Abstract
Fcp1 is a protein phosphatase that facilitates transcription elongation and termination by dephosphorylating the C-terminal domain of RNA polymerase II. High-throughput genetic screening and gene expression profiling of fcp1 mutants revealed a novel connection to Cdk8, the Mediator complex kinase subunit, and Skn7, a key transcription factor in the oxidative stress response pathway. Briefly, Skn7 was enriched as a regulator of genes whose mRNA levels were altered in fcp1 and cdk8Δ mutants and was required for the suppression of fcp1 mutant growth defects by loss of CDK8 under oxidative stress conditions. Targeted analysis revealed that mutating FCP1 decreased Skn7 mRNA and protein levels as well as its association with target gene promoters but paradoxically increased the mRNA levels of Skn7-dependent oxidative stress-induced genes (TRX2 and TSA1) under basal and induced conditions. The latter was in part recapitulated via chemical inhibition of transcription in WT cells, suggesting that a combination of transcriptional and posttranscriptional effects underscored the increased mRNA levels of TRX2 and TSA1 observed in the fcp1 mutant. Interestingly, loss of CDK8 robustly normalized the mRNA levels of Skn7-dependent genes in the fcp1 mutant background and also increased Skn7 protein levels by preventing its turnover. As such, our work suggested that loss of CDK8 could overcome transcriptional and/or posttranscriptional alterations in the fcp1 mutant through its regulatory effect on Skn7. Furthermore, our work also implicated FCP1 and CDK8 in the broader response to environmental stressors in yeast.
Keywords: transcription factor, transcription regulation, transcriptomics, yeast transcription, transcription, gene regulation, Cdk8, Fcp1, Mediator complex, Skn7, transcription repression, RNA polymerase II, oxidative stress
Introduction
In eukaryotes, RNA polymerase II (RNAPII)2 is the enzyme responsible for transcribing all protein-coding genes. Its largest subunit contains a highly conserved C-terminal domain (CTD) composed of heptapeptide repeats (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) that is essential for viability (1–3). The CTD is differentially phosphorylated throughout the transcription cycle, with each transcriptional stage characterized by a unique phosphorylation signature and the recruitment of specific regulatory, RNA-processing, and chromatin-remodeling factors (4). As such, a number of kinases (Kin28, Ctk1, Bur1, and Cdk8) and phosphatases (Fcp1, Rtr1, Ssu72, and Glc7) play key roles in transcription regulation (5).
Fcp1 (TFIIF-associating CTD phosphatase 1) was the first RNAPII-CTD phosphatase identified (6), encoded by a conserved gene that is essential for viability (7, 8). It contains a catalytic FCP homology (FCPH) domain and a single BRCA1 C-terminal (BRCT) domain, both of which are important for activity (9, 10). The latter mediates direct contacts with the RNAPII-CTD (9, 11), although its exact role within Fcp1 remains enigmatic. Functionally, Fcp1 associates along the length of genes and facilitates transcription elongation and recycling by dephosphorylating Ser2 and Thr4 residues on the RNAPII-CTD (12–15). Underscoring the significance of Fcp1 in facilitating transcription, temperature-sensitive fcp1 mutants that target the catalytic domain result in the accumulation of hyperphosphorylated RNAPII and a global shutdown of transcription at nonpermissive temperatures (6, 13, 14). Despite its importance, the full extent of Fcp1 function in the cell, as well as the exact mechanisms controlling its activity and recruitment to sites of transcription, remains poorly understood. Nevertheless, a subset of factors, including the general transcription factors TFIIF and TFIIB, have been implicated in the regulation of Fcp1 activity (7, 10, 14, 16–21). Furthermore, recent studies have ascribed Fcp1 roles beyond the RNAPII-CTD. For instance, in fission yeast, Fcp1 can dephosphorylate Spt5 (22); in mammals, mitotic exit factors (Wee1, Cdc20, USP44, Ensa and Greatwall) (23–25); and in budding yeast and mammals, RNAPI transcription initiation factors (Rrn3 and TIF-IA, respectively) (26, 27). Together, these findings are consistent with evidence positioning FCP1 earlier in evolutionary history than the gene encoding its best-known substrate, the RNAPII-CTD (28).
To better-understand the role of Fcp1 in the cell, high-throughput gene expression and genetic interaction profiling was performed on a series of fcp1 mutants, leading us to uncover a role in the response to environmental stress that was linked to CDK8, encoding the Mediator complex kinase subunit. In brief, the fcp1-594 mutant was sensitive to oxidative stress and had altered regulation of Skn7, a key transcription factor in the oxidative stress response, defects that were normalized by loss of CDK8. Strikingly, the fcp1-594 mutant had elevated mRNA levels of Skn7-dependent oxidative stress–responsive genes (TSA1 and TRX2) under basal and inducing conditions, an effect dependent on the kinase activity of Cdk8. Collectively, our work suggested that loss of CDK8 could overcome the transcriptional and posttranscriptional alterations of the fcp1 mutant by increasing Skn7 stability, protein levels, and likely the expression of target genes. In addition, our results also implicate FCP1 and CDK8 in the response to other environmental stressors, laying the foundation for future investigations.
Results
Gene expression profiles of FCP1 mutants suggested a role in the response to stress and a relationship with the gene encoding the Mediator complex kinase subunit, CDK8
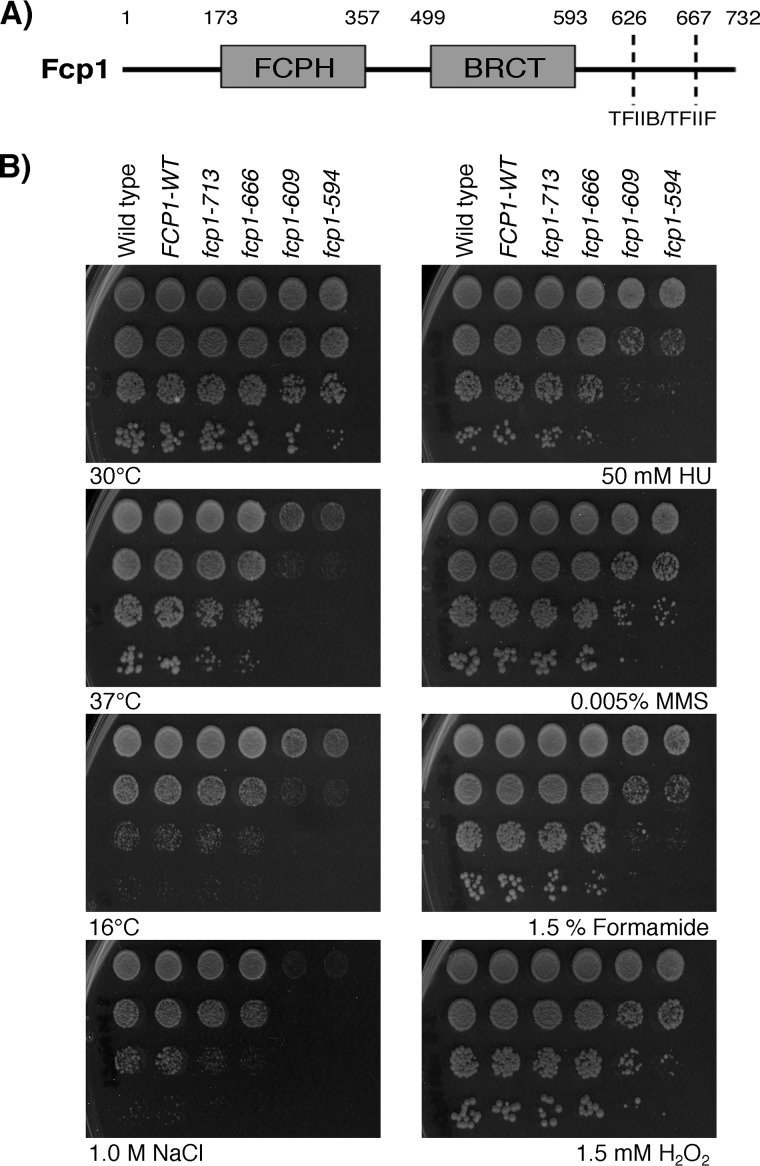
To better understand the role of FCP1 in the cell, a set of fcp1 mutants (FCP1-WT, fcp1-713, fcp1-666, fcp1-609, fcp1-594, which result in the production of C-terminally truncated Fcp1 proteins) were generated at the endogenous FCP1 locus and exposed to a variety of genotoxic agents and stress conditions (Fig. 1, A and B). These truncation alleles were described previously, and their protein products were shown to differentially disrupt the interaction with TFIIF and TFIIB while leaving the rest of Fcp1 intact (8, 10). Suggestive of a general function in the response to stress, the two shortest truncation mutants (fcp1-594 and fcp1-609) exhibited fitness defects when grown at high (37 °C) or low temperatures (16 °C) or exposed to hydroxyurea (HU), methyl methanesulfonate (MMS), formamide, osmotic stress (NaCl), or oxidative stress (H2O2) conditions (Fig. 1B). The longer fcp1-666 mutant showed an intermediate phenotype and was only sensitive to formamide and HU, whereas the fcp1-713 mutant and the FCP1-WT strain produced no growth defects.
Figure 1.
FCP1 mutants were sensitive to genotoxic agents and environmental stressors. A, schematic of FCP1 illustrating the FCPH catalytic domain, BRCT domain, and TFIIF/TFIIB-interacting region. The FCP1 mutants differentially removed the TFIIF/TFIIB-binding regions. B, the two shortest FCP1 mutants were sensitive to high (37 °C) and low (16 °C) temperatures and the indicated concentrations of hydroxyurea, methyl methanesulfonate, sodium chloride (NaCl; osmotic stress), and hydrogen peroxide (H2O2; oxidative stress). Cells with the indicated mutations were serially diluted 10-fold, spotted on YPD media with the indicated drug concentrations, and grown for 2–4 days.
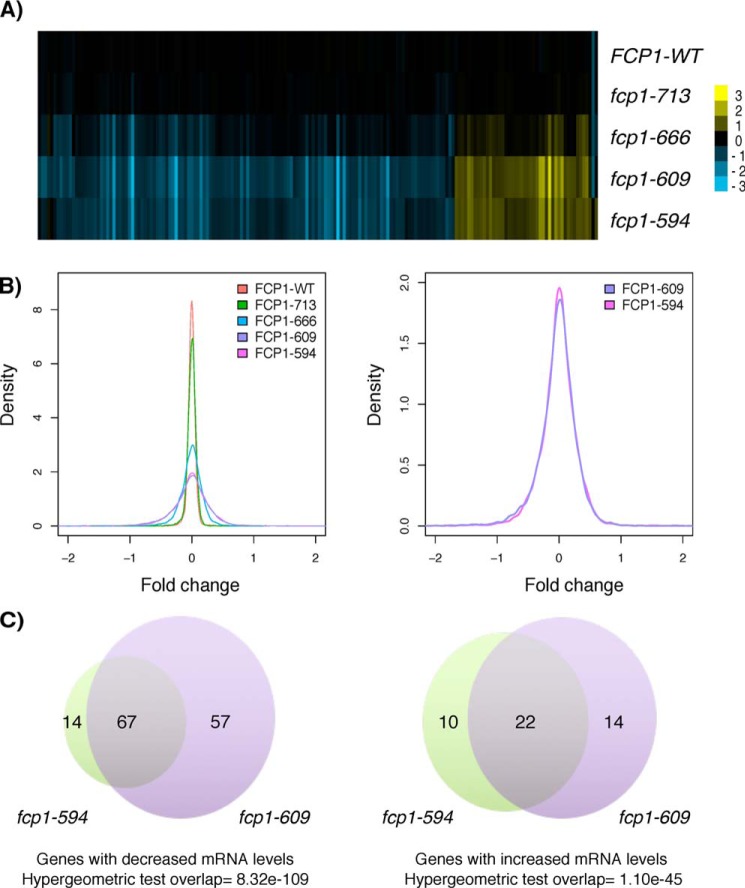
Having observed a range of fitness defects, we next determined whether these correlated with transcriptional defects. Gene expression profiles were generated using microarrays and compared with the isogenic control strain (FCP1-WT). Perhaps not surprisingly, the deletion mutants displayed milder gene expression alterations than two previously described and more severe FCP1 alleles, fcp1-1 and fcp1-2 (encoding Fcp1 R250A/R251A and L117A/L181A/H187A, respectively), which target the FCPH catalytic domain and result in a global shutdown of transcription at nonpermissive temperatures (6). Nevertheless, our gene expression profiles revealed length-dependent requirements, with the severity and number of transcriptional alterations generally increasing as the C terminus was progressively shortened (Fig. 2, A and B). However, we note that the fcp1-609 mutant, rather than the shortest fcp1-594 mutant had the greatest number of transcriptional alterations (Fig. 2C), suggesting a nuanced role for the FCP1 C terminus in transcription regulation. Despite the noted differences, there was a significant overlap between the fcp1-594 and fcp1-609 mutant expression profiles, allowing us to focus on genes whose expression was altered in both mutants (67 genes with decreased mRNA levels and 22 genes with increased mRNA levels), thus increasing confidence in our findings. Focusing on this gene set, we performed transcription factor (TF) enrichment analysis using the Yeast Promoter Atlas (29). This analysis revealed that the fcp1-dependent genes were enriched for regulation by TFs previously implicated in the response to stress (Mcm1, Ste12, Phd1, Sok2, Skn7, Sko1, Hot1, Bas1, and Cad1) (Fig. 3A), consistent with the fcp1 mutants showing growth defects across a variety of stress conditions. Follow-up targeted analysis confirmed a role for FCP1 in the regulation of a representative set of these TFs; the fcp1-594 mutant had decreased SKN7, SOK2, and MCM1 mRNA levels and Skn7, Sok2, Mcm1 and Cad1 protein levels (Fig. 3, B–E).
Figure 2.
The fcp1-594 and fcp1-609 mutants resulted in an overlapping set of gene expression alterations. A, heat map of mRNA levels showing genes differentially expressed (p < 0.01 and -fold change > 1.7) in the fcp1-594 or fcp1-609 mutant. Yellow, increased mRNA levels compared with WT; blue, decreased levels. B, left, distribution of gene expression -fold changes for the FCP1 mutants showed a length-dependent effect on gene expression. Right, focusing on the shortest mutants revealed that the fcp1-609 and not the shortest fcp1-594 mutant had the greatest number of gene expression defects. C, Venn diagram of genes differentially expressed in the fcp1-609 and fcp1-594 mutant showed a significant overlap (hypergeometric test, p < 0.01) as well as a high degree of gene-specific effects.
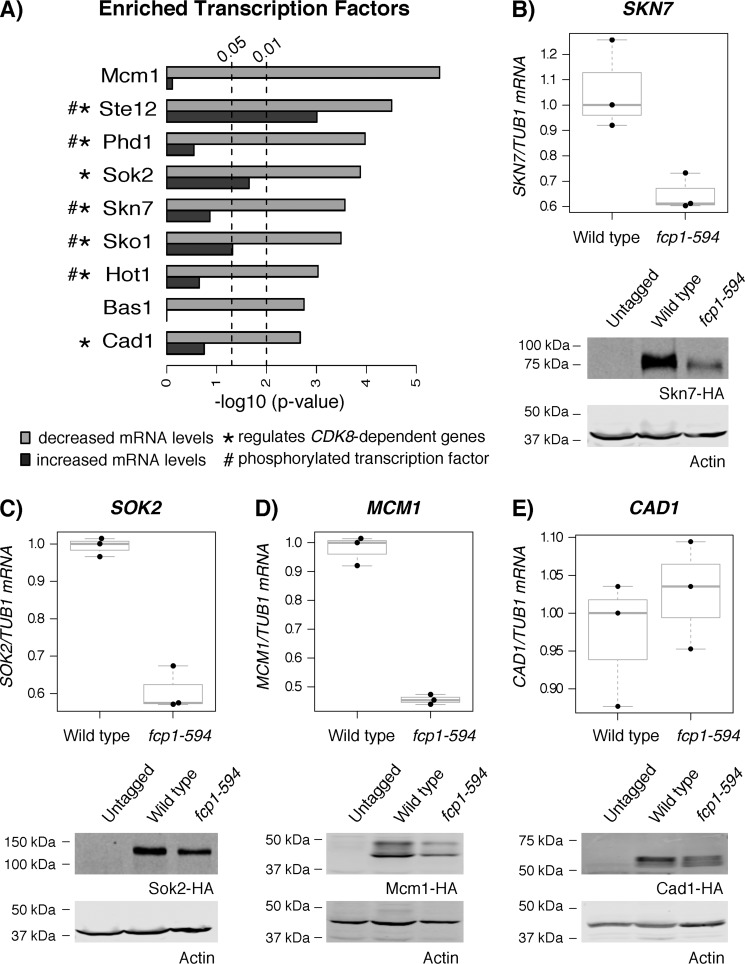
Figure 3.
FCP1 played a role in regulating the mRNA and protein levels of a subset of TFs. A, TFs enriched for regulating genes differentially expressed in the fcp1-594 and fcp1-609 mutant. Light gray bars, enrichment of TFs for genes whose mRNA levels decreased in the fcp1-594 and fcp1-609 mutant; dark gray boxes, enrichment for genes whose mRNA levels increased in the fcp1-594 and fcp1-609 mutant. Most enriched transcription factors significantly associated (p < 0.01) with genes whose mRNA levels decreased in the fcp1-594 and fcp1-609 mutant, with the exception of Ste12, which also showed a significant association with genes whose mRNA levels increased. A significant number of TFs also regulated genes whose mRNA levels are altered upon loss of CDK8 (hypergeometric test, p < 0.01 (*)) and had strong evidence of being phosphorylated proteins (hypergeometric test, p = 0.03769 (#)). B–E, top, the fcp1-594 mutant reduced SKN7, SOK2, and MCM1 mRNA levels but had no effect on CAD1. mRNA levels were normalized to TUB1 (75). mRNA levels are shown as box plots displaying the median and interquartile range, with whiskers denoting 1.5 times the interquartile range. Bottom, immunoblots of Skn7, Sok2, Mcm1, and Cad1 protein levels showed decreased levels in the fcp1-594 mutant compared with WT. Actin was used as a loading control.
Strikingly, the list of enriched TFs included two well-known substrates of the Mediator complex kinase subunit, Cdk8 (Ste12 and Phd1 (30, 31)) as well as the Phd1 ohnolog, Sok2 (32). This suggested relationship to CDK8 was further supported by a significant overlap in the TFs enriched for regulating genes whose mRNA levels were altered in the fcp1-594 and 609 mutants and those altered upon loss of CDK8 (seven of nine TFs, hypergeometric test, p = 0.0008035) (Fig. 3A (TFs marked with an asterisk) and Fig. S1A). Markedly, whereas the overlapping TFs associated with genes whose mRNA levels decreased in the FCP1 mutants (Fig. 3A, light gray bars), they related to genes whose mRNA levels increased in the cdk8Δ mutant (Fig. S1A, dark gray bars), indicating possible opposite effects on the regulation of these TFs (cdk8Δ mutant expression profile obtained from Ref. 33). To further explore the relationship of Cdk8 with these TFs, we examined available Cdk8 genome localizations derived from ChIP-chip profiles and found that Cdk8 physically associated with the promoters of a subset of genes regulated by the TFs from our analysis (Cdk8 ChIP-chip profile obtained from Ref. 34) (Fig. S1B). Finally, linking these TFs with a protein kinase and phosphatase, respectively, was intriguing, as it raised the possibility that the TFs were regulated by phosphorylation. Using the Yeast Kinase Interaction Database (35), we found a significant enrichment for proteins with strong evidence (score >6.4) of being phosphorylated in this small set of TFs (hypergeometric test, p = 0.03769) (Fig. 3A, TFs marked with a number symbol), which suggested a role for FCP1 and CDK8 in the biology of stress response TFs, perhaps through effects on their phosphorylation state.
The genetic interaction network of the FCP1 mutants supported a role in the regulation of SKN7 and SOK2
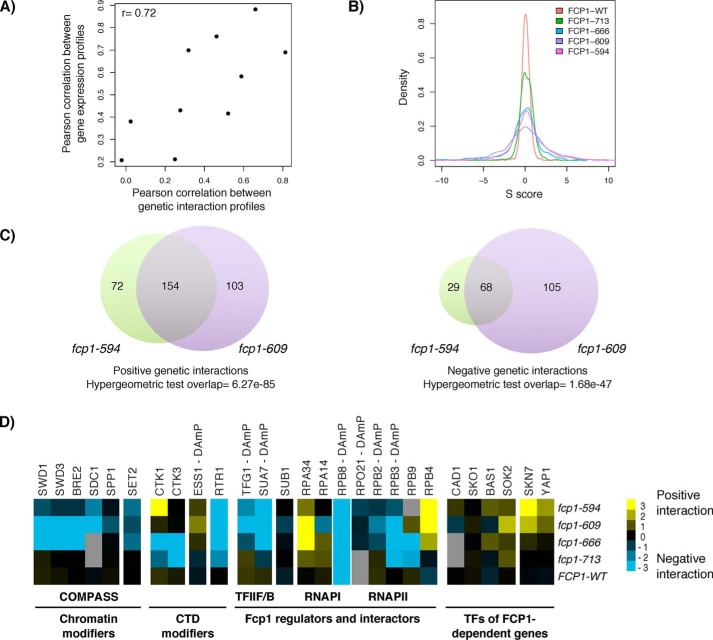
To gain insight into the cellular processes and pathways influenced by FCP1, the fcp1 truncation mutants described above were screened against a library of 1536 different mutants involved in chromatin biology, transcription, and RNA processing using epistasis miniarray profiling (E-MAP) (36). Overall, the genetic interaction profiles revealed similar functional information as the gene expression profiles (Fig. 4A). They showed that the number and strength of genetic interactions increased as Fcp1 protein was progressively shortened from the C terminus (Fig. 4B) and that the fcp1-609 mutant, and not the shortest fcp1-594 mutant, had the greatest number of genetic interactions (Fig. 4C). Furthermore, these profiles were consistent with known Fcp1 functions, including its role as an RNAPII-CTD phosphatase. For instance, they showed length-dependent genetic interactions with TFIIF and TFIIB, consistent with the differential effects of truncated Fcp1 mutant proteins on the physical interaction with both of these general transcription factors (10). This analysis also revealed significant genetic interactions with genes encoding RNAPII-CTD–modifying enzymes (CKT1, ESS1, and RTR1) (13, 37–40) (Fig. 4D) and genes encoding factors that bind the RNAPII-CTD in a phosphorylation-dependent manner, including members of the Set1-containing H3K4 methyltransferase COMPASS complex (41). Finally, the FCP1 genetic interaction profiles supported a functional relationship with the TFs identified by the gene expression analysis. Notably, both the fcp1-594 and fcp1-609 mutants showed strong positive interactions with SOK2 and SKN7 (Fig. 4D), with a link to SKN7 further supported by a positive interaction between the fcp1-594 and fcp1-609 mutants with YAP1, a transcription factor partner of Skn7 (42).
Figure 4.
Genetic interaction profiles of FCP1 mutants were consistent with its function as an RNAPII-CTD phosphatase and supported a role in transcription factor biology. A, the genetic and gene expression profiles revealed similar relationships across the various FCP1 mutants. The scatter plot of profile-paired correlations of the genetic interaction and gene expression profiles revealed a high correlation (0.72). B, distribution of S scores for the FCP1 mutants showed that the fcp1-609 mutant had the greatest number of significant genetic interactions. The S score is a modified T-statistic that captures the significance and strength of the genetic interaction. S scores greater than 2 and less than −2.5 were considered significant. C, Venn diagrams comparing the significant genetic interactions of the fcp1-609 and fcp1-594 mutants. Whereas the overlap in genetic interactions was significant (hypergeometric test, p < 0.01), there were also several genetic interactions that were unique to each mutant. D, subset of genetic interactions for the FCP1 mutants showed significant interactions with SOK2 and SKN7. Each mutant was screened in triplicate, with yellow, blue, and gray indicating alleviating, aggravating, and missing values, respectively.
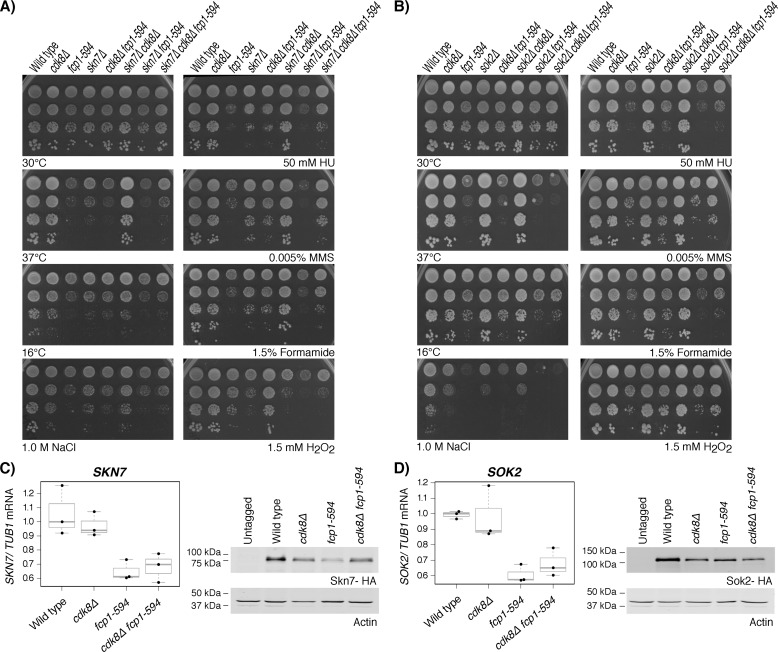
Loss of CDK8 suppressed phenotypes associated with the fcp1-594 and skn7Δ mutant
Collectively, our high-throughput data suggested a link between FCP1 and CDK8 in SKN7 and SOK2 function. To explore this relationship further, single, double, and triple mutants of fcp1-594, cdk8Δ and either skn7Δ or sok2Δ were generated, and their phenotypes were analyzed by measuring growth under the conditions described above (Fig. 5, A and B). Under these conditions, the cdk8Δ mutant generally produced phenotypes typical of the WT strain, whereas the skn7Δ mutant was sensitive to most of the conditions tested, including high temperature, H2O2, and NaCl, effects consistent with known roles for Skn7 (Fig. 5A) (43–45). Interestingly, across most conditions tested, loss of CDK8 suppressed the fcp1-594 and skn7Δ mutant growth defects, underscoring a functional relationship. We note that the suppression of skn7Δ mutant phenotypes by loss of CDK8 required FCP1. In contrast, the suppression of fcp1-594 mutant phenotypes by loss of CDK8 was generally independent of SKN7, indicating that a functional relationship between FCP1 and CDK8 likely involved multiple pathways, some of which are independent of SKN7. A notable exception to this pattern was observed under oxidative stress conditions, wherein suppression of fcp1-594 mutant phenotypes by loss of CDK8 required SKN7. In contrast, the sok2Δ mutant showed little or no sensitivity to all of the conditions examined (Fig. 5B), indicating minimal genetic relationship with FCP1 and/or CDK8.
Figure 5.
Loss of CDK8 suppressed fcp1-594 and skn7Δ mutant growth defects. Shown is the sensitivity of cdk8Δ, fcp1-594, and either skn7Δ (A) or sok2Δ (B) single, double, and triple mutants to growth under high (37 °C) and low (16 °C) temperatures, and upon exposure to the indicated concentrations of H2O2, NaCl, hydroxyurea, formamide, and methyl methanesulfonate. Loss of CDK8 suppressed the growth defects of both the fcp1-594 and skn7Δ single mutants, suggesting a shared function. Shown are RT-qPCR measurements (left) of SKN7 (C) and SOK2 (D) mRNA levels in WT or the indicated mutants normalized to TUB1 (75). mRNA levels are shown as box plots displaying the median and interquartile range, with whiskers denoting 1.5 times the interquartile range. Immunoblotting (right) of Skn7 (C) and Sok2 (D) bulk protein levels revealed that loss of CDK8 increased Skn7 protein levels in the fcp1-594 mutant background. Extracts were prepared from the indicated strains, and actin was used as a loading control.
To gain further insight into the relationship between FCP1 and CDK8 in SKN7 or SOK2 function, we next examined how these factors affected their mRNA and/or protein levels. As described above, the fcp1-594 mutant decreased the mRNA and protein levels of both SKN7 and SOK2, an effect that differed from the cdk8Δ mutant strain, which showed decreased protein levels of both TFs but no effects on their mRNA levels (Fig. 5, C and D). Consistent with the genetic dependences described above, loss of CDK8 alleviated the decreased Skn7 protein levels observed in the fcp1-594 mutant, although it did not alter Sok2 protein levels or the mRNA levels of either of these TFs. Finally, we note that the effects of FCP1 and CDK8 on Skn7 were specific, given that the trends described above were not observed on the mRNA or protein levels of Yap1, Skn7's intimate functional partner (Fig. S2).
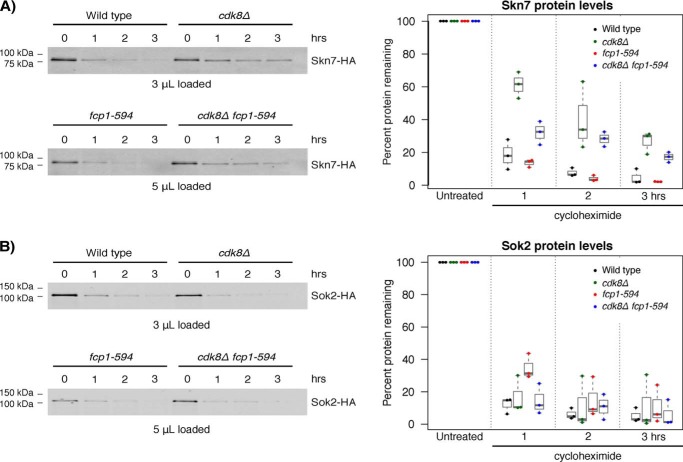
Altered CDK8 and FCP1 function led to changes in Skn7 protein stability
The discrepancy between SKN7 and SOK2 mRNA and protein levels observed in the cdk8Δ mutant suggested that CDK8 might affect Skn7 and or Sok2 protein stability, an effect reminiscent of CDK8's well-known role in phosphorylating and destabilizing Ste12, Phd1, and Gcn4 (30, 31, 46). Taking advantage of the ability to inhibit protein synthesis using cycloheximide, we measured Skn7 and Sok2 stability in WT and the fcp1-594 and cdk8Δ single and double mutants. Under WT conditions, Skn7 and Sok2 were almost completely degraded 3 h after blocking protein production, leading to estimates of half-life for both TFs at roughly 30 min (Fig. 6, A and B). Strikingly, in the cdk8Δ mutant, Skn7 protein levels decayed at a slower rate, revealing a role for CDK8 in Skn7 protein turnover. In contrast, the fcp1-594 mutant on its own had no effect on Skn7 protein turnover, but when combined with the cdk8Δ mutant, it decreased Skn7 protein stability to a level between the WT and cdk8Δ mutant, an effect most noticeable 1 and 3 h after inhibition of protein synthesis. As such, our results suggested that CDK8 promoted Skn7 turnover in a manner that was partly dependent on FCP1. Finally, no effects of the fcp1-594 or cdk8Δ mutants were seen on Sok2 protein stability; thus, it was not explored further.
Figure 6.
CDK8 and FCP1 altered Skn7 protein stability. Left, representative immunoblots of Skn7 (A) or Sok2 (B) protein levels before and after inhibition of protein synthesis by the addition of 100 μg/ml cycloheximide. As indicated, protein loading was adjusted in the fcp1-594 and fcp1-594 cdk8Δ mutant with the goal of having similar starting protein amounts. Right, quantification of Skn7 (A) or Sok2 (B) immunoblots with protein levels expressed as a percentage of the untreated control. Loss of CDK8 stabilized Skn7 protein levels in vivo. Normalized protein levels are shown as box plots displaying the median and interquartile range, with whiskers denoting 1.5 times the interquartile range.
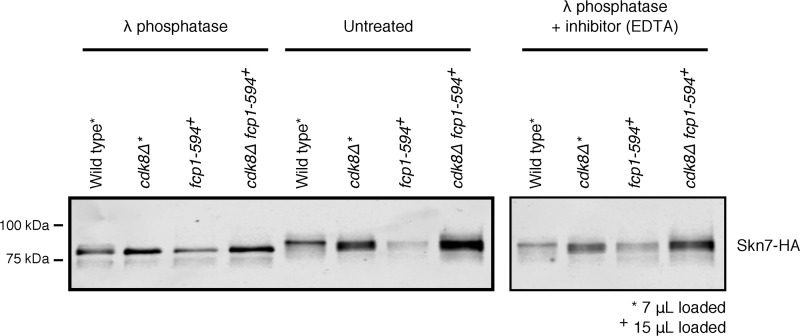
CDK8 and FCP1 regulated Skn7 phosphorylation
A role for CDK8 and FCP1 on Skn7 protein stability prompted us to determine whether this involved an effect on Skn7 phosphorylation in vivo. In the WT strain, treatment of Skn7, purified by immunoprecipitation, with λ-phosphatase resulted in faster migration compared with untreated or λ-phosphatase plus inhibitor–treated samples, indicating that Skn7 was generally phosphorylated under normal growth conditions, consistent with previous reports (Fig. 7) (42). In the fcp1-594 and cdk8Δ single and double mutants, Skn7 migrated as an intermediate species, indicating a role for these enzymes on Skn7 phosphorylation in vivo. Observing an intermediate effect for the cdk8Δ mutant was consistent with evidence that other kinases are involved in Skn7 phosphorylation (47).
Figure 7.
Phosphorylation of Skn7 depended on CDK8 and FCP1. Skn7 purified by immunoprecipitation was untreated or treated with 200 units of λ-phosphatase for 1 h in the presence or absence of 100 mm EDTA inhibitor, as indicated. Slower Skn7 migration was observed in untreated samples and in samples treated with λ-phosphatase plus EDTA phosphatase inhibitor, indicating the presence of phosphorylation. Reduced but not completely abolished phosphorylation was observed in the fcp1-594, cdk8Δ, and fcp1-594 cdk8Δ mutants compared with WT. Protein loading was adjusted as indicated.
A role for CDK8 in regulating the levels of Skn7 phosphorylation, combined with previous reports of a direct physical interaction between Cdk8 and Skn7 (48), suggested that Cdk8 may directly phosphorylate Skn7. To test this hypothesis, in vitro kinase reactions were performed with recombinant Skn7 as substrate for WT or kinase-inactive Cdk8 (D290A (49)) recovered from yeast by immunoprecipitation. Under these conditions, WT Cdk8, but not the kinase inactive mutant, robustly phosphorylated the RNAPII-CTD, a previously reported substrate (Fig. S3) (49–51). Under the same conditions, we detected no phosphorylation of Skn7 by Cdk8, suggesting that its effect in vivo may reflect an indirect mechanism or that Skn7 phosphorylation requires an additional co-factor that is absent from the in vitro reactions.
FCP1 and CDK8 functioned in the regulation of Skn7-dependent oxidative stress–responsive genes
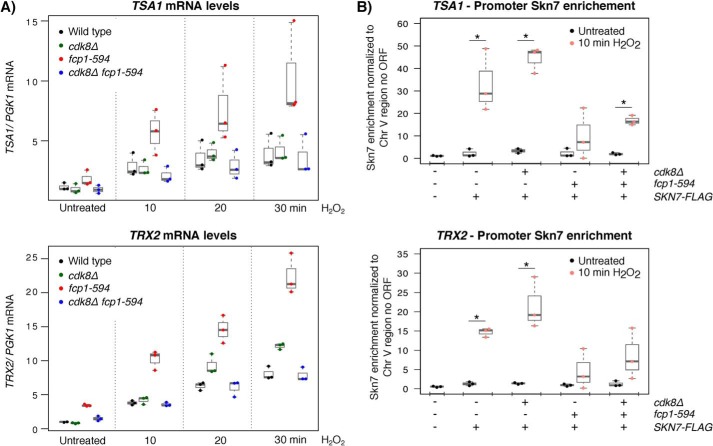
The results presented so far revealed a role for CDK8 and FCP1 in the regulation of Skn7, leading us to examine whether these factors also functioned in the transcriptional regulation of oxidative stress–responsive genes. To this end, we focused on two representative and well-established Skn7-dependent oxidative stress-induced genes, TRX2 and TSA1, and measured their mRNA levels using RT-qPCR in untreated cells and following exposure to 0.2 mm H2O2. Consistent with previous findings, the expression of TRX2 and TSA1 was induced in response to oxidant (52, 53), an effect also seen in the fcp1-594 and cdk8Δ single and double mutants (Fig. 8A). However, we note that the fcp1-594 mutant robustly increased the basal and induced mRNA levels of both TSA1 and TRX2, an effect not easily reconcilable with the decreased levels of Skn7 observed in the same strain. Under oxidative stress conditions, the robust increases of TRX2 and TSA1 mRNA levels observed in the fcp1-594 mutant were not a result of bypassing the requirement for SKN7, as no increase was observed in a strain carrying a skn7 deletion in combination with the fcp1-594 mutant or any of the other mutant backgrounds (Fig. S4A). Most interestingly and consistent with shared roles for FCP1 and CDK8 in the regulation of Skn7, loss of CDK8 in an fcp1-594 mutant background normalized the elevated TRX2 and TSA1 mRNA levels observed in the fcp1-594 mutant under basal and induced conditions (Fig. 8A and Fig. S4B). Furthermore, the effect of CDK8 on TSA1 and TRX2 mRNA levels depended on its kinase activity, given that introduction of a catalytically inactive CDK8D290A allele decreased the induced mRNA levels of TSA1 and TRX2 in the fcp1-594 cdk8Δ mutant compared with a WT CDK8 allele (Fig. S4B).
Figure 8.
FCP1 altered expression of TSA1 and TRX2 in a CDK8-dependent manner. A, RT-qPCR analysis of TSA1 (top) and TRX2 (bottom) mRNA levels with and without H2O2 treatment normalized to PGK1 mRNA levels. The elevated TRX2 and TSA1 mRNA levels detected in the fcp1-594 mutant were normalized by loss of CDK8. B, ChIP-qPCR analysis of Skn7 at the promoter of TSA1 (top) and TRX2 (bottom). Skn7 levels were normalized to an intergenic region of chromosome V (72). The fcp1-594 mutant reduced Skn7 enrichment at the promoter of TRX2 and TSA1 upon induction. *, p < 0.05 using a two-tailed Student's t test. RT-qPCR and ChIP-qPCR results are shown as box plots displaying the median and interquartile range, with whiskers denoting 1.5 times the interquartile range.
The unexpected effect of the fcp1-594 mutant on the expression of Skn7-dependent genes led us to examine Skn7 protein levels under oxidative stress conditions. Here, we found that oxidative stress recapitulated the observations under normal growth conditions, where the fcp1-594 mutant showed decreased Skn7 protein levels under basal and oxidative stress conditions that were normalized by loss of CDK8. Thus, in the fcp1-594 mutant, elevated TSA1 and TRX2 mRNA levels were dependent on SKN7, despite its protein levels being decreased in the fcp1-594 mutant under basal and oxidative stress conditions.
To understand the discrepancy between Skn7 protein levels and expression of its target genes in the fcp1-594 mutant, we measured RNAPII and Skn7 occupancy at these loci via ChIP of endogenous Rpb3 and Skn7-FLAG, respectively. Consistent with the increase in TRX2 and TSA1 mRNA levels upon exposure to 0.2 mm H2O2 and Skn7's role as a transcriptional activator (52, 54), we observed increased RNAPII and Skn7 occupancy at these loci upon treatment in the WT strain, an effect most striking at the TSA1 locus (Fig. 8B and Fig. S5 (B and C)). For RNAPII, the fcp1-594 and cdk8Δ single and double mutants also showed increased RNAPII occupancy upon exposure to H2O2, indicating increased transcription in response to stress. However, we note that whereas the cdk8Δ and fcp1-594 cdk8Δ mutants tended to increase RNAPII occupancy at the promoter and along the length of TRX2 and TSA1, the fcp1-594 mutant showed a more pronounced effect along the length of these genes compared with the promoters. For Skn7, a significant increase was also observed in the cdk8Δ mutant upon exposure to H2O2, an effect that tended to be higher compared with WT and was consistent with the observed stabilizing effect of the cdk8Δ mutant on Skn7. Most unexpected was the effect of the fcp1-594 mutant, which despite robustly increasing TRX2 and TSA1 mRNA levels in a SKN7-dependent manner, showed no significant increase in Skn7 occupancy when comparing basal and inducing conditions. Loss of CDK8 in the fcp1-594 mutant background had limited effects on Skn7 occupancy at both the TSA1 and TRX2 promoters. Here, Skn7 occupancy remained low upon exposure to H2O2 compared with WT. However, we note that at the TSA1 locus, loss of CDK8 in the fcp1-594 mutant background led to a significant difference between the uninduced and induced conditions.
The low Skn7 occupancy at the TSA1 and TRX2 promoter that we observed in the fcp1-594 mutant prompted us to examine whether the gene expression alterations were linked to defects in transcription, which have been reported to increase the mRNA levels of stress-responsive genes through a combination of transcriptional and posttranscriptional effects (55). Here, we showed that inhibiting transcription by the addition of 100 μg/ml 1,10-phenanthroline increased TSA1 and TRX2 mRNA levels in the WT strain to levels similar to the untreated fcp1-594 mutant (Fig. S6). Thus, defects in transcription in the fcp1-594 mutant may in part underpin the gene expression alterations observed in this mutant under basal conditions. Comparing the effect of inhibiting transcription on the fcp1-594 and cdk8Δ single and double mutants revealed that TSA1 mRNA levels decreased in the fcp1-594 mutant, increased in the cdk8Δ mutant, and more closely resembled the WT in the fcp1-594 cdk8Δ double mutant. Different patterns were observed for TRX2, suggesting that the effect of FCP1 and CDK8 in regulating the balance of transcriptional and posttranscriptional events was gene-specific.
Discussion
This study describes an unexpected role for FCP1 and CDK8 in the regulation of Skn7 and the expression of its target genes. We show that the genetic relationship between FCP1 and CDK8 upon exposure to oxidant depended on SKN7 and that these factors modulated Skn7 protein levels, stability, and expression of target genes. Paradoxically, whereas the fcp1-594 mutant increased expression of Skn7-dependent oxidative stress-induced genes, it decreased Skn7 promoter occupancy; the former was overcome by loss of CDK8 likely through the stabilization of Skn7. As such, by probing the genes most sensitive to altered Fcp1 activity, our findings underscored a novel connection to CDK8 in the regulation of Skn7 and the expression of its target loci.
Most defects of Skn7 biology observed in the fcp1-594 mutant were normalized by loss of CDK8, revealing a shared function in the regulation of Skn7 and expanding well-known activities in the regulation of TFs and the transcriptional response to environmental stress (30, 31, 46, 56). Here, we observed that loss of CDK8 normalized the growth defects of both the fcp1-594 and skn7Δ mutants across a range of stress conditions, an effect that may be the result of the reported activation of stress-responsive genes upon loss of CDK8 under basal conditions (57). Our findings also complemented previous work implicating CDK8 in the biology of Skn7 as well as the oxidative stress response (48, 58–60). For instance, it has been shown that loss of CDK8 can normalize the increased mRNA levels of OCH1, a Skn7-dependent osmotic stress-response gene, observed in a not4Δ mutant (which encodes a ubiquitin ligase subunit of the Ccr4-Not complex) (48). However, we note that whereas loss of NOT4 increased Skn7 occupancy at the OCH1 gene promoter (48), we observed that the fcp1-594 mutant decreased Skn7 occupancy at the TRX2 and TSA1 promoters, suggesting that loss of CDK8 may suppress Skn7-associated phenotypes in the not4Δ and fcp1-594 mutant through distinct functional pathways. Cdk8 has also been implicated in the oxidative stress response by priming the degradation of Med13/Srb9, a Mediator kinase domain subunit (58). In fact, many Mediator subunits, including components of the kinase module (e.g. Med13 and CycC) have been implicated in the oxidative stress response and shown to be degraded upon oxidant exposure (58–60). Given that Cdk8 stability does not seem to be altered by exposure to oxidant, we hypothesize that the role described here may underscore a Mediator-independent activity for Cdk8.
Here, we report a role for CDK8 in influencing Skn7 phosphorylation, protein levels, promoter occupancy, and expression of oxidative stress–induced genes, findings that are remarkably similar to CDK8's effect on its reported direct TF substrates (30, 31, 46, 56, 61). Despite the similarities, our kinase assays did not support a direct kinase–substrate relationship between Cdk8 and Skn7, suggesting that either our kinase assays were missing important factors required for Skn7 phosphorylation or that the effects may be through indirect mechanisms. Consistent with the former, we note that Skn7 phosphorylation is complex, requiring Sln1 for phosphorylation by Ypd1 in vitro (47) and Yap1 for phosphorylation upon oxidative stress in vivo (62). Although some questions remain, Skn7 phosphorylation clearly is important for its activity as a transcriptional activator (62). At osmotic stress genes, phosphorylation of Asp-427 is required for their activation (45). During oxidative stress, Asp-427 phosphorylation is dispensable (52); although there is evidence that a number of Skn7 residues are phosphorylated under these conditions (62), their exact identity and responsible kinases remain unknown. Intriguingly, here we linked Skn7 phosphorylation and function in the oxidative stress response to Cdk8, a kinase that can bind Skn7 (48), and Fcp1, a protein phosphatase. However, their effects on Skn7 biology were not easily explained by direct phosphorylation and dephosphorylation by these factors. Therefore, it is formally possible that the effects of FCP1 and CDK8 on Skn7 function may be independent of its phosphorylation state. However, given that many sites on Skn7 have been suggested to undergo phosphorylation, it is also possible that the effect of FCP1 and CDK8 on Skn7 phosphorylation differ in a manner not readily detected by bulk shifts in migration patterns but that nonetheless result in functional differences to Skn7.
FCP1 has been previously implicated in the response to other environmental stressors. In fly-derived cell lines, FCP1 knockdown leads to decreased expression and RNAPII association at heat shock genes (63), effects that contrasted the increased mRNA levels and RNAPII occupancy at oxidative stress response genes observed in yeast for the fcp1-594 mutant. Although potentially indicative of species-specific activities, our yeast growth assays supported a distinct role for Fcp1 in the oxidative and heat shock response, showing that the sensitivity of the fcp1-594 mutant to oxidative stress but not heat shock was suppressed by loss of CDK8. Altogether, here we show a role for Fcp1 in the transcriptional response to other environmental stressors while also revealing an ability to function as a transcriptional inhibitor, a particularly unexpected finding, given its well-established roles in facilitating transcription (6, 10).
Experimental procedures
Yeast strains
All yeast strains are listed in Table 1. They were primarily generated in the W303 background, with the exception of strains used for microarray expression profiling and E-MAP analysis, which were derived from a BY4742 background, the standard genotype for those assays. Complete or partial gene deletions and the integration of the 3XFLAG or HA tags were done using the one-step gene replacement method to integrate PCR-amplified segments (64). All double and triple mutant strains were generated via mating and tetrad dissection. All plasmids used are listed in Table 2.
Table 1.
Strains used in this study
| Strain | Genotype | Background | Source |
|---|---|---|---|
| 1810 | Mata fcp1–594-flag::nat his3Δ1 leu2Δ0 ura3Δ0 | BY4742 | This study; gene expression |
| 1811 | Mata fcp1–609-flag::nat his3Δ1 leu2Δ0 ura3Δ0 | BY4742 | This study; gene expression |
| 1812 | Mata fcp1–666-flag::nat his3Δ1 leu2Δ0 ura3Δ0 | BY4742 | This study; gene expression |
| 1813 | Mata fcp1–713-flag::nat his3Δ1 leu2Δ0 ura3Δ0 | BY4742 | This study; gene expression |
| 1814 | Mata FCP1-WT-flag::nat his3Δ1 leu2Δ0 ura3Δ0 | BY4742 | This study; gene expression |
| 1815 | Matα fcp1–594-flag::nat his3Δ 1 leu2Δ 0 LYS2+ met15Δ0 ura3Δ0 Δcan1::MATaPr-HIS3 Δlyp1::MATα Pr-LEU2 | BY4742 | This study; E-MAP |
| 1816 | Matα fcp1–609-flag::nat his3Δ 1 leu2Δ 0 LYS2+ met15Δ0 ura3Δ0 Δcan1::MATaPr-HIS3 Δlyp1::MATα Pr-LEU2 | BY4742 | This study; E-MAP |
| 1817 | Matα fcp1–666-flag::nat his3Δ 1 leu2Δ 0 LYS2+ met15Δ0 ura3Δ0 Δcan1::MATaPr-HIS3 Δlyp1::MATα Pr-LEU2 | BY4742 | This study; E-MAP |
| 1818 | Matα fcp1–713-flag::nat his3Δ 1 leu2Δ 0 LYS2+ met15Δ0 ura3Δ0Δcan1::MATaPr-HIS3 Δlyp1::MATα Pr-LEU2 | BY4742 | This study; E-MAP |
| 1819 | Matα FCP1-WT-flag::nat his3Δ 1 leu2Δ 0 LYS2+ met15Δ 0 ura3Δ 0 Δcan1::MATaPr-HIS3 Δlyp1::MATα Pr-LEU2 | BY4742 | This study; E-MAP |
| 1820 | Matα fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1821 | Matα fcp1-609-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1822 | Matα fcp1-666-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1823 | Matα fcp1-713-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1824 | Matα FCP1-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1825 | Matα Skn7-HA::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1826 | Matα Skn7-HA::kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1827 | Matα Skn7-HA::kan fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1828 | Matα Skn7-HA::kan cdk8::his fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1829 | Matα Sok2-HA::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1830 | Matα Sok2-HA::kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1831 | Matα Sok2-HA::kan fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1832 | Matα Sok2-HA::kan cdk8::his fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1833 | Matα Cad1-flag::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1834 | Matα Cad1-flag::kan fcp1-594-HA::hygro ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1835 | Matα Mcm1-flag::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1836 | Matα Mcm1-flag::kan fcp1-594-HA::hygro ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1837 | Matα Yap1-HA::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1907 | Matα Yap1-HA::kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1838 | Matα Yap1-HA::kan fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1845 | Matα Yap1-HA::kan cdk8::his fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1839 | Matα skn7::Kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1840 | Matα skn7::Kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1841 | Matα skn7::Kan fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1842 | Matα skn7::Kan cdk8::his fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1901 | Matα sok2::Kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1902 | Matα sok2::Kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1903 | Matα sok2::Kan fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1904 | Matα sok2::Kan cdk8::his fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1905 | Matα cdk8::His ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1906 | Matα cdk8::His fcp1-594-flag::nat ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 5 | Matα ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | |
| 1908 | Matα Skn7-flag::kan ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1909 | Matα Skn7-flag::kan cdk8::his ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1910 | Matα Skn7-flag::kan fcp1-594-HA::hygro ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| 1911 | Matα Skn7-flag::kan cdk8::his fcp1-594-HA::hygro ade2-1 can1-100 his3-11 leu2-3, 112 trp 1-1 ura3-1 LYS2 | W303 | This study |
| YCN44 | Mata ura3-52 cdk8::URA3 | Σ1278b | Ref. 74 |
Table 2.
Plasmids used in this study
Growth assays
Overnight cultures grown in YPD were diluted to 0.5 OD600, 10-fold serially diluted, and spotted onto YPD plates with or without the indicated amount of HU (Sigma), MMS (Sigma), formamide (Sigma), hydrogen peroxide (H2O2) (Sigma), or sodium chloride (NaCl) (Bioshop). Plates were incubated at the indicated temperatures for 3–5 days.
Microarray experiments
Microarray expression profiling was performed in duplicate as described previously (65, 66). Cultures were grown in a 24-well plate incubator/reader to mid-log phase, with spiked-in RNA controls to monitor global changes in mRNA levels. Because no global changes in mRNA levels were detected, expression values were normalized to total mRNA levels. -Fold changes were determined by comparing mutant with WT profiles, with differentially expressed genes having a p value < 0.01 and absolute -fold change > 1.7 (67).
Data analysis
TF enrichment was done using the Yeast Promoter Atlas database and a hypergeometric test with Bonferroni correction (adjusted p value of 0.05) (29). The gene expression profile for the cdk8Δ mutant was reported previously (33). TFs with a score greater than 6.4 in the YeastKID database were categorized as phosphorylated, and a hypergeometric test was used to test for significance (p = 0.03355) (35). Complete gene expression profiles can be found in GSE128936.
Protein extraction and protein blotting
Overnight cultures were diluted to 0.3 OD600 and grown to 1.0 OD600. Whole-cell extracts were prepared by glass bead lysis in the presence of TCA. Immunoblotting was done using anti-HA (Sigma), anti-FLAG (Sigma), or actin (ICN) antibodies. Immunoblots were scanned with the Odyssey IR imaging system (LI-COR).
RT-qPCR
Overnight cultures were diluted to 0.15 OD600 and grown to 0.5 OD600. RNA was extracted and purified using the Qiagen RNeasy minikit. cDNA was generated using the Qiagen QuantiTect reverse transcription kit and analyzed using the PerfeCTa SYBR Green FastMix (VWR) and a Rotor-Gene 6000 (Qiagen). All strains were grown in YPD, except for strains carrying pRS314-based vectors, which were grown in −TRP medium. 0.2 mm H2O2 or 100 μg/ml 1,10-phenanthroline were used to treat cells for the indicated amount of time. TUB1, PKG1, RIB5, and SNR19 were used as control genes as indicated. The choice of control gene depended on the stability of the gene in the condition tested. PKG1 was used for experiments involving oxidative stress, and a composite of RIB5 (68) and SNR19 (69) was used for experiments involving transcriptional inhibition. mRNA levels were quantified from three independent biological replicates. All primer sequences used are listed in Table 3.
Table 3.
Primers used in this study
| Primer name | Forward sequence | Reverse sequence | Source or reference |
|---|---|---|---|
| SKN7 RT-qPCR | GCGACGGTCTTTCAGCTATC | AAATCATCCCTCGTGAATGG | This study |
| SOK2 RT-qPCR | GGGTGTCTGGATACCGTTTG | ATAGGTACGCGGACTGATGG | This study |
| MCM1 RT-qPCR | CATCCTCGACCACAGCAAG | GGCAAAAGCCTGTTGGTG | This study |
| CAD1 RT-qPCR | CTTTATTCCCCAGCGTGCTT | GGAAGCACCGAAGCTAGAGA | This study |
| TUB1 RT-qPCR | TCTTGGTGGTGGTACTGGTT | TGGATTTCTTACCGTATTCAGCG | Ref. 75 |
| TSA1 3′ and RT-qPCR | GGCTGACACCAACCACTCTT | TTGGAAGGCTTCAACCAATC | This study |
| TRX2 3′ and RT-qPCR | AGCTGAAGTTTCTTCCATGCCT | TTGATAGCAGCTGGGTTGGC | This study |
| PGK1 RT-qPCR | CAAGAGCTCTGCTGCTGGTA | AAAGCAACACCTGGCAATTC | This study |
| TSA1 promoter | TGGCAACAAACCAGGACATA | TGTGGTTGGTTGTGAGCAAT | This study |
| TRX2 promoter | CCATTCGGGGGATGAAAAG | TCGTAGACTCTCGTGTATGTGTG | This study |
| Chr V | GGCTGTCAGAATATGGGGCCGTAGTA | CACCCCGAAGCTGCTTTCACAATAC | Ref. 72 |
| RIB5 RT-qPCR | CGCAAGGTGGAGCCTTCTAT | TGCTTCTCAATAATCTTCCCAGT | This study |
| SNR19 RT-qPCR | ACTTTTCTCTAGCGTGCCAT | CTTCAAACTACAATCCCGACCA | This study |
E-MAP
E-MAP screens were performed as described previously (36). Briefly, FCP1 mutants were crossed, using a Singer robot, to a library of 1536 mutants, including full gene deletions or DAmP (decrease abundance by mRNA perturbation) alleles. Mating, diploid selection, sporulation, haploid selection, and double mutant selection were done by replicate plating on selective media, as described previously. All strains were screened in triplicate, and normalization was performed as described previously (36). S scores ≥2 and ≤−2.5 were considered significant.
Protein stability assay
Overnight cultures were diluted to 0.3 OD600 and grown to 1.0 OD600. 10 OD600 units were collected prior to the addition of 100 μg/ml cycloheximide (Sigma). Following protein shutdown, 10 OD600 units were collected at the indicated times. All proteins were extracted using TCA as described above. Due to the differences in starting protein levels in the FCP1 mutant, protein loading was adjusted as indicated in each figure. Signal intensities from each lane were used to calculate percentage protein levels relative to the untreated control.
Immunoprecipitation (IP) and phosphatase treatment
The phosphatase treatment protocol was adapted from Flott and Rouse (70). Overnight cultures were diluted to 0.3 OD600 and grown to 1.0 OD600. Samples were mechanically lysed in TAP-IP buffer (50 mm Tris (pH 7.8), 150 mm NaCl, 1.5 mm MgAc, 10% glycerol, 0.15% Nonidet P40, 1 mm DTT) with phosphatase inhibitors (10 mm NaPPi, 5 mm EGTA, 5 mm EDTA, 0.1 mm Na3VO4, and 5 mm NaF) and Complete EDTA-free protease inhibitor mixture (Roche Applied Science). Lysates were split equally into three tubes containing 40 μl of HA beads (Sigma) and incubated for 1.5 h. Samples were washed twice with TAP-IP buffer with phosphatase and protease inhibitors and twice with TAP-IP buffer lacking phosphatase and protease inhibitors prior to resuspending in 1× NEBuffer for protein metallophosphatases (50 mm HEPES, 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 1 mm MnCl2). 200 units of λ-phosphatase (New England BioLabs) and 100 mm EDTA was added as indicated and incubated for 60 min at 30 °C. Samples were washed twice using Tandem Affinity Purification (TAP)-IP buffer without phosphatase and protease inhibitors and resuspended in 2× SDS buffer prior to analysis by SDS-PAGE. Protein loading was adjusted as indicated in an effort to ensure that the differences in protein migration were not influenced by changes in the amount of protein in each sample.
Recombinant protein expression
An N-terminally 6×HIS-tagged SKN7 was cloned into a pET28a derivative and expressed from Escherichia coli BL21 (DE3)-RIL cells. Cells were induced at 0.6 OD600 for 4 h at 37 °C using 1.0 mm isopropyl 1-thio-β-d-galactopyranoside, harvested by centrifugation, and lysed in buffer containing 25 mm Tris-Cl (pH 7.5), 0.5 m NaCl, 30 mm imidazole, and 1 mg/ml lysozyme. Skn7 protein was purified by passing the supernatant over a HisTrap FF (5 ml) column (GE Healthcare) and eluted using 0.5 m imidazole.
In vitro kinase assays
The in vitro kinase assay protocol was adapted from Hirst et al. (56). Strains from a Σ1278b background, expressing either WT or kinase-impaired FLAG-tagged CDK8 (D290A), were grown to 1.0 OD600. Cells were collected and mechanically lysed in kinase lysis buffer (KLB) (50 mm Tris, 200 mm NaCl, 5 mm EDTA, 0.1% Nonidet P40, and protease inhibitors). Cdk8 was immunoprecipitated by incubating cell lysates with anti-FLAG M2 beads for 60 min, followed by two washes with KLB and two washes with kinase buffer (10 mm MgCl2, 50 mm Tris (pH 7.5), 1 mm DTT, and protease inhibitors). Kinase reactions were performed for 20 min at 30 °C in kinase buffer, with 3.33 pmol of [γ-32P]ATP (PerkinElmer Life Sciences) and 1 μg of the indicated substrate. Reactions were run on a 10% SDS-polyacrylamide gel and exposed on Kodak Biomax film.
ChIP
Overnight cultures were diluted to 0.15 OD600, grown to 0.5 OD600, and cross-linked or induced using 0.2 mm H2O2 for 10 min prior to cross-linking with 1% formaldehyde for 20 min. Chromatin was prepared as described previously (71). Anti-FLAG antibody (4.2 μl) (Sigma) or anti-Rpb3 antibody (7 μl) (Neoclone) was coupled to 60 μl of Protein A Dynabeads (Invitrogen) overnight. Following cross-linking reversal and DNA purification, both the immunoprecipitated and input DNA were analyzed by qPCR using a Rotor-Gene 6000 and PerfeCTa SYBR Green FastMix (Quanta Biosciences). All samples were analyzed from three independent biological replicates and normalized to an intragenic region of Chromosome V (72). Primers used are listed in Table 3.
ChIP-on-chip analysis
Cdk8 ChIP-on-chip profiles were reported previously (35). Average scores were obtained by averaging all probes mapping to 500 bp upstream of the ORF. TF target gene pairs were obtained from the YEASTRACT database using the DNA binding and expression evidence category (73). Genes that are regulated by more than one TF were included in the respective box plots.
Author contributions
M. J. A., K. D., and M. S. K. conceptualization; M. J. A. data curation; M. J. A., G. L. N., and J. J. B. formal analysis; M. J. A., F. C. H., N. J. K., I. S., and M. S. K. supervision; M. J. A. and M. S. K. funding acquisition; M. J. A., K. D., M. S., N. H., J. J. B., F. C. H., N. J. K., and I. S. investigation; M. J. A. visualization; M. J. A., K. D., F. C. H., N. J. K., and I. S. methodology; M. J. A. and K. D. writing-original draft; M. J. A., G. L. N., F. C. H., N. J. K., I. S., and M. S. K. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Emanuel Rosonina for insightful discussions, Dr. Alexandre Lussier for critical reading of the manuscript, Alyssa Kirlin for assistance with the purification of Skn7 and critical reading of the manuscript, Marian Groot Koerkamp for help with the microarray data submission, and Konstantin Mestnikov for help with the 1,10-phenanthroline exposure experiment.
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant RPGIN-2016–04297 (to M. S. K.), a PDF fellowship (to M. J. A.), and a USRA studentship (to M. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
Microarray data were submitted to the Gene Expression Omnibus under accession number GSE128936.
- RNAPII
- RNA polymerase II
- CTD
- C-terminal domain
- FCPH
- FCP-homology domain
- BRCT
- BRCA1 C terminus domain
- TFIIF and TFIIB
- general transcription factor IIF and IIB, respectively
- MMS
- methyl methanesulfonate
- HU
- hydroxyurea
- E-MAP
- epistasis mini-assay profiling
- TF
- transcription factor
- qPCR
- quantitative PCR
- HA
- hemagglutinin
- OD
- optical density
- IP
- immunoprecipitation
- KLB
- kinase lysis buffer
- TAP
- tandem affinity purification
- IQR
- interquantile range.
References
- 1. Allison L. A., and Ingles C. J. (1989) Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc. Natl. Acad. Sci. U.S.A. 86, 2794–2798 10.1073/pnas.86.8.2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nonet M., Sweetser D., and Young R. A. (1987) Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50, 909–915 10.1016/0092-8674(87)90517-4 [DOI] [PubMed] [Google Scholar]
- 3. Corden J. L., Cadena D. L., Ahearn J. M. Jr., and Dahmus M. E. (1985) A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 82, 7934–7938 10.1073/pnas.82.23.7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harlen K. M., and Churchman L. S. (2017) The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- 5. Zaborowska J., Egloff S., and Murphy S. (2016) The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 23, 771–777 10.1038/nsmb.3285 [DOI] [PubMed] [Google Scholar]
- 6. Kobor M. S., Archambault J., Lester W., Holstege F. C. P., Gileadi O., Jansma D. B., Jennings E. G., Kouyoumdjian F., Davidson A. R., Young R. A., and Greenblatt J. (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4, 55–62 10.1016/S1097-2765(00)80187-2 [DOI] [PubMed] [Google Scholar]
- 7. Kimura M., Suzuki H., and Ishihama A. (2002) Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell Biol. 22, 1577–1588 10.1128/MCB.22.5.1577-1588.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Archambault J., Chambers R. S., Kobor M. S., Ho Y., Cartier M., Bolotin D., Andrews B., Kane C. M., and Greenblatt J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 14300–14305 10.1073/pnas.94.26.14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghosh A., Shuman S., and Lima C. D. (2008) The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell 32, 478–490 10.1016/j.molcel.2008.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobor M. S., Simon L. D., Omichinski J., Zhong G., Archambault J., and Greenblatt J. (2000) A motif Shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 7438–7449 10.1128/MCB.20.20.7438-7449.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu X., Chini C. C., He M., Mer G., and Chen J. (2003) The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 10.1126/science.1088753 [DOI] [PubMed] [Google Scholar]
- 12. Zhang D. W., Mosley A. L., Ramisetty S. R., Rodríguez-Molina J. B., Washburn M. P., and Ansari A. Z. (2012) Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 287, 8541–8551 10.1074/jbc.M111.335687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho E.-J., Kobor M. S., Kim M., Greenblatt J., and Buratowski S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15, 3319–3329 10.1101/gad.935901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bataille A. R., Jeronimo C., Jacques P.-É., Laramée L., Fortin M.-È., Forest A., Bergeron M., Hanes S. D., and Robert F. (2012) A Universal RNA Polymerase II CTD Cycle Is Orchestrated by Complex Interplays between Kinase, Phosphatase, and Isomerase Enzymes along Genes. Molecular Cell 45, 158–170 10.1016/j.molcel.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 15. Hsin J.-P., Xiang K., and Manley J. L. (2014) Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol. Cell Biol. 34, 2488–2498 10.1128/MCB.00181-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calvo O., and Manley J. L. (2005) The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24, 1009–1020 10.1038/sj.emboj.7600575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suh M.-H., Ye P., Zhang M., Hausmann S., Shuman S., Gnatt A. L., and Fu J. (2005) Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc. Natl. Acad. Sci. U.S.A. 102, 17314–17319 10.1073/pnas.0507987102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kops O., Zhou X. Z., and Lu K. P. (2002) Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. 513, 305–311 10.1016/S0014-5793(02)02288-3 [DOI] [PubMed] [Google Scholar]
- 19. Chambers R. S., Wang B. Q., Burton Z. F., and Dahmus M. E. (1995) The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270, 14962–14969 10.1074/jbc.270.25.14962 [DOI] [PubMed] [Google Scholar]
- 20. Garavís M., González-Polo N., Allepuz-Fuster P., Louro J. A., Fernández-Tornero C., and Calvo O. (2017) Sub1 contacts the RNA polymerase II stalk to modulate mRNA synthesis. Nucleic Acids Res. 45, 2458–2471 10.1093/nar/gkw1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang B., Yang G., Chen Y., Zhao Y., Gao P., Liu B., Wang H., and Zheng Z.-L. (2016) C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast. Proc. Natl. Acad. Sci. U.S.A. 113, E8197–E8206 10.1073/pnas.1605871113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwer B., Ghosh A., Sanchez A. M., Lima C. D., and Shuman S. (2015) Genetic and structural analysis of the essential fission yeast RNA polymerase II CTD phosphatase Fcp1. RNA 21, 1135–1146 10.1261/rna.050286.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visconti R., Palazzo L., Della Monica R., and Grieco D. (2012) Fcp1-dependent dephosphorylation is required for M-phase-promoting factor inactivation at mitosis exit. Nat. Commun. 3, 894 10.1038/ncomms1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hégarat N., Vesely C., Vinod P. K., Ocasio C., Peter N., Gannon J., Oliver A. W., Novák B., and Hochegger H. (2014) PP2A/B55 and Fcp1 regulate Greatwall and Ensa dephosphorylation during mitotic exit. PLoS Genet. 10, e1004004 10.1371/journal.pgen.1004004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Della Monica R., Visconti R., Cervone N., Serpico A. F., and Grieco D. (2015) Fcp1 phosphatase controls Greatwall kinase to promote PP2A-B55 activation and mitotic progression. eLife 4, e10399 10.7554/eLife.10399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bierhoff H., Dundr M., Michels A. A., and Grummt I. (2008) Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol. Cell Biol. 28, 4988–4998 10.1128/MCB.00492-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fath S., Kobor M. S., Philippi A., Greenblatt J., and Tschochner H. (2004) Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J. Biol. Chem. 279, 25251–25259 10.1074/jbc.M401867200 [DOI] [PubMed] [Google Scholar]
- 28. Guo Z., and Stiller J. W. (2005) Comparative genomics and evolution of proteins associated with RNA polymerase II C-terminal domain. Mol. Biol. Evol. 22, 2166–2178 10.1093/molbev/msi215 [DOI] [PubMed] [Google Scholar]
- 29. Chang D. T.-H., Huang C.-Y., Wu C.-Y., and Wu W.-S. (2011) YPA: an integrated repository of promoter features in Saccharomyces cerevisiae. Nucleic Acids Res. 39, D647–D652 10.1093/nar/gkq1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raithatha S., Su T.-C., Lourenco P., Goto S., and Sadowski I. (2012) Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol. Cell Biol. 32, 664–674 10.1128/MCB.05420-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nelson C., Goto S., Lund K., Hung W., and Sadowski I. (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421, 187–190 10.1038/nature01243 [DOI] [PubMed] [Google Scholar]
- 32. Byrne K. P., and Wolfe K. H. (2005) The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15, 1456–1461 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T. J. P., van Leenen D., and Holstege F. C. P. (2005) Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19, 511–522 10.1016/j.molcel.2005.06.033 [DOI] [PubMed] [Google Scholar]
- 34. Aristizabal M. J., Negri G. L., Benschop J. J., Holstege F. C. P., Krogan N. J., and Kobor M. S. (2013) High-throughput genetic and gene expression analysis of the RNAPII-CTD reveals unexpected connections to SRB10/CDK8. PLoS Genet. 9, e1003758 10.1371/journal.pgen.1003758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharifpoor S., Nguyen Ba A. N., Young J.-Y., van Dyk D., Friesen H., Douglas A. C., Kurat C. F., Chong Y. T., Founk K., Moses A. M., and Andrews B. J. (2011) A quantitative literature-curated gold standard for kinase-substrate pairs. Genome Biol. 12, R39 10.1186/gb-2011-12-4-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Collins S. R., Roguev A., and Krogan N. J. (2010) Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 470, 205–231 10.1016/S0076-6879(10)70009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu P. L., Yang F., Smith-Kinnaman W., Yang W., Song J.-E., Mosley A. L., and Varani G. (2014) Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J. Mol. Biol. 426, 2970–2981 10.1016/j.jmb.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mosley A. L., Pattenden S. G., Carey M., Venkatesh S., Gilmore J. M., Florens L., Workman J. L., and Washburn M. P. (2009) Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34, 168–178 10.1016/j.molcel.2009.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu X., Wilcox C. B., Devasahayam G., Hackett R. L., Arévalo-Rodríguez M., Cardenas M. E., Heitman J., and Hanes S. D. (2000) The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 19, 3727–3738 10.1093/emboj/19.14.3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee J. M., and Greenleaf A. L. (1991) CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1, 149–167 [PMC free article] [PubMed] [Google Scholar]
- 41. Ng H. H., Robert F., Young R. A., and Struhl K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 10.1016/S1097-2765(03)00092-3 [DOI] [PubMed] [Google Scholar]
- 42. Mulford K. E., and Fassler J. S. (2011) Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryotic Cell 10, 761–769 10.1128/EC.00328-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krems B., Charizanis C., and Entian K.-D. (1996) The response regulator-like protein Pos9/Skn7 ofSaccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29, 327–334 10.1007/BF02208613 [DOI] [PubMed] [Google Scholar]
- 44. Raitt D. C., Johnson A. L., Erkine A. M., Makino K., Morgan B., Gross D. S., and Johnston L. H. (2000) The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11, 2335–2347 10.1091/mbc.11.7.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ketela T., Brown J. L., Stewart R. C., and Bussey H. (1998) Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259, 372–378 10.1007/s004380050824 [DOI] [PubMed] [Google Scholar]
- 46. Chi Y., Huddleston M. J., Zhang X., Young R. A., Annan R. S., Carr S. A., and Deshaies R. J. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15, 1078–1092 10.1101/gad.867501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li S., Ault A., Malone C. L., Raitt D., Dean S., Johnston L. H., Deschenes R. J., and Fassler J. S. (1998) The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17, 6952–6962 10.1093/emboj/17.23.6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lenssen E., Azzouz N., Michel A., Landrieux E., and Collart M. A. (2007) The Ccr4-Not complex regulates Skn7 through Srb10 kinase. Eukaryot. Cell 6, 2251–2259 10.1128/EC.00327-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hengartner C. J., Myer V. E., Liao S.-M., Wilson C. J., Koh S. S., and Young R. A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2, 43–53 10.1016/S1097-2765(00)80112-4 [DOI] [PubMed] [Google Scholar]
- 50. Liu Y., Kung C., Fishburn J., Ansari A. Z., Shokat K. M., and Hahn S. (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell Biol. 24, 1721–1735 10.1128/MCB.24.4.1721-1735.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liao S.-M., Zhang J., Jeffery D. A., Koleske A. J., Thompson C. M., Chao D. M., Viljoen M., van Vuuren H. J. J., Young R. A. (1995) A kinase–cyclin pair in the RNA polymerase II holoenzyme. Nature 374, 193–196 10.1038/374193a0 [DOI] [PubMed] [Google Scholar]
- 52. Morgan B. A., Banks G. R., Toone W. M., Raitt D., Kuge S., and Johnston L. H. (1997) The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16, 1035–1044 10.1093/emboj/16.5.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee J., Godon C., Lagniel G., Spector D., Garin J., Labarre J., and Toledano M. B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16046 10.1074/jbc.274.23.16040 [DOI] [PubMed] [Google Scholar]
- 54. He X.-J., and Fassler J. S. (2005) Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 58, 1454–1467 10.1111/j.1365-2958.2005.04917.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., and Hughes T. R. (2004) Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell Biol. 24, 5534–5547 10.1128/MCB.24.12.5534-5547.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hirst M., Kobor M. S., Kuriakose N., Greenblatt J., and Sadowski I. (1999) GAL4 is regulated by the RNA polymerase II holoenzyme–associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3, 673–678 10.1016/S1097-2765(00)80360-3 [DOI] [PubMed] [Google Scholar]
- 57. Holstege F. C. P., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., and Young R. A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 10.1016/S0092-8674(00)81641-4 [DOI] [PubMed] [Google Scholar]
- 58. Willis S. D., Stieg D. C., Ong K. L., Shah R., Strich A. K., Grose J. H., and Cooper K. F. (2018) Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress. Microbial Cell 5, 357–370 10.15698/mic2018.08.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cooper K. F., Scarnati M. S., Krasley E., Mallory M. J., Jin C., Law M. J., and Strich R. (2012) Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 125, 1015–1026 10.1242/jcs.096479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krasley E., Cooper K. F., Mallory M. J., Dunbrack R., and Strich R. (2006) Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 172, 1477–1486 10.1534/genetics.105.052266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosonina E., Duncan S. M., and Manley J. L. (2012) Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev. 26, 350–355 10.1101/gad.184689.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. He X.-J., Mulford K. E., and Fassler J. S. (2009) Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot. Cell 8, 768–778 10.1128/EC.00021-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fuda N. J., Buckley M. S., Wei W., Core L. J., Waters C. T., Reinberg D., and Lis J. T. (2012) Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes. Mol. Cell Biol. 32, 3428–3437 10.1128/MCB.00247-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 65. Lenstra T. L., Benschop J. J., Kim T., Schulze J. M., Brabers N. A. C. H., Margaritis T., van de Pasch L. A. L., van Heesch S. A. A. C., Brok M. O., Groot Koerkamp M. J. A., Ko C. W., van Leenen D., Sameith K., van Hooff S. R., Lijnzaad P., et al. (2011) The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell 42, 536–549 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Wageningen S., Kemmeren P., Lijnzaad P., Margaritis T., Benschop J. J., de Castro I. J., van Leenen D., Groot Koerkamp M. J. A., Ko C. W., Miles A. J., Brabers N., Brok M. O., Lenstra T. L., Fiedler D., Fokkens L., et al. (2010) Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143, 991–1004 10.1016/j.cell.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kemmeren P., Sameith K., van de Pasch L. A. L., Benschop J. J., Lenstra T. L., Margaritis T., O'Duibhir E., Apweiler E., van Wageningen S., Ko C. W., van Heesch S., Kashani M. M., Ampatziadis-Michailidis G., Brok M. O., Brabers N. A. C. H., et al. (2014) Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157, 740–752 10.1016/j.cell.2014.02.054 [DOI] [PubMed] [Google Scholar]
- 68. Molin C., Jauhiainen A., Warringer J., Nerman O., and Sunnerhagen P. (2009) mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA 15, 600–614 10.1261/rna.1403509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Molina-Navarro M. M., Castells-Roca L., Bellí G., García-Martínez J., Marín-Navarro J., Moreno J., Pérez-Ortín J. E., and Herrero E. (2008) Comprehensive transcriptional analysis of the oxidative response in yeast. J. Biol. Chem. 283, 17908–17918 10.1074/jbc.M800295200 [DOI] [PubMed] [Google Scholar]
- 70. Flott S., and Rouse J. (2005) Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem. J. 391, 325–333 10.1042/BJ20050768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., Haug J., Johnston M., Jaspersen S. L., Kobor M. S., and Shilatifard A. (2009) Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35, 626–641 10.1016/j.molcel.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Keogh M.-C., and Buratowski S. (2004) Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol. Biol. 257, 1–16 10.1385/1-59259-750-5:001 [DOI] [PubMed] [Google Scholar]
- 73. Teixeira M. C., Monteiro P. T., Palma M., Costa C., Godinho C. P., Pais P., Cavalheiro M., Antunes M., Lemos A., Pedreira T., and Sá-Correia I. (2018) YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 46, D348–D353 10.1093/nar/gkx842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pan X., and Heitman J. (2002) Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell Biol. 22, 3981–3993 10.1128/MCB.22.12.3981-3993.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu P. Y. T., and Kobor M. S. (2014) Maintenance of heterochromatin boundary and nucleosome composition at promoters by the Asf1 histone chaperone and SWR1-C chromatin remodeler in Saccharomyces cerevisiae. Genetics 197, 133–145 10.1534/genetics.114.162909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.