Abstract
Recombinant inbred lines (RILs) are an important resource for mapping genes controlling complex traits in many species. While RIL populations have been developed for maize, a maize RIL population with multiple teosinte inbred lines as parents has been lacking. Here, we report a teosinte nested association mapping (TeoNAM) population, derived from crossing five teosinte inbreds to the maize inbred line W22. The resulting 1257 BC1S4 RILs were genotyped with 51,544 SNPs, providing a high-density genetic map with a length of 1540 cM. On average, each RIL is 15% homozygous teosinte and 8% heterozygous. We performed joint linkage mapping (JLM) and a genome-wide association study (GWAS) for 22 domestication and agronomic traits. A total of 255 QTL from JLM were identified, with many of these mapping near known genes or novel candidate genes. TeoNAM is a useful resource for QTL mapping for the discovery of novel allelic variation from teosinte. TeoNAM provides the first report that PROSTRATE GROWTH1, a rice domestication gene, is also a QTL associated with tillering in teosinte and maize. We detected multiple QTL for flowering time and other traits for which the teosinte allele contributes to a more maize-like phenotype. Such QTL could be valuable in maize improvement.
Keywords: RIL, TeoNAM, JLM, GWAS, maize, domestication, multiparental populations; MPP
RECOMBINANT inbred line (RIL) populations are powerful tools for investigating the genetic architecture of traits and identifying the causal genes that underlie trait variation. RIL populations have been widely used in many organisms. In mammals, the well-known Collaborative Cross, consisting of a large panel of mouse multiparental RILs, has been specifically designed for the analysis of complex traits (Churchill et al. 2004). Similarly, the Drosophila Synthetic Population Resource (DSPR), which consists of two sets of RILs, has been designed to combine the high mapping resolution offered by multiple generations of recombination with the high statistical power afforded by a linkage-based design (King et al. 2012). In plants, the maize nested association mapping (NAM) population, which crossed 25 founders to a common parent in maize (Yu et al. 2008), has been successfully applied to a large number of traits (Buckler et al. 2009; Kump et al. 2011; Tian et al. 2011). The NAM design has also been utilized in other crops such as barley (Maurer et al. 2015; Nice et al. 2016), rice (Fragoso et al. 2017), sorghum (Bouchet et al. 2017), wheat (Jordan et al. 2018), and soybean (Xavier et al. 2018). In Arabidopsis, another design, called the Multiparent Advanced Generation Intercross (MAGIC) population, provides high precision for the detection of QTL (Kover et al. 2009; Huang et al. 2011). This design has also been used in wheat (Huang et al. 2012; Mackay et al. 2014), rice (Bandillo et al. 2013), and maize (Dell’Acqua et al. 2015).
For the study of maize domestication, many new discoveries were made using a biparental maize–teosinte BC2S3 RIL population. Shannon (2012) performed QTL mapping for 16 traits and examined the genetic architecture of domestication at the whole-genome level. This RIL population has also been widely used to fine-map QTL, and identify causal or candidate genes for many traits, including seed shattering (Lin et al. 2012), leaf number (Li et al. 2016), kernel row number (KRN) (Calderón et al. 2016), shoot apical meristem morphology (Leiboff et al. 2016), vascular bundle number (Huang et al. 2016), tassel-related traits (G. Xu et al. 2017), nodal root number (Zhang et al. 2018), and leaf morphological traits (Fu et al. 2019). Using this population, several QTL have been fine-mapped to single genes including grassy tillers1 (gt1) for controlling prolificacy (PROL) (Wills et al. 2013), prolamin box-binding factor1 (pbf1) for kernel weight (Lang et al. 2014), glossy15 (gl15) for vegetative phase changes (D. Xu et al. 2017), UPA1 (Upright Plant Architecture1) and UPA2 for leaf angle (Tian et al. 2019), as well as several genes regulating flowering time: ZmCCT10 (Hung et al. 2012), Zea Agamous-like1 (zagl1) (Wills et al. 2017), ZmCCT9 (Huang et al. 2018), Zea mays CENTRORADIALIS8 (ZCN8) (Guo et al. 2018), and MADS-box transcription factor69 (ZmMADS69) (Liang et al. 2019). In addition to phenotypic traits, the maize–teosinte BC2S3 RIL population was used for comprehensive genome-wide expression QTL (eQTL) analysis to study the changes in gene expression during maize domestication (Wang et al. 2018), and metabolite QTL analysis to study metabolic divergence between maize and teosinte (Xu et al. 2019).
Despite its utility, the maize–teosinte BC2S3 RIL population has three limitations. First, there is only a single teosinte parent, which cannot broadly represent the diversity of teosinte. Second, this population had two generations of backcrossing, which has produced a background in which some teosinte traits are suppressed and do not segregate among the RILs. Third, the teosinte parent was a wild outcrossed individual which, unlike an inbred line, could not be maintained as a permanent resource.
In this paper, we report the development of a teosinte NAM (TeoNAM) population of 1257 BC1S4 RILs using five teosinte inbred parents crossed with a common maize parent (W22) for mapping QTL for domestication and agronomic traits. We have genotyped the RILs with 51,544 genotype-by-sequencing (GBS) markers that provide a high-density genetic map. The TeoNAM population captures a large number of recombination events for localizing QTL to genomic locations and the single generation of backcross allows enhanced expression of teosinte traits as compared to the BC2S3 RIL population. We report data for 22 traits but focus our discussion on nine traits to illustrate the utility of TeoNAM, including the identification of candidate genes. TeoNAM will be a valuable resource for dissecting the genetic basis of domestication and agronomic traits.
Materials and Methods
Population development
The TeoNAM population was designed as a genetic resource for studying maize genetics and domestication. Five wild teosinte parents were chosen, with four teosinte inbred lines that capture some diversity of Z. mays ssp. parviglumis (TIL01, TIL03, TIL11, and TIL14) and one teosinte inbred line of Z. mays ssp. mexicana (TIL25). The common parent is a modern maize inbred line (W22) that has been widely used in maize genetics. The five teosinte parents were crossed to W22, and followed by one generation of backcrossing and four generations of selfing (Supplemental Material, Figure S1). We obtained 1257 BC1S4 RILs with 223, 270, 219, 235, and 310 lines for W22 × TIL01, W22 × TIL03, W22 × TIL11, W22 × TIL14, and W22 × TIL25, respectively.
Marker data
All DNA samples of 1257 lines were genotyped using GBS technology (Elshire et al. 2011). The genotypes were called from GBS raw sequencing reads using the TASSEL5-GBS Production Pipeline based on 955,690 SNPs in the ZeaGBSv2.7 Production TagsOnPhysicalMap file (Glaubitz et al. 2014). Then, the raw GBS markers were filtered in each RIL subpopulation using the following steps. We first removed sites with minor allele frequencies (MAFs) < 5% and thinned sites 64-bp apart using “Thin Sites by Position” in TASSEL5 (Bradbury et al. 2007), and then we ran FSFHap Imputation in TASSEL5 separately for each chromosome using the following parameters: backcross, Phet = 0.03125, Fillgaps = TRUE, and the default settings for other features. The imputed parental call files from the 10 chromosomes were then combined together and passed to R/qtl (Broman et al. 2003) to estimate the genetic map. The B73 reference genome v2 was used to determine marker order, and genetic distances between markers were calculated using the Haldane mapping function as part of the est.map command with an assumed genotyping error rate of 0.001, taking the BC1S4 pedigree of the RIL into consideration (Shannon 2012). Bad genetic markers were identified by visual inspection of the genetic map and removed, then we repeated all filtering steps. Finally, an average of 13,733 high-quality SNPs was obtained for each subpopulation (Table 1).
Table 1. TeoNAM genetic map statistics.
| Population | No. RILs | No. Markers | Length (cM) | No. XOs | cM/Mb | W22 (%) | Heterozygous (%) | Teosinte (%) |
|---|---|---|---|---|---|---|---|---|
| W22 × TIL01 | 223 | 13,088 | 1457 | 6,291 | 0.71 | 75.8 | 7.7 | 16.0 |
| W22 × TIL03 | 270 | 16,109 | 1596 | 8,505 | 0.78 | 75.5 | 8.1 | 16.2 |
| W22 × TIL11 | 219 | 13,187 | 1398 | 5,745 | 0.68 | 76.3 | 7.6 | 15.6 |
| W22 × TIL14 | 235 | 11,395 | 1348 | 6,462 | 0.65 | 75.7 | 9.4 | 14.6 |
| W22 × TIL25 | 310 | 14,884 | 1506 | 8,877 | 0.73 | 77.6 | 8.0 | 14.2 |
| Composite | 1257 | 51,544 | 1540 | 35,880 | 0.75 | 76.6 | 8.1 | 15.0 |
No., number; RIL, recombinant inbred line; XO, crossover.
Field design and phenotyping
The TeoNAM population was planted using a randomized complete block design at the University of Wisconsin West Madison Agricultural Research Station in different years. The subpopulations W22 × TIL01, W22 × TIL03, and W22 × TIL11 were grown in summer 2015 and 2016, subpopulation W22 × TIL14 was grown in summer 2016 and 2017, and subpopulation W22 × TIL25 was grown in summer 2017 with two blocks. We planted one subpopulation within each block and all lines were randomized within each block. Each row had 16 seeds planted 1-ft apart, and spacing between any two rows was 30 in.
Twenty-two traits were scored (Table 2): days to anthesis (DTA) (number of days between planting and when at least one-half of the plants in a plot were shedding pollen); days to silk (DTS) (number of days between planting and when at least one-half of the plants in a plot were showing silk); anthesis–silk interval (ASI) (number of days between anthesis and silk); tassel branch number (TBN) (number of tassel branches on the main stalk); culm diameter (CULM) (diameter of the narrowest plane of the main stalk right above the ground); plant height (PLHT) (distance from the ground to the topmost node on the main stalk); leaf length (LFLN) (length of a well-developed leaf, usually fourth to sixth from top); leaf width (LFWD) (width of a well-developed leaf, usually fourth to sixth from top); tiller number (TILN) (number of tillers surrounding main stalk); PROL (0 vs. 1 for absence/presence of secondary ears at the topmost branch-bearing node on the main stalk); ear branch number (EB) (number of branches on the primary lateral inflorescence); staminate spikelet (STAM) (0–3 scale for spikelet sex on the primary lateral inflorescence, where 0 indicates completely feminized and 3 indicates completely staminate); KRN (number of internode columns on the primary lateral inflorescence); ear length (EL) (length of the primary lateral inflorescence); ear diameter (ED) (diameter of the primary lateral inflorescence); kernel weight (KW) (average weight of 50 random kernels from five ears); shattering (SHN) (number of pieces into which an ear shatters when dropped to the floor from a height of ∼1.8 m); barren ear base (BARE) (0–2 scale for lack of kernels at the base of the ear, where 0 indicates kernels present at the base and 2 indicates no developed kernels at the base of the ear); glume score (GLUM) (0–3 scale for glume size, where 0 indicates small and 3 indicates large); glume color (GLCO) (0–4 scale of glume color for white through brown); red pericarp (REPE) (0–2 scale for colorless to red pericarp); and yellow pericarp (YEPE) (0–2 scale for dull yellow to bright yellow pericarp). The average trait values from 2 years were used for QTL analysis.
Table 2. List of 22 domestication and agronomic traits scored.
| Trait | Abbreviation | Units | Category |
|---|---|---|---|
| Anthesis–silk interval | ASI | count | Agronomic |
| Barren ear base | BARE | score | Agronomic |
| Days to anthesis | DTA | count | Agronomic |
| Days to silk | DTS | count | Agronomic |
| Plant height | PLHT | cm | Agronomic |
| Tassel branch number | TBN | count | Agronomic |
| Yellow pericarp | YEPE | score | Agronomic |
| Culm diameter | CULM | mm | Domestication |
| Ear branch number | EB | count | Domestication |
| Ear diameter | ED | mm | Domestication |
| Ear length | EL | cm | Domestication |
| Glume color | GLCO | score | Domestication |
| Glume score | GLUM | score | Domestication |
| Kernel row number | KRN | count | Domestication |
| Kernel weight | KW | g | Domestication |
| Leaf length | LFLN | cm | Domestication |
| Leaf width | LFWD | cm | Domestication |
| Prolificacy | PROL | binary | Domestication |
| Red pericarp | REPE | score | Domestication |
| Shattering | SHN | count | Domestication |
| Staminate spikelet | STAM | score | Domestication |
| Tiller number | TILN | count | Domestication |
Genetic map construction and marker imputation
A composite genetic map was constructed for the TeoNAM population. The markers from the five RIL subpopulations were combined together into 51,544 unique SNPs, and the missing genotypes were imputed according to the flanking markers. If the flanking markers had same genotypes, the missing genotype was imputed as the same with flanking markers, or otherwise left as missing. The imputed genotypes were then passed to R/qtl software to estimate the genetic map.
Since stepwise regression cannot use individuals with missing marker data, we performed a further step to impute missing data around breakpoint as previously described (Tian et al. 2011). First, we transformed genotypes to a numeric format, in which markers with a homozygous W22 parent were coded as 0, markers with a homozygous non-W22 parent were coded as 2, and markers with heterozygous genotypes were coded as 1. Markers within breakpoint were imputed according to the genetic distance of flanking two markers. Because stepwise regression is computationally intensive, we thinned SNPs within 0.1 cM. We finally obtained 4578 markers for subsequent joint linkage analysis.
Simple QTL mapping
QTL mapping was carried out using a modified version of R/qtl (Broman et al. 2003), which takes into account the BC1S4 pedigree of the RILs (Shannon 2012). For each trait, a total of 1000 permutation tests were used to determine the significance threshold levels for claiming QTL. After permutation, an approximate LOD score of 4.0 at P < 0.05 was obtained across all traits. With the LOD threshold, simple interval mapping was first fitted using Haley–Knott regression implemented in the scanone command of R/qtl. The multiple QTL model was then applied to search for additional QTL, and accurately refine QTL positions using refineqtl and addqtl in R/qtl. The entire process was repeated until significant QTL could no longer be added. The total phenotypic variation explained by all QTL was calculated from a full model that fitted all QTL terms in the model using the fitqtl function. The percentage of phenotypic variation explained by each QTL was estimated using a drop-one ANOVA analysis implemented with the fitqtl function. The C.I. for each QTL was defined using a 1.5-LOD support interval. To make results comparable among five subpopulations, the composite genetic map was used for QTL mapping.
Joint linkage mapping
To map QTL in the TeoNAM population, a joint linkage mapping (JLM) procedure was performed as previously described (Buckler et al. 2009; Tian et al. 2011). First, a total of 1000 permutations were performed to determine the significance cutoff for each trait. JLM was performed using the stepwise linear regression fixed model implemented by the PROC GLMSELECT procedure in SAS software. The family main effect was fitted first, and then marker effects nested within families were selected to enter or leave the model based on the permutated P-value using a marginal F-test. After the model was fitted with stepwise regression, each marker was dropped from the full model one at a time and a single best marker was refitted to improve the overall fit of the model. A threshold of α = 0.05 was used to declare significant allele effects across families within each QTL identified by stepwise regression. The QTL support interval was calculated by adding each marker from the same chromosome of that QTL at a time to the full model. If the P-value of the marginal F-test of the QTL was not significant at the 0.01 level, the flanking marker should be in the support interval for the QTL, as the new flanking marker explained the QTL as well as the original marker.
Genome-wide association study
A genome-wide association study (GWAS) approach was also used to map QTL in the TeoNAM population. Since GBS produces relatively low-density markers, the 955,690 raw SNPs from the GBS pipeline were filtered using less-conservative criteria: MAF > 0.01, missing rate < 0.75, and heterozygosity rate < 0.1. After this filtering, 181,404 GBS SNPs were used to run FSFHap Imputation in TASSEL5 separately for each chromosome and subpopulation using the following parameters: backcross, Phet = 0.03125, Fillgaps = TRUE, and the default settings for other features. Imputed genotypes were then combined together, and SNPs with missing rate > 0.2 and MAF < 0.05 across 1257 RILs were removed, and a total of 118,838 SNPs were kept and used for GWAS. GWAS was performed using a linear mixed model accounting for population structure (Q) and kinship matrix (K), where Q was computed as the first five principle components and K was calculated using a centered Identity-By-State (IBS) method as implemented in TASSEL (Bradbury et al. 2007). P-values < P = 0.00001 (LOD = 5) were considered to be the significance threshold, following a previous study (Kremling et al. 2018).
QTL candidate analysis
To report the QTL position following the latest genomic version, we used the CrossMap (Zhao et al. 2014) software to uplift the GBS SNP positions from maize B73 reference AGPv2 coordinates to AGPv4 coordinates. QTL candidates were analyzed by checking the gene annotations of genes within QTL support intervals.
Data availability
Seeds for all 1257 RILs in TeoNAM are available at the Maize Genetics Cooperative Stock Center. The SNP genotypes for TeoNAM are available at the Cyverse Discovery Environment under the directory: http://datacommons.cyverse.org/browse/iplant/home/shared/panzea/genotypes/GBS/TeosinteNAM. The genotypic data were uploaded with AGPv2 position in the marker name. The raw phenotypic data for TeoNAM are available at Figshare database: https://doi.org/10.6084/m9.figshare.9820178. The seed information, SNP genotypes, and phenotypic data for the BC2S3 maize–teosinte RIL population can be found in Shannon et al. (2019). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9250682.
Results
Characterization of a TeoNAM population
We developed a TeoNAM population, which was constructed by crossing five teosinte inbred lines to a maize inbred line (W22), followed by one generation of backcross to the common recurrent maize parent and four generations of selfing (Figure S1). The teosinte parents include four Z. mays ssp. parviglumis lines and one Z. mays ssp. mexicana line. As such, TeoNAM encompasses five biparental families, each with 219–310 BC1S4-derived RILs for a total of 1257 RILs. The number of segregating SNP markers ranges from 11,395 to 16,109 per family, with over 51,000 total SNP markers (Table 1).
The expected segregation for a BC1S4 population is 73.44% homozygous recurrent, 3.13% heterozygous, and 23.44% homozygous donor parent. Overall, the percentage of genotypes observed were 76.6% W22 homozygous, 15% teosinte homozygous, and 8.1% heterozygous across all SNP sites in the TeoNAM population (Table 1). The percentage of teosinte varied among subpopulations from 14.2 to 16.2% (Table 1), and also varied across the genome in all subpopulations (Figure S2). The observed higher than expected heterozygosity may be due to unconscious selection for more heterozygous plants that had hybrid vigor. The chromosomal region of highest heterozygosity is on the short arm of chromosome 4 near teosinte glume architecture1 (tga1) (Wang et al. 2005). Selection against homozygotes for the teosinte allele of tga1, which have poor ear and kernel quality, may be the cause. For a BC1S4, the expected frequency of the maize allele is 75%. All subpopulations deviate from this with an excess of maize alleles (Table 1) and the amount of excess varies across the genome (Figure S3).
We constructed genetic linkage maps for each family and a composite linkage map based on all RILs across all families, and identified and annotated 51,544 high-confidence SNPs that were used to impute the SNP alleles in the RILs. The composite genetic map based on these markers is 1540 cM in length including 35,880 crossovers. We examined the relationship between genetic distance in centimorgans and physical distance in megabases based on the composite genetic map. The mean value is 0.75 cM/Mb. However, there is wide deviation from the mean across the genome (0–5.52 cM/Mb). As expected, there is suppressed recombination near the centromeres (Figure S2) and frequent recombination near the telomeres, where gene density is high as well (Figure S2).
We scored 22 traits for the TeoNAM lines of which 15 traits are domestication related, including vegetative gigantism (CULM, LFLN, and LFWD), prolificacy (PROL), tillering (TILN), ear SHN, conversion of the inflorescence from staminate to pistillate (STAM), multiple ear-related traits (EB, ED, EL, KRN, and KW), glume traits (GLCO and GLUM), and REPE (Table 2). Additionally, several agronomic traits were scored including flowering (ASI, DTA, and DTS), plant architecture (PLHT and TBN), BARE, and YEPE. Most traits (ASI, CULM, DTA, DTS, ED, EL, KRN, KW, LFLN, LFWD, PHLT, and TBN) follow approximately normal distributions, suggesting oligo- or polygenic genetic control of these traits, but other traits (BARE, EB, GLCO, GLUM, PROL, REPE, SHN, STAM, TILN, and YEPE) exhibited a skewed or nonnormal distribution. Some of these traits are meristic or discrete traits (e.g., PROL or TILN). A few traits, like STAM, show a two-part distribution with a spike at 0 plus a continuous range of values from 0 to 2, which suggests that they may be polygenic threshold traits (Figure S4). There are also substantial differences in trait means among the five subpopulations, indicating underlying differences in genetic architecture among the five teosinte inbreds (Figure S5).
Validation of the power of TeoNAM
QTL mapping:
We used both JLM and the GWAS method as two complementary approaches for QTL detection. We also used basic interval QTL mapping for the five individual subpopulations to provide a guide for future work to fine-map the genes underlying the QTL. We detected 255 QTL for 22 traits by JLM, which combines information across all families (Figure 1 and Table S1). We detected a total of 150 QTL by GWAS, among which 57 QTL overlapped with QTL by JLM (Table S2). Separate QTL mapping for each subpopulation detected 464 QTL in total, among which 293 QTL overlapped with QTL by JLM (Figures S6–S27 and Table S3). Below, we focus on QTL detected by JLM for our characterization of the genetic architecture and the distribution of QTL allelic effects.
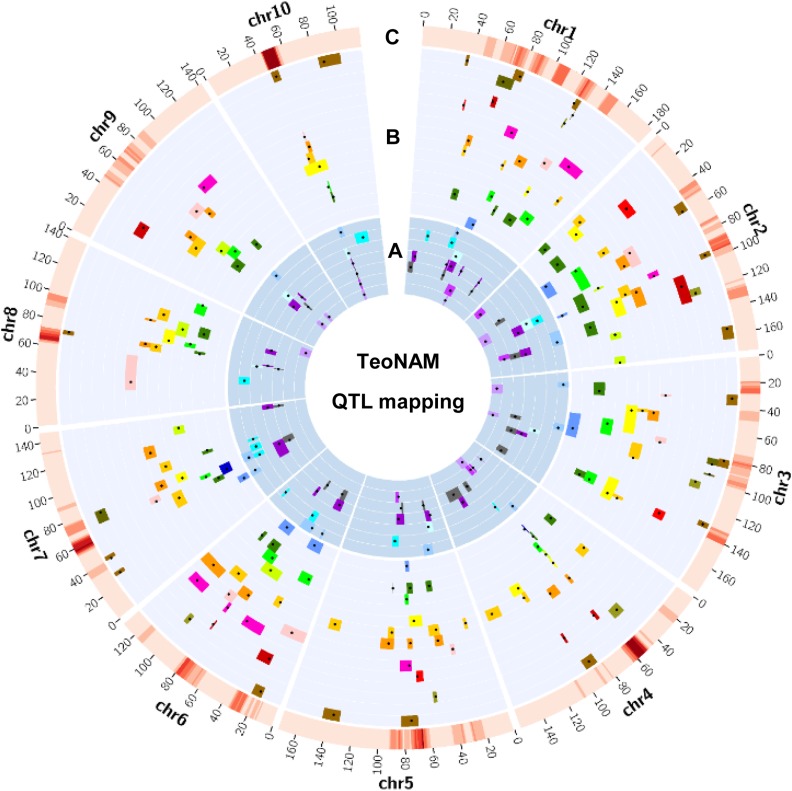
Figure 1.
Genomic distribution of QTL for all 22 traits in TeoNAM. The 22 agronomic (A) and domestication (B) traits are plotted in layers with different background colors, following the order of ASI, BARE, DTA, DTS, PLHT, TBN, YEPE, CULM, EB, ED, EL, GLCO, GLUM, KRN, KW, LFLN, LFWD, PROL, REPE, SHN, STAM, and TILN outward. Black dots indicate QTL peaks detected by JLM and colored bars indicate the support interval of QTL for different traits. The heat map in the outmost layer (C) shows the number of QTL peaks using a sliding window of 10- and 1-cM steps, where low-to-high densities of QTL (0–13) are shown in light-to-dark red, respectively. ASI, anthesis–silk interval; BARE, barren ear base; DTA, days to anthesis; DTS, days to silk; CULM, culm diameter; EB, ear branch number; ED, ear diameter; EL, ear length; GLCO, glume color; GLUM, glume score; KRN, kernel row number; KW, kernel weight; LFLN, leaf length; LFWD, leaf width; PLHT, plant height; PROL, prolificacy; REPE, red pericarp; SHN, shattering; STAM, staminate spikelet; TBN, tassel branch number; TILN, tiller number; YEPE, yellow pericarp. chr, chromosome; JLM, joint linkage mapping; TeoNAM, teosinte nested association mapping.
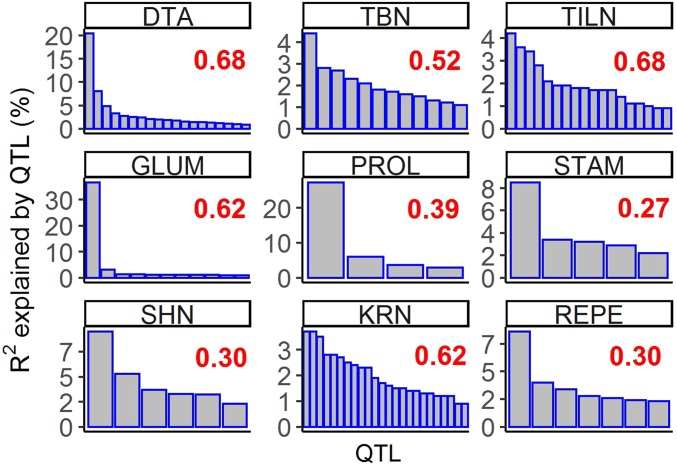
Among 22 traits, the numbers of QTL range from 2 to 24; the trait with the most QTL is KRN. Genetic architecture varies considerably among traits (Figure 2 and Figure S28). Several traits—including BARE, GLCO, GLUM, PROL, REPE, and STAM—had relatively simple genetic architectures, with a single QTL of very large effect plus a few (3–10) QTL of small effect. The largest QTL for each of these traits has between 2.1 and 11.7 times the size of the effect of the second largest QTL. A second class of traits has a genetic architecture that is either more polygenic (ED, KRN, KW, LFLN, TBN, and TILN) or has only a few QTL of small effect (ASI, CULM, EB, LFWD, PLHT, and SHN). For these traits, there was no single large-effect QTL that accounts for the majority of the explainable variation. The largest effect QTL for each of these traits has between 1 and 1.8 times the size of the effect of the second largest QTL. A final class of traits has a genetic architecture with both a single QTL of moderately large effect plus multiple (11–19) QTL of small effect. These traits include DTA, DTS, EL, and YEPE. The largest effect QTL for each of these traits is between 2.1 and 3.7 times the size of the effect of the second largest QTL.
Figure 2.
Distinct genetic architectures for different traits. The nine traits that we focused on in the main text are shown. The x-axes indicate QTL and the y-axes indicate the phenotypic variation explained by each QTL (R2). Red numbers indicate variance explained by the QTL model for each trait. The R2 distribution for 13 additional traits can be found in Figure S28. DTA, days to anthesis; GLUM, glume score; KRN, kernel row number; PROL, prolificacy; REPE, red pericarp; SHN, shattering; STAM, staminate spikelet; TBN, tassel branch number; TILN, tiller number.
QTL for agronomic traits:
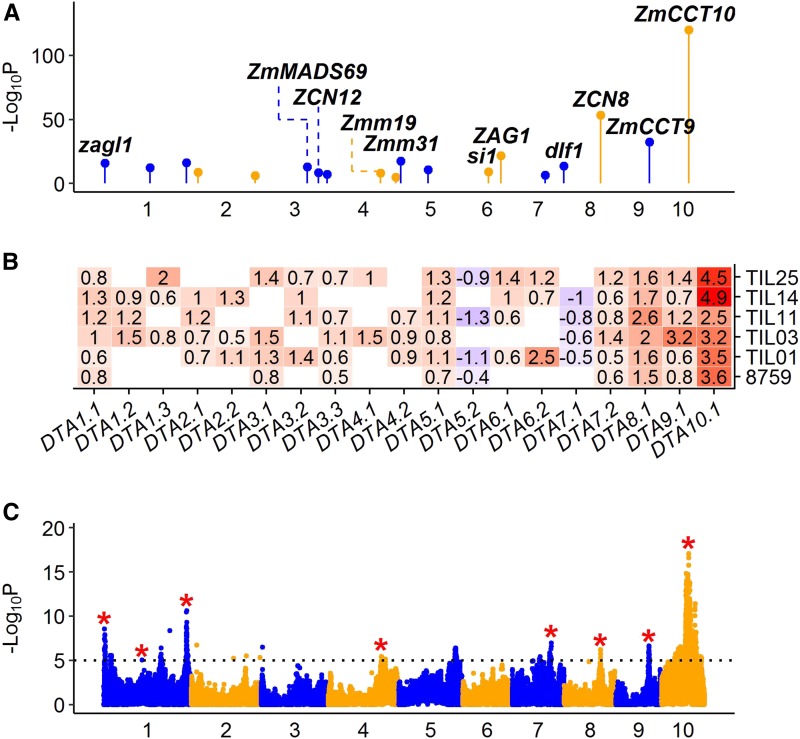
DTA is a classical quantitative trait for maize, and in TeoNAM, it is controlled by a large-effect QTL plus many small-effect QTL from JLM results. We detected 19 QTL that explained 68% of the total variance for DTA (Figure 3). Among them, several recently cloned flowering time genes were detected. For example, zagl1 was included within QTL DTA1.1, which affects flowering time as well as multiple traits related to ear size, with the maize allele conferring larger ears with more kernels (Wills et al. 2017). ZmMADS69 was included within QTL DTA3.1, which functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module, and contributes to both long-day and short-day adaptation (Liang et al. 2019). ZCN8 was included within QTL DTA8.1, which is the maize florigen gene and has a central role in mediating flowering (Meng et al. 2011; Guo et al. 2018). ZmCCT9 was included within QTL DTA9.1, in which a distant Harbinger-like transposon acts as a cis-regulatory element to repress its expression to promote flowering under the long days of higher latitudes (Huang et al. 2018). ZmCCT10 was included within QTL DTA10.1, which is a known gene involved in the photoperiod response in maize (Hung et al. 2012; Yang et al. 2013).
Figure 3.
QTL characterization for agronomic trait DTA. (A) Genomic distribution of 19 QTL for DTA detected by JLM. The known candidate genes are shown above the corresponding QTL in bold italic. (B) Heat map shows additive allele effects of teosinte relative to maize in number of days for 19 QTL detected by JLM. The allele effect of teosinte parent 8759 was estimated from the 866 maize–teosinte BC2S3 RILs (Shannon 2012). Insignificant effects are shown as blank. Red and blue colors indicate that the teosinte alleles delay or promote flowering time, respectively. (C) Manhattan plot shows QTL detected by GWAS. The significance threshold at LOD = 5 is indicated by black dotted line. The red stars indicate GWAS signals overlapping with QTL by JLM. In (A) and (C), odd and even numbered chromosomes are shown in blue and orange colors, respectively. DTA, days to anthesis; GWAS, genome-wide association study; JLM, joint linkage mapping; RIL, recombinant inbred line.
In addition to these genes, we also identified several other candidate genes for DTA that have not previously been characterized as genes underlying a QTL. Z. mays CENTRORADIALIS12 (ZCN12) was included within QTL DTA3.2, which is a potential floral activator (Meng et al. 2011). Z. mays MADS19 (Zmm19) was included within QTL DTA4.1 and Z. mays MADS31 (Zmm31) was included within DTA5.1. silky1 (si1) was included within QTL DTA6.1, which is also a MADS-box gene required for lodicule and stamen identity (Ambrose et al. 2000). Zea AGAMOUS1 (ZAG1) was included within QTL DTA6.2, which is known to affect maize flower development (Schmidt et al. 1993). It is well known that MADS-box genes encode transcription factors that are key regulators of plant inflorescence and flower development (Theissen et al. 2000). Other than MADS genes, delayed flowering1 (dlf1), a floral activator gene downstream of ZCN8 (Meng et al. 2011), was included within QTL DTA7.2.
As expected, the teosinte alleles delayed flowering for the above QTL that mapped to candidate genes (Figure 3). We plotted the phenotypic difference in DTA between teosinte and maize across the whole genome, and the teosinte genotype was associated with late flowering over most of the genome, even where no QTL were detected, suggesting that there are many additional minor-effect QTL that were not detected due to insufficient statistical power (Figure S29). Interestingly, chromosomes 5 and 7 are exceptions to this pattern, with the teosinte genotype being associated with early flowering at most sites (Figure S29). Results for DTS were similar to those for DTA, as expected (Figure S30).
TBN is the only tassel trait that we scored. We detected 12 QTL of small effect that explained 52% of the total variance for TBN (Figure S31). Among them, several classical genes were identified. fasciated ear4 (fea4) was included within QTL TBN6.1, which is a bZIP transcription factor with fasciated ears and tassels, as well as greatly enlarged vegetative and inflorescence meristems (Pautler et al. 2015). tasselsheath1 (tsh1) was included within QTL TBN6.2, which is a GATA class transcription factor that promotes bract growth and reduces branching (Whipple et al. 2010). ramosa1 (ra1) was included within QTL TBN7.1, which is a C2H2 zinc finger transcription factor that has tassels with an increased number of long branches as well as branched ears (Vollbrecht et al. 2005). tasselsheath4 (tsh4) was included within QTL TBN7.2, which is a SQUAMOSA promoter-binding protein (SBP)-box transcription factor that functions to repress lateral organ growth and also affects phyllotaxy, axillary meristem initiation, and meristem determinacy within the floral phase (Chuck et al. 2010). Barren inflorescence1 (Bif1) was included within QTL TBN8.1, which shows decreased production of branches and spikelet pairs (Barazesh and McSteen 2008). Zea floricaula leafy1 (zfl1) was included within QTL TBN10.1, which together with its homolog zfl2 leads to a disruption of floral organ identity and patterning, as well as to defects in inflorescence architecture and in the vegetative-to-reproductive phase transition (Bomblies et al. 2003).
QTL for domestication traits:
TILN is a classical domestication trait that measures the difference in plant architecture between maize and its wild relative teosinte, i.e., the low apical dominance of a highly branched teosinte plant as compared to the less-branched maize plant. We detected 18 small-effect QTL that explained 68% of the total variance for TILN (Figure S32). Among them, teosinte branched1 (tb1) was included within QTL TILN1.3, which is a TCP family of transcriptional regulators contributing to the increase in apical dominance during maize domestication (Doebley et al. 1997). Additionally, Zea AGAMOUS2 (ZAG2) was included within QTL TILN3.2, which is a MADS-box gene recently found to be downstream of tb1 (Studer et al. 2017). Zmm20 and Zmm26 were included within QTL TILN1.1 and TILN5.2, respectively, which are two other MADS-box genes that were possible targets of selection during domestication (Zhao et al. 2011). PROSTRATE GROWTH1 (PROG1) was included within QTL TILN7.1, which is a C2H2 zinc finger protein controlling a key change during rice domestication from prostrate to erect growth, and also affecting plant architecture and yield-related traits (Jin et al. 2008; Tan et al. 2008). There are 13 genes in the support interval and the QTL peak is closest to PROG1, being ∼14 kb 5′ of the start site (Figure S32). This is the first evidence that PROG1 may have had a role in maize domestication.
GLUM is a classical maize domestication trait that measures the dramatic change from the fruitcase-enveloped kernels of the teosinte ear to the naked grains of maize ear. Previously, this trait was shown to be largely controlled by a single gene that is known as teosinte glume architecture1 (tga1) (Wang et al. 2005, 2015). Interestingly, tga1 is a direct target of tb1. We detected 11 QTL that explained 62% of the total variance for GLUM. These QTL include tga1 itself within a large-effect QTL GLUM4.1 plus several small-effect QTL (Figure S33). Among the small-effect QTL, two MADS genes (Zmm27 and Zmm7) were included within GLUM2.2 and GLUM7.1, respectively. In this regard, Studer et al. (2017) recently defined a maize domestication gene network in which tga1 regulates multiple MADS-box transcription factors.
PROL is also an important domestication trait that measures the difference between the many-eared plants of teosinte and the few-eared (one or two) plants of maize. Previously, a large-effect QTL was fine-mapped to a region 2.7 kb upstream of gt1 (Wills et al. 2013). Interestingly, gt1 is a known target of tb1 (Whipple et al. 2011). We detected four QTL that explained 39% of the total variance for PROL, which included a single large-effect QTL plus three small-effect QTL (Figure S34). Concordantly, gt1 was included within QTL PROL1.1. The QTL PROL3.1 support interval included sparse inflorescence1 (spi1), which affects the initiation of axillary meristems and lateral organs during vegetative and inflorescence development in maize (Gallavotti et al. 2008).
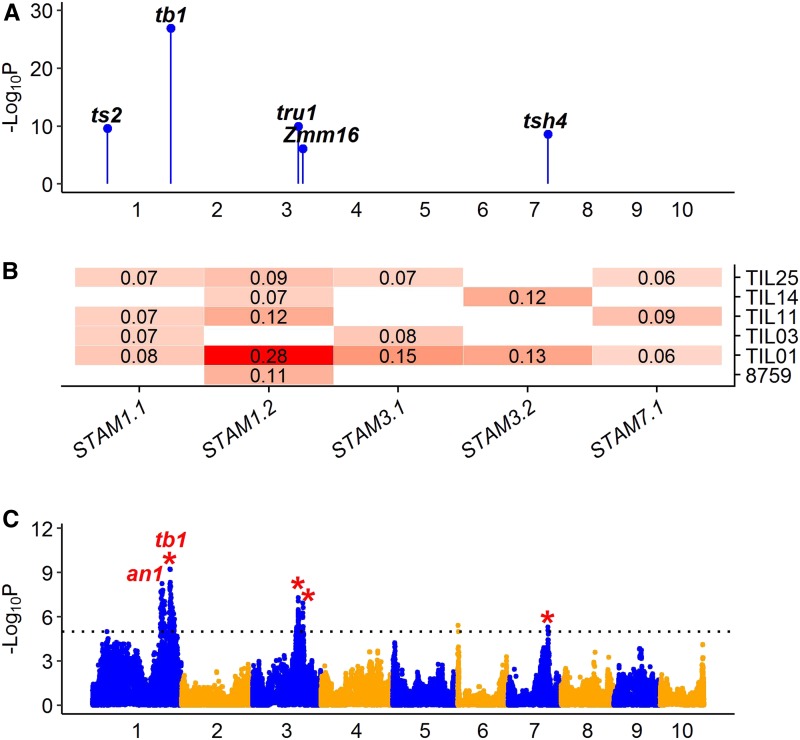
STAM measures the proportion of the terminal lateral inflorescence on the uppermost lateral branch that is staminate. Relative to domestication, this trait represents the sexual conversion of the terminal lateral inflorescence from tassel (staminate) in teosinte to ear (pistillate) in maize. Currently, tb1 and tassels replace upper ears1 (tru1) are the only two genes that have been shown to regulate this sexual difference. We detected five QTL that explained 27% of the total variance for STAM (Figure 4). QTL STAM1.2 mapped upstream of tb1, which is an important domestication gene known for various traits (Doebley et al. 1995). tru1 was included within QTL STAM3.1, which is a direct target of tb1 (Dong et al. 2017). tassel seed2 (ts2) was included within QTL STAM1.1, which is a recessive mutant that produces pistillate spikelets in the terminal inflorescence (tassel) (Irish and Nelson 1993). Z. mays MADS16 (Zmm16) was included within QTL STAM3.2. tassel sheath4 (tsh4) was included within QTL STAM7.1, which is an SBP-box transcription factor that regulates the differentiation of lateral primordia (Chuck et al. 2010). In addition to these QTL, two other STAM QTL were detected by GWAS. Notably, a QTL on chromosome 1 (AGPv4 chromosome 1:234.4–249.9 Mb) is located upstream of tb1 and colocalized with STAM1.1 from a recent study (Yang 2018). The known gene anther ear1 (an1) is a strong candidate gene for this QTL since loss of an1 function results in the development of staminate flowers in the ears (Bensen et al. 1995). The tb1 QTL region was also detected by GWAS with a strong signal (AGPv4 chromosome 1:264.1–283.1 Mb), which includes tb1.
Figure 4.
QTL characterization for domestication trait STAM. (A) Genomic distribution of five QTL for STAM detected by JLM. The known candidate genes are shown above the corresponding QTL in bold italic. (B) Heat map shows additive allele effects of teosinte relative to maize for five QTL detected by JLM. The allele effect of teosinte parent 8759 was estimated from the 866 maize–teosinte BC2S3 RILs (Shannon 2012). Insignificant effects are shown as blank. The teosinte genotypes at all QTL consistently contribute to a staminate lateral inflorescence. (C) Manhattan plot shows QTL detected by GWAS. The significance threshold at LOD = 5 is indicated by black dotted line. The red stars indicate GWAS signals overlapping with QTL by JLM. In (A) and (C), odd and even numbered chromosomes are shown in blue and orange colors, respectively. GWAS, genome-wide association study; JLM, joint linkage mapping; RIL, recombinant inbred line; STAM, staminate spikelet.
SHN measures ear shattering, the loss of which is a key step during crop domestication (Doebley et al. 2006). Teosinte ears have abscission layers between the fruitcases (modified internodes) that allow the ear to shatter into single-seed units (fruitcase) at maturity. The maize ear lacks abscission layers and remains intact at maturity. Currently, only two maize orthologs (ZmSh1-1 and ZmSh1-5.1+ZmSh1-5.2) of sorghum and rice Shattering1 (Sh1) have been verified for seed SHN (Lin et al. 2012). We detected six QTL that explained 30% of the total variance for SHN (Figure S35). Sh1-1 and Sh1-5.1/5.2 were included within QTL SHN1.1 and SHN5.1, respectively, confirming prior identification of these maize paralogs of the sorghum SHN gene as strong candidates for our QTL.
KRN is a domestication trait measuring the dramatic change from the two-ranked teosinte ear to a multiple-ranked (four or more) maize ear. We detected 24 small-effect QTL that explained 62% of the total variance for KRN (Figure S36). Among them, indeterminate spikelet1 (ids1) was included within QTL KRN1.3, which is an APETALA2-like transcription factor that specifies determinate fates by suppressing indeterminate growth within the spikelet meristem (Chuck et al. 1998). A previous fine-mapping study of KRN using a maize–teosinte BC2S3 RIL population also identified ids1 as a strong candidate for KRN (Calderón et al. 2016). unbranched3 (ub3) was included within QTL KRN4.2, which is an SBP transcription factor that has been shown to regulate KRN in both mutant and QTL studies (Chuck et al. 2014; Liu et al. 2015). ramosa1 (ra1) was included within KRN7.1, which is a C2H2 zinc finger transcription factor controlling inflorescence architecture (Vollbrecht et al. 2005).
REPE, which concerns a reddish–brownish pericarp, is a trait that distinguishes teosinte kernels from those of most maize. The role of pigmentation in domestication is complex in that pigments can provide defenses against molding and bird predation, but can also impart bitterness and astringency (Morohashi et al. 2012). The red (or reddish–brownish) pigmentation often results from the accumulation of phlobaphenes, which are flavonoid pigments (Morohashi et al. 2012). In the absence of the reddish–brownish pigment, the kernels are white unless anthocyanins (blue–purple) or carotenoids (yellow–orange) are present. Our results show that Pericarp color1 (P1) was included within QTL REPE1.1 (Figure S37), which encodes an R2R3 Myb-like transcription factor that governs the biosynthesis of brick-red flavonoid pigments (Grotewold et al. 1994).
Results for 13 additional traits (ASI, BARE, CULM, DTS, EB, ED, EL, GLCO, KW, LFLN, LFWD, PLHT, and YEPE) are reported in the supplemental figures and tables (Figures S30 and S38–S49, and Table S1).
New discoveries and avenues for future research
QTL hotspots:
To evaluate whether QTL detected in TeoNAM are overrepresented in specific genomic regions, we counted and plotted the number of QTL peaks using sliding windows of 10- and 1-cM steps (Figure 1). The windows with at least five QTL were considered as QTL hotspots under 1000 permutation tests (P < 0.001) and continuous windows were merged together. We totally identified 15 QTL hotspots, ranging from 5 to 13 QTL (Table S4). The two largest hotspots (hs10-1 and hs4-1) were located on chromosomes 10 and 4, respectively. A close view of genes under the hotspots shows that hs10-1 and hs4-1 are around ZmCCT10 and tga1, respectively. These results suggest that these two regulatory genes may have pleiotropic effects on multiple traits.
QTL detection and effects:
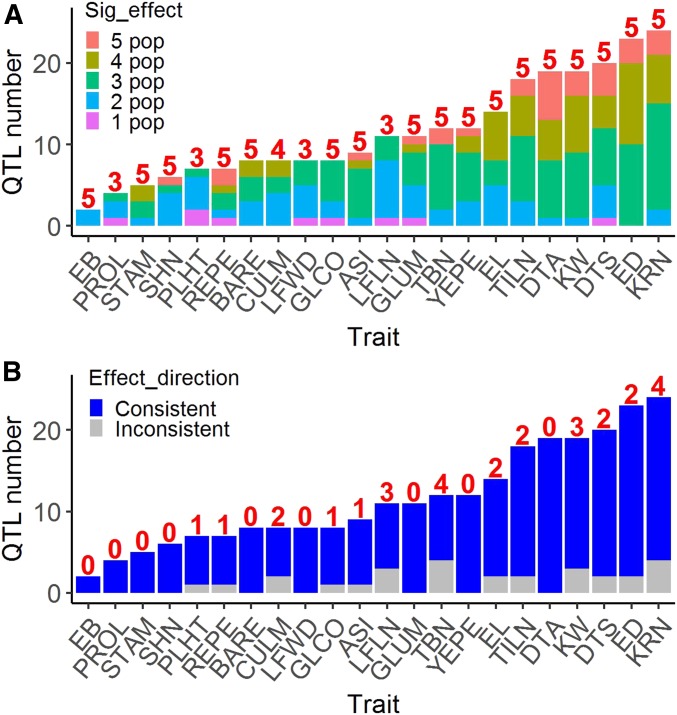
To evaluate the power of QTL mapping using TeoNAM, we summarized the distribution of QTL detected with significant effects in the different subpopulations. Among 255 QTL for 22 traits, 246 QTL (96%) were detected in two or more subpopulations, 186 QTL (73%) were detected in three or more subpopulations, 83 QTL (33%) were detected in four or more subpopulations, and 29 QTL (11%) were detected in all five subpopulations (Figure 5A). These percentages are conservative as not all traits were scored in all five subpopulations. If one considers whether the QTL was detected in subpopulations in which it was scored, then 205 QTL (80%) were detected in at least one-half of the subpopulations and 39 QTL (15%) were detected in all subpopulations.
Figure 5.
QTL detection and effects for all 22 traits. (A) Summary of QTL detection for all 22 traits. The number above the bar indicates the number of subpopulations in which the trait was scored. (B) Summary of QTL effect directions for all 22 traits. The number above the bar indicates the number of QTL within which a teosinte allele associated with the maize phenotype was detected.
The allelic effects from different teosinte parents were estimated simultaneously by JLM. For most QTL, the allelic effects from different subpopulations are in the same direction (Figure 5B). For seven traits (EB, GLUM, LFWD, PROL, SHN, STAM, and YEPE), the teosinte genotypes were consistently associated with a teosinte phenotype and the W22 allele with a maize phenotype at all QTL. For all other traits, there were cases in which a teosinte allele was associated with the maize phenotype. For example, the teosinte genotype is associated with late flowering at most QTL for DTA except DTA5.2 and DTA7.1, for which the teosinte genotype consistently contributes to early flowering in at least three subpopulations (Figure 2). Similar results were observed for KRN and EL. The teosinte genotype is associated with lower KRN at most QTL, but there is one QTL (KRN5.1) for which the teosinte genotype is consistently associated with higher KRN in four subpopulations and also in the BC2S3 population (Figure S36). The teosinte genotype is associated with shorter EL at most QTL, but there are two QTL (EL4.1 and EL9.1) for which the teosinte genotype is consistently associated with longer EL in four and two subpopulations, respectively (Figure S43). These QTL might be worth exploring further for use in maize improvement.
We also observed notable results for different teosinte parents. For KW, the teosinte genotype from different subpopulations is associated with reduced kernel weight at most QTL. Only three QTL (KW5.3, KW6.2, and KW9.1) are exceptions, with one teosinte allele conferring heavier kernels. Interestingly, for these three QTL, the teosinte alleles with effects in the opposite direction are all from the TIL14 subpopulation (Figure S45). Similar results were observed for ED, where the teosinte genotype is associated with a decrease in ED at most QTL, but the teosinte allele from TIL03 at two QTL (ED2.1 and ED6.1) is associated with the increase of ED (Figure S42). These results suggest that there are beneficial alleles from teosinte that could be utilized for maize improvement.
Comparing and combining TeoNAM with BC2S3:
We compared TeoNAM with our previous maize–teosinte BC2S3 RIL population. The composite genetic map for TeoNAM is 1540 cM in length. The individual genetic maps based on the five subpopulations have an average length of 1461 cM with a range of 1348–1596 cM. The genetic map for BC2S3 RIL population is 1478 cM in length. Thus, the TeoNAM subpopulations are similar to the BC2S3 RIL population in genetic map length. The median length of homozygous teosinte segments in TeoNAM is 7.5 Mb. The median length of homozygous teosinte segments in BC2S3 population is 4.8 Mb. The longer segment length for TeoNAM is expected given it had one fewer generations of backcrossing and less opportunity for recombination. The mean number of homozygous teosinte segments in TeoNAM is 2414 and the number of homozygous teosinte segments in BC2S3 is 5745. The total length of teosinte segments for the five subpopulations is 67 gigabases (Gb) (W22 × TIL01), 87 Gb (W22 × TIL03), 66 Gb (W22 × TIL11), 59 Gb (W22 × TIL14), and 79 Gb (W22 × TIL25), and BC2S3 (W22 × 8759) exceeds this range with 110 Gb.
Previously, Shannon (2012) performed a comprehensive interval QTL analysis for 16 agronomic traits in the BC2S3 population and identified 218 QTL for 16 traits. Among these traits, 14 traits were also scored in TeoNAM population. For the common 14 traits, 168 and 179 QTL were detected for the TeoNAM and BC2S3 populations, respectively. The mean 1.5-LOD QTL support interval across 14 traits for BC2S3 is 5.7 Mb, which is significantly smaller than the TeoNAM interval of 17.2 Mb determined by JLM (P = 2.6E–08) (Figure S50). Among these QTL, 50 overlapped between the two populations. For the common QTL, the mean variance explained by QTL is 3.4 and 2.9% for BC2S3 and TeoNAM, respectively. Thus, there is no significant difference in QTL effect size (P = 0.3) (Figure S51).
Among the 50 common QTL between TeoNAM and BC2S3, 12 QTL were detected with significant effects and consistent effect directions in all five subpopulations of TeoNAM and BC2S3 (Table 3). Another 23 QTL were not significant in all five subpopulations of TeoNAM but with consistent effect direction (Table 3). Among these QTL, known candidates include zagl1 for DTA1.1, Zmm31 for DTA5.1, dlf1 for DTA7.2, ZCN8 for DTA8.1, ZmCCT9 for DTA9.1, ZmCCT10 for DTA10.1, tga1 for GLUM4.1, ids1 for KRN1.3, ra1 for KRN7.1, P1 for REPE1.1, ZmSh1-1 for SHN1.1, ZAG2 for TILN3.2, and tb1 for ED1.3 and STAM1.2. There are also novel candidates: smk1 (small kernel1) for ED2.4 and GIF1 (GRAIN INCOMPLETE FILLING 1) for EL10.1. smk1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 (NADH dehydrogenase subunit7) transcript editing, and seed development in maize and rice (Li et al. 2014). GIF1 encodes a cell wall invertase required for carbon partitioning during early grain filling in rice (Wang et al. 2008). Among the candidates, causative sites have been reported for only three: ZCN8, ZmCCT10, and tga1. Therefore, there are many new candidates for future fine-mapping of their causative polymorphisms.
Table 3. QTL with same effect direction between TeoNAM and BC2S3.
| Trait | QTL | Chr | QTL peak (V4) (Mb) | Support interval (V4) (Mb) | QTL peak (cM) | Support interval (cM) | P-value | TIL01 | TIL03 | TIL11 | TIL14 | TIL25 | 8759a | Candidate gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BARE | BARE4.3 | 4 | 55.9 | 39.0–124.3 | 61.5 | 58.2–66.8 | 4.72E–07 | 0.19 | 0.25 | 0.19 | 0.14b | 0.09b | 0.63 | tga1 |
| DTA | DTA1.1 | 1 | 4.5 | 2.8–5.6 | 9.0 | 3.4–13.2 | 1.26E–16 | 0.59 | 1.02 | 1.23 | 1.31 | 0.77 | 0.81 | zagl1 |
| DTA | DTA3.3 | 3 | 231.4 | 226.3–233.4 | 167.2 | 156.7–170.0 | 4.54E–08 | 0.64 | 1.05 | 0.68 | 0.13b | 0.73 | 0.49 | |
| DTA | DTA5.1 | 5 | 6.9 | 6.7–7.2 | 30.7 | 29.2–31.5 | 2.62E–18 | 1.11 | 0.80 | 1.11 | 1.24 | 1.30 | 0.66 | Zmm31 |
| DTA | DTA7.2 | 7 | 180.4 | 180.1–180.8 | 140.7 | 139.3–141.8 | 2.92E–14 | 0.53 | 1.37 | 0.77 | 0.58 | 1.16 | 0.59 | dlf1 |
| DTA | DTA8.1 | 8 | 126.9 | 126.6–126.9 | 68.3 | 68.2–68.3 | 4.05E–54 | 1.64 | 2.00 | 2.63 | 1.72 | 1.59 | 1.54 | ZCN8 |
| DTA | DTA9.1 | 9 | 117.5 | 115.7–118.8 | 72.9 | 72.4–74.6 | 4.76E–33 | 0.65 | 3.16 | 1.24 | 0.74 | 1.39 | 0.79 | ZmCCT9 |
| DTA | DTA10.1 | 10 | 95.4 | 94.9–99.4 | 54.7 | 54.3–55.1 | 9.29E–121 | 3.47 | 3.22 | 2.46 | 4.87 | 4.54 | 3.61 | ZmCCT10 |
| EB | EB7.1 | 7 | 121.5 | 103.9–132.7 | 64.8 | 60.1–72.7 | 5.18E–14 | 0.02b | 0.01b | 0.06 | 0.05 | 0.01b | 0.05 | |
| ED | ED1.3 | 1 | 277.1 | 265.1–283.1 | 134.7 | 127.6–141.3 | 6.45E–06 | −1.02 | −0.28b | −1.38 | −0.25b | −0.64 | −1.87 | tb1 |
| ED | ED2.4 | 2 | 217.2 | 211.9–223.4 | 123.3 | 116.8–133.5 | 1.22E–06 | −0.46b | −0.79 | −0.39b | −0.72 | −0.85 | −0.88 | smk1 |
| ED | ED3.2 | 3 | 218.4 | 214.8–219.8 | 140.1 | 133.8–142.3 | 1.34E–13 | −1.18 | −0.45 | −0.72 | −0.34b | −1.33 | −0.89 | |
| ED | ED4.1 | 4 | 7.8 | 6.8–11.1 | 23.8 | 21.5–30.1 | 1.50E–11 | −1.59 | −0.48 | −0.01b | −0.64 | −0.25b | −0.74 | |
| ED | ED7.2 | 7 | 163.6 | 160.4–164.2 | 98.0 | 94.1–98.4 | 2.47E–19 | −0.67 | −0.94 | −1.59 | −1.30 | −0.78 | −0.81 | |
| EL | EL7.1 | 7 | 125.5 | 123.7–130.3 | 66.8 | 65.7–70.5 | 4.64E–17 | −0.82 | −0.34 | −0.56 | −0.21b | −0.81 | −0.97 | |
| EL | EL8.1 | 8 | 156.5 | 150.4–157.6 | 82.5 | 78.1–83.4 | 2.84E–11 | −0.19b | −0.48 | −0.15b | −0.21b | −0.68 | −0.66 | |
| EL | EL10.1 | 10 | 115.0 | 115.0–118.9 | 58.5 | 58.8–60.0 | 6.87E–40 | −0.12b | −0.88 | −0.85 | −1.68 | −1.17 | −1.34 | GIF1 |
| GLCO | GLCO4.1 | 4 | 42.1 | 39.0–42.1 | 59.6 | 58.2–59.6 | 5.98E–24 | 0.01b | 0.12 | 0.08b | 0.31 | 0.37 | 0.27 | |
| GLUM | GLUM4.1 | 4 | 42.1 | 40.4–55.9 | 59.6 | 59.1–61.5 | 2.57E–168 | 0.74 | 0.77 | 0.73 | 0.93 | 1.06 | 0.8 | tga1 |
| KRN | KRN1.3 | 1 | 298.5 | 298.1–299.6 | 165.7 | 164.9–168.2 | 2.73E–16 | −0.51 | −0.58 | −0.35 | −0.09b | −0.26 | −0.63 | ids1 |
| KRN | KRN4.3 | 4 | 243.5 | 241.6–244.8 | 145.6 | 137.4–152.1 | 8.27E–07 | −0.22 | −0.41 | −0.18 | −0.23 | −0.23 | −0.55 | |
| KRN | KRN5.1 | 5 | 2.4 | 2.3–2.9 | 12.7 | 11.7–15.0 | 8.72E–14 | 0.36 | 0.51 | 0.12b | 0.40 | 0.24 | 0.3 | |
| KRN | KRN7.1 | 7 | 95.2 | 21.5–123.7 | 58.6 | 49.6–65.7 | 7.00E–11 | −0.36 | −0.18 | −0.54 | −0.06b | −0.42 | −0.26 | ra1 |
| KRN | KRN8.2 | 8 | 171.3 | 169.6–173.4 | 106.8 | 100.8–113.0 | 6.70E–13 | −0.49 | −0.40 | −0.38 | −0.04b | −0.15b | −0.25 | |
| KW | KW1.2 | 1 | 234.3 | 222.8–252.5 | 110.3 | 106.5–116.4 | 6.01E–10 | −0.01 | −0.01 | −0.01 | −0.00b | −0.01 | −0.01 | |
| KW | KW2.1 | 2 | 53.3 | 36.9–57.5 | 74.3 | 64.3–76.1 | 1.32E–11 | −0.01 | −0.00b | −0.01 | −0.01 | −0.00b | −0.01 | |
| KW | KW4.2 | 4 | 170.1 | 156.8–174.8 | 82.8 | 73.6–84.2 | 1.69E–08 | −0.01 | −0.01 | −0.01 | −0.00b | −0.00b | −0.01 | |
| KW | KW8.1 | 8 | 137.7 | 132.5–145.9 | 72.7 | 70.4–75.4 | 1.31E–12 | −0.00b | −0.01 | −0.01 | −0.01 | −0.01 | −0.01 | |
| REPE | REPE1.1 | 1 | 47.0 | 46.5–51.8 | 64.9 | 64.4–68.3 | 2.71E–11 | −0.10 | −0.10 | −0.06 | −0.10 | −0.10 | −0.10 | P1 |
| SHN | SHN1.1 | 1 | 265.1 | 264.1–265.1 | 127.6 | 127.2–127.6 | 3.21E–16 | 0.08 | 0.09 | 0.18 | 0.17 | 0.11 | 0.07 | ZmSh1-1 |
| SHN | SHN6.1 | 6 | 162.5 | 162.2–163.6 | 98.2 | 97.4–101.1 | 3.49E–11 | 0.06 | 0.04b | 0.17 | 0.03b | 0.10 | 0.06 | |
| STAM | STAM1.2 | 1 | 270.5 | 269.8–270.5 | 130.2 | 129.5–130.2 | 1.23E–27 | 0.28 | 0.02b | 0.12 | 0.07 | 0.09 | 0.11 | tb1 |
| TILN | TILN3.2 | 3 | 138.3 | 137.6–158.6 | 80.5 | 80.2–86.2 | 5.22E–24 | 0.00b | 0.42 | 0.33 | 0.08b | 0.20 | 0.14 | ZAG2 |
| TILN | TILN5.1 | 5 | 69.0 | 36.0–140.7 | 75.4 | 69.9–82.7 | 6.66E–10 | 0.18 | 0.24 | 0.14 | 0.14 | 0.16 | 0.10 | |
| TILN | TILN10.1 | 10 | 62.5 | 25.3–82.1 | 47.7 | 45.0–51.1 | 4.10E–13 | 0.15 | 0.24 | 0.06b | 0.08b | 0.28 | 0.12 |
BARE, barren ear base; DTA, days to anthesis; EB, ear branch number; ED, ear diameter; EL, ear length; GLCO, glume color; GLUM, glume score; KRN, kernel row number; KW, kernel weight; REPE, red pericarp; SHN, shattering; STAM, staminate spikelet; TILN, tiller number. Chr, chromosome; JLM, joint linkage mapping; TeoNAM, teosinte nested association mapping.
The allele effect of 8759 relative to maize was estimated in BC2S3 by Shannon (2012).
Indicates that the additive allele effect of teosinte relative to maize estimated from JLM in TeoNAM is not significant.
To maximize the power to detect QTL, we combined TeoNAM and BC2S3 for eight traits (DTA, ED, EL, KRN, KW, GLCO, GLUM, and TILN) that were measured in all six subpopulations via the exact same method used to perform JLM. Before analysis, we imputed the genotype for BC2S3 at 4578 TeoNAM SNPs according to the flanking markers using the same procedure as for TeoNAM and permuted a new P-value cutoff for statistical significance for each trait. The Least Squared Means (LSMs) from previous analysis (Shannon 2012) were used for JLM. With the combined TeoNAM–BC2S3 data, we detected 184 QTL for these eight traits, which included 109 QTL overlapping with TeoNAM, 80 QTL overlapping with the BC2S3, and 32 novel QTL that were not detected in either TeoNAM or BC2S3 (Table S5). The QTL with significant allele effects in multiple subpopulations will be good targets for fine-mapping. For future analysis of additional traits, one could combine TeoNAM and the BC2S3 together. The value of this combination is that there is one additional teosinte allele and increased QTL detection power, but the downside is that one would need to assay the BC2S3 population using 866 RILs plus TeoNAM with 1257 RILs.
Discussion
RILs are powerful tools for dissecting the complex genetic architectures of different traits and for gene discovery. RILs such as the maize NAM population have been successfully used for the genetic dissection of many traits (Buckler et al. 2009; Kump et al. 2011; Tian et al. 2011). RILs with multiple parents greatly increase the power and precision to identify QTL compared to the traditional biparent RIL population. Multiparent RILs also enable the estimation of allele effects simultaneously from each inbred parent. Our TeoNAM RILs were created by crossing five teosinte inbred parents with a maize inbred parent, but differ from MaizeNAM in that we applied a generation of backcrossing to the maize parent before four generations of selfing. The power and precision of TeoNAM can be shown with several traits. For example, we detected 19 QTL for DTA, among which many QTL mapped to recently cloned genes such as ZmCCT10, ZmCCT9, ZCN8, zagl1, and ZmMADS69. QTL also mapped to some novel candidates such as dlf1, si1, ZAG1, ZCN12, Zmm19, and Zmm31, which may have an important role in flowering time regulation.
For RIL populations, both JLM and GWAS are common methods for QTL detection. In this study, we identified 255 QTL for 22 traits by JLM, and significant peaks were detected at 57 QTL by GWAS, which suggests that GWAS is less powerful than JLM for mapping QTL in TeoNAM. Nevertheless, there were a few instances in which GWAS gave evidence of closely linked QTL that were not separated by JLM. For example, we did not identify an1, a strong candidate for STAM QTL on chromosome 1, with JLM possibly because it is closely linked to tb1 (candidate of QTL STAM1.2), but we detected distinct and significant peaks at both an1 and tb1 through GWAS as it tests each SNP independently.
TeoNAM reveals that there are distinct genetic architectures for different traits. Traits like PROL and GLUM are controlled by a major effect QTL plus several QTL of very small effect, while traits like DTA and KRN show more classic polygenic inheritance. Domestication traits controlled by single major genes have frequently been reported in a variety of crops (Doebley et al. 2006). There is less emphasis in the literature on polygenic inheritance of domestication traits, although a well-known example is fruit size in tomato (Frary et al. 2000). Our results highlight the importance of these two modes of inheritance during the domestication process.
A relatively large phenotypic data set of 22 traits scored in TeoNAM allowed us to evaluate QTL hotspots. We detected 15 QTL hotspots, in which the largest hotspot could affect 13 traits. However, this should be treated with caution regard whether these hotspots were caused by a single master gene with pleiotropic effects or several genes that are tightly linked together, as domestication may favor the selection of tightly linked combinations of genes (Le Thierry d’Ennequin et al. 1999).
In our study, a total of 15 domestication traits and 7 agronomic traits were analyzed. Further fine-mapping and gene cloning will be required to find the causal genes underlying QTL for these traits. TeoNAM should also be useful for investigating the genetic control of many new traits that we did not assay. Morphological traits such as root architecture, shoot apical meristem size, vasculature, pollen size, and kernel shape can be explored. Also, molecular traits such as gene expression (eQTL) (Wang et al. 2018), alternative splicing (Chen et al. 2018), grain protein content (Cook et al. 2012), and metabolites (Xu et al. 2019) can also be explored to better understand the full spectrum of changes that occurred during maize domestication. In this context, we note that TeoNAM was developed as part of an undergraduate research project and phenotyping performed over three field seasons, but the entire set of lines was never grown in a single season. Increased power beyond that which we report here can be gained by growing the entire set of lines each season for multiple seasons to obtain better phenotypic data, and more power in QTL detection.
Acknowledgments
We thank Karl Broman for suggestions on the analyses and Jesse Rucker, Elizabeth Buschert, Eric Rentmeester, Adam Mittermaier, David Sierakowski, and Brian Schaeffer for assistance with field work and phenotyping. This research was supported by the US National Science Foundation (grant IOS 1238014) and the China Postdoctoral Science Foundation (2018M640204).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9250682.
Communicating editor: T. Juenger
Literature Cited
- Ambrose B. A., Lerner D. R., Ciceri P., Padilla C. M., Yanofsky M. F. et al. , 2000. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579. 10.1016/S1097-2765(00)80450-5 [DOI] [PubMed] [Google Scholar]
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A., Lobina I. T. et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice (N Y) 6: 11 10.1186/1939-8433-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazesh S., and McSteen P., 2008. Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 179: 389–401. 10.1534/genetics.107.084079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen R. J., Johal G. S., Crane V. C., Tossberg J. T., Schnable P. S. et al. , 1995. Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84. 10.1105/tpc.7.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Wang R. L., Ambrose B. A., Schmidt R. J., Meeley R. B. et al. , 2003. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395. 10.1242/dev.00457 [DOI] [PubMed] [Google Scholar]
- Bouchet S., Olatoye M. O., Marla S. R., Perumal R., and Tesso T., 2017. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 206: 573–585. 10.1534/genetics.116.198499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y. et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen Ś., and Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J. et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. 10.1126/science.1174276 [DOI] [PubMed] [Google Scholar]
- Calderón C. I., Yandell B. S., and Doebley J. F., 2016. Fine mapping of a QTL associated with kernel row number on chromosome 1 of maize. PLoS One 11: e0150276 10.1371/journal.pone.0150276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Han Y., Liu H., Wang X., Sun J. et al. , 2018. Genome-wide association analyses reveal the importance of alternative splicing in diversifying gene function and regulating phenotypic variation in maize. Plant Cell 30: 1404–1423. 10.1105/tpc.18.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Meeley R. B., and Hake S., 1998. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12: 1145–1154. 10.1101/gad.12.8.1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Whipple C., Jackson D., and Hake S., 2010. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137: 1243–1250. 10.1242/dev.048348 [DOI] [PubMed] [Google Scholar]
- Chuck G. S., Brown P. J., Meeley R., and Hake S., 2014. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 111: 18775–18780. 10.1073/pnas.1407401112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D. et al. , 2004. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. 10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- Cook J. P., McMullen M. D., Holland J. B., Tian F., Bradbury P. et al. , 2012. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 158: 824–834. 10.1104/pp.111.185033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua M., Gatti D. M., Pea G., Cattonaro F., Coppens F. et al. , 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol. 16: 167 10.1186/s13059-015-0716-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., and Gustus C., 1995. Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., and Hubbard L., 1997. The evolution of apical dominance in maize. Nature 386: 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., and Smith B. D., 2006. The molecular genetics of crop domestication. Cell 127: 1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Dong Z., Li W., Unger-Wallace E., Yang J., Vollbrecht E. et al. , 2017. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA 114: E8656–E8664. 10.1073/pnas.1714960114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K. et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso C. A., Moreno M., Wang Z., Heffelfinger C., Arbelaez L. J. et al. , 2017. Genetic architecture of a rice nested association mapping population. G3 (Bethesda) 7: 1913–1926. 10.1534/g3.117.041608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A., Nesbitt T. C., Frary A., Grandillo S., Van Der Knaap E. et al. , 2000. fw2. 2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. 10.1126/science.289.5476.85 [DOI] [PubMed] [Google Scholar]
- Fu Y., Xu G., Chen H., Wang X., Chen Q. et al. , 2019. QTL mapping for leaf morphology traits in a large maize-teosinte population. Mol. Breed. 39: 103 10.1007/s11032-019-1012-5 [DOI] [Google Scholar]
- Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D. et al. , 2008. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 105: 15196–15201. 10.1073/pnas.0805596105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J. et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B. J., Bowen B., and Peterson T., 1994. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553. 10.1016/0092-8674(94)90117-1 [DOI] [PubMed] [Google Scholar]
- Guo L., Wang X., Zhao M., Huang C., Li C. et al. , 2018. Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 28: 3005–3015.e4. 10.1016/j.cub.2018.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. E., George A. W., Forrest K. L., Kilian A., Hayden M. J. et al. , 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839. 10.1111/j.1467-7652.2012.00702.x [DOI] [PubMed] [Google Scholar]
- Huang C., Chen Q., Xu G., Xu D., Tian J. et al. , 2016. Identification and fine mapping of quantitative trait loci for the number of vascular bundle in maize stem. J. Integr. Plant Biol. 58: 81–90. 10.1111/jipb.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Sun H., Xu D., Chen Q., Liang Y. et al. , 2018. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 115: E334–E341. 10.1073/pnas.1718058115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Paulo M. J., Boer M., Effgen S., Keizer P., Koornneef M. et al. , 2011. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493. 10.1073/pnas.1100465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L. M., Tian F., Bradbury P. J., Chen C. et al. , 2012. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109: E1913–E1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish E. E., and Nelson T. M., 1993. Development of tassel seed 2 inflorescences in maize. Am. J. Bot. 80: 292–299. 10.1002/j.1537-2197.1993.tb13802.x [DOI] [Google Scholar]
- Jin J., Huang W., Gao J. P., Yang J., Shi M. et al. , 2008. Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. 10.1038/ng.247 [DOI] [PubMed] [Google Scholar]
- Jordan K. W., Wang S., He F., Chao S., Lun Y. et al. , 2018. The genetic architecture of genome-wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 95: 1039–1054. 10.1111/tpj.14009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S. et al. , 2012. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22: 1558–1566. 10.1101/gr.134031.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M. et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling K. A., Chen S. Y., Su M. H., Lepak N. K., Romay M. C. et al. , 2018. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523. 10.1038/nature25966 [DOI] [PubMed] [Google Scholar]
- Kump K. L., Bradbury P. J., Wisser R. J., Buckler E. S., Belcher A. R. et al. , 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. 10.1038/ng.747 [DOI] [PubMed] [Google Scholar]
- Lang Z., Wills D. M., Lemmon Z. H., Shannon L. M., Bukowski R. et al. , 2014. Defining the role of prolamin-box binding factor1 gene during maize domestication. J. Hered. 105: 576–582. 10.1093/jhered/esu019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiboff S., DeAllie C. K., and Scanlon M. J., 2016. Modeling the morphometric evolution of the maize shoot apical meristem. Front. Plant Sci. 7: 1651 10.3389/fpls.2016.01651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thierry d’Ennequin M., Toupance B., Robert T., Godelle B., and Gouyon P., 1999. Plant domestication: a model for studying the selection of linkage. J. Evol. Biol. 12: 1138–1147. 10.1046/j.1420-9101.1999.00115.x [DOI] [Google Scholar]
- Li D., Wang X., Zhang X., Chen Q., Xu G. et al. , 2016. The genetic architecture of leaf number and its genetic relationship to flowering time in maize. New Phytol. 210: 256–268. 10.1111/nph.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Zhang Y. F., Hou M., Sun F., Shen Y. et al. , 2014. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79: 797–809. 10.1111/tpj.12584 [DOI] [PubMed] [Google Scholar]
- Liang Y., Liu Q., Wang X., Huang C., Xu G. et al. , 2019. ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 221: 2335–2347. 10.1111/nph.15512 [DOI] [PubMed] [Google Scholar]
- Lin Z., Li X., Shannon L. M., Yeh C. T., Wang M. L. et al. , 2012. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724. 10.1038/ng.2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Du Y., Shen X., Li M., Sun W. et al. , 2015. KRN4 controls quantitative variation in maize kernel row number. PLoS Genet. 11: e1005670 10.1371/journal.pgen.1005670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I. J., Bansept-Basler P., Barber T., Bentley A. R., Cockram J. et al. , 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 (Bethesda) 4: 1603–1610. 10.1534/g3.114.012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A., Draba V., Jiang Y., Schnaithmann F., Sharma R. et al. , 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16: 290 10.1186/s12864-015-1459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Muszynski M. G., and Danilevskaya O. N., 2011. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960. 10.1105/tpc.110.081406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Casas M. I., Falcone Ferreyra M. L., Mejía-Guerra M. K., Pourcel L. et al. , 2012. A genome-wide regulatory framework identifies maize Pericarp Color1 controlled genes. Plant Cell 24: 2745–2764 (erratum: Plant Cell 24: 3853). 10.1105/tpc.112.098004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice L. M., Steffenson B. J., Brown-Guedira G. L., Akhunov E. D., Liu C. et al. , 2016. Development and genetic characterization of an advanced backcross-nested association mapping (AB-NAM) population of wild × cultivated barley. Genetics 203: 1453–1467. 10.1534/genetics.116.190736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler M., Eveland A. L., LaRue T., Yang F., Weeks R. et al. , 2015. FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27: 104–120. 10.1105/tpc.114.132506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. J., Veit B., Mandel M. A., Mena M., Hake S. et al. , 1993. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5: 729–737. 10.1105/tpc.5.7.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, L. M., 2012 The genetic architecture of maize domestication and range expansion. Ph.D. Thesis, The University of Wisconsin–Madison, Madison, WI. [Google Scholar]
- Shannon, L. M., Q. Chen, and J. F. Doebley, 2019 A BC2S3 maize-teosinte RIL population for QTL mapping. Maize Genet. Coop. News Lett. 93.
- Studer A. J., Wang H., and Doebley J. F., 2017. Selection during maize domestication targeted a gene network controlling plant and inflorescence architecture. Genetics 207: 755–765. 10.1534/genetics.117.300071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C. et al. , 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J. T. et al. , 2000. A short history of MADS-box genes in plants. Plant Mol. Evol. 42: 115–149. 10.1007/978-94-011-4221-2_6 [DOI] [PubMed] [Google Scholar]
- Tian F., Bradbury P. J., Brown P. J., Hung H., Sun Q. et al. , 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162. 10.1038/ng.746 [DOI] [PubMed] [Google Scholar]
- Tian J., Wang C., Xia J., Wu L., Xu G. et al. , 2019. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365: 658–664. 10.1126/science.aax5482 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Springer P. S., Goh L., Buckler E. S. IV, and Martienssen R., 2005. Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126. 10.1038/nature03892 [DOI] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L. et al. , 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y. et al. , 2005. The origin of the naked grains of maize. Nature 436: 714–719. 10.1038/nature03863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Studer A. J., Zhao Q., Meeley R., and Doebley J. F., 2015. Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics 200: 965–974. 10.1534/genetics.115.175752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen Q., Wu Y., Lemmon Z. H., Xu G. et al. , 2018. Genome-wide analysis of transcriptional variability in a large maize-teosinte population. Mol. Plant 11: 443–459. 10.1016/j.molp.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Whipple C. J., Hall D. H., DeBlasio S., Taguchi-Shiobara F., Schmidt R. J. et al. , 2010. A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578. 10.1105/tpc.109.073536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C. J., Kebrom T. H., Weber A. L., Yang F., Hall D. et al. , 2011. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. 10.1073/pnas.1102819108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills D. M., Whipple C. J., Takuno S., Kursel L. E., Shannon L. M. et al. , 2013. From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9: e1003604 10.1371/journal.pgen.1003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills D. M., Fang Z., York A. M., Holland J. B., and Doebley J. F., 2017. Defining the role of the MADS-box gene, Zea Agamous-like1, a target of selection during maize domestication. J. Hered. 109: 333–338. 10.1093/jhered/esx073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A., Jarquin D., Howard R., Ramasubramanian V., Specht J. E. et al. , 2018. Genome-wide analysis of grain yield stability and environmental interactions in a multiparental soybean population. G3 (Bethesda) 8: 519–529. 10.1534/g3.117.300300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Wang X., Huang C., Xu G., Liang Y. et al. , 2017. Glossy15 plays an important role in the divergence of the vegetative transition between maize and its progenitor, teosinte. Mol. Plant 10: 1579–1583. 10.1016/j.molp.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Xu G., Wang X., Huang C., Xu D., Li D. et al. , 2017. Complex genetic architecture underlies maize tassel domestication. New Phytol. 214: 852–864. 10.1111/nph.14400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Cao J., Wang X., Chen Q., Jin W. et al. , 2019. Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor, teosinte. Plant Cell 31: 1990–2009. 10.1105/tpc.19.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. J., 2018 Dissection of the genetic architecture of domestication traits in maize and its ancestor teosinte. Ph.D. Thesis, The University of Wisconsin–Madison, Madison, WI. [Google Scholar]
- Yang Q., Li Z., Li W., Ku L., Wang C. et al. , 2013. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 110: 16969–16974. 10.1073/pnas.1310949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., and Buckler E. S., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. 10.1534/genetics.107.074245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang X., Lin Z., Wang J., Xu M. et al. , 2018. The genetic architecture of nodal root number in maize. Plant J. 93: 1032–1044. 10.1111/tpj.13828 [DOI] [PubMed] [Google Scholar]
- Zhao H., Sun Z., Wang J., Huang H., Kocher J. P. et al. , 2014. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30: 1006–1007. 10.1093/bioinformatics/btt730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Weber A. L., McMullen M. D., Guill K., and Doebley J., 2011. MADS-box genes of maize: frequent targets of selection during domestication. Genet. Res. (Camb) 93: 65–75. 10.1017/S0016672310000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Seeds for all 1257 RILs in TeoNAM are available at the Maize Genetics Cooperative Stock Center. The SNP genotypes for TeoNAM are available at the Cyverse Discovery Environment under the directory: http://datacommons.cyverse.org/browse/iplant/home/shared/panzea/genotypes/GBS/TeosinteNAM. The genotypic data were uploaded with AGPv2 position in the marker name. The raw phenotypic data for TeoNAM are available at Figshare database: https://doi.org/10.6084/m9.figshare.9820178. The seed information, SNP genotypes, and phenotypic data for the BC2S3 maize–teosinte RIL population can be found in Shannon et al. (2019). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9250682.