Abstract
The large state space of gene genealogies is a major hurdle for inference methods based on Kingman’s coalescent. Here, we present a new Bayesian approach for inferring past population sizes, which relies on a lower-resolution coalescent process that we refer to as “Tajima’s coalescent.” Tajima’s coalescent has a drastically smaller state space, and hence it is a computationally more efficient model, than the standard Kingman coalescent. We provide a new algorithm for efficient and exact likelihood calculations for data without recombination, which exploits a directed acyclic graph and a correspondingly tailored Markov Chain Monte Carlo method. We compare the performance of our Bayesian Estimation of population size changes by Sampling Tajima’s Trees (BESTT) with a popular implementation of coalescent-based inference in BEAST using simulated and human data. We empirically demonstrate that BESTT can accurately infer effective population sizes, and it further provides an efficient alternative to the Kingman’s coalescent. The algorithms described here are implemented in the R package phylodyn, which is available for download at https://github.com/JuliaPalacios/phylodyn.
Keywords: Bayesian nonparametrics, coalescent, effective population size, Tajima coalescent
MODELING gene genealogies from an alignment of sequences (timed and rooted bifurcating trees reflecting the ancestral relationships among sampled sequences), is a key step in coalescent-based inference of evolutionary parameters such as effective population sizes. In the neutral coalescent model without recombination, observed sequence variation is produced by a stochastic process of mutation acting along the branches of the gene genealogy (Watterson 1975; Kingman 1982a), which is modeled as a realization of the coalescent point process at a neutral nonrecombining locus. In the coalescent point process, the rate of coalescence (the merging of two lineages into a common ancestor at some time in the past) is a function that varies with time, and it is inversely proportional to the effective population size at time t, (Kingman 1982b; Slatkin and Hudson 1991; Donnelly and Tavaré 1995). Our goal is to infer , which we will refer to as the “effective population size trajectory.”
Multiple methods have been developed to infer using the standard coalescent model with or without recombination. Some of these methods infer from summary statistics such as the sample frequency spectrum (SFS) (Bhaskar et al. 2015; Terhorst et al. 2017); however, the SFS is not a sufficient statistic for inferring (Sainudiin et al. 2011). Other methods have been proposed that directly use molecular sequence alignments at a completely linked locus, i.e., without recombination (Griffiths and Tavaré 1996; Kuhner and Smith 2007; Minin et al. 2008; Li and Durbin 2011; Drummond et al. 2012; Gill et al. 2013; Palacios and Minin 2013). Our approach is of this type. Still other methods account for recombination across larger genomic segments (Li and Durbin 2011; Sheehan et al. 2013; Schiffels and Durbin 2014; Palacios et al. 2015). In spite of their variety, all these methods must contend with two major challenges: (1) choosing a prior distribution or functional form for , and (2) integrating over the large hidden state space of genealogies. For example, several previous approaches have assumed exponential growth (Griffiths and Tavaré 1996; Kuhner et al. 1998; Kuhner and Smith 2007), in which case the estimation of is reduced to the estimation of one or two parameters. In general, the functional form of is unknown and needs to be inferred. A commonly used naive nonparametric prior on is a piecewise linear or constant function defined on time intervals of constant or varying sizes (Heled and Drummond 2008; Sheehan et al. 2013; Schiffels and Durbin 2014). The specification of change points in such time-discretized effective population size trajectories is inherently difficult because it can lead to runaway behavior or large uncertainty in . These difficulties can be avoided by the use of Gaussian process priors in a Bayesian nonparametric framework, allowing accurate and precise estimation (Gill et al. 2013; Palacios and Minin 2013; Lan et al. 2015; Palacios et al. 2015). More precisely, the autocorrelation modeled with the Gaussian process avoids runaway behavior and large uncertainty in .
The second challenge for coalescent-based inference of is the integration over the hidden state space of genealogies for large sample sizes. Given molecular sequence data at a single nonrecombining locus and a mutation model with parameter , current methods rely on calculating the marginal likelihood function by integrating over all possible coalescence and mutation events. Under the infinite sites mutation model without intralocus recombination (Watterson 1975), this integration requires a computationally expensive importance sampling technique or Markov Chain Monte Carlo (MCMC) techniques (Griffiths and Tavaré 1994a; Stephens and Donnelly 2000; Hobolth et al. 2008; Wu 2010; Gronau et al. 2011). Moreover, a maximum likelihood estimate of cannot be explicitly obtained; instead, it is obtained by exploring a grid of parameter values (Tavaré 2004). For finite sites mutation models, current methods approximate the marginal likelihood function by integrating over all possible genealogies via MCMC methods [Equation 1; Kuhner (2006); Drummond et al. (2012)]. In both cases, the marginal likelihood may be expressed as
| (1) |
in which Pr is used to denote both the probability of discrete variables and the density of continuous variables. The integral above involves an -dimensional integral over coalescent times and a sum over all possible tree topologies with n leaves. Therefore, these methods require a very large number of MCMC samples, and exploration of the posterior space of genealogies continues to be an active area of research (Kuhner et al. 1998; Rannala and Yang 2003; Drummond et al. 2012; Whidden and Matsen 2015; Aberer et al. 2016).
Current methods rely on the Kingman n-coalescent process to model the sample’s ancestry. However, the state space of genealogical trees grows superexponentially with the number of samples, making inference computationally challenging for large sample sizes. In this study, we develop a Bayesian nonparametric model that relies on Tajima’s coalescent, a lower-resolution coalescent process with a drastically smaller state space than that of Kingman’s coalescent. In particular, we approximate the posterior distribution , where corresponds to the Tajima’s genealogy of the sample (see Figure 1A and Tajima’s genealogies), has a Gaussian process prior with precision hyperparameter τ that influences the smoothness of the resulting trajectory, and mutations occur according to the infinite sites model of Watterson (1975). This results in a new efficient method for inferring called Bayesian Estimation by Sampling Tajima’s Trees (BESTT), with a drastic reduction in the state space of genealogies. Using simulated data, we show that BESTT can accurately infer effective population size trajectories and that it provides a more efficient alternative than Kingman’s coalescent models.
Figure 1.
For a sample of size n, the number of Tajima’s genealogies is superexponentially fewer compared to the number of Kingman’s genealogies. (A) A Kingman’s genealogy and a Tajima’s genealogy for . A Kingman’s genealogy (left) comprises a vector of coalescent times and the labeled topology; the number of possible labeled topologies for a sample of size n is . A Tajima’s genealogy (right) comprises a vector of coalescent times and a ranked tree shape. In both cases, coalescent events are ranked from 2 at time to n at time . Coalescent times are measured from the present (time 0) back into the past. (B) The numbers of labeled topologies and ranked tree shapes (formulas provided in Tajima’s genealogies) for different values of the sample size, n.
Next, we start with an overview of BESTT, detail our representation of molecular sequence data, and define the Tajima coalescent process. We then introduce a new augmented representation of sequence data as a directed acyclic graph (DAG). This representation allows us to both calculate the conditional likelihood under the Tajima coalescent model and to sample tree topologies compatible with the observed data. We then provide an algorithm for likelihood calculations and develop an MCMC approach to efficiently explore the state space of unknown parameters. Finally, we compare our method to other methods implemented in Bayesian Evolutionary Analysis Sampling Trees (BEAST) (Drummond et al. 2012) and estimate the effective population size trajectory from human mitochondrial DNA (mtDNA) data. We close with a discussion of possible extensions, and limitations of the proposed model and implementation.
Materials and Methods
Overview of BESTT
Our objective in the implementation of BESTT is to estimate the posterior distribution of model parameters by replacing Kingman’s genealogy with Tajima’s genealogy . A Tajima’s genealogy does not include labels at the tips (Figure 1): we do not order individuals in the sample but label only the lineages that are ancestral to at least two individuals (that is, we only label the internal nodes of the genealogy). Replacing Kingman’s genealogy by Tajima’s genealogy in our posterior distribution exponentially reduces the size of the state space of genealogies (Figure 1B). To compute , the conditional likelihood of the data conditioned on a Tajima’s genealogy, we assume the infinite sites model of mutations, and leverage a DAG representation of sequence data and genealogical information. Note that the overall likelihood, Equation 1, will differ only by a combinatorial factor from the corresponding likelihood under the Kingman coalescent. Our DAG represents the data with a gene tree (Griffiths and Tavaré 1994a), constructed via a modified version of the perfect phylogeny algorithm of Gusfield (1991). This provides an economical representation of the uncertainty, and conditional independences induced by the model and the observed data.
Under the infinite sites mutation model, there is a one-to-one correspondence between observed sequence data and the gene tree of the data (Gusfield 1991) (Summarizing sequence data as haplotypes and mutation groups and Representing as a gene tree). We further augment the gene tree representation with the allocation of the number of observed mutations along the Tajima’s genealogy to generate a DAG (An augmented data representation using directed acyclic graphs). The conditional likelihood is then calculated via a recursive algorithm that exploits the auxiliary variables defined in the DAG nodes, marginalizing over all possible mutation allocations (Computing the conditional likelihood). We approximate the joint posterior distribution via an MCMC algorithm using Hamiltonian Monte Carlo for sampling the continuous parameters of the model and a novel Metropolis–Hastings algorithm for sampling the discrete tree space.
Summarizing sequence data as haplotypes and mutation groups
Let the data consist of n fully linked haploid sequences or alignments of nucleotides at s segregating sites sampled from n individuals at time (the present). Note that any labels we affix to the individuals are arbitrary in the sense that they will not enter into the calculation of the likelihood. We further assume the infinite sites mutation model of Watterson (1975) with mutation parameter μ and known ancestral states for each of the sites. Then, we can encode the data into a binary matrix of n rows and s columns with elements , where 0 indicates the ancestral allele.
To calculate the Tajima’s conditional likelihood , we first record each haplotype’s frequency and group repeated columns to form mutation groups; a mutation group corresponds to a shared set of mutations in a subset of the sampled individuals. We record the cardinality of each mutation group (i.e., the number of columns that show each mutation group). In Figure 2A, there are two columns labeled “b,” corresponding to two segregating sites that have the exact same pattern of allelic states across the sample. Further, two individuals carry the derived allele of mutation group b, so in this case the frequency of haplotype 7 and the cardinality of mutation group b are both equal to 2. Likewise, haplotype 4 has frequency 1 and carries five mutations that are split into mutation groups “a,” “f,” and “g” (the latter is not shown in Figure 2A, but appears in Figure 2B) of respective cardinalities 1, 3, and 1. We denote the number of haplotypes in the sample as h, the number of mutation groups as m, and the representation of as haplotypes and mutation groups as .
Figure 2.
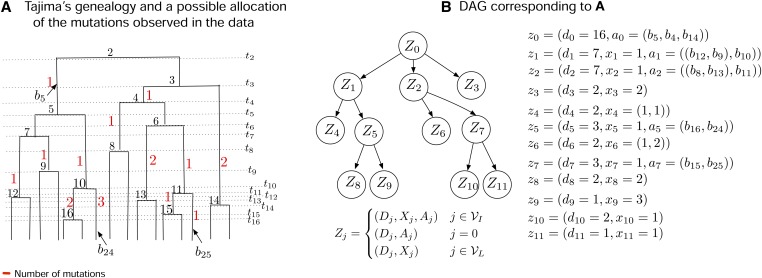
Data structures employed by our method, Bayesian Estimation of population size changes by Sampling Tajima’s Trees (BESTT), for calculating the conditional likelihood of the data. (A) Compressed data representation of sequences and (columns, only the first 10 of which are shown), comprised of nine haplotypes and 13 mutation groups. Rows correspond to haplotypes and each polymorphic site is labeled by its mutation group . (B) Gene tree representation of the data in (A). Red numbers indicate the cardinality of each mutation group [number of columns with the same label in (A)]. Black letters indicate the mutation group [column labels in (A)], and black numbers indicate the frequency of the corresponding haplotype. (C) A Tajima’s genealogy compatible with the gene tree in (B). Internal nodes are labeled according to order of coalescent events from the root to the tips. Coalescent event i happens at time and branches are labeled (see An augmented data representation using directed acyclic graphs for details). (D) A Directed Acyclic Graph (DAG) representation of the gene tree in (B) together with allocation of mutation groups along the branches of the Tajima’s genealogy in (C). denotes the set of internal nodes and the set of leaf nodes. A detailed description of the DAG is given in An augmented data representation using directed acyclic graphs.
Representing as a gene tree
(Figure 2A) can alternatively be represented as a gene tree or perfect phylogeny (Gusfield 1991; Griffiths and Tavaré 1994b). This representation relies on our assumption of the infinite sites mutation model in which, if a site mutates once in a given lineage, all descendants of that lineage also have the mutation and no other individuals carry that mutation. The gene tree is a graphical representation of the haplotypes (as tips) arranged by their patterns of shared mutations. The haplotype data summarized in Figure 2A correspond to the gene tree given in Figure 2B. Details of the correspondence between haplotype data and gene tree are listed below, and an additional example is given in Figure E1 (Appendix E).A gene tree for a matrix of h haplotypes and m mutation groups is a rooted tree with h leaves and at least m edges, such that (Figure 2B):
Each row of corresponds to exactly one leaf of . The black numbers at leaf nodes in Figure 2B are the haplotype frequencies.
Each mutation group of is represented by exactly one edge of , which is labeled accordingly (letters in Figure 2, A and B). The red numbers along edges in Figure 2B give the cardinality of each mutation group (i.e., the number of segregating sites in this group; see Figure 2A). Some external edges (edges subtending leaves) may not be labeled, indicating that they do not carry additional mutations to their parent edge. This happens when the other edges emanating from the parent node necessarily correspond to other mutation groups.
Edges are placed in the gene tree in such a way that each “path” from the root to a leaf fully describes a haplotype. Edges corresponding to shared mutations between several haplotypes are closest to the root. For example, in Figure 2B, haplotype 4 corresponds to the leaf at which one arrives starting from the root and going along edges a, g, and f; in contrast, haplotype 7 corresponds to the leaf at which one arrives going from the root along edge b. Thus, the labels and the numbers associated with the edges along the unique path from the root to a leaf exactly specify a row of .
Dan Gusfield’s perfect phylogeny algorithm (Gusfield 1991) transforms the sequence data into a gene tree and this transformation is one-to-one. We note that the perfect phylogeny or gene tree is not the same as the genealogy . While a genealogy is a bifurcating tree of individuals of the sample, the gene tree is a multifurcating tree of haplotypes.
Tajima’s genealogies
Our method of computing the probability of the recoded data, , uses ranked tree shapes rather than fully labeled histories. We refer to these ranked tree shapes as Tajima’s genealogies but note they have also been called unlabeled rooted trees (Griffiths and Tavaré 1995) and evolutionary relationships (Tajima 1983). In Tajima’s genealogies, only the internal nodes are labeled and they are labeled by their order in time. Tajima’s genealogies encode the minimum information needed to compute the probability of data , which consists of nested sets of mutations, without any approximations. In Figure 1A for example, it matters only that mutation group e occurs on a subgroup of the individuals who carry mutation group “a,” and that this is different from the subgroups carrying c, d, and f. No other labels matter because individuals are exchangeable in the population model we assume.
This represents a dramatic coarsening of tree space compared to the classical leaf-labeled binary trees of Kingman’s coalescent. The number of possible ranked tree shapes for a sample of size n corresponds to the n-th term of the sequence A000111 of Euler zig-zag numbers (Disanto and Wiehe 2013) whereas the number of labeled binary tree topologies is . As can be seen from Figure 1B, this provides a much more efficient way to integrate over the key hidden variable, the unknown gene genealogy of the sample, when computing likelihoods.
We model this hidden variable using the vintaged and sized coalescent (Sainudiin et al. 2015), which corresponds exactly to this coarsening of Kingman’s coalescent. As can be seen in Figure 1A, we assign vintages/labels 2 through n starting at the root of the tree and moving toward the present, so that the node created by the final splitting event, which is also the first coalescence event looking back in the ancestry of the sample, is labeled n. We write for the time of node k, measured from the present back into the past. We set to be the present time. Then during the interval the sample has exactly k extant ancestors, for .
The coarsening of the tree topology does not change the law of the times between two coalescence events. Thus, conditional on the effective population size trajectory and the time at which the number of ancestors to the sample decreases to k, the distribution of the time during which the sample has k ancestors is given by
| (2) |
(Slatkin and Hudson 1991), where . Writing the density at of the vector of coalescence times as a product of conditional densities, we obtain
| (3) |
We use a lower triangular matrix denoted to represent Tajima’s genealogies; see Appendix A. The probability of a ranked tree shape was derived independently in Sainudiin et al. (2015) and Palacios et al. (2015). Specifically, for every ranked tree shape with n leaves,
| (4) |
where c is the number of cherries in (i.e., nodes subtending two leaves; in Figure A1A). Note that this probability is independent of the effective population size trajectory since the choice of the pair of lineages that coalesce during an event is independent of (recall that in Kingman’s coalescent, the coalescing pair is chosen uniformly at random among all possible pairs). Since the distribution of Tajima’s genealogies conditional on can be factored as the product of the probability of the ranked tree shape and the coalescent times density, we arrive at
| (5) |
An augmented data representation using directed acyclic graphs
A key component of BESTT is the calculation of the conditional likelihood . We compute the conditional likelihood recursively over a DAG, . Our DAG exploits the gene tree representation of the data (Figure 2B), incorporates the branch length information of the Tajima’s genealogy (Figure 2C), and facilitates the recursive allocation of mutations to the branches of . Here, we detail the construction of the DAG.
We construct the DAG using three pieces of information: the observed gene tree , a given Tajima’s genealogy and a latent allocation of mutations along the branches of the Tajima’s genealogy (Figure 3). An allocation refers to a possible mapping (compatible with the data) of the observed numbers of mutations (red numbers in Figure 2B) to branches in the Tajima’s genealogy. Figure 3A shows one possible mapping for the Tajima’s genealogy in Figure 2C; usually this mapping is not unique. Our construction of enables an efficient recursive consideration of all possible allocations of mutations along when computing the conditional likelihood .
Figure 3.
Directed Acyclic Graph (DAG) construction. (A) A Tajima’s genealogy from Figure 2C with added allocation of mutations shown in red. (B) The corresponding augmented DAG with allocation of mutations. At the root , there are no mutations by convention. Node has 16 descendants across three subtrees of and two descendants, corresponding to nodes . These three subtrees subtend from , and , respectively, in (A). Node corresponds to the tree subtending from of size 7 with mutation along , and subtends three subtrees from and . Subtrees subtending from are grouped together in leaf node because they both have two descendants and have the same parent node. When leaf nodes represent more than one tree, such as in Figure 4B, the random variable is the vector that denotes the number of mutations along the branches that subtends from the tree node j that have descendants, and is the number of edges subtending from .
Constructing the DAG :
The graph structure of our DAG (Figure 2D) with nodes and edges E is constructed from a gene tree . The number of internal nodes in the DAG is the same as the number of internal nodes in . However, sister leaf nodes in with the same number of descendants are grouped together in and leaf nodes descending from edges with no mutations are treated as singletons grouped together in . For example, the leaves in Figure 2B subtending from edges i and j are grouped into in Figure 2D, as they both have haplotype frequency 2. However, the leaves subtending from the e and f edges are not grouped (and correspond to and in the DAG Figure 2D) since they have respective haplotype frequencies 2 and 1. We label the root node of as and increase the index i of each node from top to bottom, moving left to right. For , we assign a directed edge if the node in corresponding to is connected to the node in corresponding to . The index set of internal nodes in is denoted by and the index set of leaf nodes is denoted by .
Information carried by the nodes in :
Each node in represents a vector, , which includes number of descendants, number of mutations, and latent allocation of mutations. Although the number of descendants and number of mutations are part of the observed data, the allocation of mutations can be seen as a random variable; for ease of exposition, we use capital letters to denote all three types of information. We define the vector as follows:
where denotes the number of descendants of (i.e., of sampled sequences subtended by) node , denotes the number of mutations separating from its parent node, and denotes the allocation of mutations along (described in detail below). The number of descendants is thus the number of individuals/sequences descending from node (this information is part of ). For internal nodes, records the cardinality of a mutation group, represented as a red number along the edge of in Figure 2B, where i is the index of the parent node of . Leaf nodes in may correspond to more than one leaf node in , namely any sister nodes with the same number of descendants. In this case, is a vector with the cardinalities of the corresponding mutation groups (see for example node in Figure 3B). To keep the DAG construction simple, we only allow groupings of leaf nodes and not of internal nodes with identical descendants carrying identical numbers of mutations. We note that, in principle, it would be possible to compress the number of internal nodes of the DAG by exploiting all the symmetries observed in the data.
Allocation of mutation groups along :
The latent allocation variables determine a possible correspondence between subtrees in and nodes in ; in particular, indicates the branches in that subtend the subtrees corresponding to nodes if are child nodes of .
Allocations of mutations to branches are usually not unique and computation of the conditional likelihood requires summing over all possible allocations. In Figure 3A we show one such possible allocation of the mutation groups of the gene tree in Figure 2B along the Tajima’s genealogy in Figure 2C. For example, mutation group a in Figure 2B with cardinality 1 (number in red) is a mutation observed in seven individuals (sum of black numbers of leaves descending from edge marked a). This same mutation group, a, is shown as a red number 1 in Figure 3A allocated to branch . If is an internal node, the number of mutations is denoted as a vector of length 1. If is a leaf node, can be a vector of length >1. Details on notation for allocations can be found in Appendix B.
Computing the conditional likelihood
Under the infinite sites mutation model, mutations are superimposed independently on the branches of as a Poisson process with rate μ. To compute , we marginalize over the latent allocation information in the DAG ; that is, we sum over all possible mappings of mutations in to branches in as follows:
where , , denotes the index of the parent of node i in , and we set because it is assumed that there are no mutations above the root node and the length of the root branch . Writing for the tree length of (i.e., the sum of the lengths of all branches of ) and factoring out a global factor (due to the Poisson distribution of mutations across the genealogy) from each of the above products over , we have
where is the set of all permutations of divided into groups of different sizes. The number of different permutations of the k values of divided into groups of sizes is
| (6) |
For example, assume that and with branch lengths . In this case, because there will be three branches with two mutations, because there will be one branch with no mutations, and because there will be two branches with three mutations. The number of permutations of mutations groups divided into groups with cardinalities of sizes is .
The conditional likelihood is calculated via a backtracking algorithm (Appendix C). The algorithm marginalizes the allocations by traversing the DAG from the tips to the root. The pseudocode and an example can be found in Appendix C.
The case of unknown ancestral states
Up to now, we have assumed that the ancestral state was known at every segregating site. The representation of the data that we use in this case records the cardinalities of each mutation group and the genealogical relations between these groups, but does not assign labels to the sequences. Hence, in the terminology of Griffiths and Tavaré (1995), our data corresponds to an unlabeled rooted gene tree.
When the ancestral types are not known, the data (now denoted ) may be represented as an unlabeled unrooted gene tree. By the remark following Equation 1 in Griffiths and Tavaré (1995), if s is the number of segregating sites, then there are at most unlabeled rooted gene trees that correspond to the unrooted gene tree of the observed data . By the law of total probability [see also Equation 10 in Griffiths and Tavaré (1995)], the conditional likelihood of can be written as the sum over all compatible unlabeled rooted gene trees of the probability of conditionally on . That is:
| (7) |
where each of the corresponds to a unique unlabeled rooted gene tree compatible with the unrooted gene tree and denotes the number of those unlabeled rooted gene trees. In the following sections, we shall assume that the ancestral type at each site is known.
Bayesian inference of the effective population size trajectory
Our posterior distribution of interest is
| (8) |
where has a Gaussian process prior with mean 0 and covariance function (Rasmussen and Williams 2006). This specification ensures is nonnegative. In our implementation, we assume a regular geometric random walk prior, that is, at B regularly spaced time points in with
The parameter τ is a length-scale parameter that controls the degree of smoothness of the random walk. We place a γ prior with parameters and on τ, reflecting our lack of prior information in terms of high variance about the smoothness of the logarithm of the effective population size trajectory.
We approximate the posterior distribution of model parameters via an MCMC sampling scheme. Model parameters are sampled in blocks within a random scan Metropolis-within-Gibbs framework. Our algorithm initializes with the corresponding Tajima genealogy of the UPGMA estimated tree implemented in phangorn (Schliep 2011). Given an initial genealogy, our algorithm initializes and τ with the method of Palacios and Minin (2012) implemented in phylodyn (Karcher et al. 2017). We then proceed to generate: (1) a sample of the vector of effective population sizes and precision parameter as described in Split Hamiltonian Monte Carlo updates of , (2) a sample of the vector of coalescent times as described in Hamiltonian Monte Carlo updates of coalescent times and Local updates of coalescent times where we modify a single coalescent time, and (3) a sample of ranked tree shape as described in Metropolis–Hastings updates for ranked tree shapes in each iteration. To summarize the effective population size trajectory, we compute the posterior median and 95% credible intervals pointwise at each grid point in , were is the maximum time to the most recent common ancestor sampled.
Metropolis–Hastings updates for ranked tree shapes:
There is a large body of literature on local transition proposal distributions for Kingman’s topologies (Kuhner et al. 1998; Rannala and Yang 2003; Drummond et al. 2012; Whidden and Matsen 2015; Aberer et al. 2016). In this paper, we adapted the local transition proposal of Markovtsova et al. (2000) to Tajima’s topologies. We briefly describe the scheme below and provide a pseudocode algorithm in Appendix C (Algorithm 3).
Given the current state of the chain , we propose a new ranked tree shape in two steps. For step 1, we first sample a coalescent interval uniformly at random, where . Note that we will never select the interval at the top of the tree (see Figure A1A). Given k, we focus solely on the coalescent events at times and . For step 2, there are two possible scenarios. Case A: the lineage created at time , labeled k, coalesces at time (first row of Figure 4A). Case B: lineage k does not coalesce at time (Figure 4B). In Case A, we choose a new pair of lineages at random to coalesce at time from the three lineages subtending k and (excluding k), and we coalesce the remaining lineage with k at ( and in Figure 4). In Case B, we invert the order of the coalescent events; that is, the two lineages descending from k are set to coalesce at time and lineages descending from are set to coalesce at time ( a in Figure 4). Note that the numerical labels are included to clarify the picture: lineages subtending both Case A and Case B can be either labeled (if there is a vintage subtending that lineage) or not (if there is a singleton). The transition probability is given by the product of the probabilities of the two steps. The new ranked tree shape is accepted with probability given by the Metropolis–Hastings ratio defined below:
| (9) |
We note that our proposal can result in the same ranked tree shape. However, we tested alternative proposals that precluded this event and we did not find any notable difference in the overall performance of the MCMC algorithm.
Figure 4.
Markov moves for topologies. First row: possible coalescent patterns (Case A or Case B) for a given topology . Second row: possible Markov moves in Case A ( and ) and Case B . k indexes the coalescent interval sampled. Numerical labels at the tips are added for convenience: conditionally on a given , tips can be labeled (vintage) or not (singleton). Figure is adapted from Figures 2–4 of Markovtsova et al. (2000).
Split Hamiltonian Monte Carlo updates of :
To make efficient joint proposals of and τ, we use the Split Hamiltonian Monte Carlo method proposed by Lan et al. (2015). Conditioned on , the target density becomes . This is the same target density implemented in Karcher et al. (2017) for fixed coalescent times .
Hamiltonian Monte Carlo updates of coalescent times:
Given the current state , we propose a new vector of coalescent times with target density by numerically simulating a Hamilton system with Hamiltonian
| (10) |
where is the momentum vector assumed to be normally distributed. The system evolves according to:
| (11) |
We use the leapfrog method (Neal 2011) with step size ϵ and a p Poisson with mean 10 distributed number of steps to simulate the dynamics from time to . Each leapfrog step of size ϵ follows the trajectory:
| (12) |
For our implementation, we set the mass matrix , the identity matrix. We simulate the Hamiltonian dynamics of the logarithm of times to avoid proposals with negative values. Solving the equations of the Hamilton system requires calculating the gradient of the logarithm of the target density with respect to the vector of log coalescent times. The gradient of the log conditional likelihood (score function) is calculated at every marginalization step in the algorithm for the likelihood calculation.
At the beginning of Bayesian inference of the effective population size trajectory, we described how we assume a regular geometric random walk prior on at B regularly spaced time points in . Ideally, the window size T must be at least , the time to the most recent common ancestor (TMRCA). However, is not known. Our initial values of coalescent times are obtained from the UPGMA implementation in phangorn (Schliep 2011) with times properly rescaled by the mutation rate, and we set . We initially discretize the time interval into B intervals of length . As we generate new samples of , we expand or contract our grid according to the current value of by keeping the grid interval length fixed to , effectively increasing or decreasing the dimension of .
Local updates of coalescent times:
In addition to Hamiltonian Monte Carlo (HMC) updates of coalescent times, we propose a move of a single coalescent time (excluding the TMRCA ) chosen uniformly at random and sampled uniformly in the intercoalescent interval; that is, we choose and . This is a symmetric proposal and the corresponding Metropolis–Hastings acceptance probability is
| (13) |
While these updates may seem unnecessary in light of the Hamiltonian updates of coalescence times (Hamiltonian Monte Carlo updates of coalescent times), we observed better performance of our MCMC sampler by including this additional proposal. One reason may be our choice of in Hamiltonian Monte Carlo updates of coalescent times that does not account for the geometric structure of the posterior distribution of coalescent times. However, a better choice of comes with higher computational burden than a simple local update of coalescent times.
Multiple independent loci:
Thus far, we have assumed our data consist of a single linked locus of s segregating sites. We can extend our methodology to l independent loci with segregating sites for . In this case, our data consist of l aligned sequences with elements , where 0 indicates the ancestral allele as before. We then jointly estimate the Tajima’s genealogies , precision parameter τ, and vector of log effective population sizes through their posterior distribution:
| (14) |
In Equation 14, we enforce that all loci follow the same effective population size trajectory but every locus can have its own mutation rate .
Data availability
The methods presented in this article are implemented in the open source R package phylodyn (https://github.com/JuliaPalacios/phylodyn). The data files necessary to reproduce both the simulated and real data analyses are provided with the R package.
Results
The performance of BESTT in applications to simulated data
We tested our new method, BESTT, on simulated data under four different demographic scenarios. Note that in this section, is rescaled to the coalescent timescale, meaning that is the pairwise rate of coalescence at time t in the past relative to the rate at the present time zero. We simulated genealogies under four different population size trajectories:
- A period of exponential growth followed by constant size:
(15) - A trajector with instantaneous growth:
(16) Exponential growth: .
A constant trajectory: .
Given a genealogy of length , where is the intercoalescent length while there are j lineages, we drew the total number of mutations (segregating sites) s according to a Poisson distribution with parameter . We then placed the mutations uniformly at random along the branches of the genealogy. For each of the s mutations, we assigned the mutant type to individuals descending from the branch where the mutation occurred and the ancestral type otherwise.
We summarize our posterior inference by the posterior median and 95% Bayesian credible intervals (BCIs) after 200 thousand iterations, and thinned every 10 iterations with 100 iterations of burn in. Our initial number of change points for was set to 50 over the time interval , where is the initialized time to the most recent common ancestor; however, over the course of MCMC iterations, this number could increase or decrease according to the posterior distribution of .
We assess the accuracy and precision of our estimates using the sum of relative errors (SRE)
| (17) |
where is the estimated effective population size trajectory at time . Second, we computed the mean relative width (MRW) as
| (18) |
where corresponds to the 97.5% upper limit and corresponds to the 2.5% lower limit of the estimated posterior distribution of . In addition, we measured how well the 95% credible intervals cover the truth and compute the envelope measure, :
| (19) |
We first simulated three data sets of individuals with an average number of 100 segregating sites under different types of population size trajectories: constant, exponential growth, and instantaneous growth. Results are depicted in the first column of Figure 8. Posterior medians and 95% credible intervals are shown as black curves and gray shaded areas, respectively. The trajectories used to simulate the data are depicted as dashed lines. Figure 8 shows that our BESTT method recovers the constant and exponential growth trajectories very well, but the instantaneous growth scenario is less accurate and with high uncertainty (wide credible intervals). In all three cases, our envelope measure is > 95%. Performance measures on all simulations are summarized in Table 1.
Figure 8.
Bayesian Estimation of population size changes by Sampling Tajima’s Trees (BESTT) and BEAST comparison. Posterior inference from simulated data of sequences under constant, exponential, and instantaneous growth trajectories (rows) obtained from our method BESTT (first column), BEAST Extended Bayesian Skyline Plot (EBSP) (second column), and BEAST Skygrid (third column). Shaded areas correspond to 95% credible intervals, solid black lines show posterior medians, and dashed lines show the simulated truth. In the fourth column, we show the posterior density of the time to the most recent common ancestor (TMRCA) from the three methods: BESTT (black), BEAST EBSP (blue), and BEAST Skygrid (red). The true value of the TMRCA is depicted as a vertical dashed line.
Table 1. Empirical measures of performance in the simulations described in the text.
| Simulation | % ENV | SRE | MRW |
|---|---|---|---|
| Instantaneous growth (n = 10,s = 31) | 96 | 5.87 | 124352 |
| Instantaneous growth (n = 10,s = 63) | 100 | 2.15 | 2296 |
| Instantaneous growth (n = 10,s = 120) | 98 | 0.53 | 80 |
| Instantaneous growth (n = 25) | 90 | 0.40 | 3.43 |
| Instantaneous growth (n = 35) | 92 | 0.31 | 3.16 |
| Constant | 100 | 0.30 | 1.16 |
| Exponential | 100 | 0.35 | 5.45 |
| Exponential and constant (n = 10, 1 locus) | 100 | 4.31 | 22608 |
| Exponential and constant (n = 10, 5 loci) | 100 | 2.37 | 309.1 |
| Exponential and constant (n = 10, 10 loci) | 100 | 0.16 | 4.19 |
% ENV, % envelope measure; MRW, mean relative width; SRE, sum of relative errors.
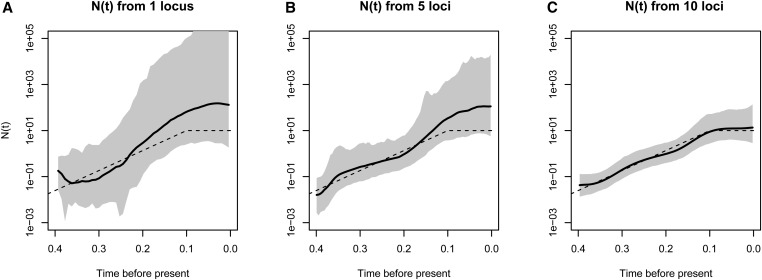
We analyzed the effect of increasing the number of segregating sites, the number of samples, and the number of independent genealogies on posterior inference with BESTT. In all three cases, we expect our method to better recover the truth. Figure 5 shows our results on simulated data under a population size trajectory with instantaneous growth (Equation 16) of individuals with 31, 63, and 120 segregating sites. As expected, our method recovers the truth with higher precision (MRW) and accuracy (SRE) when we increase the number of segregating sites. Increasing the number of segregating sites may result in more constraints in the gene tree. For , there are 7936 possible ranked tree shapes; however, for the data sets simulated with 31, 63, and 102 segregating sites, there are only , , and ranked tree shapes compatible with their corresponding gene trees. These numbers were estimated by importance sampling (Cappello and Palacios 2019).
Figure 5.
Varying the number of segregating sites. Posterior inference from simulated data of sequences under a population size trajectory with instantaneous growth (dashed lines). s is the number of segregating sites. Posterior medians are depicted as solid black lines and 95% Bayesian credible intervals are depicted by shaded areas.
As another performance assessment, we simulated data sets from a population size trajectory with instantaneous growth with varying numbers of samples. We simulated data sets with , 25, and 35 samples with 215 expected numbers of segregating sites. Our results depicted in Figure 6 show that our method performs better in terms of SRE and MRE when the number of samples increases. Similarly, precision (MRW) and accuracy (SRE) increases when inference is done from a larger number of independent data sets. Finally, Figure 7 shows our results from 1, 5, and 10 data sets simulated from 1, 5, and 10 independent genealogies of 10 individuals with a population size trajectory of growth followed by a constant period (Equation 15). As expected, our method’s performance substantially increases by increasing the number of independent data sets.
Figure 6.
Varying the number of samples under a population size trajectory with instantaneous growth. Posterior inference from simulated data of , 25, and 35 sequences under the population size trajectory with instantaneous growth. Shaded areas correspond to 95% credible intervals, solid lines to posterior medians, and dashed lines to the truth.
Figure 7.
Multiple independent data sets. Posterior inference from simulated data of sequences under exponential followed by constant trajectory (Equation 15). (A) Inference from a single simulated data set, (B) from five independently simulated data sets, and (C) from 10 independently simulated data sets. Shaded areas correspond to 95% credible intervals, solid black lines show posterior medians, and dashed lines show the simulated truth.
Comparison to other methods
To our knowledge, there is no other method for inferring (variable) effective population size over time from haplotype data that assumes the infinite sites mutation and a nonparametric prior on ; therefore, we cannot directly compare our method to others. Moreover, our method is the only one that explicitly averages over Tajima genealogies instead of Kingman genealogies. BEAST (Drummond et al. 2012) is a program for analyzing molecular sequences that uses MCMC to average over the Kingman tree space and it is therefore a good reference for comparison to our method. We compared our results to the Extended Bayesian Skyline Plot method (EBSP) (Heled and Drummond 2008) and the Skygrid method (Gill et al. 2013) implemented in BEAST.
Since the infinite sites mutation model is not implemented in BEAST, we first converted our simulated sequences of 0s and 1s to sequences of nucleotides by sampling s ancestral nucleotides uniformly on and assigning one of the remaining three types uniformly at random to be the mutant type. This corresponds to a simulation of the Jukes–Cantor mutation model (Jukes and Cantor 1969) that is currently implemented in BEAST.
We compare the results of BESTT to those of BEAST EBSP and Skygrid (Drummond et al. 2005, 2012) in Figure 8. We note that results from BEAST are generated from 10 million iterations and thinned every 1000 iterations, while results from BESTT are generated from 200 thousand iterations.
We compared our point estimates from all methods to the ground truth for each simulation (Table 2). In two cases, BESTT has better envelope than BEAST. For the exponential growth simulation (Figure 8, second row) the BEAST EBSP result has better SRE and MRW, however, the credible intervals are uneven with very wide intervals at the ends. In all cases, the BEAST Skygrid results have wider credible intervals. For the instantaneous growth simulation (Figure 8, third row), BEAST EBSP did not generate many simulations beyond the time point 0.06; for this reason, we recomputed the performance statistics for the overlapping time interval . In this interval, BESTT outperforms both methods implemented in BEAST in terms of envelope and SRE. The last column of Figure 8 shows the posterior distribution of the TMRCA. For the case of constant population size, the true value of the TMRCA is contained in the 95% BCI estimated with BESTT but it is not contained in the 95% BCIs estimated with the two methods implemented in BEAST. In the exponential growth simulation, the true TMRCA is contained in the 95% BCIs estimated with the three methods and the instant growth method; the true TMRCA is not contained in the 95% BCIs of the three methods.
Table 2. Performance comparison between BESTT and BEAST in simulations.
| % ENV | SRE | MRW | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Simulation | BESTT | EBSP | Skygrid | BESTT | EBSP | Skygrid | BESTT | EBSP | Skygrid |
| Constant | 100 | 100 | 100 | 0.30 | 0.24 | 0.24 | 1.16 | 1.49 | 4.41 |
| Exponential | 100 | 97 | 100 | 0.35 | 0.26 | 0.29 | 5.45 | 2.56 | 46.13 |
| Inst. growth | 97 | 94 | 100 | 0.61 | 2.65 | 22.60 | 105.60 | 14.95 | > 1000 |
BESTT, Bayesian Estimation of population size changes by Sampling Tajima’s Trees; % ENV, % envelope measure; EBSP, Extended Bayesian Skyline Plot; MRW, mean relative width; SRE, sum of relative errors. Bold values indicate best performance.
We note that BEAST Bayesian Skygrid (Gill et al. 2013) is a more comparable method to BESTT since it assumes Gaussian process priors on log like BESTT.
Computational performance of BESTT
BESTT approximates the posterior distribution (a) , where is a Tajima’s genealogy instead of (b) , where is a Kingman’s genealogy and is the labeled data, to estimate . These two posterior distributions are the same when every individual of the sample has its own private mutation group and no shared mutation groups. Otherwise, the number of Tajima’s trees compatible with observed data , i.e., Tajima’s trees such that , is smaller than the number of Kingman’s trees compatible with observed labeled data (Cappello and Palacios 2019). That is, we are required to estimate the posterior of a smaller number of trees. For this reason, we argue that Tajima’s coalescent is a more efficient model than Kingman’s coalescent for estimating the posterior distribution of . However, a single conditional likelihood calculation requires the sum over all possible allocation of mutation groups to branches of . Our algorithm only accounts for allocations constrained by the DAG and the ranked tree shape of . For the data depicted in Figure 2, A and B and of Figure 2C, there are only eight different possible allocation paths of all mutation groups to branches. In Appendix C, we detail how our algorithm finds these paths. The number of paths depends on the number of subtrees with the same family size path in the DAG and in the ranked tree shape. In the best case, our algorithm will find a path in , where is the number of nodes in the gene tree. In general, the number of allocation paths will be much smaller than the number of labeled trees compatible with a ranked tree shape. In our implementation, we estimate posterior (a) with MCMC. The main difference between our MCMC algorithm and the MCMC algorithm implemented in BEAST is the tree topology sampler. While our MCMC algorithm explores the space of ranked tree shapes with local move proposals of ranked tree shapes, BEAST explores the space of labeled, ranked tree shapes with local move proposals of labeled trees. A formal assessment of the efficiency of our MCMC algorithm and its comparison to the MCMC implementation in BEAST is beyond the scope of this manuscript and subject of future research.
Inferring human population demography from mtDNA
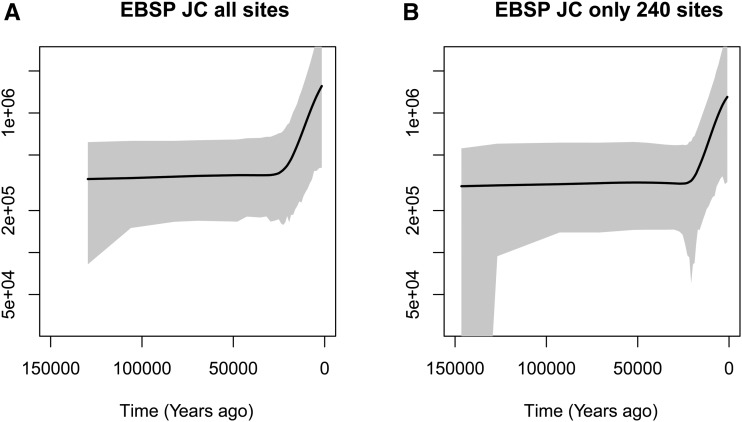
We selected samples of mtDNA at random from 107 Yoruban individuals available from the 1000 Genomes Project phase 3 (1000 Genomes Project Consortium et al. 2015). We retained the coding region, bp, according to the rCRS reference of human mtDNA (Anderson et al. 1981; Andrews et al. 1999) and removed 38 insertions/deletions. Of the 260 polymorphic sites, we retained 240 sites compatible with the infinite sites mutation model. The final file is available in https://github.com/JuliaPalacios/phylodyn. To encode our data as 0s and 1s, we use the inferred root sequence RSRS of Behar et al. (2012) to define the ancestral type at each site. To rescale our results in units of years, we assumed a mutation rate per site per year of (Rebolledo-Jaramillo et al. 2014). We compare our results with the Extended Bayesian Skyline method (Drummond et al. 2012) implemented in BEAST in Figure 9. When applying BEAST, we assumed the Jukes–Cantor mutation model. Both methods detect an inflection point ∼20 KYA followed by exponential growth. The mean TMRCA inferred for these YRI mtDNA samples with BESTT is ∼170 KYA with a 95% BCI of , while the mean TMRCA inferred with BEAST is ∼160 KYA with a 95% BCI of . In Appendix D, we include two more comparisons of BESTT and BEAST.
Figure 9.
Posterior inference of female effective population size from 35 mitochondrial DNA (mtDNA) samples from Yoruban individuals in the 1000 Genomes Project using our method Bayesian Estimation of population size changes by Sampling Tajima’s Trees (BESTT) (first plot) and the BEAST Extended Bayesian Skyline Plot (second plot). Posterior median curves are depicted as solid black lines and 95% credible intervals by shaded regions.
Discussion
The size of emergent sequencing data sets prohibits the use of standard coalescent modeling for inferring evolutionary parameters. The main computational bottleneck of coalescent-based inference of evolutionary histories lies in the large cardinality of the hidden state space of genealogies. In the standard Kingman coalescent, a genealogy is a random labeled bifurcating tree that models the set of ancestral relationships of the samples. The genealogy accounts for the correlated structure induced by the shared past history of organisms and explicit modeling of genealogies is fundamental for learning about the past history of organisms. However, the genomic era is producing large data sets that require more efficient approaches that efficiently integrate over the hidden state space of genealogies.
In this manuscript, we show that a lower-resolution coalescent model on genealogies, Tajima’s coalescent, can be used as an alternative to the standard Kingman coalescent model. In particular, we show that the Tajima coalescent model provides a feasible alternative that integrates over a smaller state space than the standard Kingman model. The main advantage in Tajima’s coalescent is modeling of the ranked tree topology as opposed to the fully labeled tree topology, as in Kingman’s coalescent.
A priori, the cardinality of the state space of ranked tree shapes is much smaller than the cardinality of the state space of labeled trees. However, in this manuscript we show that when the Tajima coalescent model is coupled with the infinite sites mutation model, the space of ranked tree shapes is constrained by the data and the reduction on the cardinality of the hidden state space of Tajima’s trees is even more pronounced than expected.
To leverage the constraints imposed by the data and the infinite sites mutation model, we apply Dan Gusfield’s perfect phylogeny algorithm (Gusfield 1991) to represent sequence alignments as a gene tree. We exploit the gene tree representation for conditional likelihood calculations and for exploring the state space of ranked tree shapes.
For the calculation of the likelihood of the data conditioned on a given Tajima’s genealogy, we augment the gene tree representation of the data with the Tajima’s genealogy and map observed mutations to branches. We define a DAG with the augmented gene tree. This new representation as a DAG allows for calculating the likelihood as a backtracking algorithm that transverses the gene tree from the leaves to the root. Our implementation’s computational bottleneck lies in the likelihood calculation. Given a Tajima’s genealogy, our likelihood algorithm sums over all possible allocations of mutation groups to branches. Although this number is generally much smaller than the number of labeled genealogies, our algorithm can be further optimized. In future studies, we will explore a sum-product type of algorithm for the likelihood calculation. In the present implementation, we are able to infer effective population size trajectories from samples of size on a regular personal laptop computer within few hours.
Our statistical framework draws on Bayesian nonparametrics. We place a flexible geometric random walk process prior on the effective population size that allows us to recover population size trajectories with abrupt changes in simulations. The inference procedure proposed in this manuscript relies on MCMC methods with three large Gibbs block updates of: coalescent times, effective population size trajectory, and ranked tree shape topology. We use Hamiltonian Monte Carlo updates for continuous random variables—coalescent times and effective population size—and a Metropolis–Hastings sampler for exploring the space of ranked tree shapes. For exploring the genealogical space, Markovtsova et al. (2000) suggest a joint local proposal for both coalescent times and topology. Here, we restrict our attention to the topology alone. A future line of research includes the development of a joint local proposal of coalescent times and ranked tree shapes. We also envision that a joint sampler of coalescent times and effective population size trajectories should improve mixing and convergence.
Our method does not model recombination, population structure, or selection. It assumes completely linked and neutral segments from individuals from a single population, and the infinite sites mutation model. While this model is a good approximation for some human molecular data, it is not appropriate for modeling molecular data from other organisms such as pathogens and viral populations. Finally, haplotype data of many organisms are usually sparse with few unique haplotypes presented at high frequencies. Since our algorithm exploits molecular data at the haplotype level, our proposed method is ideally suited for this scenario where the space of ranked tree shapes is drastically smaller than the space of labeled topologies.
Acknowledgments
We acknowledge the developers of R-ape, R-phangorn, and R-phylodyn, which facilitated our implementations, and thank the editor and two anonymous referees whose suggestions considerably improved the manuscript. This research was supported in part by National Institutes of Health grant R01 GM-131404 and funding from the Alfred P. Sloan Foundation to J.A.P, and by National Science Foundation CAREER award DBI-1452622 to S.R. A.V. was supported in part by the Chaire Modélisation Mathématique et Biodiversité program of VEOLIA Environnement, École Polytechnique, Museum National d’Histoire Naturelle, Fondation X. A.V. and J.A.P. were supported by the France-Stanford Center for Interdisciplinary Studies.
Appendix
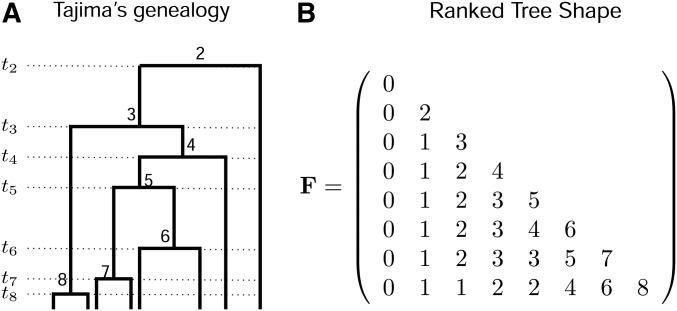
Appendix A: Matrix Representation of a Ranked Tree Shape
Our algorithms exploit the following encoding of a ranked tree shape by a triangular matrix of size , which we denote by (Figure A1). Recall that, by convention, and .
First, we declare that if . Next, the number of lineages through time is encoded on the diagonal of : for i in . Finally, for , the entry denotes the number of lineages that do not coalesce in the time interval ; in particular, and for every i in , denotes the number of singletons (i.e., external branches that have not coalesced) in the time interval (Figure A1). Other statistics of the ranked tree shape can be expressed in terms of the corresponding matrix . Among them, the number c of cherries is equal to the number of times that the number of singletons decreases by two between lines i and , since such an event means that the coalescence separating these two epochs was that of two external branches. That is,
Appendix B
Detailed allocation of mutation groups along
The latent allocation random variables are constrained by the information in the Tajima’s genealogy . In a given , every subtree is labeled by its ranking from past to present (Figure A1). Subtree i is subtended by branch with length , for . We will assume that , the length of the root branch, is 0. Let c be the number of cherries (nodes with two leaves) in ; the two branches of a given cherry share the same label . The actual label of external branches is arbitrary but, for ease of exposition in our figures, we first label the cherries’ branches from left to right by ; singleton branches are labeled from left to right by (Figure 2C). As mentioned before, the length of is the number of the corresponding sister nodes in that were grouped together in forming node . In this case, denotes a collection of vectors of branch labels in subtending the child node subtrees of node . corresponds to the branch subtending from the leftmost child node of on , corresponds to the branch subtending from the next child node of , etc., and corresponds to the branch subtending from the rightmost child node of on . Observe that, since we group some of the leaf nodes in into a single node in , any may be a vector of branch labels; for example and in Figure 3B.
Appendix C
Algorithms for conditional likelihood calculation
The following two algorithms detail the calculation of . is encoded in , the observed data as a tree structure. Each node in has number of descendants (or lineages) and mutation information attached to it. Tajima’s genealogy is encoded as , which contains the ranked tree shape , and , which contains the vector of coalescent times multiplied by the mutation rate μ.
Algorithm 1 Calculate Likelihood procedure
Input: ,
Output: Log Likelihood
1: Initiate to be the set of leaf nodes of with at least one descendant. Initiate and to be zero. Initiate current_path to be empty.
2: Call CalcLL_recursive(LL, index, current_path, Fpath, times, Genetree).
3: return
Algorithm 2 CalcLL_recursive(, , current_path, , , ) procedure
1: if {When a complete path node is found} then
2: for in do
3: Calculate log likelihood based on and number of mutations of in current_path.
4: Accumulate to total log likelihood
5: end for
6: else
7: for in do
8: Check compatibility of the , according to the given .
9: if is compatible with then
10: Update by assigning it to the current step in
11: Update . If a has been mapped entirely, remove from pool, update its parent , and potentially add parent node to if parent node has not been entirely assigned.
12: Append this to current_path
13: Call CalcLL_recursive(LL, index + 1, current_path, Fpath, Genetree)
14: Restore previous , , and current_path
15: end if
16: end for
17: end if
To illustrate algorithms 1 and 2, we use our examples of Figure 2 and Figure 3. Algorithm 1 initiates the pool with nodes . Then, Algorithm 2 cycles through this list. Assume the first node is . This node has descendants and the ancestry is with sizes: . On the other hand, the first coalescent event (from present to past) labeled 16 in (Figure 3A) has ancestry with sizes: . Therefore, this node is compatible. The node is removed from the pool, its parent node added to the pool, and is assigned to the path. At this time current_path = Z8 and the pool becomes: . The algorithm then cycles through this list and picks . This node has size ancestry . On the other hand the second coalescent event labeled 15 has size ancestry: . Since the node size ancestry is contained in the second coalescent event’s size ancestry, this node is compatible. The current path becomes current_path = Z8 − Z10 and the pool becomes . We continue this procedure until we reach the path current_path = .
Once a path is found, the algorithm backtracks the path until there is one compatible node and the path continues to grow. A sequence of backtracking and growing is the following:
In the first sequence of steps , the path decreases. This happens because there are no alternative compatible paths at the point when the sequence starts to grow until step 10. At step 10, the algorithm does not find a compatible way to keep growing the path so the algorithm starts to backtrack again until step 12. From steps 12 to 18, the algorithm grows the path until a new complete path has been found. A complete path has the correspondence of coalescent events to nodes in the gene tree. The first element of the path corresponds to the coalescent event at time , the second element of the path corresponds to the second coalescent event at time . The last element of the path is when all sequences coalesce at time . In this example, the algorithm finds eight paths. Once the paths are found, the algorithm computes the likelihood and the result is the sum of the likelihoods of the eight paths.
Markovian proposal of ranked tree shapes
The following algorithm generates a new ranked tree shape from a Markovian proposal and outputs the corresponding transition probabilities. This proposal is used in Metropolis–Hastings updates for ranked tree shapes.
Algorithm 3 Transition proposals for ranked tree shapes
-
Input: Output: , ,
1. Set .
2. Sample with uniform discrete probability a coalescent event k from the set Set .
3. If lineage k coalesces at time (Figure 4, Case A)
-
If the lineages coalescing at time are singletons (Figure 4, Case A, lineages 1 and 2 in )
No sampling required to distinguish between two singletons. Set .
Update : merge one singleton with the lineage coalescing at (excluding lineage k) in , then merge the second singleton at time with lineage k.
Compute the probability of restoring the ordering of to .
-
Else
Sample one of the lineages coalescing at time with uniform discrete probability. Set .
Update : merge the sampled lineage with the one coalescing at time in . At time , merge the lineage not sampled with the new lineage k.
Compute the probability of restoring the ordering of to .
Else (Figure 4, Case B)
Swap the coalescent events. Lineages descending from k are now set to coalesce at time and lineages previously descending from are now set to coalesce at time . Set and .
4. , .
Appendix D
We replicated the BEAST EBSP analysis of the 35 Yoruban individuals from the 1000 Genomes Project phase 3 using the whole mtDNA coding region consisting of 15,409 sites. In both cases, we assumed the Jukes–Cantor mutation model (Jukes and Cantor, 1969). Figure D1 shows the comparison between EBSP inference from the 240 segregating sites retained in Inferring human population demography from mtDNA that are compatible with the infinite sites mutation model assumption. In both cases, we recover very similar trajectories.
In addition, we compared our results with BEAST Bayesian Skyline Plot (BSP) (Drummond and Rodrigo, 2000). For our reduced data set of 240 segregating sites, we could not generate valid inference of with Metropolis–Hastings acceptance probability > 0. Instead, we were able to generate results with BEAST BSP from the complete data set of 15,409 sites. The comparison of our method from 240 segregating sites to BEAST BSP from 15,409 sites is depicted in Figure D2.
Appendix E
In Figure E1A, we show the data from Figure 2A with an additional haplotype (10) with frequency 1 and an additional column grouped with mutation group h (not shown in the table). In Figure E1B we show the corresponding perfect phylogeny. This new perfect phylogeny has a new tip with black label 1 (frequency) subtending from a branch with 0 mutations (red label). The path from the leaf to the root shows that this haplotype has a unique mutation corresponding to mutation group a. We note that mutation group labels carry no information. We incorporate the labels in the figure for ease of exposition. Since mutation group h has now multiplicity 2, the branch labeled h has now a red label 2.
Figure A1.
Ranked tree shape. Left: Example of a Tajima’s genealogy (redrawn from Figure 1A) with coalescent events ranked from 2 at time to n at time . Right: The corresponding matrix, with , that encodes the ranked tree shape information of the Tajima’s genealogy on the left. denotes the number of lineages that do not coalesce in the time interval . In particular, for i in denotes the number of singletons (external branches that have not coalesced) in the time interval .
Figure D1.
Posterior inference of female effective population size from 35 mtDNA samples from Yoruban individuals in the 1000 Genomes Project using BEAST EBSP (first plot) from all 15,409 sites and the BEAST EBSP (second plot) from the 240 segregating sites retained. In both cases, the mutation model assumed is Jukes Cantor (JC). Posterior median curves are depicted as solid black lines and 95% credible intervals by shaded regions.
Figure D2.
Posterior inference of female effective population size from 35 mtDNA samples from Yoruban individuals in the 1000 Genomes Project using our BESTT (first plot) from only 240 segregating sites and the BEAST BSP (second plot) from all the 15,409 sites. Posterior median curves are depicted as solid black lines and 95% credible intervals by shaded regions.
Figure E1.
Second example of Perfect Phylogeny. (A). Compressed data representation of sequences and (columns, only the first 10 of which are shown), comprised of 10 haplotypes and 13 mutation groups. This data table has one more haplotype (10) and one more mutation labeled h than the example of Figure 2. (B). Gene tree representation of the data in (A). Red numbers indicate the cardinality of each mutation group (number of columns with the same label in (A). Black letters indicate the mutation group (column labels in (A), and black numbers indicate the frequency of the corresponding haplotype.
Footnotes
Communicating editor: G. Coop
Literature Cited
- 1000 Genomes Project Consortium, Auton A., Abecasis G. R., Altshuler D. M, Durbin R. M. et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberer A. J., Stamatakis A., and Ronquist F., 2016. An efficient independence sampler for updating branches in Bayesian Markov chain Monte Carlo sampling of phylogenetic trees. Syst. Biol. 65: 161–176. 10.1093/sysbio/syv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R. et al. , 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- Andrews R. M., Kubacka I., Chinnery P. F., Lightowlers R. N., Turnbull D. M. et al. , 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23: 147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Behar D. M., Van Oven M., Rosset S., Metspalu M., Loogväli E.-L. et al. , 2012. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90: 675–684. 10.1016/j.ajhg.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A., Wang Y. R., and Song Y. S., 2015. Efficient inference of population size histories and locus-specific mutation rates from large-sample genomic variation data. Genome Res. 25: 268–279. 10.1101/gr.178756.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello L., and Palacios J. A., 2019. Sequential importance sampling for multi-resolution Kingman-Tajima coalescent counting. arXiv. Available at: https://arxiv.org/abs/1902.05527. [DOI] [PMC free article] [PubMed]
- Disanto F., and Wiehe T., 2013. Exact enumeration of cherries and pitchforks in ranked trees under the coalescent model. Math. Biosci. 242: 195–200. 10.1016/j.mbs.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Donnelly P., and Tavaré S., 1995. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 29: 401–421. 10.1146/annurev.ge.29.120195.002153 [DOI] [PubMed] [Google Scholar]
- Drummond A., and Rodrigo A., 2000. Reconstructing genealogies of serial samples under the assumption of a molecular clock using serial-sample UPGMA. Mol. Biol. Evol. 17: 1807–1815. 10.1093/oxfordjournals.molbev.a026281 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., Shapiro B., and Pybus O. G., 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22: 1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D., and Rambaut A., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. S., Lemey P., Faria N. R., Rambaut A., Shapiro B. et al. , 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol. Biol. Evol. 30: 713–724. 10.1093/molbev/mss265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. C., and Tavaré S., 1994a Simulating probability distributions in the coalescent. Theor. Popul. Biol. 46: 131–159. 10.1006/tpbi.1994.1023 [DOI] [Google Scholar]
- Griffiths R. C., and Tavaré S., 1994b Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 344: 403–410. 10.1098/rstb.1994.0079 [DOI] [PubMed] [Google Scholar]
- Griffiths R. C., and Tavaré S., 1995. Unrooted genealogical tree probabilities in the infinitely-many-sites model. Math. Biosci. 127: 77–98. 10.1016/0025-5564(94)00044-Z [DOI] [PubMed] [Google Scholar]
- Griffiths R. C., and Tavaré S., 1996. Monte Carlo inference methods in population genetics. Math. Comput. Model. 23: 141–158. 10.1016/0895-7177(96)00046-5 [DOI] [Google Scholar]
- Gronau I., Hubisz M. J., Gulko B., Danko C. G., and Siepel A., 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43: 1031–1034. 10.1038/ng.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield D., 1991. Efficient algorithms for inferring evolutionary trees. Networks 21: 19–28. 10.1002/net.3230210104 [DOI] [Google Scholar]
- Heled J., and Drummond A. J., 2008. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8: 289. 10.1186/1471-2148-8-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A., Uyenoyama M. K., and Wiuf C., 2008. Importance sampling for the infinite sites model. Stat. Appl. Genet. Mol. Biol. 7: Article32. 10.2202/1544-6115.1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., and Cantor R. C., 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, edited by Munro H. N. Academic Press, New York: 10.1016/B978-1-4832-3211-9.50009-7 [DOI] [Google Scholar]
- Karcher M. D., Palacios J. A., Lan S., and Minin V. N., 2017. phylodyn: an r package for phylodynamic simulation and inference. Mol. Ecol. Resour. 17: 96–100. 10.1111/1755-0998.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J., 1982a The coalescent. Stochastic Process. Appl. 13: 235–248. 10.1016/0304-4149(82)90011-4 [DOI] [Google Scholar]
- Kingman J. F. C., 1982b Exchangeability and the evolution of large populations, pp. 97–112 in Exchangeability in Probability and Statistics, edited by Koch G. and Spizzichino F.. North-Holland, Amsterdam. [Google Scholar]
- Kuhner M. K., 2006. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22: 768–770. 10.1093/bioinformatics/btk051 [DOI] [PubMed] [Google Scholar]
- Kuhner M. K., and Smith L., 2007. Comparing likelihood and Bayesian coalescent estimation of population parameters. Genetics 175: 155–165. 10.1534/genetics.106.056457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner M. K., Yamato J., and Felsenstein J., 1998. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan S., Palacios J. A., Karcher M., Minin V., and Shahbaba B., 2015. An efficient Bayesian inference framework for coalescent-based nonparametric phylodynamics. Bioinformatics 31: 3282–3289. 10.1093/bioinformatics/btv378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. 10.1038/nature10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsova L., Marjoram P., and Tavaré S., 2000. The age of a unique event polymorphism. Genetics 156: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V. N., Bloomquist E. W., and Suchard M. A., 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25: 1459–1471. 10.1093/molbev/msn090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal, R. M., 2011 MCMC using Hamiltonian dynamics, pp.113–162 in Handbook of Markov Chain Monte Carlo, edited by S. Brooks, A. Gelman, G. L. Jones, and X.-L. Meng. CRC Press, Boca Raton, FL. [Google Scholar]
- Palacios, J. A., and V. N. Minin, 2012 Integrated nested Laplace approximation for Bayesian nonparametric phylodynamics, pp. 762–735 in Proceedings of the Twenty-Eighth Conference on Uncertainty in Artificial Intelligence. Association for Uncertainty in Artificial Intelligence Press, Arlington, VA. [Google Scholar]
- Palacios J. A., and Minin V. N., 2013. Gaussian process-based Bayesian nonparametric inference of population size trajectories from gene genealogies. Biometrics 69: 8–18. 10.1111/biom.12003 [DOI] [PubMed] [Google Scholar]
- Palacios J. A., Wakeley J., and Ramachandran S., 2015. Bayesian nonparametric inference of population size changes from sequential genealogies. Genetics 201: 281–304. 10.1534/genetics.115.177980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B., and Yang Z., 2003. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. E., and Williams C. K. I., 2006. Gaussian Processes for Machine Learning. MIT Press, Cambridge, MA. [Google Scholar]
- Rebolledo-Jaramillo B., Su M. S., Stoler N., McElhoe J. A., Dickins B. et al. , 2014. Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 111: 15474–15479. 10.1073/pnas.1409328111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainudiin R., Thornton K., Harlow J., Booth J., Stillman M. et al. , 2011. Experiments with the site frequency spectrum. Bull. Math. Biol. 73: 829–872. 10.1007/s11538-010-9605-5 [DOI] [PubMed] [Google Scholar]
- Sainudiin R., Stadler T., and Véber A., 2015. Finding the best resolution for the Kingman-Tajima coalescent: theory and applications. J. Math. Biol. 70: 1207–1247. 10.1007/s00285-014-0796-5 [DOI] [PubMed] [Google Scholar]
- Schiffels S., and Durbin R., 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. 10.1038/ng.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep K. P., 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S., Harris K., and Song Y. S., 2013. Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194: 647–662. 10.1534/genetics.112.149096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., and Hudson R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., and Donnelly P., 2000. Inference in molecular population genetics. J. R. Stat. Soc. Series B Stat. Methodol. 62: 605–635. 10.1111/1467-9868.00254 [DOI] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., (2004). Part I: ancestral inference in population genetics, pp 1–188 in Lectures on Probability Theory and Statistics, Volume 1837 of Lecture Notes in Mathematics. Springer-Verlag, New York. [Google Scholar]
- Terhorst J., Kamm J. A., and Song Y. S., 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49: 303–309. 10.1038/ng.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. 10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Whidden C., and Matsen F. A. IV, 2015. Quantifying MCMC exploration of phylogenetic tree space. Syst. Biol. 64: 472–491. 10.1093/sysbio/syv006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., 2010. Exact computation of coalescent likelihood for panmictic and subdivided populations under the infinite sites model. IEEE/ACM Trans. Comput. Biol. Bioinformatics 7: 611–618. 10.1109/TCBB.2010.2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The methods presented in this article are implemented in the open source R package phylodyn (https://github.com/JuliaPalacios/phylodyn). The data files necessary to reproduce both the simulated and real data analyses are provided with the R package.