Abstract
To identify genes involved in cerebral infarction, we have employed a forward genetic approach in inbred mouse strains, using quantitative trait loci (QTL) mapping for cerebral infarct volume after middle cerebral artery occlusion. We had previously observed that infarct volume is inversely correlated with cerebral collateral vessel density in most strains. In this study, we expanded the pool of allelic variation among classical inbred mouse strains by utilizing the eight founder strains of the Collaborative Cross and found a wild-derived strain, WSB/EiJ, that breaks this general rule that collateral vessel density inversely correlates with infarct volume. WSB/EiJ and another wild-derived strain, CAST/EiJ, show the highest collateral vessel densities of any inbred strain, but infarct volume of WSB/EiJ mice is 8.7-fold larger than that of CAST/EiJ mice. QTL mapping between these strains identified four new neuroprotective loci modulating cerebral infarct volume while not affecting collateral vessel phenotypes. To identify causative variants in genes, we surveyed nonsynonymous coding SNPs between CAST/EiJ and WSB/EiJ and found 96 genes harboring coding SNPs predicted to be damaging and mapping within one of the four intervals. In addition, we performed RNA-sequencing for brain tissue of CAST/EiJ and WSB/EiJ mice and identified 79 candidate genes mapping in one of the four intervals showing strain-specific differences in expression. The identification of the genes underlying these neuroprotective loci will provide new understanding of genetic risk factors of ischemic stroke, which may provide novel targets for future therapeutic intervention of human ischemic stroke.
Keywords: ischemic stroke, wild-derived mouse strains, neuroprotection, quantitative trait mapping
STROKE, the sudden death of the brain tissue occurring when the blood flow to the brain is lost by blockage or rupture, is a leading cause of death in the United States (the fourth) and the world (the second) (Roger et al. 2012; Johnson et al. 2016). The majority (over 80%) of strokes are ischemic in origin and results in irreversible death of brain tissue (infarction). Studies of genetic risk factors for stroke susceptibility in humans have employed both family-based linkage (Gretarsdottir et al. 2003; Helgadottir et al. 2004) and population-based genome-wide association studies (GWAS) (Matarín et al. 2007; Matarin et al. 2008a,b). Unfortunately, these studies have not yet uncovered druggable targets to treat stroke and modify clinical outcomes. These studies were designed to identify stroke susceptibility risk factors, and not factors that modulate infarct size or neurological outcomes once a stroke has occurred. More relevant to this goal would be a study of anatomic or neurological outcomes in ischemic stroke patients. A GWAS has recently been published on neurological or behavioral outcomes of 6165 ischemic stroke patients (Söderholm et al. 2019), but only a single variant passed the threshold of genome-wide significance. Furthermore, despite an explosion of the human GWAS for disease phenotypes, no published GWAS for infarct volume in ischemic stroke has been published. The paucity of human genetic variants shown to modulate stroke outcomes is perhaps not surprising. Genetic studies in the human related to infarct volume or neurological outcomes are intrinsically problematic due to uncontrollable variation in the extent and location of the occluded vessel, and especially, variation in the critical time between first recognized symptoms of stroke and medical intervention. Instead, current understanding of the mechanisms underlying infarct damage is primarily based on experimental animal models.
Animal models of focal cerebral ischemia have been established to investigate the pathophysiologic events occurring after ischemic stroke. Most stroke models induce cerebral ischemia within the middle cerebral artery (MCA) territory as most relevant for thrombo-embolic stroke. MCA occlusion models vary both in the extent of occlusion (permanent vs. transient occlusion) and the site of occlusion (proximal vs. distal portion of the vessel). Several reports have determined that the permanent occlusion of distal MCA (pMCAO) method produces more restricted and reproducible damage to the cerebral hemisphere (Majid et al. 2000; Lambertsen et al. 2002; Carmichael 2005). Furthermore, previous studies have also demonstrated that different inbred mouse strains show robust differences in stroke outcomes, providing evidence that the innate response to permanent focal cerebral ischemia is under strong genetic control (Barone et al. 1993; Majid et al. 2000; Lambertsen et al. 2002; Sugimori et al. 2004).
Therefore, we have taken a forward genetic approach using quantitative trait loci (QTL) mapping to identify novel genes/pathways (genetic components) involved in modulating infarct volume in ischemic stroke, using the well-established model of pMCAO. Previously, we uncovered several genetic loci that are involved in regulating ischemic stroke and identified several genes modulating infarct volume within the genetic loci (Keum and Marchuk 2009; Chu et al. 2013; Keum et al. 2013; Lee et al. 2016, 2018).
One of these loci located on distal chromosome 7 (cerebral infarct volume QTL 1; Civq1) is the strongest and most significant locus modulating infarct volume, found in multiple pairwise crosses of inbred mouse strains (Keum and Marchuk 2009; Keum et al. 2013). Intriguingly, Civq1 overlaps with a locus on chromosome 7 that modulates cerebral collateral vessel number (collateral artery number QTL 1; Canq1) (Keum and Marchuk 2009; Zhang et al. 2010; Keum et al. 2013; Lee et al. 2016). We and others have noted a strong inverse correlation between the volume of the infarct after pMCAO and the number of collateral vessel connections in the cerebral vasculature (Keum and Marchuk 2009; Zhang et al. 2010). These vessels connect portions of the same or different arteries, and upon occlusion of an artery, provide a circulatory shunt, enabling reperfusion of the ischemic cerebral territory. Canq1 was recently shown to be due to variation in the Rabep2 (Lucitti et al. 2016), encoding a protein that modulates endosomal recycling of VEGFR2, a receptor for vascular endothelial growth factor (Kofler et al. 2018). This locus and gene exert an overwhelming effect on the size of the infarct after vessel occlusion across a wide spectrum of inbred mouse strains.
Given the strong influence that the collateral circulation plays in the modulation infarct volume across most inbred mouse strains, we sought strains that break the inverse correlation between collateral vessel density and infarct volume. Previously, we found one such strain, C3H, which breaks this inverse correlation. Genome-wide QTL mapping for infarct volume after pMCAO in F2 progeny of a cross between B6 and C3H identified a neuroprotective locus, Civq4, on chromosome 8 (Chu et al. 2013). Civq4 was identified in a cross between two classical inbred mouse strains that overall exhibit relatively low-sequence variation compared to that found within the entire species. These and other commonly used inbred mouse strains contain 94% of their genetic background from Mus musculus domesticus, 5% from M. m. musculus, and <1% from M. m. castaneus (Yang et al. 2011). Thus, genetic mapping using these inbred mouse strains may systemically miss potential candidate loci that might cause phenotypic variation within the entire species. Therefore, in this study, we utilize the eight founder mouse strains of the Collaborative Cross (CC) mouse to expand the scope of genetic variation that we survey in the mouse genome. We use this information to select strains for QTL mapping that will uncover novel loci that modulate infarct size via a collateral-independent mechanism.
Materials and Methods
Animals
All inbred mouse strains were obtained from the Jackson Laboratory (Bar Harbor, ME), and then bred locally to obtain mice used in all experiments. Mice (both male and female animals) were age-matched (P21 for collateral vessel perfusion and 12 ± 1 week for pMCAO) for all experiments. All animal procedures were conducted under protocols approved by the Duke University Institutional Animal Care and Use Committee in accordance with National Institutes of Health guidelines.
Collateral vessel density measurement
As collateral vessel traits are determined by 3 weeks of age and remain constant for many months (Clayton et al. 2008), the collateral vessel phenotype was measured at P21 as previously described (Lee et al. 2016, 2018). Mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), and the ascending thoracic aorta was cannulated. The animals were perfused with freshly made buffer (1 mg/ml adenosine, 40 g/ml papaverine, and 25 mg/ml heparin in PBS) to remove the blood. The pial circulation was then exposed after removal of the dorsal calvarium and adherent dura mater. The cardiac left ventricle was cannulated and a polyurethane solution with a viscosity sufficient to minimize capillary transit (1:1 resin to 2-butanone, PU4ii; VasQtec) was slowly infused; cerebral circulation was visualized under a stereomicroscope during infusion. The brain surface was topically rinsed with 10% PBS-buffered formalin and the dye solidified for 20 min. After postfixation with 10% PBS-buffered formalin, pial circulation was imaged. All collaterals interconnecting the anterior cerebral artery (ACA) and MCA trees of both hemispheres were counted.
pMCAO
Focal cerebral ischemia was induced by direct pMCAO as previously described (Lee et al. 2016, 2018). Briefly, adult mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), and then 0.5% bupivacaine (5 mg/ml) was also administrated by injection at the incision site. The right MCA was exposed by a 0.5 cm vertical skin incision midway between the right eye and ear under a dissecting microscope. After the temporalis muscle was split, a 2-mm burr hole was made with a high-speed microdrill at the junction of the zygomatic arch and the squamous bone through the outer surface of the semitranslucent skull. The MCA was clearly visible at the level of the inferior cerebral vein. The inner layer of the skull was removed with fine forceps, and the dura was opened with a 32-gauge needle. While visualizing under an operating microscope, the right MCA was electrocauterized. The cauterized MCA segment was then transected with microscissors to verify permanent occlusion. The surgical site was closed with 6–0 sterile nylon sutures. The temperature of each mouse was maintained at 37° with a heating pad during the surgery, and then placed in an animal recovery chamber until the animal was fully recovered from the anesthetic. Mice were then returned to their cages and allowed free access to food and water in an air-ventilated room with the ambient temperature set to 25°.
Infarct volume measurement
Cerebral infarct volumes were measured 24 hr after surgery because the size of the cortical infarct is largest and stable at 24 hr after pMCAO (Lambertsen et al. 2005). Twenty-four hours after pMCAO surgery, the animals were killed, and the brains were carefully removed. The brains were placed in a brain matrix and sliced into 1-mm coronal sections after being chilled at −80° for 4 min to slightly harden the tissue. Each brain slice was placed in 1 well of a 24-well plate and incubated for 20 min in a solution of 2% 2,3,5-triphenyltetrazolium chloride in PBS at 37° in the dark. The sections were then washed once with PBS and fixed with 10% PBS-buffered formalin at 4°. Then, 24 hr after fixation, the caudal face of each section was scanned using a flatbed color scanner. The scanned images were used to determine infarct volume (Wexler et al. 2002). Image-Pro software (Media Cybernetics) was used to calculate the infarcted area of each slice by subtracting the infarcted area of the hemisphere from the noninfarcted area of the hemisphere to minimize error introduced by edema. The total infarct volume was calculated by summing the individual slices from each animal.
Flow cytometry
Adult (8–12 weeks old) mouse brain cortex of three CAST/EiJ (CAST) and six WSB/EiJ (WSB) mice were digested with collagenase A (1.5 mg/ml) and DNase I (0.4 mg/ml) in Hank’s Balanced Salt Solution containing 5% fetal bovine serum and 10 mM HEPES at 37° for 45 min. Pelleted postdigested brain preparations were suspended in 30% Percoll and spun at room temperature to separate cells from myelin. Isolated cells were stained with fluorophore-conjugated antibodies for 30 min at room temperature in PBS containing 3% fetal bovine serum, 5 μg/ml of anti-CD16/32, 5% normal rat serum, 5% normal mouse serum, and 10 mM EDTA. The antibodies were obtained from BioLegend (San Diego, CA) [anti-CD3-AF488 (145-2C11), anti-CD49b-PE (HMα2), anti-F4/80-PE-Cy7 (BM8), anti-B220-AF647 (RA3-6B2), anti-CD64-BV421 (X54-5/7.1), anti-CD103-BV510 (2E7), anti-CD45-BV605 (30-F11), anti-IA/IE-BV650 (M5/114.15.2), and anti-CD11c-BV785 (N418)], BD Biosciences (San Jose, CA) [anti-CD31-PerCP-Cy5.5 (MEC13.3), anti-SiglecF-PE-CF594 (E50-2440), anti-Ly6G-AF700 (1A8), anti-CD11b-APC-Cy7 (M1/70), and anti-CD24-BV711 (M1/69)], or Invitrogen (Carlsbad, CA) [anti-Ly6C-PerCP-Cy5.5 (HK1.4)]. Dead cells were positively stained with LIVE/DEAD Fixable Yellow Dead Cell Stains (Molecular Probes, Waltham, MA). Flow cytometric analysis was performed using a LSRII flow cytometer (BD Biosciences) and results analyzed using FlowJo software (BD Biosciences, Franklin Lakes, NJ).
SNP genotyping
Genomic DNA was isolated from tails of F2 intercross between CAST and WSB mice using DNeasy Tissue kit (QIAGEN, Hilden, Germany). Genome-wide SNP genotyping was performed with a Mouse Universal Genotyping Array (MUGA; 8K SNPs). Array hybridization including sample preparation was performed by Neogen/GeneSeek (Lincoln, NE).
Linkage (QTL) analysis
Genome-wide scans were performed using R/qtl software. Genotypes from MUGA were prepared for QTL mapping as follows. A total of 2460 informative markers for CAST and WSB across the mouse genome were used for genetic mapping. The significance thresholds for LOD scores were determined by 1000 permutations using all informative markers. Each QTL was indicated as a significant when its LOD score exceeded 95% (P < 0.05) of the permutation distribution and QTL intervals exceeded 99% (P < 0.01) of the permutation distribution were considered as candidate intervals. The 95% confidence interval of each peak was determined by 1.5-LOD support interval. The physical (megabase) map positions based on the genomic sequence from the GRCm38/mm10 were calculated using Mouse Map Converter tool of the Jackson Laboratory (http://cgd.jax.org/mousemapconverter/).
Haplotype analysis
For the intervals of the identified four neuroprotective loci (Civq8 on chromosome 1, Civq9 on chromosome 6, Civq10 on chromosome 13, and Civq11 on chromosome 17), SNP data were obtained from the Mouse Phenome Database (http://phenome.jax.org/). An amino acid change affected detrimental to a protein was examined by three independents in silico prediction algorithms, Polymorphism Phenotyping v2 (PolyPhen-2; http://genetics.bwh.harvard.edu/pph2/index.shtml), Sorting Intolerant From Tolerant (SIFT; http://sift.jcvi.org), and Protein Variation Effect Analyzer (PROVEAN; http://provean.jcvi.org).
RNA-sequencing analysis
Paired-end, 150-bp sequencing reads were generated from adult (8–12 weeks old) brain tissue mRNA of six CAST (three males and three females) and six WSB (three males and three females) mice on the Illumina HiSeq 2500 platform. Adapters for all paired-end sequencing reads were trimmed using Cutadapt (Martin 2011). Differential gene expression analysis is occasionally confounded by differences in alignment rate of reads obtained from strains that are highly diverged from the reference genome. Furthermore, there are millions of variant sites between the CAST and WSB strains, which would make these effects substantially pronounced. To alleviate these effects, we performed our alignment to pseudo-reference genomes (http://csbio.unc.edu/CCstatus/index.py?run= Pseudo), produced by altering the mouse reference genome (GRCm38.p4) using variant calls for the CAST and WSB strains. Reads from each strain were aligned to their respective pseudoreference genome using TopHat (v. 2.1.1) (Trapnell et al. 2009) under default settings, and remapped to reference coordinates using the Lapels pipeline (Holt et al. 2013; Huang et al. 2014).
Differential gene expression
Gene expression counts for each sample were obtained using HTseq (Anders et al. 2015) under default settings and subsequently imported into DESeq2 (Love et al. 2014), where gene counts for each mouse were normalized for differences in sequencing effort and dispersions were calculated for each sample. Differential expression was determined using a two-sided Wald test comparing two negative binomial distributions. P-values were adjusted using a false discovery rate of 5% by Benjamini–Hochberg correction (Benjamin and Hochberg 1995).
Statistical analysis
Results were represented as the mean ± SEM. Significant differences between data sets were determined using P-values < 0.05.
Data availability
Strains used in this study are available through the Jackson Laboratory catalog. Supplemental data contain two figures (Supplemental Material, Figures S1 and S2) and three tables (Tables S1–S3). Figure S1 shows flow cytometry data used to determine the expression level of CD45+ hematopoietic cells from brain cortex of CAST and WSB mice. Figure S2 shows both the number of collateral vessel connections and infarct volume after pMCAO for all individual F2 animals. Table S1 contains genotype and phenotype information of 251 F2 (CAST × WSB) animals used for QTL mapping analysis. Table S2 contains detailed information of all coding SNPs including in silico prediction of three independent algorithms. Table S3 contains detailed information of RNA-sequencing analysis within identified neuroprotective loci. RNA-sequencing data are available at Gene Expression Omnibus under the accession number GSE129379. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8984114.
Results
Analysis of collateral vessel density and infarct volume after pMCAO for all eight founder strains of the CC
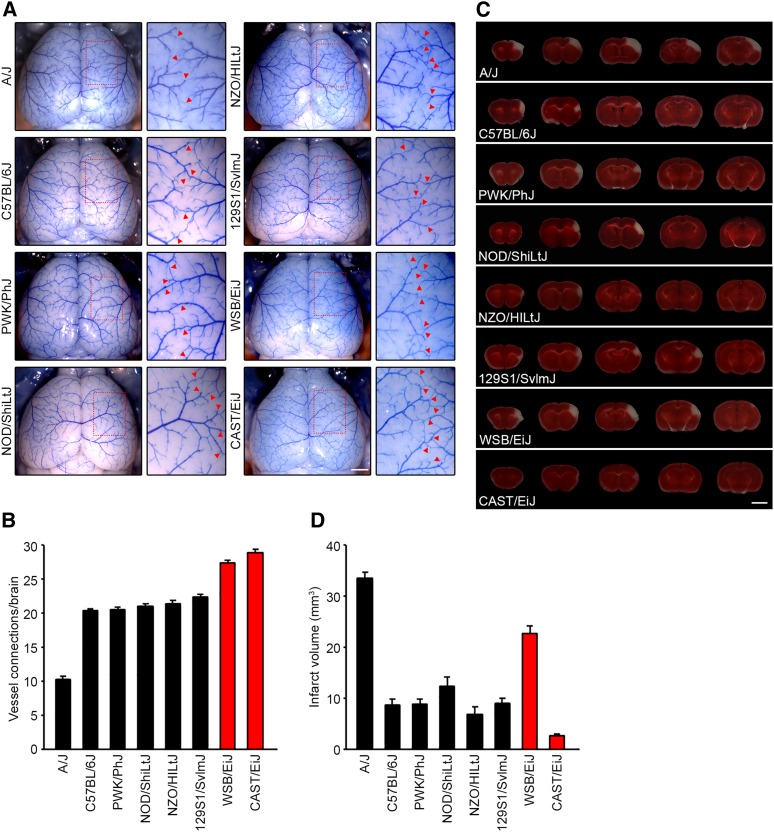
Using the eight founder mouse strains of the CC, we first examined the number of collateral vessel connections between the ACA and MCA (Figure 1, A and B). Classical inbred mouse strains B6, NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), and 129S1/SvlmJ (129S1), and the wild-derived strain PWK/PhJ (PWK) show similar level of vessel connections [B6 (20.4), NOD (21.0), NZO (21.4), 129S1 (22.4), and PWK (20.4)], whereas A/J (10.2) showed a lower level. Interestingly, two wild-derived strains, CAST and WSB, showed the highest number of collateral vessel connections (CAST, n=28.8 and WSB, n=27.3) that we have ever observed in any inbred strain.
Figure 1.
Collateral vessel density and infarct volume after pMCAO in eight founder strains of the Collaborative Cross (CC) mouse. (A) Representative images of the brains for the eight founder strains of the CC recombinant inbred mapping panel: A/J, B6, PWK, NOD, NZO, 129S1, WSB, and CAST. For each strain, the area outlined in a red box in the leftmost image is threefold magnified in the image to the right, where the red arrowheads indicate collateral vessel connections between the ACA and the MCA. Bar, 1 mm. (B) The graph indicates the average number of collateral vessel connections between ACA and MCA in the brain. The total number of animals for A/J, B6, PWK, NOD, NZO, 129S1, WSB, and CAST were 21, 37, 24, 9, 14, 31, 13, and 32 mice, respectively. Data represent the mean ± SEM. (C) Serial brain sections (1 mm) for each founder strain of the CC mouse 24 hr after pMCAO. The infarct appears as white tissue after 2% triphenyltetrazolium chloride staining. Bar, 5 mm. (D) The graph shows the infarct volume for each founder strain of the CC mouse. The total number of animals for A/J, B6, PWK, NOD, NZO, 129S1, WSB, and CAST were 19, 32, 27, 9, 10, 32, 18, and 32 animals, respectively. Data represent the mean ± SEM.
To determine whether collateral vessel density is inversely correlated with infarct volume in these new strains, we next examined ischemic infarct volume in the pMCAO model. We performed pMCAO and measured infarct volume for the eight CC founder mouse strains (Figure 1, C and D). Infarct volume in these mouse strains displayed various levels after ischemic stroke induction [A/J (33.4 mm3), B6 (8.6 mm3), PWK (8.7 mm3), NOD (12.3 mm3), NZO (6.8 mm3), 129S1 (9.0 mm3), WSB (22.6 mm3), and CAST (2.6 mm3]. As expected, most CC founder mouse strains showed an inverse correlation between collateral vessel connections and infarct volume. However, one strain, WSB, breaks this inverse correlation. Although two wild-derived mouse strains, CAST and WSB, exhibit the highest number of vessel connections we have seen in any inbred strain (Figure 1B), WSB consistently displayed a dramatically larger infarct volume (8.7-fold) after pMCAO, compared to CAST (22.6 mm3 vs. 2.6 mm3) (Figure 1D). This suggests that, at least in part, WSB modulates infarct volume after cerebral ischemia through a collateral-independent mechanism.
It is conceivable that the strain-dependent infarct volume differences are caused by different composition of cells involved in host defense mechanisms in the brains of the two strains (e.g., CD45+ hematopoietic cells). Thus, we performed flow cytometric analysis of CD45+ hematopoietic cells in the cerebral cortex of the two strains. We found no difference in the fraction of microglia, lymphocytes (B and T cells), or CD11b+ myeloid cells (macrophages) in the brain tissue of the two strains (Figure S1, A and B).
Four novel loci contribute infarct volume differences between CAST and WSB
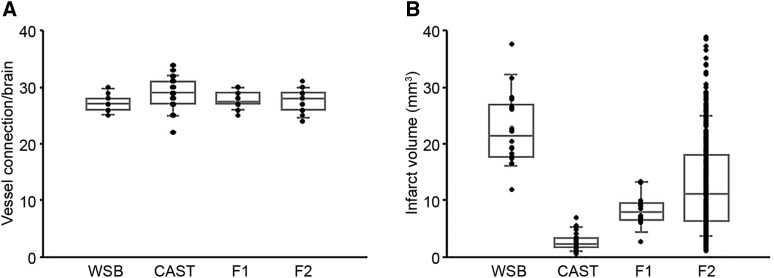
To identify the genetic loci modulating infarct volume between these two strains, we generated F1 and F2 progeny between CAST and WSB and examined both collateral vessel density and infarct volume. Consistent with the number of collateral vessel connections between CAST and WSB, collateral vessel connections in the F1 and F2 generations exhibited a very tight distribution [WSB (27.3), CAST (28.8), F1 (27.7), and F2 (27.6)] (Figure 2A). Moreover, the number of collateral vessel connections in individual F2 animals falls within a rather tight range (from 25 to 31 connections between the ACA and MCA), similar in number and range to the two parental strains (Figure S2A). Thus, genetic variation within the two strains does not appear to modulate the collateral vessel phenotype. Next, using F1 and F2 progeny, we measured infarct volume after pMCAO. In contrast to the vascular phenotype, infarct volume in the F2 generation was widely distributed, ranging from 1.1 to 38.9 mm3 (Figure 2B and Figure S2B) and covering nearly the entire phenotypic spectrum observed across all inbred strains (Keum and Marchuk 2009; Keum et al. 2013).
Figure 2.
F2 intercross animals between CAST and WSB exhibit a tight distribution of collateral vessel connections but a wide distribution for pMCAO-induced infarct volume. (A) The dot plot graph shows the number of collateral vessel connections between the ACA and MCA for parental strains, WSB and CAST, and their F1 and F2 intercross progeny. The box plot indicates the degree of dispersion and skewness. The number of animals for WSB, CAST, F1, and F2 was 13, 32, 18, and 24 animals, respectively. Data represent the mean ± SEM. (B) Dot and box plot graphs show the distribution of pMCAO-induced infarct volume for WSB, CAST, and their F1 and F2 intercross progeny. The number of animals for the infarct volume measurements was 18, 32, 14, and 251 animals, respectively. Data represent the mean ± SEM.
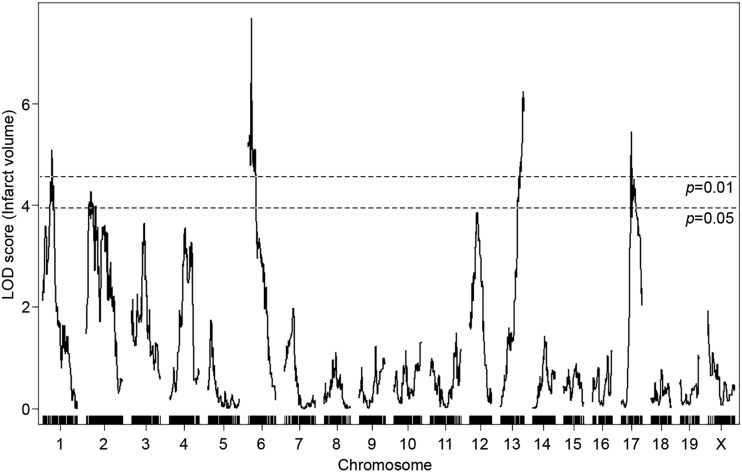
To discover genetic loci that modulate infarct volume we performed genome-wide QTL mapping analysis. A total of 251 F2 mice (Figure 2B and Figure S2B) were used for genome-wide SNP genotyping using a MUGA (8K SNPs). For genome-wide QTL mapping analysis, 2460 informative SNP markers were selected across the mouse genome (Table S1), and we identified four new QTL peaks that display highly significant linkage to the infarct volume trait. These were located on chromosomes 1, 6, 13, and 17 (Figure 3). These new loci were designated as Civq8 through Civq11 based on our previous work that identified seven distinct loci for this trait (Keum and Marchuk 2009; Chu et al. 2013). The novel loci are Civq8 [chromosome 1, LOD 5.09 (JAX00250952)], Civq9 [chromosome 6, LOD 7.68 (UNC_rs51474193)], Civq10 [chromosome 13, LOD 6.15 (UNC130410312)], and Civq11 [chromosome 17, LOD 5.45 (UNC170802188)] (Table 1).
Figure 3.
Identification of four novel Civq loci modulating infarct volume in the F2 intercross between CAST and WSB. The graph represents the analysis of a genome-wide QTL mapping scan for infarct volume measured 24 hr after pMCAO using 251 F2 progeny (CAST and WSB). Chromosomes 1 through X are indicated numerically on the x-axis. The y-axis shows an LOD score and the significant levels (P < 0.05 and 0.01) were determined by 1000 permutation tests. Four regions of the genome mapping to chromosomes 1, 6, 13, and 17 display highly significant QTL mapping to the infarct volume trait with LOD scores of 5.09, 7.68, 6.15, and 5.45, respectively.
Table 1. Characteristics of four novel QTL for ischemia-induced infarct volume.
| QTL | Location | LOD score | 1.5-LOD interval (Mb) (GRCm38/mm10) | Marker at peak (peak position) | Additive effecta | Dominance effectb | Protective allele |
|---|---|---|---|---|---|---|---|
| Civq8 | Chr1 | 5.09 | 1: 40.66–68.27 (27.61 Mb) | JAX00250952 | 6.25 | −0.77 | WSB |
| 53.22 Mb | |||||||
| Civq9 | Chr6 | 7.68 | 6: 22.41–27.88 (5.47 Mb) | UNC_rs51474193 | −4.2 | −2.74 | CAST |
| 26.49 Mb | |||||||
| Civq10 | Chr13 | 6.15 | 13: 106.02–120.42 (14.40 Mb) | UNC130410312 | 4.17 | −0.35 | WSB |
| 118.60 Mb | |||||||
| Civq11 | Chr17 | 5.45 | 17: 56.20–72.82 (16.62 Mb) | UNC170802188 | −3.93 | −1.7 | CAST |
| 61.66 Mb |
Direction of additive effect corresponds to additive effect of CAST allele.
Direction of dominance effect corresponds to deviation from additive relationship between CAST/CAST and WSB/WSB genotypes.
New loci uncover potential candidate genes
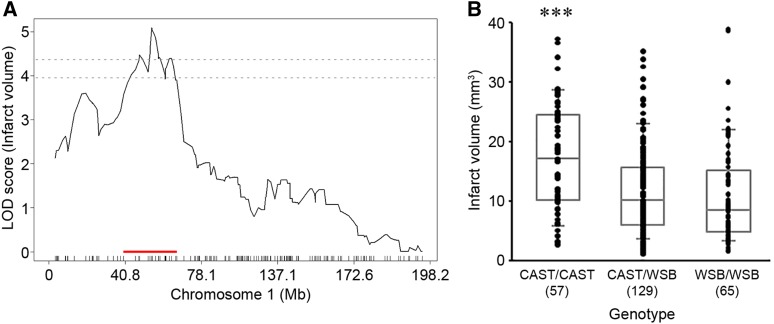
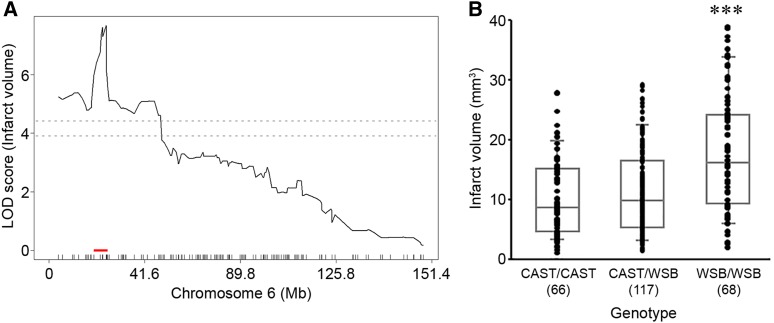
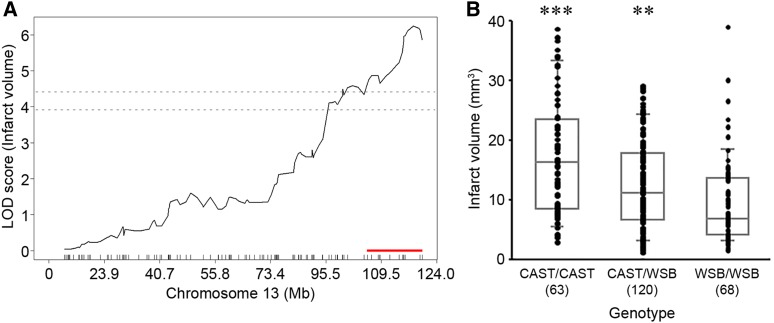
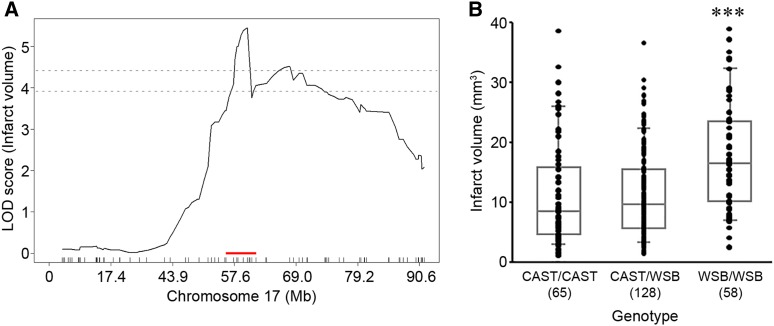
For the precise definition of the candidate interval for each locus, we extended each interval an additional 1.5 Mb flanking each arm of 1.5-LOD support interval. A total of 173 informative SNP markers were used for the definition of Civq8 mapping to chromosome 1. The resulting candidate interval for Civq8 is from 40.66 to 68.27 Mb (Figure 4A and Table 1). Interestingly, Civq8 harbors a transgressive allele (or alleles) that modulates the phenotype in the opposite direction of the parental strains. As shown in Figure 4B, the animals with the CAST allele are more sensitive to infarction, while those with the WSB allele are more resistant to infarction. For Civq9, mapping to chromosome 6, 144 informative SNP markers were used and its candidate interval is from 22.41 to 27.88 Mb (Figure 5A and Table 1). At Civq9, the CAST allele exhibits a protective effect on infarct volume in agreement with the phenotype of the parental CAST strain (Figure 5B). For Civq10, mapping to chromosome 13, 120 informative SNP markers were used and its candidate interval is from 106.02 to 120.42 Mb (Figure 6A and Table 1). Civq10 also harbors a transgressive allele (or alleles) since the WSB allele at Civq10 is protective, in contrast to the overall phenotype of the parental strain (Figure 6B). For Civq11, mapping to chromosome 17, 78 informative SNP markers were used and its candidate interval is from 56.20 to 63.71 Mb (Figure 7A and Table 1). The CAST allele in Civq11 exhibits a protective effect in agreement with the parental strain (Figure 7B).
Figure 4.
The Civq8 locus mapping to chromosome 1. (A) The graph shows the QTL mapping across chromosome 1 using 173 informative SNP markers, highlighting the Civq8 locus. The LOD score at the peak is 5.09 (JAX00250952), and the 1.5-LOD support interval is from 40.66 to 68.27 Mb, indicated by the red bar on the graph. (B) Genotype-phenotype correlation of the F2 cohort at JAX00250952. The alleles are transgressive with the CAST allele conferring increased susceptibility to infarction and the WSB allele conferring protection. Data represent the mean ± SEM. *** P < 0.001 vs. CAST/WSB and WSB/WSB, one-way ANOVA followed by Scheffe’s test.
Figure 5.
The Civq9 locus mapping to chromosome 6. (A) The graph shows the QTL mapping across chromosome 6 using 144 informative SNP markers, highlighting the Civq9 locus. The LOD score at the peak is 7.68 (UNC_rs51474193), and the 1.5-LOD support interval is from 22.41 to 27.88 Mb, indicated by the red bar on the graph. (B) Genotype-phenotype correlation of the F2 cohort at UNC_rs51474193. The CAST allele at Civq9 is protective for infarction. Data represent the mean ± SEM. *** P < 0.001 vs. CAST/CAST and CAST/WSB, one-way ANOVA followed by Scheffe’s test.
Figure 6.
The Civq10 locus mapping to chromosome 13. (A) The graph shows the QTL mapping across chromosome 13 using 120 informative SNP markers, highlighting the Civq10 locus. The LOD score at the peak is 6.15 (UNC130410312), and the 1.5-LOD support interval is from 106.02 to 120.42 Mb, indicated by the red bar on the graph. (B) Genotype-phenotype correlation of the F2 cohort at UNC130410312. The alleles are transgressive with the CAST allele conferring increased susceptibility to infarction and the WSB allele conferring protection. Data represent the mean ± SEM. *** P < 0.001 vs. CAST/WSB and WSB/WSB, ** P < 0.01 vs. WSB/WSB, one-way ANOVA followed by Scheffe’s test.
Figure 7.
The Civq11 locus mapping to chromosome 17. (A) The graph shows the QTL mapping across chromosome 17 using 78 informative SNP markers, highlighting the Civq11 locus. The LOD score at the peak is 5.45 (UNC170802188), and the 1.5-LOD support interval is from 56.20 to 63.71 Mb, indicated by the red bar on the graph. (B) Genotype-phenotype correlation of the F2 cohort at UNC170802188. The CAST allele at Civq11 is protective for infarction. Data represent the mean ± SEM. *** P < 0.001 vs. CAST/CAST and CAST/WSB, one-way ANOVA followed by Scheffe’s test.
To identify possible candidate gene(s) modulating infarct volume in these loci, we first surveyed candidate genes within the candidate interval of each QTL and then sought the presence of nonsynonymous coding SNPs (hereafter, coding SNP) in these genes. We identified a total of 330 coding SNPs in 90 coding genes in Civq8, 77 coding SNPs in 20 coding genes in Civq9, 109 coding SNPs in 34 coding genes in Civq10, and 157 coding genes in 47 coding genes in Civq11. Within these intervals, the vast majority of these genes harbor multiple coding SNPs (Table S2). To determine whether any of these nonsynonymous amino acid substitutions might affect protein function, all the coding SNPs were subjected to three independent in silico algorithms that predict their functional consequences: SIFT, PolyPhen-2, and PROVEAN. In Civq8, 46 genes have a coding SNP (or SNPs) predicted to be damaging by at least one of the prediction algorithms. However, only 11 genes harbor coding SNPs predicted to be damaging by all three programs and another 15 harbor damaging SNPs if the threshold is relaxed that only two of the three algorithms predict damaging consequences. In Civq9, seven genes have coding SNPs predicted to be damaging by at least one algorithm, with only one predicted to be damaging by all three programs and another three genes by only two algorithms. In Civq10, a total 16 genes have coding SNPs predicted to be damaging with only four predicted to be damaging by all three programs and another one gene by only two algorithms. In Civq11, a total 27 genes have coding SNPs predicted to be damaging with only three predicted to be damaging by all three programs and another seven genes by only two algorithms (Table 2 and Table S2). Although genes containing coding SNPs predicted to be damaging by all three independents in silico algorithms are the best candidate genes, even genes containing coding SNPs predicted to be damaging by only one in silico algorithm are still potential candidates. The details of all coding SNPs and functional predictions are listed in Table S2.
Table 2. Candidate genes harboring coding SNPs that are predicted to be damaging by three different in silico algorithms.
| QTL | Predicted damaging by all three in silico algorithms | Predicted damaging by only two in silico algorithms | Predicted damaging by only one in silico algorithm | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Civq8 | Aox2 | Aox3 | Aox4 | Aox1 | Bmpr2 | Ccdc150 | Boll | Carf | Ccdc168 |
| Casp8 | Crygb | Dnah7a | Ercc5 | Gm973 | Hibch | Clfar | Col5a2 | Cyp20a1 | |
| Dytn | Il1r1 | Mpp4 | Il18rap | Il1r2 | Il1rl1 | D630023F18Rik | Dnah7b | Il18r1 | |
| Nabp1 | Nbeal1 | Pard3b | Pms1 | Sgol2a | Il1rl2 | Map4k4 | Mett21c | ||
| Timem237 | Trak2 | Wdr75 | Mettl21e | Mfsd9 | Plcl1 | ||||
| Rftn2 | Slc39a10 | Spag16 | |||||||
| Spats2l | Zdbf2 | ||||||||
| Civq9 | Iqub | Hyal4 | Pax4 | Ptprz1 | Cadps2 | Cped1 | Pot1a | ||
| Civq10 | Gzma | Itga1 | Itga2 | Mrps30 | 4833420G17Rik | Cdc20b | Dhx29 | ||
| Map3k1 | Emb | Gapt | Hcn1 | ||||||
| Il31ra | Mcidas | Nnt | |||||||
| Pde4d | Tcstv3 | ||||||||
| Civq11 | Adgre1 | Arrdc5 | Ticam1 | Cntnap5c | Dennd1c | Pdzph1 | 1700061G19Rik | 4930583I09Rik | Acer1 |
| Plin4 | Pspn | Shd | Adgre4 | Chaf1a | Clpp | ||||
| Zfp119b | Ebi3 | Fer | Fsd1 | ||||||
| Mpnd | Plin5 | Safb2 | |||||||
| Stap2 | Tnfsf9 | Vmn2r120 | |||||||
| Zfp119a | Zfp959 | ||||||||
Detailed coding SNP information for each gene is available in Table S2.
Strain-specific differential gene expression discovers candidate genes in the four new loci
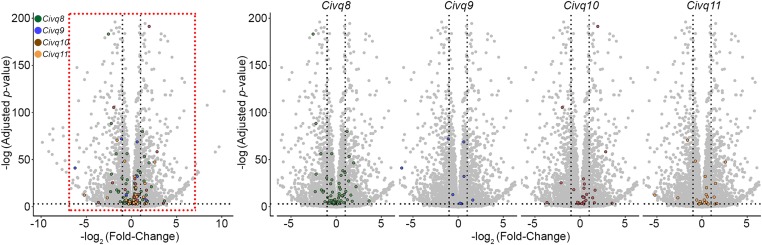
To identify other candidate genes underlying these loci, we sought evidence of strain-specific differential transcript levels between two wild-derived mouse strains, CAST and WSB. Strain-specific differences in transcript levels could potentially be caused by regulatory sequence variation acting in cis by any number of potential molecular mechanisms. We employed RNA-sequencing data analysis using adult brain tissues between CAST and WSB. As shown in Figure 8, a total 9563 genes were differentially expressed between strains but only 220 of these genes are located within one of the four new QLTs, with 135 in Civq8, 10 in Civq9, 47 in Civq10, and 28 in Civq11. Assuming that the most likely candidate genes would show at least a twofold difference in strain-specific expression reduces the overall list to only 79 candidate genes: 47 genes in Civq8, 3 genes in Civq9, 19 genes in Civq10, and 10 genes in Civq11 (Table 3 and Table 4). All 220 genes are displayed in Table S3, which includes information on direction of difference, that is, which inbred strain of the pair exhibits the higher expression. Genes harboring either coding SNPs predicted to be damaging or those showing differential expression between CAST and WSB are potential candidate genes that modulate infarct volume after ischemic stroke via a collateral vessel-independent mechanism.
Figure 8.
RNA-sequencing data identifies genes showing strain-specific differences in cerebral cortex gene expression. The volcano plot shows differential gene expression between CAST and WSB from brain cortex determined by RNA-sequencing analysis. Each dot represents a different gene with the log2 fold-change plotted against log10 P-value. The total number of significantly differential expressed genes across the mouse genome is 9563 genes but only 220 genes map within one of the four QTL intervals. Differentially expressed genes in the red box in the leftmost plot are separated for each interval on the right; green dots indicate 135 genes mapping within Civq8, blue dots indicate 10 genes mapping within Civq9, brown dots indicate 47 genes mapping within Civq10, and orange dots indicate 28 genes mapping within Civq11.
Table 3. Differential gene expression between CAST and WSB determined by RNA-sequencing analysis.
| QTL | Symbol | Description | Gene type | Log2 fold-change |
|---|---|---|---|---|
| Civq8 | Gm37373 | Predicted gene 37373 | TECa | −4.36 |
| Gm15759 | Predicted gene 15759 | antisense_RNA | −3.07 | |
| Gm28802 | Predicted gene 28802 | unprocessed_pseudogene | −2.75 | |
| Als2cr12 | Amyotrophic lateral sclerosis 2 chromosome region 12 | protein_coding | −2.55 | |
| 4930558J18Rik | RIKEN cDNA 4930558J18 gene | lincRNA | −2.31 | |
| Gm37915 | Predicted gene 37915 | antisense_RNA | −2.28 | |
| Dnah7a | Dynein, axonemal, heavy chain 7A | protein_coding | −2.24 | |
| Aox2 | Aldehyde oxidase 2 | protein_coding | −2.23 | |
| Gm26813 | Predicted gene 26813 | lincRNA | −1.80 | |
| Gm8241 | Predicted gene 8241 | processed_pseudogene | −1.72 | |
| Boll | Boule homolog, RNA binding protein | protein_coding | −1.69 | |
| Gm8419 | Predicted gene 8419 | processed_pseudogene | −1.65 | |
| Cps1 | Carbamoyl-phosphate synthetase 1 | protein_coding | −1.64 | |
| C2cd6b | C2 calcium dependent domain containing 6B | lincRNA | −1.60 | |
| E330011M16Rik | RIKEN cDNA E330011M16 gene | TECa | −1.59 | |
| Gm27607 | Predicted gene 27607 | snoRNA | −1.55 | |
| Osgepl1 | O-sialoglycoprotein endopeptidase-like 1 | protein_coding | −1.52 | |
| Stat4 | Signal transducer and activator of transcription 4 | protein_coding | −1.46 | |
| Mstn | Myostatin | protein_coding | −1.40 | |
| Gm35801 | Predicted gene 35801 | antisense_RNA | −1.28 | |
| Tmem182 | Transmembrane protein 182 | protein_coding | −1.24 | |
| Gm11600 | Predicted gene 11600 | processed_pseudogene | −1.24 | |
| Aox3 | Aldehyde oxidase 3 | protein_coding | −1.05 | |
| Civq9 | Hyal5 | Hyaluronoglucosaminidase 5 | protein_coding | −6.24 |
| Iqub | IQ motif and ubiquitin domain containing | protein_coding | −1.10 | |
| Civq10 | Gm47776 | Predicted gene 47776 | processed_pseudogene | −3.67 |
| Gm21188 | Predicted gene 21188 | protein_coding | −3.66 | |
| Mcidas | Multiciliate differentiation and DNA synthesis associated cell cycle protein | protein_coding | −2.10 | |
| Itga2 | Integrin alpha 2 | protein_coding | −1.97 | |
| Gm6270 | Predicted gene 6270 | lincRNA | −1.84 | |
| Gm15290 | Predicted gene 15290 | lincRNA | −1.69 | |
| Gm15488 | Predicted gene 15488 | unitary_pseudogene | −1.69 | |
| Gm15285 | Predicted gene 15285 | processed_pseudogene | −1.25 | |
| Civq11 | Vmn2r120 | Vomeronasal 2, receptor 120 | protein_coding | −5.23 |
| Nudt12os | Nucleoside diphosphate linked moiety X-type motif 12, opposite strand | antisense_RNA | −2.98 | |
| Cd70 | CD70 antigen | protein_coding | −2.68 | |
| Catsperd | Cation channel sperm associated auxiliary subunit delta | protein_coding | −1.59 |
The table lists all genes showing at least twofold differential expression where the CAST allele is higher than WSB allele. Additional information for all 220 genes is available in Table S3.
To be experimentally confirmed.
Table 4. Differential gene expression between CAST and WSB determined by RNA-sequencing analysis.
| QTL | Symbol | Description | Gene type | Log2 fold-change |
|---|---|---|---|---|
| Civq8 | Rps27a-ps1 | Ribosomal protein S27A, pseudogene 1 | processed_pseudogene | 3.82 |
| 1700066B17Rik | RIKEN cDNA 1700066B17 gene | lincRNA | 3.64 | |
| Gm553 | Predicted gene 553 | lincRNA | 3.07 | |
| Gm29018 | Predicted gene 29018 | antisense_RNA | 2.77 | |
| Gm28177 | Predicted gene 28177 | processed_transcript | 2.49 | |
| BC055402 | CDNA sequence BC055402 | lincRNA | 2.19 | |
| Il1rl2 | Interleukin 1 receptor-like 2 | protein_coding | 1.94 | |
| D930019O06Rik | RIKEN cDNA D930019O06 | antisense_RNA | 1.91 | |
| Sgo2a | Shugoshin 2A | protein_coding | 1.78 | |
| Rfx8 | Regulatory factor X 8 | protein_coding | 1.63 | |
| 9530026F06Rik | RIKEN cDNA 9530026F06 gene | lincRNA | 1.60 | |
| 4933402D24Rik | RIKEN cDNA 4933402D24 gene | protein_coding | 1.43 | |
| Dnah7c | Dynein, axonemal, heavy chain 7C | protein_coding | 1.39 | |
| Gm15832 | Predicted gene 15832 | processed_transcript | 1.39 | |
| Hibch | 3-hydroxyisobutyryl-Coenzyme A hydrolase | protein_coding | 1.30 | |
| Myl1 | Myosin, light polypeptide 1 | protein_coding | 1.24 | |
| 1700019D03Rik | RIKEN cDNA 1700019D03 gene | protein_coding | 1.22 | |
| Gm28826 | Predicted gene 28826 | lincRNA | 1.21 | |
| Il1r2 | Interleukin 1 receptor, type II | protein_coding | 1.07 | |
| Gm28449 | Predicted gene 28449 | lincRNA | 1.07 | |
| Gm28982 | Predicted gene 28982 | lincRNA | 1.05 | |
| 2810408I11Rik | RIKEN cDNA 2810408I11 gene | processed_transcript | 1.04 | |
| Gm18849 | Predicted gene 18849 | unprocessed_pseudogene | 1.03 | |
| C230029F24Rik | RIKEN cDNA C230029F24 gene | lincRNA | 1.02 | |
| Civq9 | Asb15 | Ankyrin repeat and SOCS box-containing 15 | protein_coding | 1.63 |
| Civq10 | BC147527 | CDNA sequence BC147527 | protein_coding | 3.53 |
| Gzmk | Granzyme K | protein_coding | 2.83 | |
| Gm15323 | Predicted gene 15323 | lincRNA | 2.28 | |
| 4833420G17Rik | RIKEN cDNA 4833420G17 gene | protein_coding | 1.96 | |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | protein_coding | 1.66 | |
| Cdc20b | Cell division cycle 20B | protein_coding | 1.57 | |
| Gm47914 | Predicted gene 47914 | lincRNA | 1.52 | |
| 3110070M22Rik | RIKEN cDNA 3110070M22 gene | protein_coding | 1.39 | |
| Gm15327 | Predicted gene 15327 | processed_transcript | 1.16 | |
| 4930467J12Rik | RIKEN cDNA 4930467J12 gene | lincRNA | 1.07 | |
| Gm15287 | Predicted gene 15287 | antisense_RNA | 1.03 | |
| Civq11 | Vmn2r118 | Vomeronasal 2, receptor 118 | protein_coding | 2.58 |
| Acsbg2 | Acyl-CoA synthetase bubblegum family member 2 | protein_coding | 1.62 | |
| Prr22 | Proline rich 22 | protein_coding | 1.49 | |
| Gm16712 | Predicted gene 16712 | lincRNA | 1.44 | |
| Plin4 | Perilipin 4 | protein_coding | 1.34 | |
| Gm17168 | Predicted gene 17168 | antisense_RNA | 1.13 |
The table lists all genes showing at least twofold differential expression where the WSB allele is higher than CAST allele. Additional information for all 220 genes is available in Table S3.
Discussion
The long-term objective of this work is to identify novel targets for stroke therapy, focusing on those factors that modulate the size of the cerebral infarct in a collateral vessel-independent manner. In support of this goal, a recent study of a meta-analysis of multiple clinical stroke cohorts showed that collateral vessel anatomy did not correlate with neurological outcomes (de Havenon et al. 2019). Furthermore, the collateral vasculature is developmentally established and consequently, may be difficult to modify therapeutically.
Here, to increase the genetic diversity of our mapping approach, we utilized eight founder mouse strains of the CC recombinant inbred mapping panel. These strains include five classical inbred strains (A/J, B6, NOD, NZO, and 129S1) and three wild-derived strains from Mus musculus subspecies (PWK, WSB, and CAST). Specifically, these wild-derived strains, PWK (M. m. musculus), WSB (M. m. domesticus), and CAST (M. m. castaneous), are more genetically diverse than the classical inbred strains (Churchill et al. 2004; Threadgill 2006; Aylor et al. 2011; Iraqi et al. 2012). Using all eight founder strains of the CC mouse, we surveyed both the number of collateral vessel connections as well as infarct volume after pMCAO. The number of collateral vessel connection of two wild-derived strains, CAST and WSB, were the highest we have observed in any inbred mouse strains. Consistent with the role of collateral vessels in reperfusion of the ischemic territory following pMCAO, CAST mice exhibit the lowest infarct volume that we have ever measured. However, although CAST and WSB have the largest number of collateral vessel connections, unlike CAST, upon pMCAO, WSB exhibits a large infarct volume. Therefore, this wild-derived WSB strain also breaks the inverse correlation between collateral vessel connections and infarct volume. Supportive evidence for a noncollateral mechanism is the observation that infarct volume in the F2 animals shows a broad range of values while the collateral vessel connections are nearly invariant among F2 animals. Based on the ability to reperfuse the ischemic tissue due to more than sufficient collateral vessel connections, but displaying a much larger infarct volume than would be predicted, WSB likely contains loci that upon ischemic insult, contribute to neuronal death. One compelling hypothesis is that most strains contain natural neuroprotective loci that are defective in WSB. In this F2 cross, we identified the most significant four novel neuroprotective loci (Civq8 on chromosome 1, Civq9 on chromosome 6, Civq10 on chromosome 13, and Civq11 on chromosome 17). Importantly, these loci identified in an F2 cross between CAST and WSB did not overlap with other loci that we previously identified with other F2 crosses. More importantly, the Civq1/Canq1 locus, the strongest locus controlling collateral vessel number, is absent in this cross. Interestingly but not surprisingly, we found transgressive alleles among the loci, alleles that operate in the opposite direction of the parental strain phenotypes. Transgressive segregation is not uncommon in intraspecific crosses (Palijan et al. 2003; Roper et al. 2003), and one of our other, previously mapped neuroprotective loci, Civq4, also harbors a transgressive allele (Chu et al. 2013).
From the QTL mapping analysis for infarct volume, we identified four candidate intervals regulating infarct volume through a collateral-independent mechanism. To identify candidate genes within the 1.5-LOD support intervals, we searched for coding SNP differences between CAST and WSB focusing on genes where the functional consequence of the variants were predicted using three different in silico prediction algorithms (Ng and Henikoff 2003; Adzhubei et al. 2010; Choi et al. 2012). Coding SNPs predicted as “damaging” by all three algorithms were considered the highest priority candidates, with full realization that any of the other genes harboring coding SNPs cannot be ignored.
As the genes underlying these four loci might harbor cis-regulatory sequence variation modulating differences in mRNA levels, we determined strain-specific differential transcript levels between CAST and WSB using RNA-sequencing analysis. Genes exhibiting a statistically significant, twofold or greater difference in transcript levels are considered our highest priority candidate genes.
Through these two independent approaches, we have narrowed our list of candidate genes for all four novel loci to 96 genes harboring potentially damaging coding SNP variation and 79 genes exhibiting greater than twofold, strain-specific differential gene expression between CAST and WSB. To identify the causative genes, further functional studies are required. However, with additional bioinformatics analyses and a search of the relevant literature, two genes stand out.
Civq8 harbors a protective WSB allele. Casp8 (Caspase 8), mapping within the interval, harbors multiple coding SNPs for which the WSB allele of one SNP in particular (rs226995171) is predicted to be damaged by all three algorithms used in our analysis. Moreover, among 37 inbred mouse strains (Sanger4: Sanger SNP and indel data, ≥89 million locations, 37 inbred strains of mice (phenome.jax.org/projects/Sanger4)), only the CAST and SPRET/EiJ strains harbor this rare allele. Casp8 initiates an extrinsic pathway during apoptotic cell death (Kischkel et al. 1995; Thornberry and Lazebnik 1998) and it is possible that a poorly functioning or nonfunctional Casp8 would protect brain tissue against ischemia-induced apoptotic cell death.
The other gene is Map3k1, a member of the mitogen-activated protein kinase kinase kinase (MAP3K) superfamily controlling the MAPKK-MAPK signaling cascade. Although MAP3K1 plays an important role in multiple aspects of cell physiology, MAP3K1 also regulates apoptosis in response to multiple cellular stresses (Xia et al. 1995; Cardone et al. 1997; Deak et al. 1998; Widmann et al. 1998). Similar to Civq8, Civq10 is also a transgressive locus where the WSB allele is protective. One of the coding SNPs (rs257092960) in Map3k1 is predicted to be damaged by all three algorithms and WSB is the only strain harboring this allele among 37 mouse inbred strain (Sanger4). As with Casp8, it is possible that a nonfunctional Map3k1 protects ischemia-induced apoptotic cell death.
In summary, we have identified a wild-derived mouse strain, WSB, which breaks the inverse correlation between collateral vessel connections and infarct volume after pMCAO. By selecting two wild-derived mouse strains, CAST and WSB, that contain similarly high numbers of collateral vessel connections but display large differences in infarct volume, we identified a strain pair that might uncover novel loci involved in neuroprotection. In a cross between these strains, we discovered four novel neuroprotective genetic loci. Using RNA-sequencing of brain tissue and in silico analyses of coding SNPs, we further prioritized the genes mapping within these intervals. The identification of such neuroprotective genes may provide novel targets for future therapeutic intervention for human ischemic stroke.
Acknowledgments
The authors thank Sena Bae and Daniel A. Snellings for helpful discussion concerning data sorting and analysis. This work was supported by grants from National Institutes of Health (5R01HL097281) and the Foundation Leducq Transatlantic Network of Excellence in Neurovascular Disease (17 CVD 03).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8984114.
Communicating editor: A. Palmer
Literature Cited
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A. et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A. et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222. 10.1101/gr.111310.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone F. C., Knudsen D. J., Nelson A. H., Feuerstein G. Z., and Willette R. N., 1993. Mouse strain differences in susceptibility to cerebral ischemia are related to cerebral vascular anatomy. J. Cereb. Blood Flow Metab. 13: 683–692. 10.1038/jcbfm.1993.87 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Cardone M. H., Salvesen G. S., Widmann C., Johnson G., and Frisch S. M., 1997. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell 90: 315–323. 10.1016/S0092-8674(00)80339-6 [DOI] [PubMed] [Google Scholar]
- Carmichael S. T., 2005. Rodent models of focal stroke: size, mechanism, and purpose. NeuroRx 2: 396–409. 10.1602/neurorx.2.3.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R., and Chan A. P., 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7: e46688 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P. L., Keum S., and Marchuk D. A., 2013. A novel genetic locus modulates infarct volume independently of the extent of collateral circulation. Physiol. Genomics 45: 751–763. 10.1152/physiolgenomics.00063.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D. et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. 10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- Clayton J. A., Chalothorn D., and Faber J. E., 2008. Vascular endothelial growth factor-A specifies formation of native collaterals and regulates collateral growth in ischemia. Circ. Res. 103: 1027–1036. 10.1161/CIRCRESAHA.108.181115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium , 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401. 10.1534/genetics.111.132639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak J. C., Cross J. V., Lewis M., Qian Y., Parrott L. A. et al. , 1998. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc. Natl. Acad. Sci. USA 95: 5595–5600. 10.1073/pnas.95.10.5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Havenon A., Mlynash M., Kim-Tenser M. A., Lansberg M. G., Leslie-Maazwi T. et al. , 2019. Results from DEFUSE 3: good collaterals are associated with reduced ischemic core growth but not neurologic outcome. Stroke 50: 632–638. 10.1161/STROKEAHA.118.023407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S., Thorleifsson G., Reynisdottir S. T., Manolescu A., Jonsdottir S. et al. , 2003. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 35: 131–138 [corrigenda: Nat. Genet. 37: 555 (2005)]. 10.1038/ng1245 [DOI] [PubMed] [Google Scholar]
- Helgadottir A., Manolescu A., Thorleifsson G., Gretarsdottir S., Jonsdottir H. et al. , 2004. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat. Genet. 36: 233–239. 10.1038/ng1311 [DOI] [PubMed] [Google Scholar]
- Holt J., Huang S., McMillan L., and Wang W., 2013. Read annotation pipeline for high-throughput sequencing data, pp. 605–612 in Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics, ACM, New York. [Google Scholar]
- Huang S., Holt J., Kao C. Y., McMillan L., and Wang W., 2014. A novel multi-alignment pipeline for high-throughput sequencing data. Database (Oxford) 2014 10.1093/database/bau057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W., Onuma O., Owolabi M., and Sachdev S., 2016. Stroke: a global response is needed. Bull. World Health Organ. 94: 634–634A. 10.2471/BLT.16.181636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum S., and Marchuk D. A., 2009. A locus mapping to mouse chromosome 7 determines infarct volume in a mouse model of ischemic stroke. Circ Cardiovasc Genet 2: 591–598. 10.1161/CIRCGENETICS.109.883231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum S., Lee H. K., Chu P. L., Kan M. J., Huang M. N. et al. , 2013. Natural genetic variation of integrin alpha L (Itgal) modulates ischemic brain injury in stroke. PLoS Genet. 9: e1003807 10.1371/journal.pgen.1003807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel F. C., Hellbardt S., Behrmann I., Germer M., Pawlita M. et al. , 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14: 5579–5588. 10.1002/j.1460-2075.1995.tb00245.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler N., Corti F., Rivera-Molina F., Deng Y., Toomre D. et al. , 2018. The Rab-effector protein RABEP2 regulates endosomal trafficking to mediate vascular endothelial growth factor receptor-2 (VEGFR2)-dependent signaling. J. Biol. Chem. 293: 4805–4817. 10.1074/jbc.M117.812172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen K. L., Gregersen R., and Finsen B., 2002. Microglial-macrophage synthesis of tumor necrosis factor after focal cerebral ischemia in mice is strain dependent. J. Cereb. Blood Flow Metab. 22: 785–797. 10.1097/00004647-200207000-00004 [DOI] [PubMed] [Google Scholar]
- Lambertsen K. L., Meldgaard M., Ladeby R., and Finsen B., 2005. A quantitative study of microglial-macrophage synthesis of tumor necrosis factor during acute and late focal cerebral ischemia in mice. J. Cereb. Blood Flow Metab. 25: 119–135. 10.1038/sj.jcbfm.9600014 [DOI] [PubMed] [Google Scholar]
- Lee H. K., Keum S., Sheng H., Warner D. S., Lo D. C. et al. , 2016. Natural allelic variation of the IL-21 receptor modulates ischemic stroke infarct volume. J. Clin. Invest. 126: 2827–2838. 10.1172/JCI84491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Koh S., Lo D. C., and Marchuk D. A., 2018. Neuronal IL-4Rα modulates neuronal apoptosis and cell viability during the acute phases of cerebral ischemia. FEBS J. 285: 2785–2798. 10.1111/febs.14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucitti J. L., Sealock R., Buckley B. K., Zhang H., Xiao L. et al. , 2016. Variants of Rab GTPase-effector binding protein-2 cause variation in the collateral circulation and severity of stroke. Stroke 47: 3022–3031. 10.1161/STROKEAHA.116.014160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid A., He Y. Y., Gidday J. M., Kaplan S. S., Gonzales E. R. et al. , 2000. Differences in vulnerability to permanent focal cerebral ischemia among 3 common mouse strains. Stroke 31: 2707–2714. 10.1161/01.STR.31.11.2707 [DOI] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matarín M., Brown W. M., Scholz S., Simon-Sanchez J., Fung H. C. et al. , 2007. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 6: 414–420. 10.1016/S1474-4422(07)70081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarin M., Brown W. M., Singleton A., Hardy J. A., and Meschia J. F., 2008a Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 39: 1586–1589. 10.1161/STROKEAHA.107.502963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarin M., Simon-Sanchez J., Fung H. C., Scholz S., Gibbs J. R. et al. , 2008b Structural genomic variation in ischemic stroke. Neurogenetics 9: 101–108. 10.1007/s10048-008-0119-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., and Henikoff S., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palijan A., Dutil J., and Deng A. Y., 2003. Quantitative trait loci with opposing blood pressure effects demonstrating epistasis on Dahl rat chromosome 3. Physiol. Genomics 15: 1–8. 10.1152/physiolgenomics.00084.2003 [DOI] [PubMed] [Google Scholar]
- Roger V. L., Go A. S., Lloyd-Jones D. M., Benjamin E. J., Berry J. D. et al. , 2012. Heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation 125: e2–e220 (erratum: Circulation 125: e1002). 10.1161/CIR.0b013e31823ac046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R. J., McAllister R. D., Biggins J. E., Michael S. D., Min S. H. et al. , 2003. Aod1 controlling day 3 thymectomy-induced autoimmune ovarian dysgenesis in mice encompasses two linked quantitative trait loci with opposing allelic effects on disease susceptibility. J. Immunol. 170: 5886–5891. 10.4049/jimmunol.170.12.5886 [DOI] [PubMed] [Google Scholar]
- Söderholm M., Pedersen A., Lorentzen E., Stanne T. M., Beven S. et al. , 2019. Genome-wide association meta-analysis of functional outcome after ischemic stroke. Neurology 92: e1271–e1283. 10.1212/WNL.0000000000007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori H., Yao H., Ooboshi H., Ibayashi S., and Iida M., 2004. Krypton laser-induced photothrombotic distal middle cerebral artery occlusion without craniectomy in mice. Brain Res. Brain Res. Protoc. 13: 189–196. 10.1016/j.brainresprot.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Thornberry N. A., and Lazebnik Y., 1998. Caspase: enemies within. Science 281: 1312–1316. 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., 2006. Meeting report for the 4th annual Complex Trait Consortium meeting: from QTLs to systems genetics. Mamm. Genome 17: 2–4. 10.1007/s00335-005-0153-5 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler E. J., Peters E. E., Gonzales A., Gonzales M. L., Slee A. M. et al. , 2002. An objective procedure for ischemic area evaluation of the stroke intraluminal thread model in the mouse and rat. J. Neurosci. Methods 113: 51–58. 10.1016/S0165-0270(01)00476-9 [DOI] [PubMed] [Google Scholar]
- Widmann C., Gerwins P., Johnson N. L., Jarpe M. B., and Johnson G. L., 1998. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol. Cell. Biol. 18: 2416–2429. 10.1128/MCB.18.4.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Dickens M., Raingeaud J., Davis R. J., and Greenberg M. E., 1995. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270: 1326–1331. 10.1126/science.270.5240.1326 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A. et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. 10.1038/ng.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Prabhakar P., Sealock R., and Faber J. E., 2010. Wide genetic variation in the native pial collateral circulation is a major determinant of variation in severity of stroke. J. Cereb. Blood Flow Metab. 30: 923–934. 10.1038/jcbfm.2010.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains used in this study are available through the Jackson Laboratory catalog. Supplemental data contain two figures (Supplemental Material, Figures S1 and S2) and three tables (Tables S1–S3). Figure S1 shows flow cytometry data used to determine the expression level of CD45+ hematopoietic cells from brain cortex of CAST and WSB mice. Figure S2 shows both the number of collateral vessel connections and infarct volume after pMCAO for all individual F2 animals. Table S1 contains genotype and phenotype information of 251 F2 (CAST × WSB) animals used for QTL mapping analysis. Table S2 contains detailed information of all coding SNPs including in silico prediction of three independent algorithms. Table S3 contains detailed information of RNA-sequencing analysis within identified neuroprotective loci. RNA-sequencing data are available at Gene Expression Omnibus under the accession number GSE129379. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8984114.