Abstract
Protamines are small, highly-specialized, arginine-rich, and intrinsically-disordered chromosomal proteins that replace histones during spermiogenesis in many organisms. Previous evidence supports the notion that, in the animal kingdom, these proteins have evolved from a primitive replication-independent histone H1 involved in terminal cell differentiation. Nevertheless, a direct connection between the two families of chromatin proteins is missing. Here, we primarily used electron transfer dissociation MS-based analyses, revealing that the protamines in the sperm of the liverwort Marchantia polymorpha result from post-translational cleavage of three precursor H1 histones. Moreover, we show that the mature protamines are further post-translationally modified by di-aminopropanelation, and previous studies have reported that they condense spermatid chromatin through a process consisting of liquid-phase assembly likely involving spinodal decomposition. Taken together, our results reveal that the interesting evolutionary ancestry of protamines begins with histone H1 in both the animal and plant kingdoms.
Keywords: 14-3-3 protein, mass spectrometry (MS), histone, electron microscopy (EM), chromatography, di-aminopropanelation, histone H1, Marchantia, mass spectrometry, protamines
Introduction
Protamines are a group of highly-specialized chromosomal sperm nuclear basic proteins (SNBPs)3 that are present in many organisms (1–4). They replace histones during the last stages of male germ terminal differentiation of spermiogenesis (5). They are small (25–100 amino acids), highly arginine-rich proteins (6, 7) that belong to the broad family of intrinsically-disordered proteins (8). Despite their disorganized structure in solution, protamines from both chordate and nonchordate organisms are able to adopt secondary structure in the presence of helicogenic buffers (9) and upon binding to DNA in sperm (10). The functional role of these proteins appears to be multifaceted (11), but their ultimate biological significance is not yet properly understood. In addition to tightly compacting the sperm genome and protecting against DNA damage (11, 12) during its exit from the body, protamines may also assist in erasing the somatic epigenetic contribution of histones. Protamines are sporadically but nonrandomly distributed (13, 14) throughout the animal and plant kingdoms (3). Fish are a good example of this (13, 15). Although the species and families of some orders contain only protamines (P-type), sperm from other orders might contain different SNBP types such as histones (H-type) or protamine-like (PL-type) proteins (4, 16).
Despite the heterogeneity in SNBP types, the PL-type, in addition to their highly-basic amino acid (Lys/Arg) composition, contains a distinctive winged helix domain (WHD) that is evolutionarily related to the distinctive WHD of somatic linker H1 histones (17). Also, in those organisms that retain the full histone complement in their sperm-like echinoderms or in zebrafish, they either contain an unusually-long sperm-specific histone H1 (18) or elevated levels of histone H1 (19), which in both instances assist in maintaining a highly-compacted chromatin organization.
Therefore, a structural (20) and evolutionary (16) role of the histone H1 family of chromosomal proteins (21) in shaping sperm chromatin organization is not surprising. However, although a connection between protamines (lacking the WHD) and histone H1 has been demonstrated, and organisms possessing a mixture of related PL-type and protamines have been described (22, 23), this is the first observation of the exclusive presence of histone H1 ontogenically-related protamines. The results described below conclusively show that in Marchantia the post-translational processing of three highly-specialized H1 histones of the PL type, which are expressed in antheridiophores, gives rise to the protamines that are found in the sperm of this organism.
Results
Initial evidence for the presence of protamines in Marchantia polymorpha sperm
The first evidence of protamines in M. polymorpha comes from a study published in 1978 by Reynolds and Wolfe (24). Fig. 1D of that paper shows an acetic acid urea-PAGE depicting a pattern of four small molecular mass proteins that is almost identical to that shown in Fig. 1a, lane 1. In both instances, the electrophoretic pattern is characterized by the complete absence of histones, and as it was initially concluded:“ … histones are completely lost and replaced by proteins … resembling animal protamines.” (see Ref. 24). Coarse electrophoretic fractionation of these proteins revealed a composition enriched in basic amino acids that was also important in reaching this conclusion. In a subsequent paper, the same authors extended their study to a broader spectrum of plants, including algae, bryophytes, and ferns, for which representative species containing protamines were also identified.
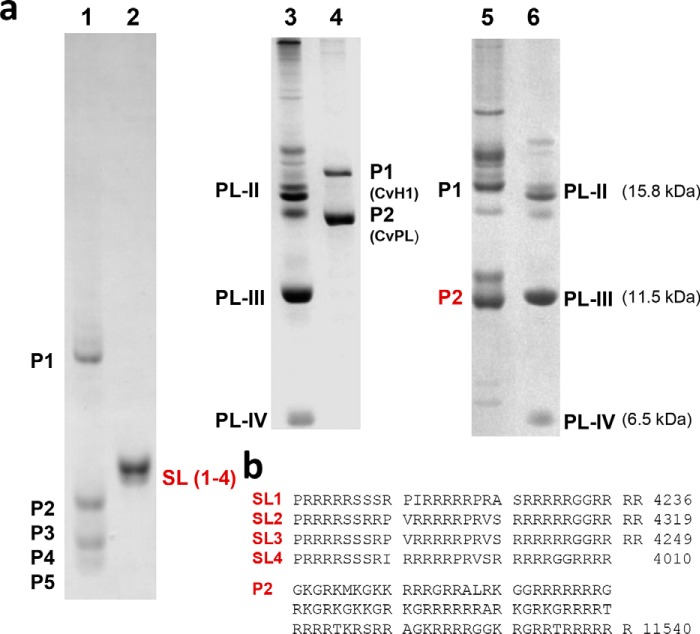
Figure 1.
Comparison of the protamines of the liverwort M. polymorpha to other nonchordate and chordate protamines. a, electrophoresis (acetic acid urea-PAGE) of the SNBPs from the following: lane 1, M. polymorpha (26); lane 2, Oncorhynchus keta (salmon, salmine (SL)) (84); lanes 3 and 6, M. californianus (California mussel) used as a marker (85, 86); lane 4, Chaetopterus variopedatus (parchment worm) 23); and lane 5, Styela montereyensis (Monterey stalked tunicate) (22). b, amino acid sequences of the O. keta salmine (SL1–4) protamines (84) and the P2 protamine of Styela plicata (sea squirt) (22).
Fig. 1 shows the electrophoretic pattern observed in the sperm of M. polymorpha SNBPs compared with those of some representative nonchordate and chordate animals. The amino acid sequence of the protamines from these organisms is arginine-rich (Fig. 1b), and in the nonchordate organisms a marine worm, Chaetopterus variopedatus (lane 4) (23), and a tunicate, Styela montereyensis (lane 5) (22), a WHD-containing PL structurally related to chordates exists (P1 in Fig. 1a, lanes 4 and 5). The high electrophoretic mobility of M. polymorpha SNBPs resembles that of the small protamines present in salmon sperm (Fig. 1a, lane 2). A very similar arginine-rich composition is observed in protamines of other nonchordate and chordate organisms which, like in salmon, exclusively contain P-type SNBPs in their sperm (4, 7, 25).
Marchantia genome and male transcriptome indicate a relation of its sperm protamines to histone H1
In 2014, we used SNBPs isolated from M. polymorpha sperm and Edman degradation N-terminal amino acid sequencing to publish a partial amino acid sequence corresponding to protamine P5 (26) (Fig. 1a, lane 1). This sequence information was more recently used to identify a histone H1 gene of the SNBP PL-type (termed MpPRM) from the analysis of the male gametogenesis transcriptome of this organism which is expressed during spermiogenesis (27). The protein contains the characteristic histone H1 WHD (Fig. 2, a, PL-1, and b) in addition to several arginine-rich clusters at its C-terminal end.
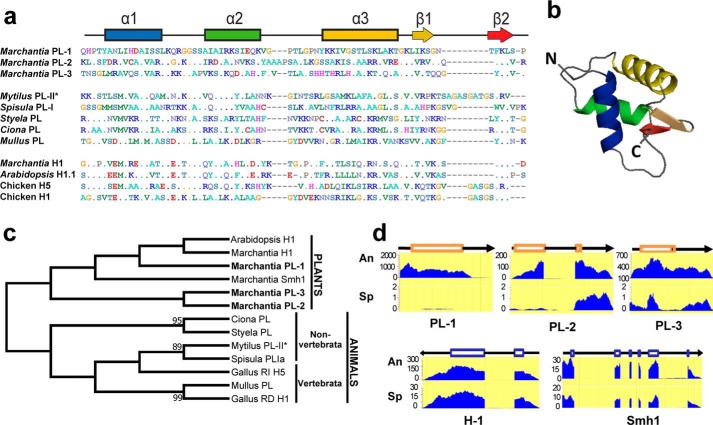
Figure 2.
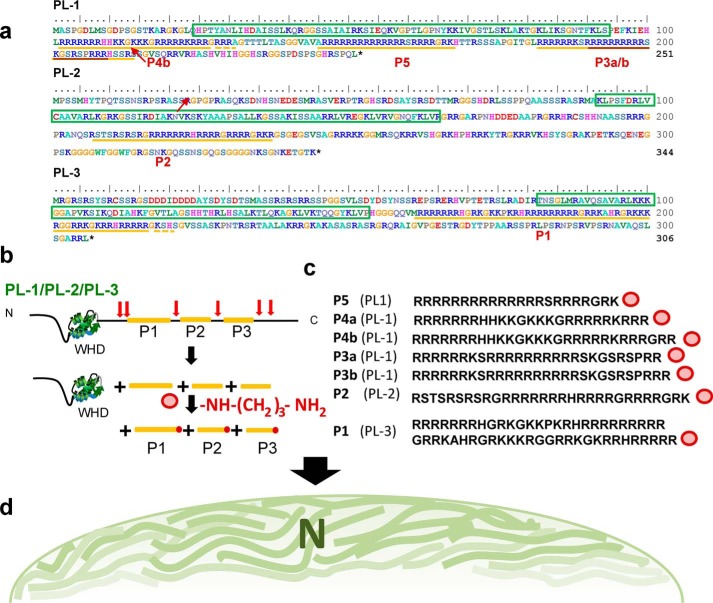
Genome of M. polymorpha reveals the presence of three histone H1-related PL proteins. a, amino acid sequence alignment of the protein regions corresponding to the WHD of M. polymorpha PL1–PL3 compared with the amino acid sequences corresponding to the WHD of nonchordate and chordate organisms as well as the WHD of some representative somatic H1 histones, including the H5 histone, which is in the terminally differentiated erythrocytes of nonmammalian vertebrates: M. californianus PL-II* (86); S. solidissima (surf clam) PL-I (87); S. plicata PL (22); C. intestinalis (transparent sea squirt) PL (22); and M. surmuletus (striped ret mullet) PL (88). b, predicted tertiary structure organization of the M. polymorpha PL-1 WHD. c, phylogenetic analysis of the proteins shown in a. d, levels of gene expression in antheridiophores (An) and in sporophytes (Sp) of M. polymorpha PL1–PL3. In the schematic representation, the boxes indicate exon domains. Smh1, single myb histone 1 (first identified in maize) that binds to telomere DNA repeats in plants (41).
The more recent availability of the M. polymorpha genome (28) has allowed us to identify two additional arginine-rich PL proteins also containing a WHD (Fig. 2a, PL-2/PL-3). The phylogenetic relationship of these three PL proteins and their evolutionary connection to other M. polymorpha histone H1s, as well as to animal PLs, is shown in Fig. 2c. The PLs identified in this way are mainly expressed in antheridiophores (the M. polymorpha male sex organ), in contrast to the histone H1 counterparts, which are more similarly expressed in both antheridiophores and sporophytes (Fig. 2d).
Determination of the amino acid sequence of M. polymorpha SNBPs provides evidence for their PL origin
The sequences for the rest of the M. polymorpha SNBPs (P1–P4 in Fig. 1a) were determined using HPLC-coupled MS. The HPLC-fractionated proteins were fragmented using electron transfer dissociation (ETD) and collisionally-activated dissociation (CAD) (Fig. 3, a–e, see also Table 1 and Table S1) (29–31). The sequences obtained (see Table 1), primarily using ETD, confirmed the arginine-rich protamine nature of these proteins, and they revealed their structural relation to the three PL proteins that were bioinformatically identified above. These results conclusively show that M. polymorpha SNBPs are the post-translational cleavage products of three independent, histone H1–related, and protamine-like precursors.
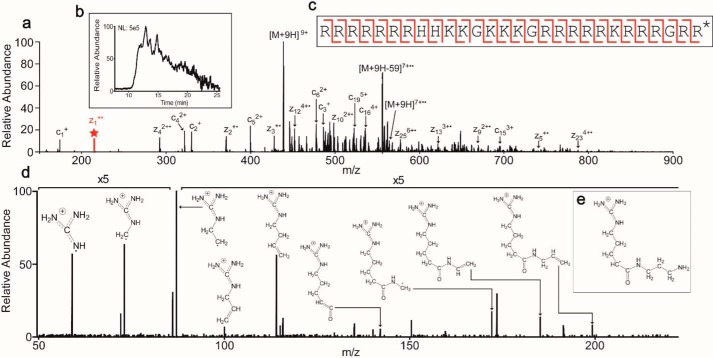
Figure 3.
Detection and fragmentation of the protamine RRRRRRRHHKKGKKKGRRRRRKRR-RGRR* (where * = di-aminopropanelated C terminus) (this peptide corresponds to amino acids 102–129 of Marchantia PL-I (MpPRM) NCBI accession number BAU71552). a, precursor ions at 438.07 m/z (z = 9) were fragmented by ETD to produce the MS2. Selected abundant ions are labeled (M = precursor mass; 59 = mass of guanidinium group). The peak in red with a star above represents the z1+· ion. b, extracted chromatogram of the protamine. Because the peak elutes for over 10 min, multiple MS2 scans were averaged to produce the final scan shown in a. c, sequence coverage by ETD. The cleavages denoted in red depict the c and z• ions observed giving unambiguous sequence coverage across the length of the protamine. d, HCD MS3 fragmentation spectrum of the z1+· ion at 215.1740 m/z (z = 1). Proposed structures for the major ions are depicted. e, proposed structure of the z1+· fragment ion.
Table 1.
Identified protamines
Asterisk indicates the di-aminopropanylated C terminus. The lower part of the table corresponds to protamines of lower abundance. Amino acids in parentheses do not contain fragment ions confirming their location. These residues can be interchanged within the parentheses.
| M + H | Sequence | Protein name | First amino acid residue no. | Last amino acid residue no. | NCBI accession no. |
|---|---|---|---|---|---|
| 3955.50 | RRRRRRKSRRRRRRRRRRSKGSRSPRRR* | PL-1 (MpPRM) | 182 | 209 | BAU71552 |
| 3799.41 | RRRRRRKSRRRRRRRRRRSKGSRSPRR* | PL-1 (MpPRM) | 182 | 208 | BAU71552 |
| 3942.56 | RRRRRRRHHKKGKKKGRRRRRKRRRGRR* | PL-1 (MpPRM) | 102 | 129 | BAU71552 |
| 3313.16 | RRRRRRRRRRRRRRSRRRRGRK* | PL-1 (MpPRM) | 146 | 167 | BAU71552 |
| 7466.75 | RRRRRRRHGRKGKKPKRHRRRRRRRRRGRRKAHRGRKKKRG-GRRKGKRRHRRRRR* | PL-3 | 162 | 216 | PTQ32223 |
| 4082.51 | RSTSRSRSRGRRRRRRRHRRRRGRRRRGRK* | PL-2 | 207 | 236 | PTQ35141 |
| 3780.36 | RRRRRRKSRRRRRRRRRR(SKGSH)SPRR* | PL-1 (MpPRM) | 182 | 208 | BAU71552 |
| 4139.33 | RRRRRRKSRRRR(RRTVDRRRRRRRRRRR) | PL-1 (MpPRM) | 182 | 209 | BAU71552 |
| 4366.65 | RRRRRRKSRRRRRRRRRRSKGSRSPRRRHSSR | PL-1 (MpPRM) | 182 | 213 | BAU71552 |
| 7819.01 | RRRRRRRHGRKGKKPKRHRRRRRRRRRGRRKAHRGRKKKRGGRRKGKRRHRRRRR(GKSH) | PL-3 | 162 | 220 | PTQ32223 |
M. polymorpha protamines are di-aminopropanelated at their C termini
Interestingly, in multiple ETD MS2 scans, a c-ion series could be deduced that covered the majority of the protein. However, there was a strong ion at 215.1740 m/z (z = 1) present in multiple species denoted by the star in Fig. 3a. From this ion, a series of amino acids was determined that complemented the c-ion series. Therefore, the ion at 215.1740 m/z must be a z• ion. Based on the sequence deduced from the c-ion series, the database BLAST search provided the protein from which the observed species originated as well as the final residue in the protein. This ion corresponded to an arginine with an unknown modification of 56.0740 Da greater than an unmodified arginine. This unknown modification was also observed on C-terminal lysines.
A theoretical elemental composition was calculated to assist in the determination of the modification present on the z1+· ion. The molecular formula C9H21N5O was determined to be the closest combination based on parts/million error. Interestingly, there was only one oxygen atom present in the theoretical composition. This would place the modification on the C terminus because an unmodified C terminus would have two oxygens. After accounting for the arginine side chain (C4H11N3) and the protein backbone (C2HO), the elements C3H9N2 remain.
To determine the structure of the modification, the ion at 215.1740 m/z was first formed by ETD and then re-isolated for fragmentation by high-energy collisional dissociation (HCD) to give the MS3 scan shown in Fig. 3d. Because of the guanidino group present on the arginine side chain, the charge should be stabilized within this group. Therefore, the structure can be determined from additions to the arginine side chain. As shown in Fig. 3d, the ions present confirm the structure shown in Fig. 3e. All identified protamines contained this modification. The modification was absent in other observed proteins from Marchantia (results not shown), indicating that this modification is specific to protamines.
Chromatin organization during M. polymorpha spermiogenesis
Organisms where SNBPs of the protamine type undergo a post-translational processing involving extensive protein cleavage often exhibit a characteristic nuclear organization during this process. A transition from lamello-fibrillar chromatin organization to a complete electron-dense highly-compacted organization in their mature sperm is observed (7). Such a transition has been observed both in internally fertilizing invertebrate (26, 32) and vertebrate organisms (33–35).
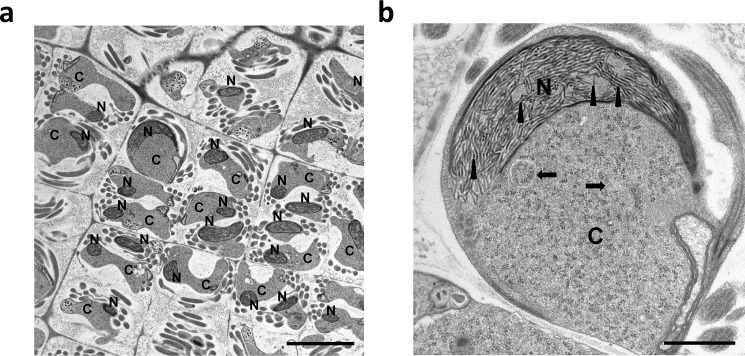
Fig. 4 shows an electron microscopic image of spermatids in developing M. polymorpha antheridia (36) (the haploid structure producing male gametes). These spermatids have been shown to contain PL-1–type protamines until the late stages of spermiogenesis (27). As can be seen in Fig. 4, chromatin adopts a distinct fibrillar organization with dispersed uniform fibers of 24 ± 3-nm thickness. The fibers appear clearly dispersed and further coalesce into a uniform, highly-condensed, and electron-dense opaque nucleus in the mature sperm (37). This type of chromatin organization has also been observed in the liverwort Blasia pusilla, a bryophyte (26). The peculiar transitional chromatin organization before complete condensation observed in these organisms has been proposed to be mediated by the liquid-liquid phase separation mechanism of spinodal decomposition (26). The process is framed on a physicochemical model that involves “kinetic, equilibrium, and structural aspects of a system in route to equilibrium.” In it, the less electron-dense nucleoplasm and the chromatin appear to be continuous rather than one disperse and one continuous phase (7).
Figure 4.
Electron micrographs of M. polymorpha spermatids. a, chromatin condensing nucleus (N) exhibiting a shrinking cytoplasm (C). The scale bar is 5 μm. b, electron micrograph (greater magnification) of a condensing spermatid showing the characteristic 24 ± 3-nm fibers (arrowheads). The arrows point to autophagosomes. The scale bar is 1 μm.
Discussion
Protamines in plants
The occurrence of protamines in the sperm of Marchantia may have broader implications for the overall evolution of SNBPs. As pointed out in earlier work, the shift to the convergent presence of SNBPs in the sperm of both plants and animals might have involved selection pressure to reduce sperm nuclear weight and volume for a swimming male gamete (24) in internally fertilizing organisms. Indeed, protamines appear to be present in more primitive lower plants with motile sperm and are much less ubiquitous in the male nuclei of the pollen grain (3). In this regard, it is interesting that while in animals protamines may be present in both internal and external fertilizers, the last type of fertilization represents an SNBP evolutionary bottleneck. Once organisms from a clade have acquired this fertilization mode, there is no return to the histone or PL SNBP types (38, 39). Moreover, evolutionarily-driven interspecific changes in the regulation of protamine genes and amino acid sequence in mice and mammals have been shown to confer inter-specific sperm competition (fertility) advantage (40, 42, 43). Thus, in addition to providing genome compaction/protection against DNA damage and somatic histone epigenetic clearance (6, 11), protamines might have evolved to fine-tune the structural chromatin features of the nucleus (44). These changes optimize the swimming potential of internal fertilizers, not only in the animal kingdom but also in plant internal fertilizers, such as in the case of bryophites, including liverworts, mosses, and hornworts (45).
Protamines and histone H1
Fig. 5 summarizes the relationship between the SNBPs of M. polymorpha (Fig. 1a, lane 1) and their histone H1–related PL precursors. Once more, these results emphasize the evolutionary relatedness of protamines to histone H1, not only in animals (4, 16) but also in plants (Fig. 2c). Although protamines are the only SNBPs found in M. polymorpha mature sperm, their PL histone H1–related precursors are present in the spermatids in the early stages of spermiogenesis (27). Moreover, M. polymorpha protamines present in the final sperm are the product of post-translational cleavage of their precursors (Fig. 5b).
Figure 5.
Protamines of M. polymorpha sperm are the post-translational cleavage products from three PL histone H1 precursors Pl-1, PL-2, and PL-3. a, amino acid sequences of the three PL precursors. The boxes with green borders indicate the WHD, which is characteristic of H1 histones. The regions underlined in yellow correspond to the peptides identified in mature sperm. The region of Pl-1 underlined in brown represents the first M. polymorpha protamine sequence determined using conventional Edman degradation sequencing (26). b, schematic representation of the post-translational cleavage undergone by M. polymorpha PLs. Precursor SNBP histone H1-related PLs consisting of an N-terminal (N) winged helix domain (WDH) and an arginine-rich C-terminal domain (C) are post-translationally cleaved (red arrows) into smaller fragments (yellow) and further C-terminally di-aminopropanelated (dark red dots). c, amino acid sequence of the protamines (Fig. 1a, lane 1) identified by MS. d, schematic representation of the intermediate liquid phase chromatin condensation stage observed in spermatids (Fig. 4).
Post-translational cleavage of protamine precursors is quite a general occurrence both in protostome and deuterostome species, which appears to be independent of their histone H1 origin, and it also appears to have been the product of evolutionary convergence (46). For instance, the protamines of cephalopods (Sepia officinalis (cuttlefish) (47)/Loligo opalescens (squid) (46)) and the protamine P2 in mammals (mouse (48) and human (49)) are the products of a gene encoding a protein with an N-terminal leading domain consisting of a mixture of neutral/polar amino acids and a highly-arginine–rich C-terminal end. The N-terminal leading sequence is gradually removed during the late stages of spermiogenesis following DNA binding (6, 48), and hence it is hypothesized to play an important role in proper chromatin deposition and condensation.
Protamine post-translational processing and sperm chromatin condensation
The post-translational modifications (PTMs) undergone by M. polymorpha protamine precursors include cleavage as well as C-terminal di-aminopropanelation (Fig. 5, b and c). Di-aminopropane is a product of the oxidation of spermine and spermidine that is catalyzed by the FAD-dependent polyamine oxidases commonly found in monocotyledonous plants (50, 51) and that in A. thaliana have been shown to participate in polyamine metabolism with important involvement in plant development (50). Upon activation of the arginine α-carboxyl group in the M. polymorpha protamine, it can readily acylate 1,3-di-aminopropane to produce the PTM observed here. Of note, oxidation of spermine by diamine oxidase occurs in human seminal plasma and is related to sperm fertility (52, 53).
The occurrence of PTMs in mammalian protamines has been well-documented (6), where phosphorylation, acetylation, and methylation of different residues have been described (54). Although the functional role of these PTMs is not well-understood, serine phosphorylation was initially proposed to regulate the proper interaction of the highly positively–charged protamines with DNA, therefore ensuring a proper chromatin assembly (55–57). In those instances where cleavage of a protamine precursor takes place, like in mammalian protamine P2, phosphorylation does not occur until the initiation of the cleavage process (56).
There is not much information about C-terminal PTMs due mainly to the experimental difficulty of their analysis (58), and hence there is not much information about their functional involvement. However, half of the biologically active peptides and peptide hormones are α-amidated at their C termini where the neutralization of the negative charge enhances their hydrophobicity and improves their receptor-binding activity (59). In this regard, the role of the carboxyl end di-aminopropanelation, observed for the first time here in chromosomal proteins, remains elusive. The possibility exists that as in the case of phosphorylation, it has an important role in the chromatin transitions undergone during spermiogenesis. In particular, it might play a role during the PL to protamine transition as it might neutralize the negative charge of the carboxyl end which, like in the hormone peptides (59), might otherwise interfere with proper protamine deposition onto DNA from a complex heterogeneous mixture of very small arginine-rich proteins (Figs. 1a, lane 1, and 5c).
Protamine precursor trimming and phosphorylation have been involved in chromatin patterning of the developing spermatid nucleus of those organisms undergoing liquid-phase condensation (60) through a process of spinodal decomposition (7, 32, 61). These events precede the final highly-compacted chromatin structure that is characteristic of mature sperm. The protein processing observed in M. polymorpha appears to play a similar role as its spermatids exhibit this pattern type (Figs. 4 and 5d), in which the intrinsically disordered nature of the proteins (8) involved (histone H1 and protamines) play a critical role in this process (10, 62–64). This is very similar to the phase separation process recently described for the heterochromatin domain formation by Drosophila HP1a (65) and other chromatin subcompartments (66). Indeed, sperm chromatin is a salient example of fully heterochromatinized nuclei.
Paraphrasing the title of the paper reporting the genome of M. polymorpha (28), the analysis of its SNBP composition and transitions, made possible through the analysis of its genome, provides a powerful evolutionary insight that transcends that of plants and has implications for the evolution of these protamines and their conserved chromatin organization encompassing the plant and animal kingdoms.
Experimental procedures
Materials
Male accession of M. polymorpha, Takaragaike-1 (Tak-1) (67), was grown and induced to sexual reproduction as described in Minamino et al. (36).
LCMS-grade water and LCMS-grade acetonitrile were purchased from VWR Scientific. LCMS-grade 2-propanol and formic acid were purchased from Thermo Fisher Scientific. Acetic acid, vasoactive intestinal peptide, and angiotensin I were purchased from Sigma.
Protein extraction
M. polymorpha sperm from antheridiophores was resuspended in 100 mm Tris-HCl (pH 7.5), 0.5% Triton X-100 containing 10 μg/ml tosyl lysine chloromethyl ketone buffer (at a ratio of 20 μl/liter antheridiophore) and centrifuged at maximum speed in an Eppendorf microcentrifuge. The pellet obtained in this way was homogenized in 0.4 n HCl (at a ratio of 10 μl/1 antheridiophore) and homogenized in a small Dounce with 10 strokes. The HCl extract was precipitated overnight with 6 volumes of acetone at −20 °C, and the next day it was centrifuged at maximum speed in an Eppendorf microcentrifuge. The protein pellet was washed with acetone at room temperature, centrifuged again under the same conditions, and dried in a speedvac. The protein pellet thus obtained was resuspended in water for further use.
Gel electrophoresis
Gel electrophoresis was carried out on acetic acid (5%), urea (2.5 m), and (15%) PAGE-polymerized using thiourea and hydrogen peroxide (68), as described previously (69).
Genome screening
M. polymorpha proteins containing a histone H1 globular domain were retrieved from the M. polymorpha Genome Database (http://marchantia.info/4 (28)). Gene structure and levels of expression were analyzed using the Genome Browser implemented in this database. PL proteins from the mussel M. californianus (PL-II*; accession no. AAB24707), the surf clam Spisula solidissima (PLIa; accession no. AAT45384), the tunicates S. montereyensis (P1; accession no. AAQ01227) and Ciona intestinalis (PL; accession no. XP_002130983), the fish Mullus surmuletus (PL; accession no. Q08GK9), histone H1.1 from Arabidopsis thaliana (accession no. AAF63139), and histones H1 and H5 from the chicken Gallus gallus (accession nos. P09987 and AAA48798, respectively) were retrieved from the GenBankTM database.
Multiple sequence alignments
These were performed using MAFFT version 7.310 (70) in the Jalview2 package (71). The evolutionary history of the winged-helix globular domains (WHD) was inferred by using the maximum likelihood method based on the Le Gascuel model (72) in the software MEGA version 7 (73). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (only bootstrap values higher than 80% are shown). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 2.3546)). The analysis involved 13 amino acid sequences. All positions with less than 95% site coverage were eliminated. That means fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 65 positions in the final dataset.
Tertiary structure prediction
The tertiary structures of the globular domains of histone H1 and PL proteins were modeled in the SWISS-MODEL server (74) using the most closely-related structure, automatically searched by the software, as a template in each case. The obtained structures were rendered using the PyMOL Molecular Graphics System version 1.2r1.
Sample preparation
One μl of the acetone-precipitated protein-HCl extract suspension was diluted 10-fold with 0.1% acetic acid in water. 300 nl of the dilution, corresponding to ∼0.2% of the total suspension, was pressure-loaded onto a reversed-phase column containing 10 cm of 3-μm diameter, 300 Å PLRP-s packing material (Polymer Laboratories) within a 360 × 75-μm fused silica capillary integrated with an electrospray tip (75, 76). Additionally, 100 fmol of both vasoactive intestinal peptide and angiotensin I were loaded onto the column as internal standards.
Chromatography and mass spectrometry
An Agilent Technologies (Palo Alto, CA) 1100 Series binary HPLC system coupled to a Thermo Fisher Scientific Orbitrap Fusion Tribrid mass spectrometer (San Jose, CA) operating in standard pressure mode was used to characterize the proteins in the sample (77). Proteins that were unable to be retained on the column due to high hydrophilicity were identified using an isocratic elution at 100% solvent A (0.3% formic acid in water) for 25 min at a flow rate of ∼50 nl/min (Fig. S1). Following the identification of proteins not retained on the column, the column was washed with solvent A for an additional 25 min at ∼100 nl/min to remove any additional salts. The remaining proteins were then eluted using a gradient of 0–10−30–70–100% solvent B (72% acetonitrile, 18% isopropenyl acetate, 10% water, and 0.3% formic acid) in 0–10−20–30–35 min at a flow rate of ∼100 nl/min (Fig. S2).
Proteins were selected for fragmentation based on a 60,000 resolution Orbitrap MS1 scan. Using top speed mode with a 3-s cycle time, proteins with a charge state of 4 and higher were isolated by the quadrupole with a width of 1.5 m/z and fragmented by ETD and CAD (29–31). All MS2 scans were analyzed in the Orbitrap at 60,000 resolution with an AGC target of 2.0e5 and a maximum injection time of 100 ms. ETD was performed on ions with m/z 300–700 using calibrated reaction times (78, 79) (Table S2). CAD was performed on ions with m/z 400–1500 at 30% normalized collision energy. Ions were placed on an exclusion list for 30 s after three scans in 30 s. For only the isocratic identification, all scans included an in-source dissociation energy of 40 to break up any adducts that may be present due to the acetone precipitation.
Determination of C-terminal di-aminopropanelation by MS3
A targeted MS3 method was used to determine the structure of the z1+· ion present in many species because it did not correspond to a known amino acid (80, 81). First, the precursor mass of 439.18 m/z (z = 9) was isolated with the quadrupole at a width of 1.5 m/z, an AGC target of 5.0e5, and a maximum injection time of 300 ms. These ions were then fragmented by ETD using 2.0e5 reagent ions and a reaction time of 12 ms. The fragment ion at 215.174 m/z (z = 1) was then isolated with a width of 1.5 m/z and fragmented by HCD using 30% HCD collisional energy. The resulting fragment ions were analyzed in the Orbitrap at 60,000 resolution and a scan range of 50–220 m/z.
MS data analysis
The data files were manually inspected for the species present. All fragmentation scans from the major species were manually sequenced de novo by averaging all MS2 scans under the peak using Qual Browser version 4.0.27.10 (Thermo Fisher Scientific). Sequenced proteins were identified by using the NCBI Protein Blast Nonredundant protein sequences database (as of 03/22/2019) against M. polymorpha) (82, 83).
EM
EM was carried out as described previously (36). Briefly, M. polymorpha antheridia were fixed overnight with paraformaldehyde (2%) and glutaraldehyde (2%) in 0.05 m cacodylate buffer (pH 7.4) at 4 °C. The samples were next washed three times with cacodylate buffer and post-fixed with osmium tetroxide (2%) in the same buffer for 3 h at 4 °C. The samples were dehydrated by rinsing them for 30 min with increasing concentrations (50, 70, and 90%) of ethanol at 4 °C, and this was repeated four times with 100% ethanol at room temperature. The samples were finally left overnight at room temperature in 100% ethanol. The samples were subsequently embedded twice in propylene oxide (PO) for 30 min and then transferred to a 70:30 mixture of PO and Quetol-651 resin (Nisshin EM Co.) for 1 h. After evaporation of PO, the samples were transferred to 100% resin, and polymerization was allowed to proceed for 48 h at 60 °C. Ultra-thin sections were stained with 2% uranyl acetate and lead stain solution (Sigma) visualized on a JEM1400 Plus (Jeol Ltd.) transmission electron microscope.
Author contributions
R. A. D., D. F. H., and J. A. conceptualization; R. A. D. and J. A. resources; R. A. D., M. S. C., D. L. B., J. S., D. F. H., and J. A. methodology; N. M. and J. A. validation; C. R.-C. visualization; M. S. C., D. F. H., and J. A. project administration; H. E. K., J. M. E.-L., T. U., D. F. H., and J. A. writing-review and editing; J. A. formal analysis; J. A. supervision; J. A. funding acquisition; J. A. writing-original draft.
Supplementary Material
Acknowledgment
We thank Katrina V. Good for carefully proofreading the manuscript.
This work was supported by Natural Science and Engineering Research Council of Canada (NSERC) Grant RGPIN-2017-04162 (to J. A.), National Institutes of Health Grant GMO37537 (to D. F. H.), and by the Model Plant Facility, NIBB BioResource Center. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SNBP
- sperm nuclear basic protein
- ETD
- electron transfer dissociation
- CAD
- collisionally-activated dissociation
- PO
- propylene oxide
- WHD
- winged helix domain
- HCD
- high-energy collisional dissociation
- PTM
- post-translational modification
- PL
- protamine-like.
References
- 1. Bloch D. P. (1969) A catalog of sperm histones. Genetics 61, Suppl. 93–111 [PubMed] [Google Scholar]
- 2. Bloch D. P. (1976) Histones of Sperm (King R. C., ed) pp. 139–167, Plenum Press, New York [Google Scholar]
- 3. Kasinsky H. E. (1989) Specificity and Distribution of Sperm Basic Proteins (Hnilica L. S., ed) pp. 73–163, CRC Press Inc., Boca Raton, FL [Google Scholar]
- 4. Ausió J. (1999) Histone H1 and evolution of sperm nuclear basic proteins. J. Biol. Chem. 274, 31115–31118 10.1074/jbc.274.44.31115 [DOI] [PubMed] [Google Scholar]
- 5. Oliva R., and Dixon G. H. (1991) Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucleic Acids Res. Mol. Biol. 40, 25–94 10.1016/S0079-6603(08)60839-9 [DOI] [PubMed] [Google Scholar]
- 6. Balhorn R. (2007) The protamine family of sperm nuclear proteins. Genome Biol. 8, 227 10.1186/gb-2007-8-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kasinsky H. E., Eirín-López J. M., and Ausió J. (2011) Protamines: structural complexity, evolution and chromatin patterning. Protein Pept. Lett. 18, 755–771 10.2174/092986611795713989 [DOI] [PubMed] [Google Scholar]
- 8. Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausió J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., et al. (2001) Intrinsically-disordered protein. J. Mol. Graph. Model. 19, 26–59 10.1016/S1093-3263(00)00138-8 [DOI] [PubMed] [Google Scholar]
- 9. Verdaguer N., Perelló M., Palau J., and Subirana J. A. (1993) Helical structure of basic proteins from spermatozoa. Comparison with model peptides. Eur. J. Biochem. 214, 879–887 10.1111/j.1432-1033.1993.tb17991.x [DOI] [PubMed] [Google Scholar]
- 10. Roque A., Ponte I., and Suau P. (2011) Secondary structure of protamine in sperm nuclei: an infrared spectroscopy study. BMC Struct. Biol. 11, 14 10.1186/1472-6807-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliva R. (2006) Protamines and male infertility. Hum. Reprod. Update 12, 417–435 10.1093/humupd/dml009 [DOI] [PubMed] [Google Scholar]
- 12. Bao J., and Bedford M. T. (2016) Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 151, R55–R70 10.1530/REP-15-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saperas N., Lloris D., and Chiva M. (1993) Sporadic appearance of histones, histone-like proteins and protamines in sperm chromatin of bony fish. J. Exp. Zool. 265, 575–586 10.1002/jez.1402650514 [DOI] [Google Scholar]
- 14. Saperas N., Ausio J., Lloris D., and Chiva M. (1994) On the evolution of protamines in bony fish: alternatives to the “retroviral horizontal transmission” hypothesis. J. Mol. Evol. 39, 282–295 10.1007/BF00160152 [DOI] [PubMed] [Google Scholar]
- 15. Herraez M. P., Ausió J., Devaux A., Gonzalez-Rojo S., Fernandez-Diez C., Bony S., Saperas N., and Robles V. (2017) Paternal contribution to development. Sperm genetic damage and repair in fish. Aquaculture 472, 45–59 10.1016/j.aquaculture.2016.03.007 [DOI] [Google Scholar]
- 16. Eirín-López J. M., and Ausió J. (2009) Origin and evolution of chromosomal sperm proteins. Bioessays 31, 1062–1070 10.1002/bies.200900050 [DOI] [PubMed] [Google Scholar]
- 17. Cheema M. S., and Ausió J. (2015) The structural determinants behind the epigenetic role of histone variants. Genes 6, 685–713 10.3390/genes6030685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poccia D. L., and Green G. R. (1992) Packaging and unpackaging the sea urchin sperm genome. Trends Biochem. Sci. 17, 223–227 10.1016/0968-0004(92)90382-J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu S. F., Zhang H., and Cairns B. R. (2011) Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578–589 10.1101/gr.113167.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis J. D., and Ausió J. (2002) Protamine-like proteins: evidence for a novel chromatin structure. Biochem. Cell Biol. 80, 353–361 10.1139/o02-083 [DOI] [PubMed] [Google Scholar]
- 21. Cole R. D. (1984) A minireview of microheterogeneity in H1 histone and its possible significance. Anal. Biochem. 136, 24–30 10.1016/0003-2697(84)90303-8 [DOI] [PubMed] [Google Scholar]
- 22. Lewis J. D., Saperas N., Song Y., Zamora M. J., Chiva M., and Ausió J. (2004) Histone H1 and the origin of protamines. Proc. Natl. Acad. Sci. U.S.A. 101, 4148–4152 10.1073/pnas.0308721101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fioretti F. M., Febbraio F., Carbone A., Branno M., Carratore V., Fucci L., Ausió J., and Piscopo M. (2012) A sperm nuclear basic protein from the sperm of the marine worm Chaetopterus variopedatus with sequence similarity to the arginine-rich C termini of chordate protamine-likes. DNA Cell Biol. 31, 1392–1402 10.1089/dna.2011.1547 [DOI] [PubMed] [Google Scholar]
- 24. Reynolds W. F., and Wolfe S. L. (1978) Changes in basic proteins during sperm maturation in a plant, Marchantia polymorpha. Exp. Cell Res. 116, 269–273 10.1016/0014-4827(78)90448-2 [DOI] [PubMed] [Google Scholar]
- 25. Lewis J. D., Song Y., de Jong M. E., Bagha S. M., and Ausió J. (2003) A walk though vertebrate and invertebrate protamines. Chromosoma 111, 473–482 10.1007/s00412-002-0226-0 [DOI] [PubMed] [Google Scholar]
- 26. Kasinsky H. E., Ellis S., Martens G., and Ausió J. (2014) Dynamic aspects of spermiogenic chromatin condensation patterning by phase separation during the histone-to-protamine transition in charalean algae and relation to bryophytes. Tissue Cell 46, 415–432 10.1016/j.tice.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 27. Higo A., Niwa M., Yamato K. T., Yamada L., Sawada H., Sakamoto T., Kurata T., Shirakawa M., Endo M., Shigenobu S., Yamaguchi K., Ishizaki K., Nishihama R., Kohchi T., and Araki T. (2016) Transcriptional framework of male gametogenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 57, 325–338 10.1093/pcp/pcw005 [DOI] [PubMed] [Google Scholar]
- 28. Bowman J. L., Kohchi T., Yamato K. T., Jenkins J., Shu S., Ishizaki K., Yamaoka S., Nishihama R., Nakamura Y., Berger F., Adam C., Aki S. S., Althoff F., Araki T., Arteaga-Vazquez M. A., et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 10.1016/j.cell.2017.09.030 [DOI] [PubMed] [Google Scholar]
- 29. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., and Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 10.1073/pnas.0402700101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Earley L., Anderson L. C., Bai D. L., Mullen C., Syka J. E., English A. M., Dunyach J. J., Stafford G. C. Jr, Shabanowitz J., Hunt D. F., and Compton P. D. (2013) Front-end electron transfer dissociation: a new ionization source. Anal. Chem. 85, 8385–8390 10.1021/ac401783f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anderson L. C., English A. M., Wang W., Bai D. L., Shabanowitz J., and Hunt D. F. (2015) Protein derivatization and sequential ion/ion reactions to enhance sequence coverage produced by electron transfer dissociation mass spectrometry. Int. J. Mass. Spectrom. 377, 617–624 10.1016/j.ijms.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harrison L. G., Kasinsky H. E., Ribes E., and Chiva M. (2005) Possible mechanisms for early and intermediate stages of sperm chromatin condensation patterning involving phase separation dynamics. J. Exp. Zool. A Comp. Exp. Biol. 303, 76–92 10.1002/jez.a.115 [DOI] [PubMed] [Google Scholar]
- 33. Gusse M., and Chevaillier P. (1978) Ultrastructural and chemical study of the chromatin during spermiogenesis of the fish Scyliorhinus caniculus (L.) (author's transl). Cytobiologie 16, 421–443 [PubMed] [Google Scholar]
- 34. Gusse M., and Chevaillier P. (1980) Molecular structure of chromatin during sperm differentiation of the dogfish Scyliorhinus caniculus (L.). Chromosoma 77, 57–68 10.1007/BF00292041 [DOI] [PubMed] [Google Scholar]
- 35. Gusse M., and Chevaillier P. (1981) Microelectrophoretic analysis of basic protein changes during spermiogenesis in the dogfish Scylliorhinus caniculus (L). Exp. Cell Res. 136, 391–397 10.1016/0014-4827(81)90018-5 [DOI] [PubMed] [Google Scholar]
- 36. Minamino N., Kanazawa T., Nishihama R., Yamato K. T., Ishizaki K., Kohchi T., Nakano A., and Ueda T. (2017) Dynamic reorganization of the endomembrane system during spermatogenesis in Marchantia polymorpha. J. Plant Res. 130, 433–441 10.1007/s10265-017-0909-5 [DOI] [PubMed] [Google Scholar]
- 37. Kreitner G. L. (1977) Transformation of the nucleus in Marchantia spermatids: morphogenesis. Am. J. Botany 64, 464–475 10.1002/j.1537-2197.1977.tb12370.x [DOI] [Google Scholar]
- 38. Kasinsky H. E., Mann M., Huang S. Y., Fabre L., Coyle B., and Byrd E. W. Jr. (1987) On the diversity of sperm basic proteins in the vertebrates: V. Cytochemical and amino acid analysis in Squamata, Testudines, and Crocodylia. J. Exp. Zool. 243, 137–151 10.1002/jez.1402430116 [DOI] [PubMed] [Google Scholar]
- 39. Kasinsky H. E. (1995) Evolution and Origins of Sperm Basic Proteins. (Jamiesson B. G. M., Ausió J., and Justine J. L., eds) pp. 447–462, Memoires du Museum d'Histoire Naturelle, Paris [Google Scholar]
- 40. Martin-Coello J., Dopazo H., Arbiza L., Ausió J., Roldan E. R., and Gomendio M. (2009) Sexual selection drives weak positive selection in protamine genes and high promoter divergence, enhancing sperm competitiveness. Proc. Biol. Sci. 276, 2427–2436 10.1098/rspb.2009.0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marian C. O., Bordoli S. J., Goltz M., Santarella R. A., Jackson L. P., Danilevskaya O., Beckstette M., Meeley R., and Bass H. W. (2003) The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro. Plant Physiol. 133, 1336–1350 10.1104/pp.103.026856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ausió J., Eirín-López J. M., and Frehlick L. J. (2007) Evolution of vertebrate chromosomal sperm proteins: implications for fertility and sperm competition. Soc. Reprod. Fertil. Suppl. 65, 63–79 [PubMed] [Google Scholar]
- 43. Lüke L., Tourmente M., and Roldan E. R. (2016) Sexual selection of protamine 1 in mammals. Mol. Biol. Evol. 33, 174–184 10.1093/molbev/msv209 [DOI] [PubMed] [Google Scholar]
- 44. Stephens A. D., Banigan E. J., and Marko J. F. (2019) Chromatin's physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 58, 76–84 10.1016/j.ceb.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Niklas K. J. (1997) The Evolutionary Biology of Plants. Ph.D. thesis, The University of Chicago Press, Chicago, IL [Google Scholar]
- 46. Lewis J. D., de Jong M. E., Bagha S. M., Tang A., Gilly W. F., and Ausió J. (2004) All roads lead to arginine: the squid protamine gene. J. Mol. Evol. 58, 673–680 10.1007/s00239-004-2589-8 [DOI] [PubMed] [Google Scholar]
- 47. Wouters-Tyrou D., Martin-Ponthieu A., Ledoux-Andula N., Kouach M., Jaquinod M., Subirana J. A., and Sautière P. (1995) Squid spermiogenesis: molecular characterization of testis-specific pro-protamines. Biochem. J. 309, 529–534 10.1042/bj3090529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elsevier S. M., Noiran J., and Carre-Eusebe D. (1991) Processing of the precursor of protamine P2 in mouse. Identification of intermediates by their insolubility in the presence of SDS. Eur. J. Biochem. 196, 167–175 10.1111/j.1432-1033.1991.tb15800.x [DOI] [PubMed] [Google Scholar]
- 49. Martinage A., Arkhis A., Alimi E., Sautière P., and Chevaillier P. (1990) Molecular characterization of nuclear basic protein HPI1, a putative precursor of human sperm protamines HP2 and HP3. Eur. J. Biochem. 191, 449–451 10.1111/j.1432-1033.1990.tb19142.x [DOI] [PubMed] [Google Scholar]
- 50. Fincato P., Moschou P. N., Spedaletti V., Tavazza R., Angelini R., Federico R., Roubelakis-Angelakis K. A., and Tavladoraki P. (2011) Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 62, 1155–1168 10.1093/jxb/erq341 [DOI] [PubMed] [Google Scholar]
- 51. Paschalidis K. A., and Roubelakis-Angelakis K. A. (2005) Sites and regulation of polyamine catabolism in the tobacco plant. Correlations with cell division/expansion, cell cycle progression, and vascular development. Plant Physiol. 138, 2174–2184 10.1104/pp.105.063941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hölttä E., Pulkkinen P., Elfving K., and Jänne J. (1975) Oxidation of polyamines by diamine oxidase from human seminal plasma. Biochem. J. 145, 373–378 10.1042/bj1450373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maayan R., Zukerman Z., and Shohat B. (1995) Oxidation of polyamines in human seminal plasma: a possible role in immunological infertility. Arch. Androl. 34, 95–99 10.3109/01485019508987836 [DOI] [PubMed] [Google Scholar]
- 54. Brunner A. M., Nanni P., and Mansuy I. M. (2014) Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin 7, 2 10.1186/1756-8935-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Louie A. J., and Dixon G. H. (1972) Kinetics of enzymatic modification of the protamines and a proposal for their binding to chromatin. J. Biol. Chem. 247, 7962–7968 [PubMed] [Google Scholar]
- 56. Green G. R., Balhorn R., Poccia D. L., and Hecht N. B. (1994) Synthesis and processing of mammalian protamines and transition proteins. Mol. Reprod. Dev. 37, 255–263 10.1002/mrd.1080370303 [DOI] [PubMed] [Google Scholar]
- 57. Kennedy B. P., and Davies P. L. (1981) Phosphorylation of a group of high molecular weight basic nuclear proteins during spermatogenesis in the winter flounder. J. Biol. Chem. 256, 9254–9259 [PubMed] [Google Scholar]
- 58. Marino G., Eckhard U., and Overall C. M. (2015) Protein termini and their modifications revealed by positional proteomics. ACS Chem. Biol. 10, 1754–1764 10.1021/acschembio.5b00189 [DOI] [PubMed] [Google Scholar]
- 59. Prigge S. T., Mains R. E., Eipper B. A., and Amzel L. M. (2000) New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell. Mol. Life Sci. 57, 1236–1259 10.1007/PL00000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shin Y., and Brangwynne C. P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 61. Martens G., Humphrey E. C., Harrison L. G., Silva-Moreno B., Ausió J., and Kasinsky H. E. (2009) High-pressure freezing of spermiogenic nuclei supports a dynamic chromatin model for the histone-to-protamine transition. J. Cell Biochem. 108, 1399–1409 10.1002/jcb.22373 [DOI] [PubMed] [Google Scholar]
- 62. Watson M., and Stott K. (2019) Disordered domains in chromatin-binding proteins. Essays Biochem. 63, 147–156 10.1042/EBC20180068 [DOI] [PubMed] [Google Scholar]
- 63. Borgia A., Borgia M. B., Bugge K., Kissling V. M., Heidarsson P. O., Fernandes C. B., Sottini A., Soranno A., Buholzer K. J., Nettels D., Kragelund B. B., Best R. B., and Schuler B. (2018) Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 10.1038/nature25762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wright P. E., and Dyson H. J. (2015) Intrinsically-disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 10.1038/nrm3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., and Karpen G. H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Erdel F., and Rippe K. (2018) Formation of chromatin subcompartments by phase separation. Biophys. J. 114, 2262–2270 10.1016/j.bpj.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ishizaki K., Chiyoda S., Yamato K. T., and Kohchi T. (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091 10.1093/pcp/pcn085 [DOI] [PubMed] [Google Scholar]
- 68. Hurley C. K. (1977) Electrophoresis of histones: a modified Panyim and Chalkley system for slab gels. Anal. Biochem. 80, 624–626 10.1016/0003-2697(77)90687-X [DOI] [PubMed] [Google Scholar]
- 69. Ausio J. (1992) Presence of a highly specific histone H1-like protein in the chromatin of the sperm of the bivalve mollusks. Mol. Cell. Biochem. 115, 163–172 [DOI] [PubMed] [Google Scholar]
- 70. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Le S. Q., and Gascuel O. (2008) An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 73. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Martin S. E., Shabanowitz J., Hunt D. F., and Marto J. A. (2000) Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 72, 4266–4274 10.1021/ac000497v [DOI] [PubMed] [Google Scholar]
- 76. Udeshi N. D., Compton P. D., Shabanowitz J., Hunt D. F., and Rose K. L. (2008) Methods for analyzing peptides and proteins on a chromatographic time scale by electron-transfer dissociation mass spectrometry. Nat. Protoc. 3, 1709–1717 10.1038/nprot.2008.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Senko M. W., Remes P. M., Canterbury J. D., Mathur R., Song Q., Eliuk S. M., Mullen C., Earley L., Hardman M., Blethrow J. D., Bui H., Specht A., Lange O., Denisov E., Makarov A., et al. (2013) Novel parallelized quadrupole/linear ion trap/Orbitrap tribrid mass spectrometer improving proteome coverage and peptide identification rates. Anal. Chem. 85, 11710–11714 10.1021/ac403115c [DOI] [PubMed] [Google Scholar]
- 78. Compton P. D., Strukl J. V., Bai D. L., Shabanowitz J., and Hunt D. F. (2012) Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Anal. Chem. 84, 1781–1785 10.1021/ac202807h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rose C. M., Rush M. J., Riley N. M., Merrill A. E., Kwiecien N. W., Holden D. D., Mullen C., Westphall M. S., and Coon J. J. (2015) A calibration routine for efficient ETD in large-scale proteomics. J. Am. Soc. Mass. Spectrom. 26, 1848–1857 10.1007/s13361-015-1183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Olsen J. V., and Mann M. (2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U.S.A. 101, 13417–13422 10.1073/pnas.0405549101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Macek B., Waanders L. F., Olsen J. V., and Mann M. (2006) Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol. Cell. Proteomics 5, 949–958 10.1074/mcp.T500042-MCP200 [DOI] [PubMed] [Google Scholar]
- 82. Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., Madden T. L., Matten W. T., McGinnis S. D., Merezhuk Y., Raytselis Y., Sayers E. W., Tao T., Ye J., and Zaretskaya I. (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., and Madden T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hoffmann J. A., Chance R. E., and Johnson M. G. (1990) Purification and analysis of the major components of chum salmon protamine contained in insulin formulations using high-performance liquid chromatography. Protein Expr. Purif. 1, 127–133 10.1016/1046-5928(90)90005-J [DOI] [PubMed] [Google Scholar]
- 85. Carlos S., Hunt D. F., Rocchini C., Arnott D. P., and Ausio J. (1993) Post-translational cleavage of a histone H1-like protein in the sperm of Mytilus. J. Biol. Chem. 268, 195–199 [PubMed] [Google Scholar]
- 86. Carlos S., Jutglar L., Borrell I., Hunt D. F., and Ausio J. (1993) Sequence and characterization of a sperm-specific histone H1-like protein of Mytilus californianus. J. Biol. Chem. 268, 185–194 [PubMed] [Google Scholar]
- 87. Lewis J. D., McParland R., and Ausió J. (2004) PL-I of Spisula solidissima, a highly elongated sperm-specific histone H1. Biochemistry 43, 7766–7775 10.1021/bi0360455 [DOI] [PubMed] [Google Scholar]
- 88. Saperas N., Chiva M., Casas M. T., Campos J. L., Eirín-López J. M., Frehlick L. J., Prieto C., Subirana J. A., and Ausió J. (2006) A unique vertebrate histone H1-related protamine-like protein results in an unusual sperm chromatin organization. FEBS J. 273, 4548–4561 10.1111/j.1742-4658.2006.05461.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.