Abstract
Sequential activation of DNA replication origins is precisely programmed and critical to maintaining genome stability. RecQL4, a member of the conserved RecQ family of helicases, plays an essential role in the initiation of DNA replication in mammalian cells. Here, we showed that RecQL4 protein tethered on the pre-replicative complex (pre-RC) induces early activation of late replicating origins during S phase. Tethering of RecQL4 or its N terminus on pre-RCs via fusion with Orc4 protein resulted in the recruitment of essential initiation factors, such as Mcm10, And-1, Cdc45, and GINS, increasing nascent DNA synthesis in late replicating origins during early S phase. In this origin activation process, tethered RecQL4 was able to recruit Cdc45 even in the absence of cyclin-dependent kinase (CDK) activity, whereas CDK phosphorylation of RecQL4 N terminus was required for interaction with and origin recruitment of And-1 and GINS. In addition, forced activation of replication origins by RecQL4 tethering resulted in increased replication stress and the accumulation of ssDNAs, which can be recovered by transcription inhibition. Collectively, these results suggest that recruitment of RecQL4 to replication origins is an important step for temporal activation of replication origins during S phase. Further, perturbation of replication timing control by unscheduled origin activation significantly induces replication stress, which is mostly caused by transcription-replication conflicts.
Keywords: DNA replication, cell cycle, checkpoint control, cyclin-dependent kinase (CDK), cell division cycle 7-related protein kinase (Cdc7), chromatin immunoprecipitation (ChiP), origin activation, RecQL4, replication stress, replication timing, transcription-replication conflicts, replication initiation, pre-replicative complex
Introduction
RecQL4 is a member of conserved RecQ family helicases that play important roles in the maintenance of genome integrity by acting in various DNA metabolic processes, such as DNA repair, DNA recombination, and DNA replication. In humans, five RecQ helicases, RecQL1, WRN, BLM, RecQL4, and RecQL5 have been identified. Mutations in WRN, BLM, and RecQL4 genes have been shown to be associated with human genetic disorders such as Werner's, Bloom, and Rothmund-Thomson syndromes, which are characterized by increased genome instability, resulting in early aging symptoms and cancer predisposition (1–3). Although all these proteins contain a well-conserved helicase domain and contribute to genome maintenance, the roles of each protein in DNA metabolism are very different.
RecQL4 is known to be involved in many DNA metabolic processes. RecQL4 plays important roles in DNA double strand break (DSB)2 repair, such as homologous recombination and nonhomologous end joining (4–6) and has also been shown to be involved in the activation of ataxia telangiectasia mutated (ATM), a major checkpoint kinase against DNA DSBs (7). The conserved helicase domain in RecQL4 and its helicase activity are required for these cellular responses to DNA DSBs (5, 7). In addition, RecQL4 has a unique N-terminal domain showing limited homology to Sld2, which is an essential replication initiation factor in yeast cells (8), and this N-terminal domain has been shown to be essential for the initiation of DNA replication in vertebrates (9, 10).
Precise control of the initiation of DNA replication is important for faithful duplication of the genome in eukaryotes, which contains multiple replication origins. For complete duplication of the whole genome once in a cell cycle, pre-replicative complexes (pre-RCs) are assembled on replication origins in G1 phase and sequentially activated during S phase in eukaryotes (11). Origin recognition complexes (ORC) bind to replication origins after mitosis. Next, Cdc6, Cdt1, and the minichromosome maintenance2–7 (Mcm2–7) are recruited to form the pre-RC in G1 phase. For the onset and progression of S phase, pre-RCs on individual replication origins are sequentially activated by forming the active replicative helicase complex containing Cdc45, Mcm2–7, and GINS, which is called the CMG complex. In yeast systems, this activation process requires many origin firing factors, including Dpb11, Sld2, Sld3, and Mcm10, and two S phase–promoting kinases, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) (8, 12). DDK phosphorylates Mcm4 and Mcm6, leading to recruitment of Cdc45 and Sld3 (13–15). CDK phosphorylates Cdc45, Sld2, Sld3, and Sld7, and together with Dpb11 facilitates recruitment of a protein complex composed of GINS, Sld2, and Pol ϵ. Association of these proteins together with Mcm10 and the action of these two kinases trigger assembly and activation of the CMG complex (11, 16, 17).
The process of origin activation appears to be fairly conserved in all eukaryotic systems and human homologues of yeast proteins play similar roles during the initiation process. Treslin/Ticrr, a mammalian homologue of yeast Sld3 has been shown to be required for origin binding of Cdc45 (18), and Mcm10 in human cells has been shown to be required for assembly and activation of the CMG complex and recruitment of DNA polymerase α (19, 20). RecQL4, a homologue of yeast Sld2 in human cells, also plays essential roles in the assembly and activation of the CMG complex. RecQL4 interacts with other essential initiation factors, such as Mcm10 and And-1/Ctf4/Wdhd1, and is required for origin association of these proteins and components of the CMG complex, Cdc45 and GINS (21, 22). Origin association of RecQL4 and interaction of RecQL4 with Mcm10 and And-1 on replication origins depend on CDK and DDK (22), and CDK phosphorylation of the RecQL4 N terminus has been shown to affect the physical interaction with Mcm10 in vitro (21). However, roles of RecQL4 and other initiation proteins leading to origin activation and control of this process by CDK and DDK during S phase are not well-understood in mammalian cells.
In this study, we forced the binding of WT or phospho-mutant RecQL4 proteins to replication origins by expressing RecQL4 proteins fused to Orc4, a component of ORC, in HeLa cells. We then examined the recruitment of initiation factors on replication origins and the time of origin activation during S phase. We found that the tethered RecQL4 on the pre-RC led to early activation of late replicating origins and increased replication stress, which was alleviated by transcription inhibition. Conversely, phospho-deficient RecQL4 protein for CDK phosphorylation failed to induce early activation of late origins because of a lack of And-1 and GINS binding. Our results indicate that recruitment of RecQL4 on replication origins is an important step for temporal activation of replication origins during S phase. Furthermore, these results provide an insight for CDK control of origin activation and replication timing control during S phase in mammalian cells.
Results
Recruitment of RecQL4, Mcm10, and And-1 on late replicating origins occurs in late S phase
Activation of the replicative helicase on individual replication origins should occur at the time of origin activation. Because replication initiation factors such as RecQL4, Mcm10, and And-1 are required for CMG assembly in mammalian cells, we investigated when these factors were recruited to late replicating origins during S phase. To do this, HeLa cells were synchronized at early or late S phase using double thymidine block and release, and the binding of these proteins to the replication origin in the β-globin locus (β-globin origin), a well-characterized late replicating origin in HeLa cells (23, 24), was examined by in vivo crosslinking followed by chromatin immunoprecipitation (ChIP). As shown in Fig. 1, binding of the pre-RC component proteins, Orc2 and Mcm4, was observed in both early S (released for 1 h from double thymidine block) and late S (released for 6 h) phases, and loading of Cdc45 and Sld5, components of the CMG complex, was very low in early S phase and significantly increased in late S phase. Binding pattern of the RecQL4, Mcm10, and And-1 proteins to the β-globin origin was similar to that of the CMG component proteins (Fig. 1), indicating that these proteins were recruited to the late replicating origin only at late S phase, but not early S phase. These results suggest that their binding to individual replication origins is possibly regulated by the replication timing control program in cells, and might be an important control step to activate late replicating origins.
Figure 1.

Recruitment of replication factors for origin activation at the late replicating origin occurs in late S phase. HeLa cells were synchronized at the G1/S boundary and released for 1 h (early S phase) or 6 h (late S phase). Cells were harvested, and ChIP assays were performed using the described antibodies at the indicated times. Relative binding of proteins to the β-globin origin compared with the non-origin region (−21 kb distal region from the β-globin origin) was examined by quantitative PCR analysis. The mean values ± S.D. with p values are shown. ns denotes not significant. ***, p < 0.001.
Tethering RecQL4, Mcm10, or And-1 on the pre-RC is sufficient to activate late replicating origins in early S phase
Because RecQL4, Mcm10, and And-1 proteins are recruited to the replication origin at the time of its activation, we tested whether their aberrant binding to replication origins affected the timing of origin activation during S phase. RecQL4 proteins fused to the N terminus of Orc4, a component of the ORC, were expressed in HeLa cells to force tethering of RecQL4 proteins on the pre-RC, and then the nascent DNA synthesis at early or late replicating origins was determined by bromodeoxyuridine-immunoprecipitation (BrdU-IP) followed by quantitative PCR analysis. In HeLa cells, nascent DNA synthesis at the lamin B2 origin, a typical early replicating origin in human cells (25, 26), was dominant in early S phase, whereas replication at late replicating origins such as GRM8 (27) and β-globin origins was almost negligible during early S phase and occurred mostly in late S phase (Fig. 2, A and D). However, expression of RecQL4-Orc4 fusion proteins in HeLa cells (Fig. 2B) clearly increased the amount of RecQL4 proteins loaded onto the late replicating origin (Fig. 2C) and nascent DNA synthesis at the late replicating origins (Fig. 2E; GRM8 and β-globin origins) in early S phase. The level of nascent DNA synthesis at the β-globin origin observed in these cells was comparable with the level of DNA synthesis observed in the late S phase in HeLa cells (Fig. 2A). In addition, this nascent DNA synthesis depended on two S phase promoting kinases, CDK and DDK (Fig. 2F), similar to the nascent DNA synthesis observed during normal DNA replication. Therefore, increases in nascent DNA synthesis observed in these cells appeared to be caused by early activation of the late replicating origins. Increases in nascent DNA synthesis at the late replicating origins in early S phase were also observed in the cell line stably expressing RecQL4-Orc4 proteins from the Tet-inducible promoter (Fig. S1, A and B).
Figure 2.

Tethering replication factors for CMG assembly on the pre-RC induces early activation of late replicating origins. A, replication timing of origins in HeLa cells. Cells were synchronized at G1/S and released into S phase. Nascent DNA synthesis was examined by BrdU-IP. B and C, expression and origin binding of RecQL4-Orc4 fusion proteins. HeLa cells transfected with RecQL4-Orc4 (R-O4) were synchronized at early S phase, and the expression levels (B) and origin binding of RecQL4 (C) were determined by Western blotting and ChIP, respectively. R4, RecQL4; O4, Orc4; GDH, GAPDH. D, the cell cycle profile of cells prepared in (A) and (C). E, RecQL4 tethered on the pre-RC induces origin activation. Cells transfected with R-O4 were synchronized at early S phase, and nascent DNA synthesis was determined by BrdU-IP. F, origin activation by tethered RecQL4 depends on both DDK and CDK. Cells transfected with R-O4 were treated with 10 μm roscovitine (Ros), Nu6140 (Nu), or PHA-767491 (PHA) for 1 h at early S phase, and nascent DNA synthesis was examined. G, tethering Mcm10 or And-1 on the pre-RC triggers early activation of the β-globin origin. Nascent DNA synthesis at the β-globin origin (left panel) was examined in cells expressing Mcm10-Orc4 (M-O4), Mcm10 (M10), Orc3-And-1 (O3-A), or And-1 (A1) at early S phase. Middle and right panels show expression levels of these proteins, and arrowheads indicated M-O4 and O3-A proteins. LB1, lamin B1. p values: ns, not significant; **, p < 0.01; ***, p < 0.001.
As RecQL4 proteins loaded onto replication origins interact with Mcm10 and And-1 proteins, and all these proteins are required for assembly of the CMG complex and origin activation in human cells (22), we tested whether early activation of the late replicating origins also occurred by tethering Mcm10 or And-1 on the pre-RC. Although the levels of nascent DNA synthesis were different, tethering Mcm10 or And-1 on the pre-RC by expressing Mcm10-Orc4 or Orc3–And-1 fusion proteins significantly increased nascent DNA synthesis at the β-globin locus in early S phase (Fig. 2G), suggesting that tethered Mcm10 or And-1 proteins also induced activation of late replicating origins in early S phase. Therefore, loading of factors required for CMG complex assembly on the replication origins seems to be sufficient to induce activation of replication origins during S phase. Any one of these proteins tethered on the pre-RC may recruit two other proteins functioning together for CMG assembly and origin activation. Consistent with this notion, RecQL4 tethering on the pre-RC did not force early activation of the late replicating origin if Mcm10 or And-1 was depleted (Fig. S2).
Recruitment of Cdc45 to replication origins by tethered RecQL4 on the pre-RC does not depend on CDK activity
In mammalian cells, concerted action of two S phase–promoting kinases, CDK and DDK, is required for recruitment of replication initiation factors on the replication origins and their action. We examined whether recruitment of replication initiation factors forced by the tethered RecQL4 depended on these two kinase activities. When we examined the amount of replication initiation proteins on the late replicating origin in early S phase by in vivo crosslinking followed by ChIP analyses, expression of RecQL4-Orc4 proteins clearly increased binding of factors required for CMG assembly (Mcm10 and And-1) as well as CMG component proteins (Cdc45 and Sld5), as expected (Fig. 3). Treatment of the DDK inhibitor (PHA767491) in these cells reduced origin binding of Mcm10, And-1, Cdc45, and Sld5, suggesting that binding of Mcm10, And-1, Cdc45, and the GINS complex forced by the tethered RecQL4 on the pre-RC depended on DDK activity (Fig. 3). The binding of Mcm10, And-1, and Sld5 also depended on CDK activity, which was judged by the decrease in origin binding after CDK inhibitor (NU6140) treatment. Conversely, origin binding of Cdc45 was still observed in cells treated with CDK inhibitor, suggesting that origin binding of Cdc45 forced by tethered RecQL4 on the pre-RC did not depend on CDK activity. Furthermore, CDK-independent recruitment of Cdc45 on replication origins forced by the tethered RecQL4 could occur in the absence of recruitment of other essential initiation factors such as Mcm10, And-1, and Sld5 (Fig. 3), suggesting that RecQL4 may play its role by directly recruiting either Cdc45 or factors required for Cdc45 recruitment, such as Treslin.
Figure 3.

Influence of CDK and DDK on origin association of replication proteins induced by RecQL4 tethering on the pre-RC. HeLa cells expressing RecQL4-Orc4 (R-O4) were synchronized at G1/S and released to S phase in the presence of 10 μm roscovitine (Ros), Nu6140 (Nu), or PHA-767491 (PHA). After 1 h, ChIP assays for the β-globin origin were carried out using antibodies targeting Mcm10, And-1, Cdc45, or Sld5. Lanes: −, mock transfection; M, mock treatment. p values: ns, not significant; ***, p < 0.001.
CDK phosphorylation of N-terminal Sld2 homology domain in RecQL4 protein plays essential role for activation of replication origins
Although CDK phosphorylation of replication factors is essential to initiate DNA replication, the exact role of individual protein phosphorylation during the initiation processes is not yet elucidated in human cells. Therefore, we decided to determine the role of RecQL4 phosphorylation by CDK during the initiation process using this tethering system. Because there are many candidates for essential CDK phosphorylation sites in RecQL4 proteins, we first determined the minimal region of RecQL4 proteins to force early activation of late replicating origins by tethering onto the pre-RC. As shown in Fig. 4, A–D, overexpression of RecQL4 alone or tethering N terminus–deleted fragment (ND1) on the pre-RC did not induce β-globin origin activation. On the other hand, tethering N-terminal fragments CD1 (amino acids 1–241) or CD2 (amino acids 1–427) on the pre-RC forced early activation of β-globin origins, although the level of nascent DNA synthesis was somewhat lower than the level of synthesis caused by tethering of full-length RecQL4 proteins (Fig. 4, A–D). This result suggests that the N terminus of the RecQL4 protein (CD1), including the Sld2 homology domain, contains minimal regions required for initiation of DNA replication. To determine the role of CDK phosphorylation, we created alanine substitution mutants of all possible CDK phosphorylation sites in the CD1 region and determined their activity to support early activation of late replicating origins by tethering on the pre-RC. When serine or threonine at any CDK phosphorylation site in the CD1 region was individually substituted with alanine, early activation of the β-globin origins was still observed by tethering these mutant proteins (S89A, T93A, and T139A) on the pre-RC, although the levels of nascent DNA synthesis were relatively low (Fig. 4E). In contrast, alanine substitution of all three phosphorylation sites (3A) almost completely abolished the ability to induce early activation of the β-globin origins. Further, the phosphomimetic mutant with glutamic acid substitution of all phosphorylation sites (3E) showed the ability to support early activation of the β-globin origins (Fig. 4E). These results suggested that CDK phosphorylation of all three sites in CD1 plays important roles to activate the replication origins. To confirm the role of these three phosphorylation sites in the context of the full-length RecQL4 protein, we constructed the phospho-deficient (3A) or phosphomimetic (3E) mutant of the full-length RecQL4 protein containing alanine or glutamate substitution of all three CDK phosphorylation sites in the CD1 region and examined its ability to induce early activation of late replicating origins by tethering onto the pre-RC. Tethering 3A mutant of the RecQL4 protein on the pre-RC barely induced activation of the β-globin origins, whereas tethering 3E mutant of the RecQL4 protein on the pre-RC resulted in a significant increase in β-globin origin activation, which is comparable with tethering of the WT RecQL4 protein (Fig. 4F). Collectively, these results suggested that CDK phosphorylation of the three phosphorylation sites in the Sld2 homology domain of RecQL4 proteins plays essential roles for activation of replication origins in human cells.
Figure 4.

CDK phosphorylation of RecQL4 N terminus is essential for origin activation. A, schematic diagrams of RecQL4-Orc4 variants. All RecQL4 variants contained 2 FLAG tags at the N terminus. Sld2, a yeast Sld2 homology domain; FL, full-length; CD, C-terminal deleted; ND, N-terminal deleted; 3A, alanine substitutions of three CDK phosphorylation sites (S89, T93, and T139); 3E, glutamic acid substitutions of three CDK phosphorylation sites. B–D, HeLa cells were transfected with plasmids for indicated RecQL4 variants-Orc4 and synchronized at early S phase. The level of expressed RecQL4 variants was determined by Western blotting (B), and nascent DNA synthesis at the β-globin origin was examined by BrdU-IP (C). The cell cycle profiles of these cells were shown by flow cytometry analysis (D). E and F, all three phosphorylation sites at the N terminus of RecQL4 are important for origin activation. HeLa cells were transfected with plasmids for various RecQL4 CD1-Orc4 (E) or full-length RecQL4-Orc4 (F) containing the indicated mutations and synchronized at early S phase. Nascent DNA synthesis at the β-globin origin (left panel) and expression levels of individual proteins (right panel) were examined by BrdU-IP and Western blotting, respectively. p values: ns, not significant; **, p < 0.01; ***, p < 0.001.
CDK phosphorylation of RecQL4 N-terminal domain is required for And-1 and GINS recruitment onto replication origin
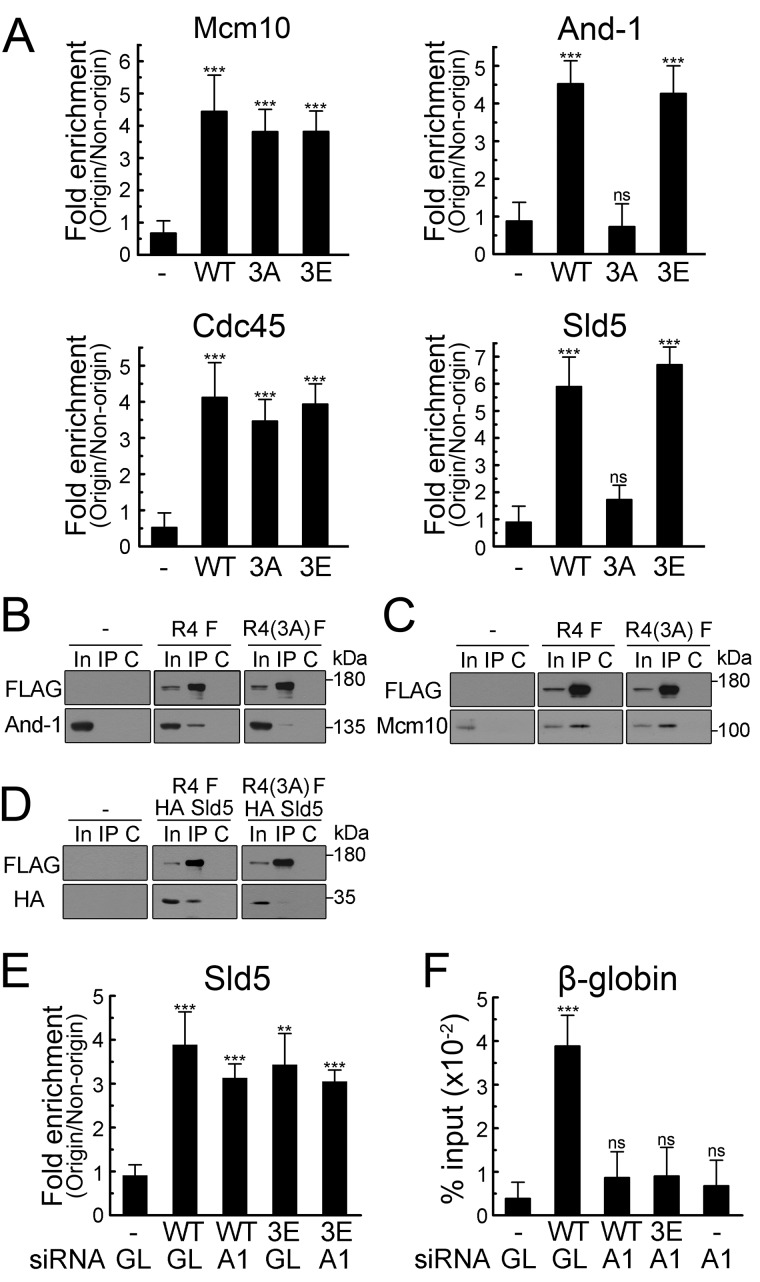
To explore the exact role of RecQL4 N-terminal phosphorylation by CDK for origin activation, the recruitment of replication initiation factors on the late replicating origin in early S phase was examined after tethering phospho-deficient (3A) or phosphomimetic (3E) mutants of full-length RecQL4 proteins on the pre-RC. As expected, Cdc45 recruitment onto the replication origin, which did not depend on CDK activity as shown in Fig. 3, occurred by tethering phospho-deficient, phosphomimetic, or WT RecQL4 proteins (Fig. 5A). The recruitment of Mcm10, which was shown to be CDK dependent (Fig. 3), was still observed by tethering phospho-deficient RecQL4 proteins (Fig. 5A). On the other hand, recruitment of And-1 or Sld5, which also depended on CDK activity (Fig. 3), was not observed by tethering phospho-deficient mutant RecQL4 proteins (Fig. 5A). These results suggested that CDK phosphorylation of the RecQL4 N terminus is essential for recruitment of the And-1 and GINS complex, but not for Mcm10. Consistent with this notion, co-immunoprecipitation (co-IP) analysis showed that the phospho-deficient form of RecQL4 stably interacted with Mcm10, but not with And-1 or Sld5; in contrast, WT RecQL4 stably interacted with And-1 and Sld5 as well as Mcm10 in HeLa cells (Fig. 5, B–D). Therefore, CDK phosphorylation of the RecQL4 N terminus was not required for RecQL4 interaction with Mcm10, but appeared to play its role in the recruitment of And-1 and Sld5 on replication origins by increasing the interaction of RecQL4 with And-1 and Sld5. However, the absence of Sld5 recruitment could be indirectly caused by lack of And-1 recruitment because And-1 has been shown to interact with GINS and is required for CMG complex assembly (20, 28). To test this possibility, we tethered RecQL4 proteins on the pre-RC in the absence of And-1 and examined Sld5 recruitment onto the replication origin. As shown in Fig. 5E, the recruitment of Sld5 proteins onto β-globin origins still occurred in the absence of And-1 by tethering WT or phosphomimetic RecQL4 proteins, although replication at the β-globin origin was not induced by tethering in the absence of And-1 proteins (Fig. 5F). Therefore, phosphorylation of the RecQL4 N terminus by CDK appears to be responsible for the interaction and recruitment of both And-1 and the GINS complex.
Figure 5.
CDK phosphorylation of RecQL4 N terminus is required for origin association of And-1 and Sld5. A, origin association of replication proteins induced by tethered RecQL4 with mutations in CDK phosphorylation sites. HeLa cells transfected with WT, phospho-deficient (3A), or phosphomimetic (3E) forms of full-length RecQL4-Orc4 were synchronized at early S phase. Then, the association of replication proteins on the β-globin origin was analyzed by ChIP. B–D, physical interaction of WT or phospho-deficient RecQL4 with And-1, Sld5, or Mcm10. HeLa cells were transfected with FLAG-tagged WT or phospho-deficient (3A) RecQL4 (B and C), or transfected with indicated RecQL4 and HA-tagged Sld5 (D). Cells were synchronized at early S phase, and the interaction between indicated proteins was analyzed by co-IP with anti-FLAG antibodies. Lanes: In, 5% of input for IP; C, control IP in the presence of FLAG peptides (0.2 mg/ml). E and F, mock-depleted (GL) or And-1 (A1) depleted HeLa cells were transfected with WT or phosphomimetic (3E) RecQL4-Orc4 and synchronized at early S phase. Then, the association of Sld5 proteins with β-globin origins (E) and nascent DNA synthesis at β-globin origins (F) were determined by ChIP and BrdU-IP analyses, respectively. p values: ns, not significant; **, p < 0.01; ***, p < 0.001.
Perturbation of replication timing control programs by forced activation of late replicating origins induces replication stress that is relieved by inhibiting transcription
Because RecQL4 tethering on the pre-RC induces unscheduled activation of late replicating origins, we investigated how cells responded to the perturbation of replication timing control. At first, we examined whether replication stress was induced by activation of late replicating origins in early S phase. In U2OS cells stably expressing RecQL4-Orc4 proteins from the Tet-inducible promoter, increases in phosphorylated ATR and phosphorylated RPA, a well-known target of ATR kinase (29, 30), were observed after addition of doxycycline (Fig. 6A), suggesting that the unscheduled activation of late replicating origins induced replication stress, which resulted in increased ssDNAs and activation of ATR, a master kinase for the replication checkpoint response. Immunostaining of RPA proteins in these cells clearly showed accumulation of ssDNAs on chromatins after induction of RecQL4-Orc4 proteins (Fig. 6C). However, phosphorylated histone H2AX (γH2AX), a marker of DNA double strand breaks, was not significantly increased during this condition (Fig. 6A), indicating that stress induced by forced activation of late replicating origins can be overcome by cellular responses to replication stress, such as the ATR pathway.
Figure 6.

Unscheduled activation of replication origins induces replication stress that is relieved by transcription inhibition. A, U2OS cells stably expressing RecQL4-Orc4 proteins from the doxycycline-inducible promoter (U2OS-RO4) were cultured in the absence (−) or presence (+) of 3 μg/ml doxycycline for 48 h. Then, the levels of p-ATR (S428), p-RPA (S33), and γH2AX were examined by Western blotting. M, Mock treatment. B and C, U2OS-RO4 cells were grown in the absence (−) or presence (+) of 3 μg/ml doxycycline for 44 h. Cells were treated with 75 μm cordycepin (Co) or 40 μg/ml α-amanitin (α-ama) for 4 h. The level of indicated proteins was examined by Western blotting (B) and ssDNA accumulation was analyzed by staining with anti-RPA32 antibody (C). D, U2OS-RO4 cells were cultured in the absence (−) or presence (+) of 3 μg/ml doxycycline and 50 μm nucleosides (Nuc) for 48 h. RPA foci formation was examined by staining with anti-RPA32 antibody. The numbers at the bottom of each image indicate the percentage of RPA foci positive cells. As a positive control, cells were treated with 30 μg/ml bleomycin (Bl) for 2 h or 10 μm etoposide (Eto) for 1 h. Scale bars: 10 μm.
Over-replication induced by expression of limiting replication factors or oncogene activation has been shown to increase replication stress by depletion of dNTP pools in some cells (31, 32). However, RPA foci were still observed after addition of nucleosides (Fig. 6D), suggesting that depletion of dNTPs was not the main reason for the accumulation of ssDNAs in cells expressing RecQL4-Orc4. On the other hand, the treatment of transcription inhibitors such as cordycepin or α-amanitin almost completely reduced RPA phosphorylation (Fig. 6B) and RPA foci (Fig. 6C). Therefore, unscheduled activation of replication origins in cells expressing RecQL4-Orc4 proteins seemed to induce replication stress by increasing transcription-replication conflicts.
Discussion
In this study, RecQL4 was tethered on replication origins by expressing RecQL4-Orc4 fusion proteins, and we found that the tethered RecQL4 protein on the pre-RC induced recruitment of replication initiation factors, assembly of the CMG complex, and nascent DNA synthesis during S phase in human cells. Because overexpression of RecQL4 alone did not induce late origin activation (Fig. 4, B–D), association of RecQL4 with replication origins rather than increase in RecQL4 protein level appeared to be critical for forced activation of late replicating origins. The process of late origin activation in early S phase by RecQL4 tethering was summarized in Fig. 7.
Figure 7.

Diagram showing the forced activation of late replicating origins by RecQL4 tethering. In early S phase, late origin activation is prevented by Rif1-dependent PP1 action. Tethered RecQL4 somehow overcomes PP1 action and induces recruitment of other initiation factors in a manner dependent upon DDK (for Cdc45) or DDK and CDK (for Mcm10, And-1, and GINS). This leads to CMG assembly and late origin activation in early S phase. Tethering of phospho-deficient RecQL4 defective in CDK phosphorylation failed to induce origin activation because of lack of origin binding of And-1 and GINS.
Although the timing of origin activation in mammalian cells appears to be governed by characteristics of the replication origins and replication timing control program during S phase (33), tethered RecQL4 on the late replicating origin somehow overcame those controls to activate late replicating origins in early S phase. RecQL4 was shown to be required for origin binding of Mcm10 and And-1, and co-dependent binding of RecQL4 and Mcm10 on replication origins is an early event that occurs before the recruitment of And-1 and the CMG assembly (20, 22). All known initiation factors tested in this study were recruited to the late replicating origin in early S phase by tethered RecQL4 (Fig. 3) or RecQL4 N terminus (Fig. 5A), and the nascent DNA synthesis induced by tethered RecQL4 depended upon other initiation factors, such as Mcm10 and And-1 (Fig. S2), and both CDK and DDK activities (Fig. 2F). Because all of these results are consistent with previous observations for the initiation of DNA replication in eukaryotes (20–22, 34, 35), origin activation induced by RecQL4 tethering seems to follow conserved mechanism of origin activation after RecQL4 binding to replication origins. Therefore, we believe that this system would be useful to determine the role of RecQL4 protein and its modification by CDK in the initiation process of DNA replication in human cells.
In the present study, we were able to determine the mechanism of origin activation in detail using this system. We found that Cdc45 was recruited to replication origins by the tethered RecQL4 even in the absence of CDK activity, whereas other initiation factors such as Mcm10, And-1, and Sld5 were not (Fig. 3). This result suggested that the RecQL4 on the pre-RC was directly responsible for the origin association of Cdc45 or Treslin, a factor required for Cdc45 recruitment. Further, the CDK dependence for origin binding of Cdc45 observed in previous studies (18, 20, 36) appeared to be caused by failure of RecQL4 loading onto the replication origin, and the interaction of RecQL4 with Cdc45 or Treslin may not depend on CDK activity. Consistent with this notion, RecQL4 loading onto the replication origin requires both CDK and DDK activities (22), and RecQL4 was shown to interact with Cdc45 proteins in cells (21, 37).
Using this system, we also found that CDK phosphorylation of the RecQL4 N terminus was essential for origin activation (Fig. 4) and origin recruitment of And-1 and Sld5, but not for Mcm10 recruitment (Fig. 5). Although origin association of essential initiation factors, except for Cdc45, depended on both CDK and DDK activities (Fig. 3), the phospho-deficient form of RecQL4 still interacted with Mcm10 (Fig. 5D) and induced recruitment of Mcm10 on origins, as well as the WT or phosphomimetic form of RecQL4 (Fig. 5A). Therefore, our results clearly showed that phosphorylation of the RecQL4 N terminus was not required for its interaction with or recruitment of Mcm10 on the replication origins. Phosphorylation of other initiation factors, including Mcm2–7 complex and/or Mcm10 itself, might be responsible for the CDK dependence of Mcm10 recruitment on replication origins. On the other hand, phospho-deficient RecQL4 proteins failed to induce origin recruitment of And-1 and Sld5, and did not show any interaction with And-1 or Sld5 proteins (Fig. 5, A–C). Because the phosphomimetic form of RecQL4 is capable of recruiting both And-1 and Sld5 on replication origins, and origin association of Sld5 during this condition did not depend on the presence of And-1 proteins (Fig. 5E), CDK phosphorylation of the RecQL4 N terminus appears to be directly responsible for the recruitment of both And-1 and the GINS complex including Sld5.
In yeast cells, DDK phosphorylation of the Mcm2–7 complex is important for recruitment of Cdc45 with Sld3 on replication origins, and no CDK phosphorylation is required for Cdc45 recruitment (38). CDK phosphorylation of several initiation factors, including Sld2 and Sld3, has been shown to facilitate recruitment of a complex that includes GINS and polymerase ϵ (8, 16). Accordingly, the roles of CDK and DDK in yeast systems appear to be conserved in human cells. Once RecQL4 is loaded onto replication origins, origin binding of Cdc45 depends on DDK activity, but not on that of CDK (Fig. 3). In addition, CDK phosphorylation of the RecQL4 N terminus containing the Sld2 homology domain is required for origin binding of the GINS complex, including Sld5 (Fig. 5). However, RecQL4 is not only required for origin association of the GINS complex as shown in yeast systems, but also required for recruitment of other essential initiation factors, including Mcm10, And-1, and Cdc45. Therefore, RecQL4 in human cells appears to play more diverse roles than yeast Sld2 during the initiation process.
RecQL4 protein tethered on the pre-RC somehow overcomes the replication timing control program and induces early activation of late replicating origins in human cells (Fig. 2). Eukaryotic cells have multiple replication origins, and the sequential activation of replication origins during S phase is governed by the replication timing control program, which appears to be important to maintain genome integrity (33). In yeast (S. cerevisiae), availability of limiting initiation factors such as Cdc45, Dbf4, Sld2, and Sld3 has been shown to be important for the control of replication timing, and their overexpression induces premature activation of late replicating origins (12). Although Cdc45 was shown to be a limiting initiation factor and its overexpression affected origin activation in human cells (32), replication timing control program in human cells appeared to be more influenced by architectural features of chromosomes, such as cis-elements in chromosomal DNAs, chromatin structure, and trans-acting factors associated with chromatin (33). In mammals, replication timing domains that contain multiple replication origins and replicate concomitantly within a short time window highly co-localize with topologically associated domains (33). Further, cis-elements influencing origin firing and chromatin structure and trans-acting factors such as Rif1 were shown to affect the replication timing control during S phase (33, 39, 40). Rif1 is a chromatin associated protein modulating organization of chromatin and has been shown to recruit protein phosphatase 1 (PP1) to prevent DDK phosphorylation of the Mcm2–7 complex. We still do not clearly understand how RecQL4 on the pre-RC overcame the replication timing control program; however, it seems important to note that both RecQL4 and Rif1 can interact with G-quadruplex (G4) structures. G4 structures have been shown to be associated with replication origins (41), and Rif1 binds to G4 structures and suppresses replication by recruiting PP1 (33, 42–46). Because RecQL4 N terminus has a strong affinity to G4 structures (47), RecQL4 may antagonize the activity of Rif1 by competing for G4 binding or resolving G4 structures. It may also be possible that RecQL4 or factors directly recruited by RecQL4 influence chromatin structure to increase accessibility to limiting initiation factors. Consistent with this notion, And-1 was shown to interact with and stabilize Gcn5, a histone acetyltransferase, in human cells (48), but its involvement in the replication process has yet to be determined.
Sequential activation of replication origins in mammalian cells is precisely programmed, and the perturbation of this program appears to increase genome instability (49). Increased replication by overexpression of limiting replication initiation proteins such as Cdc45 results in more origin firing and increases in γH2AX (32). Oncogene activation was also shown to increase replication initiation and replication stress (49). Because RecQL4 tethering results in more unscheduled origin activation, it may be possible to increase replication stress, including transcription-replication conflicts, as shown in cells with replication stress induced by cyclin-E expression (50). Consistent with this notion, RecQL4 tethering on the pre-RC increased the accumulation of ssDNAs, and their accumulation was almost completely reversed by inhibition of transcription (Fig. 6). In eukaryotic cells, several proteins have been shown to be responsible for resolving replication stress caused by transcription-replication conflicts. ATR is a major checkpoint kinase that responds to replication stress, and Mec1, a yeast homolog of ATR, has been shown to play important roles in limiting transcription-replication conflicts during replication stress (51). Active roles of p53 in replication fork processivity and in preventing transcription-replication conflicts have also been reported (52, 53). FancD2, a component of the FA pathway, plays roles in responding to replication stress and in removal of R-loops (DNA:RNA hybrids) (54, 55). Although it is not clear whether replication stress induced by tethering RecQL4 on the pre-RC is solely caused by transcription-replication conflicts, it would be interesting to see whether the proteins known to be involved in transcription-replication conflicts play any roles in response to this replication stress.
Experimental procedures
Cell culture and synchronization
HeLa cells and HeLa or U2OS cells stably expressing RecQL4-Orc4 from a doxycycline inducible promoter (HeLa-RO4 or U2OS-RO4) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) or tetracycline-free FBS and antibiotics. To synchronize the cell cycle, cells were treated with 2 mm thymidine for 18 h, incubated in fresh medium without thymidine for 12 h, and then incubated for an additional 13 h in medium containing 2 mm thymidine. For ectopic protein expression in synchronized cells, transfection was carried out at the time of release from the first thymidine block. Cell cycle profiles were monitored by flow cytometry analysis after staining with propidium iodide using FACS Calibur (BD Biosciences).
Plasmids and siRNA transfection
For expression of WT or various mutant RecQL4 proteins fused to Orc4 (R-O4), cDNAs encoding WT RecQL4 (amino acid residues 1–1209), C-terminal truncated proteins (1–241 for CD1 and 1–427 for CD2), and N-terminal truncated protein (ND, 248–1209) were amplified by PCR and inserted into the N terminus of the ORC4 gene, which was cloned into the pCDNA3.1(−) plasmid. For alanine or glutamic acid substitution of the CDK phosphorylation sites of RecQL4 (S89, T93 and T139), site-directed mutagenesis was conducted using PCR (56, 57). Mcm10 fused to the N terminus of Orc4 and Orc3 fused to the N terminus of And-1 were also generated by PCR and subcloned into the pCDNA3.1(−) plasmid. To generate a HeLa or U2OS cell line stably expressing RecQL4-Orc4 protein from a doxycycline-inducible promoter, the RecQL4-Orc4 expression vector was constructed using the pTRE plasmid and transfected into HeLa or U2OS Tet-On cells. Subsequently, a cell line stably expressing the RecQL4-Orc4 fusion protein was isolated by screening for hygromycin resistance and Western blot analysis. Transfection of plasmid DNAs was performed with Polyfect (Qiagen, Hilden, Germany), and transfection of siRNAs were performed using the Neon transfection system or Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer's instruction. All siRNAs were chemically synthesized from Bioneer (Daejeon, Korea), and sense-strand sequences of siRNAs used in this study were as follows: GL-2, 5′-AACGUACGCGGAAUACUUCGA-3′; RecQL4, 5′-GACUGAGGACCUGGGCAAA-3′; Mcm10, 5′-AGAGUUGCAAGAGGAAUUA-3′; And-1, 5′-GAUCAGACAUGUGCUAUUA-3′.
IP and Western blot analysis
To prepare whole cell extracts for IP, cells were lysed in a buffer (25 mm Tris-HCl, pH 7.5, 2.5 mm MgCl2, 1 mm DTT, 0.2% IGEPAL CA-630, 10 mm sodium fluoride, 200 μm sodium orthovanadate, and protease inhibitors) containing 150 mm NaCl or 150 mm Na-acetate. Cell lysates were sonicated, treated with 0.25 units/μl benzonase (Enzynomics, Daejeon, Korea) at 4 °C for 1 h, and cleared by centrifugation at 13,000 rpm for 10 min. The protein concentration in the lysate was measured by Bradford assay. For Western blot analysis, cells were re-suspended in a buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% IGEPAL CA-630, 0.25% sodium deoxycholate, 0.1% SDS, 10 mm sodium fluoride, 200 μm sodium orthovanadate, and protease inhibitors). The cells were disrupted by sonication, and 20 μg of proteins was subjected to SDS-PAGE. Anti-RecQL4 antibody (AbFrontier, San Diego, CA) was prepared by immunizing rabbits with recombinant proteins of the RecQL4 N terminus (amino acid residues 1–241). Anti-Orc2, anti-WDHD1/And-1, anti-phospho RPA32 (S33), and anti-γH2AX were purchased from Bethyl Laboratories (Montgomery, TX). Anti-Mcm4, anti-GAPDH, anti-Orc4, and anti-ATR antibodies were from Santa Cruz Biotechnology (Dallas, TX), and anti-Lamin B1 and anti-Mcm10 antibodies from Abcam (Cambridge, MA). Anti-HA, anti-phospho ATR (S428), and anti-Cdc45 antibodies were from Cell Signaling Technology (Danvers, MA). Anti-GINS4 (Sld5) was from GeneTex (Irvine, CA). Anti-FLAG, anti-RPA32, and anti-BrdU antibodies were supplied by Sigma-Aldrich.
Immunocytochemistry
For immunostaining, cells were grown on coverslips and pre-extracted in CSK buffer (10 mm PIPES, pH 7.0, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, and 0.5% Triton X-100) on ice for 5 min. Subsequently, cells were fixed with 3.7% paraformaldehyde, washed with PBS containing 0.25% Triton X-100, and incubated in a blocking buffer (PBS containing 5% BSA and 0.1% Tween 20). Primary and fluorescent-conjugated secondary antibodies were sequentially treated in the blocking buffer for 1 h. Cell nuclei were stained with 0.2 μg/ml 4′-6′-diamidino-2-phenylindole (DAPI) in PBS for 3 min. The images of stained cells were obtained by fluorescent microscopy.
BrdU-IP, ChIP, and quantitative PCR analysis
BrdU-IP assays were performed to measure nascent DNA synthesis at replication origins as described previously (58) with some modifications. Cells were pulse-labeled for 30 min with BrdU (150 μm) and harvested. Genomic DNA was extracted and sonicated to the average size of about 300 nucleotide pairs. After DNA denaturation by boiling and rapid cooling, the nascent DNA was immunoprecipitated with anti-BrdU antibody and Dynabeads protein G (Thermo Fisher Scientific) in an immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3% BSA, 0.1% Triton X-100). The beads were washed four times with washing buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100) and once with TE (10 mm Tris-HCl, pH 8.0, 1 mm EDTA). DNA bound to the beads was eluted by 100 mm glycine-HCl (pH 1.9) and immediately neutralized by adding Tris-HCl buffer (pH 8.5).
ChIP assays were performed as described (22), except for using Dynabeads protein G. Primer sets for the origin in Lamin B2 locus (LB2 Ori), metabotropic glutamate receptor 8 isoform locus (GRM8 Ori), β-globin locus (β-globin Ori), and the −21 kb distal region of β-globin Ori (β-globin −21 kb) were as follows: LB2 Ori-F, 5′-AATAAACTCAGAGGCAGAACC-3′; LB2 Ori-R, 5′-AGAAGATGCATGCCTAATGTG-3′; GRM8 Ori-F, 5′-GGGAAGGAAATGCAAGACAA-3′; GRM8 Ori-R, 5′-AATTTGGCTGCTTAGCATGG-3′; β-globin Ori-F, 5′-CTATTGCTTACATTTGCTTCTG-3′; β-globin Ori-R, 5′-CTTCATCCACGTTCACCTTG-3′; β-globin −21 kb-F, 5′-TGGCCACCAATTGAGTCATC-3′; β-globin −21 kb-R, 5′-AGTCAGGCTGGAAATGAGAG-3′.
Precipitated DNAs were analyzed by quantitative real-time PCR analysis using the Rotor-gene Q real-time PCR cycler (Qiagen, Hilden, Germany). The percent enrichment of specific origin DNAs in BrdU-IP compared with input DNA was calculated for nascent DNA synthesis, and enrichment of β-globin origin DNA compared with the 21 kb distal region DNA was calculated for ChIP analyses. Data represent the mean value ± S.D. for more than three independent experiments. Statistical significance was assessed using the two-tailed unpaired Student's t test.
Author contributions
G. S., J.-S. I., and J.-K. L. conceptualization; G. S., D. J., and H. K. data curation; G. S. and D. J. formal analysis; G. S., D. J., and H. K. investigation; G. S., J.-S. I., and J.-K. L. writing-original draft; J.-S. I. and J.-K. L. methodology; J.-K. L. supervision; J.-K. L. funding acquisition; J.-K. L. project administration; J.-K. L. writing-review and editing.
Supplementary Material
This work was supported by the National Research Foundation of Korea (NRF) Grants NRF-2014R1A1A2058846 and NRF-2017R1A2B2010660 funded by the Korea government (to J.-K. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- DSB
- double strand break
- pre-RC
- pre-replicative complex
- CDK
- cyclin-dependent kinase
- ssDNA
- single-stranded DNA
- ORC
- origin recognition complex
- CMG complex
- Cdc45–Mcm2–7–GINS
- DDK
- Dbf4-dependent kinase
- IP
- immunoprecipitation
- ATR
- ATM and Rad3-related
- RPA
- replication protein A
- γH2AX
- phosphorylated histone H2AX
- PP1
- protein phosphatase 1
- G4
- G-quadruplex.
References
- 1. Monnat R. J., Jr. (2010) Human RECQ helicases: Roles in DNA metabolism, mutagenesis and cancer biology. Semin. Cancer Biol. 20, 329–339 10.1016/j.semcancer.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singh D. K., Ghosh A. K., Croteau D. L., and Bohr V. A. (2012) RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 736, 15–24 10.1016/j.mrfmmm.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larizza L., Magnani I., and Roversi G. (2006) Rothmund-Thomson syndrome and RECQL4 defect: Splitting and lumping. Cancer Lett. 232, 107–120 10.1016/j.canlet.2005.07.042 [DOI] [PubMed] [Google Scholar]
- 4. Shamanna R. A., Singh D. K., Lu H., Mirey G., Keijzers G., Salles B., Croteau D. L., and Bohr V. A. (2014) RECQ helicase RECQL4 participates in non-homologous end joining and interacts with the Ku complex. Carcinogenesis 35, 2415–2424 10.1093/carcin/bgu137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu H., Shamanna R. A., Keijzers G., Anand R., Rasmussen L. J., Cejka P., Croteau D. L., and Bohr V. A. (2016) RECQL4 promotes DNA end resection in repair of DNA double-strand breaks. Cell Rep. 16, 161–173 10.1016/j.celrep.2016.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu H., Shamanna R. A., de Freitas J. K., Okur M., Khadka P., Kulikowicz T., Holland P. P., Tian J., Croteau D. L., Davis A. J., and Bohr V. A. (2017) Cell cycle-dependent phosphorylation regulates RECQL4 pathway choice and ubiquitination in DNA double-strand break repair. Nat. Commun. 8, 2039 10.1038/s41467-017-02146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park S.-Y., Kim H., Im J.-S., and Lee J.-K. (2019) ATM activation is impaired in human cells defective in RecQL4 helicase activity. Biochem. Biophys. Res. Commun. 509, 379–383 10.1016/j.bbrc.2018.12.151 [DOI] [PubMed] [Google Scholar]
- 8. Muramatsu S., Hirai K., Tak Y.-S., Kamimura Y., and Araki H. (2010) CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ϵ, and GINS in budding yeast. Genes Dev. 24, 602–612 10.1101/gad.1883410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sangrithi M. N., Bernal J. A., Madine M., Philpott A., Lee J., Dunphy W. G., and Venkitaraman A. R. (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121, 887–898 10.1016/j.cell.2005.05.015 [DOI] [PubMed] [Google Scholar]
- 10. Abe T., Yoshimura A., Hosono Y., Tada S., Seki M., and Enomoto T. (2011) The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta 1813, 473–479 10.1016/j.bbamcr.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 11. Parker M. W., Botchan M. R., and Berger J. M. (2017) Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 52, 107–144 10.1080/10409238.2016.1274717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka S., Nakato R., Katou Y., Shirahige K., and Araki H. (2011) Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21, 2055–2063 10.1016/j.cub.2011.11.038 [DOI] [PubMed] [Google Scholar]
- 13. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., and Arai K. (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281, 39249–39261 10.1074/jbc.M608935200 [DOI] [PubMed] [Google Scholar]
- 14. Deegan T. D., Yeeles J. T., and Diffley J. F. (2016) Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 35, 961–973 10.15252/embj.201593552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larasati, and Duncker B. (2017) Mechanisms governing DDK regulation of the initiation of DNA replication. Genes 8, E3 10.3390/genes8010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heller R. C., Kang S., Lam W. M., Chen S., Chan C. S., and Bell S. P. (2011) Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146, 80–91 10.1016/j.cell.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeeles J. T., Deegan T. D., Janska A., Early A., and Diffley J. F. (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435 10.1038/nature14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumagai A., Shevchenko A., Shevchenko A., and Dunphy W. G. (2010) Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140, 349–359 10.1016/j.cell.2009.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chattopadhyay S., and Bielinsky A.-K. (2007) Human Mcm10 regulates the catalytic subunit of DNA polymerase-α and prevents DNA damage during replication. Mol. Biol. Cell 18, 4085–4095 10.1091/mbc.e06-12-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Im J.-S., Ki S.-H., Farina A., Jung D.-S., Hurwitz J., and Lee J.-K. (2009) Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 10.1073/pnas.0908039106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu X., Rochette P. J., Feyissa E. A., Su T. V., and Liu Y. (2009) MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J. 28, 3005–3014 10.1038/emboj.2009.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Im J. S., Park S. Y., Cho W. H., Bae S. H., Hurwitz J., and Lee J. K. (2015) RecQL4 is required for the association of Mcm10 and Ctf4 with replication origins in human cells. Cell Cycle 14, 1001–1009 10.1080/15384101.2015.1007001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitsberg D., Selig S., Keshet I., and Cedar H. (1993) Replication structure of the human β-globin gene domain. Nature 366, 588–590 10.1038/366588a0 [DOI] [PubMed] [Google Scholar]
- 24. Goren A., Tabib A., Hecht M., and Cedar H. (2008) DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes Dev. 22, 1319–1324 10.1101/gad.468308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., and Falaschi A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287, 2023–2026 10.1126/science.287.5460.2023 [DOI] [PubMed] [Google Scholar]
- 26. Todorovic V., Giadrossi S., Pelizon C., Mendoza-Maldonado R., Masai H., and Giacca M. (2005) Human origins of DNA replication selected from a library of nascent DNA. Mol. Cell 19, 567–575 10.1016/j.molcel.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 27. Cadoret J.-C., Meisch F., Hassan-Zadeh V., Luyten I., Guillet C., Duret L., Quesneville H., and Prioleau M.-N. (2008) Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl. Acad. Sci. U.S.A. 105, 15837–15842 10.1073/pnas.0805208105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moiseeva T., Hood B., Schamus S., O'Connor M. J., Conrads T. P., and Bakkenist C. J. (2017) ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat. Commun. 8, 1392 10.1038/s41467-017-01401-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vassin V. M., Anantha R. W., Sokolova E., Kanner S., and Borowiec J. A. (2009) Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122, 4070–4080 10.1242/jcs.053702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu S., Opiyo S. O., Manthey K., Glanzer J. G., Ashley A. K., Amerin C., Troksa K., Shrivastav M., Nickoloff J. A., and Oakley G. G. (2012) Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 40, 10780–10794 10.1093/nar/gks849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bester A. C., Roniger M., Oren Y. S., Im M. M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D. S., and Kerem B. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Köhler C., Koalick D., Fabricius A., Parplys A. C., Borgmann K., Pospiech H., and Grosse F. (2016) Cdc45 is limiting for replication initiation in humans. Cell Cycle 15, 974–985 10.1080/15384101.2016.1152424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu H., Baris A., and Aladjem M. I. (2018) Replication timing and nuclear structure. Curr. Opin. Cell Biol. 52, 43–50 10.1016/j.ceb.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., and Dutta A. (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 21, 2288–2299 10.1101/gad.1585607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang Y. H., Farina A., Bermudez V. P., Tappin I., Du F., Galal W. C., and Hurwitz J. (2013) Interaction between human Ctf4 and the Cdc45/Mcm2–7/GINS (CMG) replicative helicase. Proc. Natl. Acad. Sci. U.S.A. 110, 19760–19765 10.1073/pnas.1320202110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumagai A., Shevchenko A., Shevchenko A., and Dunphy W. G. (2011) Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 193, 995–1007 10.1083/jcb.201102003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kliszczak M., Sedlackova H., Pitchai G. P., Streicher W. W., Krejci L., and Hickson I. D. (2015) Interaction of RECQ4 and MCM10 is important for efficient DNA replication origin firing in human cells. Oncotarget 6, 40464–40479 10.18632/oncotarget.6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Araki H. (2016) Elucidating the DDK-dependent step in replication initiation. EMBO J. 35, 907–908 10.15252/embj.201694227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., and Masai H. (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 26, 137–150 10.1101/gad.178491.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamazaki S., Ishii A., Kanoh Y., Oda M., Nishito Y., and Masai H. (2012) Rif1 regulates the replication timing domains on the human genome. EMBO J. 31, 3667–3677 10.1038/emboj.2012.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hänsel-Hertsch R., Di Antonio M., and Balasubramanian S. (2017) DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279 10.1038/nrm.2017.3 [DOI] [PubMed] [Google Scholar]
- 42. Sukackaite R., Cornacchia D., Jensen M. R., Mas P. J., Blackledge M., Enervald E., Duan G., Auchynnikava T., Köhn M., Hart D. J. and Buonomo S. B. C. (2017) Mouse Rif1 is a regulatory subunit of protein phosphatase 1 (PP1). Sci. Rep. 7, 2119 10.1038/s41598-017-01910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hiraga S. I., Ly T., Garzón J., Hořejší Z., Ohkubo Y. N., Endo A., Obuse C., Boulton S. J., Lamond A. I., and Donaldson A. D. (2017) Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 18, 403–419 10.15252/embr.201641983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alver R. C., Chadha G. S., Gillespie P. J., and Blow J. J. (2017) Reversal of DDK-mediated MCM phosphorylation by Rif1-PP1 regulates replication initiation and replisome stability independently of ATR/Chk1. Cell Rep. 18, 2508–2520 10.1016/j.celrep.2017.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanoh Y., Matsumoto S., Fukatsu R., Kakusho N., Kono N., Renard-Guillet C., Masuda K., Iida K., Nagasawa K., and Shirahige K. (2015) Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 22, 889 10.1038/nsmb.3102 [DOI] [PubMed] [Google Scholar]
- 46. Hiraga S.-I., Alvino G. M., Chang F., Lian H.-Y., Sridhar A., Kubota T., Brewer B. J., Weinreich M., Raghuraman M., and Donaldson A. D. (2014) Rif1 controls DNA replication by directing protein phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 28, 372–383 10.1101/gad.231258.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keller H., Kiosze K., Sachsenweger J., Haumann S., Ohlenschläger O., Nuutinen T., Syväoja J. E., Görlach M., Grosse F., and Pospiech H. (2014) The intrinsically disordered amino-terminal region of human RecQL4: multiple DNA-binding domains confer annealing, strand exchange and G4 DNA binding. Nucleic Acids Res. 42, 12614–12627 10.1093/nar/gku993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y., Jaramillo-Lambert A. N., Yang Y., Williams R., Lee N. H., and Zhu W. (2012) And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene 31, 643–652 10.1038/onc.2011.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blumenfeld B., Ben-Zimra M., and Simon I. (2017) Perturbations in the replication program contribute to genomic instability in cancer. Int. J. Mol. Sci. 18, 1138 10.3390/ijms18061138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jones R. M., Mortusewicz O., Afzal I., Lorvellec M., García P., Helleday T., and Petermann E. (2013) Increased replication initiation and conflicts with transcription underlie cyclin E-induced replication stress. Oncogene 32, 3744–3753 10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- 51. Hamperl S., and Cimprich K. A. (2016) Conflict resolution in the genome: How transcription and replication make it work. Cell 167, 1455–1467 10.1016/j.cell.2016.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yeo C. Q. X., Alexander I., Lin Z., Lim S., Aning O. A., Kumar R., Sangthongpitag K., Pendharkar V., Ho V. H. B., and Cheok C. F. (2016) p53 Maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 15, 132–146 10.1016/j.celrep.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 53. Klusmann I., Rodewald S., Müller L., Friedrich M., Wienken M., Li Y., Schulz-Heddergott R., and Dobbelstein M. (2016) p53 Activity results in DNA replication fork processivity. Cell Rep. 17, 1845–1857 10.1016/j.celrep.2016.10.036 [DOI] [PubMed] [Google Scholar]
- 54. Schwab R. A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.-C., Cohn M. A., Gibbons R. J., Deans A. J., and Niedzwiedz W. (2015) The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 60, 351–361 10.1016/j.molcel.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. García-Rubio M. L., Pérez-Calero C., Barroso S. I., Tumini E., Herrera-Moyano E., Rosado I. V., and Aguilera A. (2015) The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 11, e1005674 10.1371/journal.pgen.1005674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 57. Reikofski J., and Tao B. Y. (1992) Polymerase chain reaction (PCR) techniques for site-directed mutagenesis. Biotechnol. Adv. 10, 535–547 10.1016/0734-9750(92)91451-J [DOI] [PubMed] [Google Scholar]
- 58. Azuara V. (2006) Profiling of DNA replication timing in unsynchronized cell populations. Nat. Protoc. 1, 2171–2177 10.1038/nprot.2006.353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.