Abstract
F1 hybrids between mouse inbred strains PWD and C57BL/6 represent the most thoroughly genetically defined model of hybrid sterility in vertebrates. Hybrid male sterility can be fully reconstituted from three components of this model, the Prdm9 gene, intersubspecific homeology of Mus musculus musculus and Mus musculus domesticus autosomes, and the X-linked Hstx2 locus. Hstx2 modulates the extent of Prdm9-dependent meiotic arrest and harbors two additional factors responsible for intersubspecific introgression-induced oligospermia (Hstx1) and meiotic recombination rate (Meir1). To facilitate positional cloning and to overcome the recombination suppression within the 4.3 Mb encompassing the Hstx2 locus, we designed Hstx2-CRISPR and SPO11/Cas9 transgenes aimed to induce DNA double-strand breaks specifically within the Hstx2 locus. The resulting recombinant reduced the Hstx2 locus to 2.70 Mb (chromosome X: 66.51–69.21 Mb). The newly defined Hstx2 locus still operates as the major X-linked factor of the F1 hybrid sterility, and controls meiotic chromosome synapsis and meiotic recombination rate. Despite extensive further crosses, the 2.70 Mb Hstx2 interval behaved as a recombination cold spot with reduced PRDM9-mediated H3K4me3 hotspots and absence of DMC1-defined DNA double-strand-break hotspots. To search for structural anomalies as a possible cause of recombination suppression, we used optical mapping and observed high incidence of subspecies-specific structural variants along the X chromosome, with a striking copy number polymorphism of the microRNA Mir465 cluster. This observation together with the absence of a strong sterility phenotype in Fmr1 neighbor (Fmr1nb) null mutants support the role of microRNA as a likely candidate for Hstx2.
Keywords: Speciation, Hybrid sterility X2, Prdm9, Bionano optical mapping, SPO11Cas9 transgene, Fmr1nb
REPRODUCTIVE isolation is a basic prerequisite of speciation implemented by a range of prezygotic and postzygotic mechanisms under complex genetic control (Dobzhansky 1951; Dion-Côté and Barbash 2017). Hybrid sterility, one of the reproductive isolation mechanisms, appears in the early stages of speciation and shares common features in many animal and plant species hybrids. They include preferential involvement of the heterogametic sex (XY or ZW), known as Haldane’s rule (Haldane 1922), or the large X effect (Coyne’s rule), referring to disproportionate engagement of X chromosome compared to autosomes (Dobzhansky 1951; Forejt 1996; Coyne and Orr 2004; Good et al. 2008; Presgraves 2018). The first hypothesis on genetic control of hybrid sterility, known as Dobzhansky–Muller epistatic incompatibility, refers to a dysfunction caused by the independent divergence of mutually interacting genes (Dobzhansky 1951). More recently, an interaction between meiotic drive and its suppressors has been implicated in some instances of reproductive isolation (Orr 2005; Zhang et al. 2015; Patten 2018). However, despite extensive genetic studies in organisms of various species such as yeast, fruit fly, or house mouse, the underlying genetic architecture and molecular mechanisms of hybrid sterility remain elusive [reviewed in Maheshwari and Barbash (2011); Phifer-Rixey and Nachman (2015); Dion-Côté and Barbash (2017); Mack and Nachman (2017); Payseur et al. (2018)].
The first hybrid sterility genetic factor to be identified in vertebrate, the hybrid sterility 1 (Hst1), was described in hybrids between laboratory and wild mice (Forejt and Ivanyi 1974; Gregorová et al. 1996; Trachtulec et al. 1997) and identified as the Prdm9 gene encoding PR/SET domain-containing nine protein (Mihola et al. 2009). The PRDM9 binds genomic DNA by a zinc finger domain at allele-specific sites and trimethylates lysine 4 and lysine 36 of histone 3. In mice, humans, and other mammalian species, Prdm9 mediates meiotic recombination by determining the genomic localization of the recombination hotspots (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). In a mouse model of intersubspecific hybrids where Mus musculus domesticus subspecies is represented by inbred strain C57BL/6J (hereafter B6) and Mus musculus musculus by PWD/Ph (hereafter PWD) (Gregorova and Forejt 2000) Prdm9 causes early meiotic arrest and complete male sterility by interaction with the X-linked Hstx2 locus. Hybrids between laboratory strains PWD and B6 serve as a robust, reproducible and genetically well-defined model of hybrid sterility [reviewed in Forejt (1996); Forejt et al. (2012)]. Specific allelic combinations of the Prdm9 gene (Prdm9PWD/B6) and Hstx2 locus (Hstx2PWD) were shown necessary but not sufficient to fully explain the meiotic arrest in hybrids. Initially, three or more additional hybrid sterility genes of small effect complementing the Prdm9 and Hstx2 major hybrid sterility genes had been considered (Dzur-Gejdosova et al. 2012). Later, we identified chromosome-autonomous meiotic asynapsis of homeologous chromosomes [homologous chromosomes from related (sub)species] as the third requirement for meiotic arrest (Bhattacharyya et al. 2013, 2014). The chromosomal, nongenic effects of homeologous chromosomes in (PWD × B6) F1 hybrids, manifested as a failure of meiotic chromosome synapsis, is most likely a consequence of evolutionary erosion of PRDM9 binding sites in each subspecies, resulting in asymmetry of DNA double-strand-break (DSB) hotspots (Davies et al. 2016). The explanation of hybrid sterility by expected shortage of symmetric DNA DSBs was supported by improvement of chromosome pairing and fertility after experimentally increasing the number of symmetric DNA DSBs by random stretches of a homozygous PWD sequence (Gregorova et al. 2018). Moreover, partial improvement of meiotic chromosome synapsis in hybrid males was achieved by addition of exogenous DSBs generated by a single cisplatin injection (Wang et al. 2018).
The PWD allele of the Hstx2 locus (Hstx2PWD) is indispensable for full sterility of (PWD × B6) F1 hybrids, while the Hstx2B6 allele attenuates the phenotype to partial spermatogenesis arrest in reciprocal (B6 × PWD) F1 males (Dzur-Gejdosova et al. 2012; Flachs et al. 2012; Forejt et al. 2012). Admittedly, the mechanism of action of the Hstx2 locus in meiotic arrest of F1 hybrids remains elusive. Previously, the Hstx2 locus was mapped to a 4.7 Mb region on X chromosome [chromosome X (Chr X): 64.9–69.6 Mb] (Bhattacharyya et al. 2014). The interval that encompasses 10 protein-coding genes and a cluster of microRNA (miRNA) genes is still too large to identify the true Hstx2 candidate. The Hstx2 locus (Chr X: 64.9–69.6 Mb) harbors two additional meiosis-related genetic factors, the hybrid sterility X1 (Hstx1) locus, manifested by sperm head malformations after Hstx2PWD sequence introgression into the B6 genome (Storchová et al. 2004), and meiotic recombination 1 (Meir1), which controls meiotic recombination rate (Balcova et al. 2016). Since these factors have not yet been genetically separated, their phenotypes may represent a pleiotropic effect of the same gene.
In an attempt to reduce the size of Hstx2, we constructed an SPO11-driven CRISPR-Cas9 system to target meiotic recombination to a particular genomic locus within the Hstx2 recombination cold spot. Although the method did not work as predicted, we recovered a single recombinant, thus reducing the Hstx2 locus to 2.70 Mb. We show that the shortened version of Hstx2 still carries the genetic factors or genes responsible for hybrid sterility, meiotic chromosome asynapsis, and genome-wide control of meiotic recombination rate. Using Bionano Optical mapping technology, we show high incidence of subspecies-specific insertion/deletion variants inside and outside the Hstx2 locus. Furthermore, we interrogate the Fmr1nb gene as a possible Hstx2 candidate gene.
Materials and Methods
Animals and ethics statement
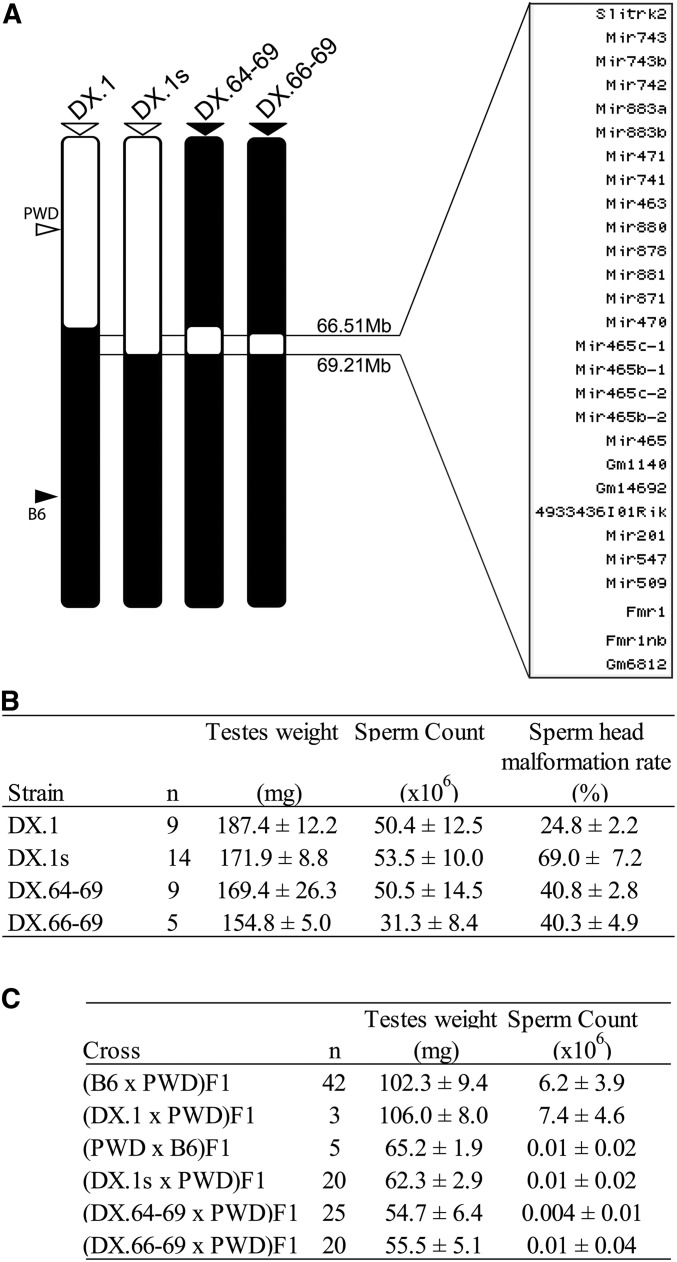
The mice were maintained in the Pathogen-Free Facility of the Institute of Molecular Genetics (Czech Academy of Sciences in Prague). The project was approved by the Animal Care and Use Committee of the Institute of Molecular Genetics AS CR and by the Czech Central Committee for Animal Welfare, and ethically reviewed and performed in accordance with European Directive 86/609/EEC. Subconsomic mouse strains C57BL/6J-ChrX.1PWD/Ph (abbreviated B6.DX.1) and C57BL/6J-ChrX.1sPWD/Ph (B6.DX.1s) were described earlier (Storchová et al. 2004). The C57BL/6J-ChrX.64-69PWD/Ph/ForeJ (B6.DX.64-69) congenic strain was established by backcrossing B6.DX.1s to the B6 strain (Figure 1). The congenic strain C57BL/6J-ChrX.66-69PWD/Ph (B6.DX.66-69) was prepared by the new CRISPR/ Cas9 Hstx2-targeting method. The PWD/B6 composition of the Chr X is depicted schematically in Figure 1 for each consomic strains.
Figure 1.
Mapping of hybrid male sterility Hstx1 and Hstx2 loci in subconsomic and congenic strains. (A) Schematic view of the chromosome X architecture in subconsomic and congenic strains B6.DX.1, B6.DX.1s, B6.DX.64-69, and B6.DX.66-69. The PWD and B6 origin of chromosomal intervals is depicted in white and black. The list of protein coding genes, noncoding RNAs, and miRNAs spanning the interval of the newly defined Hstx2 locus (66.51–69.21 Mb) is shown. (B) Hstx1 locus mapping. Fertility parameters of subconsomic and congenic males; the testes weight (weight of wet testes pair in milligrams), the sperm count (number of sperms in millions per pair of epididymes), and frequency of malformed sperm heads (in percent). (C) Hstx2 locus mapping. Fertility parameters of the (B6 × PWD) F1 and the reciprocal (PWD × B6) F1 hybrid males, and F1 male progeny of crosses of B6.DX.1, B6.DX.1s, B6.DX.64-69, and B6.DX.66-69 congenic females with PWD males are presented as mean ± SD; n, number of analyzed males.
Genotyping, fertility parameters, and histology
Genomic DNA was prepared from tails by NaOH method (Truett et al. 2000). The X chromosome recombinants in the backcross 1 (BC1) populations were genotyped by PWD/B6 allele-specific microsatellite markers (Supplemental Material, Table S1). Recombination breakpoints were determined by Sanger-DNA sequencing of the PCR amplicons carrying informative PWD/B6 SNP polymorphism(s). Genotyping of the new B6.DX.66-69 strain by microsatellite markers, Sanger DNA sequencing, and next-generation sequencing showed the maximum and minimum extent of the PWD sequence on Chr X. The Fmr1nb deletion was confirmed using primers: forward 5′CAGGAGGTTCTGGACTGCTC 3′ and reverse 5′TGAAGTCCAGAAGCCAAACC 3′. All experiments were performed with at least three animals per group. Cytological and histological experiments were performed on males between 8 and 10 weeks of age, with the exception of the males after fertility test.
Quantitative Reverse Transcription-PCR (RT-qPCR) analysis
Total RNA was extracted from testes by TRI reagent #T9424 (Sigma, St. Louis, MO) according to manufacturer’s instructions. The RNA was reverse transcribed using MuMLV-RT (28025-013; Invitrogen, Carlsbad, CA). Quantitative real-time PCR was performed with the Light Cycler DNA Fast Start Master SYBR Green I kit (Roche) in a Light Cycler 480 Instrument II at Tm = 60°. The sequences of primers for Fmr1nb were: Fmr1nb-F – 5′-TCCTGGGATTTCTGCCTATG-3′, Fmr1nb-R – 5′-CCTTCAACATCCTGTTCATCC-3′; and the primers for Actin-b were: Actb-F – 5′-CTAAGGCCAACCGTGAAAAG-3′, Actb-R – 5′-ACCAGAGGCATACAGGGACA-3′. The Fmr1nb expression values were normalized to Actin-b expression.
Western blotting
Whole testes were snap-frozen in liquid nitrogen before extraction buffer with protease inhibitors (1836153; Roche) and bensonase (1.01654.0001; Merck) was used to homogenize the tissue (see supplementary Reagent Table). After a 30 min incubation, 2% SDS was added and the mixture was heated at 95° for 20 min. Total protein concentration was measured using the Pierce BCA Protein Assay Kit (#23225; Thermo Scientific). The protein samples were then size-separated by electrophoresis on a gradient Bolt 4–12% Bis-Tris plus gel (NW04120BOX; Invitrogen), transferred onto a polyvinylidene difluoride membrane, and blocked with TBST with 5% BSA overnight. Primary antibodies against FMR1NB (sc-246953, goat polyclonal; Santa Cruz Biotechnology) and alpha-tubulin (66031-1-Ig, mouse monoclonal; Proteintech) were used at the 1:1000 and 1:2000 dilutions, respectively. Secondary antibodies (a donkey anti-goat IgG-HRP antibody, sc-2020; Santa Cruz Biotechnology, and a horse anti-mouse IgG-HRP antibody, 7076; Cell Signaling Technology) conjugated to HRP were used at 1:10000 dilution. Western Blotting Substrate (#32106; Pierce ECL Plus) was used for detection of HRP enzyme activity. Images were captured using the Bio-Rad ChemiDoc MP Imaging System and processed with ImageLab software (Bio-Rad, Hercules, CA).
Immunofluorescence microscopy
Meiotic chromosome spreads were performed as previously described (Anderson et al. 1999) with minor modifications. Briefly, the testes were dissected and transferred to 1 ml of RPMI (Sigma). Sucrose (0.1 M) was used as a hypotonic solution and cells were dropped onto a slide with 1% paraformaldehyde containing protease inhibitors (1836153; Roche). After 3 hr at 4° slides were washed and blocked with 0.5 × blocking buffer (1.5% BSA, 5% goat serum, 0.05% Triton X-100) containing protease inhibitors (1836153; Roche) for 1 hr at 4°. Primary antibodies (listed in supplementary Reagent Table) were added and the slides were incubated overnight in a humid chamber at 4°. The slides were then incubated with secondary antibodies conjugated to fluorophores (supplementary Reagent table) for 1 hr at 4°. The slides were mounted with Vectashield mounting medium containing DAPI (H1200). The immunofluorescence images were observed by Nikon Eclipse ×400 epifluorescence microscope with single band-pass filters for excitation and emission of infrared, red, blue, and green fluorescence (Chroma Technologies) and ×60 Plan Fluor objective (MRH00601; Nikon, Garden City, NY). The images were captured using a DS-QiMc monochrome CCD camera (Nikon) and NIS Elements processing program (NIS-Elements Microscope Imaging Software). The images were adjusted using Adobe Photoshop (Adobe Systems).
Construction of Fmr1nb-specific TALEN and generation of transgenic mice
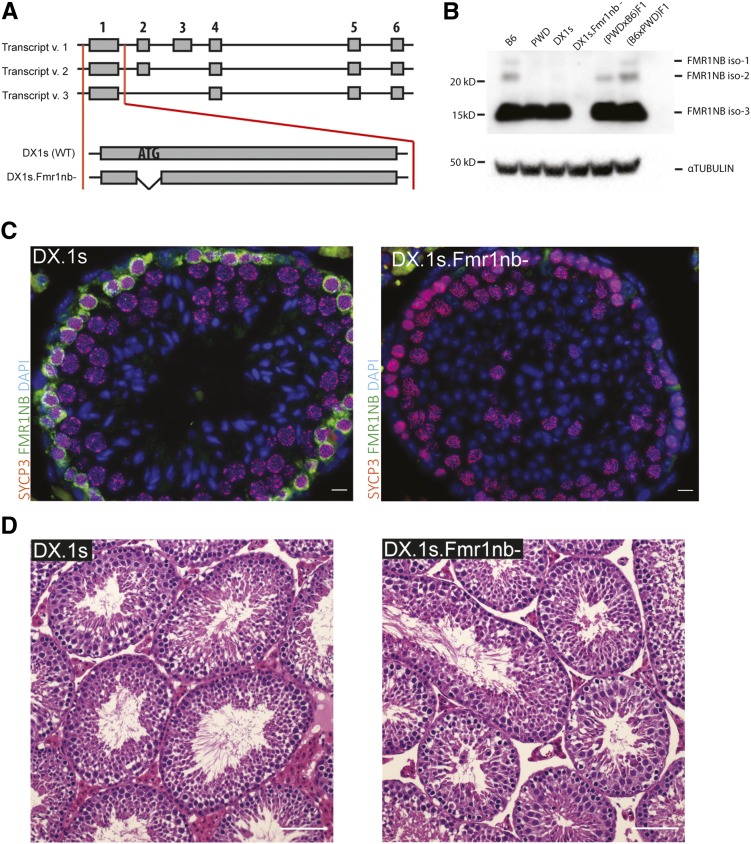
TALEN nucleases were designed using TAL Effector Nucleotide Targeter 2.0 (https://tale-nt.cac.cornell.edu/), assembled using the Golden Gate Cloning system (https://international.neb.com/applications/cloning-and-synthetic-biology/dna-assembly-and-cloning/golden-gate-assembly), and cloned into the ELD-KKR backbone plasmid. TALEN containing repeats NN-NN-HD-NG-NN-NN-NG-NG-NI-NN-NI-NN-NI-HD-HD-NG-HD-HD (for 5′ site) and NG-HD-NG-HD-NG-NN-NI-HD-NG-NG-NN-NN-HD-HD-NG-NG (for 3′ site) recognized a locus close to the ATG start codon of Fmr1nb. Each TALEN plasmid was linearized with NotI and transcribed using the mMESSAGE mMACHINE T7 Kit (Ambion). Polyadenylation of resulting messenger RNAs (mRNAs) was performed using the Poly(A) Tailing Kit (Ambion); the mRNA was purified with RNeasy Mini columns (Qiagen, Valencia, CA). TALEN mRNAs were diluted in nuclease free water and kept at −80°. Transgenic mice were generated in the transgenic facility of the Institute of Molecular Genetics by injecting purified mRNA of Fmr1nb-specific TALEN into male pronuclei of one-cell embryos of C57BL/6N or B6.DX.1s origin. Mice positive for mutations were identified by PCR reaction with Fmr1nb2outF and Fmr1RightBsrI primers followed by NspI digestion. Specific genome mutations were identified by PCR fragment sequencing. Twenty-three mouse founders (F0), each carrying a mutated allele of the Fmr1nb gene, were generated. After outcrossing the F0 mice to C57BL/6N or to B6.DX.1s we obtained five B6.Fmr1nb− mouse strains and three B6.DX.1s.Fmr1nb− strains with stable deletion mutations. Here we used two lines, the B6.Fmr1nbem1ForeJ line carrying 236 bp long deletion over the ATG start codon of the Fmr1nbB6 allele, and the B6.DX.1s.Fmr1nbem1ForeJ line carrying 19 bp long deletion over the ATG start codon of the of Fmr1nbPWD allele.
Preparation of CRISPR-Hstx2 and SPO11-Cas9 constructs, and generation of transgenic mice
To place the Cas9 nuclease under the control of the SPO11 promoter, the SPO11 coding region was replaced by a mouse codon-optimized Cas9 open reading frame in an SPO11-carrying bacterial artificial chromosome (BAC) clone (RP23-20N4, distributed by BACPAC Resources, Oakland, CA) by a marker-less GalK double-selection system via liquid culture recombineering as described (Sharan et al. 2009). Homology arms for the SPO11 BAC were introduced by PCR with Phusion polymerase (New England Biolabs, Frankfurt am Main, Germany). The 1.3 kbp PCR product was purified with a Gel Extraction Kit (QIAGEN) and confirmed by Sanger sequencing. The Cas9 cassette was produced by excision from plasmid MLM3613 (#42251; Addgene, Watertown, MA) by enzymes SacII and MssI (Thermo Fisher Scientific, Schwerte, Germany) and purified by gel extraction. The homology arms were added by PCR amplification and Phusion polymerase. The CRISPR plasmid pX260 was obtained (#42229, Addgene plasmid, a gift from Feng Zhang; Cong et al. 2013) and the CRISPR protospacers corresponding to the Hstx2 loci were cloned according to instructions from the Zhang Laboratory (https://media.addgene.org/cms/filer_public/e6/5a/e65a9ef8-c8ac-4f88-98da-3b7d7960394c/zhang-lab-general-cloning-protocol.pdf). Briefly, long oligonucleotides were ordered as Ultramers (oligos 20–21; Integrated DNA Technologies, Coralville, IA) for the following three target regions flanking the Hstx2 locus: a sequence 2.2 Mb upstream of the Ctag2 gene (Chr X: 65,069,229–65,069,258); an intergenic sequence between the Mir465 cluster and Gm1140 predicted protein coding gene (Chr X: 67,052,342–67,052,371); and a sequence 4 kbp upstream of the Aff2 gene (Chr X: 69,356,143–69,356,172). After phosphorylation (T4 Polynucleotide Kinase, New England Biolabs) and annealing by temperature ramping from 95° to 30 sec by −0.1°/min increments, the duplexes were ligated into the BbsI site of the cut pX260 plasmid (New England Biolabs) and transformed into DH5-Alpha Escherichia coli cells. The protospacer-containing plasmids were further modified by excising the Cas9 open reading frame with PstI (New England Biolabs). Each final plasmid contains the U6 promoter, protospacer, the 1H promoter, and the trans-activating CRISPR RNA. These were sequence-verified before transgenic injection. The CRISPR constructs and SPO11-Cas9-BAC construct were generated in Tubingen by the laboratory of Y.F.C. The BAC transgene was injected to the pronuclei of 1-day-old mouse embryos and the founders were generated in the laboratory of R.S. in Vestec.
Bionano optical mapping
We generated optical maps for two markers (BspQ1 and DLE-1) across the whole genome of five different mice, from two mouse subspecies: C57BL/6J (B6) and C57Bl6Crl (B6N) of M. m. domesticus and PWD/Ph (PWD) and PWK/Ph (PWK) of M. m. musculus origin. Two females were from the congenic C57BL/6J-ChrX.64-69PWD/Ph strain (B6.DX64-69), carrying a small portion of Chr X including the hybrid sterility Hstx2 locus from PWD/Ph on C57BL/6 background. First, megabase-scale high-molecular-weight (HMW) DNA was extracted according to the Saphyr Bionano Prep Animal Tissue DNA Isolation Soft Tissue Protocol (Document Number: 30077, Revision B). Briefly, cell nuclei were isolated from splenic tissue and embedded in agarose plugs. DNA in plugs was purified with Proteinase K and RNAse, then HMW genomic DNA was extracted from the agarose plugs using agarase, and purified by drop dialysis. HMW DNA was resuspended overnight before quantification with the Qubit BR dsDNA assay, then kept at 4° until labeling. Each sample was labeled at the recognition sites NtBspQ1 (GCTCTTC) and DLE-1 (CTTAAG), respectively, using two different methylation insensitive assays. The Bionano nicking, labelling, repairing, and staining protocol was used to label NtBspQ1 (Document Number: 30206, Revision C), and was performed on 900 ng of purified HMW DNA for each mouse. The Bionano direct labelling and staining protocol (Document Number: 30024, Revision I) was performed on 750 ng of DNA to label all DLE-1 recognition sites. After an initial clean-up step, the labeled HMW DNA was prestained, homogenized, and quantified with the Qubit HS dsDNA assay, before using an appropriate amount of backbone stain YOYO-1. The molecules were then imaged using the Bionano Saphyr System (Bionano Genomics, San Diego, CA). We obtained high-quality optical reads for both labeling techniques. For example, for the nicking, labelling, repairing, and staining labeling produced an average of 437 Gbps of reads, which were longer than 150 kbps and have a minimum of nine label. It achieved an average N50 length of 0.3137 Mbp with an average label density of 14.82 labels per 100 kbp. Similarly, the direct labelling and staining labeling achieved an average output of 389 Gbps (≥150 kbp and minSites ≥9), an average N50 length of 0.2663 Mbp and an average label density of 13.72/100 kbp. (Individual outputs were collected for each animal and labeling technique in Table S2). The presence of in silico recognition sites for each enzyme recognition site in the genome was used to compute separate in silico optical maps for each labeling enzyme, for the mm10 genome (Table S3).
Detection and quantification of apoptotic cells: terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay
The males were killed and the testes dissected from, and fixed in 4% paraformaldehyde overnight at 4°. Testes were dehydrated and embedded in paraffin. Paraffin sections at 3 μm thick were deparaffinized. To perform antigen retrieval for immunohistochemistry, the slides were incubated in Citrate Antigen Retrieval solution for 15 min at pH 6.0. The slides were processed as for immunofluorescence. The apoptotic cells in the tissue sections were determined by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL), using in situ DeadEnd Fluorometric detection kit (G3250-PROMEGA, Madison, WI) according to technical protocol (#TB235). TUNEL-treated testicular sections were mounted in Vectashield with DAPI to watch the nuclei. Images were captured from a Nikon E-400 Eclipse fluorescence microscope and captured with a Ds-Qi_Mc1 CCD camera (Nikon). The images were processed and TUNEL-positive cells counted by the NIS Elements picture analyzer, and processed using Photoshop (Adobe).
Fertility test
Each male was mated with one 8-week-old C57BL/6J virgin female for 3 months, during which the numbers of neonatal pups sired by B6.DX.1s.Fmr1nb− and B6.DX.1s males were recorded.
Data availability and statistics
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and in the supplemental material. The optical mapping data sets are available from L.O.-H. or K.K.U. upon reasonable request. Statistical analyses were performed by unpaired two-tailed t-test, if not indicated otherwise. Statistical significance was set at P values of * 0.05,** 0.01, and *** 0.005. Data were processed and plotted by GraphPad Prism version 6.00 (GraphPad Software, San Diego, CA; www.graphpad.com). Other types of statistical analyses are described within the text and in the corresponding figure legends. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9874460.
Results
Hstx2 locus is a recombination cold spot
The Hstx2 locus was initially defined as a 4.7 Mb PWD interval present in B6.PWD-Chr X.1s (B6.DX.1s), but absent in the partially overlapping B6.PWD-Chr X.1 (abbreviated B6.DX.1) congenic strain. (Storchová et al. 2004; Bhattacharyya et al. 2014) (Figure 1A).
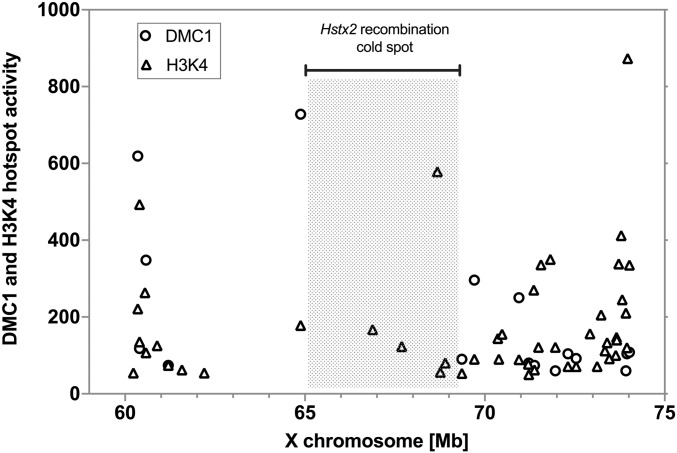
Here, we specified the PWD/B6 distal border of B6.PWD-Chr X.1s by next-generation sequencing to Chr X: 69.21 Mb narrowing the Hstx2 locus to 4.3 Mb of the PWD sequence (Figure 1A). Admittedly, such subtraction mapping could not exclude the possibility that some additional genetic information in the proximal 64.9 Mb of the PWD sequence may contribute to the genetic factors situated within Hstx2 locus. To reduce the size of Hstx2 locus and to check the possible role of the proximal region of the XPWD sequence, 52 new recombinant X chromosomes were generated in three BC1 populations (Table 1). Genotyping of 168 (B6.DX.1s × B6) × B6 BC1 mice yielded 51 recombinants with crossovers spanning the proximal region of Chr X. A new C57BL/6J-ChrX.64-69PWD/Ph congenic strain (abbreviated B6.DX.64-69) derived from this backcross carried only 4.34 Mb of the PWD sequence (Chr X: 64.87–69.21 Mb; mouse genome assembly GRCm38.p6) (Figure 1A). However, not a single recombination occurred in the Hstx2 locus tracked by markers at Chr X: 65.10 and 69.08 Mb (Table 1). In the second backcross experiment, the B6.DX.51-69 subconsomic, which carries PWD sequence in the interval 51–69 Mb was used, but again no recombinant among 111 BC1 animals was found within the Hstx2 locus. Finally, in an attempt to change the pattern of the recombination hotspots, the B6.Prdm9Hu strain carrying the “humanized” PRDM9 with ZnF array from the human PRDM9A allele (Davies et al. 2016) was used in (B6.Prdm9Hu × B6.DX.64-69) × B6 backcross. No recombinant was found within the Hstx2 locus among 369 BC1 animals. The absence of crossovers could occur due to the lack or inaccessibility of PRDM9 binding sites, the failure of SPO11 protein to target these sites and induce DNA DSBs, or because the repair of such DSBs is implemented exclusively by noncrossovers. The available data on female B6 meiosis (Brick et al. 2018) showed reduced occurrence of PRDM9-dependent H3K4me3 hotspots and absence of DMC1 hotspots within the Hstx2 locus (Figure 2), suggesting the virtual disappearance of SPO11-generated DNA DSBs as a mechanism of recombination suppression. Remarkably, in male meiosis the strong suppression of DMC1 hotspots [data from Davies et al. (2016)] over the Hstx2 locus observed in (PWD × B6) and (B6 × PWD) reciprocal F1 hybrids was attenuated in PWD and B6 parental strains (Figure S1). To conclude, no recombinant in the Hstx2 region was found among 648 BC1 mice, although 15 recombinants would be expected (P = 2.495 × 10−7, binomial test) based on the 0.526 cM/Mb mean recombination rate in the adjacent Chr X: 7.36–65.10 Mb proximal region. The recombination cold spot overlaps with the interval of low PRDM9 histone methyltransferase activity and strong suppression of DNA DSB hotspots.
Table 1. Localization of PWD/B6 recombination events on the X chromosome.
| Number of recombination events (N) in the specific X chromosome intervals / recombination ratea,b (cM/Mb) | ||||||
|---|---|---|---|---|---|---|
| Backcross (BC1) | Number of BC1 (n) | X:7.36–65.10 Mb | X:7.36–36.20 mb | X:36.20–59.66 Mb | X:59.66–65.10 Mb | X:65.10–69.08 Mb |
| DX.1s × B6) × B6 | 168 | 51 / 0.526a | 17 / 0.351a | 30 / 0.761a | 4 / 0.438a | 0 |
| (DX.51-69 × B6) × B6 | 111 | N.D. | N.D. | N.D. | 1 / 0.166a | 0 |
| (DX.64-69 × B6.P9Hu/Hu) × B6c | 369 | N.D. | N.D. | N.D. | N.D. | 0 |
The recombination rate (cM/Mb) was calculated from the number of recombination events (N) and the number of BC1 animals tested (n) using the length (L) of a specific region on the X chromosome.
Microsatellite PCR primer sequences used for genotyping are listed in Table S1.
The B6.Prdm9Hu/Hu mouse strain carries Prdm9Hu/Hu on a B6 background, which was engineered by replacing the PRDM9B6 zinc-finger array with the human “B-allele” zinc finger array (Davies et al. 2016). B6.Prdm9Hu/Hu was crossed with B6.DX.64-69, and the female progeny was backcrossed with B6 males.
Figure 2.
Activity of PRDM9-dependent H3K4 methylation and DMC1-marked DNA DSBs in female meiosis. The DMC1 and H3K4me3 hotspots plotted within the Hstx2 locus and the adjacent regions of chromosome X (mm10 genome). The strong DMC1 hotspots coupled with H3K4 methylation lie outside the Hstx2 region (shaded), which contains only H3K4 methylation marks. Data extracted from Brick et al. (2018); visualized are hotspots with activity >50.
Targeting homologous recombination to Hstx2 by CRISPR/Cas9
Because the Hstx2 locus behaved as a cold spot of recombination, we attempted to bring the recombination machinery to this region by means of Cas9 endonuclease-induced DSBs. Two transgenic lines were prepared, the first carrying Cas9 endonuclease under the control of SPO11 genomic region to ensure exclusive expression of Cas9 at early prophase I of meiosis. The second transgenic strain was generated with the U6-promoter driven CRISPR cassette targeted to three sites within the Hstx2 locus (see Materials and Methods). Next, the double transgenic F1 females (B6.DX.1s.TgSPO11-Cas9 × B6.TgCRISPR-Hstx2) were mated to B6 males to generate the BC1 population. This approach allows the generation of targeted DSB by means of a transgene that can be removed through selective breeding in a B6 backcross design. We found that double transgenic F1 females yielded a 15-fold higher frequency of recombination in the interval spanning 64.8-65.1 Mb immediately adjacent to the Hstx2 locus (10 recombinants in 181 BC1 offspring, 18.42 cM/Mb) compared to previous classical backcrosses (one recombination event in 279 BC1 offspring, 1.19 cM/Mb). However, only one homologous recombination event inside the Hstx2 locus was detected, giving rise to congenic strain B6.PWD-Chr X.66-69 (abbreviated B6.DX.66-69). The new congenic restricts the PWD sequence on Chr X to 2.70 Mb in the 66.51–69.21 Mb interval. Admittedly, all these recombinants occurred within the range bracketed by the guide RNAs but at some distance away from the sites targeted. At this point, we have not determined what may have caused the increase in recombination rate close to but not involving the targeted sites.
Phenotypes of newly defined Hstx1, Hstx2, and Meir1 loci
Hstx1 fertility phenotype:
To check the Hstx1 phenotype the fertility parameters of B6.DX.64-69 and B6.DX66-69 congenic males carrying the shortened 4.34 Mb (Chr X: 64.87–69.21 Mb) and 2.70 Mb (Chr X: 66.51–69.21 Mb) of PWD sequence were compared to B6.DX.1 and B6.DX.1s males carrying 64.9 and 69.2 Mb of proximal PWD sequence (Figure 1, A and B). Both shortened intervals of the PWD sequence reduced testes weight (P < 0.05, t-test) and caused higher frequency of morphologically malformed sperm heads compared to B6.DX.1 (P < 0.01, t-test, Figure 1B). However, compared to B6.DX.1s, the level of teratozoospermia controlled by the 4.34 and 2.70 Mb stretches of PWD sequence was significantly lower (40.8% vs. 69%, P < 0.01, t-test, Figure 1B). Thus, some additional genetic information proximal to the Chr X: 64.87–69.21 Mb interval is necessary to fully reconstruct the Hstx1 phenotype.
Hstx2 fertility and meiotic chromosome asynapsis phenotypes:
To verify the presence of Hstx2 in the newly derived congenic strains, testes weight and sperm count were compared in F1 hybrid males from crosses of PWD males and B6.DX.1, B6.DX.1s, B6.DX.64-69, and B6.DX66-69 females. The quasi-fertile phenotype of (B6.DX.1 × PWD) F1 hybrids contrasted with full sterility of the remaining three types of hybrids as shown by low testes weight (P < 0.0001, t-test) and sperm count (P < 0.0001, t-test, Figure 1C). Thus in contrast to the Hstx1 locus, the shortest version of Hstx2 (Chr X: 66.51–69.21 Mb) was necessary as well as sufficient to fully reconstruct the (PWD × B6) F1 male hybrid sterility phenotype.
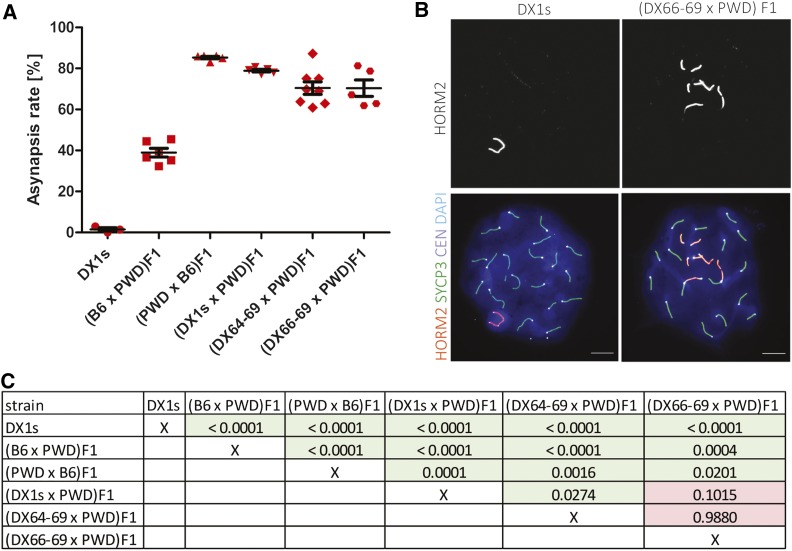
Recently, we have found out that meiotic asynapsis of homeologous chromosomes (homologs from different subspecies) in (PWD × B6) F1 hybrids depends on their subspecific origin and can be abolished by introduction a short stretches (27 Mb or more) of consubspecific homology into a given chromosome pair (Gregorova et al. 2018). Contrary to this chromosome-autonomous cis-control, the substitution of the Hstx2PWD allele for Hstx2B6 in (B6 × PWD) F1 hybrids significantly reduces meiotic asynapsis in trans, while the Prdm9PWD/Prdm9B6 genotype remains the same as in sterile hybrids (Bhattacharyya et al. 2014). To evaluate meiotic chromosome synapsis we visualized the axial elements of partially or fully asynapsed chromosomes by co-immunostaining of HORMA domain-containing protein-2, HORMAD2 (Wojtasz et al. 2012) and synaptonemal complex protein 3, SYCP3, in pachynemas of F1 hybrids carrying different intervals of XPWD (Figure 3). The highest proportion, 85.3 ± 1.3%, of pachynemas affected by asynapsis was observed in the (PWD × B6) F1 hybrid males with intact XPWD chromosome. The frequencies of pachynemas with asynapsis rates 78.9 ± 1.4%, 70.5 ± 8.6%, and 70.49% in three subconsomic F1 hybrids (B6.DX.1s × PWD) F1, (B6.DX.64-69 × PWD) F1, and (B6.DX66-69 × PWD) F1 did not differ from each other, but were significantly lower than in (PWD × B6) F1s (Figure 3A). Importantly, the XB6 chromosome in (B6 × PWD) F1 did not completely eliminate the Prdm9 controlled asynapsis, which reached 38.9 ± 5.2% in (B6 × PWD) F1 hybrid males (Figure 3A). It appears that in (B6 × PWD) F1 hybrid genomic background this level of asynapsis rate could indicate a threshold of azoospermia because (B6 × PWD) F1 hybrid males with <40% asynapsis rate showed 7.2 ± 4.2 × 106 epididymal sperm count, while males of the same genotype with >40% asynapsis were virtually azoospermic (0.12 ± 0.1 × 106 sperm count).
Figure 3.
Pivotal role of the Hstx2 locus in the pachytene asynapsis rate of male F1 hybrids. (A) The mean values of asynapsis rate (± SD) in F1 and B6.DX.1s hybrid males carrying different portions of XPWD. Autosomal asynapsis (frequency of pachynemas with one or more asynapsed autosomes) was examined in 5–8 animals of a given genotype, scoring at least 50 pachytene nuclei per one male. (B) Representative immunofluorescence micrographs show the HORMAD2-positive XY pair in a pachytene spermatocyte of B6.DX.1s congenic male and asynapsed autosomes in (DX.66-69 × PWD) F1 hybrids. Asynapsed chromosome axes are immunostained by HORMAD2 antibody. SYCP3 visualizes lateral elements of synaptonemal complexes. CEN labels centromeric heterochromatin, and DAPI labels nuclear DNA. Bar, 10 µm. (C) Comparisons of the asynapsis rates between individual animal groups were performed by two-tailed t-test, and the P-values are displayed in the table.
To conclude, ∼three-quarters of the Hstx2 effect on Prdm9-controlled asynapsis rate is preserved in the newly reduced 2.70 Mb PWD sequence version (Chr X: 66.51–69.21 Mb); the remaining effect either maps elsewhere on the X chromosome or is the consequence of a hypothetical position effect of the M. m. domesticus genome on the introgressed M. m. musculus sequence.
Meir1 control of global meiotic recombination rate:
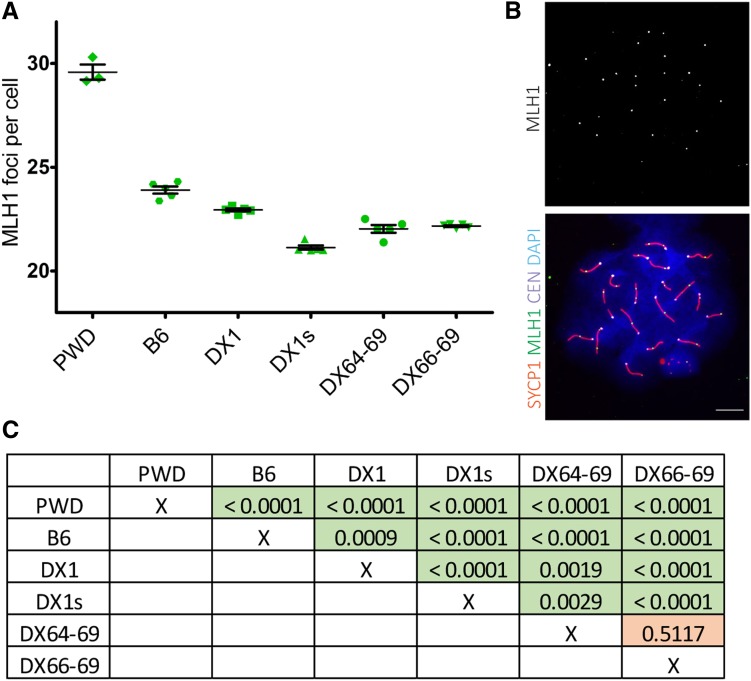
The Meir1 was localized in the Hstx2 interval as the strongest transgressive modifier of the meiotic recombination rate in B6.DX.1s males. The Meir1PWD coming from the high-recombination rate PWD strain lowered crossover frequency in a transgressive manner when introgressed into the B6 genome (Balcova et al. 2016). The crossover frequency determined by counting the MLH1 foci per pachytene spermatocyte revealed that both the 4.34 and 2.70 Mb PWD interval reduced recombination compared to B6 and B6.DX.1, thus behaving as Meir1, but the reduction did not reach the level seen in B6.DX.1s (Figure 4). We conclude that similarly as in the case of the newly defined Hstx1 locus some additional genetic information in the proximal PWD sequence besides the 2.70 Mb interval is necessary to fully reconstruct the Meir1 phenotype (Figure 4, A and B).
Figure 4.
Transgressive effect of the Hstx2PWD allele on crossover rate. (A) The mean crossover rate values (± SD) are shown for the subconsomic and congenic males carrying different portions of the chromosome XPWD on the B6 genetic background. (B) Representative immunofluorescence micrograph visualizing MLH1 foci (green), synaptonemal complex protein 1, SYCP1 (red), centromeric proteins, CEN (white), and nuclear DNA (blue) in the B6.DX.1s late pachytene spermatocyte. Bar, 10 μm. (C) Summary of comparisons of the recombination rates between individual animal groups are shown in the table as P-values analyzed by unpaired two-tailed t-test.
Optical mapping of intersubspecific structural variation within and outside the Hstx2 locus
One possible cause of the recombination cold spot overlapping the Hstx2 locus could be a structural rearrangement, typically an inversion that prevents recovery of viable recombinants. Such structural variants acting as recombination suppressors often enforce reproductive isolation between species, especially when situated on sex chromosomes (Kirkpatrick 2010; Hooper et al. 2018). To elucidate the physical structure of Hstx2 locus we analyzed the region by optical mapping using the Bionano Saphyr platform, a further development of the technique described by (Chan et al. (2018)). As a proof of concept, we examined the Hstx2PWD M. m. musculus introgression in the M. m. domesticus Chr XB6. Indeed, the 64–69 Mb interval of Chr X was easily recognizable in two optical maps from biological replicas of B6.DX.64-69 mice when matched with the reference B6/J in silico map and with the map of a female from the C57BL/6Crl substrain. The structure of the 64–69 Mb interval of Chr X matched most closely the PWD and PWK optical maps, while the flanking intervals matched the B6 optical map. (Figure 5, A–C). To inquire into the overall divergence of the Hstx2 locus as a possible cause of recombination suppression, optical maps of the region of the same size outside the recombination cold spot (Chr X: 59.6–64.0 Mb) was compared to the Hstx2 region (Chr X: 64.8–69.2 Mb) from four mouse strains (B6/N, B6.DX.64-69, PWD, and PWK) by alignment to the mm10 in silico reference (Table 2). Although only 0.08% of the control locus sequence was involved in deletions or insertions in B6/N and B6.DX.64-69, the same 4.3 Mb control interval included 6.92% of deleted or inserted sequence in PWD and PWK. In comparison, the Hstx2 locus (Chr X: 64.8–69.2 Mb) displayed 2 insertions of 8.7 kb and no deletion in the B6/N, representing 0.02% of the sequence, while 4.71% of sequence was either inserted or deleted in B6.DX.64-69, 4.58% in PWD, and 5.90% in PWK. Intraspecific comparison of the same Hstx2 interval yielded 1.11% and 2.40% of sequence involved in PWK and PWD specific inversions and deletions. To conclude, the overall structural dissimilarity is surprisingly high between M. m. musculus and M. m. domesticus subspecies, but unlikely to explain the Hstx2 recombination cold spot.
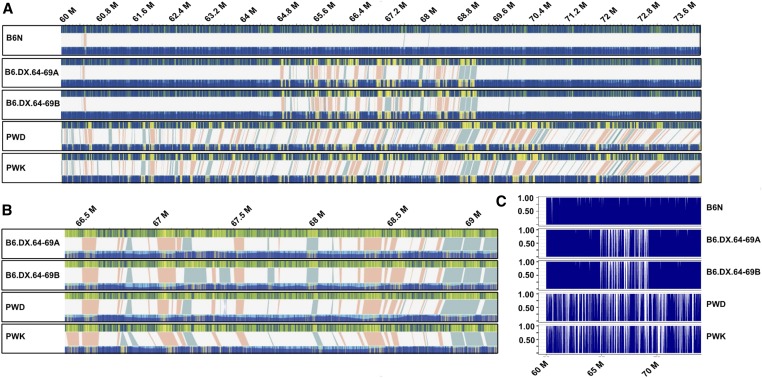
Figure 5.
Structural variants (SVs) in the Hstx2 locus and in flanking regions. Each box contains a comparative analysis of a de-novo optical map (bottom), and the mm10 in-silico reference B6 map (top) of a given individual. (A) Five maps of B6N, B6.DX.64-69A, B6.DX.64-69B, PWD, and PWK spanning Chr X 60–74 Mb (images extracted from Bionano Solve version 3.3_10252018 at maximum resolution). At this overview, individually-labeled restriction sites are not visible. However, matching intervals appear blue on both the reference and de novo map, as labeled restriction sites matching their predicted position in the reference are depicted as blue lines. In contrast, labels found in either the reference or de novo map, but not both, are marked by yellow lines. Therefore, clusters of mismatched labels become visible as yellow blocks. Label patterns are used to predict SVs by the Bionano Solve software. Putative SVs are depicted as shaded areas, connecting the upper reference and lower de novo map. Light red areas represent putative deletions, where labels present in the in silico reference, are absent in the de novo map. In contrast, light blue shaded areas depict putative insertions, where additional labels were found in the de novo map, but not the in silico reference. (B) The same optical maps for B6.DX.64-69A, B6.DX.64-69B, PWD, and PWK, zoomed in to Hstx2 position X: 66.51–69.21 Mb, which is an apparent recombination cold spot. All putative SVs are shown at higher resolution, with deletions in red and insertions in blue. Neither large inversions nor translocations have been predicted for this interval. (C) To quantify the number of labels matching between in silico map and each of the five de novo maps, we counted all labels across Chr X 60–74 M (see Table 2). Proportions of matching labels are plotted per 10 kb nonoverlapping window.
Table 2. Insertions and deletions in the Hstx2 locus compared to control intervals on chromosomes X.
| Mouse strain | Control Chr X coordinates (Mb) | Insertions, n /(kb) | Deletions, n /(kb) | Hstx2 locus coordinates (Mb) | Insertions, n /(kb) | Deletions. n /(kb) |
|---|---|---|---|---|---|---|
| B6.N | Chr X: 59.6–64.0 | 1 / 0.9 | 1 / 2.8 | Chr X: 64.8–69.2 | 2 / 8.8 | 0 |
| B6.DX64-69_A | Chr X: 59.6–64.0 | 1 / 0.9 | 1 / 2.6 | Chr X: 64.8–69.2 | 14 / 94.1 | 26 / 116.3 |
| B6.DX64-69_B | Chr X: 59.6–64.0 | 1 / 0.9 | 1 / 2.8 | Chr X: 64.8–69.2 | 15 / 92.6 | 26 / 111.6 |
| PWD | Chr X: 59.6–64.0 | 22 / 105.7 | 21 / 174.4 | Chr X: 64.8–69.2 | 12 / 85.8 | 29 / 116.4 |
| PWK | Chr X: 59.6–64.0 | 21 / 113.4 | 24 / 192.1 | Chr X: 64.8–69.2 | 14 / 140.3 | 26 / 119.4 |
Optical maps over the Hstx2 region (Chr X: 64.8–69.2 Mb) and the control Hstx2-adjacent interval of the same size (Chr X: 59.6–64.0Mb) from five mouse genome DNA samples, representing four mouse strains, were generated and aligned to the mm10 in silico reference map. Coordinates are given with respect to the position in the mouse genome reference mm10 (Mb), n / (kb) numbers and cumulative sizes of structural variants within the intervals of the same extent in the X chromosome.
Fine-scale screen for the Hstx2-specific structural variants
A structural variant within the Hstx2 locus could be a marker of the Hstx2 candidate gene. Thus we screened for PWD-specific structural variations within the Hstx2 locus because the Hstx2 alleles differ between M. m. musculus PWK and PWD and M. m. domesticus B6 mice (Flachs et al. 2014). We first aligned de novo maps of B6.DX.64-69, PWD, PWK, and C57BL/6Crl to the C57BL6/J in silico reference, generating a quadruple assembly (Figure 5). We then screened for structural variants that occur in B6.DX.64-69 and PWD but not in C57BL/6Crl or PWK. This had to be done semimanually, as due to the large genetic divergence in this interval, relying only on Bionano’s automated algorithms was insufficient. A fine-scale characterization of the refined Hstx2 interval by manual label matching revealed three high-confidence structural variants. The first locus, found between Chr X positions 66.756–66.797 Mb, contains two long terminal repeats (LTRs) in the B6 reference. While PWD and PWK both possess a 4.7 kb deletion of the first LTR, the second LTR locus downstream harbors a 3.1 kb deletion in PWD, also deleting miRNA Mir883b. In contrast, PWK shows a large overlapping 45.0 kb insertion (Figure 6A). The second significant structural variation is located between chromosomal positions 66.819–66.840 Mb, and includes the Mir465 cluster, which appears differentially duplicated in PWK and PWD (Figure 6B). In PWD we observed an insertion of 22.9 ± 4 kb, while the PWK map revealed a shorter insertion of only 16.3 kb. Previously, we found overexpression of the Hstx2 miRNA cluster, particularly of Mir465 in sterile hybrids (Bhattacharyya et al. 2014). A differential duplication could therefore harbor subspecies-specific differences in Mir465 expression, which may confer dosage effects on the regulation of downstream target genes.
Figure 6.
Detailed examination of polymorphic structural variation in the Hstx2 locus. Blue vertical lines represent perfect matches to the predicted B6 in silico optical map (mm10), while yellow vertical lines are additional detected labels that do not match the reference. Structural variants (SVs) between the B6 reference and respective de novo optical map are depicted as colored triangles, deletions in orange, and insertions in blue. At the bottom of the panel, the ENSEMBL tracks for LTRs and genes are shown, with vertical lines representing the interval affected by the SVs depicted in the top panel. (A) The optical maps zoomed to interval at Chr X: 66.75–66.80 Mb, revealing a polymorphic LTR region. Here, PWD possesses two deletions while PWK displays only one deletion, plus an insertion. (B) The optical map zoomed in at interval Chr X: 66.76–66.84 Mb. PWD and PWK both bear insertions, which duplicate the locus containing the Mir465 miRNA cluster, compared to the orthologous region in B6J. These insertions are polymorphic between the two M. m. musculus chromosomes, spanning only 16.2 kb in PWK but 23.3 kb in PWD. (C) Optical map zoomed in at interval Chr X: 67.75–67.81 Mb, which possesses a deletion in PWD only. However, the deletion does not appear to disrupt any known gene.
The third is a homozygous deletion of 4574 ± 9 bp situated at Chr X: 67,787,047–67,795,903. However, this deletion neither interrupts nor deletes any known gene, mRNA/miRNA sites, or transcripts in the available testis transcriptomics data sets (Margolin et al. 2014; Harr et al. 2016; Jung et al. 2019) (Figure 6C). This structural variant is thus an unlikely candidate for harboring Hstx2.
Probing Fmr1nb as an Hstx2 candidate gene
The newly reduced Hstx2 genomic interval incorporates eight protein-coding genes, of which the Fmr1 neighbor (Fmr1nb) appeared as the best potential candidate for the Hstx2 gene.
The priority was based on Fmr1nb expression at early meiotic prophase I (Margolin et al. 2014; Ball et al. 2016; Jung et al. 2019; Ernst et al. 2019) and two nonsynonymous single nucleotide polymorphisms between PWD and B6 parental strains (Table S5). We confirmed almost exclusive expression of Fmr1nb in the testis, with only traces in the spleen and heart (Figure S2A) and found 2.5-fold higher expression in sterile (PWD × B6) F1 adult testis compared to the PWD and B6 parental strains (P < 0.001, P < 0.001; t-test) (Figure S2B). A continuous increase of the mRNA level of Fmr1nb was found in juvenile males at 10, 12, 14, and 20 days of postnatal development; however, all three genotypes showed a similar expression pattern (Figure S2C). The predicted structure of the FMR1NB protein (Figure S3) consists of two cytosolic N- and C-terminal domains, two transmembrane domains, and an extracellular part containing a P-type trefoil domain. The mouse Fmr1nb transcripts occur in three splice variants (ENSMUSG00000062170.12, ENSEMBL) corresponding to three isoforms of FMR1NB protein (Q80ZA7, UniProt) comprising 238, 192, and 166 amino acids, respectively. We found that in the testis, the most abundant is isoform 3 (Figure S3), made up of 166 amino acids. It lacks the complete P-type trefoil domain and most of the extracellular domain. Two FMR1NB nonsynonymous substitutions create exchanges of 31 argininePWD for threonineB6 and 162 leucinePWD for isoleucineB6.
Using fluorescence immunolabeling, we detected the FMR1NB protein on histological sections of the testis of adult B6 males in the cytoplasm and spermatocyte cell membranes. The strongest FMR1NB expression was found at the leptotene and zygotene stages of the first meiotic prophase. The signal decreased in pachynemas, and disappeared in the round and elongated spermatids (Figure S2D).
Fertility phenotypes of Fmr1nb null mutants:
To test the effect of the Fmr1nb null allele on the Hstx1/2 phenotypes, two mouse lines carrying Fmr1nb deletion mutants were generated by TALEN nuclease method (see Material and Methods and Figure 7A). The coisogenic mouse line B6.Fmr1nbem1ForeJ carried 236 bp deletion within the first exon and B6.DX.1s.Fmr1nbem2ForeJ displayed a 19 bp deletion over the ATG start codon (these lines are henceforth called B6.Fmr1nb- and B6.DX.1s.Fmr1nb−). The Fmr1nb mRNA was detectable by quantitative Reverse Transcription-PCR in both transgenic lines as expected because the transcription start site was not affected (not shown). Three FMR1NB isoforms were identified by Western blotting in males carrying Fmr1nbB6 and Fmr1nbPWD alleles, while the FMR1NB protein was missing in the mutant testes (Figure 7B). Intriguingly, the most truncated isoform 3 of FMR1NB was expressed most strongly in the testes of all four genotypes, whereas the longer isoforms iso-1 and iso-2 showed low expression in B6 and (B6 × PWD) F1, and even lower in (PWD × B6) F1 sterile hybrids and no expression in PWD and B6.DX.1s (Figure 7B). Immunohistochemistry of testes of adult wild type males showed high expression of FMR1NB in spermatogenic cells in early stages of meiotic prophase I. The protein was missing in histological sections from the B6.DX.1s.Fmr1nb− knockout males (Figure 7C), but the overall composition of testicular tubules did not show any apparent changes (Figure 7D).
Figure 7.
Generation of Fmr1nb null allele. (A) Transcript variants of Fmr1nb are shown, comprising six, five, and four exons. Deletion mutants of B6 and PWD alleles of Fmr1nb were generated by TALEN nuclease pair constructs targeted to the ATG start codon of Fmr1nb in C57BL/6N (B6N) laboratory strain and C57BL/6J-ChrX.1sPWD/Ph (B6.DX.1s) subconsomic strain, respectively. (B) FMR1NB protein levels in the testes of males of indicated genotypes were assessed by Western blot. None of the three isoforms of FMR1NB was detectable in the Fmr1nb-deficient strain. Loading control was alpha-tubulin. (C) Immunolabeling of FMR1NB and SYCP3 in histological sections of testis of B6.DX.1s and B6.DX.1s.Fmr1nb. FMR1NB is shown in green, SYCP3 is shown in violet, and DAPI is shown in blue. Bar, 10 μm. (D) The histological sections of testes of the B6.DX.1s and B6.DX.1s.Fmr1nb- genotype stained with hematoxylin and eosin displayed no changes in morphology and occurrence of the meiotic cells. Bar, 100 μm.
The Fmr1nb− males bred successfully, but their mean litter size was significantly lower than litter size of males carrying the wild-type alleles (Figure S4). The B6.Fmr1nb− and B6 males did not differ significantly in the testes weight (165.8 ± 22.2 vs. 180 ± 16.8 mg; P = 0.133, t-test) or in the sperm count (54.5 ± 18.2 × 106 vs. 73.3 ± 17.5 × 106; P = 0.073, t-test) (Figure S5, A and B), but B6.Fmr1nb− displayed a significantly higher proportion of malformed sperm heads (32.9 ± 8.6 vs. 19.8 ± 4.2%; P < 0.05, t-test) (Figure S5C).
The effect of the Fmr1nbPWD null allele was stronger on the B6.DX.1s genetic background. Testes weight of the B6.DX.1s.Fmr1nb− males was significantly lower than in B6.DX.1s (148.1± 16.1 vs. 171.9 ± 8.8; P < 0.001, t-test) (Figure S5D) and the sperm count was lower in B6.DX.1s.Fmr1nb− than in B6.DX.1s males (44.2 ± 12.8 vs. 53.5 ± 10 × 106; P < 0.05, t-test) (Figure S5E). Furthermore, the B6.DX.1s.Fmr1nb− males showed significantly higher proportion of malformed sperm heads than B6.DX.1s control males (76.9 ± 8 vs. 69 ± 7.3%; P < 0.05, t-test) (Figure S5F). The frequency of apoptotic cells in seminiferous tubules assessed by fluorescence TUNEL labeling of histological sections was higher in the B6.DX.1s.Fmr1nb− males (3.36 ± 0.23) compared to B6.DX.1s males (1.46 ± 0.39, P < 0.005; Figure S6, A and B).
To inquire whether Fmr1nb interacts with the Hstx2 phenotype, the hybrid males were analyzed for the testes weight and sperm count. Neither the Fmr1nbB6 nor Fmr1nbPWD null allele rescued hybrid sterility; on the contrary, the Fmr1nbPWD null allele in (B6.DX.1s.Fmr1nb− × PWD) F1 hybrid males significantly reduced the testes weight when compared to (B6.DX.1s × PWD) F1 control males (59.3 ± 4.1, and 67.7 ± 3.5 mg; P < 0.001, t-test, see Table S6).
To conclude, the Fmr1nb on the B6 genetic background is necessary for the normal course of spermiogenesis, with stronger effects in the PWD context of B6.DX.1s Fmr1nb congenic males. In intersubspecific F1 hybrids, however, the absence of FMR1NB modifies neither the intrameiotic arrest nor hybrid sterility.
Discussion
Two-gene architecture of hybrid sterility
Our model of hybrid sterility based on (PWD × B6) F1 hybrids is composed of three main components: the Prdm9 gene, subspecific divergence of homeologous autosomes, and the Hstx2 locus. It differs in its simplicity from the complex genetic control reported by other studies using the same combination of house mouse subspecies (Tucker et al. 1992; Payseur et al. 2004; Macholán et al. 2007, 2011; Duvaux et al. 2011; Janoušek et al. 2012; Turner et al. 2012).
PRDM9 protein activates a high number of asymmetric DNA DSBs in prophase I of (PWD × B6) F1 primary spermatocytes, so that PRDM9B6-determined hotspots occur mostly on the PWD chromosome and vice versa (Davies et al. 2016; Smagulova et al. 2016; Hinch et al. 2019). The main reason of hotspot asymmetry is the evolutionary erosion of the PRDM9 DNA binding sites (Baker et al. 2015). The predominant role of PRDM9-induced DSB asymmetry in this model of hybrid sterility was emphasized by complete recovery of spermatogenesis and fertility of the (PWD × B6) F1 hybrids when the zinc-finger array of PRDM9B6 was replaced with the human orthologous sequence (Davies et al. 2016). The hotspot erosion and meiotic failure disappeared because PRDM9Hum, in contrast to PRDM9B6, has never before been in contact with mouse genome. Full recovery can be also achieved by homozygosity for the Prdm9PWD allele (Dzur-Gejdosova et al. 2012).
The importance of cis-interaction between homeologous chromosomes was shown in intersubspecific backcross males where asymmetry disappeared in conspecific autosomal intervals (PWD/PWD or B6/B6) (Gregorova et al. 2018), which initially had been misinterpreted as multiple hybrid sterility QTL (Dzur-Gejdosova et al. 2012). The major meiotic consequences of DSB hotspot asymmetry include persistent DNA DSBs and meiotic asynapsis, both leading to apoptosis (Davies et al. 2016; Gregorova et al. 2018; Wang et al. 2018).
The role of Hstx2 is apparent from attenuated manifestation of the Prdm9-driven asynapsis phenotype and subsequent meiotic arrest in the reciprocal (B6 × PWD) F1 hybrids. Previously we excluded mitochondrial inheritance, the Y chromosome, and genomic imprinting as a cause and identified the Hstx2 locus on Chr X to be the culprit (Dzur-Gejdosova et al. 2012; Bhattacharyya et al. 2014). We have not yet identified the genetic factor behind the Hstx2 locus, so it is difficult to guess why the same pair of homeologous autosomes with the same ratio of asymmetric/novel DMC1 hotspots (Davies et al. 2016; Smagulova et al. 2016) differs so strongly in DSB repair and meiotic synapsis in the reciprocal hybrids. Three main options can be considered: Hstx2 could extend the time window necessary to accomplish the repair of mutated PRDM9 binding sites, it could reduce the sensitivity of putative mismatch repair anticrossover activity to sequence heterology (Spies and Fishel 2015), or it may facilitate the switch of repair partner bias by sister chromatid homologous recombination (Garcia-Muse et al. 2019).
A recombination cold spot overlaps the Hstx2 locus
Empirical results from rabbits and mice strongly indicate that genomic regions with suppressed recombination are more differentiated and tend to accumulate reproductive isolation genes (Nachman and Payseur 2012). Ortiz-Barrientos et al. (2016) predicted that “…regions of low recombination will tend to harbor genes for various forms of reproductive isolation, as well as modifiers of recombination during the early stages of speciation…” Indeed, the hybrid sterility genetic locus Hstx2 meets both of these predictions since it is situated in a recombination cold spot and carries Meir1, an underdominant modifier of meiotic recombination rate. Moreover, Hstx2 operates at early stage of speciation when reproductive isolation of Mus musculus subspecies is still incomplete. In an attempt to reduce the size of the Hstx2 locus by genetic recombination, we used three genetic backcrosses, one of them employing the “humanized” Prdm9Hu allele known to determine a DSB hotspots landscape entirely different from the Prdm9dom2 allele. However, none of these crosses was able to break the 4.3 Mb cold spot. The only recombinant which reduced Hstx2 to 2.7 Mb was obtained in a backcross where SPO11-driven Cas9 nuclease was targeted by CRISPR to Hstx2 interval in female meiotic prophase. Because the recombination breakpoint lies outside the targeted sites and outside SPO11-oligo hotspots (Lange et al. 2016), the possibility that this unorthodox crossover arose by repairing a Cas9-generated DSB seems unlikely.
The cold spots of recombination are often caused by heterozygosity for large structural variations, often inversions, and these “frozen” blocks can harbor genetic factors important for reproductive isolation (Coyne and Orr 2004; Fuller et al. 2018). In contrast to inversions, large copy number variants can be associated with closed chromatin and reduced gene expression in germ cells, suggesting a constitutive effect on recombination by altering chromatin structure (Morgan et al. 2017). A constitutive cold spot model seems to better fit to the Hstx2 locus based on the low histone methyltransferase activity of PRDM9 and strong depression DNA DSB hotspots in the Hstx2 region in female meiosis (Brick et al. 2018). The conclusion is also supported by recombination data from 73 sequenced inbred strains of the Collaborative Cross project (Collaborative Cross Consortium 2012; Srivastava et al. 2017). We found that none of the sequenced strains carries a single recombination event within the 8 Mb (Chr X: 61.8–70.3Mb) interval spanning Hstx2, while 9 and 10 recombinants occurred in the adjacent 8 and 6 Mb regions (http://csbio.unc.edu/CCstatus/index.py?run=CCV). In the Diversity Outbred project that used the same eight parental strains strong association between copy number variants regions and recombination cold spots was found (Morgan et al. 2017).
The present results based on optical mapping of a single genomic region indicate that genome-wide optical mapping can greatly contribute to elucidating the “fluidity” of noncoding sequences between related species as well as to clarify the greater differentiation of X chromosome compared to the autosomes (Hammer et al. 2008; Presgraves 2018). The optical mapping enabled unprecedentedly high resolution of the Hstx2 locus physical map in the M. m. musculus (PWD) and M. m. domesticus (B6) genome, but did not provide evidence of an inversion that could explain the recombination cold spot. Provided that the Hstx2 phenotype is associated with a structural variant, then it should be visible in the PWD sequence, but not in PWK or B6. Three such PWD-specific variants have been found, but only one of them, including a cluster of miRNA genes, can directly implicate functional consequences related to Hstx2. To conclude, these results together with the recombination data from the Collaborative Cross project show that the Hstx2 locus is located within a constitutive recombination cold spot with the chromatin structure poorly accessible to the recombination machinery.
Hstx1 and Meir1 genetic factors located in the newly defined Hstx2 locus
The Hstx1 was mapped on Chr X as a QTL common for several male fertility phenotypes following the transgression of Chr XPWD into the B6 genome. In the same experiment the suppression of recombination in the Chr X: 59.65–72.41 Mb interval (DXMit140–DXMit199) was noticed for the first time and the QTL for number of offspring, testes weight and sperm morphology was mapped to the interval near the DXMit199 marker (Storchová et al. 2004). Later, the X-linked Hstx2 locus controlling the early meiotic arrest in (PWD × B6) F1 hybrids was localized in the same area (Bhattacharyya et al. 2014).
The effect of Meir1 genetic factor on meiotic recombination is paralleled by the male-limited transgressive/underdominant effect of Hstx2 on hybrid sterility, since the Meir1PWD allele of the high recombination rate PWD parent causes downregulation of crossover rate after introgression in the low recombination rate B6 strain. Thus the localization of Meir1 within the Hstx2 locus indicates a link between meiotic recombination and hybrid sterility (Balcova et al. 2016).
In the course of positional cloning of QTL in mice and other organisms, the QTL effect sometime weakens or even disappears with narrowing down the critical region. In most instances the weakening of QTL’s effect was explained by several physically linked small effects (Flint et al. 2005). We have seen some weakening of all three genetic factors mapping to the 2.70 Mb interval, which can be explained in the same manner. Alternatively, an epigenetic positional cis-effect could be involved.
The role of the Fmr1 neighbor (Fmr1nb) gene in male fertility
In the present study, we selected the Fmr1nb gene as the most promising candidate of Hstx2 based on its expression pattern during meiotic prophase I and two missense polymorphisms between PWD and B6 alleles. Although the role of Fmr1nb in male fertility was challenged in a study of 54 testis-expressed genes (Miyata et al. 2016), we showed that the Fmr1nb null allele induced apoptosis of spermatogenic cells, elevated the frequency of sperm head malformations and decreased sperm counts. A similar general function in cellular proliferation and apoptosis was described for human FMR1NB in glioma cells (Wu et al. 2018). The phenotype of Fmr1nb null mutants, in particular the occurrence of abnormal sperm heads mimics the Hstx1 effect. However, since teratozoospermia is a common pathological phenotype with many possible causes, and given that the null allele of Hstx1 does not eliminate fertility phenotype differences between B6.DX.1 and B6.DX.1s, we consider Fmr1nb an unlikely candidate for Hstx1. Moreover, since the lack of FMR1NB protein did not modulate the pachytene arrest in (PWD × B6) F1 hybrids, we also do not consider Fmr1nb as candidate of Hstx2.
miRNA cluster variation within the Hstx2 locus
The Hstx2 locus harbors an evolutionary conserved group of 12 testis specific miRNAs residing in two clusters of 19 and 3 miRNAs situated between Slitrk2 and Fmr1 protein coding genes. The conserved location of these miRNA clusters anchored between the two X-linked genes was reported in 12 mammalian species (Zhang et al. 2019). In spite of the interspecific variability in number of individual miRNA genes, the levels of testicular miRNAs are under regulatory constrains because depletion as well as overexpression of specific miRNA molecules or miRNA clusters can be deleterious for male fertility (Royo et al. 2015; Ota et al. 2019). The X-linked miRNAs are actively transcribed in spermatogonia and suppressed by meiotic sex chromosome inactivation in pachytene spermatocytes (Royo et al. 2010). Since mouse hybrid sterility is accompanied by PRDM9-controlled meiotic silencing of unsynapsed chromatin and consequent disturbance of meiotic sex chromosome inactivation (Bhattacharyya et al. 2013; Campbell et al. 2013; Larson et al. 2016), the uninhibited miRNA clusters could suppress genes necessary for meiosis, thus acting as “lethal mutants” contributing to meiotic arrest. Previously we have found overexpression in pachynemas of the miR-465 miRNA cluster in sterile (PWD × B6) F1 compared to reciprocal, quasi-fertile (B6 × PWD) F1 males (Bhattacharyya et al. 2013). Remarkably, this cluster is subjected to copy number variation between PWD, PWK, and B6 strains. Admittedly, until we identify the gene/sequence responsible for the Hstx2 phenotype, such speculations have to be taken with a grain of salt. Indeed, in reciprocal crosses between the M. m. musculus STUS strain and B6, both reciprocal hybrid males were fully sterile, showing that in this particular cross the Prdm9msc/Prdm9dom2 hybrid sterility phenotype was not dependent on Hstx2 allele (Bhattacharyya et al. 2013).
Summary
Early meiotic arrest of mouse intersubspecific hybrids depends on the interaction between the Prdm9 gene and Hybrid sterility X2 (Hstx2) locus on chromosome X. Lustyk et al. conducted high-resolution genetic and physical mapping of the Hstx2 locus, reduced it to 2.7 Mb interval within a constitutive recombination cold spot and found that the newly defined Hstx2 still operates as the X-linked hybrid sterility factor, controls meiotic chromosome synapsis, and modifies recombination rate. Optical mapping of the Hstx2 genomic region excluded inversion as a cause of recombination suppression and revealed a striking copy number polymorphism of the microRNA Mir465 cluster.
Acknowledgments
We are grateful to Vladana Fotopulosova for technical support; Inken Beck for generation of knockout mice (https://www.phenogenomics.cz/); and Lukas Cermak, Nikol Balogova, and Tomas Lidak for help with Western blotting. We thank Simon Myers for the B6.Prdm9Hu mice, Attila Toth for HORMAD2 antibody, Cornelia Burkhardt and Sven Künzel for sample preparation and Bionano optical mapping, and Emil Parvanov and Sarka Takacova for comments. This work was supported by LQ1604 project of the National Sustainability Program II from the Ministry of Education, Youth and Sports of the Czech Republic, and by Czech Science Foundation grant GA CR No. 16-01969S to J.F., and the Charles University Grant Agency, GA UK No. 22218 to D.L., L.O.-H. and Y.F.C. were supported by the Max Planck Society.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9874460.
Communicating editor: F. Cole
Literature Cited
- Anderson L. K., Reeves A., Webb L. M., and Ashley T., 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Saxl R. L., Raghupathy N. et al. , 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11: e1004916 10.1371/journal.pgen.1004916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcova M., Faltusova B., Gergelits V., Bhattacharyya T., Mihola O. et al. , 2016. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 12: e1005906 10.1371/journal.pgen.1005906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L., Fujiwara Y., Sun F., Hu J., Hibbs M. A. et al. , 2016. Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes. BMC Genomics 17: 628 10.1186/s12864-016-2865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C. et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. 10.1126/science.1183439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T., Gregorova S., Mihola O., Anger M., Sebestova J. et al. , 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc. Natl. Acad. Sci. USA 110: E468–E477. 10.1073/pnas.1219126110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T., Reifova R., Gregorova S., Simecek P., Gergelits V. et al. , 2014. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genet. 10: e1004088 10.1371/journal.pgen.1004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Thibault-Sennett S., Smagulova F., Lam K. G., Pu Y. et al. , 2018. Extensive sex differences at the initiation of genetic recombination. Nature 561: 338–342. 10.1038/s41586-018-0492-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P., Good J. M., and Nachman M. W., 2013. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193: 819–828. 10.1534/genetics.112.148635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Lam E., Saghbini M., Bocklandt S., Hastie A. et al. , 2018. Structural variation detection and analysis using Bionano optical mapping. Methods Mol. Biol. 1833: 193–203. 10.1007/978-1-4939-8666-8_16 [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium , 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401. 10.1534/genetics.111.132639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R. et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., and Orr H. A., 2004. Speciation, Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Davies B., Hatton E., Altemose N., Hussin J. G., Pratto F. et al. , 2016. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530: 171–176. 10.1038/nature16931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Côté A. M., and Barbash D. A., 2017. Beyond speciation genes: an overview of genome stability in evolution and speciation. Curr. Opin. Genet. Dev. 47: 17–23. 10.1016/j.gde.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1951. Genetics and the origin of Species, Columbia University, New York. [Google Scholar]
- Duvaux L., Belkhir K., Boulesteix M., and Boursot P., 2011. Isolation and gene flow: inferring the speciation history of European house mice. Mol. Ecol. 20: 5248–5264. 10.1111/j.1365-294X.2011.05343.x [DOI] [PubMed] [Google Scholar]
- Dzur-Gejdosova M., Simecek P., Gregorova S., Bhattacharyya T., and Forejt J., 2012. Dissecting the genetic architecture of f(1) hybrid sterility in house mice. Evolution 66: 3321–3335. 10.1111/j.1558-5646.2012.01684.x [DOI] [PubMed] [Google Scholar]
- Ernst C., Eling N., Martinez-Jimenez C. P., Marioni J. C., and Odom D. T., 2019. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10: 1251 10.1038/s41467-019-09182-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachs P., Mihola O., Simecek P., Gregorova S., Schimenti J. et al. , 2012. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS Genet. 8: e1003044 10.1371/journal.pgen.1003044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachs P., Bhattacharyya T., Mihola O., Pialek J., Forejt J. et al. , 2014. Prdm9 incompatibility controls oligospermia and delayed fertility but no selfish transmission in mouse intersubspecific hybrids. PLoS One 9: e95806 10.1371/journal.pone.0095806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Valdar W., Shifman S., and Mott R., 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6: 271–286. 10.1038/nrg1576 [DOI] [PubMed] [Google Scholar]
- Forejt J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417. 10.1016/0168-9525(96)10040-8 [DOI] [PubMed] [Google Scholar]
- Forejt J., and Ivanyi P., 1974. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24: 189–206. 10.1017/S0016672300015214 [DOI] [PubMed] [Google Scholar]
- Forejt J., Pialek J., and Trachtulec Z., 2012. Hybrid male sterility genes in the mouse subspecific crosses, pp. 482–503 in Evolution of the House Mouse, edited by Macholan M., Baird S. J. E., Muclinger P., and Pialek J.. Cambridge University Press, Cambridge: 10.1017/CBO9781139044547.021 [DOI] [Google Scholar]
- Fuller Z. L., Leonard C. J., Young R. E., Schaeffer S. W., and Phadnis N., 2018. Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 14: e1007526 10.1371/journal.pgen.1007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., Galindo-Diaz U., Garcia-Rubio M., Martin J. S., Polanowska J. et al. , 2019. A meiotic checkpoint Alters repair partner bias to permit inter-sister repair of persistent DSBs. Cell Rep. 26: 775–787.e5. 10.1016/j.celrep.2018.12.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., and Nachman M. W., 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228. 10.1534/genetics.107.085340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova S., and Forejt J., 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies–a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. (Praha) 46: 31–41. [PubMed] [Google Scholar]
- Gregorová S., Mnuková-Fajdelová M., Trachtulec Z., Capková J., Loudová M. et al. , 1996. Sub-milliMorgan map of the proximal part of mouse Chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome 7: 107–113. 10.1007/s003359900029 [DOI] [PubMed] [Google Scholar]
- Gregorova, S., V. Gergelits, I. Chvatalova, T. Bhattacharyya, B. Valiskova et al., 2018 Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice. eLife 7: e34282. 10.7554/eLife.34282 [DOI] [PMC free article] [PubMed]
- Haldane J., 1922. Sex ration and unisexual sterility in hybrid animals. J. Genet. 12: 101–109. 10.1007/BF02983075 [DOI] [Google Scholar]
- Hammer M. F., Mendez F. L., Cox M. P., Woerner A. E., and Wall J. D., 2008. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 4: e1000202 10.1371/journal.pgen.1000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr B., Karakoc E., Neme R., Teschke M., Pfeifle C. et al. , 2016. Genomic resources for wild populations of the house mouse, Mus musculus and its close relative Mus spretus. Sci. Data 3: 160075 10.1038/sdata.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch A. G., Zhang G., Becker P. W., Moralli D., Hinch R. et al. , 2019. Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science 363: eaau8861. 10.1126/science.aau8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D. M., Griffith S. C., and Price T. D., 2018. Sex chromosome inversions enforce reproductive isolation across an avian hybrid zone. Mol. Ecol. 28: 1246–1262. [DOI] [PubMed] [Google Scholar]
- Janoušek V., Wang L., Luzynski K., Dufkova P., Vyskocilova M. M. et al. , 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol. Ecol. 21: 3032–3047. 10.1111/j.1365-294X.2012.05583.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M., Wells D., Rusch J., Ahmad S., Marchini J. et al. , 2019. Unified single-cell analysis of testis gene regulation and pathology in five mouse strains. Elife. 25: 8 10.7554/eLife.43966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: e1000501. 10.1371/journal.pbio.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J., Yamada S., Tischfield S. E., Pan J., Kim S. et al. , 2016. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell 167: 695–708.e16. 10.1016/j.cell.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson E. L., Keeble S., Vanderpool D., Dean M. D., and Good J. M., 2016. The composite regulatory basis of the large X-effect in mouse speciation. Mol. Biol. Evol. 34: 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack K. L., and Nachman M. W., 2017. Gene regulation and speciation. Trends Genet. 33: 68–80. 10.1016/j.tig.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M., Baird S. J., Dufkova P., Munclinger P., Bimova B. V. et al. , 2011. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution 65: 1428–1446. 10.1111/j.1558-5646.2011.01228.x [DOI] [PubMed] [Google Scholar]
- Macholán M., Munclinger P., Sugerkova M., Dufkova P., Bimova B. et al. , 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61: 746–771. 10.1111/j.1558-5646.2007.00065.x [DOI] [PubMed] [Google Scholar]
- Maheshwari S., and Barbash D. A., 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45: 331–355. 10.1146/annurev-genet-110410-132514 [DOI] [PubMed] [Google Scholar]
- Margolin G., Khil P. P., Kim J., Bellani M. A., and Camerini-Otero R. D., 2014. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15: 39 10.1186/1471-2164-15-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., and Forejt J., 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. 10.1126/science.1163601 [DOI] [PubMed] [Google Scholar]
- Miyata H., Castaneda J. M., Fujihara Y., Yu Z., Archambeault D. R. et al. , 2016. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci. USA 113: 7704–7710. 10.1073/pnas.1608458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Gatti D. M., Najarian M. L., Keane T. M., Galante R. J. et al. , 2017. Structural variation shapes the landscape of recombination in mouse. Genetics 206: 603–619. 10.1534/genetics.116.197988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C. et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. 10.1126/science.1182363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., and Payseur B. A., 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 409–421. 10.1098/rstb.2011.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2005. The genetic basis of reproductive isolation: insights from Drosophila. Proc. Natl. Acad. Sci. USA 102: 6522–6526. 10.1073/pnas.0501893102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D., Engelstadter J., and Rieseberg L. H., 2016. Recombination rate evolution and the origin of species. Trends Ecol. Evol. 31: 226–236. 10.1016/j.tree.2015.12.016 [DOI] [PubMed] [Google Scholar]
- Ota H., Ito-Matsuoka Y., and Matsui Y., 2019. Identification of the X-linked germ cell specific miRNAs (XmiRs) and their functions. PLoS One 14: e0211739 10.1371/journal.pone.0211739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., and Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835 10.1126/science.1181495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten M. M., 2018. Selfish X chromosomes and speciation. Mol. Ecol. 27: 3772–3782. 10.1111/mec.14471 [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Krenz J. G., and Nachman M. W., 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078. 10.1111/j.0014-3820.2004.tb00490.x [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Presgraves D. C., and Filatov D. A., 2018. Introduction: sex chromosomes and speciation. Mol. Ecol. 27: 3745–3748. 10.1111/mec.14828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M., and Nachman M. W., 2015. Insights into mammalian biology from the wild house mouse Mus musculus. eLife 4: e05959. 10.7554/eLife.05959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2018. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol. Ecol. 27: 3822–3830. 10.1111/mec.14777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M. et al. , 2010. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20: 2117–2123. 10.1016/j.cub.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Royo H., Seitz H., ElInati E., Peters A. H., Stadler M. B. et al. , 2015. Silencing of X–linked MicroRNAs by meiotic sex chromosome inactivation. PLoS Genet. 11: e1005461 10.1371/journal.pgen.1005461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K., Thomason L. C., Kuznetsov S. G., and Court D. L., 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4: 206–223. 10.1038/nprot.2008.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Brick K., Pu Y. M., Camerini-Otero R. D., and Petukhova G. V., 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30: 266–280. 10.1101/gad.270009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M., and Fishel R., 2015. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 7: a022657 10.1101/cshperspect.a022657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Morgan A. P., Najarian M. L., Sarsani V. K., Sigmon J. S. et al. , 2017. Genomes of the mouse collaborative cross. Genetics 206: 537–556. 10.1534/genetics.116.198838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová R., Gregorova S., Buckiova D., Kyselova V., Divina P. et al. , 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. 10.1007/s00335-004-2386-0 [DOI] [PubMed] [Google Scholar]
- Trachtulec Z., Mnukova-Fajdelova M., Hamvas R. M., Gregorova S., Mayer W. E. et al. , 1997. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm. Genome 8: 312–316. 10.1007/s003359900430 [DOI] [PubMed] [Google Scholar]
- Truett G. E., Heeger P., Mynatt R. L., Truett A. A., Walker J. A. et al. , 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29: 52, 54 10.2144/00291bm09 [DOI] [PubMed] [Google Scholar]
- Tucker P., Sage R., Warner J., Wilson A., and Eicher E., 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163. 10.1111/j.1558-5646.1992.tb00625.x [DOI] [PubMed] [Google Scholar]
- Turner L. M., Schwahn D. J., and Harr B., 2012. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66: 443–458. 10.1111/j.1558-5646.2011.01445.x [DOI] [PubMed] [Google Scholar]
- Wang L., Valiskova B., and Forejt J., 2018. Cisplatin-induced DNA double-strand breaks promote meiotic chromosome synapsis in PRDM9-controlled mouse hybrid sterility. eLife 7: e42511. 10.7554/eLife.42511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Cloutier J. M., Baumann M., Daniel K., Varga J. et al. , 2012. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev. 26: 958–973. 10.1101/gad.187559.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wang W., Liu Y., Zhuang K., Cai T. et al. , 2018. RETRACTED: NY-SAR-35 is involved in apoptosis, cell migration, invasion and epithelial to mesenchymal transition in glioma. Biomed. Pharmacother. 97: 1632–1638. 10.1016/j.biopha.2017.11.076 [DOI] [PubMed] [Google Scholar]
- Zhang F., Zhang Y., Lv X., Xu B., Zhang H. et al. , 2019. Evolution of an X–linked miRNA family predominantly expressed in mammalian male germ cells. Mol. Biol. Evol. 36: 663–678. 10.1093/molbev/msz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Sun T., Woldesellassie F., Xiao H., and Tao Y., 2015. Sex ratio meiotic drive as a plausible evolutionary mechanism for hybrid male sterility. PLoS Genet. 11: e1005073 10.1371/journal.pgen.1005073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and in the supplemental material. The optical mapping data sets are available from L.O.-H. or K.K.U. upon reasonable request. Statistical analyses were performed by unpaired two-tailed t-test, if not indicated otherwise. Statistical significance was set at P values of * 0.05,** 0.01, and *** 0.005. Data were processed and plotted by GraphPad Prism version 6.00 (GraphPad Software, San Diego, CA; www.graphpad.com). Other types of statistical analyses are described within the text and in the corresponding figure legends. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9874460.