Abstract
The tubulin homolog FtsZ is the major cytoskeletal protein in the bacterial cell division machinery, conserved in almost all bacteria, archaea, and chloroplasts. Bacterial FtsZ assembles spontaneously into single protofilaments, sheets, and bundles in vitro, and it also accumulates at the site of division early during cell division, where it forms a dynamic protein complex, the contractile ring or Z-ring. The biochemical properties of FtsZ proteins from many bacteria have been studied, but comparable insights into FtsZs from cyanobacteria are limited. Here, using EM and light-scattering assays, we studied the biochemical and assembly properties of SyFtsZ, the FtsZ protein from the cyanobacterial strain Synechocystis sp. PCC 6803. SyFtsZ had a slow GTPase activity of ∼0.4 GTP/FtsZ molecule/min and assembled into thick, straight protofilament bundles and curved bundles, designated toroids. The assembly of SyFtsZ in the presence of GTP occurred in two stages. The first stage consisted of the assembly of single-stranded straight protofilaments and opened circles; in the second stage, the protofilaments associated into straight protofilament bundles and toroids. In addition to these assemblies, we also observed highly curved oligomers and minirings after GTP hydrolysis or in the presence of excess GDP. The three types of protofilaments of SyFtsZ observed here provide support for the hypothesis that a constriction force due to curved protofilaments bends the membrane. In summary, our findings indicate that, unlike other bacterial FtsZ, SyFtsZ assembles into thick protofilament bundles. This bundling is similar to that of chloroplast FtsZ, consistent with its origin in cyanobacteria.
Keywords: cyanobacteria, protein dynamics, electron microscopy (EM), cytoskeleton, cytokinesis, bacterial cell division, FtsZ, toroid, tubulin, Z-ring
Introduction
FtsZ, a tubulin homologue, is a crucial protein in bacterial cell division and is well-conserved in almost all bacteria, archaea, and chloroplasts. It assembles spontaneously into single protofilaments (pfs),5 sheets, and bundles in vitro, and it accumulates in vivo at the site of division early in the cell division process to form a dynamic protein complex, the contractile ring or Z-ring (1–3).
FtsZ from Escherichia coli (EcFtsZ) assembles in vitro into mostly single pfs with an average length of 200 nm (4). EcFtsZ pfs are highly dynamic, turning over with a half time of 5–8 s in vitro (5, 6) and in vivo (7). The mechanism of dynamics is now established to be treadmilling both in vitro (8, 9) and in vivo (10, 11). Treadmilling involves small patches of membrane-bound pfs adding subunits at one end and disassembling at the other.
FtsZ from different species can have different dynamic properties. The Z-ring in Bacillus subtilis has a similar dynamic turnover rate as in E. coli (7); however, the Z-ring from the slow-growing pathogen Mycobacterium tuberculosis has a slower turnover rate of 25-s half-time (12). Chloroplast Z-rings have unique and much slower dynamic properties (13–15). Chloroplasts have two copies of FtsZ, designated FtsZ1 and FtsZ2 (from green algae and plants) and FtsZA and FtsZB (from red algae). Both FtsZs co-localize in the middle of the chloroplast and are required for division (16–18). In vitro, they assemble into thick straight bundles of pfs, either separately or as a mixture (19, 20). Previous results suggested that one FtsZ might assemble as the primary scaffold and another might co-assemble to enhance polymer disassembly (20). The slow dynamics of FtsZ bundles might be related to the large diameter of the chloroplasts.
The chloroplast originated from a cyanobacterial endosymbiont and maintained FtsZ for its division. In contrast to the two FtsZs in chloroplasts, there is only one copy of FtsZ in cyanobacteria, similar to other bacteria. It should be interesting to compare the biochemical and assembly properties of cyanobacterial FtsZ with FtsZ from chloroplasts and bacteria. Here, we studied the biochemical properties of FtsZ from the cyanobacterium Synechocystis sp. PCC 6803 (SyFtsZ).
Results
SyFtsZ assembles into straight bundles of pfs and toroid-like circle bundles
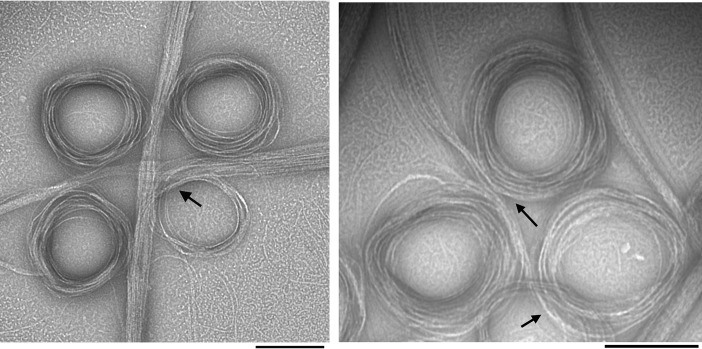
Negative-stain EM images of SyFtsZ filaments assembled in GTP showed some isolated pfs, but mostly large bundles of pfs that were either straight or curved into toroids (Fig. 1). The diameter of the toroids ranged from 200 to 300 nm. Straight bundles and toroids were mostly isolated, but occasionally, a transition between straight and curved bundles was observed (Fig. 1, arrows). In contrast to EcFtsZ, which can assemble in the absence of Mg2+ (6, 21), SyFtsZ showed no assembly without Mg2+ or when using GMPCPP to replace GTP (Fig. S1).
Figure 1.
EM images of negatively stained SyFtsZ pfs after adding 0.5 mm GTP in HMK buffer (50 mm HEPES, 100 mm KAc, 5 mm MgAc) at pH 7.5. 10 μm SyFtsZ assembled into pfs (some isolated pfs are seen in the background), which further associate into straight bundles and toroids. The diameter of the toroids is 200–300 nm. Transitions between straight bundles and toroids are indicated by arrows. Bar, 200 nm.
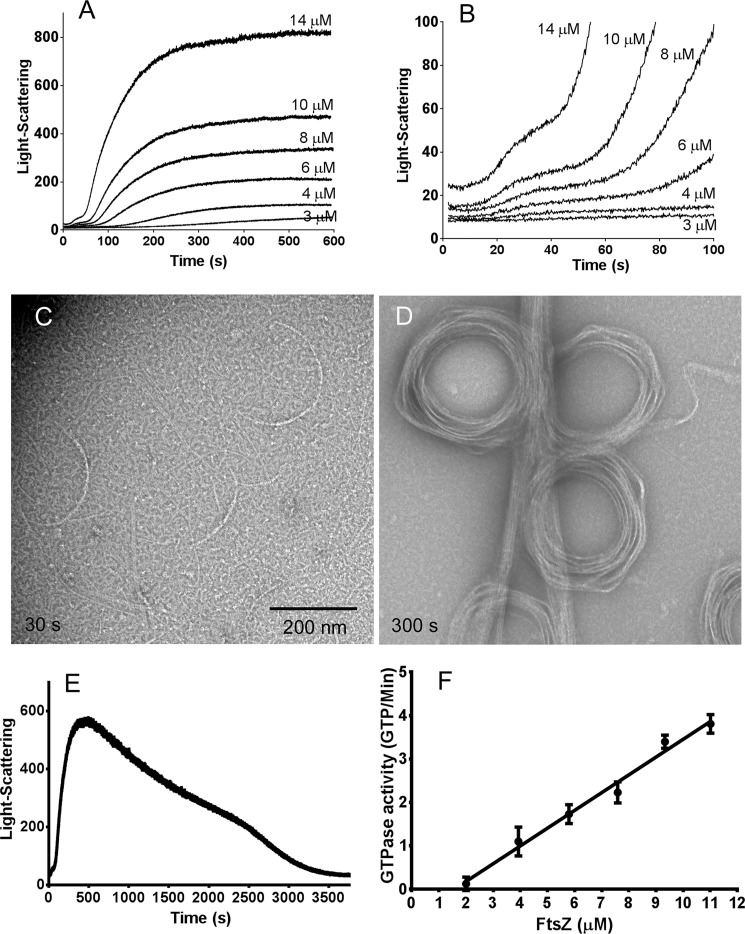
Assembly was then monitored by light scattering, which showed that it proceeded in two stages (Fig. 2). The light scattering reached a final plateau at ∼300 s (Fig. 2A). Amplification of the first 100 s (Fig. 2B) showed an early stage where a 15–20-s lag was followed by a rise to an intermediate plateau at 30–50 s. After 50 s, the second stage began as light scattering accelerated, leading to the final plateau. Negative-stain EM showed that the first stage resulted in the assembly of isolated short pfs and thin bundles of pfs curved into partial circles (Fig. 2C). The curved filaments consisted of apparently more than single filament, and their diameters were 200–300 nm, similar to toroid-like circle bundles, but mostly too short to form a complete circle. At the final plateau following the second stage of assembly, the EM showed predominantly large bundles of pfs and toroids (Fig. 2D).
Figure 2.
The assembly of SyFtsZ assayed by light scattering shows two stages in HMK buffer at pH 7.5 (A and B). A shows that light scattering reaches a final plateau at 300–600 s. B amplifies the first 100 s and shows that light scattering first approaches an intermediate plateau at 30–50 s and then accelerates to the much higher final plateau. Note that light scattering does not measure polymer, but is greatly enhanced by bundling. The signal here is primarily a measure of bundle formation. C and D, EM of 10 μm SyFtsZ shows short single pfs and partial circles at the first stage assembly, at 30 s (C), and assembly into straight bundles and toroids at the second stage of assembly, at 300 s (D). Bar, 200 nm. E, light-scattering assay of 10 μm SyFtsZ in HMK buffer assembled in 0.1 mm GTP. Light scattering reached a plateau at 500 s and then slowly decreased to the base after 1 h. F, the GTPase activity of SyFtsZ in HMK buffer was 0.41 ± 0.2 GTP/min/FtsZ above a critical concentration of 1.6 μm. Error bars, S.D.
We repeated the assembly with a limited amount of GTP (0.1 mm GTP) and found that the light-scattering signal rose to a plateau at 500 s and then slowly dropped back to the base after 1 h (Fig. 2E). This suggested that the large pf bundles disassembled after the GTP was used up. Fig. 2F shows an assay of GTPase as a function of FtsZ concentration. GTPase activity was 0.41 ± 0.2 GTP/FtsZ/min above a critical concentration around 1.6 μm (Fig. 2F). This is ∼10 times less than EcFtsZ but close to that of chloroplast FtsZ. At this rate of hydrolysis, the 10 μm SyFtsZ in Fig. 2E should consume the 100 μm GTP in about 30 min. However, the polymers in that experiment started to disassemble after 10 min, well before the GTP should be exhausted. Small and Addinall (22) documented for EcFtsZ that GDP inhibited assembly, suggesting that the early decrease in light scattering seen here was due to inhibition by the buildup of GDP.
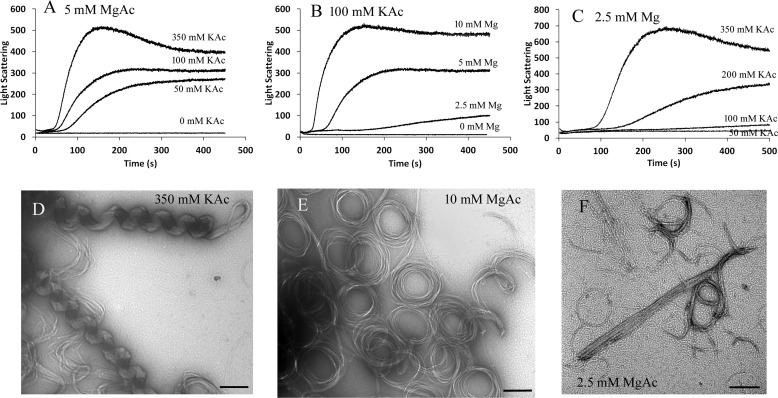
Previous results showed that high salt concentration caused an increased level of bundling of EcFtsZ (6, 23, 24). We obtained similar results with SyFtsZ. Fig. 3 shows that high concentrations of potassium and magnesium accelerated the assembly and enhanced the light scattering. Without potassium or magnesium, SyFtsZ showed no assembly in pH 7.5 buffer (Fig. 3, A and B). In most experiments, the solution we used contained 100 mm KAc and 5 mm MgAc. Increasing KAc to 350 mm increased the rate and final plateau of light scattering (Fig. 3, A and C). EM showed that in addition to straight and circular bundle structures, some helical structures could be observed. The average diameter and pitch of these helical structures were about 145 and 185 nm, respectively (Fig. 3D). Polymerization of SyFtsZ is also sensitive to MgAc concentration. When the concentration of MgAc was reduced to 2.5 mm, the light-scattering signal was much weaker and had a longer lag time (Fig. 3, B and C). EM showed that there were more single pfs and thin circles and fewer straight and circular bundles (Fig. 3F). When the concentration of MgAc increased from 5 to 10 mm, the light-scattering signal increased and the lag time decreased (Fig. 3B). EM showed more straight and circular bundles (Fig. 3E).
Figure 3.
SyFtsZ assembly in buffers of different concentration of KAc and MgAc. A–C, assembly kinetics of 5 μm SyFtsZ in 5 mm MgAc and different concentrations of KAc, from 0 to 350 mm (A); in 100 mm KAc and different concentrations of MgAc, from 0 to 10 mm (B); and in 2.5 mm MgAc and different concentrations of KAc, from 0 to 350 mm (C). D–E, negative-stain EM images of 5 μm SyFtsZ assembled in 5 mm MgAc, 350 mm KAc (D); in 10 mm MgAc, 100 mm KAc (E); and in 2.5 mm MgAc, 100 mm KAc (F). Bar, 200 nm. All buffers contain 50 mm HEPES, pH 7.5, and 0.5 mm GTP.
Assembly properties of SyFtsZ change with pH
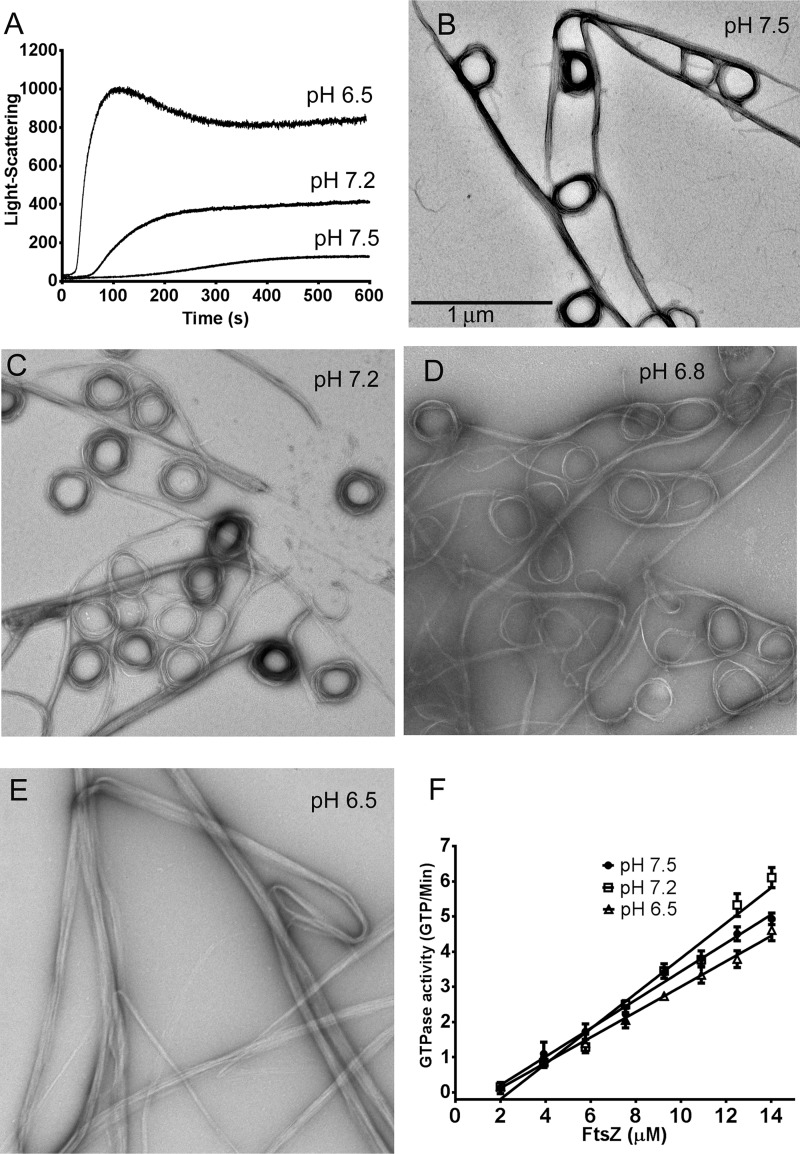
Next, we examined the assembly of SyFtsZ in buffers of different pH. The light-scattering assay showed two effects of lowering pH: an accelerated assembly and increased plateau (Fig. 4A). The assemblies displayed typical cooperative assembly kinetics with a lag time. The two-stage assemblies in pH 7.2 and pH 6.5 buffers were not as clear as we observed at pH 7.5 buffer, perhaps because the first stage was too short or too weak. EM of the pH 7.2 assembly at 40 s showed short straight bundles and toroids already assembled, mixed with short pfs and partial circles (Fig. S2). EM of assemblies at the final plateau showed a mixture of straight pf bundles and toroids at pH 7.5, pH 7.2, and pH 6.8 (Fig. 4, B–D). However, only straight bundles were observed at pH 6.5 (Fig. 4E). The increased light scattering at reduced pH is consistent with the straight bundles being larger in diameter and/or longer.
Figure 4.
SyFtsZ assembly in buffers of different pH. A, assembly kinetics of 5 μm SyFtsZ monitored by light scattering. The light-scattering signals were much higher when the pH dropped from 7.5 to 6.5. B–E, negative-stain EM images of 10 μm SyFtsZ assembled at different pH levels, from 7.5 to 6.5. Bar, 1 μm. F, GTPase activity of SyFtsZ was measured after the assembly reached a plateau. GTPase is approximately the same at pH 6.5–7.5. All buffers contained 100 mm KAc, 5 mm MgAc, and 0.5 mm GTP. pH 7.5 and 7.2 were buffered with HEPES; pH 6.8 and 6.5 were buffered with MES. Error bars, S.D.
In contrast to the large changes in light scattering and bundle morphology at lower pH, the GTPase was hardly changed. The GTPase activity of FtsZ was 0.41 ± 0.2 GTP/FtsZ/min at pH 7.5, 0.50 ± 0.1 at pH 7.2, and 0.36 ± 0.2 at pH 6.5 with critical concentrations of 1.5–2 μm (Fig. 4F).
SyFtsZ assembled into highly curved oligomers and miniring structures in the presence of GDP
Previous studies have shown that FtsZ from E. coli and Pseudomonas aeruginosa could assemble into highly curved short oligomers or miniring structures with an average diameter of 23 nm in the presence of GDP (25–28). Minirings were especially favored in the presence of ZipA protein (27). We also found highly curved oligomers and miniring structures in the EM images of SyFtsZ pfs during GTP hydrolysis (Fig. 5A) or in the presence of GDP (Fig. 5, B and C) in both pH 7.5 and pH 6.5 solutions. The diameters of these highly curved pfs or minirings were ∼20 ± 2 nm. Sedimentation results confirmed assembly of these oligomers. Using a Beckman TLA-100 rotor, we selected two centrifuge speeds: 50,000 rpm and 90,000 rpm. Only large bundles assembled in GTP could be spun down at 50,000 rpm (around 100,000 × g); in contrast, both bundles in GTP and partly short oligomers and/or minirings could be spun down at 90,000 rpm (around 310,000 × g). Fig. 5 (D and E) and Fig. S3 show the sedimentation results analyzed by SDS-PAGE. At 50,000 rpm, there was little protein in pellet with GDP. At 90,000 rpm with GDP, 21% of FtsZ was pelleted at pH 7.5 at both 5 and 10 μm FtsZ, and 49% of 10 μm FtsZ was pelleted at pH 6.5. The pellets are consistent with the short oligomers and minirings observed by EM.
Figure 5.
A–C, SyFtsZ minirings observed by negative-stain EM. A, 10 μm SyFtsZ in HMK buffer at pH 7.5 assembled in the presence of 0.5 mm GTP. In addition to straight bundles and toroids, some minirings are also observed. The diameter of miniring is ∼20 nm. Bar, 100 nm. B and C, 10 μm SyFtsZ assembled into oligomers and minirings in the presence of 0.5 mm GDP at pH 7.5 (B) or at pH 6.5 (C). Bar, 100 nm. Arrows point to minirings. D and E, SDS-PAGE analysis of sedimentation of 5 or 10 μm SyFtsZ with 1 mm GTP or 1 mm GDP centrifuged at 50,000 rpm (D) or 90,000 rpm (E) for 30 min in a Beckman TLA100 rotor. Numbers in parentheses indicate the percentage of protein in the pellet.
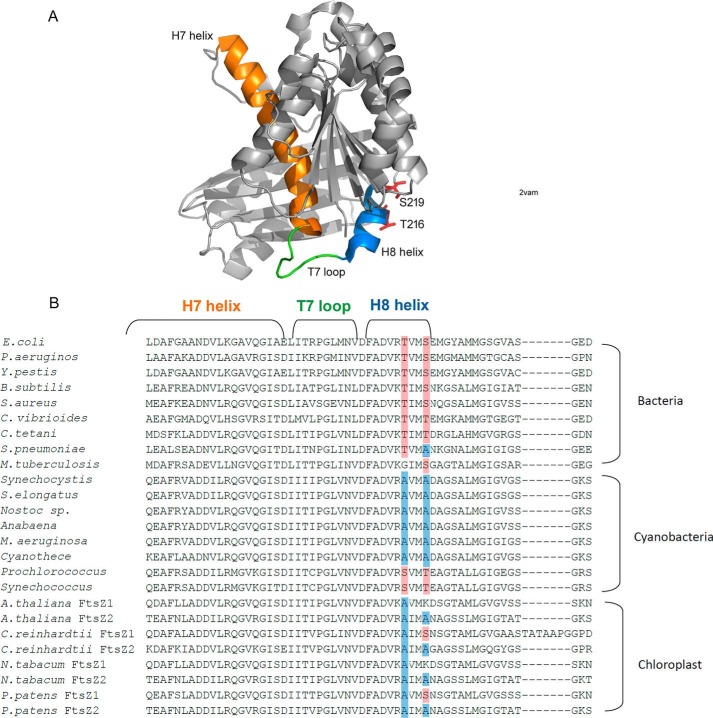
Mutant AXXA to TXXS in the H8 helix reduces bundle formation and enhances GTPase activity
FtsZ comprises two globular subdomains, which are connected by a central core helix (H7 helix) and a synergy loop (T7 loop). The T7 loop is located between the H7 helix and H8 helix, and it reaches into the active site of an adjacent subunit to activate its GTPase activity (29–31) (Fig. 6A). The amino acids Asn-207, Asp-209, and Asp-201 of EcFtsZ are highly conserved in all FtsZs. Mutation of Asp-209 in the T7 loop or Asp-212 in the H8 helix of EcFtsZ severely impaired its GTPase activity and function (32, 33). Following the very conserved sequence DFADV(R/K), we have identified two amino acids in the H8 helix that are conserved as Thr and Ser in fast-growing bacteria (Thr-215 and Ser-218 of EcFtsZ, or Thr-216 and Ser-219 of FtsZ from B. subtilis). In most cyanobacteria and chloroplast FtsZs, the corresponding amino acids are Ala (Ala-271 and Ala-274 of SyFtsZ). Interestingly, there are several cyanobacterial FtsZs, such as FtsZ from Prochlorococcus and Synechococcus, where these are still Ser and Thr. Both of these belong to marine picocyanobacteria, whose cell diameter is less than 1 μm. This size is similar to that of bacteria and much smaller than other cyanobacteria. Also, some slow-growing bacterial FtsZs have only one Thr or Ser. For example, in M. tuberculosis FtsZ (MtbFtsZ), the first Thr is changed to Gly, and in S. pneumoniae FtsZ (SpnFtsZ), the second Ser is changed to Ala. Previous results suggested that both MtbFtsZ and SpnFtsZ assembled into small bundles. MtbFtsZ slowly assembled into two-stranded pfs at pH 6.5 (12, 34), and SpnFtsZ assembled into mostly double- or multiple-strand pfs, with a high critical concentration around 3–6 μm (35, 36).
Figure 6.
A, structure of FtsZ from B. subtilis (Protein Data Bank code 2VAM). The H7 helix is shown in orange, the T7 loop is green, and the H8 helix is blue. Thr-216 and Ser-219 are shown as sticks. B, sequence alignment of FtsZ with a focus on the conserved region of H7 helix, T7 loop, and H8 helix from bacteria, cyanobacteria, and chloroplasts. Two amino acids in helix H8 are mostly conserved as Thr and Ser in bacteria (highlighted in red) and mostly Ala in cyanobacteria (highlighted in blue).
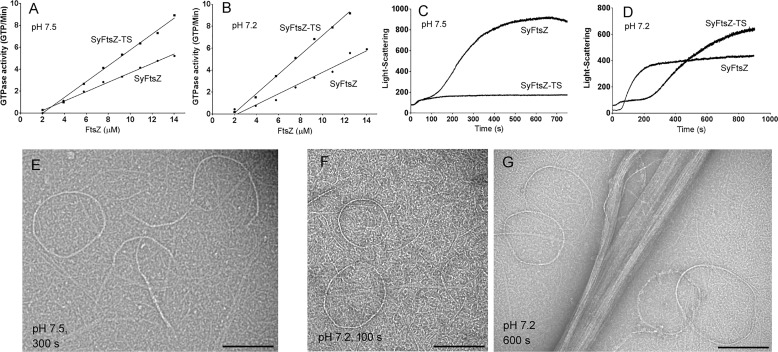
To address whether these two amino acids are related to bundle formation, we constructed the double mutant SyFtsZ-A271T/A274S (SyFtsZ-TS). The GTPase activity of this mutant increased ∼80%: from 0.4 to 0.7 GTP/FtsZ/min in pH 7.5 buffer (Fig. 7A) and from 0.5 to 0.9 GTP/FtsZ/min in pH 7.2 buffer (Fig. 7B). The increased GTPase of the mutant correlates with reduced bundle formation, suggesting that bundling results in reduced subunit turnover and GTPase. Fig. 7 (C and D) highlights the difference of their assembly properties measured by the light scattering at different pH. In pH 7.5 solution, the assembly of mutant SyFtsZ only had the fast initial stage in contrast to the two-stage assembly curves of WT SyFtsZ assembly (Fig. 7C; compare with Fig. 2B). Negative-stain EM confirmed that the assembled pfs of SyFtsZ-TS were mixtures of single pfs and thin straight and curved bundles, including some closed circles (Fig. 7E). There were no large bundles in pH 7.5 buffer for FtsZ up to 15 μm. In pH 7.2 solution, the assembly showed two stages, but they were much slower than WT (Fig. 7D). The first stage of assembly, which plateaued from 50 to 200 s, showed short pfs and circles typical of this stage (Fig. 7F). The second stage started around 200 s, rose to a much higher light-scattering signal, and approached a plateau at 900 s (Fig. 7D). EM showed mostly thick straight bundles and a few circles at pH 7.2 (Fig. 7G), and no toroid-like circular bundles were observed. Evidently, mutant proteins reduced bundle formation.
Figure 7.
Comparison of assembly properties of WT SyFtsZ and SyFtsZ-TS (SyFtsZ-A271T/A274S). A and B, the double mutant increased the GTPase somewhat at both pH 7.5 (A) and pH 7.2 (B). C, light scattering suggests that the SyFtsZ-TS at pH 7.5 assembled only to the first stage and was missing the large rise in turbidity attributed to the second stage (cf. Fig. 2). D, at pH 7.2, the first stage of assembly was prolonged, but the second stage rise in turbidity began at ∼300 s. E–G, EM confirms that SyFtsZ-TS only assembles into single straight pfs and thin curved bundles at pH 7.5, 300 s (E) and at pH 7.2, 100 s (F). These assemblies are characteristic of stage 1 (see Fig. 2). At pH 7.2, 600-s large thick bundles characteristic of stage 2 are seen (G). Bar, 200 nm. All buffers contained 50 mm HEPES, 100 mm KAc, 5 mm MgAc, and 0.5 mm GTP.
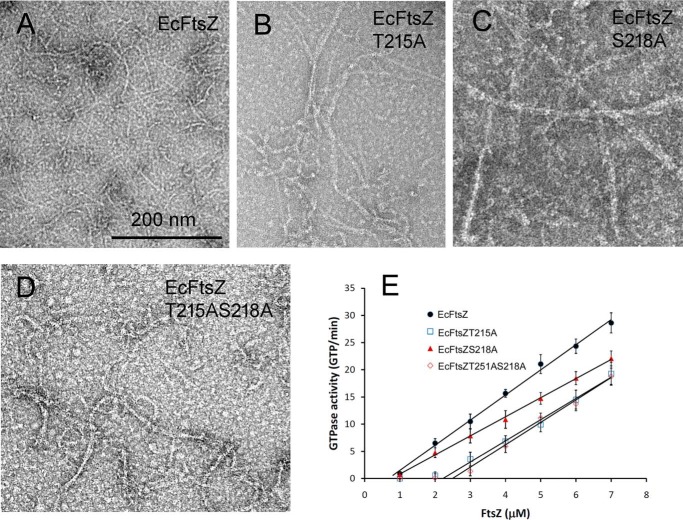
We then tested the effect on EcFtsZ of changing the Thr and Ser to Ala. We constructed two single mutants (EcFtsZ-T215A and EcFtsZ-S218A) and a double mutant EcFtsZ-T215A/S218A. Negative-stain EM showed a modest increase in bundles assembled for the mutant EcFtsZ (Fig. 8, A–D). These were mostly double pfs or small bundles, and no large bundles were seen. These modest bundling filaments were similar to the assembly of MtbFtsZ (12, 34) and SpnFtsZ (35), which had one of the two amino acids changed. Interestingly, GTPase activities of mutant EcFtsZ were only slightly changed (Fig. 8E). The GTPase was reduced from 4.6 to 3.5 GTP/FtsZ/min for EcFtsZ-S218A and little changed for EcFtsZ-T215A and EcFtsZ-T215A/S218A. The critical concentration increased from 1 to around 2.5 μm for EcFtsZ-T215A and EcFtsZ-T215A/S218A.
Figure 8.
A–D, assembly of EcFtsZ with Thr-215 and Ser-218 changed to Ala at pH 7. 5, singly or together. All mutants showed a modest increase in pf pairs and thin bundles. Bar, 200 nm. E, GTPase activity showed only small changes for the mutants, although S218A caused a significant increase in the critical concentration. Error bars, S.D.
Discussion
Perhaps the most striking observation in the present work was that SyFtsZ assembled into thick bundles of pfs that could be either straight or curved into a toroid of 200–300-nm diameter. Assembly of similar straight and curved pf bundles was reported previously for EcFtsZ and MtbFtsZ, but only in the presence of crowding agents polyvinyl alcohol or methylcellulose (37, 38). A recent study of FtsZ from the cyanobacterium Anabena showed a mixture of straight pf bundles and toroids essentially the same as those we show here for SyFtsZ. In that study, removal of the 51 N-terminal amino acids (this N-terminal peptide is much longer in cyanobacteria than the 9–15 amino acids in most bacteria) eliminated the toroids but left the thick straight pf bundles. FtsZ from the chloroplasts of Galdieria sulfuraria, GsFtsZA and GsFtsZB, also formed thick straight bundles. In this case, the assembly used the globular domains only, truncating the N- and C-terminal extensions. The morphology of SyFtsZ bundles, straight versus toroids versus helices, is sensitive to the pH value and salt concentration. Similar bundle morphologies have been observed for EcFtsZ assembled in the presence of crowding agents (38). Because the internal pH of cyanobacterial cells is above 7, SyFtsZ should assemble preferentially curved filaments and bundles in vivo.
The thick bundles seem to be a property of cyanobacterial FtsZ and the derivative chloroplast FtsZ. However, the relevance of the bundling for in vivo function is not clear. It is noted that the diameter of cyanobacterium Synechocystis sp. PCC 6803 is around 2–3 μm, and the chloroplast average of 5 μm is much larger than the ∼1 μm of the bacterial cell. These results are consistent with previous studies of chloroplast FtsZ (20), suggesting that bundling of FtsZ might benefit the cell division of large cells. The in vitro bundles appear to be three-dimensional, but because all FtsZ subunits have the C-terminal peptide that mediates membrane tethering, the in vivo bundle may be limited to a two-dimensional ribbon, with all pfs tethered to the membrane. The amount of FtsZ in cyanobacteria has not been determined, but in other bacteria, it is typically present at ∼5,000 copies (2). This could make a ribbon of 7 pfs encircling the 1-μm diameter in E. coli. Where only 30% of the FtsZ is in the Z-ring, the ribbon would average only 3 pfs wide. There is not enough FtsZ in a single cell to make a large bundle.
We found that bundling was reduced significantly when we mutated Ala-271 to Thr and Ala-274 to Ser, which were the amino acids at the corresponding positions in E. coli and other rapidly dividing bacteria. Interestingly, in the FtsZ from marine picocyanobacteria, such as Prochlorococcus and marine Synechococcus, these amino acids are still Ser and Thr, suggesting that they may reduce the bundling activity. Those marine picocyanobacteria have small sizes with diameters of less than 1 μm, compared with the 2–3-μm diameter of Synechocystis. Previous results revealed that marine Synechococcus strains and Prochlorococcus were sister clades, which are distantly related to other cyanobacteria, including freshwater Synechococcus strains, such as Synechococcus elongates (39–42). This is consistent with our analysis. One possibility is that the intrinsic bundling activity is useful for the stable Z-ring formation and benefits from the larger diameter of most cyanobacteria and chloroplasts. The biochemical properties of marine picocyanobacterial FtsZ need to be further investigated. Also, we do not have in vivo data to support this possibility, so further investigations need to be conducted.
The physiological function of the circle bundles of cyanobacterial FtsZ is still unclear. We observed that SyFtsZ assembled into both straight and circular bundles when pH levels were above 6.8; meanwhile, only straight bundles assembled when pH was 6.5. It was reported that cyanobacterial cells maintained a narrow range of survivable intracellular pH of 7.1–7.3 even when they were exposed to a low, growth-inhibiting external pH below 6 (43). This indicates that the FtsZ circles might be useful for its function. Recently, Corrales-Guerrero et al. (44) reported that deletion of N terminus of Anabaena FtsZ eliminated the circular bundles and produced only straight bundles in vitro. The mutant was nonfunctional as the sole source of FtsZ and was toxic when co-expressed with WT FtsZ. These data suggest that formation of circular bundles might be necessary for its function; however, the possibility that deletion of the N terminus affects binding of other association proteins cannot be excluded (44).
The 200–300-nm curvature of the toroids has been designated the intermediate curved conformation (45). Osawa and Erickson (46) have suggested that this intermediate curved conformation, rather than the highly curved miniring, may be responsible for generating the constriction force on the membrane. It is not clear what triggers the transformation from straight to intermediate curved. GTP hydrolysis is not required, because intermediate curved pfs can be obtained in nonhydrolyzable GMPCPP and by the EcFtsZ mutant D212A (2). Also, Popp et al. (37) observed toroids and spiral pf bundles assembled in GMPCPP and in GTP plus EDTA. Previous studies have provided evidence that a pf bundle can transition from straight to curved, in a process called spooling (47, 48). A transition from straight to curved pfs has been invoked for treadmilling pf patches (49). Fig. 1 provides some of the clearest images of the spooling transition for SyFtsZ.
Another type of curved pf is defined as highly curved pfs and minirings. The miniring with a diameter around 20–30 nm was first reported when EcFtsZ was assembled onto cationic lipid monolayers without GTP or after GTP hydrolysis (25, 50). The miniring structures of EcFtsZ could also be observed in solution (27, 28, 51), especially following the addition of ZipA (27), a FtsZ-associated protein conserved in only γ-proteobacteria. Similar miniring structures were further observed in the studies of Caulobacter crescentus FtsZ when stabilized by the regulator protein FzlA (52) and B. subtilis FtsZ with the addition of ZapA (53). It is concluded that GTP favored the assembly of straight or intermediate curved pfs, and GDP favored highly curved pfs. It is possible that these two curved conformations of FtsZ together provide a continuous force to bend and constrict the membrane at the site of cell division.
Experimental procedures
Plasmid construction and protein purification
cDNA for SyFtsZ from cyanobacterium Synechocystis sp. PCC 6803 was inserted into pET15b using PCR and engineered NdeI/BamHI digestion sites. Mutants SyFtsZ-TS, EcFtsZ-T215A, EcFtsZ-S218A, and EcFtsZ-T215A/S218A were generated by point mutagenesis.
Plasmids containing SyFtsZ or EcFtsZ were transformed into BL21 E. coli and purified. Briefly, the soluble His6 proteins were first purified using a Talon column (Clontech Lab, Inc.). After elution with 250 mm imidazole, the samples were concentrated using an Amicon Ultra-15 centrifugal filter (Millipore Sigma) and exchanged into LTK50 buffer (50 mm Tris, pH 7.9, 50 mm KCl, 1 mm EDTA, 10% glycerol). Thrombin was added to 20 units/ml and incubated at room temperature for 2–4 h to remove the His6 tag. After digestion by thrombin, three amino acid residues, Gly-Ser-His, remain at the N terminus, which should have limited impact on assembly. Further purification was over a Resource Q column (GE healthcare) with a linear 50 mm to 1 m KCl gradient in LTK buffer. Purified proteins were stored at −80 °C. Protein concentrations were determined by using a BCA assay with a corrected factor for a presumed 70% color ratio (54). Proteins were thawed and dialyzed into HMK buffer (50 mm Hepes, 100 mm KAc, 5 mm MgAc) for assembly at pH 7.5 and 7.2 or MMK buffer (50 mm MES, 100 mm KAc, 5 mm MgAc) for assembly at pH 6.8 and 6.5.
Electron microscopy
Negative-stain EM was used to visualize FtsZ polymerization. FtsZ proteins were incubated with GTP or GDP for 2–10 min. Then ∼10 μl was applied to a carbon-coated copper grid and quickly withdrawn. Grids were immediately stained with 2% uranyl acetate, and specimens were imaged with a Philips 420 electron microscope.
Light-scattering measurement
The kinetics of FtsZ assembly was measured by a light-scattering assay using a Shimadzu RF-5301 PC spectrofluorometer, with both excitation and emission at 350 nm as described previously (20, 55). The light-scattering assay is especially sensitive to the assembly of large bundles. Each measurement was repeated two or three times, with consistent results.
Sedimentation assay
Assembly of SyFtsZ was also assayed by sedimentation. 5 or 10 μm SyFtsZ was incubated with or without 1 mm GTP in HMK buffer, pH 7.5, or MMK buffer, pH 6.5, at room temperature for 10 min and centrifuged at 50,000 or 90,000 rpm for 30 min at 25 °C in a Beckman TLA100 rotor. The supernatant was carefully removed, and the pellet was resuspended in the same volume of buffer. The protein in the pellet and supernatant was assayed by 10% SDS-PAGE. The ratio of supernatant and pellet was measured using ImageJ software.
GTPase measurement
GTP turnover was measured after the assembly had reached its plateau of light scattering. The GTPase turnover was measured using a regenerative coupled GTPase assay as described previously (55, 56). Our assay mixture included 1 mm phosphoenolpyruvate, 0.8 mm NADH, 20 units/ml pyruvate kinase and lactate dehydrogenase (Sigma-Aldrich), and 0.5 mm GTP. After GTP hydrolysis by FtsZ, GDP in solution is rapidly regenerated into GTP, in a reaction that consumes one NADH per GDP. The NADH concentration was measured by the absorption at 340 nm in a Shimadzu UV-2401PC spectrophotometer, using the extinction coefficient 6,220 m−1 cm−1. The absorbance showed a linear decrease over time, giving the hydrolysis rate for each FtsZ concentration. These rates were plotted versus FtsZ concentration, and the overall hydrolysis rate in GTP/min/FtsZ was the slope of the line above the critical concentration. Each assay was repeated two or three times.
Author contributions
N. W., L. B., P. W., and Y. C. formal analysis; N. W., L. B., X. M., Y. M., C. S. C., T. Z., Z. L., P. W., and Y. C. investigation; N. W., P. W., and Y. C. writing-original draft; N. W., L. B., X. M., Y. M., C. S. C., M. u. R., T. Z., P. W., and Y. C. writing-review and editing; P. W. and Y. C. resources; P. W. and Y. C. supervision; P. W. and Y. C. project administration; Y. C. conceptualization; Y. C. data curation; Y. C. funding acquisition; Y. C. validation; Y. C. visualization; Y. C. methodology.
Supplementary Material
Acknowledgment
We thank Dr. Harold Erickson (Duke University) for providing laboratory and EM resources for some of this work and for helpful comments on the manuscript.
This work was supported by First-class University and Academic programs of Northwest University (to Y. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- pf
- protofilament
- SyFtsZ
- FtsZ from the cyanobacterium Synechocystis sp. PCC 6803
- EcFtsZ
- FtsZ from E. coli
- SyFtsZ-TS
- double mutant SyFtsZ-A271T/A274S
- MtbFtsZ
- M. tuberculosis FtsZ
- SpnFtsZ
- S. pneumoniae FtsZ
- GMPCPP
- guanylyl-(α,β)-methylene-diphosphonate.
References
- 1. Haeusser D. P., and Margolin W. (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 10.1038/nrmicro.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erickson H. P., and Osawa M. (2017) FtsZ constriction force: curved protofilaments bending membranes. Subcell. Biochem. 84, 139–160 10.1007/978-3-319-53047-5_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krupka M., and Margolin W. (2018) Unite to divide: oligomerization of tubulin and actin homologs regulates initiation of bacterial cell division. F1000Research 7, 235 10.12688/f1000research.13504.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., Bjornson K., Redick S. D., and Erickson H. P. (2005) A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88, 505–514 10.1529/biophysj.104.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y., and Erickson H. P. (2005) Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 280, 22549–22554 10.1074/jbc.M500895200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y., and Erickson H. P. (2009) FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48, 6664–6673 10.1021/bi8022653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson D. E., Gueiros-Filho F. J., and Erickson H. P. (2004) Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186, 5775–5781 10.1128/JB.186.17.5775-5781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loose M., and Mitchison T. J. (2014) The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 10.1038/ncb2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramirez-Diaz D. A., García-Soriano D. A., Raso A., Mücksch J., Feingold M., Rivas G., and Schwille P. (2018) Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 16, e2004845 10.1371/journal.pbio.2004845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bisson-Filho A. W., Hsu Y. P., Squyres G. R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M. S., Brun Y. V., and Garner E. C. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 10.1126/science.aak9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X., Lyu Z., Miguel A., McQuillen R., Huang K. C., and Xiao J. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 10.1126/science.aak9995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y., Anderson D. E., Rajagopalan M., and Erickson H. P. (2007) Assembly dynamics of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 282, 27736–27743 10.1074/jbc.M703788200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. TerBush A. D., and Osteryoung K. W. (2012) Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J. Cell Biol. 199, 623–637 10.1083/jcb.201205114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida Y., Mogi Y., TerBush A. D., and Osteryoung K. W. (2016) Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2, 16095 10.1038/nplants.2016.95 [DOI] [PubMed] [Google Scholar]
- 15. TerBush A. D., MacCready J. S., Chen C., Ducat D. C., and Osteryoung K. W. (2018) Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiol. 176, 295–306 10.1104/pp.17.00558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osteryoung K. W., Stokes K. D., Rutherford S. M., Percival A. L., and Lee W. Y. (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10, 1991–2004 10.1105/tpc.10.12.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McAndrew R. S., Froehlich J. E., Vitha S., Stokes K. D., and Osteryoung K. W. (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127, 1656–1666 10.1104/pp.010542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. TerBush A. D., Yoshida Y., and Osteryoung K. W. (2013) FtsZ in chloroplast division: structure, function and evolution. Curr. Opin. Cell Biol. 25, 461–470 10.1016/j.ceb.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 19. Olson B. J., Wang Q., and Osteryoung K. W. (2010) GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285, 20634–20643 10.1074/jbc.M110.122614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y., Porter K., Osawa M., Augustus A. M., Milam S. L., Joshi C., Osteryoung K. W., and Erickson H. P. (2017) The chloroplast tubulin homologs FtsZA and FtsZB from the red alga Galdieria sulphuraria co-assemble into dynamic filaments. J. Biol. Chem. 292, 5207–5215 10.1074/jbc.M116.767715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mukherjee A., and Lutkenhaus J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 10.1093/emboj/17.2.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Small E., and Addinall S. G. (2003) Dynamic FtsZ polymerization is sensitive to the GTP to GDP ratio and can be maintained at steady state using a GTP-regeneration system. Microbiology 149, 2235–2242 10.1099/mic.0.26126-0 [DOI] [PubMed] [Google Scholar]
- 23. Tadros M., González J. M., Rivas G., Vicente M., and Mingorance J. (2006) Activation of the Escherichia coli cell division protein FtsZ by a low-affinity interaction with monovalent cations. FEBS Lett. 580, 4941–4946 10.1016/j.febslet.2006.07.083 [DOI] [PubMed] [Google Scholar]
- 24. Mukherjee A., and Lutkenhaus J. (1999) Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erickson H. P., Taylor D. W., Taylor K. A., and Bramhill D. (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U.S.A. 93, 519–523 10.1073/pnas.93.1.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romberg L., and Mitchison T. J. (2004) Rate-limiting guanosine 5′-triphosphate hydrolysis during nucleotide turnover by FtsZ, a prokaryotic tubulin homologue involved in bacterial cell division. Biochemistry 43, 282–288 10.1021/bi035465r [DOI] [PubMed] [Google Scholar]
- 27. Chen Y., Huang H., Osawa M., and Erickson H. P. (2017) ZipA and FtsA* stabilize FtsZ-GDP miniring structures. Sci. Rep. 7, 3650 10.1038/s41598-017-03983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huecas S., Ramírez-Aportela E., Vergoñós A., Núñez-Ramírez R., Llorca O., Díaz J. F., Juan-Rodríguez D., Oliva M. A., Castellen P., and Andreu J. M. (2017) Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys. J. 113, 1831–1844 10.1016/j.bpj.2017.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Löwe J., and Amos L. A. (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 10.1038/34472 [DOI] [PubMed] [Google Scholar]
- 30. Oliva M. A., Cordell S. C., and Löwe J. (2004) Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11, 1243–1250 10.1038/nsmb855 [DOI] [PubMed] [Google Scholar]
- 31. Matsui T., Han X., Yu J., Yao M., and Tanaka I. (2014) Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 289, 3501–3509 10.1074/jbc.M113.514901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheffers D. J., de Wit J. G., den Blaauwen T., and Driessen A. J. (2002) GTP hydrolysis of cell division protein FtsZ: evidence that the active site Is formed by the association of monomers. Biochemistry 41, 521–529 10.1021/bi011370i [DOI] [PubMed] [Google Scholar]
- 33. Redick S. D., Stricker J., Briscoe G., and Erickson H. P. (2005) Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187, 2727–2736 10.1128/JB.187.8.2727-2736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White E. L., Ross L. J., Reynolds R. C., Seitz L. E., Moore G. D., and Borhani D. W. (2000) Slow polymerization of Mycobacterium tuberculosis FtsZ. J. Bacteriol. 182, 4028–4034 10.1128/JB.182.14.4028-4034.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salvarelli E., Krupka M., Rivas G., Mingorance J., Gómez-Puertas P., Alfonso C., and Rico A. I. (2015) The cell division protein FtsZ from Streptococcus pneumoniae exhibits a GTPase activity delay. J. Biol. Chem. 290, 25081–25089 10.1074/jbc.M115.650077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng Z., Zhang J., Xu D., Jiang Y. L., Zhou C. Z., and Chen Y. (2019) Multi-functional regulator MapZ controls both positioning and timing of FtsZ polymerization. Biochem. J. 476, 1433–1444 10.1042/BCJ20190138 [DOI] [PubMed] [Google Scholar]
- 37. Popp D., Iwasa M., Erickson H. P., Narita A., Maéda Y., and Robinson R. C. (2010) Suprastructures and dynamic properties of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 285, 11281–11289 10.1074/jbc.M109.084079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Popp D., Iwasa M., Narita A., Erickson H. P., and Maéda Y. (2009) FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91, 340–350 10.1002/bip.21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Honda D., Yokota A., and Sugiyama J. (1999) Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 48, 723–739 10.1007/PL00006517 [DOI] [PubMed] [Google Scholar]
- 40. Robertson B. R., Tezuka N., and Watanabe M. M. (2001) Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 51, 861–871 10.1099/00207713-51-3-861 [DOI] [PubMed] [Google Scholar]
- 41. Rocap G., Distel D. L., Waterbury J. B., and Chisholm S. W. (2002) Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68, 1180–1191 10.1128/AEM.68.3.1180-1191.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scanlan D. J., Ostrowski M., Mazard S., Dufresne A., Garczarek L., Hess W. R., Post A. F., Hagemann M., Paulsen I., and Partensky F. (2009) Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299 10.1128/MMBR.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kallas T., and Castenholz R. W. (1982) Internal pH and ATP-ADP pools in the cyanobacterium Synechococcus sp. during exposure to growth-inhibiting low pH. J. Bacteriol. 149, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corrales-Guerrero L., Camargo S., Valladares A., Picossi S., Luque I., Ochoa de Alda J. A. G., and Herrero A. (2018) FtsZ of filamentous, heterocyst-forming cyanobacteria has a conserved N-terminal peptide required for normal FtsZ polymerization and cell division. Front. Microbiol. 9, 2260 10.3389/fmicb.2018.02260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Erickson H. P., Anderson D. E., and Osawa M. (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 10.1128/MMBR.00021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Osawa M., and Erickson H. P. (2018) Turgor pressure and possible constriction mechanisms in bacterial division. Front. Microbiol. 9, 111 10.3389/fmicb.2018.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beuria T. K., Mullapudi S., Mileykovskaya E., Sadasivam M., Dowhan W., and Margolin W. (2009) Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J. Biol. Chem. 284, 14079–14086 10.1074/jbc.M808872200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Srinivasan R., Mishra M., Wu L., Yin Z., and Balasubramanian M. K. (2008) The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22, 1741–1746 10.1101/gad.1660908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holden S. (2018) Probing the mechanistic principles of bacterial cell division with super-resolution microscopy. Curr. Opin. Microbiol. 43, 84–91 10.1016/j.mib.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 50. Lu C., and Erickson H. P. (1999) The straight and curved conformation of FtsZ protofilaments-evidence for rapid exchange of GTP into the curved protofilament. Cell Struct. Funct. 24, 285–290 10.1247/csf.24.285 [DOI] [PubMed] [Google Scholar]
- 51. Romberg L., Simon M., and Erickson H. P. (2001) Polymerization of FtsZ, a bacterial homolog of tubulin: is assembly cooperative? J. Biol. Chem. 276, 11743–11753 10.1074/jbc.M009033200 [DOI] [PubMed] [Google Scholar]
- 52. Goley E. D., Dye N. A., Werner J. N., Gitai Z., and Shapiro L. (2010) Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol Cell 39, 975–987 10.1016/j.molcel.2010.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gueiros-Filho F. J., and Losick R. (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 10.1101/gad.1014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu C., Stricker J., and Erickson H. P. (1998) FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima: quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskeleton 40, 71–86 [DOI] [PubMed] [Google Scholar]
- 55. Huang H., Wang P., Bian L., Osawa M., Erickson H. P., and Chen Y. (2018) The cell division protein MinD from Pseudomonas aeruginosa dominates the assembly of the MinC-MinD copolymers. J. Biol. Chem. 293, 7786–7795 10.1074/jbc.RA117.001513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen Y., Milam S. L., and Erickson H. P. (2012) SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51, 3100–3109 10.1021/bi201669d [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.