Abstract
RNase E is a component of the RNA degradosome complex and plays a key role in RNA degradation and maturation in Escherichia coli. RNase E–mediated target RNA degradation typically involves the RNA chaperone Hfq and requires small guide RNAs (sRNAs) acting as a seed by binding to short (7–12-bp) complementary regions in target RNA sequences. Here, using recombinantly expressed and purified proteins, site-directed mutagenesis, and RNA cleavage and protein cross-linking assays, we investigated Hfq-independent RNA decay by RNase E. Exploring its RNA substrate preferences in the absence of Hfq, we observed that RNase E preferentially cleaves AU-rich sites of single-stranded regions of RNA substrates that are annealed to an sRNA that contains a monophosphate at its 5′-end. We further found that the quaternary structure of RNase E is also important for complete, Hfq-independent cleavage at sites both proximal and distal to the sRNA-binding site within target RNAs containing monophosphorylated 5′-ends. Of note, genetic RNase E variants with unstable quaternary structure exhibited decreased catalytic activity. In summary, our results show that RNase E can degrade its target RNAs in the absence of the RNA chaperone Hfq. We conclude that RNase E–mediated, Hfq-independent RNA decay in E. coli requires a cognate sRNA sequence for annealing to the target RNA, a 5′-monophosphate at the RNA 5′-end, and a stable RNase E quaternary structure.
Keywords: ribonuclease, endoribonuclease, RNA degradation, mRNA decay, RNA processing, RNA turnover, protein expression, RNA chaperone Hfq, RNase E, small guide RNA (sRNA)
Introduction
In all organisms, RNA decay is one of the important processes for overall RNA metabolism and serves to determine the intracellular RNA level. The rapid degradation of mRNA has to be precisely controlled to modulate protein expression according to the changing environment and to recycle ribonucleotides for new RNA synthesis (1, 2). In contrast, rRNA and tRNA are stably degraded only under some stress conditions or when RNA are deficient (3). RNase E, the main component of RNA degradosome in Escherichia coli, plays an important role in mRNA degradation and RNA transcript processing (4, 5). It recognizes and cuts single-stranded RNA with numerous AU bases, thereby cleaving mRNA and rRNA into their functionally active forms. This enzyme is also known to contribute to the processing of noncoding RNA, such as tRNA, 16S rRNA precursors, and RNase P (6).
RNase E is the largest RNase (118 kDa), comprising 1,061 amino acids; it is encoded by the E. coli rne gene (7). RNase E consists of two functionally distinct domains: the globular N-terminal half (NTH;2 residues 1–529) and the C-terminal half (CTH; residues 530–1,061) (8). The RNase E-NTH has been shown to contain a catalytic activity domain for RNA cleavage, including a specific RNA binding site and a cleavage site (9). Because RNase E-NTH has the RNA-processing activity, such as RNA recognition and degradation, recombinant RNase E expressing amino acids 1–529 has been shown to be sufficient for the catalytic activity (10). In contrast, the CTH contains an arginine-rich domain, which is commonly involved in protein binding (8). The RNase E-CTH provides a scaffolding core for polynucleotide phosphorylase (PNPase), RNase helicase B, enolase, polyphosphate kinase, poly(A) polymerase, GroEL, and DnaK, which cooperate together to direct RNA toward the degradosome (11, 12). RNase E and PNPase allow direct physical interactions with other components within the degradosome complex to process the degradation of mRNA (13).
Biochemical and structural analyses performed using X-ray crystallography showed that RNase E is a homotetramer constituted of a dimer of dimers, which is important in the formation of catalytic sites for RNA turnover (14, 15). The RNase E-NTH catalytic domain is divided into several subdomains consisting of a large domain (residues 1–400), Zn-link (residues 401–414), and a small domain (residues 415–529) (15). The S1 RNA-binding domain in the large domain forms a sensing pocket for the 5′-terminus of monophosphorylated RNA and an arginine-rich RNA-binding channel for single-stranded RNA (16). The RNase E selectively cleaves single-stranded RNA, but not dsRNA; it prefers specific sequences enriched in AU dinucleotides (4, 17, 18). RNase E is active toward single-stranded RNA substrates (or targets) containing a 5′-monophosphate and cleaves at the AU-rich sequences that are several nucleotides apart from the 5′-terminus (19). Bacterial RNA 5′-pyrophosphohydrolase (RppH) has been shown to readily remove pyrophosphate from the 5′-end of triphosphorylated mRNA to generate a 5′-monophosphate (20, 21). Once the 5′-end of the target RNA substrates is recognized by RNase E, conformational changes are triggered that organize active sites and stimulate cleavage rates, leading to the RNA cleavage cascade.
Small noncoding RNA have been shown to be involved in the regulation of mRNA stability and degradation of trans-encoded target mRNA in conjunction with RNase E (22, 23). Small guide RNA (sRNA) range from 50 to 300 nucleotides and contain a short segment of 7–12 bp with a cognate region in the target mRNA referred to as the “seed pairing” sequence. This is analogous to the cognate binding of microRNA in eukaryotes (10, 24). Bacterial sRNAs usually repress translation of target mRNA by base-pairing with regions adjacent to or upstream of the ribosome-binding site, resulting in the blocking of ribosome entry and concomitant stimulation of the rapid decay of mRNA by a RNA degradosome (22, 25). Bacterial sRNA also positively regulate gene expression by pairing with mRNA; several sRNAs have been shown to activate the translation of the σ factor RpoS by relieving the inhibitory secondary structure of the rpoS mRNA leader form in the presence of the RNA chaperone Hfq protein (26). The sRNA-induced mRNA decay is facilitated by the formation of the ternary complex of RNase E, sRNA, and sRNA chaperone Hfq that protects sRNA from degradation by RNase E, in which Hfq promotes the annealing of sRNA to mRNA before the involvement of RNase E (25, 27, 28). The recruited RNase E in the ternary complex can then efficiently cleave the target RNA at the preferred sites (10, 29), either proximal (30) or distal (29) to the site of sRNA pairing.
RNase E initiates RNA decay via two pathways. First, in the 5′-end–dependent pathway, the 5′-monophosphate of the RNA interacts with the 5′-sensor pocket of RNase E, which stimulates the cleavage of the substrate. Thus, RNA is cleaved more efficiently when it is 5′-monophosphorylated, a form that is often generated from primary transcripts by pyrophosphohydrolase RppH or other RNases. The second pathway, direct entry, has been suggested for sRNA-induced mRNA decay, in which mRNA with a single-stranded region and no 5′-monophosphate are degraded by RNase E stimulated with sRNA/mRNA cognate pairing (10, 31). This model envisages that specific signals invoked by a cognate sRNA seed pairing to a certain target RNA sequence would trigger RNase E to cleave at the preferred sites in the mRNA coding sequence. Several studies have shown the presence of sRNA-induced cleavage sites in the coding sequence of target RNAs, including mRNA for sodB, fumA, iscS, and ompD (29, 32). In addition, controlled cleavage of ompD mRNA by sRNA MicC has been shown to be caused by the recruitment of RNase E activated by a monophosphate group at the 5′-end of the sRNA cognate-pairing seed (10). Because RNase E contains a 5′-monophosphate–sensing pocket that can interact specifically with a 5′-monophosphorylated sRNA, binding of the sRNA to RNase E prompts the cleavage of the mRNA at the preferred sites in the coding sequence of the mRNA (33). However, recent studies have shown that RNase E can recognize specific RNA structural elements without requiring the 5′-sensing route (34).
Numerous studies have shown that RNase E and the RNA chaperone Hfq act together in many cases of sRNA-mediated mRNA decay. In contrast, the Hfq RNA chaperone has been shown to not always be required in the RNase E–mediated mRNA degradation process, as mRNA paired with sRNA is sufficient for triggering target mRNA cleavage by RNase E. By varying the pairing status as well as chemical identity of sRNA, we explored the sRNA-induced target RNA degradation by RNase E in the absence of Hfq RNA chaperone. The 5′-monophosphorylated blunt end of the sRNA/target RNA duplex can trigger RNase E to cleave target RNA at the single-stranded region at distal sites, once the sRNA paired with the target RNA is anchored at the 5′-sensing pocket. In the absence of the RNA chaperone Hfq, RNase E with an intact quaternary structure can still cleave target RNA at both the proximal and distal sites to the monophosphorylated 5′-end of sRNA that are base-paired with the target RNA.
Results
RNase E cleaves target RNA paired with sRNA in an Hfq-independent manner
It has been previously reported that RNase E cleaves single-stranded RNA (ssRNA) with 5′-monophosphate (i.e. 5′-P), but not with 5′-hydroxyl (i.e. 5′-OH) or triphosphate group (15, 19). To examine whether the group at the 5′-terminus of the RNA substrate can affect the cleavage activity of RNase E, we tested this general principle using ssRNA with different chemical groups at the 5′-end (Fig. 1A). These experiments used 47- and 25-mer ssRNA substrates containing AU-rich sequences preferable for RNA cleavage by RNase E (35). The 47- and 25-mer ssRNA substrates that were predicted to be devoid of secondary structure were 32P-labeled either at the 5′-end nucleotides as [γ-32P]ATP or at internal nucleotides as [α-32P]UMP. Radioactive phosphate labeling instead of other fluorescent modifications at the 5′-end was used to ensure that the 5′-sensing pocket in the RNase E catalytic domain was not disturbed. The ssRNAs containing triphosphates at the 5′-end were prepared using in vitro transcription, mimicking authentic mRNA in E. coli. When the 25- and 47-mer ssRNAs with 5′-monophosphate were incubated with recombinant RNase E, digestion products of similar size (shorter than 10 nt) were observed after 30 min of incubation. The substrate RNA cleavage by RNase E required Mg2+ ion, but did not need ATP or ATP hydrolysis energy (i.e. AMP-PNP, nonhydrolyzable ATP analogue). This indicates that RNase E cleaves substrate ssRNA at the AU-rich sequences close to the 5′-end monophosphate regardless of substrate RNA length. In contrast, RNA cleavage patterns were not detected when control ssRNA containing either triphosphates or hydroxyl groups at the 5′-end were used (Fig. 1A). Thus, RNase E senses the chemical status at the 5′-end of substrate RNA, and the 5′-P is a prerequisite for RNA cleavage by RNase E. When the blunt-ended duplex RNA formed by the annealing of the 5′-32P–labeled 47-mer ssRNA (R0) to complementary 5′-32P–labeled 47-mer ssRNA (cR0) was reacted with RNase E, RNA cleavage was not observed (Fig. 1A), indicating that RNase E can cleave only single-stranded regions of RNA substrates harboring 5′-P.
Figure 1.
Cleavage of RNAs by RNase E in the absence or presence of Hfq. A, cleavage of RNAs with different chemical groups at the 5′-end (monophosphorylated ssRNAs, hydroxylated ssRNA, triphosphorylated ssRNA, and blunt-ended dsRNA). Top, monophosphorylated ssRNAs of two different lengths (25 and 47 nt for 25R1 and 47R0, respectively) were reacted with RNase E under different conditions (lanes a, b, c, and d; shown in table). Each reaction was performed by incubating 10 nm 32P-labeled RNA substrate in the absence (i.e. 0 min) or presence of RNase E (0.1 μm) for 30 min. Bottom, RNase E reactions were performed by incubating 10 nm 32P-labeled RNA substrates, such as triphosphorylated ssRNA (3P-47R0), 5′-end–hydroxylated ssRNA (HO-47R0), or blunt-ended duplex RNA (47R0/47cR0) with RNase E (0.1 μm) for increasing time (0 to 30 min). RNA cleavage products were resolved on 15% native PAGE and visualized using a phosphorimager. Asterisks represent 32P-labeled nucleotides. Lane c, size control for RNA of 47 and 25 nt. B, cleavage of sRNA (25R1)-guided target RNA (47R0) by RNase E in the absence or presence of Hfq. The partial duplex RNA (47R0/25R1, 10 nm) was incubated with RNase E (0.1 μm) in the absence or presence of Hfq (0.1 μm) for 0, 2, 4, 10, 20, and 40 min. RNA cleavage products were resolved on 15% native PAGE and visualized using a phosphorimager. Kinetic analysis results of cleaved RNA products, which were quantified using densitometry from the gel, were plotted and fitted to an exponential function. Mean values of triplicate independent experiments and S.D. (error bars) are shown. The difference between RNase E plus Hfq and RNase E alone was determined to be statistically insignificant by Student's t test (p = 0.079).
In E. coli, target mRNA-paired small RNAs guide and modulate the rates of translation initiation and target mRNA degradation through base-pairing, which is likely facilitated by the RNA chaperone Hfq protein (10). To test whether 5′-monophosphorylated sRNA alone could guide recombinant RNase E to cleave a target RNA in the absence of Hfq, artificial sRNAs (25-mer R1) with 5′-P were designed and annealed to the target RNA (47-mer R0; Fig. 1B). When the duplex RNA substrate consisting of the target RNA and cognate sRNA with 5′-P was incubated with RNase E, the target RNA was readily cleaved by RNase E at the single-stranded region of R0 RNA close to the 5′-end of the sRNA in the absence or presence of Hfq (Fig. 1B). RNA cleavage products shorter than 25 nt were not observed, indicating that the artificial 5′-monophosphorylated sRNA acts in trans to stimulate RNase E–mediated cleavage of the target RNA that is tethered by base-pairing. This result suggests that radioactive bands appearing between 25- and 47-mer result from the partial cleavage of the RNA substrate by RNase E, which is stimulated by 5′-monophosphorylated sRNA, in the absence of Hfq RNA chaperone. In both the case of RNase E alone and RNase E plus Hfq, RNA cleavage products accumulated as a function of time, and product accumulation was quantified and fitted to an exponential function to compare the catalytic efficiency of RNA cleavage (graph in Fig. 1A). The rate for duplex RNA cleavage was 0.20 and 0.12 min−1 for RNase E plus Hfq and RNase E alone, respectively, indicating that there is little statistically significant difference for RNA cleavage by RNase E in the absence and presence of Hfq. Thus, we suggest that 5′-monophosphorylated sRNA and RNase E are sufficient for target RNA cleavage with moderate efficiency, and RNA chaperone Hfq is not a prerequisite for duplex RNA stabilization.
The 5′-monophosphorylated sRNA can guide RNase E to preferred site cleavage in target RNA in the absence of Hfq
Because Hfq did not markedly affect the RNA cleavage activity of RNase E, we examined the effect of Mg2+ ion or ATP when 5′-P sRNA guides target RNA cleavage by RNase E in the absence of Hfq protein. We used duplex RNA substrate (P47R0/P25R1) formed by annealing of 5′-32P–labeled target RNA to the 5′-32P–labeled sRNA for radiometric detection of cleaved target RNA using urea-PAGE. When the duplex RNA substrate was incubated with RNase E in the presence of Mg2+ ion, bands corresponding to partially cleaved duplex RNA were observed (native PAGE in Fig. 2A). Consistent with the results shown in Fig. 1A, RNase E needs neither ATP nor ATP hydrolysis energy for the cleavage of duplexed substrate RNA in the absence of Hfq. RNA products cleaved by RNase E were resolved by heating, and the reaction products were analyzed for the presence of ssRNA components (urea-PAGE in Fig. 2A). RNA cleavage products consisted of shortened target RNA (<47 nt) and 25-mer sRNA, suggesting that RNase E cleaved single-stranded regions in the target RNA complementary to the 5′-monophosphorylated sRNA in the absence of Hfq. Importantly, the sRNA (R1) used in the experiment was not cleaved and remained protected from RNase E digestion.
Figure 2.
RNase E–mediated degradation of the single-stranded region in target RNA paired with 5′-monophosphorylated sRNA. A, RNase reaction of 47-mer target RNA paired with 5′-monophosphorylated sRNA by RNase E. The partial duplex RNA substrates (10 nm) were reacted with or without RNase E (0.1 μm) under different conditions (lanes a, b, c, and d; shown in table). RNase E reaction products were simultaneously resolved on 15% native and denaturing (12% polyacrylamide in 8 m urea) PAGE, in which 32P-labeled RNA were visualized using a phosphorimager. B, mapping of RNase E cleavage sites in the single-stranded region of the partial duplex RNA substrate. The 32P-labeled dsRNA substrate (30 nm, P47R0/P25R1 shown as RNA sequences) was incubated with or without RNase E (0.25 μm, lanes 6 and 7, respectively) at 37 °C for 30 min. For comparison as RNA ladders, the 5′-end 32P-labeled single-stranded RNA (30 nm; P47R0) was mixed with alkaline hydrolysis buffer at 25 °C for 10 or 5 min (lanes 1 and 2, respectively). RNase T1 (10, 5, or 1 unit; lanes 3, 4, and 5, respectively) was incubated with P47R0 RNA for 15 min. Cleavage sites after guanine residues are indicated in the RNA sequences (underlined Gs). All reactions were quenched with an equal volume of loading buffer and analyzed on 15% denaturing (8 m urea) PAGE and visualized using a phosphorimager. RNA fragments generated using RNase E cleavage at adenine residues in single-stranded regions are indicated with A and the nucleotide position with arrows. Dashed line, spliced part of two gel images for size controls (25- and 47-mer).
To pinpoint cleavage sites for RNase E in the absence of Hfq, the preferred site for RNase E cleavage in the target RNA was identified by mapping the cleavage sites in the duplex 47-mer R0 RNA annealed to sRNA using alkaline hydrolysis and RNase T1 digestion (Fig. 2B). The duplex R0/R1 RNA was cleaved by RNase E at positions +29, ∼31, +35, and +37, which are the downstream cleavage sites proximal to the 5′-end of the sRNA. Interestingly, we observed that the “A” nucleotide is a more preferred base than the “U” nucleotide in the target RNA guided by the sRNA for RNase E cleavage. When a 47-mer target RNA (R0) is present in the reaction without sRNA, the major cleavage products are short RNAs (<10 nt) resulting from degradation at the upstream cleavage sites by RNase E stimulated by the 5′-P (Fig. 1A). Thus, the 25-mer sRNA forming a duplex region resists cleavage by RNase E, resulting in preferred cleavage patterns in the unpaired region ranging from 26 to 47 nt. Based on these results, we suggest that RNase E can only cleave the preferred sites of single-stranded regions in the target RNA that are tethered by base-pairing with 5′-monophosphorylated sRNA.
The 5′-end status and pairing region of sRNA affect target RNA cleavage by RNase E
Subunit organization in RNase E is presumed to be required for the cooperative activities of 5′-P sensing and RNA cleavage (36). The RNA-cutting site and 5′-sensing site residing at each protomer constitute the principal dimers in the RNase E homotetramer (15). Thus, two RNA cleavage sites, located at each principal dimer, exist in the interlaced quaternary structure of RNase E. Based on this conjecture of RNase E structural organization, we hypothesized that the recognition of 5′-P in sRNA might simultaneously trigger the cleavage of target RNA in trans at two active sites created by the association of four subunits of RNase E catalytic domains. To test this hypothesis, we investigated the cleavage patterns of several dsRNA substrates by RNase E in the presence of different seed regions and 5′-P and 5′-OH forms in the sRNA (Fig. 3). First, sRNA with or without 5′-P was paired with 47-mer target RNA (R0) at the 5′-site, and the resulting duplex RNA substrates were subjected to RNase E cleavage reaction (Fig. 3A). Target RNA was readily cleaved at the 3′-tail site, which was close to the phosphorylated 5′-end of the sRNA in trans (lanes for substrates 1 and 3 in Fig. 3A). When the 5′-OH sRNA was paired with the 5′-phosphorylated target RNA, the target RNA was significantly cleaved at the 3′-tail site, although to a lesser extent (substrate 2 in Fig. 3A). This result shows that RNase E can cleave the 47-mer RNA with 5′-P when it is paired with 5′-OH sRNA, which results from cleavage at the sites that are 30 nt apart from the 5′-monophosphorylated end. Thus, the RNA cleavage site is likely separated from the 5′-P pocket site, which might reside at the other subunit of the principal dimer. In contrast, RNase E cannot cleave the 47-mer RNA with the 5′-OH form when it is paired with 5′-OH sRNA (Fig. S1), indicating that the presence of 5′-P is a prerequisite for target RNA cleavage by RNase E. Thus, we suggest that the recognition of 5′-P in sRNA in trans and/or target RNA in cis can trigger the cleavage of target RNA at two active sites created by the association of the four subunits of RNase E catalytic domains. One active site close to the 5′-P pocket site (i.e. the proximal site) might cleave the target RNA with higher efficiency than the other active site that is further from the 5′-P pocket site (i.e. the distal site), likely residing at the other subunit of the principal dimer.
Figure 3.
Cleavage of various partial duplex RNA substrates with different seed regions in sRNA (3′-overhang, 5′-overhang, and double overhang duplex RNAs). Various RNA substrates were reacted with RNase E: 3′-overhang dsRNA substrates (A), 5′-overhang dsRNA substrates (B), and double overhang dsRNA substrates (C). Reactions were performed by incubating 32P-labeled duplex RNA substrates (10 nm) in the presence or absence of RNase E (0.1 μm) at 37 °C for 30 min. Cleaved RNAs were simultaneously resolved on 15% native and denaturing (8 m urea) PAGE and visualized using a phosphorimager.
When the 5′-phosphorylated 47-mer target RNA (47R0) was paired with sRNA at the 3′-terminal region, RNase E cleaved the single-stranded 5′-tail of the target RNA (substrate 1 in Fig. 3B). RNA cleavage of 5′-monophosphorylated RNA generated fragments shorter than 10 nt. Because the 5′-phosphorylated target RNA that was paired with sRNA with 5′-OH was also readily cleaved to generate the 5′-phosphorylated RNA fragments (substrate 2 in Fig. 3B), the RNA cleavage observed was likely caused by the 5′-P in the target RNA rather than by the sRNA lacking 5′-P. RNase E also cleaved the 47-mer RNA with 5′-OH when the target RNA was paired with 5′-phosphorylated sRNA (substrate 3 in Fig. 3B), which resulted from cleavages at the 5′-tail of the target RNA by RNase E. The 5′-monophosphorylated sRNA was not degraded by RNase E, indicating that it indeed guides RNase E to the cleavage sites on the target RNA, which are separated from the 5′-end of the sRNA. Thus, this result suggests that sensing of 5′-P in sRNA and cleavage of target RNA in trans occur simultaneously at the two distant sites (i.e. the 5′-P–sensing pocket and the RNA cleavage site, respectively) that are present in the RNase E homotetramer.
We also evaluated whether sRNA can guide RNase E to the preferred cleavage sites on the target RNA when it is paired in the middle of the target RNA, leaving two tails at the 5′- and 3′-ends (Fig. 3C). RNase E cleaved the 5′-terminal region of 47-mer RNA (47R0) that was paired with sRNA, in which sRNA was not degraded, generating fragments of 5′-monophosphorylated RNA shorter than 10 nt (substrates 1 and 2 in Fig. 3C). Taken together, these results suggest that RNase E can degrade single-stranded regions of a target RNA that is paired with monophosphorylated sRNA at the proximal active sites close to the 5′-P pocket site as well as at the distal active sites apart from the 5′-P pocket site.
RNase E prefers to cleave RNA at the single-stranded region proximal to the 5′-P of guide oligonucleotides
Next, we investigated the catalytic efficiency of RNA cleavage by RNase E, depending on the presence of a 5′- or 3′-tail in the target RNA (Fig. 4A). Duplex RNA substrates were designed to allow RNA cleavage at the 5′- or 3′-tail region, which was stimulated by either 5′-P sRNA in trans (substrates 1 and 3 in Fig. 4A) or 5′-P end of the target RNA in cis (substrates 2 and 4 in Fig. 4A). When the duplex RNA contained 5′-phosphorylated target RNA, the 5′-end of the sRNA was unphosphorylated. When the duplex RNA substrates had a 3′-tail (substrates 1 and 2), RNA cleavage was readily observed at the single-stranded tail regions of the 47-mer target RNA (Fig. 4A). This result indicates that RNA cleavage was stimulated by either proximal 5′-P of sRNA or distal 5′-P of target RNA itself. Similarly, duplex RNA substrates having a 5′-tail were also significantly cleaved by RNase E at the single-stranded regions of the 47-mer RNA, which was stimulated by either the distal 5′-P of sRNA (substrate 3) or proximal 5′-P of target RNA itself (substrate 4).
Figure 4.
Degradation of target RNA by RNase E with 5′-monophosphorylated guide sRNA or sDNA. The RNase E reaction was performed using target RNA (47 nt, R0) annealed with 25-mer guide oligonucleotides: sRNA (A) or sDNA (C) (indicated with red lines). RNase E (0.1 μm) was incubated with 32P-labeled dsRNA or duplex RNA/DNA substrates (10 nm; asterisks represent 32P-labeled nucleotides) at 37 °C for 2, 4, 10, 20, 40, and 60 min. The reaction mixtures were resolved on 15% native PAGE and visualized using a phosphorimager. Kinetic analysis results of RNA cleavage product accumulation from A and C were plotted and fitted to an exponential function in B and D, respectively. Mean values of triplicate independent experiments and S.D. are shown.
RNA cleavage products accumulated as a function of time, and each cleavage product was quantified and fitted to an exponential function to compare the catalytic efficiency of RNA cleavage (Fig. 4B). The accumulation of the cleaved products as a function of time was affected by the position of the single-stranded RNA region. RNase E more preferentially cleaved the 3′-tailed duplex RNA than the 5′-tailed RNA substrates (circles versus triangles in Fig. 4B). In addition, RNA cleavage was more efficient at the single-stranded RNA region proximal to the 5′-P of either sRNA or target RNA than at the single-stranded RNA region distal from the 5′-P of sRNA and target RNA (i.e. substrates 2 and 3). Thus, we suggest that the 5′-P of sRNA and 5′-P of target RNA can stimulate RNA cleavage owing to the 5′-sensor near the active site and distant from the active site, respectively, which is located at the other dimer in the RNase E homotetramer.
Several RNase H family members have been reported to cleave single-stranded RNA of RNA-DNA hybrids (37). Because RNase E contains the RNase H domain, we determined whether the 5′-monophosphorylated small complementary DNA (i.e. sDNA) can guide RNase E to cleave a target RNA that was cognately paired with sDNA. Duplex RNA/DNA hybrid substrates with a 5′- or 3′-tail were designed that were exactly the same as the duplex RNA substrates (Fig. 4A), except that the sRNA was replaced with sDNA. When the duplex RNA-DNA hybrid substrates were subjected to RNA digestion with RNase E, RNA cleavage preference was observed (Fig. 4C and Fig. S2) that was similar to the results obtained with the duplex RNA substrates. The small guide DNA with 5′-P readily stimulated the cleavage of the RNA strand at both the proximal and distal sites (substrates 5 and 7 in Fig. 4C). When the target RNA was annealed to either sRNA or sDNA, the kinetics of RNA cleavage was similar irrespective of the nucleotides in the guide oligonucleotides (Fig. 4, B and D). Thus, the digestion of an RNA strand with RNase E only requires 5′-P in either RNA or DNA beyond seed pairing of target RNA.
Tetrameric RNase E favors RNA cleavage at both the proximal and distal 5′-P–sensing sites
Maintenance of the quaternary structure of RNase E has been reported to be important for the RNA cleavage activity (15). RNase E contains a conserved cysteine motif (residues 404 and 407) that coordinates the Zn2+ ion in the Zn2+-link (residues 404–411), which is shared by two protomers at the dimer interface. In addition, small domains are responsible for maintaining the tetrameric structure of RNase E by interlacing two principal dimers (15). To determine whether the quaternary structure of RNase E is required for RNA cleavage at the sites either proximal or distal to the 5′-P in the duplex RNA substrate, we prepared two mutant constructs of RNase E that could affect the formation of the tetrameric structure. We generated and purified truncated WT RNase E, named RNase EMT(499), in which a part of the small domain (residues 500–529) was removed, and a mutant RNase E, named RNase EMT(C/S), in which two cysteines at positions 404 and 407 were exchanged with Ser to block Zn2+ ion coordination. To characterize the oligomeric state of these proteins, we determined the molecular weight of the three RNase E proteins by using analytical size exclusion chromatography (Fig. 5A). We observed significant differences in the elution profiles between WT RNase E and the mutant proteins. RNase E showed a single major peak corresponding to the molecular weight of tetrameric RNase E, whereas the two mutant proteins showed a decreased tetrameric peak and an increased monomeric peak. As shown in the elution profile of RNase EMT(C/S), Zn2+ ion coordination was likely involved in the stabilization of the tetrameric structure of RNase E. In addition, the small domain was required to maintain the tetrameric structure of RNase E, because the RNase EMT(499) mutant showed fewer tetramers and enhanced accumulation of dimers and trimers. Consistent with the result obtained for size determination in RNase E, chemical cross-linking also showed a difference in multimerization of RNase E and RNase EMT(C/S) (Fig. S3A), in which cross-linking of RNase E subunits occurred regardless of the presence or absence of the RNA substrate. RNase E protein readily formed dimers and multimers, whereas RNase EMT(C/S) could efficiently form dimers, but not multimers.
Figure 5.
Quaternary structure analysis and RNA cleavage activities of RNase E and RNase E mutants. A, size-exclusion chromatography profiles showing molecular mass distributions of WT and mutant RNase E proteins: RNase E (solid line), RNase EMT(499) (thick dashed line), and RNase EMT(C/S) (thin dashed line). The molecular weight of proteins was calculated using a standard curve obtained using size control proteins (inset graph). B, cleavage of partial duplex RNA substrates containing proximal or distal phosphate (HO47R0/P25R1 and P47R0/HO25R1, respectively) with RNase E and RNase E mutants. RNase E reactions were performed by incubating 32P-labeled 3′-overhang RNA duplex substrates (10 nm) with RNase E proteins (0.1 μm) at 37 °C for various incubation times (0, 2, 5, 10, 30, and 60 min). The reaction mixtures were resolved on 15% native PAGE and visualized using a phosphorimager. Kinetic analysis results of RNA cleavage product accumulation were plotted and fitted to an exponential function.
Next, we investigated how the tetrameric assembly of RNase E is needed for the efficient cleavage of target RNAs, which is stimulated by either the proximal 5′-P of sRNA or the distal 5′-P of target RNA (Fig. 5B and Fig. S3B). WT RNase E cleaved target RNA equally efficiently at both the proximal and distal 5′-P constructs of duplex RNA substrates. However, RNase EMT(499) mutant showed decreased RNA cleavage of the duplex RNA substrate containing the distal 5′-P of target RNA, whereas RNA cleavage with the proximal 5′-P of sRNA was not affected. In contrast, the RNase EMT(C/S) mutant showed significantly diminished RNA cleavage activity at both the proximal and distal 5′-P constructs of duplex RNA substrates. Based on these results, we concluded that tetrameric RNase E facilitates RNA cleavage at both the proximal and distal 5′-P–sensing sites. Importantly, RNA cleavage at the distal active site likely requires an intact tetrameric structure composed of two interlaced principal dimers. In addition, Zn2+ ion coordination in the tetrameric RNase E structure is necessary for efficient RNA cleavage.
The 5′-phosphorylated guide RNA attenuates target gene expression via RNA degradation by RNase E in vivo
Next, we investigated whether guide RNA-stimulated target RNA cleavage by RNase E can decrease the expression of a target gene in E. coli. To this end, rne-deleted E. coli strain, which can be complemented by plasmid-borne rne gene under the control of an arabinose gene promoter, was transformed by two plasmids encoding RNase E and GFP as a target gene. A 5′-phosphorylated guide RNA that can pair with an RNA transcript of GFP with proximal site 5′-P was designed and prepared. When the 5′-P GFP target guide RNA was transfected into E. coli expressing GFP during incubation at 37 °C, the GFP expression was significantly decreased (Fig. 6A). When the 5′-phosphorylated RNA that does not target GFP mRNA was used in the transfection, GFP expression was not decreased, and GFP fluorescence intensity was similar to that of the negative control (i.e. no transfection). The GFP fluorescence signal was normalized with the bacterial mass during the incubation, because there was not any change in bacterial growth after sRNA transfection. After a 6-h incubation of E. coli expressing GFP with the sRNA, GFP fluorescence decreased by about 40%. Next, we designed and prepared 5′-phosphorylated guide RNA targeting GFP mRNA with distal site 5′-P. After a 6-h incubation of the E. coli transfected with the sRNA with a distal 5′-site (i.e. distal GFP target RNA), the GFP fluorescence was decreased to the same extent as when the proximal GFP target RNA was used (Fig. 6B).
Figure 6.
Suppression of target gene expression through target RNA decay with RNase E and 5′-monophosphorylated guide RNA in bacteria. A, time course analysis of GFP expression in E. coli cells transformed with GFP expression vector and transfected with GFP transcripts targeting or nontargeting (scrambled sequences irrespective of GFP transcript) sRNA. The GFP-transformed bacterial cells were transfected with 5 μm phosphorylated GFP target sRNA (p-GFP target sRNA) or nontarget control sRNA (p-nontarget sRNA) by electroporation. No transfection for negative control was included. The absorbance showing cell growth and GFP fluorescence was measured every 1 h, in which GFP fluorescence was normalized to cell growth absorbance. The resulting GFP fluorescence is shown in the graph as relative to the unity value at the starting point (time 0). Data are presented as the mean ± S.D. (error bars), n = 3. B, E. coli cells transformed with GFP expression vector were transfected with the GFP transcripts targeting phosphorylated (proximal or distal site) sRNAs or nontargeting sRNA by electroporation and further incubated for 6 h. GFP fluorescence was measured and represented as shown in A. C, levels of GFP mRNA in E. coli cells that were transfected with the GFP transcripts targeting phosphorylated (proximal or distal site) sRNA or nontargeting sRNA. Bacterial RNA was extracted and reverse-transcribed. Quantitative real-time PCR was performed for the quantification of GFP transcripts. D and E, time course analysis of GFP expression in the WT (D) or Hfq deletion (E) E. coli cells transformed with GFP expression vector and transfected with GFP transcripts targeting or nontargeting (scrambled sequences irrespective of GFP transcript) sRNAs. The experiment was conducted in the same manner as in A. F, levels of GFP mRNA in the WT and Hfq deletion E. coli cells that were transfected with the GFP transcripts targeting phosphorylated (proximal or distal site) sRNAs or nontargeting sRNA. The experiment was conducted in the same manner as in C. G, primer extension assay for the measurement of GFP mRNA. GFP mRNA synthesized in vitro (100 nm) was incubated with various concentrations (0, 0.1, 1, and 10 nm) of RNase E with or without GFP targeting (proximal) p-sRNA for 30 min. After the reaction, the GFP reverse primer DNAs annealed to GFP RNA were synthesized into complementary DNA using reverse transcriptase. Urea-PAGE and subsequent SYBR Gold staining were used to resolve the synthesized cDNA that reflects the amount of GFP mRNA remaining in the reaction. H, quantitative real-time PCR was used for monitoring the levels of the remaining target GFP RNA (100 nm) after RNA decay by RNase E. GFP mRNA synthesized in vitro (100 nm) was incubated with 200 nm targeting (proximal or distal site) sRNA or nontargeting sRNA. After the reaction, cDNA was synthesized and amplified for quantitative real-time PCR. I, RNA cleavage assay with long dsRNA (72- and 47-mer RNAs annealed). The partial duplex RNA (10 nm) was incubated with RNase E (100 nm) for 0, 2, 4, 10, 20, 40, and 60 min. For comparison, partial duplex RNA harboring 25 bp was also incubated with RNase E for 30 min as a positive control in the assay. RNA cleavage products were resolved on 15% native PAGE and visualized using a phosphorimager. Dashed line, spliced part of two gel images for comparison of RNA cleavage. Mean values of triplicate independent experiments and S.D. are shown. Statistical significance was determined using paired Student's t test (**, p < 0.01).
Next, we examined steady-state mRNA levels to test whether the decrease in GFP mRNA transcript is consistent with an increase in GFP mRNA transcript decay. Real-time quantitative PCR was performed using bacterial cDNA prepared from RNA from the E. coli used in the experiment (Fig. 6B), and decreased GFP mRNA expression was observed in both the proximal and distal 5′-sites of the guide RNA targeting the GFP mRNA (Fig. 6C). Thus, attenuated GFP gene expression with low GFP fluorescence resulted from the decreased steady-state GFP mRNA transcript level, suggesting enhanced decay by RNase E with sRNA in E. coli. Furthermore, when artificial guide DNA that is equivalent to the sRNA was introduced in E. coli, the target gene repression was similarly observed (Fig. S4).
To further examine the decrease of target gene expression with sRNA-guided target RNA cleavage by RNase E in an Hfq-independent manner, we prepared the WT E. coli strain MG1655 (i.e. WT) and Hfq-deletion type MG1655 strain (i.e. ΔHfq). Each E. coli strain was transformed with plasmid DNA encoding GFP, and guide RNA transfections were performed by using the same sRNA as in Fig. 6A. As a result, GFP expression was significantly reduced when the 5′-P GFP target guide RNA was transfected into E. coli expressing GFP in both strains (Fig. 6, D and E). When the 5′-phosphorylated RNA that is not targeting the GFP mRNA was used in the transfection, GFP expression was not decreased, and GFP fluorescence intensity was similar to the control group. We next investigated whether the decreased GFP expression in both WT and Hfq deletion type E. coli was indeed caused by the decay of the GFP mRNA transcript. The experiment was conducted in the same manner as in Fig. 6C. Decreased GFP mRNA expression was observed in both the proximal and distal 5′-sites of the guide RNA targeting the GFP mRNA (Fig. 6F). These results indicate that the decrease in target gene expression is a result of the quantitative decrease in target gene mRNA transcripts by 5′-P GFP target guide RNA combined with RNase E in an Hfq-independent manner.
To further confirm that the 5′-P guide RNA attenuates GFP expression through RNA degradation by RNase E in E. coli, the in vitro synthesized target GFP RNA was paired with the guide RNA and was subsequently incubated with RNase E. When the RNase E was incubated with the guide RNA and the GFP RNA transcript, RNA cleavage was readily observed, as revealed by the decrease in the remaining RNA measured by primer extension and quantitative RT-PCR (Fig. 6, G and H). In contrast, when the GFP target transcript without guide RNA was incubated with RNase E, RNA cleavage was not observed.
The crystal structure of the tetrameric catalytic domain of RNase E has been solved (15), and the length of the tetramer comprising the dimer–dimer complex is about 125 Å. Based on the structural model of RNase E organization, we hypothesized that RNA cleavage with sRNA having the distal 5′-P is likely to occur between the 5′-phosphate–sensing site at one dimer and the remote cleavage site at the other dimer. To test this hypothesis, we prepared a partial duplex RNA with a longer duplex region (Fig. 6I); 72-mer RNA annealed to 47-mer RNA with a dsRNA region of 47 bp, which is longer than the length of tetrameric RNase E. The duplex RNA harboring a longer duplex region did not show any indication of RNA degradation after 60 min of incubation with RNase E (Fig. 6I). In contrast, when the same RNA substrate was annealed with a shorter sRNA, resulting in a short duplex RNA region about 25 bp, it was completely degraded in 30 min (Fig. S5). Thus, we suggest that RNA decay is only feasible for the target RNA with an appropriate duplex length within the distance between the 5′-P–sensing site of one dimer and the cleavage site of another dimer.
Discussion
Our experimental results showed that target RNA degradation by RNase E does not necessarily require the activity of the RNA chaperone Hfq (Fig. 1B). We observed that 5′-monophosphorylated sRNA alone could guide recombinant RNase E to cleave a target RNA that was cognately seed-paired with the sRNA. When the duplex RNA substrate consisting of target RNA and cognate sRNA with 5′-P was incubated with RNase E without other accessory proteins, the target RNA was cleaved at the single-stranded A/G-rich region close to or far from the 5′-end of the sRNA. Thus, the 5′-monophosphorylated trans-acting sRNA was sufficient to stimulate RNase E–mediated cleavage of target RNA in the absence of Hfq RNA chaperone.
RNase E degrades cis- as well as trans-acting RNA with a monophosphorylated 5′-end and readily cleaves ssRNA with 5′-P regardless of substrate RNA length (Fig. 1). When ssRNA containing a triphosphate or hydroxyl group at the 5′-end or blunt-ended duplex RNA was incubated with the enzyme, no RNA cleavage was observed. These results underline that RNase E selectively degrades single-stranded RNA with 5′-P, as is known from previous studies. Further, the RNase E catalytic activity for RNA degradation was not detected in the absence of Mg2+ ions and required neither ATP nor ATP hydrolysis energy (Fig. 1).
The RNA cleavage activation occurring via the sensing of the 5′-P in sRNA by RNase E is likely to trigger a conformational change in the enzyme, resulting in the cleavage of the target RNA within 10-mer from the 5′-P (10). Thus, a certain distance exists between the sensing pocket for the 5′-end and the RNA cleavage site in the quaternary structure of RNase E. We observed that RNase E cuts short-stranded AU-rich regions of ssRNA apart from the 5′-monophosphate by 25-mer or more as well as those within 10-mer from the 5′-P (Fig. 4A). These results suggest that the 5′-P of sRNA is recognized by one dimer of the tetramer structure of RNase E, and subsequent cleavage of RNA is likely to occur at the active site present in the other dimer that is distant from the 5′-end sensing pocket. Conversely, RNase EMT(499) and RNase EMT(C/S), RNase E mutant proteins with an incomplete tetramer structure, showed a marked decrease in cleavage activity of distal 5′-P substrates compared with that of the WT RNase E (Fig. 5B). Hence, the intact tetrameric structure of RNase E is a prerequisite for RNA cleavage of the target RNA annealed with sRNA having a distal 5′-P. However, RNase E did not cleave a partial duplex RNA with a long dsRNA region that is longer than the tetrameric RNase E (Fig. 6I). Thus, the decay of target RNA occurs at the single-stranded region present either near to or away from the RNA cleavage site residing in one dimer, in which the distance between the 5′-P and ssRNA cleavage site in the target RNA should not be longer than that between the 5′-P–sensing pocket and the RNA decay active site in RNase E.
To assess the sRNA-mediated translational control at the 5′-region of a given target mRNA in vivo, we used the GFP plasmid (Fig. 6), as previously used for the depression of target gene expression by RNase E (39). Urban and Vogel (39) introduced the plasmid encoding sRNA for suppressing GFP-tagged target gene expression in E. coli cells and showed the decreased expression level of GFP, but not the translational control. In our system, we directly transfected sRNA targeting GFP mRNA and examined both protein and mRNA levels to explore how RNase E regulates a target mRNA in vivo. We found that the amount of GFP fluorescence and GFP mRNA decreased only when the sRNA (proximal or distal P) targeting GFP mRNA was transfected. In addition, we observed that the amount of GFP fluorescence and GFP mRNA was diminished by transfection of sRNA in E. coli, and this change in fluorescence was independent of the presence of Hfq (Fig. 6). To our knowledge, this is the first attempt to suppress target gene expression in bacteria by the administration of artificial RNA. More importantly, designing the artificial guide RNA for suppressing target gene expression in bacteria is very simple. The guide RNA requires only 5′-P and cognate sequences for target RNA binding. In addition, sRNA can be alternatively substituted with DNA for target RNA decay, as shown in our study (Fig. 4B). Thus, this strategy can be used for selective knockdown of bacterial genes in vivo.
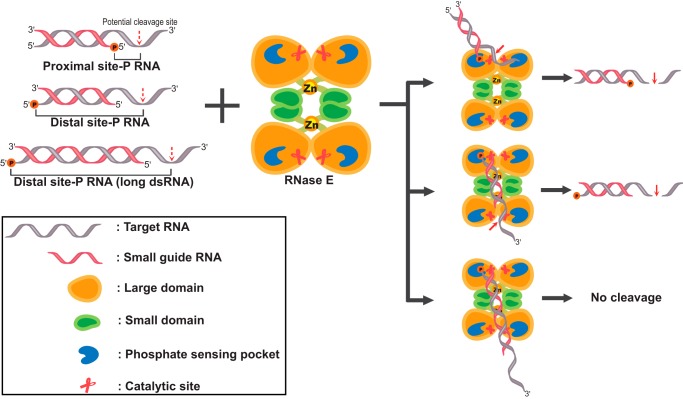
Based on the results obtained in this study, we suggest a pathway in which RNase E having a quaternary structure efficiently degrades target RNA annealed with trans-acting sRNA harboring 5′-monophosphorylated ends both proximal and distal from the cleavage site (Fig. 7). In the model, the duplex formed between trans-acting sRNA and target RNA directs the 5′-end of a trans-acting RNA into the 5′-sensing pocket, allowing the cleavable sites in the cognate single-stranded RNA formed in the duplex to be accommodated in the enzyme active site. In the dimers of RNase E, the two large domains constitute two separate enzyme active sites that interact with RNA substrates by recognizing the 5′-P of the RNA substrate and subsequently cleaving the AU-rich sequences of target RNA (15). Thus, the 5′-P–sensing pocket is necessary to increase the affinity of RNase E for RNA substrates and plays a role in distinguishing monophosphorylated RNA from triphosphorylated RNA (40, 41). A detailed structural study of this complex with a bound sRNA/mRNA pair at the atomic level would be warranted to elucidate our suggested pathway of RNA cleavage by RNase E.
Figure 7.
Models for RNase E cleavage with sRNA without the Hfq RNA chaperone. The quaternary structure of RNase E is assumed to be an intact dimer–dimer complex. RNase E can recognize and degrade target (partial duplex) RNA via the 5′-phosphate–sensing pocket and the RNA cleavage site in RNase E. When target RNA is base-paired with the proximal 5′-P sRNA, both the single-stranded RNA cleavage site and the 5′-phosphate–sensing pocket reside on the same dimer, enabling RNase E to cleave the target RNA near the 5′-P of sRNA. In the case of the distal 5′-P sRNA, target RNA base-paired with the distal 5′-P sRNA was recognized and degraded in separate dimeric sites. In contrast, RNA cleavage is not possible for target RNA with a longer duplex length exceeding the distance between the 5′-P–sensing site of a dimer and cleavage site of another dimer. The cleavage of RNA can occur both near to and distal from the 5′-P in RNA, depending on the alignment of the RNA cleavage site and the 5′-phosphate–sensing pocket on RNase E.
Our results suggest that mRNA paired with sRNA is sufficient for triggering target mRNA cleavage by RNase E without requiring an Hfq chaperone. We explored sRNA-induced target RNA degradation by RNase E in the absence of the Hfq RNA chaperone. The 5′-monophosphorylated blunt end of sRNA/target RNA duplex could trigger RNase E to cleave target RNA at the single-stranded region at distal sites. In the absence of the RNA chaperone Hfq protein, RNase E with an intact quaternary structure can cleave target RNA at sites both proximal and distal to the monophosphorylated 5′-end of sRNA that are base-paired with the target RNA (Fig. 7). Thus, we suggest that Hfq-independent RNA decay by RNase E requires these factors for efficient target RNA decay in bacteria: the 5′-P in sRNA, cognate sRNA sequence for annealing to target RNA, and a stable quaternary structure of RNase E. Given that sRNA can be designed to facilitate seed pairing, small guide oligonucleotides can be further explored for their application in selective gene knockdown in bacteria in vivo.
Experimental procedures
Purification of RNase E proteins and RNA chaperone Hfq
The rne-deleted E. coli strain KSL2000 and a previously described pNRNE4 plasmid (42) were used without further modification. The WT NTH of E. coli RNase E (residues 1–529, RNase E), mutant RNase E 499 aa (residues 1–499, RNase EMT(499)), and mutant C404S and C407S (residues 1–529, RNase EMT(C/S)), all of which contain a His6 tag at the N terminus, were expressed in BL21 (DE3) cells. Each strain was grown in Luria–Bertani (LB) medium (2.0 liters), supplemented with 100 μg/ml ampicillin, 100 μg/ml kanamycin, 5 μg/ml chloramphenicol, 5 μg/ml tetracycline, and 10 μm IPTG. Protein expression was induced by the addition of 0.1 mm IPTG, and cells were grown at 37 °C to an A600 of 0.5–0.6. After harvesting, cell pellets were resuspended in lysis buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 10 mm imidazole) supplemented with 1 mg/ml lysozyme and 10 mg/ml RNase A, and then the resuspended cells were disrupted on ice by sonication. The lysate was clarified by centrifugation, and the soluble fraction was loaded onto a nickel-nitrilotriacetic acid HisTrap column (GE Healthcare), washed extensively with washing buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 20 mm imidazole), and eluted with a gradient of elution buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, and 250 mm imidazole). The eluants were further purified using size-exclusion column chromatography (Superdex 200 column, Amersham Biosciences). Purified protein samples were concentrated with centrifuge-based ultrafiltration (Amicon® Ultra centrifugal filters, molecular weight cutoff 30,000; Merck).
The plasmid pGSO146 (43) encoding Hfq RNA chaperone protein was kindly provided by Dr. Yong-Hoon Lee (Korea Advanced Institute of Science and Technology). RNA chaperone Hfq was overexpressed in E. coli BL21 (DE3) cells harboring pGSO146 plasmid by growing the cells in LB medium (1.0 liter) supplemented with 100 μg/ml ampicillin. The overexpressed Hfq protein was purified using a nickel-nitrilotriacetic acid HisTrap column as described previously (44). Purified protein samples were concentrated using pressure-based ultrafiltration (Amicon® stirred cells, Merck). All purified proteins were >95% pure based on SDS-PAGE results. Proteins were aliquoted and mixed with 50% glycerol for storage at −20 °C. Protein concentration was determined using the Bradford method (Bio-Rad protein assay) with BSA as the standard.
Site-directed mutagenesis
The cysteine residues at positions 404 and 407 in recombinant RNase E were changed to serine residues using a Muta-DirectTM site-directed mutagenesis kit (iNtRON Biotechnology) with gene-specific primers, according to the manufacturer's protocol. The oligonucleotides used were 5′-CCA GTC ATC ACG TTA GTC CGC GTA GTT CTG GTA CTG GC-3′ and its reverse complement 5′-GCC AGT ACC AGA ACT ACG CGG ACT AAC GTG ATG ACT GG-3′ (changes introduced into serine codons are underlined). These mutations in the corresponding expression construct pNRNE4 were confirmed using DNA sequencing (Macrogen).
Oligonucleotide preparation and 32P labeling
Sequences of oligonucleotide substrates used for RNA cleavage by RNase E are shown in Table 1. Oligoribonucleotides for RNA cleavage enzymatic assays were chemically synthesized (ST Pharm Co.). For 5′-end labeling with 32P, RNA or DNA oligonucleotides were incubated with T4 polynucleotide kinase (10 units; Takara) and 1 μl of [γ-32P]ATP (3,000 Ci/mmol; GE Healthcare) in 50 μl of reaction buffer containing 50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, and 5 mm DTT at 37 °C for 1 h. The labeled RNA strands were subsequently purified using Micro Bio-SpinTM columns (Bio-Rad). Internally radiolabeled RNA was produced by performing in vitro transcription in the presence of [α-32P]UTP (3,000 Ci/mmol; GE Healthcare) according to a standard protocol. Internally 32P-incorporated RNAs were purified on 10% polyacrylamide gel with 8 m urea. RNA duplexes were prepared by annealing two RNA oligonucleotides as follows. RNA or DNA oligonucleotides were mixed with annealing buffer (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 1 mm EDTA), boiled at 85 °C for 5 min, and then annealed at 25 °C for 20 min.
Table 1.
Sequences of single-stranded oligonucleotides used in this study
| No. | Oligonucleotide name | Sequence (5′ → 3′) |
|---|---|---|
| 1 | R0/47 | GGAAAAUGUAAAUGACAUAGGCGCGCUGAAAGGGAGAAGUGAAAGUG |
| 2 | cR0/47 | CACUUUCACUUCUCCCUUUCAGCGCGCCUAUGUCAUUUACAUUUUCC |
| 3 | R1/25 | CGCGCCUAUGUCAUUUACAUUUUCC |
| 4 | R2/25 | CACUUUCACUUCUCCCUUUCAGCGC |
| 5 | R3/25 | CUCCCUUUCAGCGCGCCUAUGUCAU |
| 6 | D1/25 | CGCGCCTATGTCATTTACATTTTCC |
| 7 | D2/25 | CACTTTCACTTCTCCCTTTCAGCGC |
| 8 | GFP target sRNA (proximal) | GGCUACGUCCAGGAGCGCACC |
| 9 | GFP target sRNA (distal) | UGAAGUUCGAGGGCGACACCC |
| 10 | GFP target sDNA (proximal) | GGCTACGTCCAGGAGCGCACC |
| 11 | GFP target sDNA (distal) | TGAAGTTCGAGGGCGACACCC |
| 12 | RR0/72 | GGAAAAUGUAAAUGACAUAGGCGCGCUGAAAGGGAGAAGUGAAAGUGGGAAAAUGUAAAUGACAUAGGCGCG |
| 13 | RR1/65 | GGAAAAUGUAAAUGACAUAGGCGCGCUGAAAGGGAGAAGUGAAAGUGGGAAAAUGUAAAUGACAU |
| 14 | RNase E C404S/C407S forward primer | CCAGTCATCACGTTAGTCCGCGTAGTTCTGGTACTGGC |
| 15 | RNase E C404S/C407S reverse primer | GCCAGTACCAGAACTACGCGGACTAACGTGATGACTGG |
| 16 | GFP forward primer | AAGTGCCACCTGACGTCTAA |
| 17 | GFP reverse primer | CACAACATACGAGCCGGAAG |
| 18 | RNase E forward primer | TCCAGCGTGAAGTCCGTCTG |
| 19 | RNase E reverse primer | GTCCTGACGAGTTTCAATGG |
RNA cleavage assays
RNA cleavage assays were performed in the reaction buffer (5 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm KCl, 2 mm MgCl2, 0.2 mm DTT, and 0.2 units/μl RNase inhibitor) containing each RNA substrate (duplex or single-stranded RNA). For the RNA cleavage reaction, 10 nm RNA substrate mixed with 20 nm guide sRNA, in which RNAs were mixed with a trace amount of 32P-labeled RNA (substrate RNA and/or guide RNA), was incubated with 0.1 μm RNase E per 10-μl reaction at 37 °C for 30 min. The time course reactions were stopped with quenching buffer (0.5 mg/ml protease K in 100 mm Tris-HCl, pH 7.5, 12.5 mm EDTA, 150 mm NaCl, 0.05% bromphenol blue, 0.05% xylene cyanol FF, and 10% glycerol), and the quenched reaction mixture was loaded on a native polyacrylamide gel. For denaturing PAGE analysis, reaction quenching was performed by adding RNA-loading dye (8 m urea, 3% Ficoll 400, 0.05% bromphenol blue, and 0.05% xylene cyanol FF) in TBE buffer to the reaction mixture, and whole samples were loaded on polyacrylamide gels with 8 m urea after denaturation at 95 °C for 3 min. Radioactive RNAs in gel were visualized and quantified on a Cyclone phosphorimager (PerkinElmer Life Sciences).
Sequence analysis of cleaved RNA
The site of RNA cleavage by RNase E was identified by sequence analysis of the cleaved RNA. The 32P-labeled dsRNA substrate (30 nm, P47R0/P25R1) in the reaction buffer (50 μl) was incubated with or without RNase E (0.25 μm) at 37 °C for 30 min. The alkaline hydrolysis and RNase T1 (Ambion) cleavage assays were performed as a control for RNA-Seq. For this, 5′-end 32P-labeled single-stranded RNA (30 nm, P47R0) was mixed with alkaline hydrolysis buffer at 25 °C for 5 or 10 min. RNase T1 (1, 5, or 10 units) was incubated with P47R0 RNA at 25 °C for 15 min. All reactions were quenched with an equal volume of RNA loading dye in TBE buffer, and RNA sequences were analyzed on 15% denaturing polyacrylamide gel (8 m urea) and visualized using a phosphorimager.
Analytical size-exclusion chromatography
The oligomeric state of proteins was determined by conducting size-exclusion chromatography. Purified RNase E, RNase EMT(C/S), and RNase EMT(499) were dialyzed with gel-filtration running buffer (25 mm Tris-HCl, pH 8.0, and 300 mm NaCl) at 4 °C overnight. The proteins (typically 300 μg) were subjected to analytical size-exclusion chromatography (Superdex 200 10/300 GL; GE Healthcare). The column was equilibrated with running buffer and calibrated using a molecular weight marker kit for gel-filtration chromatography (Bio-Rad). The column was run with the same running buffer, and 0.3-ml fractions were subsequently collected and analyzed using spectrophotometry at 260-nm absorbance.
Transcription suppression by using guide sRNA in bacteria
The pNRNE4 plasmid encoding RNase E and the pUCBB-eGFP plasmid were sequentially introduced into rne-deleted E. coli strain KSL2000. The E. coli K-12 MG1655 strain and the Hfq mutant strain were kindly provided by Dr. Y. Lee (KAIST; Korea Advanced Institute of Science and Technology). The pUCBB-eGFP plasmid was introduced into these two strains. The transformed cells were further transfected with 5 μm phosphorylated GFP target sRNA (with proximal or distal site) or phosphorylated GFP nontarget control sRNA using an electroporation method (38) and Gene Pulser XcellTM electroporation systems (Bio-Rad). The sRNA-transfected cells were grown in LB medium supplemented with 100 μg/ml ampicillin, 100 μg/ml kanamycin, 5 μg/ml chloramphenicol, 5 μg/ml tetracycline, and 10 μm IPTG at 37 °C. Untransfected cells were also incubated as a negative control. Cell growth was monitored by measuring absorbance at 595 nm, and the GFP fluorescence was measured using a spectrofluorometer (excitation wavelength of 485 nm and emission wavelength of 535 nm; VICTOR X3 Multilabel Plate Reader, PerkinElmer Life Sciences) every hour. The GFP fluorescence, which was normalized to cell growth absorbance, was shown in the graph as relative to the unity value at the starting point (time 0).
Total RNA was extracted from E. coli using TRIzol (Invitrogen) according to the manufacturer's protocol at 6 h after the transfection of small guide RNA. Total RNA (1 μg) was reverse-transcribed by using a PrimeScript first strand cDNA synthesis kit (Takara). PCR amplification of cDNA was performed using 500 nm specific primers and a Rotor-Gene SYBR® Green PCR kit (Qiagen). Cycling conditions were 95 °C for 10 s and 60 °C for 45 s, and real-time amplified SYBR Green signals were observed using Rotor-Gene Q (Qiagen). The mRNA levels of GFP were normalized with those of RNase E. Data are presented as the mean ± S.D., n = 3.
Chemical cross-linking assay
The chemical cross-linking reagent dimethylsuberimidate (DMS) was used to investigate the multimerization of RNase E. DMS was dissolved in chilled triethanolamine (TEA/HCl; 0.15 m, pH 8.2) to a concentration of 5 mg/ml, and pH was adjusted to 8.2. The RNase E (2, 5, and 10 μm) was preincubated with 50 mm TEA buffer containing 50 mm NaCl and 5 mm EDTA for 10 min at 25 °C, and cross-linking was initiated by adding DMS (10 mg/ml). After incubation for 3 h at 25 °C, the cross-linking reaction was quenched by adding an equal volume of 1 m glycine. The quenched samples were analyzed using 4–12% gradient SDS-PAGE and stained with Coomassie Brilliant Blue.
Author contributions
Y. M. B. and D.-E. K. conceptualization; Y. M. B., K.-J. J., S. Y., and A. B. data curation; Y. M. B., H. L., S. Y., and D.-E. K. formal analysis; Y. M. B. and D.-E. K. validation; Y. M. B. and D.-E. K. methodology; Y. M. B. writing-original draft; Y. M. B. and D.-E. K. writing-review and editing; K. L. and D.-E. K. resources; D.-E. K. supervision; D.-E. K. funding acquisition; D.-E. K. investigation; D.-E. K. project administration.
Supplementary Material
This work was supported by research funds of Konkuk University in 2018. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- NTH
- N-terminal half
- CTH
- C-terminal half
- PNPase
- polynucleotide phosphorylase
- RppH
- 5′-pyrophosphohydrolase
- sRNA
- small guide RNA
- ssRNA
- single-stranded RNA
- nt
- nucleotides
- LB
- Luria–Bertani
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- DMS
- dimethylsuberimidate
- TEA
- triethanolamine
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1. Chen H., Shiroguchi K., Ge H., and Xie X. S. (2015) Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11, 808 10.15252/msb.20159000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kushner S. R. (2002) mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184, 4658–4665 10.1128/jb.184.17.4658-4665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deutscher M. P. (2003) Degradation of stable RNA in bacteria. J. Biol. Chem. 278, 45041–45044 10.1074/jbc.R300031200 [DOI] [PubMed] [Google Scholar]
- 4. Carpousis A. J. (2007) The RNA degradosome of Escherichia coli: an mRNA degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87 10.1146/annurev.micro.61.080706.093440 [DOI] [PubMed] [Google Scholar]
- 5. Coburn G. A., Miao X., Briant D. J., and Mackie G. A. (1999) Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 13, 2594–2603 10.1101/gad.13.19.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z., Pandit S., and Deutscher M. P. (1999) RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18, 2878–2885 10.1093/emboj/18.10.2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caruthers J. M., Feng Y., McKay D. B., and Cohen S. N. (2006) Retention of core catalytic functions by a conserved minimal ribonuclease E peptide that lacks the domain required for tetramer formation. J. Biol. Chem. 281, 27046–27051 10.1074/jbc.M602467200 [DOI] [PubMed] [Google Scholar]
- 8. Tsai Y. C., Du D., Domínguez-Malfavón L., Dimastrogiovanni D., Cross J., Callaghan A. J., García-Mena J., and Luisi B. F. (2012) Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res. 40, 10417–10431 10.1093/nar/gks739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDowall K. J., and Cohen S. N. (1996) The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255, 349–355 10.1006/jmbi.1996.0027 [DOI] [PubMed] [Google Scholar]
- 10. Bandyra K. J., Said N., Pfeiffer V., Górna M. W., Vogel J., and Luisi B. F. (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 47, 943–953 10.1016/j.molcel.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miczak A., Kaberdin V. R., Wei C. L., and Lin-Chao S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U.S.A. 93, 3865–3869 10.1073/pnas.93.9.3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leroy A., Vanzo N. F., Sousa S., Dreyfus M., and Carpousis A. J. (2002) Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 45, 1231–1243 10.1046/j.1365-2958.2002.03104.x [DOI] [PubMed] [Google Scholar]
- 13. Morita T., Kawamoto H., Mizota T., Inada T., and Aiba H. (2004) Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54, 1063–1075 10.1111/j.1365-2958.2004.04329.x [DOI] [PubMed] [Google Scholar]
- 14. Callaghan A. J., Grossmann J. G., Redko Y. U., Ilag L. L., Moncrieffe M. C., Symmons M. F., Robinson C. V., McDowall K. J., and Luisi B. F. (2003) Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry 42, 13848–13855 10.1021/bi0351099 [DOI] [PubMed] [Google Scholar]
- 15. Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., and Luisi B. F. (2005) Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191 10.1038/nature04084 [DOI] [PubMed] [Google Scholar]
- 16. Kaberdin V. R., Walsh A. P., Jakobsen T., McDowall K. J., and von Gabain A. (2000) Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli ribonuclease E. J. Mol. Biol. 301, 257–264 10.1006/jmbi.2000.3962 [DOI] [PubMed] [Google Scholar]
- 17. Chao Y., Li L., Girodat D., Förstner K. U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H. J., Luisi B. F., and Vogel J. (2017) In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65, 39–51 10.1016/j.molcel.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mackie G. A. (2013) RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11, 45–57 10.1038/nrmicro2930 [DOI] [PubMed] [Google Scholar]
- 19. McDowall K. J., Kaberdin V. R., Wu S. W., Cohen S. N., and Lin-Chao S. (1995) Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374, 287–290 10.1038/374287a0 [DOI] [PubMed] [Google Scholar]
- 20. Celesnik H., Deana A., and Belasco J. G. (2007) Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27, 79–90 10.1016/j.molcel.2007.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richards J., Luciano D. J., and Belasco J. G. (2012) Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol. Microbiol. 86, 1063–1072 10.1111/mmi.12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Desnoyers G., Bouchard M. P., and Massé E. (2013) New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 29, 92–98 10.1016/j.tig.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 23. Waters L. S., and Storz G. (2009) Regulatory RNAs in bacteria. Cell 136, 615–628 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morita T., Maki K., and Aiba H. (2005) RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186 10.1101/gad.1330405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soper T., Mandin P., Majdalani N., Gottesman S., and Woodson S. A. (2010) Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 107, 9602–9607 10.1073/pnas.1004435107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Göpel Y., Khan M. A., and Görke B. (2016) Domain swapping between homologous bacterial small RNAs dissects processing and Hfq binding determinants and uncovers an aptamer for conditional RNase E cleavage. Nucleic Acids Res. 44, 824–837 10.1093/nar/gkv1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikeda Y., Yagi M., Morita T., and Aiba H. (2011) Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 79, 419–432 10.1111/j.1365-2958.2010.07454.x [DOI] [PubMed] [Google Scholar]
- 29. Prévost K., Desnoyers G., Jacques J. F., Lavoie F., and Massé E. (2011) Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 25, 385–396 10.1101/gad.2001711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Afonyushkin T., Vecerek B., Moll I., Bläsi U., and Kaberdin V. R. (2005) Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33, 1678–1689 10.1093/nar/gki313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kime L., Jourdan S. S., Stead J. A., Hidalgo-Sastre A., and McDowall K. J. (2010) Rapid cleavage of RNA by RNase E in the absence of 5′ monophosphate stimulation. Mol. Microbiol. 76, 590–604 10.1111/j.1365-2958.2009.06935.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeiffer V., Papenfort K., Lucchini S., Hinton J. C., and Vogel J. (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16, 840–846 10.1038/nsmb.1631 [DOI] [PubMed] [Google Scholar]
- 33. Garrey S. M., Blech M., Riffell J. L., Hankins J. S., Stickney L. M., Diver M., Hsu Y. H., Kunanithy V., and Mackie G. A. (2009) Substrate binding and active site residues in RNases E and G: role of the 5′-sensor. J. Biol. Chem. 284, 31843–31850 10.1074/jbc.M109.063263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bandyra K. J., Wandzik J. M., and Luisi B. F. (2018) Substrate recognition and autoinhibition in the central ribonuclease RNase E. Mol. Cell 72, 275–285.e4 10.1016/j.molcel.2018.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McDowall K. J., Lin-Chao S., and Cohen S. N. (1994) A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269, 10790–10796 [PubMed] [Google Scholar]
- 36. Bouvet P., and Belasco J. G. (1992) Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360, 488–491, 10.1038/360488a0 [DOI] [PubMed] [Google Scholar]
- 37. Pileur F., Toulme J. J., and Cazenave C. (2000) Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA-RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res. 28, 3674–3683 10.1093/nar/28.18.3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller E. M., and Nickoloff J. A. (1995) Escherichia coli electrotransformation. Methods Mol. Biol. 47, 105–113 10.1385/0-89603-310-4:105 [DOI] [PubMed] [Google Scholar]
- 39. Urban J. H., and Vogel J. (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35, 1018–1037 10.1093/nar/gkl1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang X., and Belasco J. G. (2004) Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc. Natl. Acad. Sci. U.S.A. 101, 9211–9216 10.1073/pnas.0401382101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mackie G. A. (2000) Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 275, 25069–25072 10.1074/jbc.C000363200 [DOI] [PubMed] [Google Scholar]
- 42. Kim D., Song S., Lee M., Go H., Shin E., Yeom J. H., Ha N. C., Lee K., and Kim Y. H. (2014) Modulation of RNase E activity by alternative RNA binding sites. PLoS One 9, e90610 10.1371/journal.pone.0090610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang A., Wassarman K. M., Ortega J., Steven A. C., and Storz G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9, 11–22 10.1016/S1097-2765(01)00437-3 [DOI] [PubMed] [Google Scholar]
- 44. Santiago-Frangos A., Kavita K., Schu D. J., Gottesman S., and Woodson S. A. (2016) C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 113, E6089–E6096 10.1073/pnas.1613053113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.