Abstract
Ribonucleotide reductase (RNR) catalyzes the first committed reaction in DNA synthesis. Most of what we know about RNR regulation comes from studies with cultured cells and with purified proteins. In this study, Tran et al. use Cre-Lox technology to inactivate RNR large subunit expression in heart and skeletal muscle of mouse embryos. Analysis of these mutants paints a picture of dNTP regulation in whole animals quite different from that seen in studies of purified proteins and cultured cells.
Introduction
Several years ago, I entitled a preview article comparable with this one “The most interesting enzyme in the world” (1). That enzyme—ribonucleotide reductase (RNR)2—is of great interest because of its novel structure and regulation, its metabolic role in catalyzing the first reaction committed to DNA synthesis, its status as a potential and actual cancer drug target (2, 3), the long-range electron transfer involved in its mechanism, and its function in controlling DNA precursor pools (3). Indeed, the only other way to alter these pools is through nucleotide salvage pathways, but how de novo and salvage pathways work in tandem is poorly understood. This is partly because most of our understanding of this enzyme comes from analysis of purified proteins and cultured cells. In the accompanying paper (4), Tran et al. now expand these horizons in their analysis of the function of RNR in whole animals. The data, perhaps surprisingly, demonstrate that dNTP salvage pathways are insufficient to support rapidly proliferating cells and that nucleoside triphosphate pools in the cells of living animals, both ribo- and deoxyribo-, are quite different from those previously thought—highlighting the importance of studying metabolic regulation at whole-organism, not just cellular, levels.
RNR is the gatekeeper for de novo biosynthesis of dNTPs in animals. The pathway involves reduction of ribonucleoside diphosphates (rNDPs) to deoxyribonucleoside diphosphates (dNDPs) by RNR (see Fig. 1 and Fig. 1A of Ref. 4), followed by phosphorylation. Alternatively, deoxyribonucleosides and nucleobases, derived from DNA breakdown or transport from other tissues, can be converted to dNTPs by salvage pathways.
Figure 1.
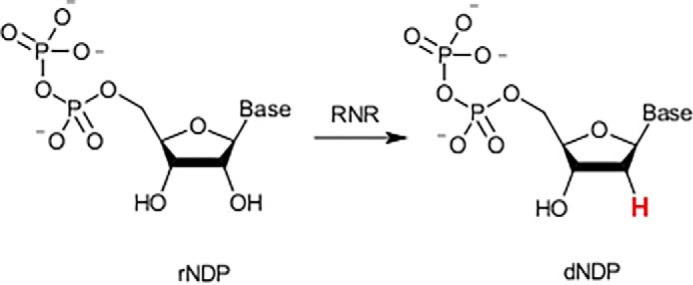
Reduction of ribonucleoside diphosphates to deoxyribonucleoside diphosphates (dNDPs) by RNR.
What Tran et al. have done is to use Cre-Lox technology to inactivate RNR large subunit expression specifically in heart and skeletal muscle of mouse embryos. Analysis of nucleoside triphosphate pools in these animals (dNTPs and rNTPs) allowed understanding of the regulation of nucleotide synthesis in living animals. Analysis of post-birth survival provided understanding of the ability of salvage pathways and import from other tissues to compensate for the loss of the main de novo route for dNTP synthesis. These latter studies involved extensive cell ultrastructural and physiological analysis.
The Cre-Lox technology used by Tran et al. inactivated synthesis of the large (R1) subunit late in embryonic development, leading to progressive loss of active de novo dNTP synthesis—specifically in heart and skeletal muscle. Postnatal survival was about 2 weeks, during which time the investigators observed multiple cell structural and physiological abnormalities in heart and skeletal muscle. The effects of R1 knockout were more dramatic in heart than in skeletal muscle. However, in either tissue, the de novo pathway cannot be bypassed during this period of extensive cell division and DNA synthesis.
From a biochemist's standpoint, perhaps the results of greatest interest pertain to the consequences of RNR R1 inactivation on nucleotide and nucleic acid metabolism, and here is where the study yielded some surprises. Extensive analysis of cell culture systems shows dGTP to be by far the least abundant of the four dNTPs in cell extracts—to the point that some have speculated that intracellular dGTP levels may be rate-limiting for DNA replication. However, in all organs analyzed in this study, dGTP was by far the most abundant dNTP—more than double the sum of the pools of dATP, dCTP, and dTTP. Quite aside from the central theme of this paper, these results indicate that studies on the effects of antimetabolites—whether nucleotide antagonists or inhibitors of other pathways—need to focus on their effects on intact animals as well as on cell culture systems. From a personal standpoint, these results were of great interest to me because my laboratory had reported dGTP levels to be exceptionally high in rat liver mitochondria (5), although the biological significance of this finding—either in cells or mitochondria—remains unknown.
Analysis of ribonucleoside triphosphates yielded a similar picture, with GTP being more abundant than CTP or UTP, trailing only ATP. The authors cogently discuss processes that involve guanine nucleotides other than as nucleic acid precursors, and that might explain the unexpected abundance of guanine ribo- and deoxyribonucleotides in living cells. Again, the results of these experiments demand caution in interpreting data from cell culture systems.
An impressive feature of this paper is the broad range of experimental techniques used—from electrophysiology to cell ultrastructure to metabolite analysis—and the care with which a complex manuscript has been assembled. The paper could serve as a model for creating other papers involving multiple techniques and approaches—from designing multipanel figures to deciding which data belong in the paper itself and which in a supplement. As papers involve more multiple-author teams and increased numbers of experimental approaches, this paper could serve as a model for constructing a manuscript using the multiple experimental approaches of today—even for those with limited interest in nucleotide metabolism.
Finally, the results of this study provide a challenge to investigators developing any metabolic antagonist for possible therapeutic purposes. It may be an obvious point, but whether a target enzyme is inactivated genetically, as in this study, or pharmaceutically, as with an enzyme inhibitor or prodrug (e.g. see Ref. 2), the consequences, in terms of interorgan metabolism, may be complex and unexpected. Multiple experimental approaches are needed to understand these effects and to use the antagonist effectively.
The author declares that he has no conflicts of interest with the contents of this article.
- RNR
- ribonucleotide reductase.
References
- 1. Mathews C. K. (2016) The most interesting enzyme in the world. Structure 24, 843–844 10.1016/j.str.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 2. Aye Y., Li M., Long M. J., and Weiss R. S. (2015) Ribonucleotide reductase and cancer: Biological mechanisms and target therapies. Oncogene 34, 2011–2021 10.1038/onc.2014.155 [DOI] [PubMed] [Google Scholar]
- 3. Nordlund P., and Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 10.1146/annurev.biochem.75.103004.142443 [DOI] [PubMed] [Google Scholar]
- 4. Tran P., Warnrooij P. H., Lorenzon P., Sharma S., Thelander L., Nilsson A. K., Olofsson A.-K., Mendini P., von Hofsten J., Stål P., and Chabes A. (2019) De novo dNTP production is essential for postnatal murine heart development. J. Biol. Chem. 294, 15889–15897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wheeler L. J., and Mathews C. K. (2011) Nucleoside triphosphate pool asymmetry in mammalian mitochondria. J. Biol. Chem. 286, 16992–16996 10.1074/jbc.M111.236968 [DOI] [PMC free article] [PubMed] [Google Scholar]


