Abstract
While Caenorhabditis elegans was originally regarded as a model for investigating determinate developmental programs, landmark studies have subsequently shown that the largely invariant pattern of development in the animal does not reflect irreversibility in rigidly fixed cell fates. Rather, cells at all stages of development, in both the soma and germline, have been shown to be capable of changing their fates through mutation or forced expression of fate-determining factors, as well as during the normal course of development. In this chapter, we review the basis for natural and induced cellular plasticity in C. elegans. We describe the events that progressively restrict cellular differentiation during embryogenesis, starting with the multipotency-to-commitment transition (MCT) and subsequently through postembryonic development of the animal, and consider the range of molecular processes, including transcriptional and translational control systems, that contribute to cellular plasticity. These findings in the worm are discussed in the context of both classical and recent studies of cellular plasticity in vertebrate systems.
Keywords: reprogramming, transdifferentiation, transdetermination, cell type conversion, stem cells, WormBook
Cellular Plasticity
Definitions
A defining property of our tissues and organs is that they remain functional over time. This implies that their main constituents, cells, persistently maintain their specialized identity. This specialized cellular identity is characterized by the combination of the cell’s morphology and function—properties that are underlined by a specific transcriptional program. A major change in the way we view the maintenance of the specialized cellular identity has occurred recently: while the identity of differentiated cells was originally thought to be forever fixed after being acquired—a view that was largely held until 2006—it has become clear that it can be altered or entirely changed, even after terminal differentiation. This ability of a cell to give rise to cell(s) with a different identity is called cellular plasticity. Cellular plasticity can occur naturally in an organism, or can be triggered exogenously, either experimentally or by the environment. Although the term has initially been used to describe a cellular path proceeding from a less differentiated to a more differentiated identity, cellular plasticity entails a variety of processes including retrodifferentiation (the reversal to a lineally related progenitor or stem cell identity, and even reversal to a pluripotent state), transdetermination (the swap in differentiation commitment), and transdifferentiation (Td, the stable switch from one differentiated identity to another). The concept of cellular plasticity does not imply any directionality, e.g., toward a more or less differentiated state, or what the final identity will be (see Box 1 Definitions).
Box 1: Definitions.
Totipotent: In reference to vertebrate development, a cell which descendants can form all embryonic and extraembryonic lineages
Pluripotent: In reference to vertebrate development, a cell which descendants can form all embryonic lineages, including germ cells.
Multipotent: A cell which descendants can give rise to several different cell types
Unipotent: A cell which descendants can form only one cell type
Cellular potential: describes the range of cell types a given cell can give rise to
Stem cell: The classical definition of a stem cell is a non-differentiated cell that can self-renew over a long period of time and give rise to daughter(s) with a different - and more differentiated - fate (i.e. exhibit cellular potential). Classically again, stem cells are viewed as discrete entities, and stemness as an intrinsic property the cells are born with. Note, however, that this definition does not fit all stem cells and that the ability to produce in vitro induced Pluripotent Stem (iPS) cells suggests that stemness is a state that can be acquired.
Blastomere: an early embryonic cell, obtained after cleavage of the zygote and before tissue germ layers are formed, that has the potential to give rise to a number of specialized cells and has a reduced capacity for self-renewal
Progenitor: A non-differentiated cell that has the potential to give rise to a number of specialized cells within a lineage and has a lower capacity for self-renewal than the stem cells. In a lineage, all cells that are in between the stem cells and the differentiated cells are called progenitors
Differentiated cell: A cell that exhibits defined specialized characteristics, morphology and behavior. Differentiated cells are conceived as discrete entities defined by intrinsic properties that ensure their function
Cellular plasticity: Describes the ability for a cell to give rise to different cell(s). No directionality - from/to non-differentiated - is implied; rather, it represents either the cellular potential of a stem cell or progenitor, or the ability of a cell to escape/change its initial identity
Reprogramming: Describes the ability for a differentiated cell to change its identity. By contrast with cellular plasticity, a directionality - starting from differentiated - is implied here
Transdifferentiation (or Td): The stable conversion of a differentiated cell into another type of differentiated cell. Both natural and induced transdifferentiation events have been described. While a direct lineal relationship must be established between the initial and final cellular identity, the original definition, as proposed by Eguchi, Kodama (1993), does not entail any specific mechanism underlying the transition. However, it does imply that only one initial inducing event is used to trigger - experimentally - induced transdifferentiation, as opposed to a succession of experimental manipulations. Aka cell type conversion, direct reprogramming
Direct reprogramming: Same as Transdifferentiation, i.e. the stable conversion of a differentiated cell into another type of differentiated cell. Although sometimes used to solely imply an experimentally triggered event, direct reprogramming can be either natural (natural direct reprogramming) or induced (induced direct reprogramming)
Pluripotent reprogramming: The conversion of a differentiated cell into a pluripotent stem cell-like state. To date, this has only been observed after experimental induction such as during the generation of iPS cells; also called nuclear reprogramming
Transdetermination: The conversion of a committed (but not differentiated) cell into another type of committed cell.
Origin of the concept, relationship with cellular potential
Cellular plasticity has classically been used as a defining property of stem cells. Stem cells self-renew and can give rise to descendants that have adopted a more differentiated identity. The number of possible alternative identities they can engender represents the cellular potential of the initial stem cell, and is often used to classify stem cells. While concepts and definitions have been largely defined and tested in vertebrate animals (see below), they are used to describe developmental events throughout the animal kingdom (see Box 1 Definitions). Thus, cells can be totipotent (i.e., can give rise to all the embryonic and extraembryonic tissues), pluripotent (i.e., can give rise to cells belonging to all three embryonic germ layers—endoderm, mesoderm, ectoderm), multipotent (i.e., can give rise to several types of cells constituting one or more tissues), or unipotent (i.e., can give rise to a specific lineage and differentiate into one cell type only) (see Box 1 Definitions).
By contrast, a differentiated cell is a specialized cell associated with a particular function in an organ or tissue (see Box 1 Definitions). In most tissues, differentiated cells ultimately become postmitotic, and the differentiated state is classically seen as an end point, a view influenced by Waddington’s epigenetic landscape, in which cells during development are represented as balls rolling down from their highest cellular potential, represented as the top of a mountain, to their final differentiated state where they rest, represented as the bottom of a valley (Waddington 1957).
Classical examples of cellular plasticity: toti-, pluri-, multi- or uni-potent cells and their demonstration
When and where are cells with broad cellular potential found in living animals? A large body of work has focused on the vertebrate embryo, in which the early embryonic cells have been shown to be totipotent, after which their cellular potential becomes more restricted as development proceeds.
Such cellular potential has been demonstrated through a variety of approaches. For example, mouse embryonic cells up to the eight-cell embryonic stage are believed to be totipotent, as they retain the ability to contribute to both embryonic and extraembryonic tissues in blastomere aggregation experiments and yield viable pups (Boroviak and Nichols 2014). One natural demonstration of totipotency at the two-cell-stage is the production of identical twins following separation of the two blastomeres in mammals.
A large proportion of embryonic cells is thought to retain the ability to form an implanting embryo until the 16-cell stage, at which stage the cells of the Inner Cell Mass (ICM) start specializing as embryonic epiblast cells or primitive endoderm cells (Morgani and Brickman 2014). Cell lines, called Embryonic Stem (ES) cells, have been derived from these ICM cells of preimplantation embryos. These ES cells express markers of the ICM cells and self-renew over a long time, in contrast with the transient ICM cells from which they are derived, and can be maintained indefinitely in culture (Boroviak and Nichols 2014). Both ICM and ES cells are thought to be pluripotent. Pluripotency is classically functionally demonstrated by testing the potential of single cells to contribute to normal development. This can be achieved by injecting them into a developing embryo, or through blastomere aggregation experiments or tetraploid complementation assays, and by testing their ability to contribute to all lineages. ES cells meet this functional definition. In addition, their differentiation potential can be tested by experimentally inducing differentiation along different lineages in vitro. While ES cells are engineered through isolation of ICM cells in very defined culture conditions and may progress to a specific pluripotent stage during derivation, it has been hypothesized that ES cells capture a naïve pluripotency state naturally found in ICM cells (Boroviak and Nichols 2014).
Early embryonic pluripotency is rapidly lost and cells later in development are thought to be multipotent, oligopotent, or unipotent. To characterize these properties, in vitro clonogenic assays, in vitro and/or in vivo phenotyping, and in vivo transplantation assays are classically used (Blanpain and Simons 2013). For instance, in vitro differentiation approaches have been performed using a variety of primary cells—or cell lines—and culture conditions, from embryoid bodies or neurospheres to single cells such as intestinal stem cells, and, more recently, through the use of 3D matrices. In addition, transplantation assays and label-retaining approaches are performed in vivo to identify and follow stem cells and their descendants in their physiological environment.
Stem cells have also been described in adults, where they are thought to contribute to homeostasis, repair, and regeneration of adult tissues. Classical examples include unipotent satellite cells, which are muscle stem cells (Sambasivan and Tajbakhsh 2015), and the multipotent intestinal crypt stem cells (van der Flier and Clevers 2009).
As highlighted for ICM cells, it should be noted that the cells exhibiting cellular plasticity during development, some with broad cellular potential, exist only transiently, by contrast with the classical definition of stem cells involving long-term self-renewal. These cells are therefore classically called blastomeres or progenitors. Thus, the notion of stem cells is, in large part, built on the ability to culture in vitro pluripotent cell lines (like ES cells), and on the description in several adult tissues of long-term resident stem cells, many of which are unipotent.
Together, this body of work has led to a hierarchical view of development in which early blastomeres in vertebrate embryos transition from totipotency to pluripotency and then continue to restrict their cellular potential as they progress along their specialization path, ultimately adopting their final differentiated and fixed identity. This notion of gradual morphing is implicitly suggested by Waddington’s epigenetic landscape graphical representation, although whether differentiation paths actually follow an incremental hierarchical process or, on the contrary, proceed through a succession of sharp transitions, remains to be determined.
Developmental Programming: Regulative vs. Mosaic Models
Are the properties of cellular potential, pluripotency, and multipotency, universal and do they apply to the worm? There have been a number of studies addressing the question of cellular potential in Caenorhabditis elegans. This section and the next will focus on those studies examining cellular plasticity in the embryo.
Programming in the worm: classical view and fixed lineage
C. elegans development has been described at the single cell level in landmark studies [Sulston and Horvitz 1977; Deppe et al. 1978; Krieg et al. 1978; Sulston et al. 1983; see Giurumescu and Chisholm (2011) for more recent automated and semi-automated lineage data]. Knowledge of its cellular lineage has revealed that development proceeds in a highly stereotyped way: a generally fixed relationship between cell ancestry and cell fate, as well as largely invariant cell positioning, has been observed throughout embryonic development.
Does the ancestry of the cells constrain their fate and, if so, how? The striking invariance of the cell lineage raised the question of whether differentiation paths are intrinsically (cell-autonomously) determined, a process called mosaic development, or whether cells in the C. elegans embryo could adapt to changes and interference, indicating that cell fates are dependent on cell–cell interactions, a process called regulative development (Conklin 1905).
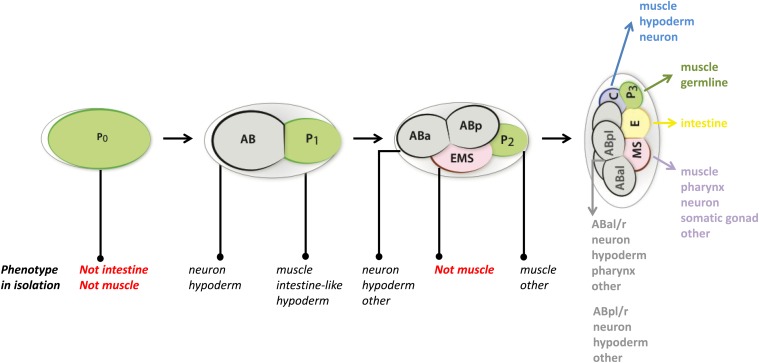
As presented in the next section, C. elegans development had initially been seen as mosaic. This view resonated with the apparent mosaic development and segregation of cytoplasmic determinants described in many invertebrates (Conklin 1905). Initial studies in C. elegans focused on the fate and determination of the early embryonic blastomeres. The fertilized egg, named P0, divides unequally into a larger anterior daughter, called AB, and a smaller posterior one called P1 (Sulston et al. 1983), which continues to divide along different division patterns and contribute to different tissues. Experiments in which the size of AB was reduced through removal of cytoplasm showed that the differential size of these two blastomeres does not substantially alter their division behavior or fate, suggesting that qualitative cytoplasmic differences dictate their fate (Schierenberg 1984, 1986; Schierenberg and Wood 1985). The AB blastomere will divide into what will become the anterior, ABa, and posterior, ABp, daughters. ABa lineage follows complicated asymmetrical patterns while ABp divides mostly through symmetrical patterns, both producing similar and unique cell types. Their descendants will be the major contributors to the cells of the hatching larva, and will generate cells of the nervous system, the hypodermis, and the pharynx. P1 continues to divide unequally to give rise to the EMS and P2 blastomeres. P2 will give rise to the C and P3 blastomeres, and P3 to the D and P4 blastomeres. EMS gives rise to MS, which produces primarily mesodermal cells and E, which generates only intestine. C generates muscles and hypodermis, and D only muscles. P4 is the precursor of the germline (Figure 1). These blastomeres are called founder cells, as they solely or mostly give rise to one tissue.
Figure 1.
Early embryonic lineage, evidence for cell-intrinsic determination and nonlinear segregation of developmental potential. The fertilized egg, named P0, divides unequally into the AB and P1 blastomeres. AB further divides into ABa and ABp, which will follow different division patterns. The tissue contribution of their descendants is indicated in gray. P1 divides unequally to give rise to the EMS and P2 blastomeres. P2 will give rise to the C and P3 blastomeres, and EMS gives rise to MS and E. The tissue contributions of the descendants of the C, P3, E, and MS blastomeres is indicated on the right. The cellular phenotype observed (in black) or not (in red) after culture in isolation, with or without cell arrest, of each blastomere up to the four-cell stage is also indicated.
Early specification suggests plasticity is lost early
A range of evidence from early embryological studies suggested that much of C. elegans embryogenesis is characterized by mosaic development, as reviewed in this section.
Founder cells are largely specified by apparent cell-intrinsic mechanisms at the time of their birth
To address the extent to which autonomous specification mechanisms act during embryogenesis, a number of approaches have been undertaken. One of these consisted of isolating specific blastomeres, culturing them in isolation, and assessing which fate they or their descendants adopted. In isolation, the AB blastomere divides and produces recognizable hypodermal and neuronal cells, similar to many of the cell types it produces in wild type embryos (Priess and Thomson 1987; Gendreau et al. 1994; Moskowitz et al. 1994). The P1 blastomere obtained after gently bursting the eggshell and eliminating the AB blastomere, is able to divide in culture and generates four cells, one of which shows characteristics of the endodermal precursor cell E, specifically the gut-specific rhabitin granules (Laufer et al. 1980). When P1 blastomeres obtained similarly are treated with a cleavage inhibitor, they also give rise to cells with gut granules (Figure 1) (Laufer et al. 1980). These experiments suggested that the ability of P1 to generate gut fate is intrinsic to the P1 cell at the two-cell-stage. This work also suggested that cell division is not a requirement for expression of at least certain differentiation characteristics. Furthermore, if isolated P1 blastomeres are allowed to divide further, they yield a partial embryo containing a few hundred cells that twitches and that contains muscle cells (Figure 1) (Laufer et al. 1980; Gossett et al. 1982). Again, this suggested that the potential to produce muscle is intrinsically present in the P1 blastomere and is segregated to its descendants. These data were interpreted as strong evidence that early embryonic specification is largely carried out autonomously and already established in the early blastomeres, consistent with a mosaic pattern of development.
One intrinsic mechanism underlying mosaic development is cell fate determination through the segregation of cytoplasmic determinants. Early observations supported such a mechanism at both the organelle and molecular levels. The germline-specific P granules are transmitted specifically to germline precursors during the early asymmetric cell divisions that establish the germline progenitor, P4 (Strome and Wood 1983). Similarly, proteins such as PIE-1 and MEX-5/6 (Mello et al. 1996; Schubert et al. 2000) are distributed unequally in the zygote and subsequent blastomeres prior to cell division (Cowan and McIntosh 1985). The levels of the SKN-1 protein, required in P1 descendants to produce pharyngeal cells, are also higher in the P1 blastomere compared to its AB sister (Bowerman et al. 1993). Caveats to these experiments exist: for instance, a role for cell–cell interactions in specification, preceding cell separation manipulations for example, cannot be definitely excluded; in addition, the impact of cell–cell signaling in rapidly dividing blastomeres may only be seen in one-to-a-few cell generations later. However, collectively, these findings made a strong case that cell fate potential is driven largely by cell-intrinsic mechanisms in early C. elegans embryos.
Cleavage-arrest experiments and exclusivity of cell fate
The law of exclusivity of differentiation postulated by Weiss (1939) posits that a single cell chooses only one path of differentiation at a time. This principle was supported in C. elegans through cleavage-arrest experiments on early embryos, which were used both to assess when the potential to differentiate particular cell types first appears, and to examine whether the potential to produce several differentiated cell types can coexist in the same blastomere (Cowan and McIntosh 1985). Cowan and McIntosh observed cleavage-arrested isolated P1 blastomeres and asked if gut, muscles, and hypodermis markers, all fates found in the P1 progeny, can be coexpressed in such setting. While they observed that all three markers were found in similar proportion in cleavage-arrested P1 blastomeres, they never found them to be coexpressed (Cowan and McIntosh 1985). These findings supported the view of exclusivity of differentiation pathways (Weiss 1939) and were interpreted as yet further evidence for mosaic development in the worm.
Importance of DNA replication in specification
An early model, called the quantal cell cycle model (Holtzer et al. 1975, 1983), posited that progression through the cell cycle, and DNA replication in particular, is a necessary requirement for cells to alter their expression profiles and change their commitment or differentiation state. The model postulates that the DNA:cytoplasm ratio controls transcription through its impact on cell cycle. In the context of embryogenesis, it implied that even when embryos are competent for transcription, they would start transcribing only once a G1 or G2 phase is added to the cell cycle.
However, cell fusion experiments to produce nondividing heterokaryons, carried out around the same time, have shown that cells in culture can alter portions of their transcriptional programs in the absence of DNA replication (Blau et al. 1983, 1985; Chiu and Blau 1984), suggesting that this model may not entirely apply, at least in in vitro settings. In an effort to decipher the relationship between DNA replication and lineage specification in an in vivo physiological setting, Edgar and McGhee (1988) tested this hypothesis in live C. elegans embryos using pulses of the drug aphidicolin, which instantaneously blocks DNA replication. Focusing on the E lineage and by comparison it to the hypodermal, and body wall muscle lineages, their experiments addressed the requirement for rounds of DNA replication in the expression of intestinal, lineage markers (Edgar and McGhee 1988).
Edgar and McGhee (1988) found that neither the number of replication rounds per se, nor the DNA:cytoplasm ratio or elongation of the cell cycle prevented expression of differentiation markers or their timing, suggesting that the quantal cell cycle model did not apply, at least for the lineages examined, in C. elegans embryo. However, their results suggest that the first round of DNA synthesis after the sublineage progenitor has been established (for instance, the E founder cell) is key. This was interpreted by the authors as a critical period during which gut genes are “activated” or licensed for later expression (Edgar and McGhee 1988). This could be achieved via the replication-dependent elimination of nucleosomes and other histone modifications, thus giving access to key determinants to target regions on the chromosomes, or it could represent a critical period when cytoplasmic gut determinants are translocated to the nucleus and thus bind to specific loci on the chromosomes. One implication of these studies is that early blastomeres, rather than being generally open to adopting different differentiation fates, must be made competent to do so in vivo through a precisely timed DNA replication period. However, such an explanation must take into account how markers of several different fates, including gut, can be observed in cleavage-arrested P1 blastomeres (which continue to undergo DNA replication and mitosis) (Laufer et al. 1980), as presented in the Cleavage-arrest experiments and exclusivity of cell fate section.
The C. elegans embryo: regulative development and early cellular plasticity clues
While much evidence points to a mosaic mode of development in C. elegans, it is also clear that regulative development also functions during early embryogenesis, and that early blastomeres likely retain wider developmental potential than they express during wild-type development.
Nonlinear segregation of developmental potential
As described above, cleavage-arrested P1 blastomeres appear to differentiate gut, based on the presence of gut granules (Laufer et al. 1980; Cowan and McIntosh 1985). However, cleavage-arrested P0 blastomeres never produce gut granules (Figure 1). This suggests that gut fate determinants are not present, or are not active, in P0, but arise in the P1 cell, perhaps as a result of its interaction with the AB blastomere or of P0 division. Similar observations have been made when segregation of muscle differentiation potential is examined: it is not seen in P0, while it is present in P1 and P2 blastomeres (Figure 1). In addition, muscle differentiation potential is not present in cleavage-arrested EMS, while it is present in the P1 mother, and in its MS daughter (Figure 1). These data suggest that different developmental potentials are not simply segregated in a linear fashion but can reappear throughout the lineage in daughters of cells that do not exhibit this potential (Cowan and McIntosh 1985).
Similarly, the lineal pattern of the worm’s tissues argues against a purely mosaic pattern of development: indeed, with the exception of the intestine and the germline, whose origin can be traced to one early ancestor, C. elegans tissues are polyclonal in origin (Sulston et al. 1983), excluding simple linear segregation of fates along lineages.
Wide developmental capacity of early blastomere nuclei
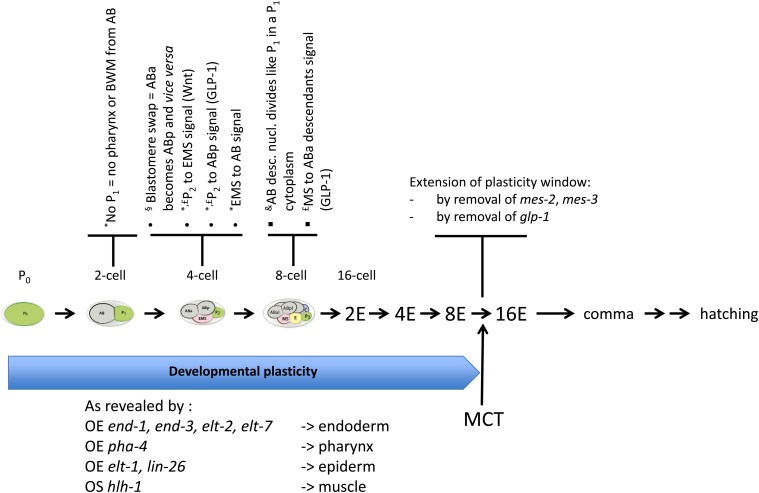
Studies have suggested that there is greater plasticity in the early blastomeres than is evident from the lineage. When the anterior cytoplasm together with the zygote nucleus are extruded from the anterior pole of wild-type 1-cell embryos, it produces an AB-like cell that can divide. If one of the nuclei obtained after two divisions is slipped back into the posterior enucleated part that had remained, the newly recreated cell then divides like a P1 blastomere that is able to produce P derivatives like muscles (Figure 2) (Schierenberg and Wood 1985; Schierenberg 1986). These experiments suggested that an AB-like nucleus still contains P1 developmental potential, even after two cell divisions, and thus could be regarded as totipotent. However, another interpretation could be that the posterior enucleated part behaves like a P1 environment that can reprogram the AB-like nucleus, in analogy to what John Gurdon observed in his landmark nuclear transfer experiments in frogs (Gurdon 1962).
Figure 2.
Evidence for the early developmental plasticity window and the control of its timing. Early blastomeres up to the 8E-cell stage can exhibit high plasticity, as revealed by blastomeres swap (§), blastomeres ablations or culture in isolation (*), nuclear transfer (&) or analysis of lineage mutants (£). In addition, overexpression (OE) of defined TFs during this time-window transforms most, if not all, blastomeres into cells bearing characteristics of the indicated tissues. Transition from the 8E to the 16E-cell stage marks the multipotency-to-commitment transition (MCT). The window can be extended by removing the polycomb PRC2 complex (mes-2, mes-3 mutants) or early Notch activity (glp-1 mutant).
Blastomere rearrangements and latent developmental potential
In another key set of experiments, displacement of blastomeres within the embryo revealed that early sister blastomeres that normally invariably adopt different fates are actually initially equipotent. For example, the AB daughters generate common but also unique cell types: only ABa generates pharyngeal muscles and inner labial neurons, while only ABp generates the GABA-containing neurons. To examine if ABp retains ABa developmental potential and vice versa, Priess and Thomson swapped their positions by micromanipulation before their separation is complete in otherwise normal embryos, and examined the identity of their descendants in embryos, larvae, and adults. All 11 such embryos developed into seemingly normal and viable larvae and adults, and the expected ABa and ABp derivatives were all found at their normal location, though likely generated by the other, interchanged, AB daughter (Figure 2) (Priess and Thomson 1987). These striking results strongly suggested that, although the AB daughters stereotypically follow different developmental paths, they in fact have a broader developmental potential than the one they normally express during development. Selective expression of a subset of this developmental potential thus relies on the environment and differential cell–cell interactions that each AB daughter experiences—a conclusion reinforced by the concomitant finding that AB blastomeres cultured in isolation do not generate all expected cell types (Priess and Thomson 1987). Together with the Schierenberg experiments described above, these studies argue that the AB blastomere at least is inherently more potent—and maybe more totipotent—than is apparent during normal development.
Early ablations also revealed instances of regulative development
Laser-mediated ablation of individual blastomeres in embryos comprising 28 cells or more revealed a restricted number of cases of regulative development, that occurred mostly during late embryogenesis (Sulston et al. 1983). These experiments initially reinforced the idea that C. elegans development and early embryogenesis in particular was largely mosaic.
However, further experiments revealed numerous cell–cell interactions that are required for the correct specification of early blastomeres as well as the complete repertoire of all cells in the C. elegans embryo. Experiments using temperature-sensitive mutants that alter the cell-cycle length of particular blastomeres, in which case the resulting embryo developed abnormally, or of all founder cells, in which case normal development was observed, suggested a requirement for timely cell–cell interactions (Schierenberg et al. 1980). Priess and Thomson (1987) further examined when descendants of the AB and P1 blastomeres are first determined using ablation experiments and monoclonal antibodies that specifically label pharyngeal or body wall muscles (contributed to by both the AB and P1 blastomeres) in late embryos. They showed that the blastomeres that generate these cells appear to be determined by the 28-cell stage. The muscles descendants of the P1 blastomere did not require AB-derived cells, as ablation of the AB blastomere at the two-cell stage did not alter the ability of the P1 descendants to produce pharyngeal and body wall muscles (Priess and Thomson 1987). These results are consistent with the observed segregation of muscle fate in P1 (Laufer et al. 1980). By contrast, removing the P1 blastomere did not allow generation of the expected pharyngeal or body wall muscles in AB descendants development (Figure 2). It was further shown that removal of EMS, a P1 daughter, led to the absence of pharyngeal muscles, while removal of P2 still allowed the generation of both body wall and pharyngeal muscles (Figure 2) (Priess and Thomson 1987). This strongly suggested that an interaction between P1 or its daughter EMS and the AB lineage is necessary for the formation of muscles in AB descendants.
Along the same lines, the role of inductive interactions during gut development has been examined more closely. The entire gut derives from a single blastomere, the founder E cell in C. elegans. Based on blastomere isolation experiments, it was initially suggested that EMS, the mother of the founder E cell, and even the P1 blastomere, have the cell-intrinsic ability to produce intestinal fate (Laufer et al. 1980). However, subsequent studies revealed that isolation of the EMS blastomere at the early four-cell stage blocked its ability to generate gut descendants. It was found that EMS requires a signal from P2 at the four-cell stage to generate gut descendants (Figure 2) (Schierenberg 1987; Goldstein 1992, 1993). During later embryogenesis, several additional cell–cell inductions were also revealed by ablation experiments, such as the regulative interaction between ABprapaapa (aka W) and ABplapaapa (aka G2) or the regulative interactions between ABplpaaaapa (aka excretory duct) and ABprpaaaapa (aka G1) (Sulston and Horvitz 1977). Likewise, several regulative interactions have also been described during larval development, including, but not restricted to, the specification of the anchor (AC) and vulval uterine (VU) cells or of the Vulval Precursor Cells (VPCs) (Greenwald 2012).
Lineage defects in mutants provide strong evidence in favor of regulative interactions
Analysis of mutations affecting embryogenesis further revealed numerous cell–cell interactions that are required for the correct specification of early blastomeres and the complete repertoire of all cells in the C. elegans embryo. We will here focus on three such examples in early embryogenesis. In particular, lineage founders were shown to be differentially specified at the four-cell stage through intercellular directional signals that depend on the geometrical arrangement of the blastomeres. This specification involves a P2 signal to ABp through the Notch extracellular receptor GLP-1 (Figure 2); both AB descendants express the maternally encoded Notch receptor GLP-1, but only the posterior one contacts P2, which expresses the Notch ligand APX-1, and, therefore, only it is induced to acquire an ABp fate (see http://www.wormbook.org/chapters/www_notchembryo/notchembryo.html and references therein). Further analysis of the glp-1 mutants also revealed a signal from the MS blastomere to the two (of the four) ABa descendants that it contacts, and which express glp-1, occurring in the 12-cell stage embryo (Figure 2) (Priess and Thomson 1987; Shelton and Bowerman 1996). This interaction is necessary to induce the ABa descendants to produce pharyngeal cells, and to break the left-right symmetry of the AB lineage (Priess and Thomson 1987; Gendreau et al. 1994; Hutter and Schnabel 1994; Mango et al. 1994; Moskowitz et al. 1994).
The blastomere isolation experiments also revealed a P2 to EMS signal that is necessary for subsequent production of endoderm (Figure 2) (Goldstein 1992, 1993). Genetic screens and mutant analyses have shown that this signal is mediated by redundant Wnt, src, and MAPK pathways, resulting in the polarization of the EMS cell, and asymmetric concentration of the TCF/POP-1 TF in the nucleus of the MS daughter vs. the E daughter (Lin et al. 1995; Thorpe et al. 1997; Bei et al. 2002).
Together, these studies showed that, even in an animal with a stereotyped cell lineage, development is not strictly driven by intrinsic factors, but also relies on timely cell–cell inductive interactions and the embryo’s cellular geometry. They also emphasize that the cellular and developmental potential of single cells should be assessed out of their normal contexts as it can be greater than their fate could predict.
Early Embryonic Cellular Plasticity is Restricted to a Defined Time Window: the Multipotency-to-Commitment Transition
Early embryonic founder cells are specified at the time of birth, but remain developmentally plastic through several divisions
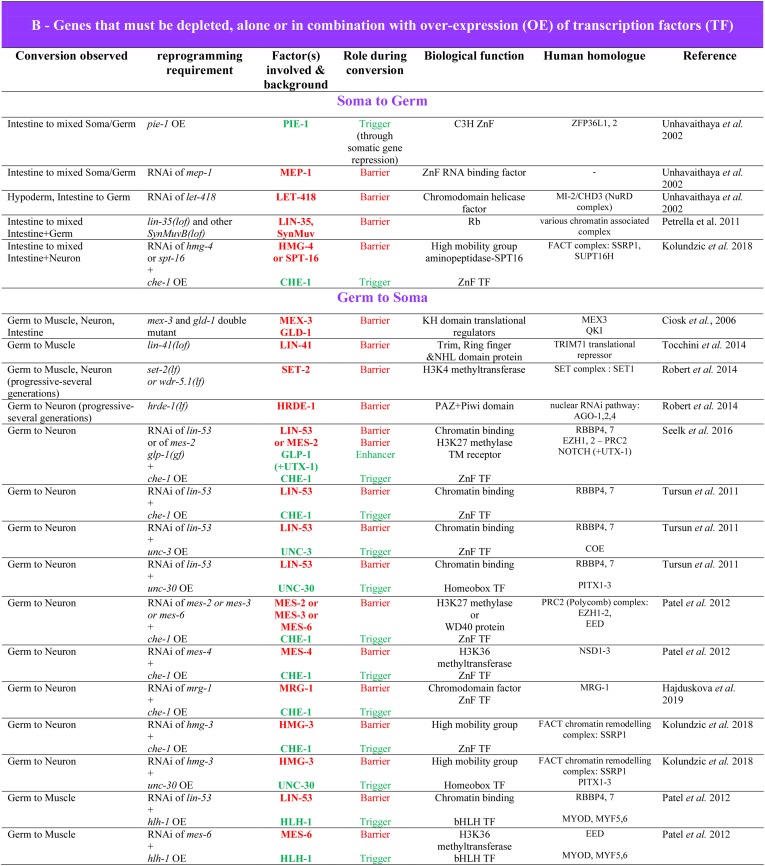
The cell-intrinsic ability of founder cells to remain developmentally plastic through several subsequent rounds of cell division was further demonstrated by the ability of them and their descendants to change fate and even lineage commitment in response to ectopically driven expression of cell fate-determining transcription factors (TFs). For example, it was found that all somatic lineages could be converted to endoderm in response to ubiquitous expression of single GATA-type TFs that function in endoderm specification and differentiation (Figure 2; END-1, END-3, ELT-2, and ELT-7) (Fukushige et al. 1998; Zhu et al. 1998; Maduro et al. 2005; Sommermann et al. 2010), resulting in embryos containing essentially only differentiated gut cells. Similarly, transcription factors for other differentiated cell types are capable of converting most or all cells of embryos into pharynx (driven by PHA-4/FoxA; Horner et al. 1998), muscle (driven by the bHLH factor HLH-1; Fukushige and Krause 2005), or epidermis (driven by the ELT-1 GATA or LIN-26 ZnF factor; Gilleard and McGhee 2001; Quintin et al. 2001) (Figure 2).
This conversion process, or transdetermination, reflects bona fide redirection of most or all somatic progenitors into the pathway for endoderm (or pharynx, muscle, or epidermis) differentiation, as other cell-type specific differentiation, characteristic of the other two germ layers, is prevented in the cells that inappropriately express markers of the differentiated gut.
Thus, several divisions after founder cell lineages are established, many or all somatic lineages of the early embryo, though normally specified to particular lineages, appear to remain largely pluripotent (capable of contributing to any of the three germ layers). These findings underscore the exclusivity of cell fates that had been originally inferred from early experiments on cleavage-arrested embryos (Cowan and McIntosh 1985): genuine acquisition of one differentiated fate by transdetermination or reprogramming precludes expression of genes characteristic of other fates.
Plasticity is retained in early blastomeres during a defined developmental window
The demonstrated developmental plasticity of early embryonic cells has provided a useful test bed for exploring in vivo mechanisms that control pluripotency and subsequent developmental commitment. Although early descendants in each founder cell lineage retain the ability to be transdetermined into any of three germ layer types, this multipotency is lost by midembryogenesis, after which embryos undergo essentially normal development and differentiation when challenged with widespread expression of cell fate-determining TFs (Zuryn et al. 2012; Spickard et al. 2018). Thus, embryos transition from a multipotent state into one in which cells resist reprogramming into other cell types. This multipotency-to-commitment transition (MCT), occurs within a few hours of fertilization, at around the 200–300 cell stage, or after the E lineage has divided to produce eight descendants (8E stage, Figure 2; the simple E lineage provides a convenient landmark for scoring various stages in embryonic development). Most, if not all, animal embryos undergo a similar transition from a plastic to a committed state of differentiation (Boroviak and Nichols 2014; Bernardo et al. 2018); hence, the MCT generally marks a major event that occurs during embryogenesis across metazoan phylogeny.
Molecular events controlling plasticity of early blastomeres and timing of the MCT
The molecular processes underlying the MCT have been investigated by analyzing genes whose loss delays its onset, resulting in extension of the period during which cells can be redirected into other developmental pathways (Spickard et al. 2018). Concomitant with this transition into commitment, nuclei undergo a change in structure that was proposed to reflect a global increase in the condensation of chromatin, suggesting conversion to heterochromatin (Yuzyuk et al. 2009). Consistent with this notion, components of the PRC2 chromatin remodeling complex, including MES-2, which encodes the H3K27 methyltransferase catalytic subunit, and MES-3, were found to be required for the normal onset of the MCT (Yuzyuk et al. 2009). Embryos deficient for this complex are delayed in commitment to normal pathways of differentiation, as evidenced by the ability to redirect normally nonmuscle cells into muscle when forced to express the muscle-determining factor HLH-1 (Figure 2) (Yuzyuk et al. 2009). The delayed transition from a multipotential to a committed state in mes-2 mutants is paralleled by a widespread change in gene expression: genes that are normally expressed specifically during early embryogenesis (including the muscle-specific unc-120 gene, the endoderm-determining end-1 gene, and the ABa lineage-expressed tbx-37 and -38 genes) and whose expression is attenuated after the 4E stage, were found to be overexpressed and maintained expression after this stage, perhaps reflecting perdurance of an early embryo-like (uncommitted) state (Yuzyuk et al. 2009).
The ability to probe developmental plasticity by challenging embryos with forced expression of TFs provided an entry point into identifying genes that, when debilitated, result in a failure of cells to commit at the normal developmental stage. A screen for such genes revealed that Notch signaling contributes to establishing the MCT in the AB lineage. In embryos lacking the Notch receptor GLP-1, the MCT is delayed to well after the 8E stage, allowing END-3 to activate widespread, ectopic gut differentiation much later than in glp-1(+) embryos (Figure 2) (Djabrayan et al. 2012).
Maternally encoded GLP-1 is expressed specifically in AB and its descendants and Notch signaling is known to specify many cell types during the first several divisions of the AB lineage in response to signals from descendants of the P1 blastomere (Mello et al. 1994; Moskowitz et al. 1994; Hutter and Schnabel 1995; Moskowitz and Rothman 1996). Thus, specification of AB descendants per se might result in commitment of these cells, consistent with temperature-shift experiments that revealed that the stage at which GLP-1 functions in several early cell fate decisions also correlated with its temporal requirement in establishing the MCT (Djabrayan et al. 2012). However, some AB-derived cells apparently never receive cell-type-specifying Notch signals during embryogenesis and yet, as with Notch-signaled cells, appear to resist reprogramming into endoderm by END-3 at the appropriate time. In fact, it was found that the requirement for GLP-1 in determining developmental plasticity is, apparently, not dependent on the AB-extrinsic specification signals from P1 descendants, but is autonomous to the AB lineage: isolated AB-derived partial embryos, which cannot receive signals from P1 descendants, continue to show GLP-1-dependent activation of the MCT at the normal time (Djabrayan et al. 2012). The role of GLP-1 in establishing the MCT in the AB lineage independently of the known signaling ligands led to the finding that two noncanonical, apparently secreted DSL-like ligands, DSL-1 and -3, show a similar requirement for establishment of the MCT (Djabrayan et al. 2012). Thus, these putative ligands may act on GLP-1 specifically in the AB lineage to direct commitment to nonendoderm fate without specifying cell identities. As elimination of either PRC2 or Notch function (in the AB lineage) results in superficially similar effects, i.e., delay of the MCT and persistence of developmental plasticity, it is conceivable that Notch acts through the PRC2 system to alter chromatin accessibility to reprogramming. Of note, a similar relationship between GLP-1/Notch and PRC2 has been unraveled in germline-to-soma conversion, as described below (Seelk et al. 2016).
It is believed that deployment of the gene regulatory networks that direct embryonic development progressively restricts the developmental fates of cells through the action of TFs and cis-regulatory sequences. Such networks establish autoregulatory “lockdown” circuits that underlie commitment to specific pathways of differentiation (Davidson and Levine 2008; Davidson 2010). The findings described above suggest that additional mechanisms operate independently of these transcriptional networks, and function to provoke developmentally plastic cells to commit to particular differentiated states. Given that mes-2(−) embryos are viable, as are dsl-1(−) and -3(−) mutants, timely activation of the MCT does not appear to be critical for successful differentiation and development, at least in conditions in which cell fate acquisition is not challenged. Thus, systems that commit initially developmentally plastic cells to differentiation may function to ensure developmental fidelity per se. As such, these systems may function independently of pathways that control the differentiated fate of cells.
Cellular Plasticity of Specialized Cells: Reprogramming and Transdifferentiation—an Overview
Is cellular plasticity a property restricted to stem cells or to progenitors found in developing embryos? Classical developmental experiments highlighted very early on that differentiated cells have an inherent cellular plasticity that is not normally expressed but that can be experimentally revealed. For instance, late 19th century studies showed that newts can regenerate their lens after complete removal, and that the likely source of the regenerated lens is pigmented epithelial cells of the dorsal iris (Collucci 1891; Wolff 1895; Kodama and Eguchi 1995). Cellular plasticity was also demonstrated for cells that are not yet fully specialized: grafting the dorsal lip of the blastopore of a frog or newt embryo onto the ventral side of a receiving embryo leads to the induction of a second body axis: the fate of the ventral cells of the receiving embryo is changed by the graft (Mangold and Spemann 1927). Later, John Gurdon found that the transplantation of a differentiated somatic nucleus into an enucleated frog egg led to the reprogramming of the differentiated nucleus, allowing the development of fertile frogs (Gurdon 1962; Gurdon and Uehlinger 1966). Thus, very early on, it appeared that, at least under specific conditions, specialized cells could exhibit cellular plasticity. This and subsequent sections will focus on the cellular plasticity of specialized cells, e.g., differentiated tissue cells and germ cells.
In fact, the ability of cells that have acquired specialized identities in multicellular organisms to undergo conversion into other cell types has been observed throughout both the plant and animal kingdoms, including members of the ecdysozoan and lophotrochozoan branches of protostomes and the deuterostomes. Cell fate conversion has been described in both natural and experimental settings outside of development. Examples include regeneration following injury, in which cells of a variety of types, and entire structures, can be provoked to develop from either differentiated cells or multipotential progenitors present in the tissue, as occurs, for example, in regenerating amphibian lens (Henry and Tsonis 2010). In the context of disease, cell type conversions and metaplasias might be at the basis of a variety of cancers (Syder et al. 2004; Means et al. 2005). While resistance to the notion that specialized cells could change their identity was initially strong, a new way to induce cellular plasticity experimentally has been described more recently in landmark experiments. Indeed, pluripotent stem cells have been engineered in vitro starting from differentiated cells (Takahashi and Yamanaka 2006) that exhibit similar properties to ES cells, and can also be grown indefinitely in culture. Such feats, called pluripotent reprogramming, necessitate the forced expression of a cocktail of transcription factors endogenously expressed in ES cells and important to maintaining their pluripotency. Although variations in the cocktail composition exist, and small molecules replacements for some of these factors have been attempted, four factors are commonly used: Oct4, Sox2, Klf4, and cMyc (Takahashi and Yamanaka 2006; Takahashi et al. 2007). It should be noted that, while all differentiated cell types used to date have been reprogrammed, the programming success rate is very low. These landmark experiments have an important conceptual implication, as they imply that pluripotency in not an inherent property of a stem cell entity, but a state that can be acquired. In addition, these studies, together with the early descriptions of unexpected cellular plasticity events, have decisively shown that the differentiated cellular identity is not fixed, and opened the door to the possibility that cellular plasticity may not be a property uniquely found in stem cells or blastomeres.
Various terms can be found in the literature to describe similar or distinct cellular plasticity events (see Box 1 Definitions). Cellular reprogramming, transdetermination, and Td all share the common characteristic that the normal fate of a cell is altered to that of another cell type, and generally involve remodeling of the transcriptional state of a nucleus, a process that translates into morphological and functional changes. Though “cellular reprogramming” can also be found in the literature with a very narrow application, that is the reprogramming of differentiated cells into an embryonic-like stem cell called induced pluripotent stem (iPS) cell, cellular reprogramming is a more generic term that does not imply the degree of specialization of the initial or final identities. We have thus used “pluripotent reprogramming” to designate reversion from a differentiated identity to an ES-like state. By contrast, transdetermination involves a switch in the fate of progenitor or stem cells from one lineage type into another, and Td applies to events in which cells that have adopted a differentiated fate are converted to another differentiated cell type [for the first use of Td in English, see Selman and Kafatos (1974); for definition and criteria, (Kodama and Eguchi 1995)]. It is important to note that the definition of these processes is based on the initial and final identities and their switch after a single inducing event, but not on the cellular or molecular mechanisms underlying the conversion itself, which are in many cases unknown and may vary in different contexts. In addition, “direct reprogramming” or “direct cell type conversion” are also found in the literature instead of Td, often to describe experimentally induced processes in cell culture assays. For clarity, we propose to use them indistinctly, but to specify, by using “natural” or “induced,” when these processes are observed in nature or have been triggered experimentally respectively (see Box 1 Definitions). While applications taking advantage of cellular plasticity have been pushed forward more recently, these concepts have deep roots in developmental biology.
Transdetermination was first used to describe the conversion of imaginal disk identity in Drosophila larvae, from leg to wing, for example (Garcia-Bellido 1966; Maves and Schubiger 1999; Worley et al. 2012) as a result of environmental changes or injury. This redirection of progenitor cells that have not yet become specialized can be contrasted with bona fide Td events. This latter process, again first described in insects (Selman and Kafatos 1974) has been observed both in dividing and nondividing cells.
The distinction between transdetermination and transdifferentation of fully differentiated cells cannot always be precisely defined, the limitation being the precision with which it is possible to define the extent of differentiation of the initial cell prior to its reprogramming. However, alteration of a lineage trajectory before cells have adopted a specific function—for example, when the fates of blastomeres in early C. elegans embryos are redirected as a result of loss of gene function or forced expression of TFs, or as occurs with homeotic mutations and when an eye is induced at the end of a leg by forced expression of eyeless in Drosophila—is definitively transdetermination. When the cell, prior to its conversion, has already performed a differentiated function—as with the rectal epithelial cell in C. elegans, which converts to a specialized neuron (Jarriault et al. 2008), or with fibroblasts that can be converted to muscle cells as a result of forced MyoD expression (Davis et al. 1987; Tapscott et al. 1988)—the event can be definitively considered Td.
Germ cell reprogramming is a special case, complicating the distinction between transdetermination and Td. Developing germ cells are a distinct population of cells, which one may view as very specialized, that are undergoing the process of later transforming into a totipotent embryo. As such, conversion of germ cells into differentiated cells may be the result of derepression of somatic differentiation that is poised to occur following fertilization. This might be exemplified by the acquisition of differentiated somatic features of several lineages by germ cells mutant for translational regulator-coding genes (Ciosk et al. 2006). Though the distinction is partly semantic, and similar molecular mechanisms might be at play, the latter process could be considered premature activation of the normal—and poised—embryonic program, rather than a de novo switch from one transcriptional program to another, as would be expected for transdetermination or Td.
The goals of studying mechanisms of developmental reprogramming in model organisms such as C. elegans are both fundamental and practical. The intensive genetic approaches available with the system can be highly informative as to the stepwise mechanisms and key molecular regulatory processes underlying conversion of one cell type to another within an in vivo context. In addition, one key aspect and nagging technical limitation in multicellular organisms is the ability to follow and unambiguously assess the cell type conversion at the single cell level. With its transparency and fully described cellular lineage (Sulston and Horvitz 1977; Sulston et al. 1983), the worm offers a unique model in this respect. Although cell identity can be changed either naturally or experimentally, it is of importance to understand at the fundamental level at what stage in the differentiation process there is a restricted or unique pathway that must be followed to acquire that identity. Moreover, it is critical to learn the extent to which the state of the nucleus is identical in two cells that ultimately adopt the same differentiated fate, either during normal development from a multipotent progenitor, or as a result of Td. This information can also guide the adaptation of reprogramming approaches in regenerative medicine. To safely replace damaged or lost cells resulting from injury or disease with genetically identical cells from a patient that have been created by inducing differentiation of stem cells or Td of a different cell type requires a complete understanding of the similarities between the natural and induced cells at the level of both gene expression and epigenetic states, and the stability of the induced state once transplanted. Such studies might also advance insights into which source of cells, whether directly from patient-harvested tissues, or from those cultured for long periods, affects the efficiency and longevity of the transdifferentiated replacement tissues.
Current Questions in the Field that can be Addressed with Studies of Plasticity in C. elegans
The ability to experimentally induce cellular reprogramming, either to another differentiated identity, or to a pluripotent state, as well as the existence of natural reprogramming events, have triggered a booming new field of investigations. Here, we summarize the main questions that have emerged, and are or can be pursued using C. elegans as a model system, with a focus on induced and natural direct reprogramming (aka Td).
What are the cellular and molecular mechanisms underlying natural and induced direct reprogramming?
The worm allows for systematic genetic analyses, a powerful tool, which, combined with candidate approaches and cellular analyses, has and further will allow the steps and the requirements to change the initial identity involved to be deciphered, including barriers and drivers, the importance of cell division, any potential requirement for transient reversal to a lineage-related progenitor/stem cell-like state, and the redifferentiation process.
Do the mechanisms of reprogramming differ between different cell types?
The field still awaits a systematic side-by-side comparison of different Td events in a given species. Several cell type-specific characteristics could impact the process. For instance, overexpression of a given TF does not lead to reprogramming of all cell types (Davis et al. 1987), possibly pointing to different mechanistic paths. In addition, some natural Td events appear to involve cellular division, while others do not. The actual requirement and contribution of cell division, or of DNA synthesis, to reprogramming and whether it reveals mechanistic differences remains to be determined. Of note, studies on the microevolution of the vulva in nematodes (Srinivasan et al. 2001; Félix 2005) have shown that different mechanistic strategies can lead to the same end point. A related question is to examine to what extent are redirection of a cell’s commitment (i.e., transdetermination) and Td mechanistically related. In addition, it may be informative to compare germ and somatic cells. Indeed, how is germline multipotency regulated and how are germ cells protected from reprogramming? Are these mechanisms similar to how the differentiated state of somatic cells is maintained?
Are the steps, molecular and cellular processes in natural vs. induced reprogramming identical, overlapping, or entirely distinct? Since the end point between different reprogramming events varies (from diverse differentiated identities to a pluripotent one), it is likely that aspects such as those involved in redifferentiation will differ between reprogramming events. However, it is conceivable that all reprogramming events, natural and induced, require the proper erasure of the initial identity and that the networks allowing this erasure share common features. In fact, the finding that homologous factors are involved in both erasure of the initial identity during natural Td in worms and in the induction of pluripotent reprogramming in mammals argues for the possibility that general principles underlying the cellular plasticity of the differentiated identity may be found (Kagias et al. 2012; Becker and Jarriault 2016). Further studies should shed light on whether shared mechanisms will be found and whether induced reprogramming events borrow from mechanisms that naturally exist.
Does reprogramming occur through gradual, continuous process or discrete steps?
We tend to view differentiation as an incremental process represented by a continuum of very close states, both at the cellular and molecular levels. This view has been very much influenced by the classical representation of differentiation along, for example, the hematopoietic lineage in vertebrates. Does reprogramming, and direct reprogramming in particular, proceed similarly? Related, is a pluri/multipotent intermediate necessary? While some studies of cell populations have suggested a gradual morphing from one identity into another, with some mixed intermediate features after direct reprogramming is triggered (Zhou et al. 2008), studies at the single-cell level in the worm suggest a process that involves clear ruptures from one state to the other (Richard et al. 2011). Subsequent transcriptomic analysis on mammalian cell populations induced to directly reprogram have given support to this hypothesis (e.g., Di Tullio et al. 2011), and single cells transcriptomic data are expected to bring more answers to this debate. Determining the cellular and molecular states and the nature of their transitions involved during direct reprogramming, may have a broader conceptual impact as it may change as well how we conceptually view the natural differentiation process.
What is the range of cell types that can be reprogrammed?
All cell types that have been used for pluripotent reprogramming have yielded the production of iPS, albeit with a very low efficiency. However, specific inducing cues used for direct reprogramming have sometimes been successful at reprogramming of given cell types but not others, suggesting that a given reprograming cue does not have the same efficiency in different cell types (Davis et al. 1987; Zhou et al. 2008; Riddle et al. 2016). This may be due to the intrinsic cellular context, the micro- and macro-environment, or a combination of both. In addition, the induced reprogramming efficiency is very low, with only some cells in the population seemingly more amenable to changing their identity while most cells do not. It is thus important to address what makes a cell amenable to reprogramming, whether some cells are more prone to it, and why, as this will determine if there are limits to the cellular repertoire that can be targeted for reprogramming, or if any cell can be converted into any other cell type.
To what extent does functional or lineal relatedness predispose cells to reprogramming into a particular cell type?
In other terms, does the initial identity of a cell influence what its final identity can be? For instance, identity swaps between different cell types within the same germ layer, or between cells that perform similar functions, such as epithelia, could be more readily implemented. Many of the described reprogramming events that occur naturally during development involve a switch of identities within one germ layer, suggesting that such events might be more easily evolvable (Selman and Kafatos 1974; Monier et al. 2005; Jarriault et al. 2008; Sprecher and Desplan 2008; Gettings et al. 2010). However, examples of trans-germ layer reprogramming have been described, albeit with low efficiency, when experimentally induced (Vierbuchen and Wernig 2011), suggesting that germ-layers boundaries can be crossed, at least experimentally. But if there are barriers to reprogramming, does the strength of these barriers increase with the degree of unrelatedness between the final identity and the initial one? Alternatively, it could be that related cell types share factors and properties that maximize the action of a given inducing cue when experimentally forcing reprogramming of one into the other. Such investigations will determine whether there are limits to the repertoire of final reprogrammed identities for each starting cell type.
Current Road Blocks, Conceptual Relevance, and Experimental Contribution of C. elegans
A number of technical road blocks exist that have slowed down obtaining answers to the questions listed in the previous section. A major limitation at the base of many controversies in the direct reprogramming field relates to the ability (or lack thereof) to trace reprogramming events at the single cell level, and unambiguously assess the lineal relationship between the initial and the final identity. Indeed, such an unambiguous relationship is required to exclude that the initial cell population was not homogenous and contained undetected progenitors of the final identity. Accurate cell-tracing is still a challenge in many models, and, when such tools are available, relies on the reliability of these tools for following precise cells over time. For instance, when based on the use of a promoter that labels a cell population, detailed knowledge is required regarding the range of cell identities, especially within a lineage, that this promoter labels. Another limitation is that in vivo Cre-Lox label-retaining approaches can be subjected to variations (Comai et al. 2014). In addition, the accessibility and transparency of the region observed to tracing and imaging of transdifferentiating cells over time are other important factors that arise in in vivo analyses. Furthermore, the efficiency of reprogramming influences the ability to predict which cells will be reprogrammed. Indeed, apart from a few instances that stand out (Xie et al. 2004; Di Tullio et al. 2011), induced direct reprogramming efficiency is usually low, and synchronization of the process can be an issue. This low efficiency precludes easily following and imaging the process at the single-cell level. Thus, while population studies can be performed, events or molecules that are unique to the few reprogramming cells in this population may be masked. It is expected that the availability of sensitive approaches that can be performed at the single cell level on multiple cells at once, such as Next Generation Sequencing, will greatly improve the ability to study direct reprogramming dynamically. These challenges have slowed down the ability to study the process, the early steps in particular, and to elucidate the molecular and cellular mechanisms underlying it. Another crucial aspect is the ability to obtain, and be able to assess the completeness of the cell type conversion, both at the genetic and epigenetic levels. This may affect aspects of the functionality of the cell made, as well as its stability over time. Finally, this stability of the final cell, in particular when in a physiological, multi-signal environment, and long after any inducing cue has been removed, is a crucial aspect to evaluate, especially when translational applications are envisioned.
C. elegans-specific features provide unique assets to study Td
Above all, the knowledge of the detailed and stereotyped somatic lineage (Sulston and Horvitz 1977; Sulston et al. 1983; White et al. 1986) of the worm, and its associated cellular anatomy, provides unique and unmatched advantages: whether natural or induced direct reprogramming is studied, the cells that have changed their identity can be unambiguously identified, as well as the variability of the process in terms of the number of these cells or their identity, from one experiment to another. The early description of the lineage subsequently made it possible to identify many cell- and tissue-specific markers, further increasing the investigator’s ability to study each identity’s characteristics. In addition, owing to the complete transparency of the worm, the process can be followed in live animals, over time, in given cells. Important questions such as the function of the naturally transdifferentiated cells and their physiological integration, or the ageing properties of the experimentally or naturally converted cell, can thus be directly examined in vivo.
One of the key advantages of the worm, besides knowledge of its cellular lineage, is its amenability to genetic studies. This enables not only the testing of candidates using existing mutants, newly engineered ones via the CRISPR-Cas9 technology, or using reverse genetic approaches including RNAi, but, importantly, it enables the identification of genes involved in the process without preconceived ideas, by conducting large-scale forward genetic screens. Such systematic, intensive screens have proven time and again their power to unravel novel concepts and molecular players (Jorgensen and Mango 2002; Pasquinelli and Ruvkun 2002; Greenwald 2012). In addition, owing to the availability of live fluorescent reporters, these screens can be designed to target one or a few specific cells, as well as to target specific steps or time points in a process. This brings an unparalleled level of precision and allows an effective mutant analysis process as relevant primary mutations directly affecting the process are more likely to be isolated.
We should bear in mind, however, that there are a number of limitations associated with the model: while C. elegans is very amenable to in vivo studies, the lack of a long-term cell culture system has limited the ability to study Td in vitro. In addition, owing to the cuticle of the worm acting as an exoskeleton, as well as to the small size of cells and nuclei, cell or nucleus transplantations are not currently possible. This limitation precludes a number of useful tests on cells’ reprogramming abilities and functionality. Nuclear transfers would be very useful to assess for instance the reprogramming capacity of a naturally transdifferentiating cell’s content. Moreover, cellular transplantation of an experimentally induced reprogrammed cell into the tissue corresponding to its final identity would be very powerful to test whether it can function as such. And transplantation of a naturally transdifferentiating cell at different locations, or at the same location but at different developmental stages would make it possible to test the impact of the micro-environment. It is hoped that the technology for such micro-transplantations will become available in the future to expand the available tools. Of note, blastomere isolation and recombination have been used (Goldstein 1992), which, combined with our increasing abilities to purify single cells from whole worms using a combination of fluorescent markers, may allow some of these tests to be performed.
Overall relevance and how C. elegans contributes new concepts and paradigms
One important question is whether the study of particular reprogramming events in the worm (or in any given organism) will yield general insights into cellular plasticity that will apply to other reprogramming events within the same species and across phyla. More than a century of work on small model organisms, such as yeast, Drosophila, and, more recently, C. elegans, have shown that the fundamental principles governing the cell’s organization, identity acquisition, and function are very similar across species, and shared with mammalian cells. As we will see below, the study of the maintenance, or of the reprogramming, of cellular identity in the worm is also very likely to highlight general cellular principles. Owing to the attributes of the worm, answers to a number of critical questions have emerged in the last decade that examine all issues related to the control of cellular potential and identity at different developmental stages: how is the latent pluripotency in the embryo controlled?
Natural Reprogramming in the Worm
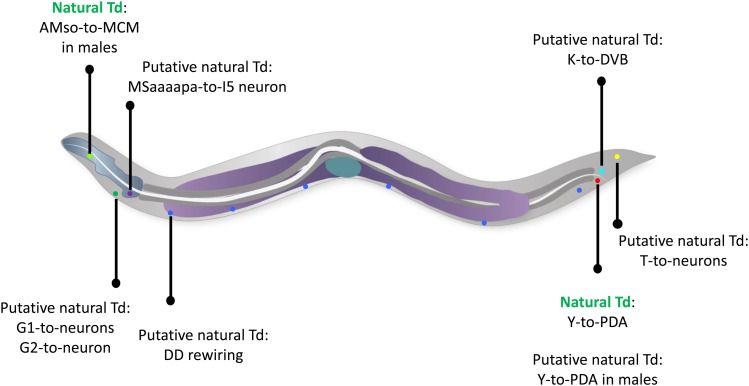
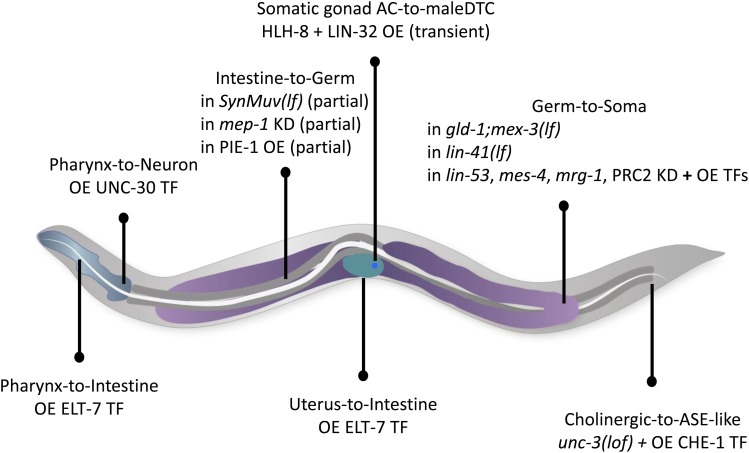
During the last decades, several instances of natural reprogramming have been described in the worm (Figure 3). These include extensive (rectal-to-neuronal, glial-to-neuronal) and possibly (neuronal-to-neuronal) mild identity changes.
Figure 3.
Schematic representation of the demonstrated and putative natural transdifferentiations in the worm. The initial and final identities, as well as their approximate localization in the worm are shown. Blue, pharynx; dark gray, intestine; purple, germline; green, uterus. Individual cells are represented as colored dots; red: rectal Y cell, green: amphid socket (AMso), in the male; dark green: G1 and G2 excretory pore cells; turquoise: rectal K cell; blue, DD neurons; dark blue: MSaaaapa cell; yellow: phasmid socket T cell. Anterior is to the left, and ventral to the bottom.
A rectal-to-neuronal cell identity conversion
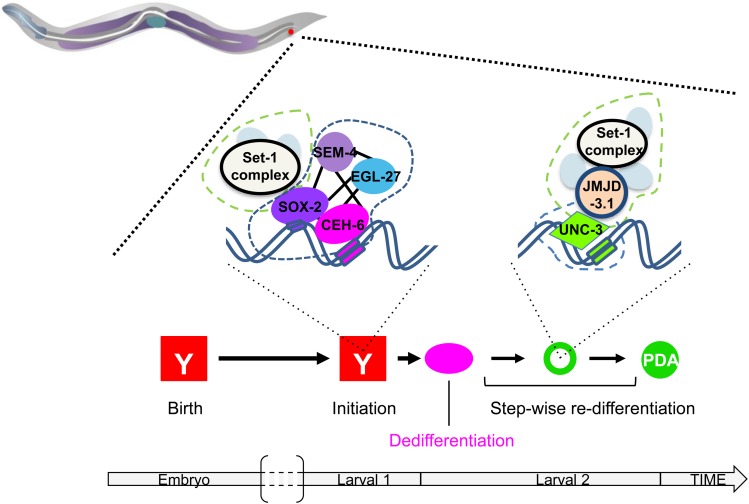
The rectum is a tube formed during embryogenesis and made of six cells, organized in three rings of two cells each (http://www.wormatlas.org/hermaphrodite/rectum/Rectframeset.html). The cells that form the rectum, named Y, B, U, F, K, and K’ (Figure 4), are born in the embryo ∼300 min after fertilization (Sulston et al. 1983). In hermaphrodites, they mostly remain rectal cells until the death of the animal. The Y cell, however, has an extraordinary behavior: by the end of the L1 larval stage, it will retract from the rectum, migrate away, and transform into a specific motoneuron, named PDA (Figure 3 and Figure 4). PDA extends a ventral axon posteriorly, and, after a commissure to the dorsal side of the animal, extends its axon toward the head of the worm and its synapses onto dorsal muscles (White et al. 1986; Jarriault et al. 2008). This transformation represents a bona fide natural Td event (Jarriault et al. 2008): their appearance as observed using Differential Interference Contrast (DIC), their electron microscopy characteristics, and the analysis of the molecular markers expressed in Y and PDA show that these represent two distinct cellular identities (Sulston et al. 1983; Jarriault et al. 2008). It is interesting to note that this direct reprogramming event occurs in absence of cell division.
Figure 4.
Rectal-to-neuron transdifferentiation. Rectal Y-to-PDA neuron Td occurs through a succession of discrete steps, including a dedifferentiation step, where the cell is stripped of its initial rectal identity without acquiring more cellular potential, followed by apparent step-wise redifferentiation into the PDA neuron. This succession of cellular steps is mirrored at the molecular level by step-specific combinations of histone modifier complexes (outlined in dashed green), such as JMJD-3.1 and the SET-1/MLL complex, and transcription factor complexes (outlined in dashed blue) such as a NODE-like + SOX-2 complex, and the UNC-3/COE TF.
Y-to-PDA Td: cellular steps, molecular processes:
To test potential external influences, the impact of the neighboring cells on this process was tested. Ablation experiments suggested that individual rectal cells did not influence the ability of the Y cell to change its identity (Jarriault et al. 2008). It was determined that migration of Y away from the rectum did not provide a needed micro-environment that would promote its cell type conversion, as inhibition of its migration did not preclude Td into a PDA neuron. In addition, while Y retraction from the rectum is concomitant with its replacement by the newly born P12.pa cell, both events are independent (Jarriault et al. 2008). In addition, the control of the timing of this Td event remains unclear, and the heterochronic pathway does not appear to be involved (Jarriault et al. 2008). Thus, no immediate external influences have been identified. To identify the intrinsic and putative extrinsic mechanisms underlying Y natural Td (Table 1), forward and reverse genetic screens have been carried out. The design of these screens has made it possible to target this event at the single cell level, and to target all possible steps of this process, by using transgenic animals where the final PDA identity is labeled, then looking for animals unable to make the PDA neuron in the progeny of mutagenized worms (Zuryn et al. 2010). Additionally, screens targeting the early events, before the Y cell leaves the rectum, have also been designed. Such screens take advantage of transgenic animals where the initial Y rectal cell is labeled, making it possible to look for animals unable to initiate Td after gene inactivation (Kagias et al. 2012).
Table 1. Necessary factors for natural transdifferentiation in C. elegans.
| Natural transdifferentiation | |||||
|---|---|---|---|---|---|
| Event | Involved factor(s) | Step | Protein domains and function | Human homolog | Reference |
| Y-to-PDA | sox-2 | Initiation of Td | Homeodomain TF | SOX2 | Kagias et al. (2012) |
| ceh-6 | Initiation of Td | Pou domain TF | POU3F4/OCT | Kagias et al. (2012) | |
| sem-4 | Initiation of Td | ZnF TF | SALL | Jarriault et al. (2008) | |
| Kagias et al. (2012) | |||||
| egl-27 | Initiation of Td | BAH, ELM2, SANT coregulator | MTA1 | Kagias et al. (2012) | |
| egl-5 | Initiation of Td | Homeobox TF | HOX | Jarriault et al. (2008) | |
| Kagias et al. (2012) | |||||
| SET1 complex | Initiation of Td | H3K4 methyltransferase complex | SET1/MLL | Zuryn et al. (2014) | |
| Redifferentiation | |||||
| unc-3 | Redifferentiation | HLH TF | COE | Richard et al. (2011) | |
| jmjd3.1 | Redifferentiation | H3K27 demethylase | JMJD3 | Zuryn et al. (2014) | |
| lin-12 | Competence to Td | EGF repeats TM receptor | NOTCH | Jarriault et al. (2008) | |
| T. Daniele and S.J., unpublished results | |||||
Transition occurs through discrete cellular steps:
In several instances of TF-induced direct reprogramming, expression of markers of the target tissue can be obtained, but the original expression program is not completely switched off (Weintraub et al. 1989; Marro et al. 2011). These incompletely reprogrammed cells suggested that transition through a mixed identity may occur. The C. elegans mutants retrieved in the first screen described above presented several phenotypes, suggesting that different steps existed that may be affected (Zuryn et al. 2010; Richard et al. 2011). By contrast, no intermediate with a mixed rectal-epithelial/neuronal identity was detected (Richard et al. 2011). A dynamic assessment of the Y cellular transition at the single cell level was further performed to elucidate the exact cellular steps of Y Td. These detailed analyses revealed that the Y cell transitions through distinct discrete identities, the first involving the complete erasure of the initial rectal identity. This apparent dedifferentiation is not coupled to an increase in cellular potential, as overexpression of cell fate determinants that reprogram the early embryo were not able to convert the Y cell at any stage in the process (Richard et al. 2011). After this dedifferentiation phase, redifferentiation takes then place in a step-wise manner that may mimic developmental neural differentiation and requires the UNC-3/EBF transcription factor (Richard et al. 2011). Thus, cell type conversion does not necessitate transition through a pluripotent ground state to be complete and no stable intermediate is generated (Figure 4).
Initiation is controlled by conserved nuclear factors:
Four conserved nuclear factors have been identified that are each crucially needed for the initiation of Y Td and are found to interact in a complex (Figure 4 and Table 1): sox-2, ceh-6/Oct, sem-4/Sall, and egl-27/Mta (Kagias et al. 2012). Their mammalian counterparts are individually known to impact on mouse ES cells pluripotency and to associate with (SOX2) or as (OCT4, SALL4, and MTA1) NODE complex members (Liang et al. 2008). Their activity in a natural cellular plasticity process in the worm suggest (i) that nuclear events are key for the dedifferentiation step, and (ii) that these four factors may together represent a conserved plasticity cassette. While strictly required for the dedifferentiation step, these factors are expressed in the other rectal cells that never change their identity, and are thus likely to provide a permissive context for Y Td rather than an inductive cue (Kagias et al. 2012). This may be true in mammals as well, as recent studies have used ectopic pluripotency factors such as OCT4 and SOX2 to increase the efficiency of the induced direct reprogramming process (Szabo et al. 2010; Efe et al. 2011; Kim et al. 2011). Taken together, these data suggest that a conserved function of the combination of these factors is to “open” the cell’s ability to initiate a reprogramming event, rather than systematically inducing reversion to a pluripotent state. It further opens the intriguing possibility that pluripotent reprogramming may highjack mechanisms that in fact naturally exist.
Invariance of natural Td is ensured by successive layers of histone modifying activities:
A striking aspect of natural Td is its associated efficiency and reproducibility. For instance, lens regeneration in newt has been shown to occur perfectly after 19 successive lensectomies over a span of 16 years (Sousounis et al. 2015). Similarly, Y Td is consistently observed in all wild-type animals, irrespective of the culture conditions (Zuryn et al. 2014). To probe the mechanisms that underlie the invariance and efficiency observed in natural conversions of one cell type into another, weakly penetrant mutants affecting Y Td were examined. The reasoning was that the low penetrance in such mutants, when not due to redundancy with other activities or to partial loss-of-functions, may point to genes involved in ensuring a perfect process every time. This way, multiple, conserved histone-modifying activities (JMJD3, SET1) acting on distinct lysines of histone H3 were found to mediate invariant cell conversion. Importantly, these activities were involved dynamically at different steps of the process, and the balance between different methylation marks appeared determinant for progression through Td. Genetic and biochemical analyses, as well as the use of a photo-convertible fluorescent protein, showed that partitioning of these histone-modifying activities is achieved through the active and timely regulation of nuclear JMJD-3.1 protein levels and phase-specific association with critical TFs that have conserved roles in cell plasticity (i.e., the NODE-like complex member and SOX-2) or terminal fate selector (e.g., UNC-3) (Zuryn et al. 2014) (Figure 4 and Table 1). It is noteworthy that these associations may represent conserved roles for these histone modifiers in mammals, as WDR5, a member of the SET1 complex, associates with OCT4 during the initial phases of pluripotent reprogramming (Ang et al. 2011); moreover, JMJD3 is required for adult neuronal differentiation, a process that resembles a Td, in line with its activity during the redifferentiation phase of Y Td (Park et al. 2014). Finally, SET1-dependent H3K4me3 methylation and JMJD-3-dependent H3K27me3/me2 demethylation were found to be not only necessary for optimal Td, but also critical to provide protection against variations that may be encountered in the worm’s natural environment, thus ensuring the robustness of cell conversion under suboptimal conditions (Zuryn et al. 2014). Together with the findings that TFs are critical for the process (Kagias et al. 2012), these data suggest a model where key TFs drive the conversion of cell identity through the different phases of dedifferentiation and redifferentiation while the interplay of different histone modifications ensures its invariance.
MCM neurons arise from the amphid socket cells in males
More recently, a second putative natural Td event has been reported in C. elegans (Figure 3), when a novel class of neurons was discovered (Sammut et al. 2015). These two neurons, named MCM, arise in males during the late L3 stage and appear to be fully differentiated in L4 larvae. Thus, the MCM neurons, which have been shown to be necessary for sex-specific learning, are made timely, when males undergo sexual maturation and their behavior changes. Examination of an S-phase reporter, together with ablation experiments. showed that the MCMs originate from the division of the two amphid socket (AMso) cells (Sammut et al. 2015). Amphid sockets, equated to glial cells, are support cells of the head sensilla, which wrap around the distal end of the latter and form the junction with the hypodermis on the left and right side of the worm (http://www.wormatlas.org/hermaphrodite/neuronalsupport/Neurosupportframeset.html, (Shaham 2015). While AMso cells do not divide in hermaphrodites, each AMso cell in males generates two daughters with asymmetric fates: one remains the adult AMso, and the other becomes an MCM neuron. Interestingly, this sex-specific process is intrinsically poised by the status of the sex-determination pathway in the AMso cells. Are AMso differentiated cells with a specialized function before they divide? Although function per se is difficult to assess, each mother AMso cell appears fully differentiated, exhibits the same morphology as in hermaphrodites or as the male adult ones, and expresses socket-specific markers. By contrast, becoming a MCM neuron, one daughter cell no longer shows expression of socket markers, and exhibits a new neuronal morphology (Sammut et al. 2015). Thus, while the mechanisms underlying this conversion, including the significance of the potentially asymmetric cell division, remain to be assessed, it appears that natural Td in C. elegans may include both events occurring in absence of cell division, and events including a cell division. Determining the impact of the cell division on the process and their possible shared mechanistic aspects will constitute future interesting questions.
Other putative plasticity events
Examination of the embryonic or larval lineage of the worm (Sulston and Horvitz 1977; Sulston et al. 1983) suggests additional putative Td events (Figure 3). For instance, the embryonic cell lineage revealed the production of neurons by the MS (mesodermal) lineage. MSaaaapa gives rise to the I4 pharyngeal neuron and its sister the pm5 pharyngeal muscle. A combination of two independent pathways, involving hlh-3 and hlh-2, is both necessary in the embryo for the anterior MSaaaapa daughter to become the I4 neuron, and sufficient to induce, at least transiently, the widespread expression of pan-neural transgenes in embryos or in other muscle and hypodermal cells of young larvae (Luo and Horvitz 2017). While the precise identity and differentiation status of the MSaaaapa mother and the extent to which its MSaaaapaa daughter is a mesodermal cell after its birth, and prior to adopting the I4 neuronal fate, remain to be determined, the formation of a neuron in this mesodermal lineage could represent an additional example of cellular plasticity in the worm. It should be noted that when the Td process includes a cell division, it will be important to rule out that asymmetric division per se, rather than Td, is enough to allow one daughter to adopt a different fate.
During larval development, a number of cells—the postembryonic “blast” cells—further divide to give rise to one or two daughter cells with a new cell type (Sulston and Horvitz 1977). Among these, a handful of blast cells appear to participate as a functioning part of an organ, indicative of a specialized function. Nonetheless, they divide, often only once, and produce cells of a different type. Of particular interest are the excretory pore G1 and G2 cells, the putative phasmid socket T cell, and the rectal K cell. The G1 pore cell is formed during embryogenesis (Sulston et al. 1983). This epithelial cell forms a small unicellular tube through a wrapping mechanism and establishment of autocellular junctions (Abdus-Saboor et al. 2011). In the middle of the L1 stage, the G1 pore delaminates to divide and give rise to two neurons (Sulston and Horvitz 1977; Parry and Sundaram 2014). The G2 cell concomitantly undergoes wrapping and establishes autocellular junctions to become the new excretory pore cell, until it divides and is replaced by its posterior daughter (Sulston and Horvitz 1977; Sundaram and Buechner 2016). The very different and specialized morphologies, functions, and marker expression of the G1, G2, and G2.p epithelial cells on one hand, and their neuronal daughters or sister on the other suggest that these cells undergo Td (Sundaram and Buechner 2016; C. Riva and S. J., unpublished results). Similarly, ultrastructural, morphological, and molecular studies suggest that the rectal K cell, which divides around the end of the first larval stage to give rise to a daughter that adopts the rectal identity and a daughter that adopts the DVB neuronal identity, also undergoes Td (Sulston and Horvitz 1977; White et al. 1986; C. Riva, C. Gally, H. Hajduskova, and S. J., unpublished results). Additional Td events include the formation of the PDA neurons in males, which occurs through the division of the Y cell, and the formation of neurons and glial cells in phasmid socket cells (T) lineage in hermaphrodites (Sulston and Horvitz 1977; C. Riva and S. J., unpublished results).
Finally, other plasticity events have been described in the worm that may represent more of a functional reprogramming than an identity change per se. Such events include the rewiring of the Dorsal D motoneurons during the L1 larval stage. During this stage, new ventral motoneurons (called VD neurons) develop, that will make synapses onto the ventral body wall muscles (BWM). Concomitantly, in a striking example of neuronal plasticity (Figure 3), extensive rewiring of the DD motoneurons occurs around the L1 molt, leading to the elimination of their ventral synapses (and input), and the formation of new synapses (and input) onto the dorsal BWM (White et al. 1978). This intralineage functional plasticity appears to be a genetically coordinated event in both presynaptic and postsynaptic partners (Gally and Bessereau 2003), and, thus, to be genetically programmed in the worm, very much in line with the swap in photoreceptor type or the switch from larval to adult cardiac myocytes that are observed during metamorphosis in the fly (Monier et al. 2005; Sprecher and Desplan 2008). Further investigations will be needed to determine if these narrower cellular plasticity events represent a simpler version of more dramatic Td events, or if they rely on fundamentally different strategies.
Induced Reprogramming in the Worm
There are documented examples in C. elegans of experimentally induced reprogramming from germline-to-soma, soma-to-germline, and soma-to-soma identities. These reveal both cytoplasmic and nuclear contributions to cellular identity maintenance and plasticity.
Germline-to-soma
The germline is the only tissue in bilaterian animals that is ultimately capable of producing all three somatic germ layers formed during embryogenesis. In C. elegans, as in other animals in which the germline is set aside starting at the earliest stages of development, it is also a bona fide immortal lineage that is passed from one generation to the next without passage through any somatic lineages. A number of maternally inherited transcripts are actively transcribed in developing germline cells. These transcripts, which will be packaged into mature oocytes, encode factors that initiate programs of somatic differentiation in the early embryo. Thus, there must exist mechanisms that ensure that germ cells do not translate these somatic differentiation factors from the accumulated maternal transcripts, thereby preventing premature somatic differentiation prior to the initiation of embryonic development by fertilization.
Cytoplasmic control of cell fate in the germline
Studies of the repression of somatic differentiation in the C. elegans germline have revealed critical regulatory mechanisms that act at the level of the cytoplasm. Indeed, the first studies to elucidate mechanisms that preclude somatic gene expression and differentiation in the germline demonstrated that translational inhibition is a major level at which somatic differentiation is blocked in the germline (Figure 5 and Figure 6).
Figure 5.
Schematic representation of the known experimentally induced transdifferentiations in the worm. The initial and final identities, as well as their approximate localization in the worm are shown. Blue, pharynx; dark gray, intestine; purple, germline; green, uterus, blue dot, somatic gonad AC cell. lf, loss-of-function; KD, RNAi-mediated knock down; OE, overexpression; TF, transcription factor. Anterior is to the left, and ventral to the bottom.
Figure 6.
Germ-to-soma transdifferentiation. Translational repressors and chromatin complexes safeguard the C. elegans germline identity: (1) In mutants for translational repressors or H3K4 methylation, listed in the blue box, germ cells (blue ovals) adopt several different somatic fates and resemble neuron (green cells) or muscle (beige cells)-like cells. Note that the germ-to-soma conversion seen in set-2 and hrde-1 mutants, members of the SET1 complex, arises progressively over several generations. (2) After knock down by RNAi of one of the following chromatin factors [lin-53, mes-2, -3, -4, -6, mrg-1, hmg-3; purple box], overexpression (OE) of a developmental regulator transcription factor (TF; white lettering) leads to specific germ-to-soma conversion: che-1 OE leads to ASE-like neuronal fate; unc-30 OE to a GABAergic neuronal-like fate; unc-3 OE to a cholinergic neuronal-like fate and hlh-1 OE leads to a muscle-like fate.
Two translational regulators, GLD-1 and MEX-3, both KH-domain-type RNA binding proteins (Draper et al. 1996; Jones et al. 1996) that function to regulate lineage-specific fates during early embryonic development (Draper et al. 1996; Mootz et al. 2004) are also key repressors of somatic differentiation in the totipotent adult germline (Figure 5, Figure 6, and Table 2) (Ciosk et al. 2006). GLD-1, which controls the transition of proliferating mitotic cells emerging from the stem cell niche into meiosis, is expressed along the distal and central region of the meiotic germline (Jones et al. 1996). MEX-3 is present in a complementary pattern proceeding from the proximal end of the GLD-1 expression domain through the proximal gonad, where it persists through the completion of oogenesis and into the early embryo (Draper et al. 1996; Ciosk et al. 2004; Mootz et al. 2004). GLD-1 functions critically in the control of the switch from mitotic germline proliferation in the distal gonad into meiosis and initiation of germ cell differentiation (Francis et al. 1995). GLD-1 represses MEX-3 expression in the germline (Ciosk et al. 2004; Mootz et al. 2004), raising the possibility that any potentially redundant role of GLD-1 and MEX-3 in other processes might be masked by derepression of MEX-3 in the absence of GLD-1. Indeed, it was found that simultaneous removal of both factors results in the appearance of a new phenotype—the presence of germline teratomas—in which clusters of differentiated cells characteristic of the three germ layers, including intestine, neurons, and both body wall and pharynx muscles, appear in the regions of the gonad in which germline meiosis and differentiation normally occur (Ciosk et al. 2006). This effect is also seen at a much lower frequency in gld-1(−) single mutants (Ciosk et al. 2006). Thus, the combined action of the MEX-3 and GLD-1 translational regulators prevents premature activation of the embryonic program of somatic differentiation during germline development. It is interesting to note that one of the known targets of the MEX-3 RNA binding protein is the worm homolog of the human pluripotency-inducing factor SOX2 (Pagano et al. 2009), providing additional support for possible conservation of mechanisms that either maintain totipotency or promote cellular plasticity across metazoans. The absence of MEX-3 and GLD-1 results in elimination of P granules in parallel with derepression of somatic differentiation in the germline, raising the possibility that the latter is attributable to loss of P granules per se, as has been reported in subsequent findings (see below).
Regulation of developmental plasticity in the germline at the cytoplasmic level also occurs during the oocyte-to-embryo transition. The heterochronic gene lin-41, which was originally identified based on its requirement in regulating the timing of stage-specific developmental events in late larval development (Slack et al. 2000; Tocchini et al. 2014), is required to prevent somatic differentiation in developing oocytes (Tocchini et al. 2014). LIN-41 is a cytoplasmic protein containing a TRIpartite Motif (TRIM) with a RING finger domain, B box, and coiled-coil domain (RBCC protein), and an NHL (NCL-1, HT2A, LIN-41) domain. In the context of germline development, lin-41 mutations were identified in a screen for mutants with proximal germline expression of vet-4/pes-2.1, a gene whose expression is normally initiated very early in embryogenesis (Seydoux et al. 1996; Biedermann et al. 2009). This phenotype appears to reflect a general derepression of RNA polymerase II transcription in oocytes, which are normally transcriptionally quiescent, as the initiation form of the enzyme (which contains a phosphorylated Ser5 on the C-terminal domain) is present in the proximal germline in lin-41 mutants, but not wild-type animals (Tocchini et al. 2014). Moreover, embryonic transcripts for a variety of cell types, including those associated with muscle, intestine, and neurons, which are normally absent in the germline, are expressed in these proximal cells, concomitant with formation of germline teratomas (Figure 5, Figure 6, and Table 2). The proximal germline cells also inappropriately switch from meiosis to mitosis and show persistent centrosomes, characteristic of early embryonic cells rather than cells undergoing oogenesis. These effects are associated with loss of germline identity, based on loss of expression of germline proteins that normally persist throughout the germline and that are not degraded until early embryogenesis.
LIN-41, and its TRIM-NHL relatives in other phyla, perform apparently evolutionarily conserved roles in controlling developmental plasticity across metazoans, in particular in regulating the balance between self-renewal and differentiation (Nimmo and Slack 2009). The Drosophila TRIM-NHL protein Mei-P26 functions in the ovarian stem cell lineage to regulate growth and proliferation and the Brat (“Brain Tumor”) gene represses self-renewal and growth during brain development in fly larvae (Betschinger et al. 2006). In both cases, removal of the protein results in tumor formation (Betschinger et al. 2006; Bowman et al. 2008; Neumüller et al. 2008). Another relative in mouse, TRIM32, also regulates self-renewal vs. differentiation in neural progenitors in part by activating expression of the murine let-7 microRNA (Schwamborn et al. 2009); however, in that context, it promotes differentiation and inhibits proliferating progenitor fate. In the C. elegans heterochronic pathway, the lin-7 microRNA inhibits LIN-41 expression, allowing for terminal differentiation (Reinhart et al. 2000; Vella et al. 2004). However, while the let-7/lin-41 relationship has been proposed to be a conserved regulatory mechanism for self-renewal vs. differentiation, let-7 does not appear to be required for LIN-41 function in preventing somatic differentiation in the germline. Although LIN-41, as with other TRIM-NHL proteins, regulates self-renewal vs. differentiation (as a heterochronic function) in somatic cells, reminiscent of its role in repressing somatic differentiation in the germline, the protein appears to act through different mechanisms in the two tissues, as mutations that interfere with its presumed RNA binding activity in the soma do not prevent its germline activity (Tocchini et al. 2014).
The acquisition of somatic differentiation in the germline is correlated with loss of the normally germline-specific P granules (Ciosk et al. 2006; Tursun et al. 2011), raising the possibility that P granules may function in repressing somatic gene expression. Indeed, depletion of PGL-1 and -3, which are required for cytoplasmic P granule nucleation, in conjunction with removal of GLH-1 and -4, which are necessary for localizing P granules to the nuclear periphery, leads to derepression of somatic cell fates in the germline, including muscle and neuronal differentiation (Updike et al. 2014). Derepression of a marker of neuronal gene expression (unc-119::GFP) in these animals suggested that P granules inhibit both transcription and translation of somatic genes, as transcripts were detectable more widely in the germline than the GFP marker. While general neural gene expression is activated in these animals, markers for a variety of specific neuronal subtypes were undetectable, suggesting that the cells did not resolve into the range of neural types that are normally made during embryogenesis. However, the neuronal cells can be induced to express ASE-type neuronal fate by forced CHE-1 expression in a lin-53 mutant background (Tursun et al. 2011); thus, the germline can be induced to become not only generic neuronal cells but also specific neuronal cell types (Figure 5, Figure 6, and Table 2). In contrast to the hermaphrodite-specific effects in gld-1(−) mex-3(−) animals (Ciosk et al. 2006), depletion of P granule function causes somatic differentiation in both hermaphrodites and males, implying their broader action in repressing somatic gene expression in the germline. It should be noted that the role of P granules in the maintenance of cell fate is not clearly established, or whether the loss of P granules causes, or is a consequence of, reprogramming. Moreover, the presence of detectable P granules per se does not block reprogramming: csr-1(−) mutants, which contain abnormally large P granules (Andralojc et al. 2017), show at least partial reprogramming (Fassnacht et al. 2018).
Nuclear control of pluripotency in the germline and germline-to-soma reprogramming:
Beyond translational control, what other molecular and cellular processes are sufficient to trigger cell type conversion? A number of studies in C. elegans have demonstrated a critical role for chromatin modification and epigenetic states, pointing to the important role of the nucleus in the regulation of germline pluripotency and repression of inappropriate somatic differentiation (Figure 5, Figure 6, and Table 2). The potential of the germline to undergo premature somatic differentiation has been demonstrated, in specific genetic backgrounds, as a result of ectopic expression of TFs that direct the differentiation of particular neuron types (Tursun et al. 2011). In embryos, forced ubiquitous expression of CHE-1—a zinc-finger transcription factor required for differentiation of the ASE gustatory neurons—results in widespread activation of a reporter under the control of CHE-1 binding sites throughout the animal, but not Td of non-neuronal cells. It is noteworthy that forced expression in the larval and adult stages led to expression of ASE reporters only in very few head sensory neurons, suggesting that the ability to challenge cell fate decreases with age (Tursun et al. 2011; Patel and Hobert 2017; Coraggio et al. 2019). However, in animals in which LIN-53—the C. elegans member of the chromatin remodeling RbAp46/48 retinoblastoma binding protein—is knocked down, CHE-1 was found to induce Td of germline cells into ASE neuron-like cells in the mitotically proliferating zone (Figure 5, Figure 6, and Table 2), as assessed using reporters and later on endogenous gene expression (Tursun et al. 2011). Knock down of LIN-53 allows germline cells to inappropriately express neuronal cell fates more broadly: UNC-3, an EBF-like transcription factor required for production of two types of cholinergic motor neurons (Prasad et al. 1998, 2008), and UNC-30, a Pitx-type transcription factor that promotes the differentiation of GABAergic motor neurons (Jin et al. 1994; Cinar et al. 2005) are both capable of activating differentiation into neuronal-like cells that express markers of their respective neuronal cell types in the germlines of lin-53(RNAi) animals (Tursun et al. 2011). The activation of somatic differentiation in the pluripotent germline may parallel the normal progression of events during embryonic neurogenesis, as the pan-neural hlh-2 gene, which is expressed throughout the developing embryonic neural tissue prior to terminal differentiation, is also expressed transiently in the germline during the germline-to-neuronal Td process (Tursun et al. 2011).
The histone chaperone family members that include LIN-53 act with several distinct complexes that remodel chromatin through distinct mechanisms (Eitoku et al. 2008). Polycomb Repressive Complex 2 (Margueron and Reinberg 2011), formed by MES-2/3/6 in worms, was also found to be required autonomously in the germline to block inappropriate neuronal differentiation activated by CHE-1 (Figure 5, Figure 6, and Table 2) (Patel et al. 2012). PRC2 directs H3K27 trimethylation, associated with repressed chromatin, and removal of mes-2, 3, or -6, or lin-53 results in loss of H3K27 trimethylation (Bender et al. 2004). Methylation of H3K27 by PRC2 is opposed by H3K36 methylation, which excludes PRC2 from autosomes, and is mediated by MES-4 (Rechtsteiner et al. 2010), a SET and PHD domain-containing histone methyltransferase (Yuan et al. 2011; Gaydos et al. 2012). As seen with removal of PRC2 components, depletion of MES-4 also allows CHE-1-mediated activation of neurons in the germline, an effect postulated to result from the observed redistribution of H3K27me3 and depletion of this modification broadly on somatically expressed gene that would otherwise be silenced by appropriate distribution of this modification (Patel et al. 2012). The potential of germline cells to undergo somatic differentiation in the absence of LIN-53/PRC2 components is not limited to neuronal differentiation, as it was found that muscle differentiation could be induced in the germline through forced expression of HLH-1 in this background. HLH-1 is a member of the MyoD transcription factor family that is required for normal muscle differentiation in the animal (Chen et al. 1994). Thus, as with the post-MCT embryo, where it acts to restrict developmental plasticity and block transdetermination (see above), PRC2 also functions to repress somatic differentiation in the germline.
The ability of PRC2 to block somatic gene expression in the mitotically proliferating (aka Mitotic Region or MR) zone of the germline is antagonized by GLP-1/Notch signaling, which is essential for proliferation of undifferentiated germ cells in the distal stem cell niche (Austin and Kimble 1987). In the absence of GLP-1/Notch function, mitotically proliferating germ cells are eliminated, and the germline cannot be provoked to undergo somatic differentiation, consistent with the observation that germline-to-neuron Td is observed in the MR (Tursun et al. 2011). In contrast, hyperactivation of the GLP-1/Notch receptor by a gain-of-function (gf) mutation, which results in excessive mitotic proliferation and germline tumors, increases the ability of these cells to undergo CHE-1-activated neuronal differentiation in the absence of LIN-53 (Seelk et al. 2016). Two lines of evidence suggest that this effect is not due merely to an increased cell number. Eliminating LAG-1, the transcription factor that transduces the Notch signal was found to abrograte the enhanced germline-to-neuronal Td in the glp-1(gf) mutant. However, the excessive mitotic proliferation that arises in a gld-1(−); gld-2(−) double mutant independent of Notch signaling did not enhance this effect (Seelk et al. 2016). Both observations suggest that this effect of Notch signaling in enhancing Td of germline nuclei is separable from its role in promoting mitotic germline proliferation. Comparing these observations with those described above, it is apparent that Notch signaling can act either to inhibit developmental plasticity, as in the embryonic MCT, or to enhance it, in the proliferating germline.
The antagonistic action of Notch and PRC2 appears to be mediated through a common set of target genes, with an enrichment of target genes on the X chromosome, as a strongly overlapping set of genes was found to be upregulated in response to Notch signaling and in the absence of PRC2 function (Seelk et al. 2016). One such gene, utx-1, which encodes a conserved H3K27me3 demethylase containing a JmjC domain, is essential for the enhancement of germline-to-neuronal Td. This finding suggests that Notch-mediated elevation of UTX-1 expression results in removal of methylation marks made by PRC2, hence relieving its repressive effects on somatic gene expression in the germline. Consistent with this possibility, LAG-1, the transcriptional mediator of Notch signaling, binds to the utx-1 gene and PRC2 represses utx-1 expression. The biological significance of this Notch activation of utx-1 is unclear as utx-1 expression is very low in the wild-type Notch-signaled mitotic zone. However, it is interesting to note that the utx-1 family member named jmjd-3.1 is necessary for natural rectal-to-neuronal Td in the worm (Zuryn et al. 2014), suggesting shared principles between these two distinct reprogramming events with an emphasis on the importance of H3K27me removal during cellular plasticity events.
Soma-to-germline reprogramming
The foregoing discussion pointed to many levels at which premature differentiation of somatic tissues is repressed in the pluripotent germline prior to fertilization of mature gametes. The mechanisms that specify germline vary widely across metazoans and in many animals (Extavour and Akam 2003). While in C. elegans, and in many other invertebrate species, the germline lineage is specified at the earliest divisions of the zygote, in other animals it arises as a result of instructive cues from cells that were otherwise destined to become somatic tissue, raising the possibility that there exist mechanisms that actively repress inappropriate germline fate specification in somatic cells. Such regulatory processes that suppress germline-like fate in somatic cells have been identified in C. elegans.
Removal of MEP-1, a Kruppel like Zn finger protein, results in arrested L1s in which somatic cells morphologically resemble germ cells and P granules accumulate in intestinal and hypodermal cells as a result of transcriptional derepression of genes encoding P granule components in the soma, beginning in late embryogenesis (Figure 5 and Table 2) (Unhavaithaya et al. 2002). This effect is not associated with detectable changes in somatic gene expression, which persists in the differentiated cells, suggesting that these cells possess a mixed soma/germline fate (Figure 5 and Table 2). This phenotype is also observed in animals lacking LET-418, which also physically interacts with MEP-1 and which is the C. elegans homolog of mammalian Mi-2/CHD3 (von Zelewsky et al. 2000)—a component of the NURD chromatin remodeling complex (Taunton et al. 1996). Loss-of-function of several mes genes, which are required for germline fate, and which encode components of the PcG and TrxG chromatin modifying complexes, suppress the ectopic expression of germline genes expression in the soma and L1 lethality of mep-1(−) animals, implying that ectopic germline fate is activated inappropriately through the normal embryonic germline regulatory system (Unhavaithaya et al. 2002). How is this implemented? MEP-1 and LET-418 form a complex with the HDA-1/HDAC-1 histone deacetylase and PIE-1, a C3H-type zinc finger protein, which is required for this interaction. PIE-1 is known to repress zygotic transcription and somatic differentiation in the early embryonic germline (Seydoux et al. 1996), and forced expression of PIE-1 in somatic cells results in derepression of germline-like fate in somatic cells, reminiscent of the phenotype of mep-1(−) (Unhavaithaya et al. 2002). Further, PIE-1 inhibits the histone deacetylase activity of HDA-1 in vitro. Thus, PIE-1 activates germline fate by antagonizing the activity of a chromatin modifying complex, including MEP-1, LET-418, and HDA-1, that promotes somatic development. When the activity of this complex is removed, somatic cells acquire a mixed soma/germline fate, which may be interpreted as incomplete Td.
MEP-1 and LET-418 are part of the so-called “SynMuv B” factors—a group of evolutionarily conserved factors involved in transcriptional repression that have been shown to be required to restrict competency to vulval induction cues during larval development (Fay and Yochem 2007). Several studies showed that the broad set of SynMuv B components also prevent germline-type differentiation in the soma (Unhavaithaya et al. 2002; Wang et al. 2005; Petrella et al. 2011). While single mutations in the SynMuv B genes (other than mep-1 and let-418) do not cause larval arrest, mutations in some, but not all, of these other SynMuv B components, like the unconditional phenotypes of mep-1(−) and let-418(−) mutations, result in partial derepression of germline-like fates in the soma (Figure 5 and Table 2), as well as enhanced RNAi sensitivity, characteristic of the germline (Cui et al. 2006). Mutations in this subset of SynMuv B genes are associated with ectopic expression of P-granule components and other germline genes but, unlike mep-1(−) and let-418(−) mutants, are viable to adulthood at 20°. However, all those mutants in which P-granule proteins are expressed in somatic cells also showed variably penetrant, and irreversible larval arrest at 26°, with a temperature-sensitive period during late embryogenesis and L1, immediately preceding the point of arrest (Petrella et al. 2011). An increase in somatically expressed P-granule components is generally, though not universally, observed in these mutants at the elevated temperature. Expression of germline components in the soma in these mutants at the restrictive temperature is associated with upregulation of many germline genes, including those involved in germline meiosis. In addition, intestine genes are also upregulated in these mutants and ectopic germline protein expression is observed most prominently in the intestine. Further, expression of the SynMuv B protein LIN-35 in the intestine was shown to be necessary to prevent expression of germline proteins, and its absence from the intestine was sufficient to cause larval arrest, suggesting that intestinal dysfunction, as a result of expression of germline factors in the organ, is responsible for the arrest. SynMuv B activities appear therefore to act broadly on repressing transcriptional programs, and may do so by modulating the chromatin landscape. In support, mutants for chromatin factors known or suggested to impact chromatin structure genetically interact with SynMuv B mutants. Indeed, as seen for the mep-1(−) and let-418(−) mutants, the L1 arrest of the SynMuv B mutants is suppressed by depletion of the germline chromatin remodeling factors encoded by several mes genes as well as by removal of MRG-1 and ISW-1, which are also required for the SynMuv phenotype and germline function. Thus, these activities may be used reiteratively in the worm to establish new stable chromatin states and reinforce the maintenance of the cellular identity.
While it is clear from these studies that germline-like characteristics can be derepressed in somatic cells, they do not demonstrate bona fide reprogramming of soma to germline per se. Rather, the observations may simply reflect expression of germline genes in an otherwise somatic cell background, resulting in a mixed cell fate (Petrella et al. 2011). A critical test of this possibility would require transplanting the cells into a normal gonad—an experiment whose technical feasibility has not been demonstrated.
Soma-to-soma: in vivo induced reprogramming of fully differentiated postmitotic somatic cells
Both natural and experimentally induced reprogramming events have been documented in C. elegans. While replication of DNA and chromatin during the cell cycle could in principle underlie “erasing” of the original differentiated state and new patterns of gene expression and reprogramming, findings in C. elegans have demonstrated at the single-cell level that even fully differentiated, postmitotic somatic cells can be reprogrammed to an entirely different cell type (Jarriault et al. 2008; Riddle et al. 2013, 2016). This section will focus on the induction of direct reprogramming of fully differentiated cells in live worms, long after the embryonic MCT has occurred, and even in fully developed adults. This feat has been achieved by forced expression of various TFs, and expression of a single TF has been shown to be sufficient to reprogram specialized, postmitotic cells, though it should be noted that the majority of cells resist efforts to reprogram them to new identities by a single specific TF.
Cells from two distinct organ types, the pharynx (the neuromuscular foregut of the digestive tract), and the somatic gonad, can be reprogrammed to a gut-like fate when forced to express the ELT-7 GATA-type transcription factor (Figure 5 and Table 2), which is redundantly required with ELT-2 (Riddle et al. 2013, 2016) at the terminal steps in the endoderm transcriptional cascade for differentiation of the intestine (McGhee et al. 2009; Riddle et al. 2013, 2016). For the pharynx, this event can occur at any stage of development, including in adults, and long after development of the pharynx has completed during the late stages of embryogenesis. In response to ectopic ELT-7 expression, the reprogrammed pharynx and gonadal cells undergo dramatic changes in morphology at the fine ultrastructural level, causing them to strongly resemble that of normal gut cells and to express molecular markers of terminal gut differentiation. In the case of the somatic gonad, the uterus appears to be converted to a gut-like organ that looks identical at the fine ultrastructural level to that of the normal gut in a process that has been called “transorganogenesis” (Riddle et al. 2016). Brief forced expression of ELT-7, and other endoderm GATA factors, not only initiates ectopic gut gene expression throughout the animal, but also leads to rapid attenuation of normal pharynx gene expression, implying that the entire gene regulatory network for pharynx differentiation is replaced by that of the intestine.
It is not clear why the somatic gonad and pharynx are specifically sensitive to postmitotic Td to gut. Both, like the gut, are tubular structures, and, thus, might share common elements across their regulatory networks (http://www.wormatlas.org/hermaphrodite/intestine/Intframeset.html; http://www.wormatlas.org/hermaphrodite/pharynx/Phaframeset.html; http://www.wormatlas.org/hermaphrodite/somatic%20gonad/Somframeset.html). FoxA-type transcription factors drive formation of tubular structures (de-Leon 2011), and both the pharynx and gonad express the PHA-4/FoxA transcription factor (Azzaria et al. 1996), which is required for pharynx organogenesis (Horner et al. 1998; Kalb et al. 1998). FoxA transcription factors, including PHA-4, have been shown to act as “pioneer” factors, which are thought to be able to access nucleosome-dense chromatin, and could prime chromatin states to make them competent for subsequent transcriptional activation (Iwafuchi-Doi and Zaret 2014; Hsu et al. 2015) Thus, expression of PHA-4 may make it possible to ELT-7 to engage its targets in nuclei that have long passed the MCT. Indeed, PHA-4 was found to be required for Td of the pharynx to endoderm (Riddle et al. 2016), suggesting that the differentiated state of the pharynx is required for this event. In contrast, no evidence was obtained for a requirement of PHA-4 in the gonad-to-gut Td event and forced expression of PHA-4 did not allow ELT-7 to promote Td in cells that normally cannot undergo this event. It appears, then, that the combination of ELT-7 and PHA-4 is insufficient to direct cells to undergo postmitotic Td to the gut in other cells, and other, as yet undefined, conditions must be met for this process to proceed.
The Td event that converts pharynx (foregut) cells to intestine (midgut) is reminiscent of a condition in humans known as Barrett’s metaplasia—a premalignant state that is associated with an aggressive and frequently fatal cancer: esophogeal adenocarcinoma (Slack et al. 2010). In this syndrome, cells of the lower esophagus (foregut) are converted from characteristic stratified squamous into those with the histological appearance of columnar goblet cells of the small intestine (midgut), a conversion that is analogous, and potentially homologous, to the Td of the C. elegans pharynx into intestine. In this regard, it is of note that the transcriptional regulatory mechanisms that specify the major divisions of the digestive tract are similar across metazoans including in C. elegans (Mango 2009; Boyle et al. 2014), suggesting that the subdivision of the alimentary canal into its major components occurs through an evolutionarily conserved process. Thus, the pharynx-to-gut Td event in worms might provide a useful model for this human condition.
Other examples of cell fate alterations following TF overexpression include the forced expression of single or pairs of bHLH factors, namely HLH-8 and LIN-32, in the AC and its precursors (L2 stage), which have been shown to alter AC fate and function (Sallee et al. 2017). This is coupled to the ectopic expression in the AC of a marker of the male DTC cells normally expressing this precise combination of bHLH factors, and a change in morphology reminiscent of the target somatic fate, suggesting a conversion from AC fate to male DTC-like features (Sallee et al. 2017). However, this possibly partial conversion appears to be only transient, perhaps because the driver used to force the bHLH cocktail expression may become less efficiently expressed over time, or because of transient superimposition of developmental programs.
Barriers to Reprogramming
Are there barriers to reprogramming? Several reports have examined this aspect during both induced pluripotent or direct reprogramming, in mammals and worms, and suggested that switching off the initial transcriptional program is step-limiting. Overcoming this limitation can be achieved via the ability of the reprogramming factors to target the active loci in the initial cell (Wapinski et al. 2013; Chronis et al. 2017; Mall et al. 2017), by removing a key transcription factor that controls the expression of many identity genes (Patel and Hobert 2017), or by using a powerful transcriptional repressor that will knock down other lineages’ transcriptional program (Mall et al. 2017). Additional genetic or epigenetic barriers that impact the ability to reprogram postmitotic cells and the efficiency of the process have yet to be discovered. In particular, if the differentiated identity is actively maintained, as proposed by Helen Blau (Blau 1992), the efficient removal of these maintenance mechanisms should enhance induced reprogramming scope.
What mechanisms might underlie these processes? Transcriptional regulatory networks have been associated with different cell states. For instance, the observation that the period of developmental plasticity correlates with the time during which restricted differentiation patterns are being specified in the embryo raises the possibility that the complex transcriptional regulatory networks activated by cell fate specification factors per se result in the pluripotency-to-commitment switch. Such gene regulatory networks are known to include positive transcriptional feedback regulatory loops that “lock down” differentiation pathways during specification (e.g., Davidson and Levine 2008; Davidson 2010), and the lockdown of one gene regulatory state might be sufficient to prevent the activation of others. If this is the case, then eliminating the function of genes essential for the specification of a cell type might be expected to cause the descendant cells to remain pluripotent. A recent study suggests that this may not be the case at least for pharyngeal cell fates: for example, elimination of the pha-4/FoxA, critical for pharynx specification, did not result in an extension of the window during which the affected embryonic cells are capable of being reprogrammed (Yuzyuk et al. 2009). Thus, there may exist global mechanisms controlling pluripotency that are independent of the known cell fate regulatory programs. It might also be that specific cell fate TFs and networks, besides endowing a cell with a specific identity, are also involved in maintaining a cell’s specialized identity, and that their removal opens up cellular plasticity without reverting to a pluripotent state. A recent study suggests that, at least for neurons, this might be the case: while ectopic expression of certain TFs have been shown to convert early embryos into balls of a given cell type (see section Early Embryonic Cellular Plasticity is Restricted to a Defined Time Window: the Multipotency-to-Commitment Transition), later overexpression of these same TFs on their own mostly failed to convert differentiated cells in late embryos or in larvae (see section Induced Reprogramming in the Worm). However, if the same experiment is performed in a mutant background in which the UNC-3 transcription factor, normally involved in specifying cholinergic motoneuron identity (and called terminal selector for its ability to induce a generic neuron to adopt a specific identity and function) is absent, a number of these cholinergic motoneurons can now be induced to express markers of other neuronal identities, such as ASE features in response to overexpression of the CHE-1 TF (Patel and Hobert 2017). A similar result has been obtained in mutants for the other “terminal selectors” ets-5, ttx-1, ceh-14, lin-11, but not when unc-30 or unc-42, or when the unc-4, unc-55 and mab-9 TFs, which control diversification into various cholinergic motoneuron subtypes, are absent (Patel and Hobert 2017). Thus, although it has not been formally demonstrated that removal of these “terminal selectors” in the neurons only after specification has the same effect, it appears that certain, but not all, TFs involved in identity-specific programs, may restrict cellular plasticity in neurons. This plasticity-restraining activity might involve control of chromatin structure, as absence of UNC-3 affects the compaction of arrays bearing CHE-1 targets, and, further, absence of enzymes depositing H3K27me and H3K9me marks also result in widened cellular plasticity (Patel and Hobert 2017).
It is possible that several layers of transcriptional control are involved in restricting cell type reprogramming. In an RNAi screen for factors that restrict the ability of a developmental TF overexpression to reprogram cells in C. elegans, several members of the FACT (“facilitates chromatin transcription”) complex were identified (Kolundzic et al. 2018). The FACT complex is involved in modulating nucleosome reorganization (Hsieh et al. 2013). Two subcomplexes were found that seem to function specifically either in the germline or in somatic cells, particularly the intestine. Elimination of the FACT complex in germ cells results in their conversion into neurons upon expression of a neuronal determinant such as CHE-1 (Kolundzic et al. 2018). This conversion is evidenced by expression of several neuronal markers (as assessed using transgenes and endogenous expression), morphological changes, and decrease of germline markers (Kolundzic et al. 2018). By contrast, elimination of the somatic FACT complex led to expression of some neuronal markers in intestinal cells, but no loss of intestinal markers or morphology. In the nucleus, loss of FACT results in mild changes in chromatin accessibility, as shown by ATAC-Seq on whole worms, and transcriptional changes, with both upregulated and downregulated genes observed (Kolundzic et al. 2018). Thus, different FACT complexes appear to act as a safeguard of cellular identity in the germ cells or in the intestine through their action on transcription. Interestingly, this activity may be evolutionarily conserved, as depletion of FACT in human fibroblasts appear to enhance both pluripotent and direct reprogramming efficiency (Kolundzic et al. 2018). Potentially representing another layer of transcriptional control, an RNAi screen targeting factors involved in chromatin regulation further showed that RNAi-mediated knock-down of mrg-1, a homolog of human MORF4L1 and MORF4L2, allows OE of the CHE-1 TF to convert germ cells into neurons expressing endogenous pan-neuronal and ASI markers (Hajduskova et al. 2019). Although the exact role of MRG-1 in this context is still unclear, it is possible that MRG-1 acts by modulating chromatin activity. Indeed, ChIP experiments against MRG-1 showed that it can be found associated to DNA, and immunoprecipitation experiments suggest an interaction with several chromatin-regulation factors, such as the H3K9 methyltransferase SET-26, the O-GlcNAc transferase OGT-1, and the HDAC repressor complex member SIN-3 (Hajduskova et al. 2019).
Additional histone modification events, as well as the nuclear RNAi pathway, have also been shown to regulate pluripotency and inhibition of somatic differentiation in the C. elegans germline (Robert et al. 2014). H3K4 methylation is associated with active promoters and enhancers (Bernstein et al. 2005; Heintzman et al. 2007) as well as transcriptional repression (Pinskaya and Morillon 2009; Margaritis et al. 2012), and changes in H3K4 methylation patterns are associated with developmental reprogramming (Gaspar-Maia et al. 2011; Koche et al. 2011). This modification is catalyzed by SET1/MLL class of histone methyltransferases that include conserved subunits SET1 (denoted SET-2 in C. elegans), WDR5 (WDR-5.1 in C. elegans), and ASH2 (ASH-2 in C. elegans), and these three components were shown to perform both overlapping and distinct functions in germline transcription (Robert et al. 2014). Removal of each of these functions results in loss of germline gene expression, progressive temperature-sensitive sterility (a mortal germline, or Mrt phenotype) and loss of germline P granules over several generations. In these animals, many normally soma-specific genes, particularly those in differentiated neurons, muscle, and intestine, are derepressed. Consistent with these observations, in set-2(−) and wdr-5.1(−) mutants, but not ash-2(−), animals, the defective germlines undergo neuronal and muscle differentiation with a progressive, cumulative effect over multiple generations, paralleling the temperature-sensitive Mrt germline phenotype. This phenotype is associated with loss and redistribution of the repressive H3K9me3 modification but an increase in H3K37me3, in contrast to the reduction seen in lin-53 mutants (see above, paragraph Nuclear control of pluripotency in the germline and germline-to-soma reprogramming). A similar temperature-sensitive Mrt phenotype is also seen in animals lacking HRDE-1 (Buckley et al. 2012) and NRDE-4 (Robert et al. 2014), which function in the nuclear RNAi pathway and associate with siRNAs that repress gene expression in the germline. Consistent with a similar mode of action, hrde-1(−) mutants, like set-2(−) mutants, also show a loss in H3K9me3, an increase in H3K27me3 in the germline, depression of somatic gene expression, and progressive increase in inappropriate somatic differentiation in the germline. Loss of SET-2/WDR-5.1 and HRDE-1/NRDE-4 show mutual enhancement for these phenotypes, implying that they act through parallel genetic pathways to repress somatic differentiation in the germline over successive generations.
While progressive loss of germline identity in the absence of SET1/WDR5 and HRDE-1/NRDE-4 is associated with a loss of germline-specific P granules, this is likely to be an effect rather than a cause of the phenotype, as changes in P granule gene expression were not observed in the transcriptional profiling experiments (Robert et al. 2014). However, other studies have shown that somatic development is indeed repressed in the germline by components of P granules (Updike et al. 2014). Thus, multiple mechanistic layers exist to protect germ cells from reprogramming, and to repress somatic differentiation in the germline. Among these, histone modifications and their likely impact on chromatin organization seem particularly crucial in the germ cells as opposed to the soma. It will be interesting to elucidate further how the regulatory machinery for the cell cycle, translation, and transcription are precisely coordinated.
Many studies described throughout this review point to the importance of chromatin alterations in establishing developmentally plastic states (Table 2). The conversion of a normally repressed, condensed, heterochromatic state into an open state that is susceptible to transcriptional activation as a result of manipulation of important chromatin regulatory factors may be analogous to the changes during normal development in what has classically been referred to as “facultative heterochromatin,” chromosomal regions in which an inactive, repressed state can be converted to actively transcribed euchromatin (Grigoryev et al. 2006; Trojer and Reinberg 2007; Wiles and Selker 2017). The genetic manipulations that allow reprogramming of cell fates might mimic this natural normal conversion of facultative heterochromatin that occurs during normal development. In this line, it is particularly striking that a crucial requirement for a change in chromatin activity/structure has been, in large part, revealed by experiments aiming at swapping from a germ to a somatic identity and vice and versa (Table 2), reinforcing the notion that chromatin organization is qualitatively different in germ cells (Schaner et al. 2003; Vágnerova et al. 2014) (http://www.wormbook.org/chapters/www_germlinechromatin/germlinechromatin.html). In addition, this may mean that the barrier mechanisms, or the relative importance of each such mechanism, may be qualitatively different in somatic vs. germ cells. Finally, experiments aiming at removing barriers to reprogram germ to somatic cells have used, for technical reasons, RNAi knock down of chromatin factors, rather than loss-of-function mutants, raising the intriguing possibility that the RNAi process per se may also facilitate the conversion.
General Principles Underlying Reprogramming
Studies in worms, as described here, and in other models strongly point to several common themes across reprogramming events. These are, at the cellular level: (i) The importance of erasing the initial identity to make way for the new one (see previous section), thereby assuring exclusivity of cell fates. This has lent additional support to the notion that cellular identities, or stable states, are reinforced by active maintenance mechanisms. (ii) The transition through discrete states in a step-wise process. The analysis of a natural Td event showed that, even in absence of cell division, Td occurs in multiple discrete steps (Richard et al. 2011). Both the step-by-step nature of the process, and the transition through a dedifferentiation state, have been observed during direct reprogramming events in other species, and thus may represent conserved aspects. Indeed, when murine primary pre-B lymphocytes are provided with exogenous C/EBPalpha, up to 100% of the cell population can be converted into macrophages (Di Tullio et al. 2011). This conversion appears to occur synchronously, allowing the authors to study the genome-wide transcriptional changes at different time points during the process. Analysis of these changes showed that the pre-B transcriptional program is switched off first, before the macrophage transcriptional program is activated (Di Tullio et al. 2011). Thus, a stage during which both transcriptional programs are off, representing a possibly dedifferentiated intermediate, can be identified. The parallel with Y Td extended further, as the order in which macrophage-lineage markers became activated was reminiscent of macrophage developmental differentiation (Di Tullio et al. 2011). A similar sequence of events is more widely observed: for instance, during newt lens regeneration following removal of the lens, pigmented epithelial cells of the iris re-enter the cell cycle and dedifferentiate—but do not revert to a pluripotent state—and lose their pigmentation before they redifferentiate into lens cells (Henry and Tsonis 2010). Thus, induced and natural reprogramming in murine cells, and newts or worms, respectively, show similar step-wise cellular transitions, raising the possibility that different direct reprogramming types could rely on similar mechanistic principles, and in widely divergent species. (iii) The uncoupling between the erasure of a cell’s identity and the reversion to a pluripotent state. As noted above, this step-wise Td process involves transition through a dedifferentiated state, in which the cell loses its initial characteristics prior to acquiring new ones. While the use of ES cells and the ability to force differentiated cells to reprogram into a pluripotent state has reinforced the implicit idea that a dedifferentiated state is equivalent to an early embryonic-like pluripotent state, studies of direct reprogramming processes have shown that this is not the case (Richard et al. 2011; Luz-Madrigal et al. 2014). (iv) The exact impact of cell division. Developmental or regenerational reprogramming and Td are commonly associated with ongoing cell division, which could represent a process for making a sufficient number of new cells. In contrast, Td in several distinct settings can occur in the absence of mitosis or cell division in C. elegans (see above). While somatic differentiation of germ cells, triggered either by CHE-1 or in P granule-depleted germlines, does appear to occur in the context of mitotically dividing cells (Updike et al. 2014), or in the case of mex-3(−) gld-1(−), in cells undergoing meiotic division (Ciosk et al. 2006), the germline-to-neuronal reprogramming mediated by CHE-1 seen in a developing germline undergoing continuous mitotic and meiotic division, can also occur even when cell division is blocked pharmacologically or genetically (Patel et al. 2012; Seelk et al. 2016). Thus, as is the case with the natural Y-to-PDA Td event (Jarriault et al. 2008), and postmitotic conversion of pharynx and uterine cells into intestine (Riddle et al. 2013, 2016), cell division is dispensable for germline-to-somatic Td. Therefore, while cell division does not represent an obligate step during direct reprogramming, it is associated with several direct reprogramming events in various species and the impact and the potential impact of DNA replication in these latter cases remains to be further explored. (v) Finally, the importance of nuclear events and the control of gene expression. These events include the activity of TFs, modulators of the chromatin structure and conformation, and, in the case of the germline, translational control.
Summary and Future Perspectives
The broad toolkit of forward and reverse genetics methods available with C. elegans will continue to advance our knowledge of the mechanisms that regulate cellular plasticity, the processes that convert multipotent cells to committed states, and the conditions that make it possible to interconvert cells between distinct differentiated states. In addition, while the expanding palette of quantitative genetics tools (Gaertner and Phillips 2010; Andersen et al. 2012) available to C. elegans researchers has not yet been applied to these problems, it may prove informative to ask whether natural genetic variation across the species, which dramatically influences many traits, also affects cellular plasticity, for example the timing of the MCT; the susceptibility to, or efficiency of, induced Td; or the specific requirements for chromatin modifiers in the process. Understanding such genetic influences may prove of particular relevance in the context of patient-specific strategies in regenerative medicine. The accessibility of in vivo analysis with the worm, providing the ability to characterize these events dynamically as they occur in individual identified cells, makes it likely that the system will continue to provide a comprehensive view of the processes in many cell contexts.
Conversion of one cell type into another most likely involves the rewiring of gene regulatory networks. Profiling of the transcriptome as cells convert from one type to another will be highly informative as to the mechanisms of this rewiring. Another unresolved question is whether the redifferentiation process mimics developmental differentiation and redeploys pathways and strategies that are similar to the developmental differentiation of lineally related cells. Together with genetic screens targeted at identifying the molecules involved, it will thus be of particular interest to assess, using transcriptomic data, the degree to which the transcriptional program during reprogramming is similar to, or the same as, what is used in the normal developmental context, and whether the fully reprogrammed differentiated cells retain marks left from the original cell state. Such conversion appears to involve switching between bistable states. Analysis of both the transcriptional, and ultimately structural, changes occurring throughout nuclei during conversion will confirm whether the switch between the two stable conditions occurs through a stereotyped stepwise series of events, progressively or not, or through a more stochastic, less determinate, progression.
As described here, most studies to date have emphasized the importance of transcriptional regulators, chromatin remodeling factors, and translational regulators in controlling plasticity and establishing commitment. However, the potential role of other major regulatory processes in regulating plasticity in C. elegans has not been illuminated. Thus, it is possible, for example, that noncoding RNAs, which play key roles both in RNA stability and expression, as well as in establishing heritable epigenetic states of gene expression (Ashe et al. 2012; Shirayama et al. 2012), might modulate cellular plasticity during natural and induced Td. The metabolic status of the animal, including its passage through developmental diapause, has long been known to influence phenotypic outcomes (Liu and Ambros 1991) and the quantitative requirement for redundant developmental regulators (Milloz et al. 2008), and it will be important to understand the relationship between metabolic or physiological conditions and cellular plasticity. Further, there is currently limited information regarding the possible influence of proteostasis, stress response, or other signaling pathways on these processes, and assessment of the roles they may play in these events should be informative.
Td of cells results not only in major changes in transcriptional states and nuclear structure, but also in the remodeling of the overall cellular structure, including cell surface components, the cytoskeleton, cell polarity, and organelle number and morphology. It will be important to dissect the detailed cellular basis for these striking remodeling processes, and to reveal how the converted cells are incorporated into new structures and form new interactions with cells in the surrounding tissues. In addition, the requirements of components involved in macromolecular turnover (e.g., proteasome, autophagic mechanisms, etc.) in these major remodeling events should prove informative as to the mechanisms of these transformations in differentiated cell function.
How related are the processes of natural Td, induced Td, and regeneration? The origins of cells involved in these events, and the mechanisms underlying regeneration, have been hotly debated, and are subject to substantial controversy. Resident adult stem cells, dedifferentiation of mature cells such as muscle fibers followed by redifferentiation into cells of the same lineage, or Td of cells adjacent to the injury site, have all been described. A survey of the literature suggests that a variety of strategies are observed that may depend on the species and complexity of the tissues or organ that are regenerated, the type and extent of the injury, and the developmental stage of the individual (Vibert et al. 2018). Detailed comparisons will make it possible to better define when, and in what conditions, Td is involved in the process, and its mechanistic relationship to the dedifferentiation of mature cells that then proliferate prior to redifferentiating. The actual cellular state of these dedifferentiated intermediates and their associated cellular potential, as well as their similarity to transient intermediate states that can be observed during injury-independent Td are all exciting problems that need to be addressed. Many of the studies of plasticity in C. elegans have pointed to evolutionarily conserved mechanisms that influence Td. With additional studies in worms and other models, a picture will emerge regarding which mechanisms underlying cellular plasticity among metazoans and between distinct processes (e.g., regeneration vs. natural Td vs. induced Td) are broadly general and which are phylogenetically restricted.
A related question for the future is whether there are important differences between in vivo and in vitro reprogramming. Indeed, studies focusing on induced events, whether in vitro or in vivo, trigger reprogramming in very different settings. What might be the impact of the cellular environment? This parameter might influence the number of cells reprogrammed, the efficiency of the process, its viability, or on the redifferentiation component, for example, by influencing the maturation and functional integration of the reprogrammed cells. In this context, the precise control of cellular environment, through defined manipulation of in vitro conditions, or in an intact organism with defined and fixed cell types and number, will likely be a great advantage to revealing potential influencing conditions that could be at work.
How does the ability of a given cell to be successfully reprogrammed change with age of cells or an organism? Induced direct reprogramming experiments in the worm suggest that, at least with certain reprogramming cues, cells in younger larvae are more prone to change their identity than in older larvae or adults (Richard et al. 2011; Tursun et al. 2011). Similarly, the success of in vivo induced direct reprogramming in the murine brain has been shown to be linked to the age of the neurons targeted (Rouaux and Arlotta 2013). Similarly, the efficiency of inducing direct reprogramming in mammals is maximized when embryonic fibroblasts are used. These observations in different species point to the influence of the age of a cell on its ability to be reprogrammed. However, specific TFs can reprogram postmitotic differentiated cells even in adult worms (Riddle et al. 2016). It will thus be important to confirm and decipher the mechanisms and context that underlie the increased resistance to reprogramming with age, and how this varies for given reprogramming cues or starting cell types.
A major feature appreciated by all C. elegans researchers is the comprehensive information available describing its development, structure, and biology at the cellular and subcellular level. Studies of cellular plasticity may ultimately reveal the instructions that make it possible to convert any one cell type into any other. It is therefore not unreasonable to imagine that the worm may be the first animal in which this is comprehensively understood, owing to its simplicity and the depth of information available. Beyond that goal, one might imagine in the far-flung future that C. elegans could be the first animal that is truly fully understood, as demonstrated by the ability to recreate it entirely in the laboratory. A detailed understanding of Td and remodeling is pivotal toward this distant goal of obtaining a complete understanding of how a complete, functioning animal self-assembles from a zygote.
Acknowledgments
We thank Sandra Bour for help with the figures, Anne-Cécile Reymann for a first reading, Lucie Laplane for discussions around stem cell concepts, and Meera Sundaram as a helpful editor. J.R. is the Wilcox Family Chair in Biotechnology and work in his laboratory is funded by grants from the National Institutes of Health (NIH 5 R01 HD081266 and NIH 5 R01 HD082347). S.J. is a research director of the Centre national de la recherche scientifique (CNRS) and work in her laboratory has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (PlastiCell CoG grant No 648960).
Footnotes
Communicating editor: M. Sundaram
Literature Cited
- Abdus-Saboor I., Mancuso V. P., Murray J. I., Palozola K., Norris C. et al. , 2011. Notch and Ras promote sequential steps of excretory tube development in C. elegans. Development 138: 3545–3555. 10.1242/dev.068148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R. et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. 10.1038/ng.1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andralojc K. M., Campbell A. C., Kelly A. L., Terrey M., Tanner P. C. et al. , 2017. ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 13: e1006611 10.1371/journal.pgen.1006611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J. et al. , 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197. 10.1016/j.cell.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E. M., Mitchell J., Bagijn M. P. et al. , 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J., and Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. 10.1016/0092-8674(87)90128-0 [DOI] [PubMed] [Google Scholar]
- Azzaria M., Goszczynski B., Chung M. A., Kalb J. M., and McGhee J. D., 1996. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev. Biol. 178: 289–303. 10.1006/dbio.1996.0219 [DOI] [PubMed] [Google Scholar]
- Becker S. F., and Jarriault S., 2016. Natural and induced direct reprogramming: mechanisms, concepts and general principles-from the worm to vertebrates. Curr. Opin. Genet. Dev. 40: 154–163. 10.1016/j.gde.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Bei Y., Hogan J., Berkowitz L. A., Soto M., Rocheleau C. E. et al. , 2002. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev. Cell 3: 113–125. 10.1016/S1534-5807(02)00185-5 [DOI] [PubMed] [Google Scholar]
- Bender L. B., Cao R., Zhang Y., and Strome S., 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14: 1639–1643. 10.1016/j.cub.2004.08.062 [DOI] [PubMed] [Google Scholar]
- Bernardo A. S., Jouneau A., Marks H., Kensche P., Kobolak J. et al. , 2018. Mammalian embryo comparison identifies novel pluripotency genes associated with the naïve or primed state. Biol. Open 7: bio033282 10.1242/bio.033282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K. et al. , 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181. 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., and Knoblich J. A., 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124: 1241–1253. 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G. et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. 10.1016/j.devcel.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Blanpain C., and Simons B. D., 2013. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 14: 489–502. 10.1038/nrm3625 [DOI] [PubMed] [Google Scholar]
- Blau H. M., 1992. Differentiation requires continuous active control. Annu. Rev. Biochem. 61: 1213–1230. 10.1146/annurev.bi.61.070192.010025 [DOI] [PubMed] [Google Scholar]
- Blau H. M., Chiu C. P., and Webster C., 1983. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32: 1171–1180. 10.1016/0092-8674(83)90300-8 [DOI] [PubMed] [Google Scholar]
- Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L. et al. , 1985. Plasticity of the differentiated state. Science 230: 758–766. 10.1126/science.2414846 [DOI] [PubMed] [Google Scholar]
- Boroviak T., and Nichols J., 2014. The birth of embryonic pluripotency. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: pii: 20130541. 10.1098/rstb.2013.0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Draper B. W., Mello C. C., and Priess J. R., 1993. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74: 443–452. 10.1016/0092-8674(93)80046-H [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G. et al. , 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14: 535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M. J., Yamaguchi E., and Seaver E. C., 2014. Molecular conservation of metazoan gut formation: evidence from expression of endomesoderm genes in Capitella teleta (Annelida). Evodevo 5: 39 10.1186/2041-9139-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A. et al. , 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Krause M., Sepanski M., and Fire A., 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120: 1631–1641. [DOI] [PubMed] [Google Scholar]
- Chiu C. P., and Blau H. M., 1984. Reprogramming cell differentiation in the absence of DNA synthesis. Cell 37: 879–887. 10.1016/0092-8674(84)90423-9 [DOI] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G. et al. , 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168: 442–459.e20. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar H., Keles S., and Jin Y., 2005. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr. Biol. 15: 340–346. 10.1016/j.cub.2005.02.025 [DOI] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., and Priess J. R., 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131: 4831–4841. 10.1242/dev.01352 [DOI] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., and Priess J. R., 2006. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311: 851–853. 10.1126/science.1122491 [DOI] [PubMed] [Google Scholar]
- Collucci V., 1891. Sulla rigenerazione parziale deell’occhio nei tritoni: isogenesi esvilluppo-Studio seprimentale. Mem. R. Accad. Sci. Ist. Bologna 5: 593–629. [Google Scholar]
- Comai G., Sambasivan R., Gopalakrishnan S., and Tajbakhsh S., 2014. Variations in the efficiency of lineage marking and ablation confound distinctions between myogenic cell populations. Dev. Cell 31: 654–667. 10.1016/j.devcel.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Conklin E. G., 1905. Mosaic development in ascidian eggs. J. Exp. Zool. 2: 145–223. 10.1002/jez.1400020202 [DOI] [Google Scholar]
- Coraggio F., Püschel R., Marti A., and Meister P., 2019. Polycomb and Notch signaling regulate cell proliferation potential during Caenorhabditis elegans life cycle. Life Sci. Alliance 2: e201800170 10.26508/lsa.201800170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan A. E., and McIntosh J. R., 1985. Mapping the distribution of differentiation potential for intestine, muscle, and hypodermis during early development in Caenorhabditis elegans. Cell 41: 923–932. 10.1016/S0092-8674(85)80073-8 [DOI] [PubMed] [Google Scholar]
- Cui M., Kim E. B., and Han M., 2006. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2: e74 10.1371/journal.pgen.0020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., 2010. Emerging properties of animal gene regulatory networks. Nature 468: 911–920. 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., and Levine M. S., 2008. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA 105: 20063–20066. 10.1073/pnas.0806007105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., and Lassar A. B., 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- de-Leon S. B., 2011. The conserved role and divergent regulation of foxa, a pan-eumetazoan developmental regulatory gene. Dev. Biol. 357: 21–26. 10.1016/j.ydbio.2010.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppe U., Schierenberg E., Cole T., Krieg C., Schmitt D. et al. , 1978. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 75: 376–380. 10.1073/pnas.75.1.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tullio A., Vu Manh T. P., Schubert A., Castellano G., Mansson R. et al. , 2011. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl. Acad. Sci. USA 108: 17016–17021 [corrigenda: Proc. Natl. Acad. Sci. USA 109: 11053 (2012)]. 10.1073/pnas.1112169108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djabrayan N. J., Dudley N. R., Sommermann E. M., and Rothman J. H., 2012. Essential role for Notch signaling in restricting developmental plasticity. Genes Dev. 26: 2386–2391. 10.1101/gad.199588.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B. W., Mello C. C., Bowerman B., Hardin J., and Priess J. R., 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216. 10.1016/S0092-8674(00)81339-2 [DOI] [PubMed] [Google Scholar]
- Edgar L. G., and McGhee J. D., 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599. 10.1016/0092-8674(88)90575-2 [DOI] [PubMed] [Google Scholar]
- Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K. et al. , 2011. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13: 215–222. 10.1038/ncb2164 [DOI] [PubMed] [Google Scholar]
- Eitoku M., Sato L., Senda T., and Horikoshi M., 2008. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 65: 414–444. 10.1007/s00018-007-7305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour C. G., and Akam M., 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130: 5869–5884. 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Fassnacht C., Tocchini C., Kumari P., Gaidatzis D., Stadler M. B. et al. , 2018. The CSR-1 endogenous RNAi pathway ensures accurate transcriptional reprogramming during the oocyte-to-embryo transition in Caenorhabditis elegans. PLoS Genet. 14: e1007252 10.1371/journal.pgen.1007252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., and Yochem J., 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306: 1–9. 10.1016/j.ydbio.2007.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., 2005. An inversion in the wiring of an intercellular signal: evolution of Wnt signaling in the nematode vulva. BioEssays 27: 765–769. 10.1002/bies.20275 [DOI] [PubMed] [Google Scholar]
- Francis R., Maine E., and Schedl T., 1995. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., and Krause M., 2005. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 132: 1795–1805. 10.1242/dev.01774 [DOI] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., and McGhee J. D., 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302. [PubMed] [Google Scholar]
- Gaertner B. E., and Phillips P. C., 2010. Caenorhabditis elegans as a platform for molecular quantitative genetics and the systems biology of natural variation. Genet. Res. 92: 331–348. 10.1017/S0016672310000601 [DOI] [PubMed] [Google Scholar]
- Gally C., and Bessereau J. L., 2003. GABA is dispensable for the formation of junctional GABA receptor clusters in Caenorhabditis elegans. J. Neurosci. 23: 2591–2599. 10.1523/JNEUROSCI.23-07-02591.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A., 1966. Changes in selective affinity following transdetermination in imaginal disc cells of Drosophila melanogaster. Exp. Cell Res. 44: 382–392. 10.1016/0014-4827(66)90444-7 [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., and Ramalho-Santos M., 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12: 36–47 (erratum: Nat. Rev. Mol. Cell Biol. 12: 273). 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., and Strome S., 2012. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2: 1169–1177. 10.1016/j.celrep.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau S. B., Moskowitz I. P., Terns R. M., and Rothman J. H., 1994. The potential to differentiate epidermis is unequally distributed in the AB lineage during early embryonic development in C. elegans. Dev. Biol. 166: 770–781. 10.1006/dbio.1994.1355 [DOI] [PubMed] [Google Scholar]
- Gettings M., Serman F., Rousset R., Bagnerini P., Almeida L. et al. , 2010. JNK signalling controls remodelling of the segment boundary through cell reprogramming during Drosophila morphogenesis. PLoS Biol. 8: e1000390 10.1371/journal.pbio.1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J. S., and McGhee J. D., 2001. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol. Cell. Biol. 21: 2533–2544. 10.1128/MCB.21.7.2533-2544.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurumescu C. A., and Chisholm A. D., 2011. Cell identification and cell lineage analysis. Methods Cell Biol. 106: 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., 1992. Induction of gut in Caenorhabditis elegans embryos. Nature 357: 255–257. 10.1038/357255a0 [DOI] [PubMed] [Google Scholar]
- Goldstein B., 1993. Establishment of gut fate in the E lineage of C. elegans: the roles of lineage-dependent mechanisms and cell interactions. Development 118: 1267–1277. [DOI] [PubMed] [Google Scholar]
- Gossett L. A., Hecht R. M., and Epstein H. F., 1982. Muscle differentiation in normal and cleavage-arrested mutant embryos of Caenorhabditis elegans. Cell 30: 193–204. 10.1016/0092-8674(82)90025-3 [DOI] [PubMed] [Google Scholar]
- Greenwald I., 2012. Notch and the awesome power of genetics. Genetics 191: 655–669. 10.1534/genetics.112.141812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev S. A., Bulynko Y. A., and Popova E. Y., 2006. The end adjusts the means: heterochromatin remodelling during terminal cell differentiation. Chromosome Res. 14: 53–69. 10.1007/s10577-005-1021-6 [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., 1962. Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 4: 256–273. 10.1016/0012-1606(62)90043-X [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., and Uehlinger V., 1966. “Fertile” intestine nuclei. Nature 210: 1240–1241. 10.1038/2101240a0 [DOI] [PubMed] [Google Scholar]
- Hajduskova M., Baytek G., Kolundzic E., Gosdschan A., Kazmierczak M. et al. , 2019. MRG-1/MRG15 is a barrier for germ cell to neuron reprogramming in Caenorhabditis elegans. Genetics 211: 121–139. 10.1534/genetics.118.301674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W. et al. , 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Henry J. J., and Tsonis P. A., 2010. Molecular and cellular aspects of amphibian lens regeneration. Prog. Retin. Eye Res. 29: 543–555. 10.1016/j.preteyeres.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer H., Rubinstein N., Fellini S., Yeoh G., Chi J. et al. , 1975. Lineages, quantal cell cycles, and the generation of cell diversity. Q. Rev. Biophys. 8: 523–557. 10.1017/S0033583500001980 [DOI] [PubMed] [Google Scholar]
- Holtzer H., Biehl J., Antin P., Tokunaka S., Sasse J. et al. , 1983. Quantal and proliferative cell cycles: how lineages generate cell diversity and maintain fidelity. Prog. Clin. Biol. Res. 134: 213–227. [PubMed] [Google Scholar]
- Horner M. A., Quintin S., Domeier M. E., Kimble J., Labouesse M. et al. , 1998. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 12: 1947–1952. 10.1101/gad.12.13.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh F. K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K. et al. , 2013. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110: 7654–7659. 10.1073/pnas.1222198110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. T., Chen H. M., Yang Z., Wang J., Lee N. K. et al. , 2015. TRANSCRIPTION. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348: 1372–1376. 10.1126/science.aab1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H., and Schnabel R., 1994. glp-1 and inductions establishing embryonic axes in C. elegans. Development 120: 2051–2064. [DOI] [PubMed] [Google Scholar]
- Hutter H., and Schnabel R., 1995. Establishment of left-right asymmetry in the Caenorhabditis elegans embryo: a multistep process involving a series of inductive events. Development 121: 3417–3424. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., and Zaret K. S., 2014. Pioneer transcription factors in cell reprogramming. Genes Dev. 28: 2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Schwab Y., and Greenwald I., 2008. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105: 3790–3795. 10.1073/pnas.0712159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Hoskins R., and Horvitz H. R., 1994. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372: 780–783. 10.1038/372780a0 [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., and Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Jorgensen E. M., and Mango S. E., 2002. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3: 356–369. 10.1038/nrg794 [DOI] [PubMed] [Google Scholar]
- Kagias K., Ahier A., Fischer N., and Jarriault S., 2012. Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc. Natl. Acad. Sci. USA 109: 6596–6601. 10.1073/pnas.1117031109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb J. M., Lau K. K., Goszczynski B., Fukushige T., Moons D. et al. , 1998. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development 125: 2171–2180. [DOI] [PubMed] [Google Scholar]
- Kiefer J. C., Smith P. A., and Mango S. E., 2007. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev. Biol. 303: 611–624. 10.1016/j.ydbio.2006.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Efe J. A., Zhu S., Talantova M., Yuan X. et al. , 2011. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA 108: 7838–7843. 10.1073/pnas.1103113108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche R. P., Smith Z. D., Adli M., Gu H., Ku M. et al. , 2011. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell 8: 96–105. 10.1016/j.stem.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama R., and Eguchi G., 1995. From lens regeneration in the newt to in-vitro transdifferentiation of vertebrate pigmented epithelial cells. Semin. Cell Biol. 6: 143–149. 10.1006/scel.1995.0020 [DOI] [PubMed] [Google Scholar]
- Kolundzic E., Ofenbauer A., Bulut S. I., Uyar B., Baytek G. et al. , 2018. FACT sets a barrier for cell fate reprogramming in Caenorhabditis elegans and human cells. Dev. Cell 46: 611–626.e12. 10.1016/j.devcel.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M., and Hobert O., 2011. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15: 205–214. 10.1038/nn.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg C., Cole T., Deppe U., Schierenberg E., Schmitt D. et al. , 1978. The cellular anatomy of embryos of the nematode Caenorhabditis elegans. Analysis and reconstruction of serial section electron micrographs. Dev. Biol. 65: 193–215. 10.1016/0012-1606(78)90190-2 [DOI] [PubMed] [Google Scholar]
- Laufer J. S., Bazzicalupo P., and Wood W. B., 1980. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19: 569–577. 10.1016/S0092-8674(80)80033-X [DOI] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H. et al. , 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10: 731–739. 10.1038/ncb1736 [DOI] [PubMed] [Google Scholar]
- Lin R., Thompson S., and Priess J. R., 1995. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83: 599–609. 10.1016/0092-8674(95)90100-0 [DOI] [PubMed] [Google Scholar]
- Liu Z., and Ambros V., 1991. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature 350: 162–165. 10.1038/350162a0 [DOI] [PubMed] [Google Scholar]
- Luo S., and Horvitz H. R., 2017. The CDK8 complex and proneural proteins together drive neurogenesis from a mesodermal lineage. Curr. Biol. 27: 661–672. 10.1016/j.cub.2017.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz-Madrigal A., Grajales-Esquivel E., McCorkle A., DiLorenzo A. M., Barbosa-Sabanero K. et al. , 2014. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 12: 28 10.1186/1741-7007-12-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M. F., Meneghini M. D., Bowerman B., Broitman-Maduro G., and Rothman J. H., 2001. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7: 475–485. 10.1016/S1097-2765(01)00195-2 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Hill R. J., Heid P. J., Newman-Smith E. D., Zhu J. et al. , 2005. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 284: 509–522. 10.1016/j.ydbio.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Mall M., Kareta M. S., Chanda S., Ahlenius H., Perotti N. et al. , 2017. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544: 245–249. 10.1038/nature21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E., 2009. The molecular basis of organ formation: insights from the C. elegans foregut. Annu. Rev. Cell Dev. Biol. 25: 597–628. 10.1146/annurev.cellbio.24.110707.175411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E., Thorpe C. J., Martin P. R., Chamberlain S. H., and Bowerman B., 1994. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development 120: 2305–2315. [DOI] [PubMed] [Google Scholar]
- Mangold O., and Spemann H., 1927. Über Induktion von Medullarplatte durch Medullarplatte im Jüngeren Keim, ein Beispiel homöogenetischer oder assimilatorischer Induktion. Wilhelm Roux Arch. Entwickl. Mech. Org. 111: 341–422. 10.1007/BF02080953 [DOI] [PubMed] [Google Scholar]
- Margaritis T., Oreal V., Brabers N., Maestroni L., Vitaliano-Prunier A. et al. , 2012. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 8: e1002952 10.1371/journal.pgen.1002952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., and Reinberg D., 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S., Pang Z. P., Yang N., Tsai M. C., Qu K. et al. , 2011. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9: 374–382. 10.1016/j.stem.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., and Schubiger G., 1999. Cell determination and transdetermination in Drosophila imaginal discs. Curr. Top. Dev. Biol. 43: 115–151. 10.1016/S0070-2153(08)60380-4 [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Fukushige T., Krause M. W., Minnema S. E., Goszczynski B. et al. , 2009. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 327: 551–565. 10.1016/j.ydbio.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A. L., Meszoely I. M., Suzuki K., Miyamoto Y., Rustgi A. K. et al. , 2005. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development 132: 3767–3776. 10.1242/dev.01925 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Draper B. W., and Priess J. R., 1994. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell 77: 95–106. 10.1016/0092-8674(94)90238-0 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Schubert C., Draper B., Zhang W., Lobel R. et al. , 1996. The PIE-1 protein and germline specification in C. elegans embryos. Nature 382: 710–712. 10.1038/382710a0 [DOI] [PubMed] [Google Scholar]
- Milloz J., Duveau F., Nuez I., and Felix M. A., 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 22: 3064–3075. 10.1101/gad.495308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier B., Astier M., Semeriva M., and Perrin L., 2005. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development 132: 5283–5293. 10.1242/dev.02091 [DOI] [PubMed] [Google Scholar]
- Mootz D., Ho D. M., and Hunter C. P., 2004. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131: 3263–3272. 10.1242/dev.01196 [DOI] [PubMed] [Google Scholar]
- Morgani S. M., and Brickman J. M., 2014. The molecular underpinnings of totipotency. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: pii: 20130549. 10.1098/rstb.2013.0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz I. P., and Rothman J. H., 1996. lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are regulated by maternal GLP-1 signaling in C. elegans. Development 122: 4105–4117. [DOI] [PubMed] [Google Scholar]
- Moskowitz I. P., Gendreau S. B., and Rothman J. H., 1994. p. 120 in Combinatorial Specification of Blastomere Identity by glp-1-Dependent Cellular Interactions in the Nematode Caenorhabditis elegans. Development, Cambridge. [DOI] [PubMed] [Google Scholar]
- Neumüller R. A., Betschinger J., Fischer A., Bushati N., Poernbacher I. et al. , 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R. A., and Slack F. J., 2009. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma 118: 405–418. 10.1007/s00412-009-0210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano J. M., Farley B. M., Essien K. I., and Ryder S. P., 2009. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. USA 106: 20252–20257. 10.1073/pnas.0907916106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H., Hong S. J., Salinas R. D., Liu S. J., Sun S. W. et al. , 2014. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 8: 1290–1299. 10.1016/j.celrep.2014.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry J. M., and Sundaram M. V., 2014. A non-cell-autonomous role for Ras signaling in C. elegans neuroblast delamination. Development 141: 4279–4284. 10.1242/dev.112045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E., and Ruvkun G., 2002. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18: 495–513. 10.1146/annurev.cellbio.18.012502.105832 [DOI] [PubMed] [Google Scholar]
- Patel T., and Hobert O., 2017. Coordinated control of terminal differentiation and restriction of cellular plasticity. eLife 6: pii: e24100. 10.7554/eLife.24100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T., Tursun B., Rahe D. P., and Hobert O., 2012. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2: 1178–1186. 10.1016/j.celrep.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V. et al. , 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M., and Morillon A., 2009. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics 4: 302–306. 10.4161/epi.4.5.9369 [DOI] [PubMed] [Google Scholar]
- Prasad B. C., Ye B., Zackhary R., Schrader K., Seydoux G. et al. , 1998. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125: 1561–1568. [DOI] [PubMed] [Google Scholar]
- Prasad B., Karakuzu O., Reed R. R., and Cameron S., 2008. unc-3-dependent repression of specific motor neuron fates in Caenorhabditis elegans. Dev. Biol. 323: 207–215. 10.1016/j.ydbio.2008.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess J. R., and Thomson J. N., 1987. Cellular interactions in early C. elegans embryos. Cell 48: 241–250. 10.1016/0092-8674(87)90427-2 [DOI] [PubMed] [Google Scholar]
- Quintin S., Michaux G., McMahon L., Gansmuller A., and Labouesse M., 2001. The Caenorhabditis elegans gene lin-26 can trigger epithelial differentiation without conferring tissue specificity. Dev. Biol. 235: 410–421. 10.1006/dbio.2001.0294 [DOI] [PubMed] [Google Scholar]
- Rechtsteiner A., Ercan S., Takasaki T., Phippen T. M., Egelhofer T. A. et al. , 2010. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 6: e1001091 10.1371/journal.pgen.1001091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C. et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Richard J. P., Zuryn S., Fischer N., Pavet V., Vaucamps N. et al. , 2011. Direct in vivo cellular reprogramming involves transition through discrete, non-pluripotent steps. Development 138: 1483–1492. 10.1242/dev.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle M. R., Weintraub A., Nguyen K. C., Hall D. H., and Rothman J. H., 2013. Transdifferentiation and remodeling of post-embryonic C. elegans cells by a single transcription factor. Development 140: 4844–4849. 10.1242/dev.103010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle M. R., Spickard E. A., Jevince A., Nguyen K. C., Hall D. H. et al. , 2016. Transorganogenesis and transdifferentiation in C. elegans are dependent on differentiated cell identity. Dev. Biol. 420: 136–147. 10.1016/j.ydbio.2016.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Mercier M. G., Bedet C., Janczarski S., Merlet J. et al. , 2014. The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Rep. 9: 443–450. 10.1016/j.celrep.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Rouaux C., and Arlotta P., 2013. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 15: 214–221. 10.1038/ncb2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee M. D., Littleford H. E. and Greenwald I., 2017. A bHLH code for sexually dimorphic form and function of the C. elegans somatic gonad. Curr. Biol. 27: 1853–1860.e5. 10.1016/j.cub.2017.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., and Tajbakhsh S., 2015. Adult skeletal muscle stem cells. Results Probl. Cell Differ. 56: 191–213. 10.1007/978-3-662-44608-9_9 [DOI] [PubMed] [Google Scholar]
- Sammut M., Cook S. J., Nguyen K. C. Q., Felton T., Hall D. H. et al. , 2015. Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526: 385–390. 10.1038/nature15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D., and Kelly W. G., 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5: 747–757. 10.1016/S1534-5807(03)00327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E., 1984. Altered cell-division rates after laser-induced cell fusion in nematode embryos. Dev. Biol. 101: 240–245. 10.1016/0012-1606(84)90136-2 [DOI] [PubMed] [Google Scholar]
- Schierenberg E., 1986. Developmental strategies during early embryogenesis of Caenorhabditis elegans. J. Embryol. Exp. Morphol. 97: 31–44. [PubMed] [Google Scholar]
- Schierenberg E., 1987. Reversal of cellular polarity and early cell-cell interaction in the embryos of Caenorhabditis elegans. Dev. Biol. 122: 452–463. 10.1016/0012-1606(87)90309-5 [DOI] [PubMed] [Google Scholar]
- Schierenberg E., and Wood W. B., 1985. Control of cell-cycle timing in early embryos of Caenorhabditis elegans. Dev. Biol. 107: 337–354. 10.1016/0012-1606(85)90316-1 [DOI] [PubMed] [Google Scholar]
- Schierenberg E., Miwa J., and von Ehrenstein G., 1980. Cell lineages and developmental defects of temperature-sensitive embryonic arrest mutants in Caenorhabditis elegans. Dev. Biol. 76: 141–159. 10.1016/0012-1606(80)90368-1 [DOI] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R. H., and Priess J. R., 2000. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell 5: 671–682. 10.1016/S1097-2765(00)80246-4 [DOI] [PubMed] [Google Scholar]
- Schwamborn J. C., Berezikov E., and Knoblich J. A., 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925. 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelk S., Adrian-Kalchhauser I., Hargitai B., Hajduskova M., Gutnik S. et al. , 2016. Increasing Notch signaling antagonizes PRC2-mediated silencing to promote reprograming of germ cells into neurons. eLife 5: e15477. 10.7554/eLife.15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman K., and Kafatos F. C., 1974. Transdifferentiation in the labial gland of silk moths: is DNA required for cellular metamorphosis? Cell Differ. 3: 81–94. 10.1016/0045-6039(74)90030-X [DOI] [PubMed] [Google Scholar]
- Seydoux G., Mello C. C., Pettitt J., Wood W. B., Priess J. R. et al. , 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382: 713–716. 10.1038/382713a0 [DOI] [PubMed] [Google Scholar]
- Shaham S., 2015. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb. Perspect. Biol. 7: a020578 10.1101/cshperspect.a020578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C. A., and Bowerman B., 1996. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122: 2043–2050. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H. C., Gu W., Ishidate T. et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R. et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Slack J. M., Colleypriest B. J., Quinlan J. M., Yu W. Y., Farrant M. J. et al. , 2010. Barrett’s metaplasia: molecular mechanisms and nutritional influences. Biochem. Soc. Trans. 38: 313–319. 10.1042/BST0380313 [DOI] [PubMed] [Google Scholar]
- Sommermann E. M., Strohmaier K. R., Maduro M. F., and Rothman J. H., 2010. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Dev. Biol. 347: 154–166. 10.1016/j.ydbio.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K., Qi F., Yadav M. C., Millan J. L., Toyama F. et al. , 2015. A robust transcriptional program in newts undergoing multiple events of lens regeneration throughout their lifespan. eLife 4: pii: e09594. 10.7554/eLife.09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickard E. A., Joshi P. M., and Rothman J. H., 2018. The multipotency-to-commitment transition in Caenorhabditis elegans-implications for reprogramming from cells to organs. FEBS Lett. 592: 838–851. 10.1002/1873-3468.12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher S. G., and Desplan C., 2008. Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature 454: 533–537. 10.1038/nature07062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Pires-daSilva A., Gutierrez A., Zheng M., Jungblut B. et al. , 2001. Microevolutionary analysis of the nematode genus Pristionchus suggests a recent evolution of redundant developmental mechanisms during vulva formation. Evol. Dev. 3: 229–240. 10.1046/j.1525-142x.2001.003004229.x [DOI] [PubMed] [Google Scholar]
- Strome S., and Wood W. B., 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. 10.1016/0092-8674(83)90203-9 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., and Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., and Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Sundaram M. V., and Buechner M., 2016. The Caenorhabditis elegans excretory system: a model for tubulogenesis, cell fate specification, and plasticity. Genetics 203: 35–63. 10.1534/genetics.116.189357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syder A. J., Karam S. M., Mills J. C., Ippolito J. E., Ansari H. R. et al. , 2004. A transgenic mouse model of metastatic carcinoma involving transdifferentiation of a gastric epithelial lineage progenitor to a neuroendocrine phenotype. Proc. Natl. Acad. Sci. USA 101: 4471–4476. 10.1073/pnas.0307983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E., Rampalli S., Risueno R. M., Schnerch A., Mitchell R. et al. , 2010. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468: 521–526 [corrigenda: Nature 560: E32 (2018)]. 10.1038/nature09591 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T. et al. , 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H. et al. , 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411. 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig C. A., and Schreiber S. L., 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411. 10.1126/science.272.5260.408 [DOI] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C., and Bowerman B., 1997. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705. 10.1016/S0092-8674(00)80530-9 [DOI] [PubMed] [Google Scholar]
- Tocchini C., Keusch J. J., Miller S. B., Finger S., Gut H. et al. , 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10: e1004533 10.1371/journal.pgen.1004533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P., and Reinberg D., 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28: 1–13. 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Tursun B., Patel T., Kratsios P., and Hobert O., 2011. Direct conversion of C. elegans germ cells into specific neuron types. Science 331: 304–308. 10.1126/science.1199082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., Shin T. H., Miliaras N., Lee J., Oyama T. et al. , 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002. 10.1016/S0092-8674(02)01202-3 [DOI] [PubMed] [Google Scholar]
- Updike D. L., Knutson A. K., Egelhofer T. A., Campbell A. C., and Strome S., 2014. Germ-granule components prevent somatic development in the C. elegans germline. Curr. Biol. 24: 970–975. 10.1016/j.cub.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vágnerova L., Gombitova A., Cmarko D., and Lanctot C., 2014. Distinct chromatin organization in the germ line founder cell of the Caenorhabditis elegans embryo. Dev. Growth Differ. 56: 605–614. 10.1111/dgd.12160 [DOI] [PubMed] [Google Scholar]
- van der Flier L. G., and Clevers H., 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71: 241–260. 10.1146/annurev.physiol.010908.163145 [DOI] [PubMed] [Google Scholar]
- Vella M. C., Choi E. Y., Lin S. Y., Reinert K., and Slack F. J., 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 18: 132–137. 10.1101/gad.1165404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibert L., Daulny A., and Jarriault S., 2018. Wound healing, cellular regeneration and plasticity: the elegans way. Int. J. Dev. Biol. 62: 491–505. 10.1387/ijdb.180123sj [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., and Wernig M., 2011. Direct lineage conversions: unnatural but useful? Nat. Biotechnol. 29: 892–907. 10.1038/nbt.1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky T., Palladino F., Brunschwig K., Tobler H., Hajnal A. et al. , 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127: 5277–5284. [DOI] [PubMed] [Google Scholar]
- Waddington C. H., 1957. The Strategy of the Genes Routlege Library Editions: 20th Centure Science. George Allen & Unwin Ltd., London. [Google Scholar]
- Wang D., Kennedy S., Conte D. Jr., Kim J. K., Gabel H. W. et al. , 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597. 10.1038/nature04010 [DOI] [PubMed] [Google Scholar]
- Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S. et al. , 2013. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155: 621–635. 10.1016/j.cell.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A. et al. , 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 86: 5434–5438. 10.1073/pnas.86.14.5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., 1939. Principles of Development. Henry Holt and Company, New York. [Google Scholar]
- White J. G., Albertson D. G., and Anness M. A., 1978. Connectivity changes in a class of motoneurone during the development of a nematode. Nature 271: 764–766. 10.1038/271764a0 [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., and Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Wiles E. T., and Selker E. U., 2017. H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43: 31–37. 10.1016/j.gde.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., 1895. Entwicklungsphyiologische studien. I. Die regeneration der Urodelenlinse. Wilhelm Roux Arch. Entw.-Mech. Org. 1: 380–390. [Google Scholar]
- Worley M. I., Setiawan L., and Hariharan I. K., 2012. Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46: 289–310. 10.1146/annurev-genet-110711-155637 [DOI] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., and Graf T., 2004. Stepwise reprogramming of B cells into macrophages. Cell 117: 663–676. 10.1016/S0092-8674(04)00419-2 [DOI] [PubMed] [Google Scholar]
- Yuan W., Xu M., Huang C., Liu N., Chen S. et al. , 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286: 7983–7989. 10.1074/jbc.M110.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T., Fakhouri T. H., Kiefer J., and Mango S. E., 2009. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell 16: 699–710. 10.1016/j.devcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Brown J., Kanarek A., Rajagopal J., and Melton D. A., 2008. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455: 627–632. 10.1038/nature07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Fukushige T., McGhee J. D., and Rothman J. H., 1998. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 12: 3809–3814. 10.1101/gad.12.24.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Le Gras S., Jamet K., and Jarriault S., 2010. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430. 10.1534/genetics.110.119230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Daniele T., and Jarriault S., 2012. Direct cellular reprogramming in Caenorhabditis elegans: facts, models, and promises for regenerative medicine. Wiley Interdiscip. Rev. Dev. Biol. 1: 138–152. 10.1002/wdev.7 [DOI] [PubMed] [Google Scholar]
- Zuryn S., Ahier A., Portoso M., White E. R., Morin M. C. et al. , 2014. Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science 345: 826–829. 10.1126/science.1255885 [DOI] [PubMed] [Google Scholar]