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noda-García L, Davidi D, Korenblum E, Elazar A, Putintseva E, Aharoni A, et al. Chance and pleiotropy dominate genetic diversity in complex bacterial environments. Nat Microbiol. 2019;4:1221–1230. doi: 10.1038/s41564-019-0412-y. [DOI] [PubMed] [Google Scholar]
- 168.Ahler Ethan, Register Ames C., Chakraborty Sujata, Fang Linglan, Dieter Emily M., Sitko Katherine A., Vidadala Rama Subba Rao, Trevillian Bridget M., Golkowski Martin, Gelman Hannah, Stephany Jason J., Rubin Alan F., Merritt Ethan A., Fowler Douglas M., Maly Dustin J. A Combined Approach Reveals a Regulatory Mechanism Coupling Src’s Kinase Activity, Localization, and Phosphotransferase-Independent Functions. Molecular Cell. 2019;74(2):393-408.e20. doi: 10.1016/j.molcel.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sample PJ, Wang B, Reid DW, Presnyak V, McFadyen IJ, Morris DR, et al. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat Biotechnol. 2019;37:803. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chang W, Cheng J, Allaire JJ, Xie Y, McPherson J. shiny: web application framework for R. 2018. https://CRAN.R-project.org/package=shiny.
- 171.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang CY, Chang PM, Ary ML, Allen BD, Chica RA, Mayo SL, et al. ProtaBank: a repository for protein design and engineering data. Protein Sci. 2018;27:1113–1124. doi: 10.1002/pro.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pires DEV, Blundell TL, Ascher DB. Platinum: a database of experimentally measured effects of mutations on structurally defined protein–ligand complexes. Nucleic Acids Res. 2015;43:D387–D391. doi: 10.1093/nar/gku966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bava KA, Gromiha MM, Uedaira H, Kitajima K, Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32:D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 177.Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Oscarson M, Ingelman-Sundberg M. CYPalleles: a web page for nomenclature of human cytochrome P450 alleles. Drug Metab Pharmacokinet. 2002;17:491–495. doi: 10.2133/dmpk.17.491. [DOI] [PubMed] [Google Scholar]
- 179.Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 180.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Matuszewski S, Hildebrandt ME, Ghenu A-H, Jensen JD, Bank C A statistical guide to the design of deep mutational scanning experiments. Genetics. 2016;204:77–87. doi: 10.1534/genetics.116.190462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rubin AF, Gelman H, Lucas N, Bajjalieh SM, Papenfuss AT, Speed TP, et al. A statistical framework for analyzing deep mutational scanning data. Genome Biol. 2017;18:150. doi: 10.1186/s13059-017-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fowler DM, Araya CL, Gerard W, Fields S. Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27:3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hietpas R, Roscoe B, Jiang L, Bolon DNA. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat Protoc. 2012;7:1382–1396. doi: 10.1038/nprot.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Python. https://www.python.org/.
- 186.Django. https://www.djangoproject.com/.
- 187.PostgreSQL. https://www.postgresql.org/.
- 188.RabbitMQ. https://www.rabbitmq.com/.
- 189.Celery. http://www.celeryproject.org/.
- 190.Saint-Andre P, Klensin J. Uniform Resource Names (URNs) 2017. [Google Scholar]
- 191.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 192.ORCID. https://orcid.org/.
- 193.D. Hardt E. The OAuth 2.0 Authorization Framework. 2012. http://www.rfc-editor.org/info/rfc6749.
- 194.Markdown. https://daringfireball.net/projects/markdown/.
- 195.Pandoc. https://pandoc.org/.
- 196.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.R-project.org.
- 197.Docker. https://www.docker.com/index.html.
- 198.Mitternacht S. FreeSASA: an open source C library for solvent accessible surface area calculations. F1000Research. 2016;5:189. doi: 10.12688/f1000research.7931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 200.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Bioinformatics. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 201.Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol Clifton NJ. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 202.Creative Commons — Attribution-NonCommercial-ShareAlike 4.0 International — CC BY-NC-SA 4.0. https://creativecommons.org/licenses/by-nc-sa/4.0/.
- 203.Creative Commons — Attribution 4.0 International — CC BY 4.0. https://creativecommons.org/licenses/by/4.0/.
- 204.Creative Commons — CC0 1.0 Universal. https://creativecommons.org/publicdomain/zero/1.0/.
- 205.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu Y, Weile J, Cote AG, Sun S, Knapp J, Verby M, et al. A web application and service for imputing and visualizing missense variant effect maps. Bioinforma Oxf Engl. 2019;35:3191–3193. doi: 10.1093/bioinformatics/btz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveDB. GitHub. (2019). https://github.com/VariantEffect/mavedb [DOI] [PMC free article] [PubMed]
- 209.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveDB. Zenodo. (2019). https://zenodo.org/record/3407951 [DOI] [PMC free article] [PubMed]
- 210.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveVis. GitHub. (2019). https://github.com/VariantEffect/mavevis [DOI] [PMC free article] [PubMed]
- 211.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveVis. Zenodo. (2019). https://zenodo.org/record/3432882 [DOI] [PMC free article] [PubMed]
- 212.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. mavedb-convert. GitHub. (2019). https://github.com/VariantEffect/mavedb-convert [DOI] [PMC free article] [PubMed]
- 213.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. mavedb-convert. Zenodo. (2019). https://zenodo.org/record/3405642 [DOI] [PMC free article] [PubMed]
- 214.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. rapimave. GitHub. (2019). https://github.com/VariantEffect/rapimave [DOI] [PMC free article] [PubMed]
- 215.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. rapimave. Zenodo. (2019). https://zenodo.org/record/3432746 [DOI] [PMC free article] [PubMed]
- 216.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. hgvsParseR. GitHub. (2019). https://github.com/VariantEffect/hgvsParseR [DOI] [PMC free article] [PubMed]
- 217.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. hgvsParseR. Zenodo. (2019). https://zenodo.org/record/3432816 [DOI] [PMC free article] [PubMed]
- 218.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveReferences. GitHub. (2019). https://github.com/VariantEffect/MaveReferences [DOI] [PMC free article] [PubMed]
- 219.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Rubin AF. MaveReferences. Zenodo. (2019). https://zenodo.org/record/3363411 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Data unavailability is a persistent feature of the MAVE literature. We compiled a list of 159 publications that contained at least one new deep mutational scanning or massively parallel reporter assay dataset and manually inspected the publication’s supplementary data and methods to determine whether counts or scores for the assayed variants were present. Refer to https://github.com/VariantEffect/MaveReferences for the full table. This figure was generated from release v0.1.1. Of the 159 total publications, 91 (57%) provide scores or counts. Figure S2. UML (Unified Markup Language) diagram of the complete MaveDB schema in PDF format. The diagram was generated using the Django Extensions package and visualized using Graphviz.
Data Availability Statement
MaveDB is hosted at https://www.mavedb.org/.
MaveVis is hosted at http://vis.varianteffect.org/.
Source code for all websites, tools, and packages is available on GitHub at https://github.com/VariantEffect/.
• MaveDB: GitHub [208] and Zenodo [209]
• MaveVis: GitHub [210] and Zenodo [211]
• mavedb-convert: GitHub [212] and Zenodo [213]
• rapimave: GitHub [214] and Zenodo [215]
• hgvsParseR: GitHub [216] and Zenodo [217]
A pre-compiled docker image for MaveVis is also available on DockerHub at https://hub.docker.com/r/jweile/mavevis/.
The full list of MAVE studies is available at GitHub [218] and Zenodo [219].