Abstract
Su(var)3–9, Enhancer-of-zeste, and Trithorax (SET) domain-containing protein 8 (SET8) is the sole enzyme that monomethylates Lys-20 of histone H4 (H4K20). SET8 has been implicated in the regulation of multiple biological processes, such as gene transcription, the cell cycle, and senescence. SET8 quickly undergoes ubiquitination and degradation by several E3 ubiquitin ligases; however, the enzyme that deubiquitinates SET8 has not yet been identified. Here we demonstrated that ubiquitin-specific peptidase 17–like family member (USP17) deubiquitinates and therefore stabilizes the SET8 protein. We observed that USP17 interacts with SET8 and removes polyubiquitin chains from SET8. USP17 knockdown not only decreased SET8 protein levels and H4K20 monomethylation but also increased the levels of the cyclin-dependent kinase inhibitor p21. As a consequence, USP17 knockdown suppressed cell proliferation. We noted that USP17 was down-regulated in replicative senescence and that USP17 inhibition alone was sufficient to trigger cellular senescence. These results reveal a regulatory mechanism whereby USP17 prevents cellular senescence by removing ubiquitin marks from and stabilizing SET8 and transcriptionally repressing p21.
Keywords: cell cycle, deubiquitylation (deubiquitination), histone methylation, posttranslational modification (PTM), senescence, p21, SET8, USP17
Introduction
Su(var)3–9, Enhancer-of-zeste, and Trithorax (SET)5 domain- containing protein 8 (SET8) (also known as SETD8, PR-Set7, and KMT5A) is the sole enzyme required to catalyze monomethylation of histone H4 lysine 20 (H4K20me1) (1–3). Besides H4K20me1, SET8 can methylate nonhistone proteins, including p53, proliferating cell nuclear antigen, Numb, and androgen receptor and estrogen receptor α (4). SET8 is involved in vital cellular processes such as DNA replication, mitosis, DNA repair, and gene transcriptional regulation (5, 6). Therefore, precise modulation of SET8 levels is important for proper cell cycle regulation, and deregulation of SET8 expression has been suggested to cause cellular transformation and contribute to cancer progression (7). Aberrant expression of SET8 has been detected in many types of tumors (8–11). High levels of SET8 are also associated with poor survival in cancer patients (9–11). Moreover, recent studies have shown that SET8 prevents cellular senescence through epigenetic regulation (12, 13). Loss of SET8 is sufficient to establish cellular senescence, and H4K20me1 modification of the cyclin-dependent kinase inhibitor p21 gene during cellular senescence is regulated by SET8 (12, 13). Because knockdown of p21 alleviates the senescence state of SET8 knockdown cells, SET8 suppresses induction of cellular senescence by repressing p21 transcription (13).
SET8 is regulated at several levels, including the transcriptional level (14), posttranscriptional level (15), and posttranslational level (7). Some E3 ubiquitin ligases have been shown to induce SET8 ubiquitination and degradation, which regulate cell cycle progression (7). The anaphase-promoting complex APC/CCdh1 induces ubiquitination and degradation of SET8 during G1 phase (16). In addition, Cullin-RING ubiquitin ligase 4Cdt2 (CRL4Cdt2) and Skp1–Cullin-1–F-box protein (SCF)–Skp2 (SCFSkp2) mediate the degradation of SET8 in S phase (17–20). SCFβ-TRCP also promotes cell growth by targeting SET8 for degradation (21). Thus, the ubiquitination machinery plays an important role in regulating SET8 protein turnover and its activity.
On the other hand, ubiquitination is a reversible reaction, and ubiquitin is removed by deubiquitinases (DUB). DUBs are classified as ubiquitin C-terminal hydrolases, Mpr1, Pad1 N-terminal (MPN) domain–containing metalloenzymes, ubiquitin-specific processing proteases (USPs), ovarian tumor (OTU) domain ubiquitin-aldehyde–binding proteins, and the motif interacting with the Ub-containing novel DUB family (22, 23). DUBs control the stability and activity of multiple proteins that are crucial for cellular proliferation and survival, including p53, Mdm2, c-Myc, and histones (24). However, the mechanisms by which SET8 is deubiquitinated and stabilized remain unclear. Here we report that USP17 is a novel SET8 deubiquitinase. Overexpression of WT USP17, but not its catalytically inactive mutant (C89S), stabilized SET8. USP17 interacted with SET8 and removed polyubiquitin chains from SET8. Furthermore, we found that knockdown of USP17 not only decreased SET8 protein levels and H4K20me1 but also increased p21 levels. As a result, knockdown of USP17 suppressed cell proliferation. USP17 was down-regulated in replicative senescence, and inhibition of USP17 alone was sufficient to trigger cellular senescence. These results reveal a regulatory mechanism whereby USP17 removes ubiquitin marks to prevent cellular senescence, stabilizing SET8 and repressing p21.
Results
USP17 stabilizes the SET8 protein
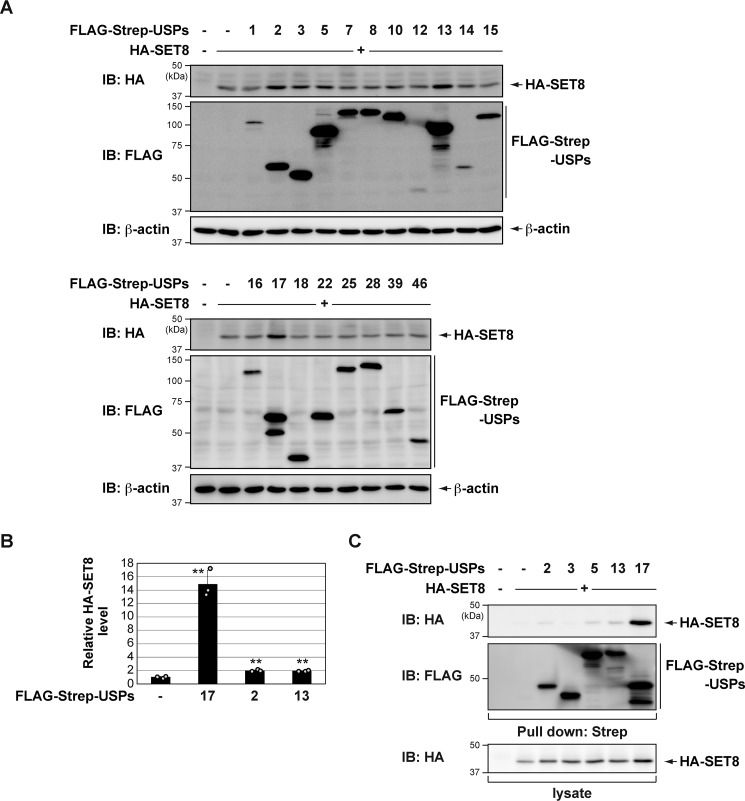
To identify the DUBs that stabilize the SET8 protein, we expressed HA-SET8 and several DUBs in COS7 cells. Cell lysates were subjected to immunoblotting using anti-HA antibodies. We found that USP17 significantly increased SET8 protein levels (Fig. 1, A and B, and Fig. S1). USP17 robustly interacted with SET8 (Fig. 1C). Thus, we focused on USP17 in subsequent functional studies.
Figure 1.
Identification of USP17 as a novel deubiquitinase of SET8. A, COS7 cells were transiently transfected with HA-SET8 in the presence of FLAG-Strep-USPs. After 24 h, cell lysates were immunoblotted (IB) with the indicated antibodies. B, quantitative analysis of the HA-SET8 protein bands shown in Fig. S1. Data in the bar graph are the means and standard deviations (n = 3). **, p < 0.01. C, COS7 cells were transiently transfected with the indicated constructs. After 24 h, cell lysates were subjected to Strep-Tactin pulldown and immunoblotting with an anti-HA antibody.
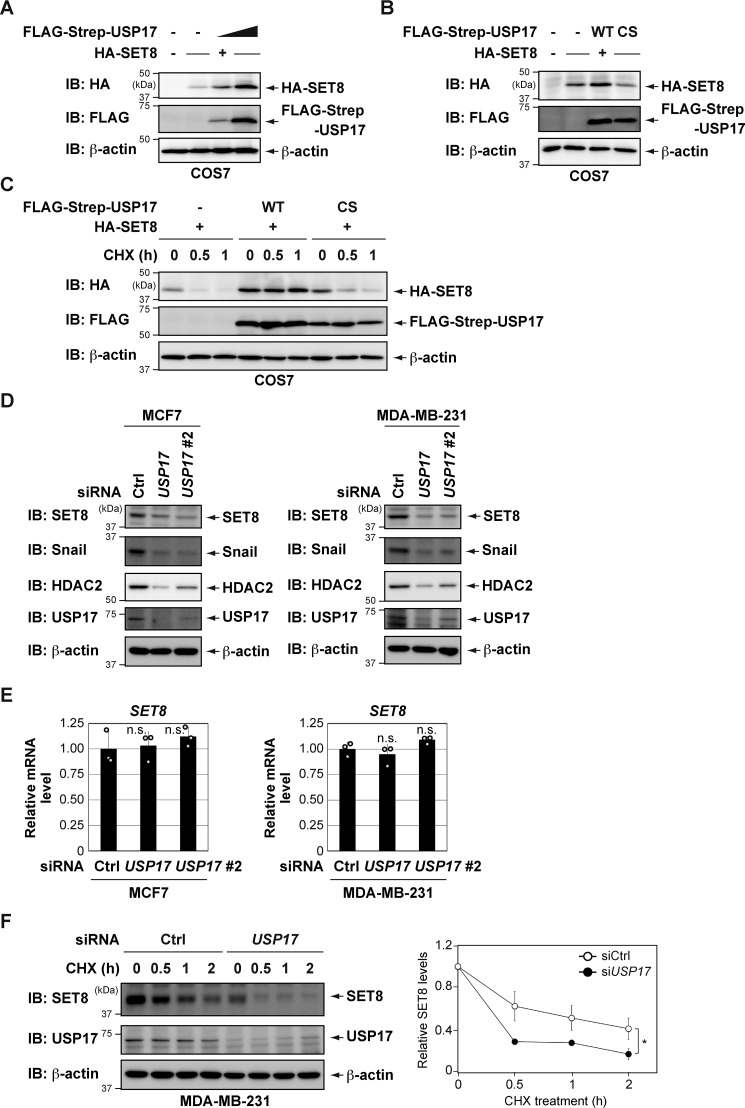
Overexpression of USP17 led to increased SET8 protein levels in a dose-dependent manner (Fig. 2A). However, expression of the mutant USP17, in which the catalytic cysteine was replaced by serine (C89S) (25), failed to stabilize the SET8 protein (Fig. 2B). Overexpression of WT USP17, but not the C89S mutant, markedly prolonged the half-life of the SET8 protein (Fig. 2C). These results suggest that the enzymatic activity of USP17 is required for SET8 protein stabilization.
Figure 2.
USP17 regulates the stability of SET8 protein. A, HA-SET8 was coexpressed with increasing amounts of FLAG-Strep-USP17 in COS7 cells. After 24 h, cell lysates were immunoblotted (IB) with the indicated antibodies. B, HA-SET8 was coexpressed with FLAG-Strep-USP17 (either the WT or the catalytically inactive mutant (C89S)) in COS7 cells. After 24 h, cell lysates were immunoblotted with the indicated antibodies. CS, C89S. C, USP17 (WT), but not USP17 (C89S), delayed SET8 protein turnover. COS7 cells were transiently transfected with the indicated constructs. After 24 h, cells were treated with 25 μg/ml CHX for the indicated periods and harvested for immunoblotting with the indicated antibodies. D, depletion of USP17 decreases SET8 protein levels. Cells were transiently transfected with the indicated siRNAs. After 72 h, cell lysates were immunoblotted with the indicated antibodies. Ctrl, control. E, cells were transiently transfected with the indicated siRNAs for 72 h. The expression of each gene was assessed by qPCR. The expression level of SET8 was normalized to that of β-actin mRNA. Results are shown as mean ± S.D. (n = 3). F, time course of CHX treatment of MDA-MB-231 cells transiently transfected with the indicated siRNAs. The protein stability of SET8 was analyzed at the indicated times by immunoblot analysis (left panel). Quantification of the SET8 protein level was normalized to β-actin. Data represent the means and S.D. of three independent experiments (right panel). *, p < 0.05; n.s., not significant.
We then knocked down endogenous USP17 expression in several cancer cell lines and examined the effects on the endogenous SET8 protein. As shown in Fig. 2, D and E, and Fig. S2, depletion of USP17 decreased SET8 protein levels without affecting SET8 mRNA levels. Other known USP17 substrates (Snail and HDAC2) (26–28) were also reduced by USP17 knockdown (Fig. 2D). Furthermore, SET8 protein was less stable in MDA-MB-231 cells depleted of USP17 in a CHX half-life assay (Fig. 2F). Collectively, these results suggest that USP17 plays a role in maintaining the abundance of the SET8 protein.
USP17 deubiquitinates SET8
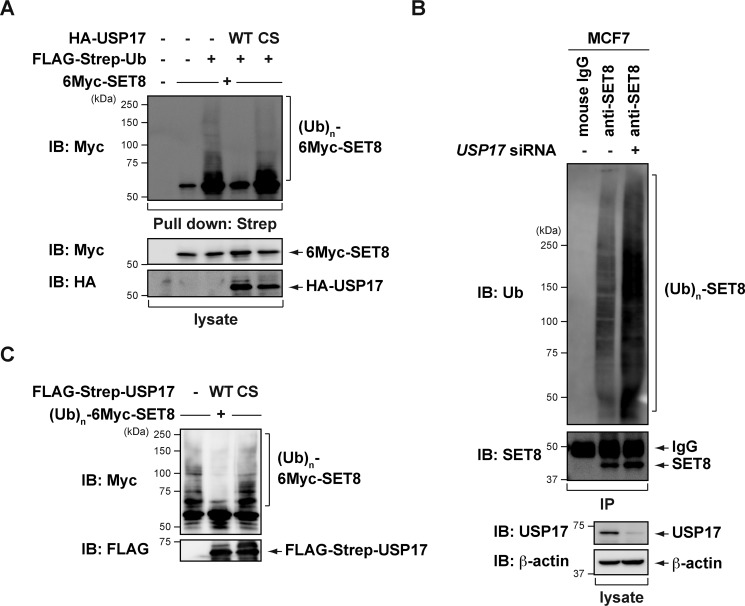
We then examined whether USP17 regulates SET8 protein stability through deubiquitination. We coexpressed 6Myc-SET8 and FLAG-Strep-ubiquitin (Ub) with the WT or C89S mutant of USP17 in COS7 cells. After precipitating ubiquitin-conjugating proteins from cells treated with the proteasome inhibitor MG132, we found that SET8 was heavily ubiquitinated, as reported previously (16–21). However, coexpression of WT USP17, but not the C89S mutant, significantly reduced the ubiquitinated species of SET8 (Fig. 3A). Conversely, SET8 ubiquitination increased in USP17-knockdown MCF7 cells (Fig. 3B). To clarify whether USP17 directly deubiquitinates SET8 in vitro, we purified ubiquitinated SET8 proteins from COS7 cells transfected with 6Myc-SET8 and Strep-Ub using affinity purification with Strep-Tactin–Sepharose and desthiobiotin elution. We incubated ubiquitinated SET8 proteins with purified USP17 (WT or C89S mutant) or control buffer. As shown in Fig. 3C, WT USP17, but not the C89S mutant, specifically reduced the ubiquitinated species of SET8. These results indicate that USP17 stabilizes SET8 by directly removing its ubiquitination.
Figure 3.
USP17 mediates SET8 deubiquitination. A, an empty vector or a plasmid expressing USP17 WT or C89S was coexpressed with 6Myc-SET8 and a FLAG-Strep-Ub expression plasmid into COS7 cells. Cells were treated with 20 μm MG132, and cell lysates were subjected to Strep-Tactin pulldown and immunoblotting (IB) with the indicated antibodies. CS, C89S. B, MCF7 cells transfected with control or USP17 siRNA were treated with 10 μm MG132 for 6 h. Cell lysates were subjected to immunoprecipitation (IP) with anti-SET8 antibody, and the polyubiquitination of SET8 was assessed by immunoblotting using an anti-ubiquitin antibody. C, USP17 deubiquitinates SET8 in vitro. Ubiquitinated SET8 was purified from MG132-treated COS7 cells expressing 6Myc-SET8 and FLAG-Strep-Ub and then incubated with purified USP17 WT or C89S in a deubiquitination assay as described under “Experimental procedures.” The polyubiquitinated state of SET8 was examined by immunoblotting with an anti-Myc antibody.
Mapping of binding regions between USP17 and SET8 proteins
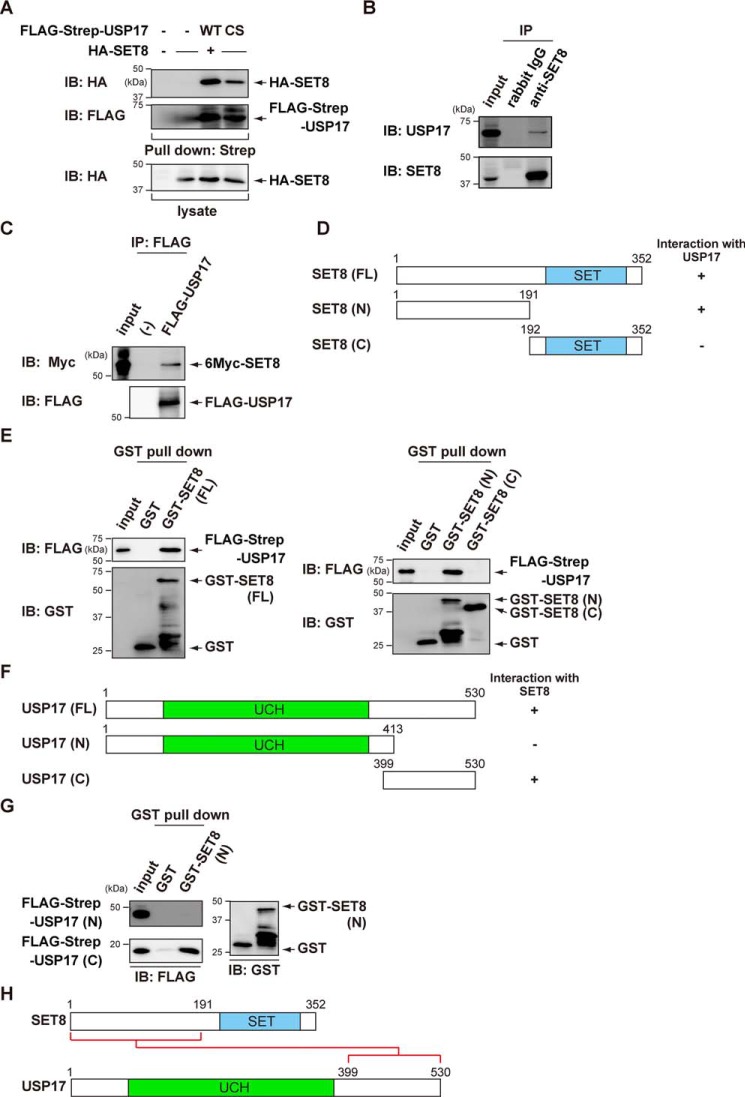
As shown in Figs. 1C and 4A, ectopic HA-SET8 was pulled down with FLAG-Strep-USP17 in COS7 cells. The USP17 C89S mutant also interacted with SET8, indicating that the catalytic activity of USP17 was not required for the interaction with SET8 (Fig. 4A). Furthermore, endogenous USP17 was immunoprecipitated by anti-SET8 antibodies (Fig. 4B). The results of the immunoprecipitation assay revealed that recombinant FLAG-USP17 and recombinant 6Myc-SET8 bound to each other in vitro, suggesting a direct interaction (Fig. 4C).
Figure 4.
USP17 interacts with SET8. A, COS7 cells were transiently transfected with the indicated constructs. After 24 h, cell lysates were subjected to Strep-Tactin pulldown and immunoblotting (IB) with an anti-HA antibody. B, endogenous SET8 was immunoprecipitated (IP) from MCF7 cells, and bound endogenous USP17 was examined by immunoblotting. C, in vitro binding assay for recombinant FLAG-USP17 and 6Myc-SET8. In vitro translated FLAG-USP17 and 6Myc-SET8 were used for the in vitro binding assay. D, schematic of full-length and deletion mutants of SET8. E, mapping of the binding region of USP17 on SET8 using SET8 deletion mutants. COS7 cells were transfected with FLAG-Strep-USP17. After 24 h, cell lysates were incubated with GST or GST-SET8 and subjected to GST pulldown, followed by immunoblotting with an anti-FLAG antibody. F, schematic of full-length and deletion mutants of USP17. UCH, ubiquitin C-terminal hydrolase G, mapping of the binding region of SET8 on USP17 using USP17 deletion mutants. COS7 cells were transfected with FLAG-Strep-USP17 (N) or FLAG-Strep-USP17 (C). After 24 h, cell lysates were incubated with GST or GST-SET8 (N) and subjected to GST pulldown, followed by immunoblotting with an anti-FLAG antibody. K, schematic of protein–protein interactions between SET8 and USP17.
We then investigated the interaction between SET8 and USP17 in more detail using a GST pulldown assay. SET8 bound USP17 through its N-terminal domain (1–191) (Fig. 4, D and E). In addition, the C-terminal domain (399–530) of USP17 was specifically pulled down with GST-SET8N (1–191) (Fig. 4, F–H).
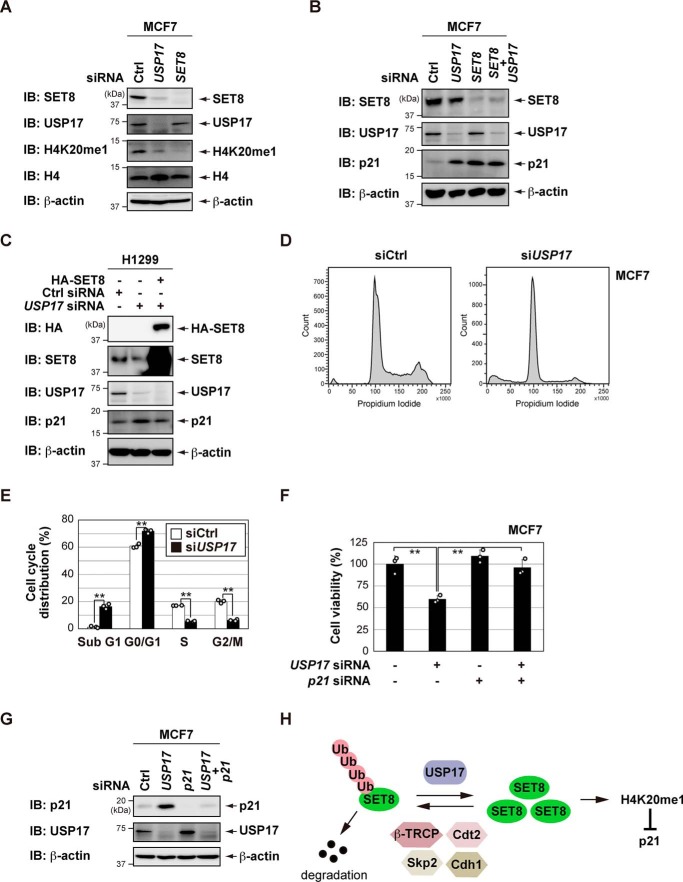
USP17 knockdown not only decreases H4K20 monomethylation but also increases p21 levels
As described above, SET8 is the sole enzyme required to catalyze H4K20me1 (1–3). To clarify the role of endogenous USP17 in the regulation of SET8, we investigated whether knockdown of USP17 affected H4K20me1 levels. As shown in Fig. 5A, knockdown of SET8 was associated with a reduction in H4K20me1 levels in MCF7 cells. Knockdown of USP17 partially but significantly decreased H4K20me1 levels in MCF7 cells. Previous studies have shown that knockdown of SET8 increases p21 levels (12, 13). Therefore, we also investigated whether knockdown of USP17 increased the expression of p21 by down-regulating SET8 expression. Similar to knockdown of SET8, knockdown of USP17 increased the expression of p21 in MCF7 cells (Fig. 5B). However, combined knockdown of USP17 and SET8 showed that p21 expression could not be increased further compared with knockdown of SET8. We also demonstrated that USP17 knockdown-induced p21 up-regulation was suppressed by SET8 overexpression in H1299 cells (Fig. 5C). These results suggest that SET8 is a downstream effector of USP17 in the regulation of p21.
Figure 5.
USP17 knockdown not only decreases H4K20 monomethylation but also increases p21 levels. A, B, and G, MCF7 cells were transiently transfected with the indicated siRNAs. After 72 h, an immunoblot (IB) analysis was performed with the indicated antibodies. Ctrl, control. C, H1299 cells were transfected with the indicated constructs and siRNAs. The cell lysates were analyzed by immunoblotting with the indicated antibodies. D and E, MCF7 cells were transiently transfected with the indicated siRNAs. After 72 h, a FACS analysis was performed (D). The percentages of cells in each phase of the cell cycle are indicated (E). Data are the means and standard deviations of three independent experiments. F, MCF7 cells were transiently transfected with the indicated siRNAs. After 72 h, cell viability was measured by a WST-8 cell proliferation assay. Data are the means and standard deviations of three independent experiments. **, p < 0.01. H, a proposed model illustrating how USP17 induces SET8 protein stabilization through deubiquitination.
A flow cytometry analysis showed that knockdown of USP17 induced G1 phase arrest and apoptosis in MCF7 cells (Fig. 5, D and E). As a result, proliferation of MCF7 cells was suppressed by knockdown of USP17, and the effects were simultaneously decreased by knockdown of p21 (Fig. 5, F and G). These results suggest that USP17 affects the status of H4K20me1 and the expression of p21 by regulating the stability of SET8 (Fig. 5H).
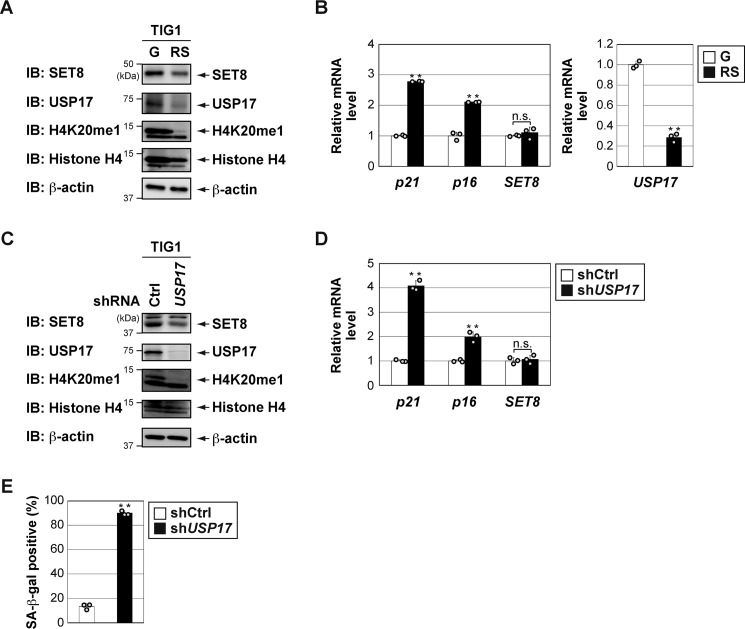
USP17 is down-regulated in senescent cells, and loss of USP17 induces senescence in human fibroblasts
Tanaka et al. (12) showed that SET8 is down-regulated in senescent cells, induced by oncogenic and replicative stress. Depletion of SET8 induces senescence in human fibroblasts (12, 13). We also found that SET8 protein levels decreased (Fig. 6A), whereas SET8 mRNA levels did not vary (Fig. 6B) in the late passage of human diploid fibroblast TIG1 cells. mRNA expression of p21 and p16 was up-regulated in the late passage of TIG1 cells (Fig. 6B). We also found a discernable decrease in USP17 expression in a prolonged culture of TIG1 cells (Fig. 6, A and B). These results suggest that the reduction in SET8 protein levels associated with cellular senescence may be due to decreases in USP17 protein levels. To investigate this possibility, we used lentiviral shRNA to knock down USP17 in proliferating TIG1 cells. Similar to the results shown in Fig. 5A, knockdown of USP17 decreased SET8 protein and H4K20me1 levels (Fig. 6C). Knockdown of USP17 was sufficient to trigger cellular senescence in TIG1 cells (Fig. 6, D and E). As reported previously (12, 13), we found that the knockdown of SET8 induced senescence of TIG1 cells (Fig. S3). However, additional depletion of USP17 did not affect senescence induction. Collectively, our results suggest that USP17 regulates cellular senescence by controlling SET8 stability and transcriptional repression of p21.
Figure 6.
USP17 is down-regulated in senescent cells, and loss of USP17 induces senescence in human fibroblasts. A, USP17 was down-regulated in replicative senescent TIG1 cells. Total protein was extracted from TIG1 cells with PD30 (growing (G)) or PD65 (replicative senescence (RS)), and an immunoblot (IB) analysis was performed with the indicated antibodies. B, total RNA was extracted from TIG1 cells with PD30 (G) or PD65 (RS), and the expression of each gene was assessed by quantitative RT-PCR. mRNA levels of the indicated genes were normalized with β-actin mRNA. Results are shown as means ± S.D. (n = 3). C–E, TIG1 cells were infected with lentivirus vectors containing shRNA for USP17 (shUSP17) or the control (shCtrl) for 144 h. C, total cell lysates were harvested for an immunoblot analysis with the indicated antibodies. D, the expression of each gene was assessed by qPCR, and the mRNA levels of the indicated genes were normalized with β-actin mRNA. Results are shown as means ± S.D. (n = 3). E, cells were subjected to senescence-associated β-gal (SA–β-gal) staining. Bar graphs below show the percentage of β-gal–positive cells in the indicated culture. **, p < 0.01; n.s., not significant.
Discussion
SET8 is the sole enzyme required to catalyze H4K20me1. In addition to histones, a number of nonhistone proteins are also methylated by SET8, leading to changes in their function or stability. Previous studies reported that overexpression of SET8 contributes to cancer tumorigenesis (15). Consequently, the mechanisms responsible for precise modulation of SET8 levels for proper cell cycle progression have been attracting increasing attention. Protein ubiquitination and deubiquitination are involved in the regulation of virtually all aspects of cellular biology, with effects ranging from altered protein stability to changes in protein function. Several E3 ligases have been reported to promote the ubiquitination and degradation of SET8 (7). In contrast, the DUB(s) of SET8 require further investigation. In this study, we identified USP17 as a novel SET8 DUB. USP17 interacts with SET8 and deubiquitinates SET8 in cells and in vitro. We also demonstrated that knockdown of USP17 not only decreased SET8 protein levels but also those of H4K20me1. These results contribute to our knowledge of the diverse mechanisms regulating SET8.
Humans have about 100 DUBs, divided into six different subfamilies (22, 23). Numerous DUBs are associated with cancer development and progression, some of which have been reported to be involved in epigenetic regulation (30). Although we have identified USP17 belonging to the USP family of DUBs as a SET8 DUB, other subfamilies of DUBs (e.g. OTU DUBs and MPN DUBs) may also regulate SET8. We tested whether other subfamilies of DUBs stabilize SET8 proteins. As shown in Fig. S4, only USP17 increased SET8 protein levels. However, the possibility that other DUBs may contribute to the regulation of SET8 protein under diverse cellular conditions cannot be ruled out. Further investigation is needed to clarify these concerns.
USP17 is an immediate-early gene and induced by the cytokines IL-4 and IL-6 (22, 31). USP17 has been reported to play an important role in tumor progression, such as cell proliferation and migration (31, 32). For example, USP17 exhibits oncogenic activity by stabilizing Cdc25A and contributes to the maintenance of pluripotency by controlling Cdc25A protein abundance in mouse embryonic stem cells (25). McFarlane et al. (32) also showed that depletion of USP17 blocks translocation and proper activation of Ras and RhoA in G1 phase, resulting in accumulation of the cyclin-dependent kinase inhibitors p21 and p27. We observed that knockdown of USP17 increased the expression of p21 in MCF7 and TIG1 cells. Previous studies showed that depletion of SET8 up-regulates p21 by reducing deposition of H4K20me1 at its gene loci (12, 13). Therefore, we speculated that knockdown of USP17 results in a decrease in SET8 protein levels, which induces p21 by decreasing H4K20me1 on the p21 gene. Furthermore, proliferation of MCF7 cells was suppressed by knockdown of USP17, and the effects were simultaneously decreased by knockdown of p21. USP17 appears to promote cell proliferation by decreasing p21 expression levels via regulation of SET8.
Cellular senescence is a tumor suppressor mechanism that prevents proliferation of precancerous cells. It is established and maintained by two major tumor suppressor pathways: the p53/p21 and p16/pRB pathways (33). We found that the expression levels of USP17 and SET8 declined with cellular senescence. USP17 repression is not only associated with the senescence response but also contributes to its induction. Previous studies revealed that loss of SET8 is sufficient to establish cellular senescence (12, 13). Based on these findings and our results, an USP17–SET8 axis suppresses p21 expression and blocks the onset of cellular senescence.
USP17 has been found to regulate breast cancer metastasis through Snail deubiquitination and stabilization (26, 27). In addition, USP17 has been shown to function as a deubiquitinating enzyme of Slug and Twist (34). Based on these findings, USP17 stimulates not only cell proliferation but also cell migration and invasion, resulting in cancer progression and metastasis. Up-regulated expression of USP17 has been detected in many types of tumors (32). On the other hand, SET8 is also overexpressed in multiple tumor types and has been implicated in cancer progression and metastasis (35). Our results revealed a novel link between USP17 and SET8. When considering USP17 as a SET8 stabilization factor, it appears to facilitate cancer progression and metastasis by stabilizing SET8. Future studies are needed to clarify this issue. Based on the important roles of SET8 in cancer, the contribution of USP17 to these pathological and physiological processes warrants further investigation.
Experimental procedures
Cell lines, plasmids, and transfections
MCF7, HCT116, MDA-MB-231, TIG-1, and COS7 cells were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 4.5 g/liter glucose, 10% FBS (Sigma), 100 units/ml penicillin G, and 100 μg/ml streptomycin. H1299 cells were cultured in Roswell Park Memorial Institute 1640 medium (Sigma) containing 10% FBS and penicillin/streptomycin. Cells were grown in a 5% CO2 atmosphere at 37 °C.
Human DUBs were cloned from the cDNA of MCF7 cells or H1299 cells into pcDNA3/FLAG-Strep or pCMV5/FLAG-Strep. HA-SET8, 6Myc-SET8, and FLAG-Strep or Strep-Ub were also inserted into pcDNA3. USP17 (C89S) was generated using the QuikChange mutagenesis kit (Agilent Technologies, La Jolla, CA). In shRNA-mediated gene silencing, gene-specific hairpin oligonucleotides were ligated into the pLKO vector (Sigma). The sequence for USP17 shRNA was 5′-GCAGGAAGATGCCCATGAATT-3′ (26). The SET8 Mission shRNA plasmid (TRCN0000130139) was obtained from Sigma. All constructs were verified by sequencing.
For DNA transfection, plasmids were transiently transfected with PEI (Polysciences, Warrington, PA) or Lipofectamine 2000 (Invitrogen). For siRNA transfection, siRNAs were transfected using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol. Human USP17 siRNA (sense, 5′-GCAGGAAGAUGCCCAUGAA-3′) (36) and human USP17 #2 siRNA (sense, 5′-GAAUGUGCAAUAUCCUGAG-3′) (37) were purchased from FASMAC (Kanagawa, Japan). siRNA targeting human SET8 (sense, 5′-CGAGGAACAGAAGAUCAAA-3′) was purchased from Sigma (38). The siRNA oligo targeting human p21 RNA has been described previously (39). Stealth RNAiTM siRNA Negative Control Med GC Duplex was obtained from Invitrogen.
RNA extraction, reverse transcription, and quantitative PCR
Total RNA was extracted using Sepasol-RNAI Super G (Nacalai Tesque, Kyoto, Japan) according to the manufacturer's protocol. First-strand cDNA was synthesized with the PrimeScript first-strand cDNA Synthesis Kit (TaKaRa Bio Inc., Shiga, Japan) as described previously (39). Quantitative PCR was performed as described previously (40). The following primer sequences were used: human USP17, 5′-CAGAAGACACAGACAGGCGA-3′ (forward) and 5′-GCTCTTTCCACCAAGTGCTC-3′ (reverse); human SET8, 5′-AAGGTGGACTTGAACAGATG-3′ (forward) and 5′-ACCTGTGCTGAGTCTTTGAC-3′ (reverse); human β-actin, 5′-TGGCACCCAGCACAATGAA-3′ (forward) and 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ (reverse); human p21, 5′-GATTTCTACCACTCCAAACGCC-3′ (forward) and 5′-AGAAGATGTAGAGCGGGC-3′ (reverse) (39); human p16, 5′-GAAGGTCCCTCAGACATCCCC-3′ (forward) and 5′-CCCTGTAGGACCTTCGGTGAC-3′ (reverse) (41). The specificities of the detected signals were confirmed by a dissociation curve, which consisted of a single peak. Values were normalized by β-actin.
Immunochemical methods and antibodies
Immunoblotting, immunoprecipitation, and Strep-Tactin pulldown were performed as described previously (42–44). Immunoreactive proteins were visualized using enhanced chemiluminescence Western blotting detection reagents (GE Healthcare), and the light emission intensity was quantified with a Lumino-image analyzer (LAS-3000 Mini) equipped with Image Gauge software (GE Healthcare). The following commercially available antibodies were used: anti-HA (Y-11, Santa Cruz Biotechnology, Santa Cruz, CA), anti-GST (B-14, Santa Cruz Biotechnology), anti-p21 (F-5, Santa Cruz Biotechnology), anti-SET8 (D-11, Santa Cruz Biotechnology), anti-SET8 (C18B7, Cell Signaling Technology, Beverly, MA), anti-HDAC2 (3F3, Cell Signaling Technology), anti-β-actin (AC-15, Sigma), anti-FLAG (IE6, Wako, Osaka, Japan), anti-H4K20me1 (ab9051, Abcam, Cambridge, UK), anti-SET8 (AM1191a, Abgent, San Diego, CA), anti-histone H4 (MABI0400, MBL, Nagoya, Japan), anti-Snail (AF3639, R&D Systems, Minneapolis, MN), anti-ubiquitin-horseradish peroxidase (P4D1, Biolegend, San Diego, CA), and anti-USP17/DUB3 (NBP1-79745, Novus Biologicals, Littleton, CO). Anti-Myc (9E10) was obtained from the culture supernatant of MYC 1-9E10.2 cells (ATCC, Manassas, VA).
Endogenous SET8 proteins were immunoprecipitated with an anti-SET8 antibody preincubated for 2 h with Dynabeads M-280 sheep anti-mouse IgG (Invitrogen). Immunoprecipitates were washed three times with lysis buffer and then subjected to immunoblotting. Recombinant FLAG-USP17 and 6Myc-SET8 were synthesized using the TnT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI).
GST pulldown assay
cDNAs encoding full-length SET8 and its deletion mutants were cloned into pGEX6P1 and expressed in the Escherichia coli BL21 (DE3) strain, and GST fusion proteins were purified as recommended in the instructions (GE Healthcare). Cells were subjected to lysis in TNTE buffer (20 mm Tris-HCl (pH 7.5), 120 mm NaCl, 1 mm EDTA, and 0.5% Triton X-100) supplemented with protease inhibitors and pulled down with GST fusion proteins. Samples were then subjected to immunoblotting.
In vitro deubiquitination assay
6Myc-SET8 and Strep-Ub were coexpressed in COS7 cells. After cells had been treated with 20 μm MG132 for 4 h, ubiquitinated SET8 was pulled down with Strep-Tactin–Sepharose, followed by elution with 10 mm desthiobiotin. In a parallel experiment, FLAG-Strep-USP17 (WT or C89S) was expressed in COS7 cells, and FLAG-Strep-USP17 was also pulled down with Strep-Tactin–Sepharose followed by elution with desthiobiotin. In the in vitro deubiquitination assay, purified FLAG-Strep-USP17 WT or the C89S mutant was incubated with ubiquitinated 6Myc-SET8 in deubiquitination reaction buffer (150 mm Tris-HCl (pH 7.5), 50 mm NaCl, 1 mm EDTA, and 5 mm DTT) at 37 °C for 2 h. The ubiquitination status of SET8 was analyzed by immunoblotting with anti-Myc antibodies.
Cycloheximide half-life assay
Cells were transiently transfected with siRNAs. Cells were incubated for the indicated periods with 50 μg/ml of cycloheximide (CHX). Cell lysates were analyzed by immunoblotting.
Cell viability assay and SA–β-gal staining
Cell viability was assessed using WST-8 according to the manufacturer's instructions (Dojindo, Kumamoto, Japan). Cells were transfected as described above. Twenty-four hours after transfection, cells were trypsinized and reseeded at a concentration of 5 × 103 cells/well in a 96-well plate. After 48 h, the WST-8 reagent was added, and cells were incubated at 37 °C for 3 h in a humidified atmosphere of 5% CO2. The absorbance at 450 nm of the medium was measured (39). SA–β-gal staining was performed as described previously (29).
Cell cycle analysis
Cells were stained with propidium iodide using the CycletestTM Plus DNA Reagent Kit (BD Biosciences) according to manufacturer's instructions. The cell cycle distribution of nuclear DNA was assessed using a FACSVerseTM flow cytometer (BD Biosciences) by analyzing at least 20,000 cells/sample. The percentages of cells in sub G1, G0/G1, S, and G2/M were analyzed using FACSuiteTM software (BD Biosciences).
Statistical tests
The significance of differences between two groups was evaluated using two-tailed Student's t test. For multigroup analyses, significance was assessed using a one-way analysis of variance with post hoc Tukey-Kramer honestly significant difference test.
Author contributions
K. F., Y. I., S. W., and M. T. data curation; K. F., Y. I., C. M., S. W., M. T., and H. H. supervision; K. F., Y. I., C. M., S. W., and M. T. investigation; K. F., Y. I., and H. H. writing-original draft; K. F., Y. I., and H. H. writing - review and editing; Y. I. and H. H. funding acquisition; C. M., M. T., D. M., N. O., M. K., and H. H. methodology; D. M., N. O., and M. K. resources; H. H. conceptualization; H. H. project administration.
Supplementary Material
Acknowledgments
We thank the members of the Hayashi laboratory for helpful discussions. We acknowledge assistance from the Research Equipment Sharing Center at Nagoya City University.
This work was supported by Grants-in-Aid for Scientific Research (C) 24590085, 15K07936, 15K07937, and 18K06660 from the Japan Society for the Promotion of Science (JSPS); Grant-in-Aid for Young Scientists (B) 24700983 from JSPS, and a grant-in-aid for research from Nagoya City University. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- SET
- Su(var)3–9, Enhancer-of-zeste, and Trithorax
- H4K20me1
- histone H4 lysine 20 monomethylation
- SCF
- Skp1–Cullin-1–F-box
- DUB
- deubiquitinase
- USP
- ubiquitin-specific processing protease
- Ub
- ubiquitin
- CHX
- cycloheximide
- cDNA
- complementary DNA
- SA
- senescence-associated
- qPCR
- quantitative PCR.
References
- 1. Jørgensen S., Elvers I., Trelle M. B., Menzel T., Eskildsen M., Jensen O. N., Helleday T., Helin K., and Sørensen C. S. (2007) The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179, 1337–1345 10.1083/jcb.200706150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Couture J. F., Collazo E., Brunzelle J. S., and Trievel. R. C. (2005) Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 19, 1455–1465 10.1101/gad.1318405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nishioka K., Rice J. C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Lis J. T., Allis C. D., and Reinberg D. (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 10.1016/S1097-2765(02)00548-8 [DOI] [PubMed] [Google Scholar]
- 4. Liu B., Zhang X., Song F., Liu Q., Dai H., Zheng H., Cui P., Zhang L., Zhang W., and Chen K. (2016) A functional single nucleotide polymorphism of SET8 is prognostic for breast cancer. Oncotarget 7, 34277–34287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck D. B., Oda H., Shen S. S., and Reinberg D. (2012) PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 26, 325–337 10.1101/gad.177444.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jørgensen S., Schotta G., and Sørensen C. S. (2013) Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41, 2797–2806 10.1093/nar/gkt012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng N., Dai X., Wang Z., and Wei W. (2016) A new layer of degradation mechanism for PR-Set7/Set8 during cell cycle. Cell Cycle 15, 3042–3047 10.1080/15384101.2016.1234552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song F., Zheng H., Liu B., Wei S., Dai H., Zhang L., Calin G. A., Hao X., Wei Q., Zhang W., and Chen K. (2009) An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin. Cancer Res. 15, 6292–6300 10.1158/1078-0432.CCR-09-0826 [DOI] [PubMed] [Google Scholar]
- 9. Ding C., Li R., Peng J., Li S., and Guo Z. (2012) A polymorphism at the miR-502 binding site in the 3′ untranslated region of the SET8 gene is associated with the outcome of small-cell lung cancer. Exp. Ther. Med. 3, 689–692 10.3892/etm.2012.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo Z., Wu C., Wang X., Wang C., Zhang R., and Shan B. (2012) A polymorphism at the miR-502 binding site in the 3′-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int. J. Cancer 131, 1318–1322 10.1002/ijc.27352 [DOI] [PubMed] [Google Scholar]
- 11. Wang C., Guo Z., Wu C., Li Y., and Kang S. (2012) A polymorphism at the miR-502 binding site in the 3′ untranslated region of the SET8 gene is associated with the risk of epithelial ovarian cancer. Cancer Genet. 205, 373–376 10.1016/j.cancergen.2012.04.010 [DOI] [PubMed] [Google Scholar]
- 12. Tanaka H., Takebayashi S. I., Sakamoto A., Igata T., Nakatsu Y., Saitoh N., Hino S., and Nakao M. (2017) The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Rep. 18, 2148–2161 10.1016/j.celrep.2017.02.021 [DOI] [PubMed] [Google Scholar]
- 13. Shih C. T., Chang Y. F., Chen Y. T., Ma C. P., Chen H. W., Yang C. C., Lu J. C., Tsai Y. S., Chen H. C., and Tan B. C. (2017) The PPARγ-SETD8 axis constitutes an epigenetic, p53-independent checkpoint on p21-mediated cellular senescence. Aging Cell 16, 797–813 10.1111/acel.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., and Sakai J. (2009) The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell Biol. 29, 3544–3555 10.1128/MCB.01856-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milite C., Feoli A., Viviano M., Rescigno D., Cianciulli A., Balzano A. L., Mai A., Castellano S., and Sbardella G. (2016) The emerging role of lysine methyltransferase SETD8 in human disease. Clin. Epigenetics 8, 102 10.1186/s13148-016-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu S., Wang W., Kong X., Congdon L. M., Yokomori K., Kirschner M. W., and Rice J. C. (2010) Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 24, 2531–2542 10.1101/gad.1984210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abbas T., Shibata E., Park J., Jha S., Karnani N., and Dutta A. (2010) CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40, 9–21 10.1016/j.molcel.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., and Zou L. (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40, 22–33 10.1016/j.molcel.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., and Reinberg D. (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40, 364–376 10.1016/j.molcel.2010.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin Y., Yu V. C., Zhu G., and Chang D. C. (2008) SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 7, 1423–1432 10.4161/cc.7.10.5867 [DOI] [PubMed] [Google Scholar]
- 21. Wang Z., Dai X., Zhong J., Inuzuka H., Wan L., Li X., Wang L., Ye X., Sun L., Gao D., Zou L., and Wei W. (2015) SCF(β-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat. Commun. 6, 10185 10.1038/ncomms10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ventii K. H., and Wilkinson K. D. (2008) Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 10.1042/BJ20080798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McClurg U. L., and Robson C. N. (2015) Deubiquitinating enzymes as oncotargets. Oncotarget 6, 9657–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pereg Y., Liu B. Y., O'Rourke K. M., Sagolla M., Dey A., Komuves L., French D. M., and Dixit V. M. (2010) Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat. Cell Biol. 12, 400–406 10.1038/ncb2041 [DOI] [PubMed] [Google Scholar]
- 26. Liu T., Yu J., Deng M., Yin Y., Zhang H., Luo K., Qin B., Li Y., Wu C., Ren T., Han Y., Yin P., Kim J., Lee S., Lin J., et al. (2017) CDK4/6-dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat. Commun. 8, 13923 10.1038/ncomms13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y., Wang Y., Lin Y., Liu Y., Wang Y., Jia J., Singh P., Chi Y. I., Wang C., Dong C., Li W., Tao M., Napier D., Shi Q., Deng J., et al. (2017) Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat. Commun. 8, 14223 10.1038/ncomms14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song H., Tao L., Chen C., Pan L., Hao J., Ni Y., Li D., Li B., and Shi G. (2015) USP17-mediated deubiquitination and stabilization of HDAC2 in cigarette smoke extract-induced inflammation. Int. J. Clin. Exp. Pathol. 8, 10707–10715 [PMC free article] [PubMed] [Google Scholar]
- 29. Debacq-Chainiaux F., Erusalimsky J. D., Campisi J., and Toussaint O. (2009) Protocols to detect senescence-associated β-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 10.1038/nprot.2009.191 [DOI] [PubMed] [Google Scholar]
- 30. Wang J., Qiu Z., and Wu Y. (2018) Ubiquitin regulation: the histone modifying enzyme's story. Cells 7, E118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burrows J. F., McGrattan M. J., and Johnston J. A. (2005) The DUB/USP17 deubiquitinating enzymes, a multigene family within a tandemly repeated sequence. Genomics 85, 524–529 10.1016/j.ygeno.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 32. McFarlane C., Kelvin A. A., de la Vega M., Govender U., Scott C. J., Burrows J. F., and Johnston J. A. (2010) The deubiquitinating enzyme USP17 is highly expressed in tumor biopsies, is cell cycle regulated, and is required for G1-S progression. Cancer Res. 70, 3329–3339 10.1158/0008-5472.CAN-09-4152 [DOI] [PubMed] [Google Scholar]
- 33. Campisi J. (2013) Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin Y., Wang Y., Shi Q., Yu Q., Liu C., Feng J., Deng J., Evers B. M., Zhou B. P., and Wu Y. (2017) Stabilization of the transcription factors slug and twist by the deubiquitinase dub3 is a key requirement for tumor metastasis. Oncotarget 8, 75127–75140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang F., Sun L., Li Q., Han X., Lei L., Zhang H., and Shang Y. (2012) SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 31, 110–123 10.1038/emboj.2011.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim J., D'Annibale S., Magliozzi R., Low T. Y., Jansen P., Shaltiel I. A., Mohammed S., Heck A. J., Medema R. H., and Guardavaccaro D. (2014) USP17- and SCFβTrCP-regulated degradation of DEC1 controls the DNA damage response. Mol. Cell Biol. 34, 4177–4185 10.1128/MCB.00530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ducker C., Chow L. K. Y., Saxton J., Handwerger J., McGregor A., Strahl T., Layfield R., and Shaw P. E. (2019) De-ubiquitination of ELK-1 by USP17 potentiates mitogenic gene expression and cell proliferation. Nucleic Acids Res. 47, 4495–4508 10.1093/nar/gkz166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takawa M., Cho H. S., Hayami S., Toyokawa G., Kogure M., Yamane Y., Iwai Y., Maejima K., Ueda K., Masuda A., Dohmae N., Field H. I., Tsunoda T., Kobayashi T., Akasu T., et al. (2012) Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 72, 3217–3227 10.1158/0008-5472.CAN-11-3701 [DOI] [PubMed] [Google Scholar]
- 39. Inoue Y., Kawachi S., Ohkubo T., Nagasaka M., Ito S., Fukuura K., Itoh Y., Ohoka N., Morishita D., and Hayashi H. (2017) The CDK inhibitor p21 is a novel target gene of ATF4 and contributes to cell survival under ER stress. FEBS Lett. 591, 3682–3691 10.1002/1873-3468.12869 [DOI] [PubMed] [Google Scholar]
- 40. Kawarada Y., Inoue Y., Kawasaki F., Fukuura K., Sato K., Tanaka T., Itoh Y., and Hayashi H. (2016) TGF-β induces p53/Smads complex formation in the PAI-1 promoter to active transcription. Sci. Rep. 6, 35483 10.1038/srep35483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen Y., Guo X., Wang Y., Qiu W., Chang Y., Zhang A., and Duan X. (2012) Expression and significance of histone H3K27 demethylase in renal cell carcinoma. BMC Cancer 12, 470 10.1186/1471-2407-12-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inoue Y., Kitagawa M., and Taya Y. (2007) Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 26, 2083–2093 10.1038/sj.emboj.7601652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miyajima C., Inoue Y., and Hayashi H. (2015) Pseudokinase Tribbles 1 (TRB1) negatively regulates tumor-suppressor activity of p53 through p53 deacetylation. Biol. Pharm. Bull. 38, 618–624 10.1248/bpb.b15-00003 [DOI] [PubMed] [Google Scholar]
- 44. Inoue Y., Iemura S., Natsume T., Miyazawa K., and Imamura T. (2011) Suppression of p53 activity through the cooperative action of Ski and histone deacetylase SIRT1. J. Biol. Chem. 286, 6311–6320 10.1074/jbc.M110.177683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.