Abstract
While long non-coding RNAs play key roles in disease and development, few structural studies have been performed to date for this emerging class of RNAs. Previous structural studies are reviewed and a pipeline is presented to determine secondary structures of long non-coding RNAs. Similar to riboswitches, experimentally determined secondary structures of long non-coding RNAs for one species may be used to improve sequence/structure alignments for other species. As riboswitches have been classified according to their secondary structure, a similar scheme could be used to classify long non-coding RNAs.
Keywords: RNA, long non-coding RNA, non-coding RNA, RNA structure, RNA biochemistry
Long non-coding RNAs (lncRNAs) have emerged as important players in development, epigenetics, stem cell biology, plant biology, RNA processing, hormone response, cancer and brain function [1–17]. Preceded by the widespread identification of non-coding RNAs in general [18, 19] long non-coding RNAs were shown to have high specificity to tissue type and developmental stage [20, 21] (also see [22] and references therein). One of the earliest known lncRNAs is Xist (X chromosome inactivation stimulated transcript), responsible for X chromosome inactivation during development [23]. More recently, several lncRNAs have been shown to be critical in HOX gene systems during development [1]. The ½sbs-lncRNA controls mRNA decay by hybridizing with mRNA to form a platform for STAU1 protein binding, triggering degradation of mRNA [6]. Other lncRNAs are required for p21 activation [24], stem cell reprogramming [25] and stress response [26].
Although the physiological relevance of many of the reported (>20,000) lncRNAs has not been determined, many lncRNAs have been shown to possess important, visible phenotypes [27]. In addition to Xist, required for dosage compensation, the Braveheart lncRNA has been shown to be required for lineage commitment in cardiomyocytes [2]. FENDRR lncRNA is required for heart, lung and gastrointestinal development [28]. Linc-brn1b is required for neocortex development [28]. The COOLAIR lncRNA is required in A. thaliana for cold-timed flowering [4]. Additionally, the NEAT1 lncRNA has the clear phenotype of being critical for paraspeckle formation [29–31].
While lncRNAs span a wide range of physiological contexts and functions, they have several common characteristics, including length (>200 nts), alternative splicing, poly-adenylation, low abundance, lack of protein product, and low sequence identity. Many studies have been performed to identify new lncRNAs, determine their protein partners, and determine their functions (via loss of function knock down and knock out experiments). However, few studies have examined their mechanism at the atomistic level [32]. In the past few years, researchers have been laying the foundation for structure-function studies. Genome-wide studies of RNA secondary structure have been performed, revealing the lncRNAs tend to be more structured than mRNAs, but less structured than ribosomal RNAs [33–39]. Detailed secondary structure studies of complete, intact single lncRNA systems show that some lncRNAs are hierarchically structured with sub-domains containing modular RNA secondary structure motifs [40–42]. Studies of Malat-1 and related lncRNAs show that the 3’-end forms a triple helix, protecting it from RNase degradation [14, 43, 44]. Recent studies have elucidated lncRNA-protein interactions, emphasizing the need for detailed structural studies and mechanistic studies at the molecular and atomistic level [45, 46].
LncRNAs tend to have low sequence identity and are often described as non-conserved. We note that some of the most well-studied non-coding RNAs (miRNAs and rRNAs) have very high sequence identity (>78% in nucleic acid sequence identity) [47]. In contrast, many other important classes of non-coding RNAs have relatively low sequence identity (nucleic acid sequence identity of ~ 50%−65%), but secondary structures that are conserved across thousands of sequences. For example, riboswitches, which regulate metabolism in bacteria, typically have sequence identities of only 50%−65%, but have secondary structures conserved across thousands of species [47]. The U2 and U4 spliceosomal RNAs have sequence identities < 60% but secondary structures conserved for > 9000 sequences. The 5S ribosomal RNA has sequence identity of ~ 60% but secondary structure conserved over 229,000 sequences. The group I intron has decidedly low sequence identity (~ 36%) but structure conserved across 60,000 species [47].
RNAs with low sequence identity are difficult to find using conventional search algorithms such as BLAST. However, knowledge of secondary structure dramatically enhances the search success. In the case of riboswitches, the RNA secondary structure was determined for a single species using in vitro chemical probing of the RNA in cell-free reconstituted systems[48–55]. Next, this structure was used as a fingerprint to find the structure in thousands of other species, despite the low sequence identity [56]. The secondary structures determined from cell-free systems by chemical probing were verified by X-ray crystallography [57–61].
To determine the RNA secondary structure of lncRNA molecules, we follow similar strategies to those used to determine the original 16S rRNA secondary structure [62–64] and the riboswitches [65] (Fig. 1). Namely, we perform chemical probing experiments to determine nucleotides that are highly mobile and likely to reside in looping regions, as well as those nucleotides with low mobility, likely to participate in Watson-Crick base pairs. To cope with the large RNA size, we employ 3S (Shot-Gun Secondary Structure), which probes the entire RNA first and then probes shorter segments of the RNA in successive rounds of probing [40, 66]. By matching signals of short segments with full RNA experiments, we identify modular sub-domains, for which a secondary structure is often readily discernable. The resulting secondary structure can be used to improve existing phylogenetic sequence alignments, and, in principle, can be used to find instances of the lncRNA not previously found in other species. In our studies, we typically begin with either alignments generated by genome browser, or alignments using synteny. We then use the initial secondary structure to improve these sequence alignments, focusing on alignment of helical regions. Covariance analysis helps to validate each helix. Next, we use the helices with the most covariant base pairs to further improve the sequence alignment. This process can be performed iteratively, with improved or validated helices enabling improved sequence alignments, and improved sequence alignments enabling more accurate covariant measures.
Figure 1.
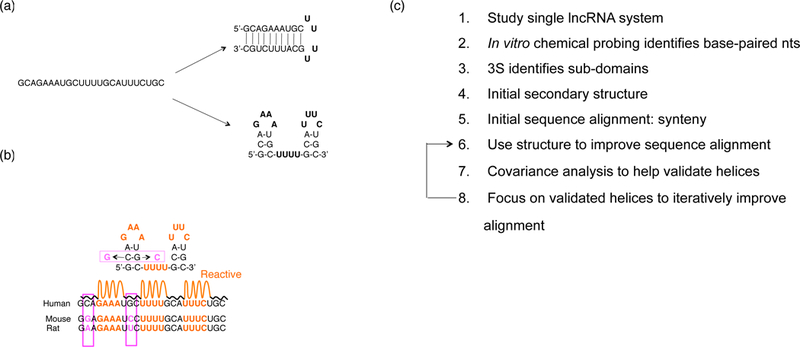
RNA secondary structure determination. (a) One sequence may be consistent with multiple folds. (b) Chemical probing reactivity data helps to lift degeneracy between folds. Multiple sequence alignment help identify covariant base pairs (pink). (c) Pipeline to determine secondary structures of long non-coding RNAs.
To demonstrate this principle, we consider the 873 nt steroid receptor RNA activator lncRNA in humans (SRA-1). This lncRNA co-activates the hormone response in human T-47D cells and co-immunoprecipitates with a large number of important proteins, including several hormone receptors (estrogen receptor, progesterone receptor, androgen receptor, glucocorticoid receptor and thyroid receptor) [67–70]. Binding assays in in vitro cell-free reconstituted systems have shown strong binding to the pseudouridinylase Pus1p, estrogen receptor, thyroid receptor, the sex reversal factor DAX-1, and the epigenetic factor SHARP. While the primary function of SRA-1 is to co-activate the hormone response, a speculated secondary function involving the binding of SRA-1 to its cognate protein SRAP has recently been shown not to occur (SRA-1 does not bind to SRAP) [71].
Our previous study demonstrated that SRA-1 contains four modular secondary structure sub-domains, each containing multiple secondary structure motifs (Fig. 2). The secondary structure was consistent with four different probing techniques (SHAPE, DMS, in-line, and RNase V1). Base pair flips with respect to species were found in the vast majority of helices. Binding studies have shown that SHARP binds to the helix 12 / helix 13 (H12/13) domain [72].
Figure 2.
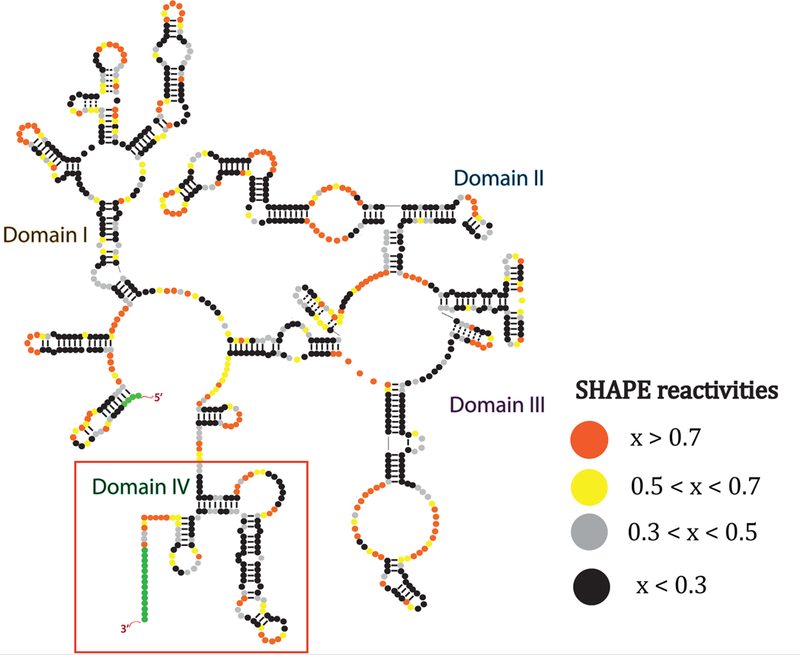
Secondary structure of steroid receptor RNA activator, as determined by 3S, consists of four sub-domains. The chemical probing reactivity data helped to improve structure/sequence alignments for domain IV.
Here, to demonstrate the utilize of secondary structure determination, we use the secondary structure of domain IV to improve the phylogenetic alignment of SRA-1 from the Ensembl dataset, in a similar manner to that used by Breaker and co-workers to improve alignments for riboswitch ncRNAs [65]. Figure 3 shows the alignment from Ensembl and the improved alignment of domain IV based on the secondary structure derived form chemical probing. The domain IV secondary structure is present in many of the sequences and covariant base pairs are observed.
Figure 3.

Example of sequence alignment for domain IV of SRA. (a) Original alignment from Ensembl database. (b) Improved alignment using knowledge of secondary structure.
This strategy can, in principle, be used to identify orthologs of lncRNAs in other species. Before classifying a lncRNA as a non-conserved RNA, we recommend that the secondary structure be studied and used to search other genomes, in addition to performing BLAST style searches. We note that in vivo probing studies provide important information validating the in vitro structures. In vitro studies are important to establish the ab initio structure because the probing signal in vivo may to be obfuscated by multiple proteins binding to the RNA, as suggested recently (Fig. 4) [11, 12]. In addition, there are few known cases where an in vitro structure of an intact, individual RNA has been shown to differ from its corresponding in vivo structure. For example, the vast majority of crystallographic structures of RNAs, which are of course determined in vitro, have either (i) been validated in vivo, or (ii) not been disproven in vivo. In the case of riboswitch RNAs, crystallographic data strongly support initial secondary structures determined by chemical probing techniques discussed above. Overall, structure-function studies of lncRNAs are in their infancy and represent an open area of research. Studies of larger lncRNAs (10–100 kB) may open a new area of structural biology and have the potential to reveal novel RNA and RNP mechanisms. Three-dimensional studies of smaller lncRNAs are also an exciting area, especially in light of the combination of specific and non-specific RNA-protein interactions thought to be involved in lncRNP complexes [12].
Figure 4.
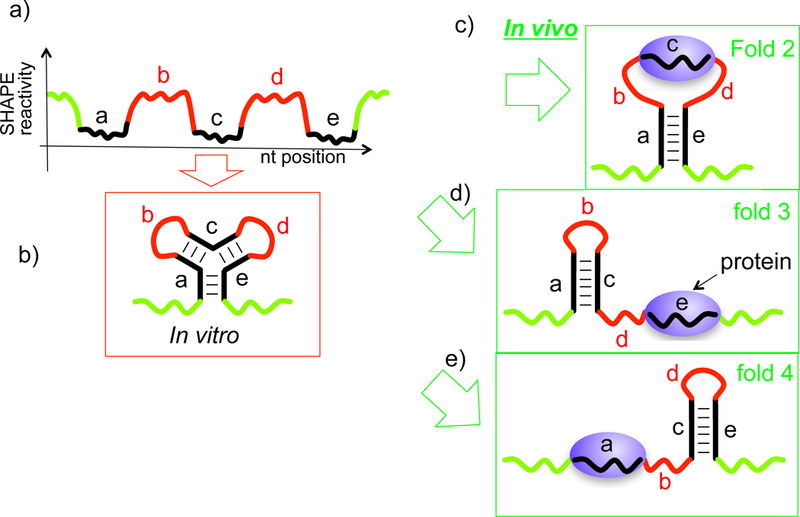
In vivo probing signals may be obfuscated by protein binding, making it more difficult to distinguish configurations. (a) Schematic of SHAPE probing signal for three-helix junction. (b)-(d) Other configurations that may be consistent with signal (a).
While there are few existing experimentally determined secondary structures of lncRNAs, the future determination of the precise and detailed secondary structure of many lncRNAs may allow immediate classification into type I: highly structured RNAs with sub-domains and complex structural motifs, such as multiway junctions, type II: loosely structured RNAs with multiple stem-loops, but lacking hierarchical domain structure and complex motifs, and type III: unstructured, disordered RNAs, which lack secondary structure. Further classification can proceed upon discovery of many lncRNAs with complex structure in terms of the specific structural motifs that organize the RNA.
Acknowledgements
The author acknowledges generous support by the LANL LDRD program.
References
- [1].Rinn JL, Chang HY, Genome regulation by long noncoding RNAs, Annual review of biochemistry, 81 (2012) 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA, Braveheart, a long noncoding RNA required for cardiovascular lineage commitment, Cell, 152 (2013) 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mercer TR, Mattick JS, Structure and function of long noncoding RNAs in epigenetic regulation, Nat Struct Mol Biol, 20 (2013) 300–307. [DOI] [PubMed] [Google Scholar]
- [4].Swiezewski S, Liu F, Magusin A, Dean C, Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target, Nature, 462 (2009) 799–802. [DOI] [PubMed] [Google Scholar]
- [5].Ulitsky I, Bartel DP, lincRNAs: genomics, evolution, and mechanisms, Cell, 154 (2013) 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gong C, Maquat LE, lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements, Nature, 470 (2011) 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D, Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin, Mol Cell, 53 (2014) 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heard E, Mongelard F, Arnaud D, Chureau C, Vourc’h C, Avner P, Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells, Proc Natl Acad Sci U S A, 96 (1999) 6841–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, Kaneko S, Helin K, Reinberg D, Stewart AF, Wutz A, Margueron R, Heard E, Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome, Mol Cell, 53 (2014) 301–316. [DOI] [PubMed] [Google Scholar]
- [10].Boumil RM, Lee JT, Forty years of decoding the silence in X-chromosome inactivation, Human molecular genetics, 10 (2001) 2225–2232. [DOI] [PubMed] [Google Scholar]
- [11].Davidovich C, Zheng L, Goodrich KJ, Cech TR, Promiscuous RNA binding by Polycomb repressive complex 2, Nat Struct Mol Biol, 20 (2013) 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR, Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA, Molecular Cell, 57 (2015) 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cech TR, Steitz JA, The noncoding RNA revolution-trashing old rules to forge new ones, Cell, 157 (2014) 77–94. [DOI] [PubMed] [Google Scholar]
- [14].Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA, Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix, Nat Struct Mol Biol, 21 (2014) 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dharap A, Nakka VP, Vemuganti R, Effect of focal ischemia on long noncoding RNAs, Stroke, 43 (2012) 2800–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ponting CP, Oliver PL, Reik W, Evolution and functions of long noncoding RNAs, Cell, 136 (2009) 629–641. [DOI] [PubMed] [Google Scholar]
- [17].Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R, The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression, Genome Res, 22 (2012) 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Numata K, Kanai A, Saito R, Kondo S, Adachi J, Wilming LG, Hume DA, Hayashizaki Y, Tomita M, Group RG, Members GSL, Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection, Genome Res, 13 (2003) 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, Consortium F, Group RGER, Genome Science G, The transcriptional landscape of the mammalian genome, Science, 309 (2005) 1559–1563. [DOI] [PubMed] [Google Scholar]
- [20].Ponjavic J, Ponting CP, Lunter G, Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs, Genome Res, 17 (2007) 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS, Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation, Genome Res, 18 (2008) 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rinn JL, Chang HY, Genome regulation by long noncoding RNAs, Annu Rev Biochem, 81 (2012) 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee JT, Jaenisch R, The (epi)genetic control of mammalian X-chromosome inactivation, Curr Opin Genet Dev, 7 (1997) 274–280. [DOI] [PubMed] [Google Scholar]
- [24].Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL, A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response, Cell, 142 (2010) 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES, lincRNAs act in the circuitry controlling pluripotency and differentiation, Nature, 477 (2011) 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP, Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor, Sci Signal, 3 (2010) ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li L, Chang HY, Physiological roles of long noncoding RNAs: insight from knockout mice, Trends in cell biology, 24 (2014) 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL, Multiple knockout mouse models reveal lincRNAs are required for life and brain development, eLife, 2 (2013) e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T, Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles, EMBO J, 31 (2012) 4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nakagawa S, Hirose T, Paraspeckle nuclear bodies--useful uselessness?, Cell Mol Life Sci, 69 (2012) 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T, MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles, Proceedings of the National Academy of Sciences of the United States of America, 106 (2009) 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY, Rise of the RNA machines: exploring the structure of long non-coding RNAs, J Mol Biol, 425 (2013) 3731–3746. [DOI] [PubMed] [Google Scholar]
- [33].Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY, Landscape and variation of RNA secondary structure across the human transcriptome, Nature, 505 (2014) 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wan Y, Qu K, Ouyang Z, Chang HY, Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing, Nat Protoc, 8 (2013) 849–869. [DOI] [PubMed] [Google Scholar]
- [35].Ouyang Z, Snyder MP, Chang HY, SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data, Genome Res, 23 (2013) 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY, Genome-wide Measurement of RNA Folding Energies, Molecular cell, 48 (2012) 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E, Genome-wide measurement of RNA secondary structure in yeast, Nature, 467 (2010) 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM, In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features, Nature, 505 (2014) 696–700. [DOI] [PubMed] [Google Scholar]
- [39].Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS, Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo, Nature, 505 (2014) 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Novikova IV, Hennelly SP, Sanbonmatsu KY, Structural architecture of the human long non-coding RNA, steroid receptor RNA activator, Nucleic Acids Res, 40 (2012) 5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, Chang HY, Akhtar A, Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila, Mol Cell, 51 (2013) 156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM, HOTAIR forms an intricate and modular secondary structure, Mol Cell, 58 (2015) 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA, A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails, Genes Dev, 26 (2012) 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wilusz JE, Freier SM, Spector DL, 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA, Cell, 135 (2008) 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY, Systematic discovery of Xist RNA binding proteins, Cell, 161 (2015) 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY, Structural imprints in vivo decode RNA regulatory mechanisms, Nature, 519 (2015) 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR, Rfam: an RNA family database, Nucleic Acids Res, 31 (2003) 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Regulski EE, Breaker RR, In-line probing analysis of riboswitches, Methods Mol Biol, 419 (2008) 53–67. [DOI] [PubMed] [Google Scholar]
- [49].Winkler W, Nahvi A, Breaker RR, Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression, Nature, 419 (2002) 952–956. [DOI] [PubMed] [Google Scholar]
- [50].Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR, Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria, Cell, 113 (2003) 577–586. [DOI] [PubMed] [Google Scholar]
- [51].Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR, A glycine-dependent riboswitch that uses cooperative binding to control gene expression, Science, 306 (2004) 275–279. [DOI] [PubMed] [Google Scholar]
- [52].Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR, Control of gene expression by a natural metabolite-responsive ribozyme, Nature, 428 (2004) 281–286. [DOI] [PubMed] [Google Scholar]
- [53].Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR, Tandem riboswitch architectures exhibit complex gene control functions, Science, 314 (2006) 300–304. [DOI] [PubMed] [Google Scholar]
- [54].Cheah MT, Wachter A, Sudarsan N, Breaker RR, Control of alternative RNA splicing and gene expression by eukaryotic riboswitches, Nature, 447 (2007) 497–500. [DOI] [PubMed] [Google Scholar]
- [55].Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR, Riboswitches in eubacteria sense the second messenger cyclic di-GMP, Science, 321 (2008) 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR, Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline, Nucleic Acids Res, 35 (2007) 4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Batey RT, Gilbert SD, Montange RK, Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine, Nature, 432 (2004) 411–415. [DOI] [PubMed] [Google Scholar]
- [58].Montange RK, Batey RT, Structure of the S-adenosylmethionine riboswitch regulatory mRNA element, Nature, 441 (2006) 1172–1175. [DOI] [PubMed] [Google Scholar]
- [59].Gilbert SD, Rambo RP, Van Tyne D, Batey RT, Structure of the SAM-II riboswitch bound to S-adenosylmethionine, Nat Struct Mol Biol, 15 (2008) 177–182. [DOI] [PubMed] [Google Scholar]
- [60].Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT, Free state conformational sampling of the SAM-I riboswitch aptamer domain, Structure, 18 (2010) 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Montange RK, Batey RT, Riboswitches: emerging themes in RNA structure and function, Annu Rev Biophys, 37 (2008) 117–133. [DOI] [PubMed] [Google Scholar]
- [62].Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF, Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence, Nucleic acids research, 8 (1980) 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Noller HF, Woese CR, Secondary structure of 16S ribosomal RNA, Science, 212 (1981) 403–411. [DOI] [PubMed] [Google Scholar]
- [64].Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Waese CR, Secondary structure model for 23S ribosomal RNA, Nucleic acids research, 9 (1981) 6167–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR, An mRNA structure that controls gene expression by binding S-adenosylmethionine, Nat Struct Biol, 10 (2003) 701–707. [DOI] [PubMed] [Google Scholar]
- [66].Novikova IV, Hennelly SP, Sanbonmatsu KY, 3S: Shotgun Secondary Structure determination for long non-coding RNAs, Methods, (2013). [DOI] [PubMed] [Google Scholar]
- [67].Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G, Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA, Genes & development, 24 (2010) 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Xu B, Yang WH, Gerin I, Hu CD, Hammer GD, Koenig RJ, Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis, Mol Cell Biol, 29 (2009) 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Colley SM, Iyer KR, Leedman PJ, The RNA coregulator SRA, its binding proteins and nuclear receptor signaling activity, IUBMB Life, 60 (2008) 159–164. [DOI] [PubMed] [Google Scholar]
- [70].Huet T, Miannay FA, Patton JR, Thore S, Steroid receptor RNA activator (SRA) modification by the human pseudouridine synthase 1 (hPus1p): RNA binding, activity, and atomic model, PLoS One, 9 (2014) e94610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].McKay DB, Xi L, Barthel KK, Cech TR, Structure and function of steroid receptor RNA activator protein, the proposed partner of SRA noncoding RNA, J Mol Biol, 426 (2014) 1766–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Arieti F, Gabus C, Tambalo M, Huet T, Round A, Thore S, The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs, Nucleic Acids Res, 42 (2014) 6742–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]


