Abstract
Bardet-Biedl syndrome (BBS) is a genetic disorder characterized by malfunctions in primary cilia resulting from mutations that disrupt the function of the BBSome, an 8-subunit complex that plays an important role in protein transport in primary cilia. To better understand the molecular basis of BBS, here we used an integrative structural modeling approach consisting of EM and chemical cross-linking coupled with MS analyses, to analyze the structure of a BBSome 2-7-9 subcomplex consisting of three homologous BBS proteins, BBS2, BBS7, and BBS9. The resulting molecular model revealed an overall structure that resembles a flattened triangle. We found that within this structure, BBS2 and BBS7 form a tight dimer through a coiled-coil interaction and that BBS9 associates with the dimer via an interaction with the α-helical domain of BBS2. Interestingly, a BBS-associated mutation of BBS2 (R632P) is located in its α-helical domain at the interface between BBS2 and BBS9, and binding experiments indicated that this mutation disrupts the BBS2-BBS9 interaction. This finding suggests that BBSome assembly is disrupted by the R632P substitution, providing molecular insights that may explain the etiology of BBS in individuals harboring this mutation.
Keywords: protein assembly, protein complex, protein cross-linking, mass spectrometry (MS), electron microscopy (EM), homology modeling, Bardet-Biedl syndrome, Cilia transport, ciliopathy, integrated modeling
Introduction
Primary cilia perform vital signaling functions in vertebrate cells, ranging from recognition of developmental cues from morphogens, such as hedgehog in the developing embryo to detection of sensory signals such as photons of light in retinal photoreceptor cells (1–4). Primary cilia are formed by the axoneme, a circularly-ordered scaffold containing nine pairs of microtubules anchored inside the cell at the basal body and protruding outward to create a finger-like projection of the plasma membrane (1, 2). Many transmembrane receptors are concentrated in this ciliary compartment, creating a type of signaling antenna for the cell (5–7). The contents of the cilium are delivered there by the intraflagellar transport (IFT)4 complexes that use kinesin motors to move cargos toward the tip of the cilium (anterograde transport) and dynein motors to move cargos toward the base of the cilium (retrograde transport) (1, 2, 4). Failure of ciliary trafficking results in diseases referred to as ciliopathies that are characterized by multiple phenotypes, including cystic kidneys, retinal degeneration, obesity, and multiple developmental disorders (8).
One of these ciliopathies is Bardet-Biedl syndrome (BBS), a disease that results from the malfunction of a large protein complex called the BBSome (9). The BBSome consists of eight subunits (named BBS1, -2, -4, -5, -7, -8, -9, and -18), and mutations in each subunit have been linked to BBS (9–12). The disease results from an inability of the BBSome to participate in ciliary transport. The proposed function of the BBSome is to act as a scaffolding complex to link protein cargos, including membrane proteins, to the IFT machinery for ciliary transport with a particularly important role in retrograde transport out of the cilium (11, 13–18). The BBSome reversibly associates with the membrane via an interaction of BBS1 with ARL6 (ADP-ribosylation factor-like protein 6), a small GTPase that interacts with the membrane in its GTP-bound form (11, 19). In this membrane-bound state, the BBSome is believed to pick up membrane proteins targeted for exit from the cilium (11, 18). Transport of these cargos occurs through association of the BBSome with the IFT-B complex via an interaction with a linker protein LZTFL1 (leucine zipper transcription factor-like 1) that binds to IFT27 in the IFT-B complex (15, 16, 20). In this manner, the BBSome collects membrane proteins for IFT- and dynein-mediated retrograde transport.
To perform its transport function, each of the 8 subunits of the BBSome must be translated on ribosomes, folded into their native state and assembled into a functional complex. Evidence suggests that BBSome assembly proceeds through several subcomplexes, with BBS9 acting as a central scaffold. A stable hexameric complex consisting of human BBS1, -4, -5, -8, -9, and -18 was recently isolated from insect cells (21). This subcomplex interacted with Arl6 as well as peptide motifs from several G protein–coupled receptor cargos. The two other subunits of the BBSome, BBS2 and BBS7, have an intricate folding and assembly process that requires a network of molecular chaperones, including the cytosolic chaperonin containing tailless polypeptide 1 (CCT; also termed TRiC) complex and three chaperonin-like proteins named BBS6, -10, and -12 (22, 23). Inactivating mutations in these chaperonin-like BBS proteins are also a major cause of BBS (24–26). The chaperonin proteins associate with BBS2 and BBS7 and assist in the formation of a BBS2/BBS7 dimer that binds BBS9 via an interaction with BBS2 (23, 27). Presumably, binding of the BBS2/BBS7 dimer with the hexameric complex completes the assembly of the BBSome octamer (21).
To better understand the mechanism of BBSome assembly, its function as a scaffold for intraflagellar transport and the molecular basis of BBS disease, we have isolated a trimeric BBSome subcomplex of BBS2, BBS7, and BBS9 and investigated its structure by electron microscopy (EM) and chemical cross-linking coupled with MS (XL-MS) using the integrative modeling platform (28). The data show that the BBS2/BBS7 dimer is stabilized by an extensive coiled-coil interaction and that BBS9 interacts with the dimer through association with the α-helical domain of BBS2. A BBS-causing mutation in BBS2 (R632P) (29–31) in this region disrupts the interaction of BBS2 with BBS9, suggesting that the inability of the BBS2/BBS7 dimer to associate with the hexameric complex is the underlying cause of BBS in patients with the R632P mutation.
Results
BBS2-7-9 purification
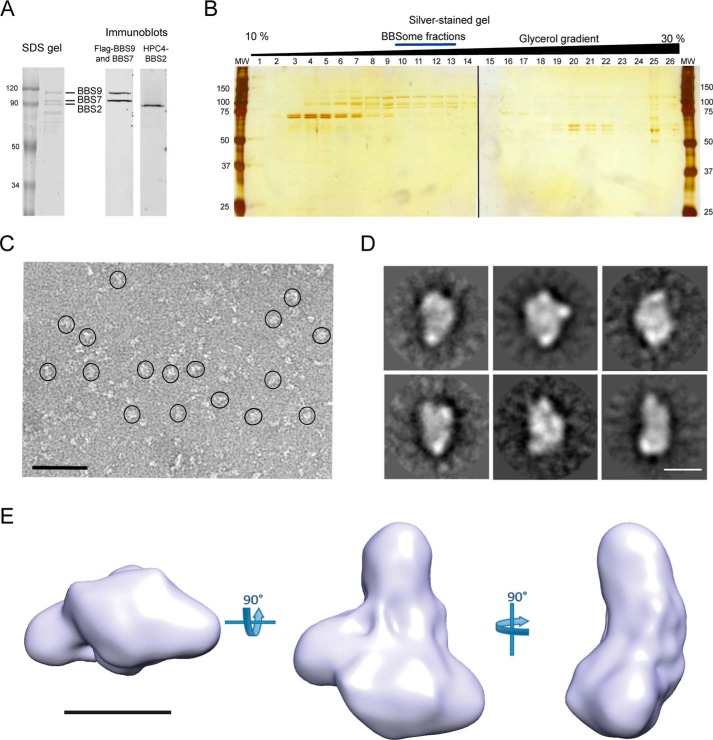
The subcomplex between BBS2, BBS7, and BBS9 is an important early intermediate in assembly of the BBSome (23). We purified this BBS2-7-9 subcomplex for structural studies by co-expressing affinity-tagged versions of each subunit in HEK-293T cells and isolating complexes containing the Strep peptide-tagged BBS7 using a Strep-Tactin column. This purification resulted in roughly equal amounts of BBS2, BBS7, and BBS9 with a 70-kDa contaminant protein corresponding to heat shock protein 70 (Hsp70) isoforms (Fig. 1A). The complex was further purified by glycerol gradient centrifugation (Fig. 1B), which separated Hsp70 and other contaminants (fractions 3–9 and 15–25) from the subcomplex (fractions 10–13).
Figure 1.
Purification of the BBS2-7-9 complex and 3D EM reconstruction. A, left, Coomassie-stained SDS gel of the BBS2-7-9 complex purified by Strep-Tactin affinity purification. FLAG-tagged human BBS9 (99.3 kDa) migrated below the 120-kDa molecular mass standard, and Strep-tagged human BBS7 (80.4 kDa) and HPC4-tagged human BBS2 (79.9 kDa) migrated near the 90-kDa molecular mass standard. The identity of these protein bands was confirmed by immunoblotting (right). B, silver-stained SDS gel of the BBS2-7-9 complex purified as in panel A and separated by a glycerol gradient. The post-centrifugation fractions were loaded from the top (left) to the bottom (right). The fractions with a blue line (fraction numbers 10–13) were used for EM analysis. C, a representative image of a negatively-stained field of BBS2-7-9 complex with the particles circled (bar = 500 Å). D, a sampling of the 2D class averages used in the reconstruction. E, three orthogonal images of the 3D reconstruction of the BBS2-7-9 complex (bar = 100 Å).
3D reconstruction of BBS2-7-9
We assessed the homogeneity of the complex by negative stain EM and found that despite the purity of the preparation, the particles were not sufficiently homogeneous for a high-resolution structural analysis by cryo-EM (Fig. 1, C and D). As a result, we carried out a low-resolution 3D reconstruction using negative-stained EM images (Table S1). The reconstruction (23 Å resolution) revealed the overall structure of the complex (Fig. 1E), which can be described as a flattened, triangular structure with a ∼200 Å height and ∼120 Å width at the base, with a small mass ∼40 Å in diameter protruding from one of the sides.
XL-MS analysis of BBS2-7-9
In the absence of high-resolution cryo-EM data, we used the low-resolution EM envelope combined with cross-link MS (XL-MS) to generate a molecular model of the BBS2-7-9 complex. XL-MS has become an effective tool to probe the structural architecture of protein complexes (32, 33). The cross-links identified by XL-MS provide distance constraints that can be combined with EM reconstructions and other structural information to define the structures of protein complexes (34–39) (Fig. 2A). We treated the purified sample with increasing amounts of the cross-linker to determine the optimal concentration of cross-linker to use (Fig. S1). Ultimately, the purified BBS2-7-9 was cross-linked with three different lysine-specific cross-linkers, disuccinimidyl suberate (DSS), disuccinimidyl glutarate (DSG), or Leiker reagent (40) and digested with proteases. The digests were analyzed by LC-MS/MS, and cross-linked peptides were identified using the cross-link search engines xQuest (41) and pLink (42). The xQuest and pLink output files are available in Table S2–S4. Only high confidence hits that satisfied the screening criteria (see “Experimental procedures”) were considered as true cross-links (Tables S2–S4). The coverage was nearly complete with 86 (BBS2), 90 (BBS7), and 87% (BBS9) of all lysine residues reacting with the cross-linking reagents. Of those, 68 (BBS2), 66 (BBS7), and 78% (BBS9) were involved in cross-links, whereas the rest formed monolinks (Table S5).
Figure 2.
Domain structural models of the components of the BBS2-7-9 complex. A, workflow for the XL-MS experiments. The purified BBS2-7-9 complex was cross-linked and digested with proteases. Cross-linked peptides were enriched and analyzed by LC-MS/MS. Cross-links were identified using the xQuest and pLink search engines. B, models of the domains of BBS2-7-9 complex components were created in I-TASSER except for the BBS9 β-propeller domain, which was from an X-ray crystal structure (PDB 4YD8). Intra-domain cross-links were mapped onto the structures. C, euclidean Cα distance distribution of the lysine cross-links based on the domain structural models. All but one cross-link (orange bar) fit inside the expected distance constraints of 35 Å for DSS, 31 Å for DSG, and 33 Å for Leiker.
Domain modeling
XL-MS generates three types of cross-links that provide different structural information: cross-links within individual domains of the BBSome subunits (intra-domain cross-links), cross-links between domains of the subunits (inter-domain cross-links), and cross-links between subunits (inter-protein cross-links). The XL-MS analysis identified 51 intra-domain cross-links (Fig. 2). These cross-links were used to validate structural models of the individual domains of the core subunits. BBS2, -7, and -9 are homologous proteins that share a well-defined domain organization with N-terminal β-propeller, followed by coiled-coil, γ-adaptin ear (GAE), platform, and C-terminal α-helical domains (11). One atomic structure of the β-propeller domain of BBS9 has recently been reported (43), and the structures of homologous domains in other proteins have been solved (11, 19). Given these homologs, we reasoned that accurate structural models of each domain could be determined, and we generated homology models of each domain using the protein structure prediction server I-TASSER (44) (Fig. 2B, Table S6). The percentage of sequence modeled in these domains for BBS2, BBS7, and BBS9 was 95, 96, and 96%, respectively (Table S7).
To assess the accuracy of the domain models, we mapped a list of all the theoretical lysine pair cross-links onto the domain models and calculated Cα-Cα distances between the lysine residues. Considering movement of the protein backbone in flexible regions, the distance constraints were <35 Å for DSS, <31 Å for the DSG, and <33 Å for Leiker (36). We found that 21% of all possible cross-links in the domain models exceeded the 35 Å DSS distance constraint (Table S8). Yet when we mapped the experimentally determined intra-domain cross-link distances on the model structures, only one DSG cross-link of the 51 intra-domain cross-links fell outside the distance constraints (Fig. 2C). This consistency between the structural models and the cross-linking distances validates the accuracy of the domain models for BBS2, -7, and -9 and the quality of the cross-linking data. These domain models were used as building blocks to assemble the BBS2-7-9 structure.
BBS2 and BBS7 coiled-coil interaction
XL-MS identified 22 inter-protein cross-links that were used to locate sites of interaction between subunits of BBS2-7-9 (Fig. 3A). Sixteen of the cross-links were between BBS2 and BBS7 and six of these were in regions predicted to form a coiled-coil (residues 334–363 of BBS2 and 339–376 of BBS7). This high density of cross-links supports the idea that BBS2 and BBS7 associate via a coiled-coil interaction in these regions. To determine whether the coiled-coil interaction was parallel or anti-parallel, we modeled the coiled-coil region to both parallel and anti-parallel templates using the Foldit program (45) (Fig. S2). When the cross-linking constraints were included in the modeling, the parallel template produced a compact coiled-coil structure with cross-link distances between 12 and 15 Å (Fig. 3B). In contrast, applying the cross-link constraints converted the anti-parallel template model into a parallel one, demonstrating that the cross-links were only compatible with a parallel model. The parallel model predicted that residues 334–363 of BBS2 participated in the coiled-coil with residues 339–363 of BBS7 and that residues 364–376 of BBS7 were not part of the coiled-coil (Fig. 3B). The interface between the two helices was populated by hydrophobic residues as expected for a coiled-coil interaction (Fig. 3C). Interestingly, no inter-protein cross-links were found in the same coiled-coil region of BBS9, indicating that BBS9 does not form a coiled-coil interaction with BBS2 or BBS7. These data are consistent with pairwise co-immunoprecipitation experiments that showed interactions between BBS2 and BBS7 as well as between BBS2 and BBS9 but not between BBS7 and BBS9 (23, 27).
Figure 3.
Coiled-coil interaction between BBS2 and BBS7. A, BBS2-7-9 complex inter-protein cross-links determined by XL-MS. Six of these interlinks were found between the coiled-coil regions of BBS2 and BBS7. B, structural model of the coiled-coil interaction between BBS2 and BBS7 with cross-links mapped onto the model. The table lists the Euclidean Cα distances of the cross-linked lysines. The region of BBS7 not involved in the coiled-coil interaction (residues 364–376) is shown in gray. C, two views of the BBS2-BBS7 coiled-coil interaction with the hydrophobic side chains at the interface shown in yellow.
Structural model of the BBSome core complex
To obtain a structural model of the entire BBS2-7-9 complex, we combined the BBS2/BBS7 coiled-coil model along with the domain models of the three BBS subunits and assembled them into the EM reconstruction using the 37 inter-domain cross-links and the 22 inter-protein cross-links identified by XL-MS. There were an additional five inter-domain cross-links that mapped to unstructured regions between the modeled domains. These cross-links could not be used for the integrated modeling. We employed the integrative modeling platform (IMP) (28), which takes structural data from the cross-links and the EM density and converts them into spatial restraints that are combined into a scoring function to rank alternative models (46). IMP iteratively searches the configurational space to generate structural models that satisfy the spatial restraints, avoid steric clashes, and retain sequence connectivity (46).
IMP iterations were run until the models converged to the point that they were producing similar structures. From these, the top 500 scoring models were compared in a distance matrix that calculates the root mean square deviation between the models (Fig. S3A). The matrix shows two clusters of models with related structures of 229 and 269 models each. Within each cluster, IMP calculates the localization density from all the models, which gives the overall shape of each protein in the complex. IMP also determines the global structural centroid for the cluster and selects the individual model whose centroid is nearest to this global centroid. The localization density and ribbon structure of the centroid model from cluster 1 is shown in Fig. 4A. Although the two clusters both fit the cross-linking and EM data comparably well, the first cluster satisfied more cross-links than the second cluster, with 89.6 and 85.4% of cross-links satisfied, respectively (Fig. 4, B and C, and Fig. S3B). The localization density of model 1 also fits the shape of the EM density well (Fig. 4D). The main differences in conformation between the two clusters is that in cluster 1, the β-propellers of BBS2 and BBS7 point out toward the short side of the EM density, whereas in cluster 2, they point out the long side. Otherwise, the inter-domain contacts between subunits in each cluster of models are the same (Fig. S3C).
Figure 4.
Structural model of the BBS2-7-9 complex. A, three orthogonal views of the IMP localization densities computed for each subunit. The ribbon structure of the centroid model is depicted. B, inter-domain and inter-protein cross-links mapped onto the centroid model ribbon structure. Some domains are labeled to provide landmarks. C, distributions of Euclidean Cα distances of the cross-linked lysines for each cross-linker. Blue bars represent cross-links within the expected distance constraints and orange bars represent cross-links outside the expected constraints (35 Å for DSS, 31 Å for DSG, and 33 Å for Leiker). D, the same views as in A docked in the EM 3D reconstruction of BBS2-7-9 complex. The reconstruction is depicted as a gray mesh surface.
An examination of the model structure shows BBS2 sitting between BBS7 and BBS9 and making close contact with both subunits (Fig. 4A). The coiled-coil is at the center of the interaction between BBS2 and BBS7, but their GAE domains are also in close proximity. The α-helical domain of BBS2 wraps around to contact the α-helical domain of BBS9, forming the primary point of contact between the two subunits. This proposed structure of the BBS2-7-9 complex is consistent with two previous studies showing binding of BBS2 to both BBS7 and BBS9 and no interaction of BBS7 with BBS9 (23, 27).
Effects of BBS-linked mutations
Multiple point mutations in BBS2, BBS7, and BBS9 have been linked to Bardet-Biedl syndrome (30, 43, 47–54). To understand how these mutations might result in BBS, we measured the effect of 14 of these mutations (7 in BBS2, 6 in BBS7, and 1 in BBS9) on the formation of the BBS2-7-9 complex. We co-expressed BBS2, BBS7, and BBS9 in HEK-293T cells and measured the interaction of BBS7 and BBS9 with BBS2 by co-immunoprecipitation. In the case of BBS7, only the BBS2 L349W mutant showed a modest 30% decrease in binding (Fig. 5A). This mutation is located in the BBS2/BBS7 coiled-coil (Fig. 3) and may weaken the coiled-coil interaction. In the case of BBS9, both the BBS2 R632P and BBS9 G141R mutants showed a marked 70% reduction in binding. The decrease observed in the BBS9 G141R mutant was a result of lower cellular expression (Fig. 5B). Quantification of the BBS9 bands from cell lysates showed that expression of the G141R mutant was diminished by 76% compared with the expression of WT BBS9. In contrast, expression of the BBS2 R632P mutant showed a slight 24% decrease that was not statistically significantly different from WT BBS2, indicating that the mutation did not significantly impair the expression of BBS2, but that it disrupted the interaction between BBS2 and BBS9. The position of the R632P mutation in the structural model suggests how the mutation would disrupt the interaction between BBS2 and BBS9. The R632P mutation is found within the α-helical domain of BBS2 and is in close proximity to the α-helical domain of BBS9 (Fig. 5, C and D). A proline substitution here would likely disrupt the formation of its helix and perturb the structure in this region, thereby interfering with the interaction between BBS2 and BBS9.
Figure 5.
Effects of mutations on BBS2-7-9 complex assembly. A, the effects of BBS-linked mutations in BBS2, -7, and -9 on BBS2-7-9 complex formation were measured. HEK-293T cells were co-transfected with c-Myc-tagged WT or mutant BBS2, FLAG-tagged WT or mutant BBS7, and FLAG-tagged WT or mutant BBS9. BBS2 was immunoprecipitated, and BBS2, -7, and -9 were detected by immunoblotting. The graph shows the quantification of the BBS7 and BBS9 bands normalized to WT. Bars represent the average ± S.D. of three experiments for each mutant and six for WT. **, p < 0.01; ***, p < 0.001. Representative immunoblots from the BBS2 immunoprecipitates are shown below the graph. B, expression levels of BBS2 L349W, R632P, and BBS9 G141R mutants in cell lysates were determined by immunoblotting. Representative blots are shown and the graph gives the quantification of the BBS2, BBS7, and BBS9 bands normalized to WT. Bars represent the average ± S.D. from six experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, map of BBS-linked mutations on the BBS2-7-9 structural model. Locations of the mutations are shown with yellow dots. D, zoomed in image shows the position of R632P in the α-helical domain of BBS2 and the cross-links (black lines) that confirm the interaction of this region with BBS9.
Discussion
To perform its essential role in ciliary transport, the eight subunits of the BBSome must be assembled into a functional complex. Our structural analysis of the BBS2-7-9 complex provides mechanistic insight into how BBSome formation could occur. BBS2 and BBS7 form a tight dimer principally via an extensive coiled-coil interaction involving residues 334–363 of BBS2 and residues 340–363 of BBS7. Previous studies indicate that BBS2 and BBS7 are brought together by chaperones, including the chaperonin-like BBS proteins (BBS6, BBS10, and BBS12) and the cytosolic chaperonin CCT (22, 23). BBS9 associates with the BBS2/BBS7 dimer through an interaction with the α-helical domain of BBS2. This interaction with BBS9 would link the BBS2/7 dimer to the hexameric BBS1, -4, -5, -8, -9, and -18 subcomplex (21). The hexameric subcomplex is stable and could be purified in milligram quantities from insect cells (21), whereas the BBS2-7-9 complex is much less stable and we could only purify microgram quantities from cultured human cells. These major differences in stability suggest that the hexameric subcomplex forms first, followed by association of the BBS2/7 dimer with the hexamer via interactions with BBS9. The instability of the BBS2/7 dimer provides an explanation why chaperonin-like proteins are needed to bring the dimer together and why mutations in the chaperonin-like proteins cause BBS.
In the BBS2-7-9 complex, the β-propeller domains of BBS2 and BBS7 are not involved in interactions that hold the complex together, leaving them accessible for other interactions. These domains might be sites of cargo binding in a manner analogous to the β-propellers of the α and β′ subunits of the COPI complex, which forms membrane vesicle coats for retrograde transport in the Golgi. These COPI β-propellers interact with membrane-associated cargos to bring them into vesicles (55–57). Similarly, the accessible β-propeller domains of the BBS2-7-9 complex may associate with membrane proteins for transport in the primary cilium.
The BBS2-7-9 structure provides insight into the molecular basis of BBS disease caused by the BBS2 R632P mutation. The interaction of this mutant with BBS9 is strongly inhibited, whereas its binding to BBS7 is not. The reason for the disruption can be seen in the predicted interaction surface between the α-helical domain of BBS2 with the α-helical domain of BBS9. Residues 628–635 of BBS2 are expected to form an α-helix that is located in the contact region between BBS2 and BBS9 (Fig. 5D). Three cross-links between the α-helical domains of BBS2 and BBS9 (Lys609–Lys687, Lys673– Lys687, Lys691–Lys821) position these two domains together in the model and indicate that these domains closely interact (Fig. 5D). The R632P mutation would disrupt this helix of BBS2, which would in turn destabilize the interactions occurring at this interface. Thus, the R632P mutation likely causes BBS because of an inability of the BBS2/7 dimer to bind to BBS9 and the rest of the BBSome.
Previous work has shown that the G141R mutation disrupts the folding of the BBS9 β-propeller and inhibits the expression of the mutant protein (43). Gly141 is found in the third β-sheet of the BBS9 β-propeller, and the Arg mutations would be predicted to disrupt the β-sheet and interfere with the folding of the β-propeller (43). We also see decreased expression of the BBS9 G141R mutant that results in decreased binding of BBS9 to BBS2. Without BBS9, the BBSome cannot assemble and the disease ensues. The other mutants found in BBS2, -7, and -9 do not disrupt formation of the BBS2-7-9 complex, suggesting that they might contribute to BBS in some other way, perhaps by interfering with the association of the other BBSome proteins or BBSome cargos.
When this work was submitted for review, a higher resolution structure of the intact BBSome containing all 8 subunits was published (58). A comparison of the two structural models shows that the overall positions of BBS2, BBS7, and BBS9 are similar in the BBS2-7-9 subcomplex and the intact BBSome (Fig. 6A). Both structures show the coiled-coil interaction between BBS2 and BBS7 as well as the helical domain interaction between BBS2 and BBS9. The positions of the β-propeller domains differ between the two structures either because of the lower resolution of the BBS2-7-9 subcomplex or because of differences resulting from interactions within the intact BBSome. As an example of the later, the BBS9 β-propeller extends away from the BBS2/7 dimer in the intact BBSome because of interactions with the other BBSome subunits. Importantly, the intact BBSome structure shows clearly how the BBS2 R632P mutation could interfere with BBS2 binding to BBS9. In the intact structure, this residue is located near a tight turn that links the two long antiparallel helices of the helical domain of BBS2. These helices constitute the interface of the helical bundle formed between BBS2 and BBS9 (Fig. 6B). The helix breaking properties of the proline mutation would disrupt the end of this helix as well as the turn, interfering with the antiparallel structure of these BBS2 helices and inhibiting their ability to form the helical bundle with BBS9.
Figure 6.
Comparison of the structural models of BBS2-7-9. A, the Cα models of BBS2, -7, and -9 from the intact BBSome (58) and the BBS2-7-9 subcomplex (this study) are shown side by side for comparison. B, a zoomed-in view of the helical bundle of BBS2 and BBS9 from the intact BBSome structure shows the position of Arg632.
In summary, this analysis reveals the structural architecture of the BBS2-7-9 complex, consisting of BBS2, BBS7, and BBS9, and identifies key interactions that bring the components of this BBSome subcomplex together. Moreover, it provides a molecular basis for Bardet-Biedl syndrome caused by the BBS2 R632P mutation. These studies show how structural information can inform our understanding of the BBSome and how its malfunction causes disease.
Experimental procedures
Purification of the BBSome 2-7-9 subcomplex
Human embryonic kidney (HEK)-293T cells cultured in 1:1 Dulbecco's modified Eagle's media (DMEM)/F-12 with 10% fetal bovine serum (FBS) in T-175 flasks were transfected at 80% confluence with a transfection mixture consisting of purified plasmids of the pcS2+ vector (Addgene) containing human BBS2 with protein C peptide (HPC4) and c-Myc tags at the N terminus, a pcDNA3.1 vector (Thermo Fisher Scientific) containing human BBS7 with a FLAG-Strep tag at the N terminus, a pcS2+ vector containing human BBS9 with a FLAG tag at the N terminus, and polyethylenimine. The DNA to polyethylenimine ratio was 1:3 by weight, and up to 90 μg of DNA was used per T-175 flask. The media was replenished with DMEM/FBS after 2–4 h, and the cells were incubated for an additional 48 h before harvesting. Cells were lysed in 3 ml of extraction buffer (20 mm HEPES, pH 7.5, 20 mm NaCl, 1% Nonidet P-40, 0.5 mm PMSF, and 6 μl/ml of protease inhibitors mixture (Sigma P8340), 24 units/ml of benzonase nuclease) per gram of cell pellet, and the lysate was cleared by centrifugation at 13,000 × g for 15 min.
The BBS2-7-9 complex was purified at 4 °C by affinity purification using 4 ml of Strep-Tactin resin (IBA Life Sciences) packed in a 2-cm diameter column and equilibrated with equilibration buffer (20 mm HEPES, pH 7.5, and 20 mm NaCl). The HEK-293T cell lysate was loaded for 1 h, washed with 5 column volumes of wash buffer (20 mm HEPES, pH 7.5, 20 mm NaCl, and 0.05% CHAPS), and then eluted with 2.5 column volumes of elution buffer (20 mm HEPES, pH 7.5, 20 mm NaCl, 2.5 mm desthiobiotin, and 0.05% CHAPS). The eluate was concentrated to ∼1 μg/μl using a 30-kDa cutoff filter (Millipore). The final protein concentration was determined by comparing the absorbance at 280 nm to a 2 μg/μl of BSA standard (Pierce) and correcting for minor buffer absorbance in a 2.5-μl nanodrop assay using a BioTek synergy H4 plate reader. The concentrated sample was flash frozen in liquid nitrogen and stored at −80 °C. The purified core complex was analyzed by 10% polyacrylamide SDS-gel electrophoresis and its purity was determined to be 80–90% by staining with Coomassie Brilliant Blue (Thermo Fisher Scientific). The composition of the core complex was confirmed by immunoblotting with anti-FLAG and anti-c-Myc antibodies. Coomassie gels and immunoblots were imaged using a LI-COR Odyssey IR scanner.
Some samples were further affinity purified using an HPC4 antibody-conjugated resin (Roche Applied Science), but yields from the HPC4 column were low, the improvements in purity were modest, and the cross-links identified were very similar to the Strep-Tactin–purified complex. Therefore, we moved away from HPC4 purifications, but included cross-linking data from both purifications in the structural analysis.
Cross-linking
The XL-MS analysis followed the protocol established by Leitner et al. (41). Approximately 200 μg of the BBS2-7-9 complex at 1 μg/μl was cross-linked in 25 mm HEPES, pH 8.0, 100 mm KCl, and 325 μm of a 50% mixture of H12/D12 DSS or H6/D6 DSG (Creative Molecules) at 37 °C for one-half hour. The reaction was quenched by adding 50 mm ammonium bicarbonate to the cross-linked sample and incubating at 37 °C for 15 min. The sample was dried by a vacuum concentrator, denatured in 100 mm Tris-HCl, pH 8.5, and 8 m urea, reduced with 5 mm tris(2-carboxyethyl)phosphine at 37 °C for 30 min, and alkylated with 10 mm iodoacetamide at room temperature in the dark for 30 min. The sample was diluted with 150 mm ammonium bicarbonate to bring the urea concentration to 4 m, and proteins were digested with 4 μg of lysyl endopeptidase (Wako Laboratories, cleaves at the C terminus of lysine) at a 1:50 enzyme to substrate ratio at 37 °C for 2 h. Subsequently, urea was diluted to 1 m, and proteins were further digested with trypsin (Wako Laboratories) and cleaved at the C terminus of lysine and arginine at a 1:50 ratio at 37 °C overnight. Peptide fragments were purified on a C18 column (Waters), dried, and reconstituted in a 35-μl size exclusion chromatography mobile phase (70:30:0.1 water:acetonitrile:TFA). Cross-linked peptide fragments were enriched by size exclusion chromatography using a Superdex Peptide PC 3.2/30 column on an AKTA pure system (GE Healthcare) at a flow rate of 50 μl/min. Fractions from the leading half of the elution peak were pooled, dried, and resuspended in 2% formic acid (41).
To increase the cross-link coverage, we also cross-linked with the Leiker trifunctional cross-linking reagent (40). We adapted the previously published protocol by first adding 50 μg of Leiker reagent to 200 μg of concentrated BBS2-7-9 complex in a final reaction volume of ∼200 μl. The reaction proceeded for 1 h and was quenched with 10 μl of 550 mm ammonium bicarbonate for 20 min. The cross-linked complex was then concentrated on a microcentrifuge filter (VWR 82031354) to <20 μl and washed three times with 800 μl of 8 m urea, 20 mm methylamine, 100 mm Tris base, pH 8.5, reconcentrating to <20 μl with each wash. The volume was brought up to 95 μl with the 8 m urea solution, reduced with 5 mm tris(2-carboxyethyl)phosphine, alkylated with 10 mm iodoacetamide, and digested by lysyl endopeptidase and trypsin as described above for the DSS and DSG cross-linking. The digested protein was then passed through the microcentrifuge filter and incubated with 40 μl of Pierce™ High Capacity Streptavidin-agarose (Thermo 20357) for 2 h. The beads where then washed, the cross-linked peptides were eluted, and the samples were prepared for MS as described (40).
Mass spectrometry
The enriched DSS and DSG cross-linked peptide samples were separated using a Thermo Fisher Scientific EASY-nLC 1000 liquid chromatograph system with a 15-cm Picofrit column (New Objective) packed with Reprosil-Pur C18-AQ of 3-μm particle size, 120-Å pore size, and gradient of 5–95% acetonitrile in 5% DMSO and 0.1% formic acid over 185 min and at a flow rate of 350 nl/min. The column was coupled via electrospray to an Orbitrap Velos Pro mass spectrometer. The resolution of MS1 was 30,000 over a scan range of 380–2000 m/z. Peptides with a charge state 3+ and greater were selected for HCD fragmentation at a normalized collision energy of 35% with 3 steps of 10% (stepped NEC) and a resolution of 7,500. Dynamic exclusion was enabled with a 10 ppm mass window and a 1-min time frame. Technical duplicates were run for each of three different DSS preparations and two DSG preparations.
The Leiker cross-linked samples were separated by LC-MS using an Easy-nLC 1200 system (Thermo Fisher Scientific) with a 75 μm × 2-cm pre-column (3 μm C18 Acclaim PepMap 100, number 164946) and a 75 μm × 25-cm analytical column (2 μm C18 3-μm C18, Acclaim PepMap 100, number 164946), using a gradient of buffer A (0.1% formic acid) and buffer B (80% acetonitrile and 0.1% formic acid) as follows: 0–6% B for 2 min, 6–35% B for 41 min, 35–100% B for 10 min, and 100% B for 12 min at a flow rate of 300 nl/min. The column was coupled via electrospray to an Orbitrap Fusion Lumos mass spectrometer run in data-dependent mode with one scan at r = 60,000 (m/z = 350–2000), followed by 10 HCD MS/MS microscans at r = 15,000 (first mass m/z = 110). The NCE was 27, with an isolation width of 2 m/z. The MS1 and MS2 scan AGC targets were 4e5 and 1e5 and the maximum injection time was 60 ms for both MS1 and MS2. Precursors of +1, +2, +7, or above, or unassigned charge states were rejected. Exclusion of isotopes was disabled. Dynamic exclusion was 30 s.
XL-MS analysis
The xProphet (version 2.5.1)/xQuest (version 2.1.2) software pipeline (41) was used to identify the DSS and DSG cross-linked peptides, the specific lysine residues involved in the cross-link, and to evaluate the quality of each hit from the MS data set. Tandem mass spectra for parent ions with a mass shift of 12.075321 Da for the DSS cross-linker and 6.04368 Da for the DSG cross-linker and a charge of +3 to +7 were classified as isotopic pairs and evaluated in ion-tag mode with the following parameters: 2 missed cleavages, 5–50 amino acid peptide length, carbamidomethyl-fixed modification (57.02146 Da mass shift), oxidation variable modification (15.99491 Da mass shift), 138.0680796 (DSS) and 96.02059 (DSG) Da mass shift for intra- and inter-protein cross-links, 156.0786442 (DSS) and 114.03115 (DSG) Da for -OH monolinks, 155.0964278 and 113.04713 for -NH2 monolinks, MS1 tolerance of 10 ppm, and MS2 tolerance of 0.2 Da for common ions and 0.3 Da for cross-link ions.
The peptide sequence database was created by xQuest based on the amino acid sequence of human BBS2, -7, and -9 (UniProt ID: Q9BXC9, Q8IW26, and Q3SYG4, respectively) and 9 common contaminant proteins (hHSPA1A, hHSPA1L, hHSPA8, hKRT1, hHSPA2, hTUBA1B, hTUBA1C, hHSPA6, hTUBA3C; UniProt ID: P0DMV8, P34931, P11142, P04264, P54652, P68363, Q9BQE3, P17066, P0DPH7, respectively). Spectra were searched against the database that covered all possible cross-link combinations of the BBSome core proteins, and any spectra that matched cross-links in the database were counted and evaluated. Cross-link hits were screened with the following xQuest criteria: 10% false discovery rate, >10% total ion counts, −4 to 7 ppm MS1 tolerance window, >20 xQuest Id-Score, and >4 fragmentation events per peptide. Any hit that met these parameters but scored below a decoy hit was not included in the cross-link list unless it was found in another run (Table S2). Those xQuest hits that satisfied these thresholds were inputted to the IMP for structural assessment (28).
All spectra were also analyzed with the pLink2 software suite (version 2.3.5). The same xQuest peptide sequence database was used and pLink added the sequences of other common contaminant proteins. The program was then run using the preset DSS and Leiker_clv linker settings and custom DSG (Linker composition: C(5)H(4)O(2), Mono composition C(5)H(6)O(3)), DSG_heavy (C(5)H(-2)2H(6)O(2), C(5)2H(6)O(3)), and DSS_heavy (C(8)H(-2)2H(12)O(2), C(5)2H(12)O(3)) linker profiles using the same cross-link and monolink masses as described for xQuest. The spectra were then analyzed using conventional cross-linking (HCD) conditions, with trypsin set as the protease and up to 3 missed cleavages allowed. Peptides were selected with a mass between 600 and 6,000 Da and a length between 6 and 60 amino acids. The precursor and fragment tolerances were ±20 ppm. The peptides were searched using carbamidomethyl (C)-fixed modifications and phospho-Tyr, -Thr, and -Ser and oxidized M variable modifications. The results were filtered with a filter tolerance of ±10 ppm and less than 5% false discovery rate (Table S3).
The residue numbers given in the xQuest and pLink output included N-terminal tag lengths of 23 for BBS2, 16 for BBS7, and 11 for BBS9. These were subtracted to obtain the correct residue number of the cross-linked amino acids. Unique cross-links were then sorted into intra-domain, intra-protein, and inter-protein data sets.
Mutagenesis
The consequences of 14 BBS-linked mutations in BBS2, -7, and -9 on the formation of BBS2-7-9 were measured by co-immunoprecipitation. The mutations were introduced into N terminally c-Myc–tagged BBS2 in pcS2+ vector, N terminally Strep, and FLAG-tagged BBS7 in pcDNA3.1 vector and N terminally FLAG-tagged BBS9 also in pcS2+ vector using mutagenic PCR primers in a conventional PCR-based cloning protocol. All constructs were sequenced to confirm that the mutations were correct. Constructs were transfected into HEK-293T cells grown in 1:1 DMEM/F-12 media with 10% FBS in 6-well plates at 80–90% confluence using Lipofectamine 2000 according to the manufacturer's protocol. Up to 3 μg of DNA was added to each well, and the relative DNA amounts were 1:1:1 for the BBS2, -7, and -9 constructs. Cells were fed with DMEM/FBS 3h after transfection and incubated at 37 °C for 2 days. Cells were washed with a PBS solution (12 mm phosphate, pH 7.4, 137 mm NaCl, 3 mm KCl) and harvested in a PBS lysis buffer (PBS plus 6 μl/ml of protease inhibitors mixture (Sigma P8340), 0.6 mm PMSF, and 1% Nonidet P-40) 48 h after transfection. Lysed cells were triturated 8–10 times with a 25-gauge needle and syringe and cleared by centrifugation at 14,800 rpm in a Sorvall Legend Micro 21 microcentrifuge for 10 min. The lysates were immunoprecipitated by incubating with an antibody to the c-Myc tag (Invitrogen 13-2500) on BBS2 as described above and immunoblotted with the c-Myc antibody for BBS2 and with the anti-FLAG antibody (Sigma F3165) for BBS7 and BBS9 also as described above.
Protein separation for EM
The BBS2-7-9 sample from the Strep affinity purification was concentrated to 1 μg/μl, and 150 μg of the sample was subjected to density gradient centrifugation to isolate the complexes. The sample was loaded onto a 10–30% glycerol gradient in a separation buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 4 mm CaCl2, 0.3 mm PMSF, 0.3% protease inhibitor mixture, 3 mm DTT) and the ultracentrifugation was performed at 34,000 rpm in a SW55Ti rotor at 4 °C for 16 h. Fractions were collected from the top to the bottom, analyzed by 10% SDS-PAGE gels, and silver stained. The fractions including pure BBS2-7-9 proteins were selected for the EM analysis.
EM grid preparation and data collection
Five-μl aliquots of the protein samples were applied to 300-mesh grids (Maxtaform Cu/Rh HR26) coated with a thin (∼8 nm) carbon layer and glow-discharged for 15 s. The grids were then stained (1 min) with 2% uranyl acetate and air-dried before transmission EM analysis. Images were taken using a Tecnai F20 transmission EM electron microscope operating at 200 kV with a 4k FEI Eagle CCD camera. Images were recorded at a sampling rate of 1.78 Å/px.
Image processing and 3D reconstruction
Contrast transfer function was corrected using the CTFFIND3 program (59), which also calculated potential astigmatism. Micrographs with visible drift and astigmatism were discarded. 8806 single particles of the BBSome core complex were selected manually, extracted from micrographs, and normalized using the XMIPP software package (60). Two types of algorithms were used to classify single images, CL2D (61) and Relion (62).
For 3D reconstruction, several initial models were tested in the first step of the 3D reconstruction procedure using EMAN software (63): artificial noise, blob, and a model created by a “common lines” algorithm based on previously obtained 2D classes. Refinement was performed until the 3D reconstructions from these initial models converged to stable, similar 3D volumes. To obtain more structural detail, the 3D reconstruction from EMAN refinement was subjected to projection matching using XMIPP. Resolution of the final 3D models was estimated based on the FSC criterion (Fourier shell correlation) (64). The spatial frequency at 0.5 correlation was taken as the resolution of the model. Visualization of the 3D models was performed using USCF Chimera (65).
Structural modeling
All three BBS2-7-9 proteins share the same domain organization with N-terminal β-propeller, coiled-coil, GAE, platform, and C-terminal α-helical domains. The secondary and tertiary structure of each domain was generated using the protein structure prediction server I-TASSER (44), and the accuracy of these domain models was confirmed using the intra-domain cross-links from the XL-MS data. A crystal structure (PDB 4YD8) was used as a domain model of the β-propeller of BBS9 (43).
Sequence analysis indicated a coiled-coil interaction between BBS2 residues 334–363 with BBS7 residues 339–376. To assess this possibility, we employed homology modeling using the parallel coiled-coil from rat PAWR protein (chains A and B from PDB ID 5fiy (66)) or the anti-parallel coiled-coil from the hantavirus nucleocapsid protein (PDB 2ic9) as templates to obtain a model structure for the BBS2/BBS7 coiled-coil. First, the coiled-coil heptad repeats were identified in the BBS2 and BBS7 sequences, revealing three clear repeats in each sequence (Fig. S2A). Similarly, three coiled-coil heptad repeats with ideal coiled-coil geometry were identified in the models and used as template structures. The program Foldit (45) was used to thread the BBS2 and BBS7 sequences into the templates. The triple heptad repeats of BBS2 and BBS7 were aligned with the heptad repeats of the templates and placed fully opposite and in register with one another. Foldit was then used to sample all side chain and backbone degrees of freedom in the resulting BBS2/BBS7 homology models. Restraints of 11.5 Å for DSS and 7.4 Å for DSG were applied between Nζ atoms of the cross-linked lysine residues identified by XL-MS. With these restraints applied, the models were again allowed to sample all side chain and backbone degrees of freedom. The region of the homology model comprising BBS7 residues 364–376 did not fit the restraints well and so Foldit was used to unfold this region and refold it using the aforementioned restraints. The result was an unstructured random coil for BBS7 residues 364–376 positioned outside the high confidence coiled-coil model for the rest of the structure.
The IMP version 2.8.0 (67) was used to predict how the individual BBSome core complex domains are oriented relative to each other. The domain models described previously were treated as rigid bodies. The parallel coiled-coil interaction of BBS2 and BBS7 was treated as a single rigid body without the unstructured residues (364–376) of BBS7. Linker regions between domains were approximated as 20 residue beads. Each of the three subunits was treated as super rigid bodies. Model restraints were generated from the cross-link data and the EM reconstruction electron density. A weight of 120 was applied to the EM restraint. 3,200,000 models were generated in runs of 100,000 frames. The top 500 scoring models were then divided into two clusters and localization densities for each subunit were calculated for each cluster. Cluster precision was then gauged by calculating the root mean square fluctuation between models in the cluster and for each residue in each subunit (Fig. S3, D and E). Model accuracy was determined by using the PDB file of the centroid model for all three subunits to calculate cross-link distances and to determine the fit within the EM density envelope.
Experimental design and statistical rationale
Three separate purifications of the BBS2-7-9 complex were cross-linked with DSS, two were cross-linked with DSG and four were cross-linked with Leiker reagent. All cross-links that met the identification requirements were included in the IMP data. Binding experiments to measure the effects of BBS-linked mutations on BBS2-7-9 interactions were repeated three times in biologically independent assays to determine whether differences from WT controls were significant. p values were calculated using a 2-tailed t test assuming a normal distribution. The raw MS spectra are deposited in the public massIVE database (http://massive.ucsd.edu)5 under the Project ID MSV000081472.
Author contributions
W. G. L., T. A., J. C., and B. M. W. conceptualization; W. G. L., T. A., J. C., M. T. B.-C., A. M., and S. F. data curation; W. G. L., J. C., M. T. B.-C., A. M., J. D. M., and J. M. V. software; W. G. L. validation; W. G. L., M. T. B.-C., J. D. M., and B. M. W. visualization; W. G. L., T. A., J. C., J. D. M., and B. M. W. methodology; W. G. L. and B. M. W. writing-original draft; W. G. L., T. A., and B. M. W. project administration; W. G. L., J. C., M. T. B.-C., J. M. V., and B. M. W. writing-review and editing; T. A. formal analysis; J. C., S. F., J. M. V., and B. M. W. resources; S. F., J. M. V., and B. M. W. supervision; J. M. V. and B. M. W. funding acquisition; B. M. W. investigation.
Supplementary Material
Acknowledgments
We thank Dr. Maxence Nachury and Dr. Thomas Walz for providing the integrative structural model of the intact BBSome prior to its release in the PDB-Dev database.
This work was supported by Spanish Ministry of Economy and Innovation Grant BFU2016-75984 (to J. M. V.) and United States National Institutes of Health Grant EY012287 (to B. M. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S2 and Tables S1–S8.
The raw MS spectra are deposited in the public massIVE database under the Project ID MSV000081472.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- IFT
- intraflagellar transport
- BBS
- Bardet-Biedl Syndrome
- BBSome
- BBS protein ciliary transport complex
- BBS2-7-9
- BBSome subcomplex containing BBS2, -7, and -9 proteins
- ARL6
- ADP-ribosylation factor-like protein 6
- LZTFL1
- leucine zipper transcription factor-like protein 1
- CCT
- cytosolic chaperonin containing tailless polypeptide 1
- XL-MS
- chemical cross-linking coupled with mass spectrometry
- HEK-293T
- human embryonic kidney 293T cells
- DMEM
- Dulbecco's modified Eagle's media
- FBS
- fetal bovine serum
- HPC4
- protein C peptide tag
- DSS
- disuccinimidyl suberate
- DSG
- disuccinimidyl glutarate
- IMP
- integrative modeling platform
- GAE
- γ-adaptin ear domain
- PDB
- Protein Data Bank
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Gerdes J. M., Davis E. E., and Katsanis N. (2009) The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goetz S. C., and Anderson K. V. (2010) The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pearring J. N., Salinas R. Y., Baker S. A., and Arshavsky V. Y. (2013) Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 36, 24–51 10.1016/j.preteyeres.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nachury M. V. (2018) The molecular machines that traffic signaling receptors into and out of cilia. Curr. Opin. Cell Biol. 51, 124–131 10.1016/j.ceb.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schou K. B., Pedersen L. B., and Christensen S. T. (2015) Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 16, 1099–1113 10.15252/embr.201540530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singla V., and Reiter J. F. (2006) The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629–633 10.1126/science.1124534 [DOI] [PubMed] [Google Scholar]
- 7. Hilgendorf K. I., Johnson C. T., and Jackson P. K. (2016) The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr. Opin. Cell Biol. 39, 84–92 10.1016/j.ceb.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lancaster M. A., and Gleeson J. G. (2009) The primary cilium as a cellular signaling center: lessons from disease. Curr. Opin. Genet. Dev. 19, 220–229 10.1016/j.gde.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., and Jackson P. K. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- 10. Jin H., and Nachury M. V. (2009) The BBSome. Curr. Biol. 19, R472–R473 10.1016/j.cub.2009.04.015 [DOI] [PubMed] [Google Scholar]
- 11. Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., and Nachury M. V. (2010) The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novas R., Cardenas-Rodriguez M., Irigoín F., and Badano J. L. (2015) Bardet-Biedl syndrome: is it only cilia dysfunction? FEBS Lett. 589, 3479–3491 10.1016/j.febslet.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 13. Lechtreck K. F., Brown J. M., Sampaio J. L., Craft J. M., Shevchenko A., Evans J. E., and Witman G. B. (2013) Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol. 201, 249–261 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liew G. M., Ye F., Nager A. R., Murphy J. P., Lee J. S., Aguiar M., Breslow D. K., Gygi S. P., and Nachury M. V. (2014) The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell 31, 265–278 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eguether T., San Agustin J. T., Keady B. T., Jonassen J. A., Liang Y., Francis R., Tobita K., Johnson C. A., Abdelhamed Z. A., Lo C. W., and Pazour G. J. (2014) IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell 31, 279–290 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Datta P., Allamargot C., Hudson J. S., Andersen E. K., Bhattarai S., Drack A. V., Sheffield V. C., and Seo S. (2015) Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E4400–E4409 10.1073/pnas.1510111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Q., Zhang Y., Wei Q., Huang Y., Li Y., Ling K., and Hu J. (2015) BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci. Rep. 5, 11855 10.1038/srep11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye F., Nager A. R., and Nachury M. V. (2018) BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847–1868 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mourão A., Nager A. R., Nachury M. V., and Lorentzen E. (2014) Structural basis for membrane targeting of the BBSome by ARL6. Nat. Struct. Mol. Biol. 21, 1035–1041 10.1038/nsmb.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seo S., Zhang Q., Bugge K., Breslow D. K., Searby C. C., Nachury M. V., and Sheffield V. C. (2011) A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 7, e1002358 10.1371/journal.pgen.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klink B. U., Zent E., Juneja P., Kuhlee A., Raunser S., and Wittinghofer A. (2017) A recombinant BBSome core complex and how it interacts with ciliary cargo. Elife 6, e27434 10.7554/eLife.27434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seo S., Baye L. M., Schulz N. P., Beck J. S., Zhang Q., Slusarski D. C., and Sheffield V. C. (2010) BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. U.S.A. 107, 1488–1493 10.1073/pnas.0910268107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q., Yu D., Seo S., Stone E. M., and Sheffield V. C. (2012) Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J. Biol. Chem. 287, 20625–20635 10.1074/jbc.M112.341487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoetzel C., Laurier V., Davis E. E., Muller J., Rix S., Badano J. L., Leitch C. C., Salem N., Chouery E., Corbani S., Jalk N., Vicaire S., Sarda P., Hamel C., Lacombe D., et al. (2006) BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 38, 521–524 10.1038/ng1771 [DOI] [PubMed] [Google Scholar]
- 25. Stoetzel C., Muller J., Laurier V., Davis E. E., Zaghloul N. A., Vicaire S., Jacquelin C., Plewniak F., Leitch C. C., Sarda P., Hamel C., de Ravel T. J., Lewis R. A., Friederich E., Thibault C., et al. (2007) Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am. J. Hum. Genet. 80, 1–11 10.1086/510256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. C., Ou Y. Y., Badano J. L., Esmail M. A., Leitch C. C., Fiedrich E., Beales P. L., Archibald J. M., Katsanis N., Rattner J. B., and Leroux M. R. (2005) MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J. Cell Sci. 118, 1007–1020 10.1242/jcs.01676 [DOI] [PubMed] [Google Scholar]
- 27. Katoh Y., Nozaki S., Hartanto D., Miyano R., and Nakayama K. (2015) Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362 10.1242/jcs.168740 [DOI] [PubMed] [Google Scholar]
- 28. Russel D., Lasker K., Webb B., Velázquez-Muriel J., Tjioe E., Schneidman-Duhovny D., Peterson B., and Sali A. (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLos Biol. 10, e1001244 10.1371/journal.pbio.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bin J., Madhavan J., Ferrini W., Mok C. A., Billingsley G., and Héon E. (2009) BBS7 and TTC8 (BBS8) mutations play a minor role in the mutational load of Bardet-Biedl syndrome in a multiethnic population. Hum. Mutat. 30, E737–E746 10.1002/humu.21040 [DOI] [PubMed] [Google Scholar]
- 30. Katsanis N., Ansley S. J., Badano J. L., Eichers E. R., Lewis R. A., Hoskins B. E., Scambler P. J., Davidson W. S., Beales P. L., and Lupski J. R. (2001) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293, 2256–2259 10.1126/science.1063525 [DOI] [PubMed] [Google Scholar]
- 31. Chen J., Smaoui N., Hammer M. B., Jiao X., Riazuddin S. A., Harper S., Katsanis N., Riazuddin S., Chaabouni H., Berson E. L., and Hejtmancik J. F. (2011) Molecular analysis of Bardet-Biedl syndrome families: report of 21 novel mutations in 10 genes. Invest. Ophthalmol. Vis. Sci. 52, 5317–5324 10.1167/iovs.11-7554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Politis A., Stengel F., Hall Z., Hernández H., Leitner A., Walzthoeni T., Robinson C. V., and Aebersold R. (2014) A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat. Methods 11, 403–406 10.1038/nmeth.2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walzthoeni T., Leitner A., Stengel F., and Aebersold R. (2013) Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 23, 252–260 10.1016/j.sbi.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 34. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., and Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 10.1073/pnas.1120559109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmström L., and Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- 36. Fernandez-Martinez J., Kim S. J., Shi Y., Upla P., Pellarin R., Gagnon M., Chemmama I. E., Wang J., Nudelman I., Zhang W., Williams R., Rice W. J., Stokes D. L., Zenklusen D., Chait B. T., Sali A., and Rout M. P. (2016) Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell 167, 1215–1228.e1225 10.1016/j.cell.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng-Elmore X., Gao X. Z., Pellarin R., Schneidman-Duhovny D., Zhang X. J., Kozacka K. A., Tang Y., Sali A., Chalkley R. J., Cote R. H., and Chu F. (2014) Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J. Mol. Biol. 426, 3713–3728 10.1016/j.jmb.2014.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leitner A., Joachimiak L. A., Bracher A., Mönkemeyer L., Walzthoeni T., Chen B., Pechmann S., Holmes S., Cong Y., Ma B., Ludtke S., Chiu W., Hartl F. U., Aebersold R., and Frydman J. (2012) The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 10.1016/j.str.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Plimpton R. L., Cuéllar J., Lai C. W., Aoba T., Makaju A., Franklin S., Mathis A. D., Prince J. T., Carrascosa J. L., Valpuesta J. M., and Willardson B. M. (2015) Structures of the Gβ-CCT and PhLP1-Gβ-CCT complexes reveal a mechanism for G-protein β-subunit folding and Gbetagamma dimer assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 2413–2418 10.1073/pnas.1419595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan D., Li Q., Zhang M. J., Liu C., Ma C., Zhang P., Ding Y. H., Fan S. B., Tao L., Yang B., Li X., Ma S., Liu J., Feng B., et al. (2016) Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife 5, e12509 10.7554/eLife.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leitner A., Walzthoeni T., and Aebersold R. (2014) Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 9, 120–137 10.1038/nprot.2013.168 [DOI] [PubMed] [Google Scholar]
- 42. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., and Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 10.1038/nmeth.2099 [DOI] [PubMed] [Google Scholar]
- 43. Knockenhauer K. E., and Schwartz T. U. (2015) Structural characterization of Bardet-Biedl syndrome 9 protein (BBS9). J. Biol. Chem. 290, 19569–19583 10.1074/jbc.M115.649202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cooper S., Khatib F., Treuille A., Barbero J., Lee J., Beenen M., Leaver-Fay A., Baker D., Popović Z., and Players F. (2010) Predicting protein structures with a multiplayer online game. Nature 466, 756–760 10.1038/nature09304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X., Cimermancic P., Yu C., Schweitzer A., Chopra N., Engel J. L., Greenberg C., Huszagh A. S., Beck F., Sakata E., Yang Y., Novitsky E. J., Leitner A., Nanni P., Kahraman A., et al. (2017) Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell Proteomics 16, 840–854 10.1074/mcp.M116.065326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laurier V., Stoetzel C., Muller J., Thibault C., Corbani S., Jalkh N., Salem N., Chouery E., Poch O., Licaire S., Danse J. M., Amati-Bonneau P., Bonneau D., Mégarbané A., Mandel J. L., and Dollfus H. (2006) Pitfalls of homozygosity mapping: an extended consanguineous Bardet-Biedl syndrome family with two mutant genes (BBS2, BBS10), three mutations, but no triallelism. Eur. J. Hum. Genet. 14, 1195–1203 10.1038/sj.ejhg.5201688 [DOI] [PubMed] [Google Scholar]
- 48. Beales P. L., Badano J. L., Ross A. J., Ansley S. J., Hoskins B. E., Kirsten B., Mein C. A., Froguel P., Scambler P. J., Lewis R. A., Lupski J. R., and Katsanis N. (2003) Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am. J. Hum. Genet. 72, 1187–1199 10.1086/375178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fauser S., Munz M., and Besch D. (2003) Further support for digenic inheritance in Bardet-Biedl syndrome. J. Med. Genet. 40, e104 10.1136/jmg.40.8.e104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deveault C., Billingsley G., Duncan J. L., Bin J., Theal R., Vincent A., Fieggen K. J., Gerth C., Noordeh N., Traboulsi E. I., Fishman G. A., Chitayat D., Knueppel T., Millán J. M., Munier F. L., et al. (2011) BBS genotype-phenotype assessment of a multiethnic patient cohort calls for a revision of the disease definition. Hum. Mutat. 32, 610–619 10.1002/humu.21480 [DOI] [PubMed] [Google Scholar]
- 51. Harville H. M., Held S., Diaz-Font A., Davis E. E., Diplas B. H., Lewis R. A., Borochowitz Z. U., Zhou W., Chaki M., MacDonald J., Kayserili H., Beales P. L., Katsanis N., Otto E., and Hildebrandt F. (2010) Identification of 11 novel mutations in eight BBS genes by high-resolution homozygosity mapping. J. Med. Genet. 47, 262–267 10.1136/jmg.2009.071365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Badano J. L., Ansley S. J., Leitch C. C., Lewis R. A., Lupski J. R., and Katsanis N. (2003) Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am. J. Hum. Genet. 72, 650–658 10.1086/368204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S. S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P., Zecha A., Mohseni M., Püttmann L., Vahid L. N., Jensen C., et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63 10.1038/nature10423 [DOI] [PubMed] [Google Scholar]
- 54. Nishimura D. Y., Searby C. C., Carmi R., Elbedour K., Van Maldergem L., Fulton A. B., Lam B. L., Powell B. R., Swiderski R. E., Bugge K. E., Haider N. B., Kwitek-Black A. E., Ying L., Duhl D. M., et al. (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum. Mol. Genet. 10, 865–874 10.1093/hmg/10.8.865 [DOI] [PubMed] [Google Scholar]
- 55. Ma W., and Goldberg J. (2013) Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32, 926–937 10.1038/emboj.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackson L. P., Lewis M., Kent H. M., Edeling M. A., Evans P. R., Duden R., and Owen D. J. (2012) Molecular basis for recognition of dilysine trafficking motifs by COPI. Dev. Cell 23, 1255–1262 10.1016/j.devcel.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dodonova S. O., Diestelkoetter-Bachert P., von Appen A., Hagen W. J., Beck R., Beck M., Wieland F., and Briggs J. A. (2015) Vesicular transport: a structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198 10.1126/science.aab1121 [DOI] [PubMed] [Google Scholar]
- 58. Chou H.-T., Apelt L., Farrell D. P., White S. R., Woodsmith J., Svetlov V., Goldstein J. S., Nager A. R., Li Z., Muller J., Dolifus H., Nudler E., Stelzl U., DiMaio F., Nachury M. V., and Walz T. (2019) The molecular architecture of native BBSome obtained by an integrated structural approach Structure 27, 1–11 10.1016/j.str.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mindell J. A., and Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 10.1016/S1047-8477(03)00069-8 [DOI] [PubMed] [Google Scholar]
- 60. Marabini R., Masegosa I. M., San Martin M. C., Marco S., Fernandez J. J., de la Fraga L. G., Vaquerizo C., and Carazo J. M. (1996) Xmipp: an image processing package for electron microscopy. J. Struct. Biol. 116, 237–240 10.1006/jsbi.1996.0036 [DOI] [PubMed] [Google Scholar]
- 61. Sorzano C. O., Bilbao-Castro J. R., Shkolnisky Y., Alcorlo M., Melero R., Caffarena-Fernández G., Li M., Xu G., Marabini R., and Carazo J. M. (2010) A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171, 197–206 10.1016/j.jsb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Scheres S. H. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 64. van Heel M., and Schatz M. (2005) Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 10.1016/j.jsb.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 65. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 66. Tiruttani Subhramanyam U. K., Kubicek J., Eidhoff U. B., and Labahn J. (2014) Cloning, expression, purification, crystallization and preliminary crystallographic analysis of the C-terminal domain of Par-4 (PAWR). Acta Crystallogr. F Struct. Biol. Commun. 70, 1224–1227 10.1107/S2053230X14014691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Webb B., Lasker K., Velázquez-Muriel J., Schneidman-Duhovny D., Pellarin R., Bonomi M., Greenberg C., Raveh B., Tjioe E., Russel D., and Sali A. (2014) Modeling of proteins and their assemblies with the integrative modeling platform. Methods Mol. Biol. 1091, 277–295 10.1007/978-1-62703-691-7_20 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.