Rowan et al. generated a dataset of over 17,000 meiotic crossovers (COs) from over 2000 F2 individuals from a single Arabidopsis thaliana cross. The unprecedented density of COs and the high-quality reference genomes of the two...
Keywords: recombination, meiosis, genetic map, structural variation, double-strand break
Abstract
Many environmental, genetic, and epigenetic factors are known to affect the frequency and positioning of meiotic crossovers (COs). Suppression of COs by large, cytologically visible inversions and translocations has long been recognized, but relatively little is known about how smaller structural variants (SVs) affect COs. To examine fine-scale determinants of the CO landscape, including SVs, we used a rapid, cost-effective method for high-throughput sequencing to generate a precise map of >17,000 COs between the Col-0 and Ler-0 accessions of Arabidopsis thaliana. COs were generally suppressed in regions with SVs, but this effect did not depend on the size of the variant region, and was only marginally affected by the variant type. CO suppression did not extend far beyond the SV borders and CO rates were slightly elevated in the flanking regions. Disease resistance gene clusters, which often exist as SVs, exhibited high CO rates at some loci, but there was a tendency toward depressed CO rates at loci where large structural differences exist between the two parents. Our high-density map also revealed in fine detail how CO positioning relates to genetic (DNA motifs) and epigenetic (chromatin structure) features of the genome. We conclude that suppression of COs occurs over a narrow region spanning large- and small-scale SVs, representing an influence on the CO landscape in addition to sequence and epigenetic variation along chromosomes.
SEXUAL reproduction generates genetic diversity from standing variation because meiotic crossovers (COs) between the maternal and paternal chromosomes provide new combinations of alleles that can be transmitted to the next generation (Barton and Charlesworth 1998). This novel variation can contribute to phenotypes upon which selection can act (Burt 2000), bring together beneficial mutations that arose independently (Muller 1932; Fisher 1999), and it can break linkage between beneficial and deleterious mutations (Peck 1994; Gray and Goddard 2012). Therefore, sexual reproduction, through the action of COs, provides a mechanism for the acceleration of adaptation. COs ensure proper chromosome segregation (Hall 1972) and are one result of several alternative outcomes of the repair of programmed DNA double-strand breaks (DSBs) that occur during meiosis, which also include non-CO (NCO) and intersister repair (Szostak et al. 1983; Keeney et al. 1997; Keeney 2001). A deeper understanding of the various influences affecting the repair of meiotic DSBs, especially those that favor the generation of COs, will strengthen our knowledge of processes that shape adaptive genetic variation.
One important factor that influences the positioning of COs along chromosomes, collinearity, has been known for almost a century. Alfred Sturtevant first proposed that a rearrangement of chromosome structure, such as an inverted segment, would suppress CO formation in the heterozygous state (Sturtevant 1921). He later confirmed that a region of chromosome III in Drosophila melanogaster that exhibited CO suppression in the heterozygous state did indeed contain an inversion (Sturtevant 1926). Such inverted regions can encompass many genes, with the consequence that alleles in the inverted segment are passed down as a single nonrecombining locus. When genes that are linked in such a way confer a particularly advantageous trait as a single unit, they form a supergene (Schwander et al. 2014; Thompson and Jiggins 2014; Charlesworth 2016), a concept that had its origins in Dobzhansky’s work and was formalized by Mather (1950). A notable example from the animal kingdom comes from ruff birds (Philomachus pugnax), where an inversion has trapped 125 genes in a nonrecombining haplotype, creating a supergene that governs both ornamental plumage and mating/social behaviors (Küpper et al. 2016). Another inversion-based supergene underlies wing pattern morphology and mimicry in butterflies (Joron et al. 2011). In plants, a chromosomal inversion is responsible for variation in life-history strategies, and adaptation to temperature and precipitation in yellow monkeyflowers (Lowry and Willis 2010; Oneal et al. 2014). Inversions are not the only type of chromosomal rearrangement resulting in CO suppression. Dobzhansky first observed reduced crossing over near interchromosomal translocations in D. melanogaster (Dobzhansky 1931), and this has been observed in other species (McKim et al. 1988; Herickhoff et al. 1993). Interestingly, some translocations can enhance recombination in “intervals” flanking the breakpoint (Sybenga 1970).
Smaller-scale structural variants (SVs) of different types have also been implicated in the suppression of meiotic recombination. For example, a 70-kb transposition on Arabidopsis thaliana chromosome 3 showed extreme local suppression of recombination at the excision site in an F2 cross between accessions BG-5 and Kro-0 (Alhajturki et al. 2018). This type of rearrangement creates an insertion/deletion (indel) polymorphism at both the original locus and the new location. It is therefore expected that other types of indels would also show local CO suppression. Indeed, transgene arrays in the order of dozens of kilobases induced local CO suppression in the nematode Caenorhabditis elegans (Hammarlund et al. 2005). In mouse, the cooccurrence of a small insertion (824 bp) and a small deletion (703 bp) was associated with a local reduction in the CO rate (Hsu et al. 2000).
Despite a large body of literature supporting a role for structural variation in determining CO positioning, there has been little systematic investigation of whether the size or the type of variant has a differential impact on the local CO rate. In Drosophila interspecies crosses, suppression of meiotic COs can extend for >1 Mb outside the borders of inversions (Kulathinal et al. 2009; Stevison et al. 2011). However, it is unclear how far beyond the variant borders the effects of CO suppression can extend in other species or for other types of SVs. In this study, we developed a new fast and cost-effective protocol for the preparation of Illumina genomic DNA sequencing libraries (Illumina, San Diego, CA). With this method, we sequenced the genomes of nearly 2000 F2 individuals from a cross between the A. thaliana accessions Col-0 and Ler-0 (hereafter referred to as Col and Ler). After including previously published sequence data for additional F2 recombinants from this cross (Choi et al. 2016; Underwood et al. 2018), we generated a set of >17,000 COs at high resolution (median interval size: 1102 bp) and examined them in the context of the precise knowledge of SVs available from high-quality reference genome sequences for these two accessions. We then compared the relationship between COs and SVs to other known influences on recombination rate.
Materials and Methods
Plant growth and DNA extraction
Col qrt1-2 CEN3 420 (Melamed-Bessudo et al. 2005; Francis et al. 2007) × Ler F2 seeds (Ziolkowski et al. 2015) were stratified for 4–7 days at 4° before sowing on soil in 40-pot trays. A portion of the seeds (∼400) had been subjected to screening for a lack of COs in the 420 CO reporter region on chromosome 3 (Melamed-Bessudo et al. 2005). Plants were cultivated in a greenhouse supplemented with additional light, and covered with a plastic dome for the first week of growth to promote germination and establishment. Leaf samples (∼0.5–1 cm long) were taken from plants between 4 and 8 weeks of age, collected in polypropylene tubes in a 96-well rack, and frozen at −80° until DNA extraction.
To extract DNA, the frozen leaf samples were ground using a TissueLyser (QIAGEN, Valencia, CA) with steel beads. A volume of 500 μl of DNA Extraction Buffer (200 mM Tris-HCl, 25 mM EDTA, 1% SDS, and 80 μg/ml RNase A) was added to the frozen powder and the tubes were mixed by gentle inversion before incubation at 37° for 1 hr. The tubes were centrifuged at 3000 × g for 5 min and a volume of 400 μl of the supernatant was transferred to each well of a 96-deep-well plate containing 130 μl of potassium acetate solution (5 M potassium acetate and 7% Tween-20); this and the following steps were performed with the aid of a Tecan Freedom EVO 150 liquid handling robot (Tecan, Männedorf, Switzerland) fitted with an eight-channel liquid handler (LiHa), a 96-channel multi-channel arm (MCA), and a gripper. Plates were covered with an adhesive seal and mixed gently by inversion, before incubation for 10 min on ice and centrifugation for 5 min at 3000 × g. A volume of 400 μl of the supernatant was transferred to a fresh 96-deep-well plate containing 400 μl of SeraPure solid-phase reverse immobilization (SPRI) beads (Rowan et al. 2017) and mixed several times by pipetting using a Tecan liquid-handling robot, before incubation for 5 min at room temperature to allow the DNA to bind to the beads. The plate was placed on a magnetic plate stand, and the supernatant was removed and discarded after all of the beads had been drawn to the magnet (∼5 min). The beads were washed three times with 900 μl of 80% EtOH. After the last wash, all traces of EtOH were removed and the beads were left to dry at room temperature for 5 min. The DNA was eluted from the beads in 100 μl of 10 mM Tris-HCl (pH 8) mixed for 1 min using a plate vortex mixer, and incubated overnight at 4°. The plates were placed on a magnet for 5 min until the samples were clear of beads, and the DNA solutions were transferred to a 96-well PCR plate and stored at −20° until library preparation.
Library preparation and sequencing using the Nextera Low Input, Transposase Enabled protocol
DNA samples were first quantified using the Quant-It kit (Thermo Fisher Scientific, Waltham, MA) with a Tecan Infinite M200 PRO fluorescence microplate reader (Tecan) and the Tecan Magellan analysis software. The samples were normalized to 2 ng/μl using a Tecan liquid-handling robot. After normalization, a subset of samples from each plate was quantified using a Qubit 2.0 fluorometer with the double-stranded DNA high-sensitivity assay kit to verify the concentration and assess the variability among samples. If the concentrations were on average 2 ng/μl with less than twofold variation among samples, then all samples in a plate were diluted uniformly to 0.25–0.5 ng/μl. Otherwise, the normalization was repeated. Once the samples were at 0.25–0.5 ng/μl, then 2 μl of DNA per sample were transferred to a fresh plate and mixed with 2.1 μl water, 0.82 μl DNA template:tagmentation (TD) buffer, and 0.08 μl TD enzyme from the Illumina Nextera 24-sample kit (catalog number FC-121-1030). Note that the TD enzyme and TD buffer are now sold separately by the manufacturer (catalog numbers 15027865 and 15027866) by special request, and the kit used to prepare these libraries has been discontinued. Plates were sealed with a Microseal B adhesive plate seal (Bio-Rad, Hercules, CA) and incubated for 10 min at 55°. After allowing the plates to cool to room temperature, the tagmented DNA was amplified by PCR using the KAPA2G Robust PCR kit (Sigma [Sigma Chemical], St. Louis, MO) using the GC buffer, along with 0.2 μM each of custom P5 and P7 indexing primers (Supplemental Material, File S1) using the following cycling conditions: 72° for 3 min, 95° for 1 min, 14 cycles of 95° for 10 sec, 65° for 20 sec, and 72° for 3 min.
Following PCR, a 5-μl sample of each library was mixed with 15 µl of 10 mM Tris-HCl (pH 8) and 5 µl of 6× loading dye, and analyzed by electrophoresis using a 2% agarose gel at 140 V for 15 min to evaluate library success and size distribution. Samples were then pooled by mixing 3 μl of each library into a single tube using a Tecan liquid-handling robot. Note that the indexing oligos in File S1 allow for 576 unique combinations of P5 and P7 indices, but additional oligos can be easily designed if higher levels of multiplexing are desired.
Size selection was performed on 100 μl of the 96-sample pool by adding 160 μl of Serapure SPRI beads (Rowan et al. 2017), which had been diluted to 0.6× strength with water, and incubating for 5 min at room temperature before placing in a magnetic 1.5-ml tube stand for 5 min. A volume of 250 μl of the supernatant was transferred to a fresh tube and 700 μl of 80% EtOH were added to the tube containing the beads (bead fraction 1). A volume of 30 μl full-strength Serapure SPRI beads was added to the supernatant and incubated for 5 min at room temperature, and then for 5 min in a magnetic 1.5-ml tube stand before transferring 280 μl of the supernatant to a fresh tube. A volume of 700 μl of 80% EtOH was added to the beads that remained after this transfer (bead fraction 2). A volume of 112 μl full-strength Serapure SPRI beads was added to the supernatant and incubated for 5 min at room temperature, and for 5 min in a magnetic 1.5-ml tube stand before removing and discarding the supernatant. A volume of 700 μl of 80% EtOH was added to the beads that remained after this transfer (bead fraction 3), and the tubes were incubated for ≥30 sec before removing the EtOH from all bead fractions and doing a second wash with 700 μl of 80% EtOH. After ≥30 sec in the second EtOH wash, all EtOH was removed from the tubes. A flash spin was performed to collect any remaining EtOH at the bottom of the tube, which was removed with a pipette. The bead samples were left to dry at room temperature for 5 min or until no EtOH remained.
The libraries for each of the three bead fractions were eluted in 26 μl of 10 mM Tris-HCl and incubated in a standard tube rack for 5 min. The tubes were then transferred back to the magnetic rack and 25 µl of the eluted libraries for each fraction were transferred to a fresh tube after all of the beads had been drawn to the magnet. Each of the three size fractions was analyzed on a Bioanalyzer to determine the size ranges and select the appropriate fraction for Illumina sequencing (often bead fraction 3, which contained 300–500-bp fragments). Samples were sequenced to an estimated depth of 1–2× average whole-genome coverage on an Illumina HiSeq 3000 instrument.
Sequence analysis and CO localization
The raw reads for all 1920 sequenced samples were demultiplexed and evaluated for quality using MultiQC v. 0.9.dev0 (Ewels et al. 2016), before adapter trimming and quality filtering using Trimmomatic v. 0.36 (Bolger et al. 2014) and sickle (https://github.com/najoshi/sickle). The reads were then aligned to the A. thaliana Col reference genome (TAIR10) with the Burrows-Wheeler Aligner (Li and Durbin 2009) v. 0.6.2 using the bwa aln algorithm with default parameters, but specifying a maximum of 10 mismatches. Demultiplexed raw reads for an additional 192 wild-type Col × Ler F2 samples were downloaded from http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4657/samples/ and for an additional 171 Col × Ler F2 samples from http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5476/samples/, and subjected to the same methods of quality filtering and alignment. Paired-end reads were aligned independently, then merged using sampe with the default parameters and specifying a maximum insert size of 500 bp.
SAMtools v.1.2 and BCFtools v.1.2 (Li et al. 2009) were used to obtain read count data for variant positions to generate the input files for the Trained Individual GenomE Reconstruction (TIGER) CO analysis pipeline (Rowan et al. 2015). A set of 545,481 total variants, including single-nucleotide polymorphisms (SNPs) and small (1–3 bp) indel polymorphisms based on assembly comparisons between Ler and Col, was filtered to SNPs only to generate the “complete” marker file (519,525 positions). The intersection between this and a set of 291,973 high-quality Col/Ler SNPs based on short-read data (kindly provided by Korbinian Schneeberger, Max Planck Institute for Plant Breeding Research, Cologne) yielded a final set of 237,288 SNPs as the “corrected” marker file.
Using this set as the input for TIGER, breakpoints were predicted on each sample independently, with the biparental mode setting and default parameters (Rowan et al. 2015). Briefly, genotypes were called for individual high-quality marker positions with the TIGER Basecaller module, and the sequence of genotypes along the chromosomes was reconstructed for each individual sample with the TIGER Hidden Markov Model, using a sample-specific error probability model generated from a β-binomial model, with which we inferred the underlying genotype probability based on the allele frequency distributions. Final recombination breakpoint resolution was refined by gathering information from additional markers (the complete marker set described above) around the breakpoints. Refined breakpoint data files for all individuals were concatenated into a single text file for downstream analysis in R. Individuals with <0.025× coverage, with excessive breakpoints, without breakpoints, or with extended marker blocks where the allele frequency was not 0, 0.5, or 1 were removed from further analysis.
For downstream analysis, a CO interval was designated as the region in between the last marker of one genotype and the first marker of the new genotype. CO positions were estimated as the midpoint between the positions of these two markers. CO positions that appeared to be double COs <500 kb apart were removed as likely false positives.
We obtained the positions and annotations of Col/Ler SVs from a high-quality Ler genome assembly (Zapata et al. 2016). The positions and annotations for disease resistance genes [including nucleotide-binding site leucine-rich repeat proteins (NLRs)] for Col were obtained from Choi et al. (2016), and we performed a comparative analysis with Ler annotations (kindly provided by Korbinian Schneeberger, Max Planck Institute for Plant Breeding Research, Cologne) to determine NLR copy number variation and SVs. The DNase I-hypersensitive (DNase I HS) hotspot data for 7-day-old seedlings (Sullivan et al. 2014) were downloaded from http://plantregulome.org/. SPO11-1-oligo and micrococcal nuclease sequencing (MNase-seq) data were obtained from Choi et al. (2016, 2018). Roger Deal and Marco Bajkic kindly provided raw Assay for Transposase Accessible Chromatin sequencing (ATAC-seq) data from Maher et al. (2018), and the data generated from meristem tissue was used in our analyses.
Statistical analysis of CO intervals and CO positions was performed using the R software environment (R Core Team 2017). Centromeres were defined as chromosome 1: 13,700,000–15,900,000; chromosome 2: 2,450,000–5,500,000; chromosome 3: 11,300,000–14,300,000; chromosome 4: 1,800,000–5,150,000; and chromosome5: 11,000,000–13,350,000 to include the regions previously defined by markers (Copenhaver et al. 1999; Arabidopsis Genome Initiative 2000). Overlaps between CO intervals and the regions of SVs, NLR loci, SPO11-1-oligo hotspots, and ATAC-seq sites were analyzed with regioneR (Gel et al. 2016). Additional plots were prepared using ggplot2 (http://ftp.auckland.ac.nz/software/CRAN/src/contrib/Descriptions/ggplot.html). Gene ontology (GO) enrichment in “CO deserts” was performed using PANTHER version 14.1 (http://pantherdb.org/). COs and SVs were grouped into 10-kb fixed windows and compared to DNA methylation data (Yelina et al. 2015) for the same genomic windows. Motif enrichment analyses were performed with Multiple Em for Motif Elicitation (MEME) (Bailey et al. 2009) after subsetting the CO data to 500 and 1000 randomly selected COs per chromosome to achieve a feasible runtime. Parameters for MEME were set to search for motifs of lengths 2–10 bp with zero or one occurrence per sequence.
Data availability
Raw read data include all 1920 individuals (including those that were later filtered out as described above) and can be found under ArrayExpress accession number E-MTAB-8165. The oligos used for the Nextera-based library preparation are available in File S1, and the full list of CO intervals and midpoints is in File S2. File S3 is a list of genes in the regions identified by 50-kb sliding window analysis with the top 5% highest CO frequencies. File S4 is a list of genes in the 95th length percentile for regions without COs in our data set. File S5 is an annotated list of NLR loci with a comparison of locus structure between Col and Ler. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9733838.
Results
Development and validation of the Nextera Low Input, Transposase Enabled protocol
We developed a fast and inexpensive library preparation protocol—Nextera Low Input, Transposase Enabled (Nextera LITE)—derived from the Illumina Nextera 24-sample reagent kit. Briefly, this protocol relies on dilution of the kit components, small reaction volumes, an optimal TD enzyme ratio, and optimized PCR amplification. For A. thaliana, we obtained fragment sizes suitable for the Illumina platform using 0.5–1 ng of DNA per 0.08 μl of TD enzyme (see Materials and Methods). Tagmented DNA fragments were directly amplified and dual-indexed in a PCR reaction without an intervening clean-up step, requiring <2 hr from start to finish and a reagent cost of $2.00–2.50 per library (Figure S1). We used a liquid-handling robot for several steps, but the entire protocol can also be easily performed by hand.
Generating a high-density genome-wide CO map
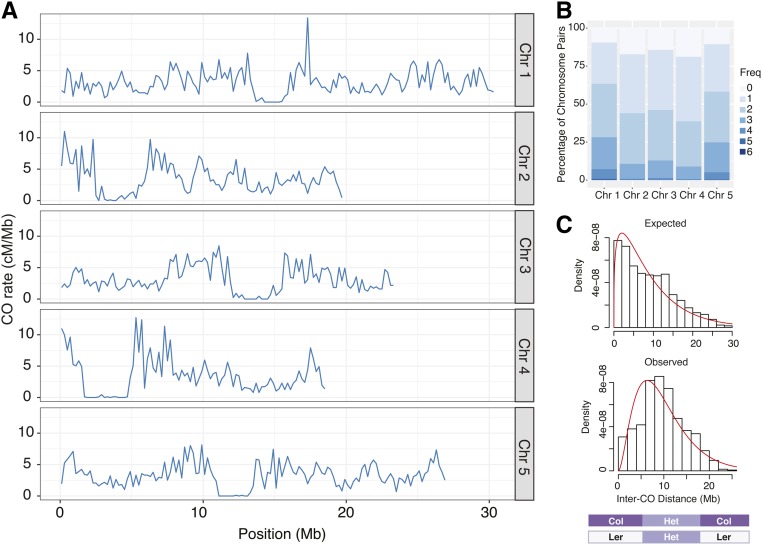
The average read depth per library gave 1–2× genome-wide coverage per individual (Figure S2). Of the 1920 Col × Ler F2 sequenced individuals, 95% had sufficient data quality and coverage (≥0.025× mean coverage of reads with quality scores ≥ 25) to call COs. We added data from 363 previously analyzed Col × Ler F2 individuals (Choi et al. 2016; Underwood et al. 2018) that were produced using another library preparation method (Rowan et al. 2015, 2017) with slightly higher average coverage. After processing and error correction for the entire data set (see Materials and Methods), we identified 17,077 COs in 2182 individuals (File S2). We observed a mean of 7.8 COs per individual, which was consistent with previous estimates from Col × Ler and other crosses (Higgins et al. 2011; Salomé et al. 2012; Wijnker et al. 2013; Rowan et al. 2015). The median resolution for CO intervals—the distance between the closest scorable markers—was 1102 bp, and three quarters of all CO positions could be estimated within an interval of 3 kb or less (Figure S3). We used the midpoints between flanking markers as estimates for CO positions in downstream analyses. In our data set, the genetic distances in specific regions that had been previously inferred by scoring recombination between fluorescent reporter genes in Col × Ler hybrids (Ziolkowski et al. 2015) were within 35% of the previously reported cM/Mb values, with the exception of a subtelomeric region on chromosome 2, which was 50% lower (Table S1). Since the recombination rate in this subtelomeric region is much higher in male than in female meiosis (Giraut et al. 2011), it is likely that this difference reflects measurement of both male and female meioses in our F2 population, while the distances estimated with fluorescent pollen markers reflect CO rates in male meiosis only (Ziolkowski et al. 2015). The distribution of CO rates in 200-kb windows along each of the five chromosomes was similar to what has been previously published for the species in general (Salomé et al. 2012) and for Col × Ler crosses specifically (Yelina et al. 2015; Choi et al. 2016), averaging 3.2 cM/Mb, with the highest rates in the pericentromeric regions and lowest rates in the centromeres (Figure 1A).
Figure 1.
CO characteristics for a population of 2182 Col × Ler F2 individuals. (A) CO rates along all five Chrs in fixed 200-kb windows. (B) Frequency of CO numbers per Chr pair for each Chr. (C) The distribution of inter-CO distances for Chrs with cis double COs, determined by the genotype transitions shown in the schematic diagram below the plot. Observed distances are compared to the distances expected using randomly selected double CO sites. The red line indicates a γ distribution fit. Chr, chromosome; CO, crossover.
To find regions with exceptionally high CO rates, we performed a sliding window analysis using 50-kb windows with 10-kb steps. Roughly 15% of the windows had CO rates that were more than about twice the genome-wide mean (>6 cM/Mb). We selected the windows with the top 5% of CO rates for further analysis. The 50-kb window with the highest rate had a CO rate of nearly 28 cM/Mb (nearly 10 times the genome-wide average), and the mean in the top 5% of windows was 11.8 cM/Mb. After merging the overlapping windows in this set we obtained 133 “hotspot regions” with a mean length of 95,714 bp and mean CO rate of 9.3 cM/Mb (File S3). GO term enrichment analysis revealed no significantly enriched categories using false discovery rate (FDR) correction, and only marginal enrichment for genes encoding transcription factors and DNA-binding proteins after Bonferroni correction (data not shown). The closest CO to each of the telomeres was most often tens of kilobases away, with the exception of those in the subtelomeres of chromosomes 4 and 5, which had COs only a few hundred to a few thousand base pairs away.
The frequencies of COs per chromosome for all five chromosomes (Figure 1B) were similar to what has been reported previously (Salomé et al. 2012). The mean number of COs per chromosome pair ranged from 1.3 to 1.9 (Table 1) and was positively correlated with chromosome length (Figure S4, R2 = 0.96), agreeing with previous reports (Giraut et al. 2011; Salomé et al. 2012). To analyze COs that must have occurred on the same chromosome, we identified chromosome pairs with two or three COs (Figure 1C). Such double COs could only be unambiguously inferred when the sequence of genotype blocks was Col-Het-Col or Ler-Het-Ler, and they therefore represent only a subset of all double COs. Using this approach, we found a total of 684 double COs with a mean inter-CO distance of 9,736,657 bp (Figure 1C and Figure S5A), which was significantly greater than the expected mean of 8,908,199 bp for a matched set of random double COs (P = 4.4 × 10−5, Mann–Whitney U-test), consistent with CO interference. We measured the strength of CO interference by fitting a γ distribution to the data and obtaining the value of the shape parameter (Figure 1C). The shape value was 2.8 for the entire data set and varied from 2.6 to 4.1 among chromosomes (Figure S5B), reflecting moderate interference (as a value of 1 would indicate no interference). The frequency of double COs was highest for the longest chromosomes, 1 and 5, which together accounted for 70% of all double COs that we could score (Table 1).
Table 1. COs per chromosome pair.
| Chr | Size (bp) | Total COs | Mean COs per pair | Double COs |
|---|---|---|---|---|
| 1 | 30,427,671 | 4,153 | 1.9 | 259 |
| 2 | 19,698,289 | 3,021 | 1.4 | 78 |
| 3 | 23,459,830 | 3,191 | 1.5 | 76 |
| 4 | 18,585,056 | 2,825 | 1.3 | 49 |
| 5 | 26,975,502 | 3,887 | 1.8 | 222 |
| TOTAL | 17,077 | 684 |
Chr, chromosome; CO, crossover.
The effects of inversions on CO positions
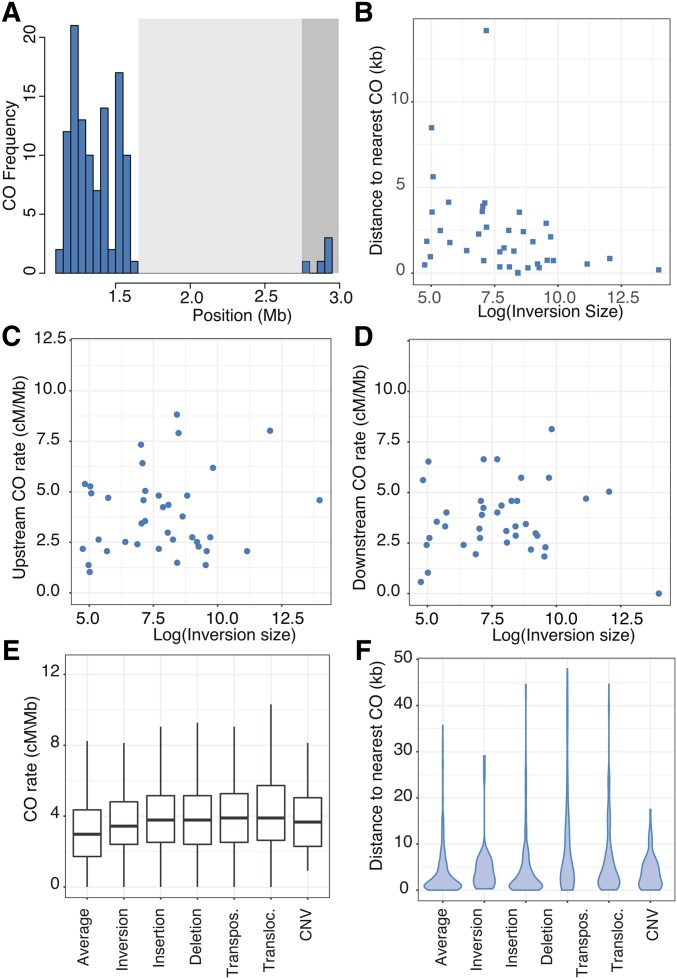
Our dense CO data set allowed us to examine in detail how SVs of different sizes affect local CO positioning. We first focused on the large paracentric inversion on chromosome 4 (Fransz et al. 2000, 2016; Zapata et al. 2016), relying on the flanking markers to establish whether a CO had occurred inside the inversion, since markers inside of SVs were filtered out during TIGER analysis. We did not find any COs within this ∼1.2-Mb inversion (Figure 2A), compared with 162 expected COs in this region when a random distribution of COs was simulated (Table 2). The closest upstream CO occurred 5425 bp from the left border and the closest downstream CO was just 1245 bp from the right border, even though it was located at the edge of the centromere. The CO rate in the 200-kb region upstream of the border was considerably higher (4.6 cM/Mb) than the genome-wide average rate of 3.0 cM/Mb and lower (0.57 cM/Mb) in the 200-kb region downstream. The low CO rate in the downstream region is likely explained by its proximity to the centromere.
Figure 2.
The effect of SVs on CO rates and positions. (A) CO frequency in 50-kb bins in a region of chromosome 4 containing “the knob,” a 1.2-Mb inversion (light gray-shaded region) that is adjacent to the centromere (dark gray-shaded region). (B) Minimum CO distance from the borders of all inversions in relation to the log10 of the inversion size. (C) CO rates (cM/Mb) in the 200 kb upstream of inversion borders. (D) CO rates (cM/Mb) in the 200 kb downstream of inversion borders. (E) CO rates (cM/Mb) in the 200 kb up- and downstream of the borders for inversions, insertions, deletions, transpos. (intrachromosomal) and transloc. (interchromosomal), and CNVs. (F) Distances to the nearest CO for inversions, insertions, deletions, transpos. (intrachromosomal) and transloc. (interchromosomal), and CNVs. CNV, copy number variations; CO, crossover; SV, structural variant; transloc., translocation; transpos., transposition.
Table 2. COs within and next to SVs.
| Variant type | N | Median length (bp) | Total COs inside | Mean distance to nearest CO (bp) | Flanking CO rate (cM/Mb) | |||
|---|---|---|---|---|---|---|---|---|
| Observed | Random | Observed | Random | Observed | Random | |||
| Inversion | 38 | 2423 | 5a | 216 | 4752a | 2276 | 3.8b | 3.2 |
| Insertion | 311 | 516 | 36 | 32 | 3969 | 3091 | 4.1b | 3.2 |
| Deletion | 426 | 605 | 0 | 0 | 4792 | 3633 | 4.1a | 3.2 |
| Transposition (intrachromosomal) | 102 | 3038 | 12a | 42 | 8446a | 2439 | 4.1a | 3.2 |
| Translocation (interchromosomal) | 271 | 996 | 30a | 87 | 8759a | 3071 | 4.4a | 3.2 |
| Copy number | 67 | 253 | 11 | 9 | 3995 | 4117 | 4.0a | 3.2 |
CO, crossover.
P < 0.05 after Mann–Whitney U-test.
P < 0.05 after Student’s t-test.
We also did not find any COs inside another large (170 kb) paracentric inversion on chromosome 3. Flanking COs were located relatively close, within 10 kb on either side of the inversion breakpoints (Figure S6). The 200-kb upstream and downstream regions of this 170 kb inversion had CO frequencies of 8.0 and 5.0 cM/Mb.
To examine CO patterns more broadly, we looked at the overlap between CO intervals and 47 inversions of different sizes (range: 115–1,178,806 bp and median: 2221 bp; (Zapata et al. 2016). Because COs inside paracentric inversions lead to the formation of dicentric and acentric products (McClintock 1931, 1933), which may be lost or lead to aneuploidy, COs inside paracentric inversions are expected to produce fewer viable offspring. A total of 13 inversions overlapped with CO intervals, which was significantly lower than expected by chance (P = 2 × 10−4, established with 5000 permutations). To further assess the impact of inversions on CO positions at a local scale, we excluded the centromeric regions where effects of inversions would be masked by the inherently low recombination rate. In a final set of 38 noncentromeric inversions and 16,776 genome-wide COs, we found only five events where the midpoint CO estimate was within an inversion, which was significantly different from 216 internal COs when a random distribution was simulated (P = 4 × 10−3, Mann–Whitney U-test). Moreover, the CO intervals spanning these five inversions were 13 kb on average, meaning that the resolution of these CO positions was much lower than the median CO resolution of the data set. Thus, it is likely that these COs occurred outside of the inversion boundaries. There was no effect (R2 = 6 × 10−4, P = 0.89) of inversion size on the distance to the nearest CO (Figure 2B) and the mean distance of an inversion breakpoint to the closest CO midpoint was 4752 ± 957 bp (mean ± SEM), which was significantly greater than the mean distance when a random CO distribution was simulated (2276 ± 430 bp, P = 7 × 10−3, Mann–Whitney U-test). The mean CO rate in the 200 kb up- and downstream of the inversions (3.8 ± 0.26) was significantly higher (P = 0.03, Student’s t-test) than the rates in a random comparison (3.2 ± 0.08). The observed CO rates in the flanking 200-kb regions of inversions (Figure 2, C and D) did not depend on inversion size (R2 = 0.013, P = 0.49). We conclude that COs are suppressed within inversions of all sizes, but that this suppression does not extend far beyond the inversion borders (<10 kb).
The effects of other SVs on CO positions
To determine whether the observed effects of inversions on COs are also seen with other SV types, we compared locations of COs in relation to insertions, deletions, tandem copy number variants (CNVs), transpositions, and translocations (Zapata et al. 2016). For all SV types except tandem CNVs, the overlap between CO intervals and the regions containing SVs was significantly lower than expected by chance (Table S2). When looking at CO positions, only transpositions and translocations had fewer internal COs and an increased distance to the nearest CO than expected from a matched random distribution of COs (Table 2). The distance to the nearest CO for all SV types was not affected by the SV length (Figure S7). CO rates in the flanking 50-, 100-, or 200-kb regions—up- and downstream of all SVs—were higher than both the observed genome-wide average and a matched random distribution of COs (Figure 2E, Table 2, Figure S8, and Table S3), and independent of the size of the variant region (Figure S9). We conclude that there is a general trend of local suppression of COs within SVs of different types and lengths, which occasionally extends several kilobases beyond the SV borders.
COs, DSBs, and SVs
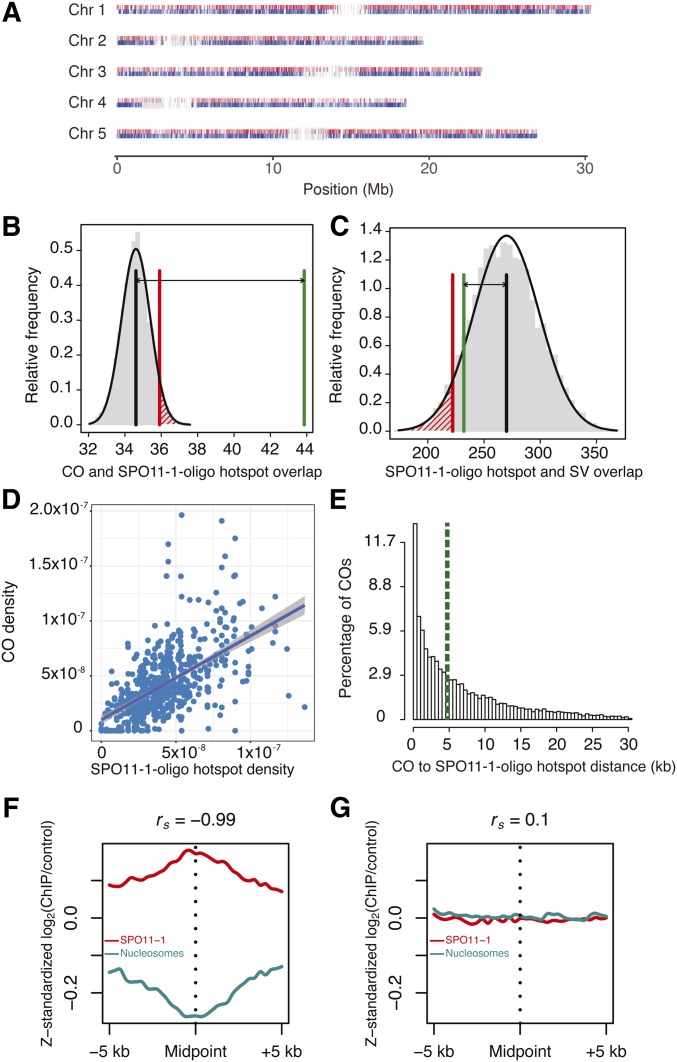
Meiotic COs occur through repair of programmed DSBs generated by SPO11 complexes during prophase I of meiosis (Szostak et al. 1983; Keeney et al. 1997; de Massy 2013). We therefore sought to examine whether suppression of COs around SVs was associated with reduced DSB formation in those regions. Meiotic DSBs have been mapped in A. thaliana via purification and sequencing of SPO11-1-oligonucleotides, which mark DSB sites (Choi et al. 2018). These data were used to define 5914 SPO11-1-oligo hotspots (Choi et al. 2018), which we compared with CO positions and the locations of SVs throughout the genome (Figure 3A). As expected, the overlap between SPO11-1-oligo hotspots and CO intervals was significantly greater than random (Figure 3B, P = 2 × 10−3 after 5000 permutations). Importantly, the overlap between SPO11-1-oligos and SVs was not significantly different from that expected by chance, indicating that the initiation of COs is not different inside and outside of SVs (Figure 3C, P = 0.09 after 5000 permutations). The density of SPO11-1-oligo hotspots and COs was also positively correlated in 200-kb windows across the genome (Figure 3D, R2 = 0.34, P < 2 × 10−16). Over one-half of all COs (53%) were within 5 kb of a SPO11-1-oligo hotspot (Figure 3E). Previous work established a relationship between low nucleosome occupancy and elevated levels of SPO11-1-oligonucleotides (Choi et al. 2018). Consistent with this, we observed that CO midpoints were associated with reduced nucleosome occupancy, measured via MNase-seq (Choi et al. 2016), and elevated SPO11-1-oligonucleotides (Figure 3, F and G). Taken together, these results suggest that suppression of COs around SVs occurs downstream of DSB formation.
Figure 3.
Coincidence of COs, SPO11-1-oligo hotspots, and SVs. (A) Locations of SPO11-1-oligo hotspots, COs, and SVs for each Chr. The position of the midpoint of the SPO11-1-oligo hotspot is shown in red, the CO midpoints in blue, and the locations of SVs in gray. Permutation tests were performed to test for overlaps between (B) SPO11-1-oligo hotspots and CO intervals, and (C) SPO11-1-oligo hotspots and SVs. In (B), the values on the x-axis are the number of overlaps (in thousands), while the values on the x-axis in (C) are the total number of overlaps. Vertical black lines indicate the mean number of overlaps expected in 5000 random permutations. The vertical red lines indicate the number of overlaps where P = 0.05. The vertical green line indicates the observed number of overlaps. The double arrow highlights the difference between the mean of 5000 permutations and the observed number. (D) Correlation between the percentage of SPO11-1-oligo hotspots and the percentage of COs in 200-kb windows. The equation of the line is 0.77x + 0.002, R2 = 0.34, P < 2 × 10−16. (E) Histogram of distances from CO breakpoints to the border of the nearest SPO11-1-oligo hotspot. The vertical green dashed line indicates the median. Outliers representing the furthest 5% of distances are not shown in the plot. (F) SPO11-1-oligo (red) or nucleosome occupancy (blue, MNase-seq) [z-score standardized log2(signal/input)] in 10-kb windows around CO midpoints. (G) As for (F), but showing analysis of the same number of randomly chosen positions. ChIP, chromatin immunoprecipitation; Chr, chromosome; CO, crossover; MNase-seq, micrococcal nuclease sequencing; SV, structural variant.
Regions devoid of COs (CO deserts)
With our ultra-dense CO data, we were able to identify large regions of the genome where CO occurrence was much lower than expected by chance. The median distance between all COs was 2610 bp and >80% of the COs were spaced within 10 kb of another CO in the data set. We selected the largest 5% of inter-CO distances (minimum 25,563 bp and maximum 1,419,083 bp, n = 839) and investigated these CO deserts to uncover genomic features that are associated with suppressed CO formation. We expected that these regions would correspond to SVs, but the overlap between CO deserts and all SVs was not significantly different from that expected by chance (P = 0.1 after 5000 permutations, Figure S10A). While only 6.3% of CO deserts were located within centromeres, these were among the longest CO deserts (top 2%). CO deserts were significantly enriched for DNA methylation at CG sites even when centromeres were excluded (Figure S10B, P < 2.2 × 10−16 with Student’s t-test or Mann–Whitney U-test). Although SVs were not more likely to be found in CO deserts, they were significantly enriched for CG DNA methylation (Figure S10C, 2.2 × 10−16 with Student’s t-test or Mann–Whitney U-test). Although there were protein-coding genes in CO deserts (File S4), GO term enrichment revealed no significantly overrepresented categories in these regions after correction for multiple hypothesis testing either by FDR or Bonferroni. We conclude that CG DNA methylation is associated with CO suppression in SVs, transposable elements, and some regions of the genome that contain protein-coding genes.
COs and NLR gene clusters
Since COs reshuffle existing genetic diversity, they represent an important mechanism for generating alleles with new functions. This is especially important for NLR genes involved in disease resistance, as COs have been implicated in the generation of new resistance specificities in plants (Richter et al. 1995; Parniske et al. 1997; McDowell et al. 1998; Noël et al. 1999). Many NLR genes in the A. thaliana genome are found in clusters that exhibit structural variation, which may locally suppress CO formation (Chin et al. 2001; Meyers et al. 2003; Alcázar et al. 2014; Chae et al. 2014; Van de Weyer et al. 2019). Indeed, a previous study showed that NLR genes in Col × Ler hybrids had variable CO rates, with some genes overlapping CO hotspots while others, including those with a high degree of structural variation, were CO coldspots (Choi et al. 2016). The dense CO data set generated in the current study enabled a deeper examination of the patterns of COs within and around NLR genes, and other genes involved in defense and immunity.
The number of overlaps between CO intervals and NLR genes, or other defense genes, was not significantly different from that expected by chance (P = 0.23, 5000 permutations, Figure S11). Considering CO positions, we found that 55 of 197 defense-related genes (28%) contained one or more CO, which was not significantly different from randomized positions (P = 0.06, Wilcoxon test). However, the mean distance from defense gene locus borders to the nearest CO was 7107 bp, which was significantly further away than expected from a random CO distribution (2212 bp, P = 7.510−6, Wilcoxon test). CO rates in the flanking 200 kb up- and downstream were slightly, but not significantly, elevated.
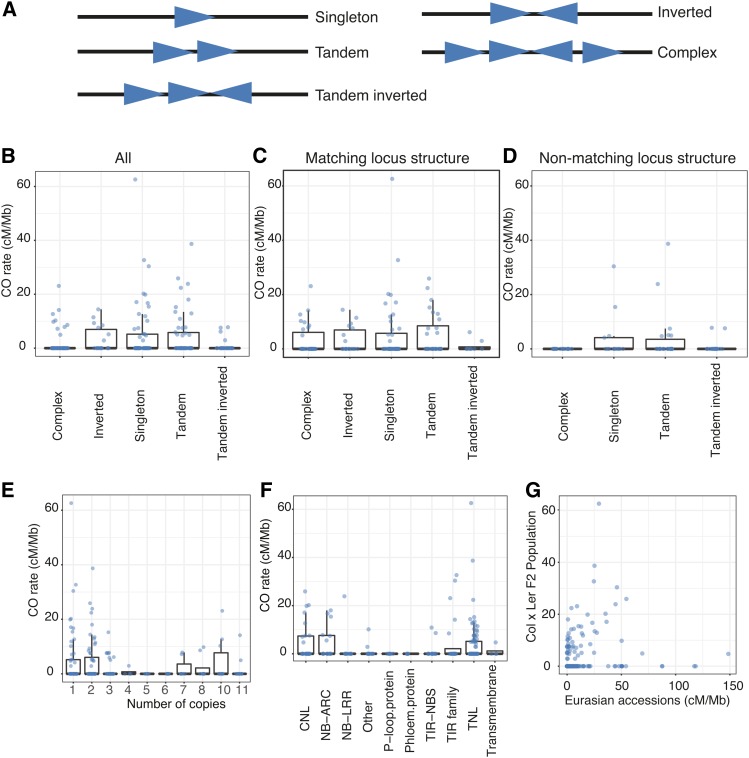
We categorized NLRs by their locus structure (Figure 4A), and found CO hotspots and coldspots among genes of all locus types. Singleton NLR genes, and those in loci with inverted and tandem repeat structures, trended toward higher proportions of CO coincidence compared with complex and tandem inverted locus structures (Figure 4B and Table 3). When Col and Ler have the same NLR locus structure (Figure 4C and File S5), the CO rates are generally higher than in NLR loci where the two parents differ in their structure (Figure 4D). However, the number of overlaps between NLR genes and CO intervals is not significantly different between the two classes (Figure S11, B and C).
Figure 4.
CO frequency in disease resistance genes. (A) Schematic drawings representing different locus structures for NLR and other disease resistance genes. (B) CO rates for all NLR loci and other disease resistance genes (File S4) based on Col locus structure. (C) CO rates for NLR loci with similar structure in Col and Ler. (D) CO rates for NLR loci that differ in structure between Col and Ler. (E) CO rates for all NLR loci as a function of copy number at the locus for Col (see Figure S12 for Col/Ler differences). (F) CO rates for NLR loci based on Col annotation. (G) CO rates in NLR genes in our Col × Ler F2 population compared with the historical recombination rate across Eurasian accessions. All boxplots (B–F) display the median, and the first and third quantiles. CNL, coiled-coil domain NLR; CO, crossover; NB–ARC, proteins containing only the nucleotide binding site domain termed ARC for its presence in APAF1, NLRs, and CED-4; NB–LRR, NLR-like proteins containing only the nucleotide-binding site and leucine-rich repeat domains; NLR, nucleotide-binding site leucine-rich repeat; TIR–NBS, NLR-like proteins containing only a TIR domain and nucelotide-binding site; TNL, TIR domain NLR.
Table 3. COs within NLR genes as a function of different structural classes.
| Locus type | Genes | Number with COs | Proportion with COs |
|---|---|---|---|
| Complex | 47 | 9 | 0.19 |
| Inverted | 18 | 7 | 0.39 |
| Singleton | 57 | 18 | 0.32 |
| Tandem | 50 | 18 | 0.36 |
| Tandem inverted | 25 | 4 | 0.16 |
CO, crossover.
COs trended toward being more prevalent in loci with only one or two genes (Figure 4E), especially when both Col and Ler had the same number of copies in a locus (Figure S12), but this was not statistically significant compared with a random CO distribution (P = 0.24, chi square test). COs showed a general trend toward exhibiting higher levels in Toll/interleukin-1 receptor (TIR) domain NLRs, coiled-coil domain NLRs, and those with only nucleotide-binding domains than in other categories of NLR genes (Figure 4F and Table S5), but this was also not statistically significant (P = 0.66, Fisher’s exact test). There was no correlation between historical recombination rates among Eurasian accessions (Choi et al. 2016) and CO rates within NLR genes among Col × Ler F2 individuals (Figure 4G). Locations of CO positions in a focal set of NLR genes that were shown to have high CO rates (Figure S13) were generally consistent with a previous study (Choi et al. 2016).
COs, chromatin, and sequence features
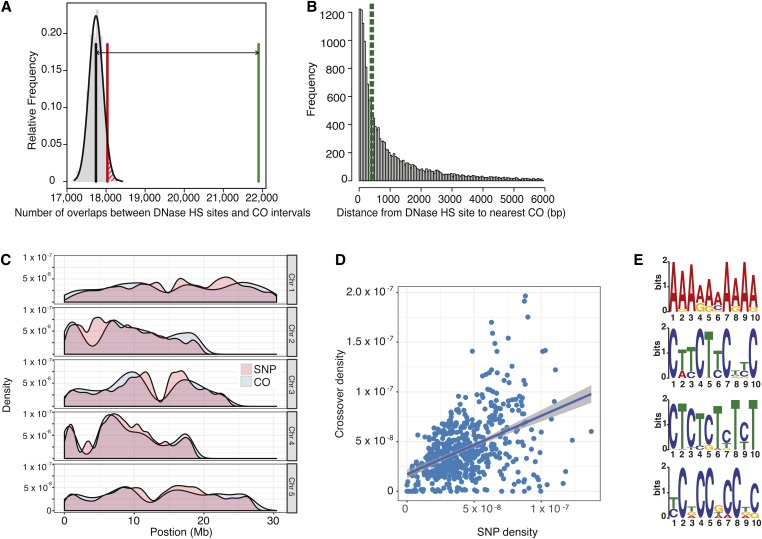
COs tend to be associated with several hallmarks of open chromatin in plants, including low levels of DNA methylation, high levels of histone H3 lysine 4 methylation (H3K4me3), and low nucleosome density (Liu et al. 2009; Yelina et al. 2012; Marand et al. 2017; Choi et al. 2018). COs have been found to be enriched close to the 5′ and 3′ flanking regions of genes, at transcription start sites, cis-regulatory regions, and regions where the histone variant H2A.Z has been deposited (Choi et al. 2013; Wijnker et al. 2013; Marand et al. 2017), which is consistent with our finding that COs were likely to be in nucleosome-depleted and SPO11-1-oligo-enriched regions of the genome (Figure 3, G and H). Therefore, we also expected to observe that COs would overlap with DNase I HS sites and ATAC-seq sites, which are two independent indicators of regions of open chromatin/cis-regulatory elements (Sullivan et al. 2014; Maher et al. 2018). We found that our CO intervals showed a significant overlap with DNase I HS sites (Figure 5A). Of 17,077 CO midpoints, 4338 (∼25%) occurred in a DNase I HS site, and a further 36% of COs occurred within 1 kb up- or downstream of a DNase I HS site. When a random distribution of COs was simulated, only 8% occurred within a DNase I HS site and 20% within 1 kb of a DNase I HS site. Over 95% of COs occurred within 6 kb of a DNase I HS site (Figure 5B), with a median distance of 558 bp to the nearest DNase I HS site. In comparison, 95% of a random set of COs were within 42 kb of a DNase I HS site and the median distance was 1 kb. Results were similar for ATAC-seq sites, where 95% of COs were within 6.5 kb of an ATAC-seq site (Figure S14), compared with 95% of a matched set of random COs occurring within 39 kb of an ATAC-seq site.
Figure 5.
Chromatin and sequence features associated with COs. (A) Permutation tests of overlaps between DNase I HS sites and CO intervals. Vertical black line indicates mean number of overlaps expected in 1000 random permutations. Vertical red line indicates the number of overlaps where P = 0.05. Vertical green line indicates the observed number of overlaps. Double arrow highlights the difference between the expected mean and the observed number. (B) Distribution of distances from CO breakpoints to the nearest DNase HS site border. The vertical dashed green line indicates the median. The top 5% of distances are omitted from this plot for ease of visualization. (C) Overlay of CO and SNP density along all five chromosomes. (D) Correlation between CO density and SNP density in 200-kb fixed windows. Line indicates best fit and gray-shaded area represents the 95% C.I. The equation of the line is 5.872e−01*x + 1.726e−08. R2 = 0.21. P < 2.2e−16. (E) Sequence motifs enriched in the 500 bp up- and downstream of the CO breakpoint site for a subset of COs (1000 per chromosome). The top four detected motifs are shown. CO, crossover; HS, hypersensitive; SNP, single-nucleotide polymorphism.
Our data set also allowed us to reexamine the question of whether recombination is itself mutagenic and would therefore be associated with genomic polymorphism (Cao et al. 2011; Long et al. 2013; Yang et al. 2015; Ziolkowski et al. 2015). However, we also note that divergence between homologs suppresses recombination (George and Alani 2012; Liu et al. 2012; Chakraborty and Alani 2016), as we observed within SVs (Figure 2). Thus, one might expect that these two opposing forces would lead to differences in the correlation between SNPs and COs over evolutionary time. We found that the pericentromeres are regions of relatively high SNP and CO density for the Col/Ler cross, despite also containing higher levels of heterochromatin (Figure 5C and Figure S15A; Yelina et al. 2012; Choi et al. 2013, 2018; Underwood et al. 2018). The densities of Col/Ler SNPs and COs among all 200-kb windows across the genome showed a moderate, but statistically significant, positive correlation (R2 = 0.21, Figure 5D), similar to that reported previously (Fernandes et al. 2018; Serra et al. 2018). In addition, the pericentromeres have been previously shown to have a general AT sequence bias (Wijnker et al. 2013). At the fine scale, several DNA sequence motifs have been associated with COs in Col × Ler crosses, including a poly(A/T) motif, a CTT/GAA repeat (Choi et al. 2013; Wijnker et al. 2013; Shilo et al. 2015), and a CCN repeat (Shilo et al. 2015). Therefore, we used MEME to identify motifs in the 500 bp up- and downstream of COs in random subsets of 500 and 1000 COs per chromosome. We found all three of these motifs (Figure 5E) in our CO data, plus a fourth motif (CT repeat). The poly(A) and CTT/GAA repeat motifs were the most common; nearly 90% of COs were within 500 bp of one or the other of these repeats (Table 4 and Table S5), while the CCN and CT repeat motifs were found in a minor fraction of the analyzed COs. Taken together, these data indicate that COs between Col and Ler are strongly associated with these four sequence motifs.
Table 4. Sequence motifs in 1-kb regions spanning of a random subset of 5000 COs.
| Motif | Occurrence | COs (%) |
|---|---|---|
| Poly(A/T) | 4314 | 86.2 |
| CTT/GAA | 1527 | 30.5 |
| CT/GA | 1721 | 34.4 |
| CCN/GGN | 901 | 18.0 |
| Poly(A/T) or CTT/GAA | 4492 | 89.8 |
| All four motifs combined | 4643 | 93.8 |
CO, crossover.
Discussion
We employed a low-cost rapid genomic DNA library preparation protocol to sequence the genomes of >1800 F2 hybrid individuals from a cross between two commonly used A. thaliana accessions, and to map COs at a very high density and precision. The availability of high-quality reference genomes for each of these accessions (Arabidopsis Genome Initiative 2000; Lamesch et al. 2012; Zapata et al. 2016) enabled a detailed analysis of the effects of SVs on CO formation, and a fine-scale examination of COs in relation to other sequence and chromatin features.
An improved method for genomic DNA library construction
Our library preparation method had a success rate of ∼95% when processing hundreds of samples in <3 hr. Furthermore, at a cost of ∼$2.50 per sample, this method is, as of writing this manuscript, highly cost-effective and compares favorably in terms of speed with similar protocols (Baym et al. 2015; Pisupati et al. 2017). A version of this protocol could also be used with homemade Tn5 transposome and buffer (Hennig et al. 2018). Combining the COs determined from 1829 F2 hybrid genomes with previously published CO data for the same cross (Choi et al. 2016; Underwood et al. 2018), we obtained a final data set of 17,077 COs. From extracted DNA to final CO data, a genetic map could be built in ∼1.5 weeks for hundreds of samples using this combination of library preparation and CO calling with TIGER analysis (Rowan et al. 2015).
Genome-wide CO distribution
The pattern of CO rates along the chromosomes (Figure 1) was in agreement with previously reported genome-wide CO data generated for A. thaliana with a variety of approaches (Giraut et al. 2011; Salomé et al. 2012; Yelina et al. 2012; Wijnker et al. 2013). Although plants, different from mammals (Tiemann-Boege et al. 2005; Paigen et al. 2008; Paigen and Petkov 2010), lack focused and narrow (1–2 kb) CO hotspots, CO rates vary along chromosomes with a broad distribution. We were able to identify 133 regions of the genome with exceptionally high CO rates (>3 times the genome-wide average cM/Mb) and CO desert regions without any COs among >2000 F2 individuals (Files S3 and S4). Hence, substantial variation in CO frequency is observed throughout the A. thaliana genome.
Insights into the mechanism for CO suppression inside SVs
Given the high density of COs in our data set, we were able to examine the relationship between COs and multiple types of SVs in fine detail. Observed COs were suppressed inside inversions and transpositions, and were slightly elevated in the flanking regions of all SV types (Figure 2, Table 2, Figure S8, Table S2, and Table S3). One interpretation for the elevated flanking CO rates is that the lack of COs inside the SVs is compensated by additional COs in the flanking regions. However, such a simple scenario would predict that larger SVs would have higher flanking CO rates, which is not what we observed (Figure S9). A more likely explanation is that SVs occur more often in regions that are already prone to high rates of recombination, as recombination is a mechanism for generating SVs (George and Alani 2012; Liu et al. 2012). In other words, COs in the vicinity of SVs are not increased because of SVs, but SVs are more likely to occur in regions with elevated CO rates. For inversions and translocations, the suppression of COs within the variant regions tends to extend moderately beyond the borders of the variant regions, as the distance to the nearest CO is slightly further than expected under a random distribution (Table 2), although this was not correlated with SV length (Figure S7). This is in contrast to Drosophila interspecific crosses, where suppression of recombination extends over 2 Mb beyond the boundaries of inversions of different sizes (Kulathinal et al. 2009; Stevison et al. 2011). Below, we discuss five possible mechanisms explaining why COs are less likely to occur inside of SVs: (1) DSB formation in variant regions is reduced; (2) reduced sequence similarity means that there is no substrate for recombinational repair; (3) COs that form within SVs create inviable gametes and thus cannot appear in progeny; (4) variant regions are physically prevented from interacting with the homologous chromosome by the structure and organization of the meiotic axis, and/or the synaptonemal complex (SC); and (5) DNA methylation suppresses COs in SVs. Below, we consider these alternative explanations in more detail.
DSB formation in variant regions is reduced. SPO11-1-oligo hotspots are as common in SVs as outside SVs (Figure 3), suggesting that meiotic DSBs that occur in these regions are repaired as NCOs if the homologous chromosome is used, or they are repaired using the sister chromatid. A caveat is that SPO11-1-oligo hotspots were mapped only in Col homozygotes (Choi et al. 2018), and there is a possibility that these hotspots are distributed differently in Ler and/or Col/Ler F1 hybrids. A recent study of inversion heterozygotes in D. melanogaster crosses suggested that the DSB number is similar in such regions, but that they are preferentially repaired as NCOs (Crown et al. 2018). While we cannot rule out a role for suppression of COs by prevention of DSBs within SVs in A. thaliana, it is likely not a major contributing factor.
Reduced sequence similarity means that there is no substrate for recombinational repair. SV types differ in their level of sequence divergence. In the case of inversions, sequence identity between the two chromosomes means that pairing is possible, but the portion of the chromosome containing the inversion must be looped so that the region is in the same orientation on both homologs. Indel polymorphisms and transpositions have no direct counterpart on the other chromosome, but the indel sequence could contain repetitive elements that occur at nearby nonallelic loci and provide a substrate for homologous recombination. Tandem CNVs should provide sufficient homology for recombination. Because inversions and tandem CNVs have homology, but exhibit reduced CO rates, this may suggest that a lack of homology alone is not sufficient to explain suppressed COs in these regions.
COs that form within SVs create inviable gametes and thus cannot appear in progeny. Although inversion loops would facilitate the alignment of homologous regions, a CO that occurs through this process is expected to generate a dicentric bridge and an acentric chromosome. This could potentially lead to inviable gametes due to aneuploidy and explain why COs in inversions are not observed in F2 progeny. A previous cytological examination of male meiotic prophase I in Col × Ler hybrids did not reveal inversion loops, and dicentric bridges were rarely observed in anaphase I (Ji 2014), but it is possible that this study did not have the power to assess whether the frequency of these events was lower than expected given the recombination rate. In hybrids of wild and domesticated maize hybrids that were heterozygous for a ∼50-Mb inversion, only 7 of 167 male meiocytes showed dicentric bridges, and pollen viability and seed set were normal (Fang et al. 2012). This suggests that there are mechanisms to prevent COs that would generate defective gametes (at least in the case of inversions), but that the frequency of deleterious COs in SVs in gametes needs to be assessed before a conclusion can be reached.
Variant regions are physically prevented from interacting with the homologous chromosome by the structure and organization of the meiotic axis, and/or the SC, or the spatiotemporal aspects of its formation. COs occur within a highly organized protein–DNA structure known as the SC, consisting of a central element flanked by two lateral elements (Zickler and Kleckner 1999). This structure of this complex is highly conserved across a broad taxonomic range and functions to hold homologous chromosomes in proximity (synapsis) during CO formation. Recombination is thought to occur between the homologs within the SC central element, where proteins that facilitate the repair of DSBs in favor of a CO outcome, such as MLH1, are recruited (Bogdanov et al. 2007). For each homolog, the rest of the chromosome that is not participating in crossing over is organized as chromatin loops tethered to the central and lateral elements, known as tethered-loop/axis architecture (Blat et al. 2002). It is possible that the portions of the chromosomes that contain SVs are prevented from interacting with the central element and the homologous chromosome. This might explain why, in some cases, the distance to the nearest CO was slightly further away than expected by chance. In A. thaliana, ZYP1, an important component of axis formation, is localized to a few foci along the chromosomes during the leptopene stage of meiosis. These foci extend and coalesce during zygotene to result in a continuous linear signal (Higgins et al. 2005; Sanchez-Moran et al. 2007). This suggests a slight asynchrony in the timing of synapsis and axis formation, which has been found to be even more pronounced in other plant species such as barley (Higgins et al. 2012). It is therefore possible that the timing of synapsis in regions containing SVs may affect the repair outcome. During meiosis in Col × Ler hybrids, the region containing the paracentric inversion on chromosome 4 remains asynapsed at pachtyene (Ji 2014). Future research, aimed toward obtaining fine-scale organization of the DNA sequences bound to the SC and more detailed examination of the spatiotemporal aspects of synapsis, will help clarify whether the formation and structure of the SC contribute to the suppression of COs in SVs.
Possible mechanism: DNA methylation prevents COs from occurring in SVs. The level of CG DNA methylation was higher in 10-kb windows that overlapped with SVs in our analysis than in 10-kb windows that did not overlap SVs (Figure S10C). We also found that regions surrounding SVs had elevated CO rates (Figure 2E). We propose that newly arising SVs are likely to occur via recombination, which, once generated, have a probability of becoming methylated. If DNA methylation in SVs prevents COs that are otherwise deleterious from occurring, then this could select for methylation around SVs. The mechanism by which DNA methylation prevents crossing over could act through organization of the SC (as proposed in point 4 above) or through other mechanisms, such as the promotion of non-CO DNA repair pathways.
Other DNA sequence features associated with COs
With this dense CO data set, we defined a more nuanced and detailed picture of CO distribution in relation to NLR and other defense genes (Choi et al. 2016), chromatin accessibility (Liu et al. 2009; Choi et al. 2013; Shilo et al. 2015; Yelina et al. 2015; Marand et al. 2017), and DNA sequence motifs (Choi et al. 2013; Wijnker et al. 2013; Shilo et al. 2015) at the fine scale. Recombination plays an important role in generating sequence diversity in NLR genes over evolutionary time (Parniske et al. 1997; Noël et al. 1999; Parniske and Jones 1999; Sun et al. 2001; Wulff et al. 2004), and it can alter resistance specificities in experimental crosses either by generating chimeric genes (Collins et al. 1999) or by altering locus copy number (Chin et al. 2001). Thus, it might be expected that CO rates in NLR genes would be higher than average. However, when examining NLR genes genome-wide in an experimental cross, NLR genes were found to be equally likely to be CO hotspots or coldspots, and this was in some cases related to variation in locus architecture (Choi et al. 2016). Roughly one-half of the NLR genes in the A. thaliana genome exist in clusters with one or more paralogs (Meyers et al. 2003). In this study, we found that tandem and inverted duplications, and singleton NLR genes, had the highest proportion of genes overlapping COs (Figure 4D and Table 3). The proportion of genes with COs was generally lower for loci that differed in structure between Col and Ler. Complex loci, defined as having at least four paralogs, only featured COs when Col and Ler had the same locus structure. CO proportions and rates were highest in loci where Col and Ler both had either only one or only two NLR gene copies. Recombination can therefore not only generate diversity in NLR genes that may alter resistance specificity, but it may also generate structural variation that suppresses recombination over time, potentially leading to resistance supergenes where several disease resistance specificities are inherited together via linkage. This would also protect newly emerged NLR genes that have not yet required a function from elimination, as long as there is a positively selected gene in such a cluster.
Methylation of H3K4me3, a hallmark of actively transcribed genes, is positively associated with COs in plants (Yelina et al. 2012, 2015; Marand et al. 2017), while dense DNA methylation, an indicator of repressed chromatin state, is negatively associated with COs (Yelina et al. 2012, 2015; Rodgers-Melnick et al. 2015; Marand et al. 2017). Chromatin accessibility, as determined by DNase I hypersensitivity, is positively associated with COs in potato (Marand et al. 2017) and maize (Rodgers-Melnick et al. 2015). Consistently, we also found COs to be highly associated with DNase I hypersensitivity (Figure 5, A and B) and with another measure of chromatin accessibility—ATAC-seq sites (Figure S14). Three previously described CO-associated sequence motifs, poly(A), CTT, and CCN repeats (Wijnker et al. 2013; Yelina et al. 2015; Melamed-Bessudo et al. 2016)—are found in regions of open chromatin (Shilo et al. 2015). At least one of these motifs, in addition to a CT repeat, was present within 1 kb of nearly all COs that were examined (Figure 5E and Table 4).
Conclusions
The ultradense catalog of COs in this study has extended and refined our knowledge of various factors that influence the position and frequency of COs along A. thaliana chromosomes. We found that all types and sizes of SVs can affect local CO positioning, not only the large-scale rearrangements of the type that have been more extensively studied in the past. Since the suppressive effect of smaller SVs does not spread far beyond the borders, only SVs that are large enough to contain multiple genes are expected to have the potential to create supergenes containing blocks of jointly inherited favorable alleles.
Acknowledgments
We thank Talia Karasov for advice in implementing the library preparation protocol; Derek Lundberg for assistance with the liquid handling robot, which Frank Chan kindly let us use for our experiments; Hans de Jong for offering helpful discussions regarding inversions; and all those who provided us with additional data (see Materials and Methods). This work was supported by the Max Planck Society.
Author contributions: B.A.R., D.W., and I.R.H. designed the study. D.H. developed the Nextera LITE library preparation protocol. B.A.R. and T.R.F. grew all of the plants and performed all of the molecular biology work. B.A.R., A.J.T., and I.R.H. analyzed the data, and B.A.R. wrote the manuscript with contributions from all authors.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9733838.
Communicating editor: J. Shendure
Literature Cited
- Alcázar R., von Reth M., Bautor J., Chae E., Weigel D. et al. , 2014. Analysis of a plant complex resistance gene locus underlying immune-related hybrid incompatibility and its occurrence in nature. PLoS Genet. 10: e1004848 10.1371/journal.pgen.1004848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhajturki D., Muralidharan S., Nurmi M., Rowan B. A., Lunn J. E. et al. , 2018. Dose-dependent interactions between two loci trigger altered shoot growth in BG-5 × Krotzenburg-0 (Kro-0) hybrids of Arabidopsis thaliana. New Phytol. 217: 392–406. 10.1111/nph.14781 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E. et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., and Charlesworth B., 1998. Why sex and recombination? Science 281: 1986–1990. 10.1126/science.281.5385.1986 [DOI] [PubMed] [Google Scholar]
- Baym M., Kryazhimskiy S., Lieberman T. D., Chung H., Desai M. M. et al. , 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10: e0128036 (erratum: PLoS One 10: e0131262). 10.1371/journal.pone.0128036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Protacio R. U., Hunter N., and Kleckner N., 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. 10.1016/S0092-8674(02)01167-4 [DOI] [PubMed] [Google Scholar]
- Bogdanov Y. F., Grishaeva T. M., and Dadashev S. Y., 2007. Similarity of the domain structure of proteins as a basis for the conservation of meiosis. Int. Rev. Cytol. 257: 83–142. [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., 2000. Perspective: sex, recombination, and the efficacy of selection–was Weismann right? Evolution 54: 337–351. [DOI] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Günther T., Bender S. et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. 10.1038/ng.911 [DOI] [PubMed] [Google Scholar]
- Chae E., Bomblies K., Kim S.-T., Karelina D., Zaidem M. et al. , 2014. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351. 10.1016/j.cell.2014.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty U., and Alani E., 2016. Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Res. 16: fow071 10.1093/femsyr/fow071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2016. The status of supergenes in the 21st century: recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. Evol. Appl. 9: 74–90. 10.1111/eva.12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. B., Arroyo-Garcia R., Ochoa O. E., Kesseli R. V., Lavelle D. O. et al. , 2001. Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157: 831–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K. A., Venn O., Higgins J. D. et al. , 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. 10.1038/ng.2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Reinhard C., Serra H., Ziolkowski P. A., Underwood C. J. et al. , 2016. Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 12: e1006179 10.1371/journal.pgen.1006179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Zhao X., Tock A. J., Lambing C., Underwood C. J. et al. , 2018. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 28: 532–546. 10.1101/gr.225599.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Drake J., Ayliffe M., Sun Q., Ellis J. et al. , 1999. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11: 1365–1376. 10.1105/tpc.11.7.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G. P., Nickel K., Kuromori T., Benito M. I., Kaul S. et al. , 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474. 10.1126/science.286.5449.2468 [DOI] [PubMed] [Google Scholar]
- Crown K. N., Miller D. E., Sekelsky J., and Hawley R. S., 2018. Local inversion heterozygosity alters recombination throughout the genome. Curr. Biol. 28: 2984–2990.e3. 10.1016/j.cub.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., 2013. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47: 563–599. 10.1146/annurev-genet-110711-155423 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1931. The decrease of crossing-over observed in translocations, and its probable explanation. Am. Nat. 65: 214–232. 10.1086/280364 [DOI] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., and Käller M., 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Pyhäjärvi T., Weber A. L., Dawe R. K., Glaubitz J. C. et al. , 2012. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191: 883–894. 10.1534/genetics.112.138578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J. B., Séguéla-Arnaud M., Larchevêque C., Lloyd A. H., and Mercier R., 2018. Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. USA 115: 2431–2436. 10.1073/pnas.1713078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1999. The Genetical Theory of Natural Selection: A Complete Variorum Edition. Oxford University Press, Oxford. [Google Scholar]
- Francis K. E., Lam S. Y., Harrison B. D., Bey A. L., Berchowitz L. E. et al. , 2007. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3913–3918. 10.1073/pnas.0608936104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., Linc G., Lee C.-R., Aflitos S. A., Lasky J. R. et al. , 2016. Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J. 88: 159–178. 10.1111/tpj.13262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P. F., Armstrong S., de Jong J. H., Parnell L. D., van Drunen C. et al. , 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376. 10.1016/S0092-8674(00)80672-8 [DOI] [PubMed] [Google Scholar]
- Gel B., Díez-Villanueva A., Serra E., Buschbeck M., Peinado M. A. et al. , 2016. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C. M., and Alani E., 2012. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit. Rev. Biochem. Mol. Biol. 47: 297–313. 10.3109/10409238.2012.675644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L., Falque M., Drouaud J., Pereira L., Martin O. C. et al. , 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7: e1002354 10.1371/journal.pgen.1002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. C., and Goddard M. R., 2012. Sex enhances adaptation by unlinking beneficial from detrimental mutations in experimental yeast populations. BMC Evol. Biol. 12: 43 10.1186/1471-2148-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. C., 1972. Chromosome segregation influenced by two alleles of the meiotic mutant c(3)G in Drosophila melanogaster. Genetics 71: 367–400. [DOI] [PubMed] [Google Scholar]
- Hammarlund M., Davis M. W., Nguyen H., Dayton D., and Jorgensen E. M., 2005. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics 171: 1047–1056. 10.1534/genetics.105.044834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig B. P., Velten L., Racke I., Tu C. S., Thoms M. et al. , 2018. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 (Bethesda) 8: 79–89. 10.1534/g3.117.300257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herickhoff L., Stack S., and Sherman J., 1993. The relationship between synapsis, recombination nodules and chiasmata in tomato translocation heterozygotes. Heredity 71: 373–385. 10.1038/hdy.1993.152 [DOI] [Google Scholar]
- Higgins J. D., Sanchez-Moran E., Armstrong S. J., Jones G. H., and Franklin F. C. H., 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. 10.1101/gad.354705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Ferdous M., Osman K., and Franklin F. C. H., 2011. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 65: 492–502. 10.1111/j.1365-313X.2010.04438.x [DOI] [PubMed] [Google Scholar]
- Higgins J. D., Perry R. M., Barakate A., Ramsay L., Waugh R. et al. , 2012. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24: 4096–4109. 10.1105/tpc.112.102483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. J., Erickson R. P., Zhang J., Garver W. S., and Heidenreich R. A., 2000. Fine linkage and physical mapping suggests cross-over suppression with a retroposon insertion at the npc1 mutation. Mamm. Genome 11: 774–778. 10.1007/s003350010134 [DOI] [PubMed] [Google Scholar]
- Ji, X., 2014 Numerical and structural chromosome aberrations in cauliflower (Brassica oleracea var. botrytis) and Arabidopsis thaliana. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands. [Google Scholar]
- Joron M., Frezal L., Jones R. T., Chamberlain N. L., Lee S. F. et al. , 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. 10.1016/S0070-2153(01)52008-6 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., and Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kulathinal R. J., Stevison L. S., and Noor M. A. F., 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5: e1000550 10.1371/journal.pgen.1000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C., Stocks M., Risse J. E., Dos Remedios N., Farrell L. L. et al. , 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48: 79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C. et al. , 2012. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Carvalho C. M. B., Hastings P. J., and Lupski J. R., 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22: 211–220. 10.1016/j.gde.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yeh C.-T., Ji T., Ying K., Wu H. et al. , 2009. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5: e1000733 10.1371/journal.pgen.1000733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A. et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. 10.1038/ng.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., and Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e10005000. [corrigenda: PLoS Biol. 10 (2012)] 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher K. A., Bajic M., Kajala K., Reynoso M., Pauluzzi G. et al. , 2018. Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30: 15–36. 10.1105/tpc.17.00581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand A. P., Jansky S. H., Zhao H., Leisner C. P., Zhu X. et al. , 2017. Meiotic crossovers are associated with open chromatin and enriched with Stowaway transposons in potato. Genome Biol. 18: 203 10.1186/s13059-017-1326-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1950. The genetical architecture of heterostyly in primula sinensis. Evolution 4: 340–352. 10.1111/j.1558-5646.1950.tb01404.x [DOI] [Google Scholar]
- McClintock B., 1931. Cytological Observations of Deficiencies Involving Known Genes, Translocations and An Inversion in Zea Mays. University of Missouri, College of Agriculture, Agricultural Experiment Station, Columbia. [Google Scholar]
- McClintock B., 1933. The association of non-homologous parts of chromosomes in the mid-prophase of meiosis in Zea mays. Z. Zellforsch 19: 191–237. 10.1007/BF02462870 [DOI] [Google Scholar]
- McDowell J. M., Dhandaydham M., Long T. A., Aarts M. G., Goff S. et al. , 1998. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874. 10.1105/tpc.10.11.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Howell A. M., and Rose A. M., 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Yehuda E., Stuitje A. R., and Levy A. A., 2005. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 43: 458–466. 10.1111/j.1365-313X.2005.02466.x [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Shilo S., and Levy A. A., 2016. Meiotic recombination and genome evolution in plants. Curr. Opin. Plant Biol. 30: 82–87. 10.1016/j.pbi.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Meyers B. C., Kozik A., Griego A., Kuang H., and Michelmore R. W., 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. 10.1105/tpc.009308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1932. Some genetic aspects of sex. Am. Nat. 66: 118–138. 10.1086/280418 [DOI] [Google Scholar]
- Noël L., Moores T. L., van Der Biezen E. A., Parniske M., Daniels M. J. et al. , 1999. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2112. 10.1105/tpc.11.11.2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oneal E., Lowry D. B., Wright K. M., Zhu Z., and Willis J. H., 2014. Divergent population structure and climate associations of a chromosomal inversion polymorphism across the Mimulus guttatus species complex. Mol. Ecol. 23: 2844–2860. 10.1111/mec.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., and Petkov P., 2010. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11: 221–233. 10.1038/nrg2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., Szatkiewicz J. P., Sawyer K., Leahy N., Parvanov E. D. et al. , 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4: e1000119 10.1371/journal.pgen.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M., and Jones J. D., 1999. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96: 5850–5855. 10.1073/pnas.96.10.5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M., Hammond-Kosack K. E., Golstein C., Thomas C. M., Jones D. A. et al. , 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832. 10.1016/S0092-8674(00)80470-5 [DOI] [PubMed] [Google Scholar]
- Peck J. R., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisupati R., Reichardt I., Seren Ü., Korte P., Nizhynska V. et al. , 2017. Verification of Arabidopsis stock collections using SNPmatch, a tool for genotyping high-plexed samples. Sci. Data 4: 170184 10.1038/sdata.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Richter T. E., Pryor T. J., Bennetzen J. L., and Hulbert S. H., 1995. New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Bradbury P. J., Elshire R. J., Glaubitz J. C., Acharya C. B. et al. , 2015. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. USA 112: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B. A., Patel V., Weigel D., and Schneeberger K., 2015. Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 (Bethesda) 5: 385–398. 10.1534/g3.114.016501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B. A., Seymour D. K., Chae E., Lundberg D. S., and Weigel D., 2017. Methods for genotyping-by-sequencing. Methods Mol. Biol. 1492: 221–242. 10.1007/978-1-4939-6442-0_16 [DOI] [PubMed] [Google Scholar]
- Salomé P. A., Bomblies K., Fitz J., Laitinen R. A. E., Warthmann N. et al. , 2012. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 108: 447–455. 10.1038/hdy.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.-L., Jones G. H., and Franklin F. C. H., 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233. 10.1101/gad.439007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T., Libbrecht R., and Keller L., 2014. Supergenes and complex phenotypes. Curr. Biol. 24: R288–R294. 10.1016/j.cub.2014.01.056 [DOI] [PubMed] [Google Scholar]
- Serra H., Lambing C., Griffin C. H., Topp S. D., Nageswaran D. C. et al. , 2018. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. USA 115: 2437–2442. 10.1073/pnas.1713071115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo S., Melamed-Bessudo C., Dorone Y., Barkai N., and Levy A. A., 2015. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27: 2427–2436. 10.1105/tpc.15.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison L. S., Hoehn K. B., and Noor M. A. F., 2011. Effects of inversions on within- and between-species recombination and divergence. Genome Biol. Evol. 3: 830–841. 10.1093/gbe/evr081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921. A case of rearrangement of genes in Drosophila. Proc. Natl. Acad. Sci. USA 7: 235–237. 10.1073/pnas.7.8.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1926. A crossover reducer in Drosophila melanogaster due to inversion of a section of the third chromosome. Biol. Zent. Bl. 46: 697–702. [Google Scholar]
- Sullivan A. M., Arsovski A. A., Lempe J., Bubb K. L., Weirauch M. T. et al. , 2014. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 8: 2015–2030. 10.1016/j.celrep.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Sun Q., Collins N. C., Ayliffe M., Smith S. M., Drake J. et al. , 2001. Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybenga J., 1970. Simultaneous negative and positive chiasma interference across the break point in interchange heterozygotes. Genetica 41: 209–230. 10.1007/BF00958908 [DOI] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., and Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. 10.1016/0092-8674(83)90331-8 [DOI] [PubMed] [Google Scholar]
- Thompson M. J., and Jiggins C. D., 2014. Supergenes and their role in evolution. Heredity 113: 1–8. 10.1038/hdy.2014.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I., Calabrese P., Cochran D. M., Sokol R. Z., and Arnheim N., 2006. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2: e70 10.1371/journal.pgen.0020070.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood C. J., Choi K., Lambing C., Zhao X., Serra H. et al. , 2018. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28: 519–531. 10.1101/gr.227116.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Weyer A.-L., Monteiro F., Furzer O. J., Nishimura M. T., Cevik V. et al. , 2019. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178: 1260–1272.e14. 10.1016/j.cell.2019.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., Velikkakam James G., Ding J., Becker F., Klasen J. R. et al. , 2013. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife 2: e01426 10.7554/eLife.01426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B. B. H., Thomas C. M., Parniske M., and Jones J. D. G., 2004. Genetic variation at the tomato Cf-4/Cf-9 locus induced by EMS mutagenesis and intralocus recombination. Genetics 167: 459–470. 10.1534/genetics.167.1.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang L., Huang J., Zhang X., Yuan Y. et al. , 2015. Parent–progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523: 463–467. 10.1038/nature14649 [DOI] [PubMed] [Google Scholar]
- Yelina N. E., Choi K., Chelysheva L., Macaulay M., de Snoo B. et al. , 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8: e1002844 10.1371/journal.pgen.1002844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina N. E., Lambing C., Hardcastle T. J., Zhao X., Santos B. et al. , 2015. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29: 2183–2202. 10.1101/gad.270876.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata L., Ding J., Willing E.-M., Hartwig B., Bezdan D. et al. , 2016. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA 113: E4052–E4060. 10.1073/pnas.1607532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., and Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
- Ziolkowski P. A., Berchowitz L. E., Lambing C., Yelina N. E., Zhao X. et al. , 2015. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 4: e03708 10.7554/eLife.03708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw read data include all 1920 individuals (including those that were later filtered out as described above) and can be found under ArrayExpress accession number E-MTAB-8165. The oligos used for the Nextera-based library preparation are available in File S1, and the full list of CO intervals and midpoints is in File S2. File S3 is a list of genes in the regions identified by 50-kb sliding window analysis with the top 5% highest CO frequencies. File S4 is a list of genes in the 95th length percentile for regions without COs in our data set. File S5 is an annotated list of NLR loci with a comparison of locus structure between Col and Ler. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9733838.