Abstract
In Alzheimer's disease (AD) and tauopathies, tau aggregation accompanies progressive neurodegeneration. Aggregated tau appears to spread between adjacent neurons and adjacent brain regions by prion-like seeding. Hence, inhibitors of this seeding offer a possible route to managing tauopathies. Here, we report the 1.0 Å resolution micro-electron diffraction structure of an aggregation-prone segment of tau with the sequence SVQIVY, present in the cores of patient-derived fibrils from AD and tauopathies. This structure illuminates how distinct interfaces of the parent segment, containing the sequence VQIVYK, foster the formation of distinct structures. Peptide-based fibril-capping inhibitors designed to target the two VQIVYK interfaces blocked proteopathic seeding by patient-derived fibrils. These VQIVYK inhibitors add to a panel of tau-capping inhibitors that targets specific polymorphs of recombinant and patient-derived tau fibrils. Inhibition of seeding initiated by brain tissue extracts differed among donors with different tauopathies, suggesting that particular fibril polymorphs of tau are associated with certain tauopathies. Donors with progressive supranuclear palsy exhibited more variation in inhibitor sensitivity, suggesting that fibrils from these donors were more polymorphic and potentially vary within individual donor brains. Our results suggest that a subset of inhibitors from our panel could be specific for particular disease-associated polymorphs, whereas inhibitors that blocked seeding by extracts from all of the tauopathies tested could be used to broadly inhibit seeding by multiple disease-specific tau polymorphs. Moreover, we show that tau-capping inhibitors can be transiently expressed in HEK293 tau biosensor cells, indicating that nucleic acid–based vectors can be used for inhibitor delivery.
Keywords: amyloid, tau protein, tauopathy, prion, fibril, crystal structure, inhibitor, protein aggregation, seeding, protein structure, neurodegeneration, structural biology, seeding, zipper interface
Introduction
Tau pathology is a marker of neurodegeneration both in AD,2 which is accompanied by deposits of aggregated β-amyloid, and pure tauopathies, which lack aggregated β-amyloid (1, 2). The spreading of minimal units of tau pathology—aggregates of tau, termed seeds—converts inert, soluble tau monomers in recipient cells into pathological aggregates through a process called prion-like seeding, in principle connecting seeding to the progressive cascade of neurodegeneration that is characteristic of tauopathies and AD (2–5). The amyloid core of tau, like other amyloidogenic proteins, is formed by steric zippers with rich β character, tightly interdigitated side chains, and strong shape complementarity (6, 7). Amyloid structures are particularly stable relative to other protein folds owing to steric zipper interactions and an extensive network of hydrogen bonds that forms along the fibril axis (8, 9), potentially explaining their ability to spread transcellularly to distant, but anatomically connected, brain regions.
Often the aggregation of amyloid proteins results in a given sequence forming different folds—structural polymorphs—that are associated with different neurodegenerative diseases. For tau, this is exemplified by cryo-EM structures of patient-derived fibrils from three different neurodegenerative diseases: two polymorphs from AD (10, 11), referred to as PHFs and SFs, which are ultrastructural polymorphs of each other, two from chronic traumatic encephalopathy (CTE) (12) and two from Pick's disease (13). Given these observations, which are supported by biological studies (3, 5, 14, 15), it has been suggested that tau adopts disease-specific fibril polymorphs. It is not known whether a single inhibitor is capable of blocking seeding by all disease-associated polymorphs or whether disease-specific inhibitors must be made to match disease-associated polymorphs.
Crystal structures of steric zippers determined from fibrils formed by short, aggregation-prone peptide segments reveal molecular interfaces that are similar to their counterparts in full-length cryo-EM structures. For α-synuclein, crystal structures of the pre-NAC and NACore homomeric steric zippers match closely the distinct interprotomer interfaces found in the “rod” and “twister” full-length cryo-EM structural polymorphs, respectively (16, 17). These studies suggest that (i) crystal structures of amyloid segments recapitulate subsets of interactions that are seen in longer fibril cores, (ii) high-resolution crystal structures can serve as viable templates for inhibitor design of full-length amyloid fibrils, and (iii) crystal structures of segments can reflect the polymorphism that is observed in full-length cryo-EM structures.
The apparent contribution of seeding to the progressive spread of tau pathology, combined with mounting evidence of disease-specific tau polymorphs, underscores the need for a diverse panel of inhibitors, with each inhibitor of the panel targeting a different polymorph of aggregated tau. The rational design of small-molecule and antibody inhibitors of seeding has proven difficult. Instead, we developed peptide-based inhibitors of seeded aggregation that work by incorporating into preformed fibrils, and “capping” fibril ends to prevent additional growth with residues that sterically block the incorporation of new molecules (18). Thus, in principle, capping inhibitors halt fibril growth both by stopping fibril extension and also by sterically interfering with and disrupting steric zipper interactions (i.e. the adhesion of one β-sheet with its partner). Using this approach, we developed capping inhibitors from crystal structures of the steric zipper segments of tau with sequences 275VQIINK280 and 306VQIVYK311 (18, 19). We found that a capping inhibitor, TLKIVW, designed to target one polymorph of VQIVYK, was a poor inhibitor of seeding by recombinant fibrils, despite strongly inhibiting 3R tau. Capping inhibitors designed using structures of three different steric zipper polymorphs of VQIINK, on the other hand, strongly inhibited seeding by recombinant tau fibrils.
In the present study, we expand our panel of capping inhibitors by designing inhibitors based on structures of five different steric zipper interfaces that are formed by tau. Four inhibitors of the panel were designed based on structural polymorphs of VQIINK, which were determined previously (19), and we introduce four new inhibitors that are designed using the crystal structure of 305SVQIVY310, which was determined as part of this work. We benchmark the efficacy of our panel by testing inhibition of seeding by pathological fibrils from 11 different donors with tau pathology and find that several inhibitors of the panel block seeding by pathological tau fibrils. Furthermore, we demonstrate that fibrils from different tau pathologies can be classified by their differential sensitivities to inhibitors within our panel, consistent with the association of particular polymorphs with particular diseases.
Results
Structure determination of steric zipper polymorphs of VQIVYK from tau
To broaden the efficacy of our panel of inhibitors against a wide range of tau polymorphs, we sought to determine structures of additional aggregation-prone interfaces of VQIVYK. As outlined in Fig. 1, we scanned the tau sequence using ZipperDB (20) and found that a segment with sequence 305SVQIVY310 has a slightly greater predicted propensity to form a steric zipper compared with the aggregation-prone parent segment with sequence 306VQIVYK311 (Fig. 1A). To determine whether SVQIVY, shifted by −1 residue compared with VQIVYK, forms a different steric zipper, we grew nanocrystals of the SVQIVY peptide (Fig. 1B) and determined the micro-electron diffraction (micro-ED) structure by direct methods at a resolution of 1.0 Å (Fig. 1C, Fig. S1, and Table S1).
Figure 1.
Top, outline of workflow used to design and test inhibitors of tau seeding. Figure citations are of figures in this paper, and lozenges are colored to correspond to A–E in this figure. A, ZipperDB (20) prediction of the aggregation-driving peptide segments in tau identifies segments VQIINK and VQIVYK. Sequences with scores exceeding an empirical threshold of −23 kcal/mol on the vertical axis (colored red) report hexapeptide segments with energetically favorable steric zipper scores. B, nanocrystals of the SVQIVY peptide segment predicted from the ZipperDB plot in A to have greater steric zipper–forming propensity compared with the parent segment, VQIVYK. C, electron diffraction collected by micro-ED from representative SVQIVY nanocrystals shown in B. D, model of a fibril-capping inhibitor (magenta) designed to block elongation by binding to the tip of a fibril (gray). The example shown is of a VQIINK capping inhibitor, WMINK, designed to inhibit aggregation from three interfaces formed by different polymorphs of the VQIINK steric zipper, labeled interface A, B, and C (19). E, seeding inhibition assay, carried out by transfecting tau biosensor cells (32) with tau fibrils (recombinant or brain-derived, as indicated) that were pretreated with capping inhibitor.
As shown in Fig. 2A, the SVQIVY structure contains two different steric zipper interfaces that belong to two different symmetry classes. A Class 1 steric zipper is formed by two copies of the peptide that are related by crystallographic symmetry and is identical to the Class 1 steric zipper that is formed by the parent VQIVYK segment that was determined previously (Fig. S1C) (7). The opposite face of SVQIVY forms a second steric zipper that is a Class 3 interface created by two molecules in the asymmetric unit. The Class 3 interface is more interdigitated than the Class 1, having a shape complementarity (Sc) of 0.87 and solvent-accessible surface area buried (Ab) of 110.9 Å2, compared with 0.79 and 106.1 Å2, respectively, for the Class 1 zipper (Fig. S1D). Class 3 steric zippers have also been observed previously in cryo-EM structures of an 11-residue segment from TDP43 (21) and a heparin-induced fibril polymorph that is formed by 2N3R tau (22).
Figure 2.
Design of SVQIVYK-based capping inhibitors and classification of seeding inhibition on recombinant tau fibrils in tau biosensor cells. A, VQIVYK Class 3 interface formed by the left and center β-sheets colored green and cyan and Class 1 interface formed by the right and center β-sheets colored orange and cyan. Lysine 311, which is absent from the SVQIVY peptide, was modeled using coordinates from the previously determined VQIVYK crystal structure (Protein Data Bank entry 2ON9). Numbering along the backbone corresponds to residues modified in B to create different capping inhibitors that are listed in Table 1. B, yellow, a composite capping inhibitor that contains tryptophan substitutions at positions 2, 3, 4, and 5 (numbering corresponding to A) to show modeled steric clashes with the VQIVYK steric zipper structure, colored gray. Steric overlaps were mapped in PyMOL (23) using the show_bumps script. Green dots and disks show favorable van der Waals contacts, and red disks show steric clashes. Larger discs represent more severe clashes. C, seeding inhibition by VQIVYK-based capping inhibitors derived from peptides with tryptophan substitutions at the indicated positions (numbering corresponding to A). Seeding (expressed as 10−3 power) and inhibition were measured using recombinant fibrils of tau40 and HEK293 tau biosensor cells that stably express P301S 4R1N tau fused to YFP (32). Seeding inhibition was determined by counting the number of fluorescent puncta as a function of inhibitor concentration. IC50 values were calculated from dose–response plots, except for cases indicated as not determined (ND). Dashed magenta lines, 50% inhibition. Inhibitor concentrations that reduce seeding by >50% are colored yellow with magenta data points. D and E, representative images of seeded tau-4R1N cells from C with no inhibitor (D) or 50 μm VY-WIW (E). Representative tau-4R1N cells containing aggregated tau puncta are marked with red arrows, and cells without are marked with white arrows. F, seeding inhibition by VQIINK-based capping inhibitors, otherwise as in C. G, seeding in tau-K18–expressing biosensor cells that were first transiently transfected with vector encoding the VY-WIW capping inhibitor peptide or a scrambled peptide as a control. Error bars, S.D.
The SVQIVY structure offers a high-resolution template for the design of inhibitors to halt seeding by tau fibrils, given its close relationship to previously determined cryo-EM structures of patient-derived tau fibrils. The Class 1 steric zipper from the peptide crystal structure is formed by a homotypic steric zipper with a mated sheet of identical sequence, whereas the VQIVYK interface observed in patient-derived fibrils forms through heterotypic steric zipper interactions (Fig. S2). Nevertheless, the features of the Class 1 interface from the peptide crystal structure, mainly the zipper class, shape complementarity, and solvent-accessible surface area buried, are highly reminiscent of those found in cryo-EM fibril structures.
We suggest that the Class 3 interface determined from the SVQIVY structure similarly represents a potential aggregation surface that contributes to fibril stability and/or seeding, although the sequences of the mated strands in the fibrillar and crystalline states are also likely to differ. The Class 3 interface, formed by Ser305, Gln307, and Val309 of the SVQIVY crystal structure, maps to a partially solvent-exposed surface in patient-derived fibrils from AD (10, 11) (Fig. S3), CTE (12), and Pick's disease (13). Importantly, the Class 1 and 3 steric zipper interfaces are not mutually exclusive, as they are both observed simultaneously in the SVQIVY crystal structure. In fact, as shown in Fig. S3, a comparison of the SVQIVY crystal structure with AD-derived fibrils reveals that unmodeled electron density overlaps with the same interface that forms the Class 3 steric zipper in both the PHF and SF (10, 11), suggesting that similar interactions could form on the surfaces of these patient-derived fibrils. Therefore, we used the high-resolution crystal structures as guides to design capping inhibitors of the Class 1 and Class 3 VQIVYK steric zipper interfaces.
Design and testing of tau inhibitors based on the structure of SVQIVY
Class 1 and Class 3 VQIVYK-based capping inhibitors were designed from the atomic-resolution SVQIVY structure by modeling single tryptophan substitutions at each position along the SVQIVY peptide to determine at which sites substitution would most potently inhibit seeding. As shown in Fig. 2B, modeling suggested that substitutions at positions 3 and 5 would disrupt the Class 3 interface, whereas substitution at position 4 would disrupt the Class 1 interface. All of the VQIVYK-based inhibitors incorporated a lysine to glutamate charge reversal at position 7 to allow for the potential formation of an electrostatic interaction between the capping peptide and the native lysine (Lys311) in the tau VQIVYK segment (Fig. 2B). For clarity, VQIVYK-based capping inhibitors are labeled with a “VY” prefix, and VQIINK-based inhibitors are labeled with an “IN” prefix.
As a first test, we measured inhibition of seeding by recombinant fibrils of full-length tau (tau40) with each of the capping inhibitors listed in Table 1. Seeding and inhibition were measured using tau biosensor cells that express a nearly full-length construct of tau, 4R1N, labeled with YFP. As shown in Fig. 2C, the Class 3 interface inhibitor VY-WIV, which has a tryptophan substitution at position 3, modestly inhibits seeding with an IC50 of 9.3 μm. The other Class 3 inhibitor VY-QIW, which has a tryptophan substitution at position 3, also exhibits some concentration-dependent inhibition of seeding, but because inhibition of seeding was relatively weak, an IC50 could not be calculated. The Class 1 capping inhibitor VY-W4, which contains a tryptophan substitution at position 4, has no measurable inhibition of seeding by recombinant tau40 fibrils, even at 50 μm. Consistent with this finding, a d-peptide inhibitor, TLKIVW, which also targets the Class 1 VQIVYK interface and potently inhibited the primary aggregation of the 3R tau (18), likewise has no measurable inhibition of seeding by full-length tau fibrils at concentrations up to 50 μm (Fig. S4).
Table 1.
Amino acid sequences of tau VQIINK and VQIVYK capping inhibitors
The top row reports the residue position number for the referenced inhibitor peptide. The second row reports the native sequence from WT human tau40 from which the respective capping inhibitor peptides were derived, and subsequent rows are inhibitor peptide sequences tested in this paper. Residues modified from the WT sequence for each capping inhibitor are highlighted in yellow and listed in red font. VQIVYK inhibitors are labeled with a “VY” prefix, and VQIINK inhibitors are labeled with an “IN” prefix.
Because both Class 3 inhibitors exhibited some inhibition of seeding, we combined the two tryptophan substitutions at positions 3 and 5 from VY-WIV and VY-QIW into a single capping inhibitor peptide referred to as VY-WIW. The IC50 of VY-WIW determined using recombinant fibrils of tau40 improved to 3.4 μm (Fig. 2, C–E). In comparison, the series of capping inhibitors IN-M4, IN-W3, and IN-R9, which target interfaces A, B, and C of the VQIINK steric zipper (Fig. 2F and Ref. 19), and WMINK, which simultaneously targets interfaces A, B, and C, all inhibited seeding by recombinant tau fibrils with IC50 values of about 1 μm.
Because capping inhibitors are composed of l-peptides, we wondered whether they could be delivered to cells by transfecting DNA that encoded the inhibitor sequence. To test this, we transfected the VY-WIW Class 3 capping inhibitor peptide into tau biosensor cells, and 24 h after transfection, biosensor cells were seeded with recombinant fibrils of tau-K18+ (residues Gln244–Glu380), which contains the entire core observed in the AD PHF and SF cryo-EM fibril structures (10, 11), and additional residues that contribute to in vitro seeding (24). Seeding inhibition was measured for cells that were transfected with inhibitor or a scrambled peptide as a negative control. As shown in Fig. 2G, cells transfected with VY-WIW resisted seeding, whereas cells seeded with vehicle only or a scrambled peptide were robustly seeded. From the data shown in Fig. 2, we conclude that (i) capping peptides either added in trans or encoded by DNA and expressed in mammalian cells inhibit aggregation that is seeded by recombinant fibrils, and (ii) VQIVYK-based capping inhibitors targeting the Class 3 interface block seeding by recombinant tau fibrils, whereas inhibitors targeting the Class 1 interface do not.
Inhibition of seeding by AD and tauopathy brain extracts
Next, we tested our panel of inhibitors on post-mortem brain tissue extracts from donors with AD. Crude AD brain extract produced strong seeding in tau-K18–expressing biosensor cells, typically 3–6 days after being transfected (Fig. S5A). Virtually no seeding was observed in cells transfected with extract from the brain of a donor with no cognitive impairment (Fig. S5A). Although seeding by crude brain extract was strong in tau-K18–expressing biosensor cells, seeding by the same specimen in 4R1N-tau–expressing biosensor cells was poor (Fig. S5B). Therefore, in all of our subsequent experiments, we use tau-K18–expressing biosensor cells, except where otherwise indicated.
As shown in Fig. 3A, seeding by crude AD brain extract in tau biosensor cells was inhibited by several peptides from our panel when 50% seeding inhibition was used as an arbitrary minimum cutoff to define inhibitor efficacy. Of the inhibitors targeting VQIVYK, only VY-WIW blocked seeding. The VQIINK inhibitor WMINK failed to block seeding by the AD-derived brain extract, despite being one of our most potent inhibitors of recombinant tau fibrils (Fig. 2D and Ref. 19). On the other hand, the three VQIINK inhibitors IN-W3, IN-M4, and IN-R9, which each target a single interface of the three that are targeted by WMINK, all blocked seeding by crude AD brain extract. Inhibition of seeding by crude brain extracts from four additional AD donors produced similar, although not identical, profiles of inhibitor sensitivity (Fig. 3E and Fig. S6). In all cases, seeding was most strongly inhibited by VY-WIW and IN-M4. Other inhibitors that were effective but exhibited greater variation were IN-W3, IN-R9, and the VQIYVK-based inhibitor TLKIVW (Fig. S6).
Figure 3.
Inhibition of seeding by AD-derived brain extracts using the VQIVYK and VQIINK panel of capping inhibitors. A and B, seeding in tau-K18 biosensor cells 6 days after the addition of crude brain extract from AD donor 1 (AD1) (A), or 1 day after seeding with fibrils purified from AD1 (B). Seeding inhibition was measured by counting the number of fluorescent puncta as a function of inhibitor. C, negative-stain electron micrograph of fibrils used for seeding in B. D, representative images from seeding inhibition experiments in A and B (seeded and inhibition of crude brain extract and purified AD fibrils). Red arrows point to representative cells containing seeded tau aggregates, and white arrows point to cells lacking aggregated tau. E, comparison of seeding by crude brain extract from AD donor 2 (AD2) with overnight versus no inhibitor preincubation. The left plot colored gray on the left shows seeding in tau-K18 biosensor cells and inhibition following overnight inhibitor preincubation. The experiment plotted on the right was performed using exactly the same conditions, except overnight inhibitor preincubation. Dashed magenta line, 50% inhibition; dotted green line, 25% inhibition. Inhibitors that reduce seeding by >50% are colored yellow with magenta data points, and inhibitors that reduce seeding by >25% but <50% are colored green with magenta data points. Error bars, S.D.
The above data establish that both VQIINK- and VQIVYK-based capping inhibitors block seeding by extracts from AD brain, although clearly the profiles of inhibition differ from recombinant fibrils, because WMINK, a strong inhibitor of recombinant tau fibrils, poorly inhibited seeding by the AD brain extracts. The latter is consistent with structural studies that show that heparin-induced recombinant tau fibrils form mixtures of polymorphs that differ from pathological fibrils (22).
As a control to ensure that our capping inhibitors target aggregated tau and not some other hypothetical seeding-competent species or cofactor in crude brain extract, we purified PHFs and SFs from brain tissue of AD donor 1 (AD1) and compared seeding inhibition of the purified fibrils and the crude brain extract. When we compared seeding by purified fibrils 1 day post-transfection (PTF) and seeding by the crude brain extract 6 days PTF, the levels of seeding without inhibitor were comparable (about 5,000 versus 6,000 aggregates per well for the crude and purified extracts). Under these conditions, we observed a similar pattern of inhibition by our panel of capping inhibitors, with VY-WIW, IN-W3, IN-M4, and IN-R9 all strongly inhibiting seeding. At 5 days PTF, seeding by purified fibrils was an order of magnitude greater than seeding by the crude brain extract, and even under these conditions, the same capping inhibitors still blocked seeding with >95% efficacy (Fig. S7), which varied among the different inhibitors from about 50 to 90%. Inhibition of purified fibrils by VY-WIV also became apparent 5 days PTF, whereas inhibition by VY-WIV was not apparent 1 day PTF. We suspect that the greater efficacy of our inhibitors toward the purified ex vivo fibrils could be due to degradation of the capping peptide and/or binding to interfering cellular components in the crude brain extracts and that, as a result, inhibition of seeding was stronger, and more apparent for purified fibrils, allowing effects to be measured from inhibitors that are typically weaker, such as VY-WIV. These data suggest that although the pattern of seeding inhibition is similar for crude and purified extracts, it is not identical. Nevertheless, these data demonstrate that seeding by both types of pathological brain extracts is strongly inhibited by capping inhibitors from our panel.
To more closely mimic a condition of therapeutic treatment, we tested the ability of our panel of capping peptides to inhibit seeding by crude brain extract without a prior overnight preincubation of the fibril and the inhibitor. As shown in Fig. 3E, most of the capping inhibitors worked better with overnight preincubation. Only IN-W3 inhibited seeding both with and without preincubation. However, IN-M4 and VY-WIW, which both inhibited seeding by >50% with overnight preincubation, also inhibited seeding without preincubation, but with moderate efficacy, as indicated with a green fill color in Fig. 3E (right), which marks inhibitors that reduce seeding by 25–50% and fall between the plotted green dotted and magenta dashed lines, which mark 25 and 50% inhibition, respectively. TLKIVW and VY-QIW, which did not inhibit seeding with overnight preincubation, also moderately blocked seeding without preincubation.
Because we measured seeding with tau-K18 biosensor cells that lack residues 373–378, which form an intramolecular heterozipper interface in the AD filament (10, 11) that is the target of VY-W4, we sought to test seeding inhibition in biosensor cells that express a tau construct that contains these additional residues to see whether VY-W4 inhibition could be detected. Because concentrated AD fibrils did not seed tau-4R1N biosensor cells at a level that was high enough to quantify, we instead created a new biosensor cell line that expresses the WT tau fragment containing 244–380, which we call tau-K18+. Tau-K18+ extends the microtubule-binding fragment (tau-K18) to include residues 373–378. Although the tau-K18+ biosensor cells could not be seeded by crude brain extract, as shown in Fig. S6C, seeding could be induced by the addition of concentrated fibrils purified from AD donor 5 (AD5). The inhibitor profile measured with tau-K18 and tau-K18+ biosensor cells differed somewhat. Both biosensor cell lines revealed seeding inhibition by VY-WIW, IN-M4, and IN-R9. K18+ cells seeded using concentrated fibrils also showed inhibition by VY-WIV and moderate inhibition by VY-QIW and TLKIVW. The tau-K18 biosensor cells that were seeded with crude brain extract, also from AD5, showed similar trends, but inhibition did not exceed 25% for these inhibitors. The only trend that did not hold between the two cell lines was inhibition by IN-W3, which showed moderate efficacy in K18 cells but appeared to stimulate seeding in K18+ biosensor cells. From these data, we conclude that a lack of inhibition by VY-W4 does not result from the lack of residues 373–378 in the tau-K18–expressing biosensor cells, because seeding inhibition by VY-W4 was also not observed with tau-K18+–expressing cells.
Next, we tested the ability of our panel of inhibitors to distinguish between a recombinant fibril polymorph and recombinant fibrils seeded with AD purified seeds. Recombinant fibrils of tau-K19+ (residues Gln244–Glu349 of 3R tau), which contains the entire sequence that was observed in the electron density maps of patient-derived AD fibrils (10, 11), were formed in the absence of heparin, with or without purified AD fibrils as a seed. As shown in Fig. 4, the morphology of recombinant tau-K19+ fibrils differs from AD brain-derived PHF, with the former forming ribbon-like fibrils (Fig. 4A) and the latter forming helical fibrils (Fig. 4B). Tau-K19+ daughter fibrils that were seeded by PHFs more closely resemble the helical morphology of the PHF parent fibril (purple arrows), although ribbon-like fibrils (orange arrows) were still observed as a minor species (Fig. 4C). We measured the response of seeded recombinant tau-K19+ daughter fibrils to our panel of VQIVYK-based capping inhibitors and found that the pattern of inhibitor sensitivity closely resembled the pattern of inhibition measured for the AD brain–derived parent fibrils (Fig. 4D). One exception was sensitivity to VY-WIW, which was more pronounced for the seeded recombinant tau-K19+ daughter fibrils and closely mirrored the acute sensitivity to VY-WIW that was observed for recombinant fibrils. These data suggest that seeding can be used to transform recombinant fibrils to a polymorph that more closely resembles the AD brain-derived parent fibril and that inhibitor profiling can be used to discern different fibril polymorphs.
Figure 4.

Seeding produces fibrils of recombinant tau that have morphologies and inhibitor sensitivities that are similar to the AD brain–derived polymorph. A–C, electron micrographs of recombinant tau-K19+ fibrils (A), AD brain-derived fibrils (B), and recombinant tau-K19+ seeded by AD brain-derived fibrils (C). Orange arrows, representative ribbon-like fibrils characteristic of the recombinant fibril polymorph; purple arrows, fibrils with helical symmetry that are characteristic of the AD brain–derived PHF polymorph. D, sensitivity of AD-derived and recombinant tau-K19+ fibrils to a panel of VQIVYK-based inhibitors. Seeding was measured in tau-K18 biosensor cells and normalized for each fibril polymorph relative to seeding without inhibitor. Note that recombinant tau-K19+ fibrils seeded by an AD brain–derived specimen exhibit inhibitor sensitivities that are more similar to AD brain–derived fibrils than to the recombinant tau-K19+ fibril polymorph. Error bars, S.D.
Fibril polymorphs are thought to vary across different tau pathologies, so we asked whether the profile of sensitivity to our panel of inhibitors differed for donors with different tauopathies. To test this, we assayed seeding inhibition by our panel of inhibitors using crude brain extract from a donor with CTE (case 2 donor from Falcon et al. (12)). Like AD-derived extracts, the CTE brain extract was well-inhibited by the VQIINK inhibitors IN-W3 and IN-M4 (Fig. 5A). By contrast, CTE brain extract was not inhibited by IN-R9 or VY-WIW. Instead, seeding was inhibited also by VY-WIV and VY-W4, two VQIVYK inhibitors that did not inhibit seeding by extracts from AD donors.
Figure 5.
Inhibitor sensitivity of CTE-derived tau seeds. A–C, seeding by crude brain extract from the temporal cortex (A), recombinant tau-K18+ fibrils that were seeded with CTE-derived tau seeds (B), or recombinant tau-K18+ fibrils that were aggregated in the presence of heparin (C). Seeding inhibition measurements for the CTE-seeded recombinant fibril polymorph in B were carried out in tau biosensor cells after three sequential rounds of in vitro seeding. Seeding inhibition was measured by counting the number of fluorescent puncta as a function of inhibitor. In A–C, magenta arrows were used to mark capping inhibitors that were effective at blocking seeding by CTE-derived tau from crude brain extracts, green arrows mark inhibitors effective at blocking seeding by recombinant tau-K18 fibrils, and the blue arrow marking IN-W3 in B emphasizes that it is the only one of the inhibitors that blocks seeding by both the CTE-derived tau and recombinant tau fibrils. D–F, representative images from A, B, and C, respectively, showing seeding and inhibition in tau-K18 biosensor cells. Red arrows, representative cells containing seeded tau aggregates; white arrows, representative cells lacking aggregated tau. Error bars, S.D.
Because CTE is a relatively rare tauopathy, we sought to amplify the CTE polymorph of tau by in vitro seeding. We carried out three sequential rounds of in vitro seeding using recombinant tau-K18+ monomer and the sarkosyl-insoluble fraction from CTE brain extract. The seeding inhibition profiles of the resulting daughter fibrils exhibited a wide range of inhibitor sensitivities (Fig. 5B), resembling the inhibitor sensitivity profile of the crude CTE brain extract. However, unlike CTE brain extract and similar to recombinant fibrils seeded with AD extract, daughter fibrils exhibited acute sensitivity to VY-WIW. We found that heparin-induced fibrils of recombinant K18+ likewise exhibit matching sensitivity to VY-WIW (Fig. 5C), suggesting that daughter fibrils inherited the inhibitor sensitivities of both the parent and recombinant fibrils. Of note, heparin was not used for in vitro seeding to amplify the CTE-derived polymorph. Collectively, these data suggest that under in vitro conditions, seeding by both AD and CTE brain–derived fibrils produces heterogeneous mixtures of fibril polymorphs that predominantly have the same characteristics of the parent fibril but also contain some fractional population that bears the hallmarks of the recombinant fibril polymorph.
We next expanded our experiments to test inhibitor efficacies on extracts from four PSP donors and one CBD donor. Contrary to the seeding inhibition profile measured for CTE-derived tau, which was inhibited by nearly half of the capping peptides in our panel, inhibition of seeding by extract from the CBD donor was poor for all of the inhibitors except IN-M4 (Fig. S8). On the other hand, the inhibitor profiles observed for the different PSP donors more closely mirrored the four AD donors, with TLKIVW and WMINK showing no inhibition and VY-W4, in some cases, stimulating seeding (Fig. 6B). The VQIVYK-based inhibitors VY-WIV, VY-QIW, and VY-WIW, produced variable effects generally ranging from no inhibition to mild inhibition (Fig. 6A), whereas the VQIINK-based inhibitors IN-W3, IN-M4, and IN-R9 were stronger inhibitors and interestingly inhibited seeding with different efficacies for the different PSP donors tested. In particular, donors 1 and 3 shared similar profiles of inhibition despite deriving from two different brain regions and were particularly sensitive to inhibition by IN-M4 and VY-WIW, whereas little to no response was seen for IN-W3 and the other two VQIVYK inhibitors, VY-WIV and VY-QIW. By contrast, seeding by extract from donor 2 was potently inhibited by all of the VQIINK-based inhibitors (IN-W3, IN-M4, and IN-R9), but vulnerability to the VQIVYK-based inhibitor VY-WIW was less pronounced.
Figure 6.
Inhibitor profiling in biosensor cells seeded by brain extract from four different PSP donors. A, tissue sections from donors 1 and 2 were harvested from the midbrain and from the locus coeruleus for donor 3. Seeding inhibition was measured by counting the number of fluorescent puncta as a function of inhibitor. VQIINK inhibitors showing >70% inhibition are highlighted on bar graphs with a red outline. B, seeding by extracts from PSP donors 1, 2, and 3 after treatment with the capping peptide VY-W4. C, representative images showing seeding and inhibition in tau biosensor cells. Red arrows, representative cells containing seeded tau aggregates; white arrows, representative cells lacking aggregated tau. D and E, as in A, except tissue sections came from two different brain regions, the cerebellum (D) or frontal cortex (E), of a fourth PSP donor. F–H, representative images from D. I–K, representative images from E. Error bars, S.D.
Because PSP donors 1–3 appeared to show variable profiles of inhibitor sensitivities, we tested the susceptibility of a fourth PSP donor using tissue sections from two different brain regions, the frontal cortex and cerebellum, using a subset of the inhibitors including IN-W3, IN-M4, and IN-R9, which proved to be the greatest discriminators observed for donors 1–3. The inhibitor profile from the cerebellum of PSP donor 4 closely matched the profiles we observed for PSP donors 1 and 3, with strong inhibitor sensitivity limited mainly to the VQIINK-based inhibitor IN-M4 (Fig. 6, D and F–H). On the other hand, the inhibitor profile we observed from a tissue section from the frontal cortex of the same donor more closely matched the profile that we observed from PSP donor 2, with the VQIINK-based inhibitors IN-W3, IN-M4, and IN-R9 all showing strong inhibition (Fig. 6, E and I–K). Unlike donor 2, however, extract from the frontal cortex of donor 4 additionally exhibited a strong sensitivity to VY-WIV.
Collectively, our data suggest that the profiles of sensitivity to our panel of inhibitors may be indicative of different fibril polymorphs, or possibly mixtures of polymorphs, and that the sensitivity profiles differ among tauopathy donors. Moreover, these data show that different profiles of inhibitor sensitivity can be found not only among donors with the same tauopathy, but, in the case of PSP donor 4, even in different brain regions of the same donor.
Discussion
From high-resolution crystal structures and moderate-resolution cryo-EM structures of patient fibrils, we have identified structures of tau aggregation interfaces to use as templates for the design of inhibitors of seeding. We find that the tau segment VQIVYK forms steric zippers belonging to two different symmetry classes. The VQIVYK Class 3 interface is partially solvent-exposed and partially occluded by islands of unknown densities in cryo-EM structures of patient-derived fibrils (10–12). The Class 1 interface is buried in the cores of the same fibrils. Capping peptides that target the Class 3 interface blocked seeding by AD extracts, whereas inhibitors of the Class 1 interface poorly inhibited seeding. Although these data suggest that stronger inhibitors of seeding can be designed by targeting a surface of the fibril that is partially solvent-exposed, it is also possible that the Class 3 inhibitors can bind to the tips of fibrils that cannot be bound by Class 1 inhibitors, perhaps because of unexpected steric clashes that prevent binding by the Class 1 inhibitors. Alternatively, we speculate that the Class 3 interface could act as a scaffold to allow for nucleation and growth of new fibrils that are transient and dynamic, owing to the partially solvent-exposed environment, and observed islands of unknown densities in cryo-EM maps of patient-derived fibrils. In either case, we demonstrate that capping inhibitors targeting the Class 3 interface block seeding by pathological extracts from donors with AD and other tauopathies.
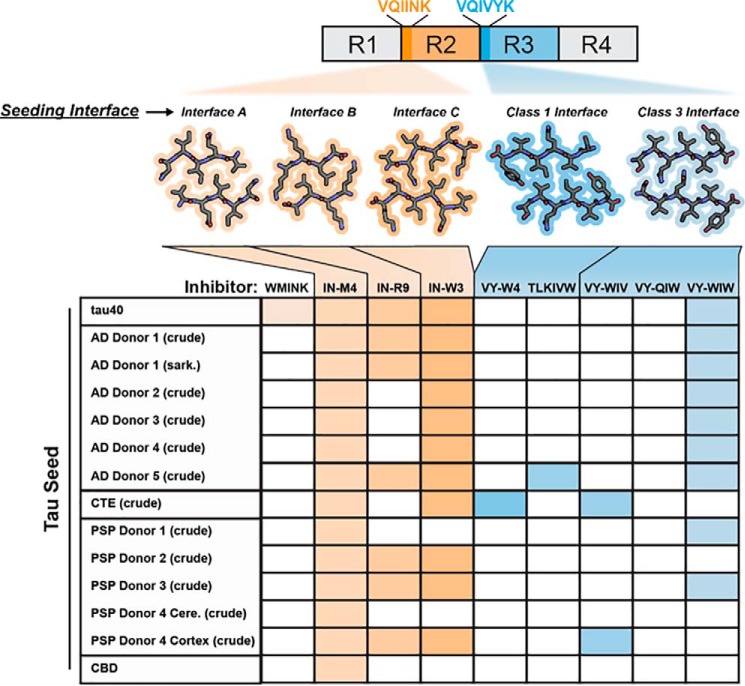
As summarized in Fig. 7, extracts from all of the autopsied diseased brains we tested were inhibited by VQIINK-based inhibitors. Thus, it is noteworthy that VQIINK localizes to the solvent-exposed fuzzy coat in structures of patient-derived fibrils. Given this observation, we speculate that VQIINK, although not resolved in the cryo-EM structure, plays a major role in seeding. In support of this, Woerman et al. (25) showed that seeding by AD and CTE extracts is most strongly transmitted in tau biosensor cells that co-express both the 3R and 4R tau isoforms. These data support the hypothesis that the 4R tau segment VQIINK plays a role in transducing seeding in pathological tissues. However, we note that the VQIVYK-based inhibitor VY-WIW is also effective at blocking seeding by AD extracts and fibrils, although epitopes in the fibril core were entirely inaccessible to antibody labeling (10, 11, 13). This apparent contradiction is explained by comparison of our SVQIVY structure with cryo-EM models of patient-derived fibrils, which show that the Class 3 interface targeted by VY-WIW also maps to a surface of the fibril that is partially solvent-exposed and is juxtaposed to the fuzzy coat.
Figure 7.
Top, locations of segments in tau targeted by different inhibitors of the panel and crystal structures of corresponding seeding interfaces. The filled boxes of the table below show efficacies of inhibitors for each donor tested in this study. For this analysis, inhibitors were scored as effective (filled box) if seeding was inhibited by 50% or more. Otherwise, inhibitors were scored as ineffective (open box).
In short, we suggest that the Class 1 interface, buried in the fibril core, is either inaccessible to Class 1 inhibitors from our panel, rendering these inhibitors ineffective, or is not as accessible to tau monomers in the cellular milieu and hence is not as effective as the Class 3 interface in seeding. On this basis, we propose that solvent-exposed aggregation-prone interfaces that decorate the outer perimeter of the fibril, including both VQIINK and the Class 3 interface of SVQIVY/VQIVYK, play a larger role in driving seeding than previously appreciated. Nevertheless, we do not dismiss the possibility that both contribute variably to seeding under different conditions that could depend on post-translational modifications, fibril size, and structure.
We find that the profile of sensitivity to our panel of inhibitors depends largely on the tauopathies of the donors, despite a shared sensitivity to IN-M4. The broad sensitivity to IN-M4 suggests that the aggregation-prone interface targeted by IN-M4 adopts a similar structure or environment in numerous tau fibril polymorphs. In support of this, it has been shown in cryo-EM studies of β2-microglobulin and α-synuclein amyloid fibrils that different polymorphs can share a common underlying subunit structure (26, 27). On the other hand, inhibitors from our panel that exhibit more variable inhibition could target other aggregation-prone interfaces that have variable accessibility in different fibrils owing to the structure of any given polymorph. Thus, these data suggest that the profiles of sensitivity to our panel of inhibitors could be diagnostic of particular disease-associated polymorphs.
The profiles of seeding inhibition using extracts from five AD donors reveal similar inhibitor sensitivity profiles, consistent with structural studies that suggest that the fibrillar polymorphs in AD could be limited to the PHF and SF. In contrast, we measured at least two different inhibitor sensitivity profiles from four different PSP donors, and in one case, we found distinct profiles from a single donor when two different brain regions were sampled. These data suggest that polymorphisms could occur among tauopathy donors and even within the brain of a given donor, although it is not clear how widespread this phenomenon is and whether it is limited to donors with specific tauopathies. To answer these questions, additional studies will focus on sampling larger numbers of patients and tauopathies and evaluating inhibitor sensitivity profiles from multiple brain regions of individual donors.
It is our hypothesis that the prion-like spread of aggregated tau drives progressive neurodegeneration, and inhibitors of seeding could thus slow or even halt disease progression. The delivery of peptides as therapeutics to the brain is liable to be challenging, and the half-lives of peptides could be short. As such, we wondered whether capping inhibitor peptides could be functionally synthesized by ribosomes following delivery to cells in a DNA vector. We found that, at least for the VY-WIW capping inhibitor peptide, transfection of plasmid DNA encoding the VY-WIW sequence into tau biosensor cells allowed cells to resist seeding with recombinant fibrils. These data suggest that capping inhibitors can be delivered to cells as nucleic acids.
The prion-like nature of amyloid aggregates is thought to perpetuate disease-specific polymorphs, and thus, it is thought that seeding produces daughter fibrils that are identical to parent fibrils. By comparing profiles of sensitivity to our panel of inhibitors for brain-derived and recombinant seeded fibrils, we found that daughter fibrils inherited specific inhibitor sensitivities that were characteristic of brain-derived fibrils. As a cautionary note, we found that daughter fibrils exhibited characteristics of the recombinant polymorph however, suggesting that in vitro seeding produces a heterogeneous mixture of fibrils and that the resulting mixture is not a perfect replicate of the tissue-derived parent polymorph. It is possible that some fibril polymorphs require mixtures of different tau isoforms to efficiently propagate (25), although in our hands, seeding mixtures of K18+ and K19+ (3R and 4R tau) did not dramatically improve the fidelity of seeding. Or, as recently proposed, amyloid formation may be steered by cellular co-factors and/or metabolites (28). In support of this, brain-derived CTE fibrils apparently enclose a hydrophobic molecule that may be necessary for faithful replication during in vitro seeding (12). Nevertheless, these data exemplify the diagnostic power of our inhibitor profiling approach by showing that the profile of inhibition of daughter fibrils is the product of its derivatives: the recombinant protein and tissue-derived fibril polymorph.
In summary, we show that the aggregation-prone segment SVQIVY forms two simultaneous steric zipper interfaces of different symmetry classes. Class 3 VQIVYK inhibitors, which target a surface that is partially solvent-exposed in patient-derived fibrils, and also VQIINK inhibitors block seeding by pathogenic tau with sensitivities that differ for extracts deriving from different tauopathies. In some cases, variations in inhibitor sensitivity were observed for donors diagnosed with the same tauopathy, and even in different brain regions of an individual donor, suggesting that fibril polymorphisms may occur in certain subsets of tauopathies. Further studies with larger sample sizes that encompass multiple brain regions will be needed to clarify how widespread the phenomenon of fibril polymorphism is among the different tauopathies. We suggest that the response of patient-derived seeds to our panel of inhibitors provides a “fingerprint” that is diagnostic of the fibrillar polymorphs it contains. This “fingerprint” might be useful for distinguishing tauopathies and detecting unique polymorphs that are associated with particular clinical characteristics.
Experimental procedures
Crystallization
305SVQIVY310 synthetic peptide was purchased from GenScript, and microcrystals were grown in batch at 3.3 mg/ml in 0.667 m dl-malic acid, pH 7.0, 8% (w/v) PEG 3350 at 18 °C.
Micro-ED data collection and processing
Crystal solution was applied to a glow-discharged Quantifoil R1/4 cryo-EM grid and plunge-frozen using a Vitrobot Mark 4. Micro-ED data were collected in a manner similar to previous studies (19). Briefly, plunge-frozen grids were transferred to an FEI Technai F20 electron microscope operating at 200 kV, and diffraction data were collected using a TVIPS F416 CMOS camera with a sensor size of 4,096 × 4,096 pixels, each 15.6 × 15.6 μm. Diffraction data were indexed using XDS, and XSCALE was used for merging and scaling together data sets from different crystals (20). Diffraction movies (numbers 1, 4, 6, 7, 8, and 9) were merged using XSCALE to produce the final data set.
Structure determination
The SHELXD macromolecular structure determination suite was used for phasing the measured intensities (21). A combination of the REFMAC, Phenix, and Buster refinement programs were used with electron scattering factors to refine the atomic coordinates determined by the direct-method protocol. Model building was performed in Coot.
Recombinant protein expression and purification
Human Tau tau-K18+ (residues Gln244–Glu380 of 4R tau) and tau-K19+ (residues Gln244–Glu349 of 3R tau) were expressed in a pNG2 vector in BL21-Gold E. coli cells grown in LB to an A600 = 0.8. Cells were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C and lysed by sonication in 20 mm MES buffer (pH 6.8) with 1 mm EDTA, 1 mm MgCl2, 1 mm DTT, and HALT protease inhibitor before the addition of NaCl (500 mm final concentration). Lysate was boiled for 20 min and then clarified by centrifugation at 15,000 rpm for 15 min and dialyzed to 20 mm MES buffer (pH 6.8) with 50 mm NaCl and 5 mm DTT. Dialyzed lysate was purified on a 5-ml HighTrap SP ion-exchange column and eluted over a gradient of NaCl from 50 to 550 mm. Protein was polished on a HiLoad 16/600 Superdex 75 pg column in 10 mm Tris (pH 7.6) with 100 mm NaCl and 1 mm DTT and concentrated to ∼20–60 mg/ml by ultrafiltration using a 3-kDa cutoff.
Human tau40 (residues 1–441) was expressed in pET28b with a C-terminal His tag in BL21-Gold E. coli cells grown in terrific broth to A600 = 0.8. Cells were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C and lysed by sonication in 50 mm Tris (pH 8.0) with 500 mm NaCl, 20 mm imidazole, 1 mm β-mercaptoethanol, and HALT protease inhibitor. Cells were lysed by sonication, clarified by centrifugation at 15,000 rpm for 15 min, and passed over a 5-ml HisTrap affinity column. The column was washed with lysis buffer and eluted over a gradient of imidazole from 20 to 300 mm. Fractions containing purified tau40 were dialyzed into 50 mm MES buffer (pH 6.0) with 50 mm NaCl and 1 mm β-mercaptoethanol and purified by cation exchange as described for K18. Peak fractions were polished on a HiLoad 16/600 Superdex 200 pg in 1× PBS (pH 7.4) and concentrated to ∼20–60 mg/ml by ultrafiltration using a 10-kDa cutoff.
Preparation of crude and purified brain-derived tau seeds
Tissue for neuropathlogically confirmed tauopathy cases from brain regions indicated in the figure legends were fresh-frozen and extracted without freeze-thaw. A discussion of the neuropathology of each case is provided in the supporting information. Tissue sections of 0.2–0.3g were excised on a block of dry ice and then manually homogenized in a 15-ml disposable tube in 1 ml of 50 mm Tris, pH 7.4, with 150 mm NaCl containing 1× HALT protease inhibitor. Samples were then aliquoted to PCR tubes and sonicated in a cup horn bath for 150 min under 30% power at 4 °C in a recirculating ice water bath, according to Ref. 29. For purification of PHFs and SFs from AD brain tissue, extractions were performed according to the previously published protocol (11) without any modifications.
Inhibitor peptides
Inhibitor peptides were designed using the native crystal structure as a starting point. Bulky side chains were modeled at sites in the VQIINK structure that were in close contact with residues in the mated sheet of the steric zipper interface. Capping residues were chosen by modeling all possible rotamers to find side chains without any compatible conformer with the steric zipper interface (i.e. side chains that clashed with the mated β-sheet at every rotamer conformer were selected). All of the inhibitor peptides shown in Table 1 were synthesized by GenScript with minimum purities of 90% and dissolved in deionized water or DMSO to a working concentration of 1.4 mm.
Tau-K18+ biosensor cells
Residues 244–380 of WT tau (referred to as tau-K18+) were fused with YFP through a linker with sequence WDPPVAT. DNA encoding tau-K18+ with a C-terminal YFP fusion was cloned into a PiggyBac vector and subsequently co-transfected into HEK293 cells with PiggyBac transposase according to the manufacturer's instructions (Systems Biosciences). Cells were selected by the addition of puromycin at a concentration of 10 μg/ml and were sorted based on FACS based on low, medium, or high YFP signal. After two rounds of puromycin selection and cell sorting, tau-K18+(YFP) cells with medium-level expression were used for experiments in this study because cells with high levels of tau-K18+ expression exhibited low background levels of spontaneous, unseeded aggregation.
Seeding in tau biosensor cells
HEK293 cell lines stably expressing tau-K18 or tau-4R1N P301S-eYFP were engineered by Marc Diamond's laboratory at the University of Texas Southwestern Medical Center (5) and used without further characterization or authentication. Tau-K18+ biosensor cells were prepared as described above. Cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Inc., catalog no. 11965092) supplemented with 10% (v/v) FBS (Life Technologies, catalog no. A3160401), 1% antibiotic-antimycotic (Life Technologies, Inc., catalog no. 15240062), and 1% Glutamax (Life Technologies, catalog no. 35050061) at 37 °C, 5% CO2 in a humidified incubator. Fibrils and patient-derived seeds were incubated for 16 h with inhibitor to yield a final inhibitor concentration of 10 μm (on the biosensor cells), except for IC50 determinations, which instead used adjustments to achieve the final indicated inhibitor concentration. For seeding, inhibitor-treated seeds were sonicated in a cup horn water bath for 3 min and then mixed with 1 volume of Lipofectamine 3000 (Life Technologies, catalog no. 11668027) prepared by diluting 1 μl of Lipofectamine in 19 μl of OptiMEM. After 20 min, 10 μl of fibrils were added to 90 μl of tau biosensor cells. The number of seeded aggregates was determined by imaging the entire well of a 96-well plate in triplicate using a Celigo image cytometer (Nexcelom) in the YFP channel. Aggregates were counted using ImageJ (30) by subtracting the background fluorescence from unseeded cells and then counting the number of peaks with fluorescence above background using the built-in particle analyzer. The number of aggregates was normalized to the confluence of each well, and dose–response plots were generated by calculating the average and S.D. values from triplicate measurements. For IC50 calculations, dose–response curves were fit by nonlinear regression in GraphPad Prism. For high-quality images, cells were photographed on a ZEISS Axio Observer D1 fluorescence microscope using the YFP fluorescence channel.
In vitro seeding
For seeding with brain-derived fibrils, seeding was carried out essentially as described by Morosova et al. (31). Briefly, either tau-K18+, tau-K19+, or a 50% mixture of both was boiled for 5 min in the presence of 100 mm 2-mercaptoethanol in the final buffer that was used for purification. Subsequently, tau monomer was diluted to 350 μm in 1× PBS (pH 7.4). Seeding was carried out by the addition of AD brain–derived fibrils that were sonicated in a cup horn water bath for 3 min at 20% power at an arbitrary concentration of 2% (v/v), and aggregation was induced by shaking at 350 rpm at 37 °C for 18 h.
Author contributions
P. M. S. and K. A. M. conceptualization; P. M. S., D. R. B., K. A. M., and T. P. Y. data curation; P. M. S., D. R. B., K. A. M., M. R. S., D. C., and C. K. W. formal analysis; P. M. S. and D. S. E. funding acquisition; P. M. S. and M. B. investigation; P. M. S. methodology; P. M. S. and D. R. B. writing-original draft; P. M. S. project administration; P. M. S., D. R. B., B. G., D. W. D., and D. S. E. writing-review and editing; G. R., C. K. W., K. L. N., B. G., M. A. D., D. W. D., H. V. V., and D. S. E. resources; D. S. E. supervision.
Supplementary Material
Acknowledgments
We thank Hilda Mirbaha for advice on conducting tau biosensor seeding experiments and Nicole Wheatley for establishing tau-K18+ biosensor cells. We also thank Tamir Gonen and Dan Shi for training and use of the FEI Technai F20 electron microscope for micro-ED data collection. Donation of CTE tissue was supported by Massachusetts Alzheimer Disease Research Center Grant P50 AG005134.
This work was supported by NIA, National Institutes of Health (NIH), Grants 1R01 AG061847 (to D. S. E.), RF1 AG054022 (to D. S. E.), and PHS P30-AG010133 (to B. G.); NINDS, NIH, Grant 1F32 NS095661 (to P. M. S.); BrightFocus Foundation Grant A2016588F (to P. M. S.); United States Public Health Service National Research Service Award 5T32GM008496 (to K. A. M.); the University of California-Caltech Medical Scientist Training Program (to K. A. M.); the National Science Foundation Graduate Research Fellowships Program (to D. R. B.); and the Howard Hughes Medical Institute. D. S. E. is SAB chair and equity holder of ADRx, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S8.
The atomic coordinates and structure factors (code 6ODG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AD
- Alzheimer's disease
- PHF
- paired helical filament
- SF
- straight filament
- CTE
- chronic traumatic encephalopathy
- micro-ED
- micro-electron diffraction
- YFP
- yellow fluorescent protein
- PTF
- post-transfection
- PSP
- progressive supranuclear palsy
- CBD
- corticobasal degeneration.
References
- 1. Goedert M., Wischik C. M., Crowther R. A., Walker J. E., and Klug A. (1988) Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc. Natl. Acad. Sci. U.S.A. 85, 4051–4055 10.1073/pnas.85.11.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mudher A., Colin M., Dujardin S., Medina M., Dewachter I., Alavi Naini S. M., Mandelkow E.-M., Mandelkow E., Buée L., Goedert M., and Brion J.-P. (2017) What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathol. Commun. 5, 99–99 10.1186/s40478-017-0488-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clavaguera F., Akatsu H., Fraser G., Crowther R. A., Frank S., Hench J., Probst A., Winkler D. T., Reichwald J., Staufenbiel M., Ghetti B., Goedert M., and Tolnay M. (2013) Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 110, 9535–9540 10.1073/pnas.1301175110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harper J. D., and Lansbury P. T. Jr. (1997) Models of amyloid seeding in Alzheimer's disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66, 385–407 10.1146/annurev.biochem.66.1.385 [DOI] [PubMed] [Google Scholar]
- 5. Sanders D. W., Kaufman S. K., DeVos S. L., Sharma A. M., Mirbaha H., Li A., Barker S. J., Foley A. C., Thorpe J. R., Serpell L. C., Miller T. M., Grinberg L. T., Seeley W. W., and Diamond M. I. (2014) Distinct Tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 10.1016/j.neuron.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., and Eisenberg D. (2005) Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 10.1038/nature03680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J. W., McFarlane H. T., Madsen A. Ø., Riekel C., and Eisenberg D. (2007) Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 10.1038/nature05695 [DOI] [PubMed] [Google Scholar]
- 8. Eisenberg D., and Jucker M. (2012) The amyloid state of proteins in human diseases. Cell 148, 1188–1203 10.1016/j.cell.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiltzius J. J. W., Landau M., Nelson R., Sawaya M. R., Apostol M. I., Goldschmidt L., Soriaga A. B., Cascio D., Rajashankar K., and Eisenberg D. (2009) Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 10.1038/nsmb.1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falcon B., Zhang W., Schweighauser M., Murzin A. G., Vidal R., Garringer H. J., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold. Acta Neuropathol. 136, 699–708 10.1007/s00401-018-1914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. W. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Falcon B., Zivanov J., Zhang W., Murzin A. G., Garringer H. J., Vidal R., Crowther R. A., Newell K. L., Ghetti B., Goedert M., and Scheres S. H. W. (2019) Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 10.1038/s41586-019-1026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falcon B., Zhang W., Murzin A. G., Murshudov G., Garringer H. J., Vidal R., Crowther R. A., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Structures of filaments from Pick's disease reveal a novel tau protein fold. Nature 561, 137–140 10.1038/s41586-018-0454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung D. C., Carlomagno Y., Cook C. N., Jansen-West K., Daughrity L., Lewis-Tuffin L. J., Castanedes-Casey M., DeTure M., Dickson D. W., and Petrucelli L. (2019) Tau exhibits unique seeding properties in globular glial tauopathy. Acta Neuropathol. Commun. 7, 36 10.1186/s40478-019-0691-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibbons G. S., Banks R. A., Kim B., Changolkar L., Riddle D. M., Leight S. N., Irwin D. J., Trojanowski J. Q., and Lee V. M. Y. (2018) Detection of Alzheimer disease (AD)-specific Tau pathology in AD and NonAD tauopathies by immunohistochemistry with novel conformation-selective Tau antibodies. J. Neuropathol. Exp. Neurol. 77, 216–228 10.1093/jnen/nly010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li B., Ge P., Murray K. A., Sheth P., Zhang M., Nair G., Sawaya M. R., Shin W. S., Boyer D. R., Ye S., Eisenberg D. S., Zhou Z. H., and Jiang L. (2018) Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609–3609 10.1038/s41467-018-05971-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez J. A., Ivanova M. I., Sawaya M. R., Cascio D., Reyes F. E., Shi D., Sangwan S., Guenther E. L., Johnson L. M., Zhang M., Jiang L., Arbing M. A., Nannenga B. L., Hattne J., Whitelegge J., Brewster A. S., Messerschmidt M., Boutet S., Sauter N. K., Gonen T., and Eisenberg D. S. (2015) Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 10.1038/nature15368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sievers S. A., Karanicolas J., Chang H. W., Zhao A., Jiang L., Zirafi O., Stevens J. T., Münch J., Baker D., and Eisenberg D. (2011) Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100 10.1038/nature10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seidler P. M., Boyer D. R., Rodriguez J. A., Sawaya M. R., Cascio D., Murray K., Gonen T., and Eisenberg D. S. (2018) Structure-based inhibitors of tau aggregation. Nat. Chem. 10, 170–176 10.1038/nchem.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldschmidt L., Teng P. K., Riek R., and Eisenberg D. (2010) Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 107, 3487–3492 10.1073/pnas.0915166107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guenther E. L., Cao Q., Trinh H., Lu J., Sawaya M. R., Cascio D., Boyer D. R., Rodriguez J. A., Hughes M. P., and Eisenberg D. S. (2018) Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471 10.1038/s41594-018-0064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang W., Falcon B., Murzin A. G., Fan J., Crowther R. A., Goedert M., and Scheres S. H. (2019) Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. eLife 8, e43584 10.7554/eLife.43584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 24. Weismiller H. A., Murphy R., Wei G., Ma B., Nussinov R., and Margittai M. (2018) Structural disorder in four-repeat Tau fibrils reveals a new mechanism for barriers to cross-seeding of Tau isoforms. J. Biol. Chem. 293, 17336–17348 10.1074/jbc.RA118.005316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woerman A. L., Aoyagi A., Patel S., Kazmi S. A., Lobach I., Grinberg L. T., McKee A. C., Seeley W. W., Olson S. H., and Prusiner S. B. (2016) Tau prions from Alzheimer's disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 113, E8187–E8196 10.1073/pnas.1616344113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iadanza M. G., Silvers R., Boardman J., Smith H. I., Karamanos T. K., Debelouchina G. T., Su Y., Griffin R. G., Ranson N. A., and Radford S. E. (2018) The structure of a β2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Commun. 9, 4517 10.1038/s41467-018-06761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guerrero-Ferreira R., Taylor N. M. I., Arteni A.-A., Kumari P., Mona D., Ringler P., Britschgi M., Lauer M. E., Verasdock J., Riek R., Melki R., Meier B. H., Böckmann A., Bousset L., and Stahlberg H. (2019) Two new polymorphic structures of α-synuclein solved by cryo-electron microscopy. bioRxiv 10.1101/654582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sade D., Shaham-Niv S., Arnon Z. A., Tavassoly O., and Gazit E. (2018) Seeding of proteins into amyloid structures by metabolite assemblies may clarify certain unexplained epidemiological associations. Open Biol. 8, 170229 10.1098/rsob.170229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaufman S. K., Del Tredici K., Thomas T. L., Braak H., and Diamond M. I. (2018) Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer's disease and PART. Acta Neuropathol. 136, 57–67 10.1007/s00401-018-1855-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morozova O. A., March Z. M., Robinson A. S., and Colby D. W. (2013) Conformational features of tau fibrils from Alzheimer's disease brain are faithfully propagated by unmodified recombinant protein. Biochemistry 52, 6960–6967 10.1021/bi400866w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holmes B. B., Furman J. L., Mahan T. E., Yamasaki T. R., Mirbaha H., Eades W. C., Belaygorod L., Cairns N. J., Holtzman D. M., and Diamond M. I. (2014) Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. U.S.A. 111, E4376–E4385 10.1073/pnas.1411649111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.