Mistranslation, incorporating an amino acid not specified by the “standard” genetic code, has applications in research and synthetic biology. Since mistranslation is toxic, its level must be modulated. Using a serine tRNA with a proline anticodon, we identify...
Keywords: serine tRNA, tRNA modifications, mistranslation, synthetic biology, statistical proteins
Abstract
Transfer RNAs (tRNAs) read the genetic code, translating nucleic acid sequence into protein. For tRNASer the anticodon does not specify its aminoacylation. For this reason, mutations in the tRNASer anticodon can result in amino acid substitutions, a process called mistranslation. Previously, we found that tRNASer with a proline anticodon was lethal to cells. However, by incorporating secondary mutations into the tRNA, mistranslation was dampened to a nonlethal level. The goal of this work was to identify second-site substitutions in tRNASer that modulate mistranslation to different levels. Targeted changes to putative identity elements led to total loss of tRNA function or significantly impaired cell growth. However, through genetic selection, we identified 22 substitutions that allow nontoxic mistranslation. These secondary mutations are primarily in single-stranded regions or substitute G:U base pairs for Watson–Crick pairs. Many of the variants are more toxic at low temperature and upon impairing the rapid tRNA decay pathway. We suggest that the majority of the secondary mutations affect the stability of the tRNA in cells. The temperature sensitivity of the tRNAs allows conditional mistranslation. Proteomic analysis demonstrated that tRNASer variants mistranslate to different extents with diminished growth correlating with increased mistranslation. When combined with a secondary mutation, other anticodon substitutions allow serine mistranslation at additional nonserine codons. These mistranslating tRNAs have applications in synthetic biology, by creating “statistical proteins,” which may display a wider range of activities or substrate specificities than the homogenous form.
MISTRANSLATION occurs when an amino acid that differs from that specified by the “standard” genetic code is incorporated into nascent proteins during translation. Although considered less frequently than protein modification, mistranslation plays a significant role in generating protein diversity. Mistranslation naturally occurs at a frequency of ∼1 in every 104–105 codons (Kramer and Farabaugh 2007; Drummond and Wilke 2009), with some conditions increasing this frequency (Santos et al. 1999; Bacher et al. 2007; Javid et al. 2014). Woese (1965) predicted that mistranslation was substantially greater during the evolution of the translational machinery, creating diversity that would allow proteins to probe phase-space. Mistranslation is also used in several systems as an adaptive response (Ling and Söll 2010; Moghal et al. 2014; Wu et al. 2014; Wang and Pan 2016). For example, in response to oxidative stress, Escherichia coli, yeast, and mammalian cells mistranslate methionine into proteins to sequester reactive oxygen species and protect cells from oxidative damage (Netzer et al. 2009; Wiltrout et al. 2012; Lee et al. 2014; Gomes et al. 2016; Schwartz and Pan 2017). In the archaeon Aeropyrum pernix, transfer RNA (tRNA)Leu is misaminoacylated by methionyl-tRNA synthetase at low temperatures, which enhances enzyme activity (Schwartz and Pan 2016). Mycoplasma species use editing-defective synthetases to generate diversity and escape the host defense systems (Li et al. 2011). In other cases, mistranslation results in nearly complete codon reassignment. Yeasts of the Candida genus naturally evolved a tRNASer variant that ambiguously decodes the leucine CUG codon mainly as serine (Massey et al. 2003; Paredes et al. 2012).
The first specificity step of translation is aminoacylation of a tRNA by its corresponding aminoacyl-tRNA synthetase [aaRS; reviewed in Pang et al. (2014)]. Each aaRS recognizes its cognate tRNAs from a pool of tRNAs with similar structure using structural elements and nucleotides within the tRNA called identity elements (Rich and RajBhandary 1976; de Duve 1988; Giegé et al. 1998). For many tRNA-aaRS interactions, the specificity is determined in large part by the anticodon. In yeast the exceptions to this are tRNASer and tRNAAla (Giegé et al. 1998). The major identity elements for tRNASer and tRNAAla are the variable arm, positioned 3′ to the anticodon stem, and a G3:U70 base pair, respectively. Because of the latter, inserting a G3:U70 base pair into other tRNAs results in misaminoacylation with alanine (McClain and Foss 1988; Francklyn and Schimmel 1989; Hoffman et al. 2017; Lant et al. 2017). In the case of tRNASer, changes to the anticodon misincorporate serine since the ribosome does not monitor the amino acid on the incoming tRNA (Chapeville et al. 1962). Post-transfer editing mechanisms exist to help maintain translation fidelity after aminoacylation. These involve editing domains that are part of the aaRS and free-standing proteins [reviewed in Ling et al. (2009)].
Mistranslation has applications in synthetic biology. tRNAs that misincorporate amino acids expand the diversity of expressed proteins, resulting in what Woese described as “statistical proteins” [Woese 1965; reviewed in Schimmel (2011)]. Statistical proteins have the potential to display a wider range of activities or substrate specificities than the homogeneous form. For example, generating antibodies that are heterogeneous mixtures, with each molecule containing one or two amino acid variants, could expand antigen recognition and be valuable for rapidly evolving antigens. Although tolerated and sometimes beneficial to cells, too much mistranslation can be lethal (Berg et al. 2017). Therefore, for mistranslation to have biological applications the activity of the mistranslating tRNA must be tuned so that it is below a toxicity threshold. Zimmerman et al. (2018) exploited the rapid tRNA decay (RTD) pathway to control tRNA levels in yeast. In this approach, mistranslating tRNAs are mutated to become substrates of the RTD pathway by destabilizing the acceptor stem (Whipple et al. 2011). The RTD pathway is controlled using an inducible MET22 gene, where repression of MET22 inhibits the RTD pathway and induces mistranslation by the mutant tRNA. Although effective, this approach also influences the levels of endogenous tRNAs.
Regulating other steps along the tRNA pathway allows the levels and/or activity of a tRNA to be controlled. These include steps during biosynthesis (e.g., transcription, processing, 3′ CCA addition, and splicing), nuclear export, aminoacylation, and interaction with the translational machinery (Nissen et al. 1996; Dreher et al. 1999; Phizicky and Hopper 2010; Hopper 2013). tRNAs are also extensively modified in both the nucleus and cytoplasm (Jackman and Alfonzo 2013). The modified bases are identity elements for aaRS enzymes (Giegé et al. 1998), regulate codon-anticodon pairing (Gustilo et al. 2008; Wei et al. 2011), maintain the reading frame during decoding (Urbonavičius et al. 2001), and regulate the tRNA structure (Lorenz et al. 2017). The final aspect of tRNA regulation is their degradation through either the RTD pathway mentioned above, which degrades hypomodified and unstable tRNAs (Chernyakov et al. 2008; Whipple et al. 2011), or the nuclear exosome, which monitors tRNA modifications and 3′ end maturation (Kadaba et al. 2004; Schneider et al. 2007; Schmid and Jensen 2008).
Because of their toxicity (Berg et al. 2017), the applications of mistranslating tRNAs to research and biotechnology requires that their activity be regulated. The goal of this work was to identify a range of base changes in tRNASer that would dampen tRNA function and fine-tune the extent of mistranslation for use in different applications and with a number of anticodon substitutions. Using a genetic suppression system that requires a proline codon be mistranslated as serine, we selected mistranslating tRNASerUGG variants with a range of activities after random mutagenesis. Many of these had increased toxicity at low temperature and upon inhibiting the RTD pathway, suggesting that they destabilize the tRNA, and enabling temperature-sensitive induction of mistranslation. Through targeted changes to predicted identity elements, we also identified substitutions in the acceptor stem and discriminator base that diminish lethality and allow mistranslation. Proteomic analysis demonstrated that tRNASer variants mistranslate to different extents with diminished growth correlating with increased mistranslation. Thus, by altering nucleotides in tRNASer it is possible to decrease tRNASer function and mistranslate with various efficiencies. In addition, we demonstrate that in combination with the correct secondary mutation, the anticodon of the tRNASer can be mutated to mistranslate arginine, glutamine, phenylalanine, and ochre stop codons, expanding the mistranslation potential of this tRNA.
Materials and Methods
Yeast strains and growth
All yeast strains are derivatives of the wild-type haploid strains BY4741 and BY4742 (Supplemental Material, Table S1; Winzeler and Davis 1997). The tti2 disruption strains covered by either TTI2 (CY6963), tti2-L187P (CY7020), or tti2-Q276TAA (CY6874) on a centromeric plasmid have been described (Hoffman et al. 2016). The tti2 disruption strain with tti2-L187R and tRNASerUCU-G26A was made by transforming tti2-L187R on an LEU2 centromeric plasmid into CY6963 along with sup17(UCU)-G26A on an HIS3 centromeric plasmid. The wild-type TTI2 on a URA3 plasmid was lost by counter selection on 5-fluoroorotic acid to generate CY8150. The met22Δ strain (CY8588) and its isogenic MET22 control (CY8589) were derived from a spore colony of the magic marker strain in the BY4743 diploid background (Tong et al. 2001).
Yeast strains were grown in yeast peptone media containing 2% glucose or synthetic media supplemented with nitrogenous bases and amino acids at 30° unless otherwise indicated. For spot assays on plates, strains were grown to saturation in selective medium, OD600 was normalized, and cultures were spotted in 10-fold serial dilutions. To quantitate growth on solid media, cells were plated after dilution to obtain single colonies. Colony size was measured using ImageJ (v1.52h; Schneider et al. 2012). Growth curves were generated by diluting saturated cultures to OD600 ∼0.1 in minimal media and incubating at 30°. OD600 was measured every 15 min using a BioTek Epoch 2 microplate spectrophotometer for 24 hr. Doubling time was calculated using the R package “growthcurver” (Sprouffske and Wagner 2016).
Plasmid constructs
SUP17 (pCB3076) and sup17(UGG) (pCB3082) expressed in YCplac33 have been previously described (Berg et al. 2017). Derivatives of YCplac33-sup17(UGG) with mutations at G9A (pCB4020), A20bG (pCB4021), G26A (pCB4023), and C40T (pCB4022) were obtained previously (Berg et al. 2017), whereas A4G (pCB4097), C5T (pCB4080), C12T (pCB4102), G15A (pCB4106), G18A (pCB4114), T20C (pCB4088), A29G (pCB4087), T33G (pCB4084), A38C (pCB4081), T39G (pCB4093), T44G (pCB4074), G45A (pCB4075), Ge23A (pCB4090), Ce21T (pCB4072), C48T (pCB4079), T51C (pCB4101), A59G (pCB4098), and T60G (pCB4089) were obtained through genetic selection.
sup17(UGG)-VAΔ was engineered using two-step mutagenic PCR with primers UG5953/VI1383 and UG5954/VI1382 (oligonucleotides are listed in Table S2) in the first round, with pCB3082 as template. Products from the first reaction were amplified with outside primers UG5953/UG5954 and cloned into YCplac33 as an EcoRI fragment after first subcloning into pGEM-Teasy (Promega, Madison, WI) to give pCB4177. sup17(UGG)-G11:C24 was cloned using the same strategy and inside primers VJ2697/VJ2698 to give pCB4191. sup17(UGG)-VAΔ1, sup17(UGG)-VA-G:C, sup17(UGG)-G73A, and sup17(UGG)-G73C were similarly engineered with inside primers VK4593/VK4594, VJ2766/VJ2767, WA5536/WA5537, or WA6571/WA6572, and cloned into YCplac33 HindIII/EcoRI to give pCB4234, pCB4206, pCB4259, and pCB4275, respectively.
Acceptor stem variants with flipped bases at positions 1:72 and 2:71 were synthesized by Life Technologies and cloned into YCplac33 as EcoRI/PstI fragments to give pCB4268 and pCB4274, respectively.
The variant with mutations at positions 3:70 was made sequentially. First, the G70C mutation was made by two-step PCR using outside primer UG5953/UG5954, inside primers VL5002/VL5003, and template pCB3082. The final product of that PCR was used as template for another two-step PCR using the same outside primers and inside primers VF8661/VF8662 to make the C3G mutation. The product was cloned into YCplac33 as an EcoRI fragment to give pCB4254. The variant with mutations at positions 4:69 was similarly made. First, the A4T mutation was made by two-step PCR using inside primers WF1163/WF1164 and template pCB3082. The final product was used as template to introduce the T69A mutation using inside primers WF1165/WF1166. The product was cloned into YCplac33 as an HindIII/EcoRI fragment to give pCB4333.
sup17(UCU) and sup17(UCU)-G9A were made by two-step mutagenic PCR with outside primers UG5953/UG5954 and inside primers VJ2409/VJ2410 from template pCB3082 or pCB4020, respectively. The products were cloned as EcoRI fragments into pRS303 (Sikorski and Hieter 1989) after first subcloning into pGEM-Teasy (Promega), giving pCB4215 or pCB4216, respectively. sup17(UCU)-G9A was also cloned into YCplac33 to give pCB4120. sup17(UCU)-G26A was similarly made using template pCB3082, and inside primers VL4943/VL4945 giving pCB4257. The following derivatives were made by two-step PCR using outside primers UG5953 and UG5954. Final products were cloned into YCplac33 as an EcoRI fragment after first subcloning into pGEM-Teasy (Promega). Template, inside primers, and construct are in parentheses: sup17(UUA) (pCB3082, VK4595/VK4596, pCB4235), sup17(UUA)-G9A (pCB4020, VK4595/VK4596, pCB4236), sup17(CUG)-G26A (pCB4023, VF7765/VF7766, pCB4311), and sup17(GAA)-G26A (pCB3082, WC8504/WC8505, pCB4358).
TTI2, tti2-L187P, and tti2-Q276TAA constructs have been described (Hoffman et al. 2016). The tti2-L187R construct was made by two-step PCR using outside primers 5693-1/5693-2, inside primers 6856-1/6856-2, and template pCB2134. The product was cloned into the wild-type DED1pr-TTI2 vector as a NotI-SacI fragment to give pCB2865.
The centromeric plasmid containing HSE-eGFP was kindly provided by Onn Brandman (Stanford University) (Brandman et al. 2012). The URA3 marker on the plasmid was switched to HIS3 using pUH7 (Cross 1997).
Selection of variants of sup17(UGG) that suppress tti2-L187P
Selection of mutant sup17(UGG) alleles that support viability and suppress tti2-L187P was performed as previously described (Berg et al. 2017). Briefly, YCplac33-sup17(UGG) were UV-irradiated and transformed into CY7020. Ura+ transformants were screened for growth on YPD containing 5% ethanol. The YCplac33 plasmids were then isolated, sequenced, and transformed back into CY7020 to analyze growth.
Fluorescence heat shock reporter
Yeast strains containing the heat shock response element (HSE)-eGFP reporter and a YCplac33-sup17(UGG) allele were grown to stationary phase in medium lacking histidine and uracil, diluted 1:20 in the same medium and grown for 6 hr at 30°. Cell densities were normalized to OD600 before measuring fluorescence. Fluorescence was measured with a BioTek Synergy H1 microplate reader at an emission wavelength of 528 nm using Gen5 2.08 software.
Mistranslation quantification using mass spectrometry
Starter cultures of yeast strains containing YCplac33-sup17(UGG) variants were grown to stationary phase in medium lacking uracil, diluted 1:20 in 10 ml of the same medium and grown for 19 hr at 30°. Cell pellets from the resulting 10 ml of yeast culture were resuspended in a denaturing lysis buffer (8 M urea, 50 mM Tris, pH 8.2, 75 mM NaCl). Cells were lysed by bead-beating with 0.5 mm glass beads at 4°. Lysates were cleared by centrifugation at 21,000 × g for 10 min at 4° and protein concentration was determined by BCA assay (Pierce, Thermo Fisher Scientific). Proteins were reduced with 5 mM dithiothreitol for 30 min at 55°, alkylated with 15 mM iodoacetamide for 30 min at room temperature, and the alkylation was quenched with additional 5 mM dithiothreitol for 15 min at room temperature. For each sample, 100 μg of protein was diluted 1:2 with 50 mM Tris, pH 8.9, and digested overnight at room temperature with 1 μg LysC (Wako Chemicals). Digestions were acidified to pH 2 with trifluoroacetic acid and desalted over Empore C18 stage tips (Rappsilber et al. 2007).
Peptide samples were resuspended in 4% acetonitrile, 3% formic acid, and subjected to liquid chromatography coupled to tandem mass spectrometry. Samples were loaded onto a fused silica capillary column packed with 1.9 μm Reprosil-Pur C18 AQ reversed-phase resin and separated using a gradient of 8–30% acetonitrile in 0.125% formic acid delivered at 250 nl/min over 95 min, with a total 120-min acquisition time. Peptides were analyzed online on the linear ion trap Orbitrap (LTQ Velos Orbitrap; Thermo Fisher Scientific) hybrid mass spectrometer using a data-dependent acquisition method. For each cycle, one full mass spectrometry scan was acquired from 350 to 1500 m/z at 60,000 resolution on the Orbitrap, with fill target of 3E6 ions and maximum injection time of 500 msec, followed by up to 20 tandem mass spectrometry on the 20 most-intense precursor ions fragmented by collision-induced dissociation and acquired in the ion trap with a 3E4 fill target and 100 msec of maximum injection time.
Raw files were converted to the open mzXML format and searched against the Saccharomyces Genome Database yeast protein sequence database (downloaded in 2014) using Comet (release 2015.01; Eng et al. 2013). The false discovery rate (FDR) was estimated using a target-decoy strategy (Elias and Gygi 2007). Data were filtered to 1% FDR at the peptide-spectrum match level using Percolator (2017; Käll et al. 2007). To identify peptides with serine substitutions at proline sites, the search was conducted with a variable modification corresponding to the mass shift of a proline to serine substitution at proline positions. A maximum of two proline to serine substitutions per peptide were allowed, as the low rate of substitution we observed was considered unlikely to produce detectable peptides with multiple substitutions. Additional search parameters were cleavage C-terminal to lysine with a maximum of two missed cleavages, constant modification of carbamidomethylation on cysteines, variable modifications of methionine oxidation and N-terminal protein acetylation, tolerance of 50 ppm for precursor masses, and 0.36 kDa with 0.11 offset for fragment ions.
To estimate the frequency of substitution, we calculated the fraction of unique peptides containing the proline sites for which the serine-substituted version of the peptide was also detected. To minimize the FDR among mistranslated peptides, we applied additional, more stringent filtering. Serine-substituted peptides with additional modifications (methionine oxidation or N-terminal acetylation) or those in which the corresponding wild-type peptide was not present in the data set were filtered out. For codon specific substitution analysis, an additional filtering step was applied. Only peptides with a single proline instance were considered so as to rule out potential site localization issues.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. File S1 contains all supplemental figures and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9742577.
Results
tRNAs that mistranslate at different codons and with a range of efficiencies will enable broad applications of mistranslation. We previously demonstrated that SUP17, the gene encoding tRNASer, is lethal when modified with a UGG anticodon and transformed into a wild-type yeast strain (Berg et al. 2017). Second-site mutations (e.g., G9A and G26A) that cripple the tRNA allow the mistranslation to be tolerated. By showing that the tRNAs suppress the stress sensitivity of a tti2-L187P mutation, we demonstrated that they mistranslate proline codons, a result confirmed by mass spectrometry. In addition, these mistranslating tRNASerUGG variants induce a cellular heat-shock response.
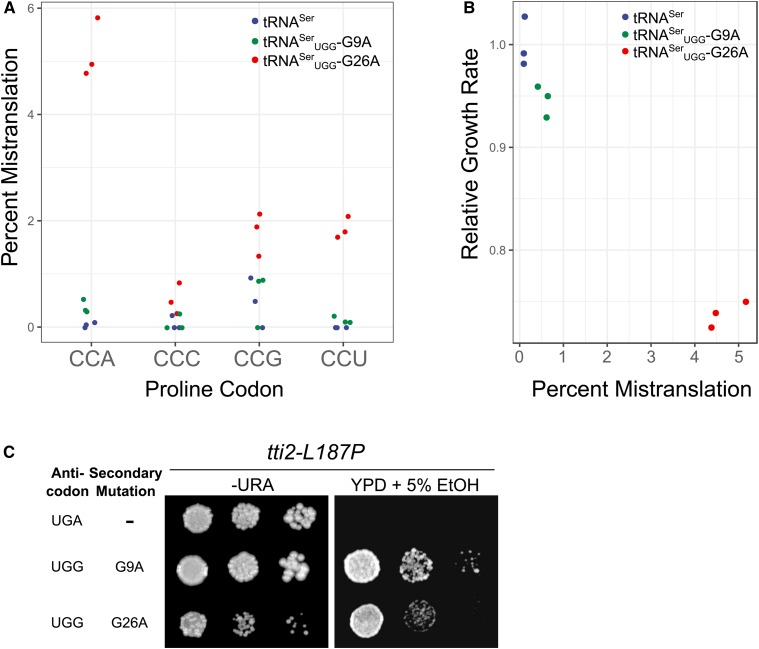
To determine the range of mistranslation induced by tRNASerUGG-G9A and -G26A, we analyzed the cellular proteome by mass spectrometry. The frequency of mistranslation detected at CCA codons in a strain containing a wild-type tRNASer was 0.04% (Figure 1A, number of peptides identified can be found in Table S3). The extent of mistranslation for tRNASerUGG-G9A and -G26A was 0.4 and 5.2%, respectively, at the CCA codon. We note that the difference between the G9A variant and the wild-type tRNA was statistically significant (Welch’s t-test; P < 0.05), although the values do approach the estimated FDR. The majority of mistranslated codons were the cognate CCA codon, but we also observed mistranslation by tRNASerUGG at the wobble codon CCG and the CCU codon. Decoding of CCU may be due to modification of tRNASerUGG, since tRNAProUGG in Salmonella enterica decodes this codon after modification of U34 to 5-oxyacetic acid (Näsvall et al. 2004). The difference in extent of mistranslation by tRNASerUGG-G9A and -G26A was consistent with the lesser effect of the G9A variant on cell growth (Figure 1B). We also observed previously that while both mistranslating variants induce a heat-shock response, G26A induces a greater heat-shock response relative to G9A (Berg et al. 2017). Although suppression of tti2-L187P is a sensitive method to detect mistranslation, it is not a quantitative measure of mistranslation (Figure 1C), likely because growth of the tti2-L187P strain under conditions of stress is a balance of both suppression by the tRNA and the toxicity caused by mistranslation.
Figure 1.
tRNASer with a proline UGG anticodon and various secondary mutations allow nonlethal levels of mistranslation. (A) Mass spectrometry analysis of the cellular proteome was performed on wild-type strain (BY4742) containing either wild-type tRNASer, tRNASerUGG-G9A or tRNASerUGG-G26A. Mistranslation of serine at proline codons was quantified at all four proline codons. (B) Growth rates for each strain in A were determined from growth curves of the strains diluted to an OD600 of ∼0.1 in media lacking uracil and grown for 24 hr. Doubling time was calculated with the R package “growthcurver” (Sprouffske and Wagner 2016), normalized to the strain containing the wild-type tRNA and plotted against the percent mistranslation at all proline codons determined through whole proteome mass spectrometry. (C) Yeast strains containing tti2-L187P (CY7020) and either wild-type tRNASer, tRNASerUGG-G9A or tRNASerUGG-G26A were grown to saturation in media lacking uracil and spotted in 10-fold serial dilutions on media lacking uracil or YPD containing 5% ethanol.
Acceptor stem and discriminator mutations can modulate tRNASer function
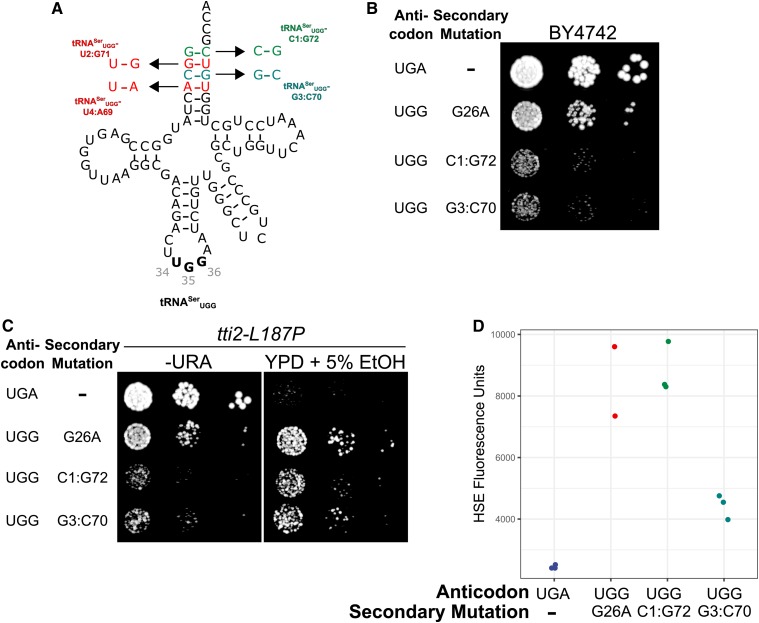
Our goal was to obtain a set of tRNA variants that mistranslate at a broader range of frequencies. Regulating the aminoacylation of a tRNA provides a possible method to modulate functionality (Giegé et al. 1998). Based on studies in E. coli and Thermus thermophilus, the identity elements for tRNASer fall within the discriminator base, first 4 bp of the acceptor stem, 1 bp in the D-arm, and unique extended variable arm (Normanly et al. 1986, 1992, Himeno et al. 1990, 1997; Sampson and Saks 1993; Asahara et al. 1994; Biou et al. 1994; Saks and Sampson 1996). We found mutations in the variable arm that either decreased the length of the arm, inverted the base pairs, or removed the arm entirely resulted in completely loss of tRNA function and no mistranslation as measured by suppression of tti2-L187P and heat-shock induction (Figure S1). The same was true when we inverted the C11:G24 base pair within the D-arm (Figure S2).
We then investigated if bases within the acceptor stem could be mutated to modulate tRNASer function and allow mistranslation. Normanly et al. (1992) converted a leucine accepting tRNA into a serine accepting tRNA in E. coli by mutating positions 2, 3, 70, 71, and 72 within the tRNALeu acceptor stem to C72, G2:C71 and A3:U70, all conserved within tRNASer, suggesting that these bases are identity elements for serylation. In addition, Saks and Sampson (1996) have shown that the charging of minihelices by SerRS requires the first 5 bp in the acceptor stem. Therefore, we constructed four alleles each inverting 1 bp in the acceptor stem (Figure 2A). Transformants were not obtained for base inversions at positions 2:71 and 4:69 (Figure S3), suggesting that these variants are highly functional and mistranslate at lethal levels. The variants with base inversions at positions 1:72 and 3:70 could be transformed. Both were partially toxic as measured by reduced growth (Figure 2B), suppressed tti2-L187P (Figure 2C) and induced a heat-shock response (Figure 2D). Together these results suggest that the C1:G72 and G3:C70 base-pair inversions result in a partial loss of tRNA function. The C1:G72 and G3:C70 mutations can thus be used to dampen the function of mistranslating tRNASer variants. Interestingly, the G3:C70 variant was more toxic than the G26A variant but the G26A variant induced a greater heat-shock response.
Figure 2.
Mutation of G1:C70 and C3:G70 reduces tRNASerUGG function allowing a nonlethal level of mistranslation. (A) Structures of four tRNASerUGG alleles created by flipping base pairs at the first four positions in the acceptor stem. (B) Wild-type strain (BY4742) expressing wild-type tRNASerUGA, tRNASerUGG-G26A or the viable tRNASerUGG-C1:G72 or -G3:C70 were grown to saturation in media lacking uracil and spotted in 10-fold serial dilutions on the same media. (C) Strains containing tti2-L187P (CY7020) and one of the tRNAs described in B were grown to saturation in media lacking uracil and spotted in 10-fold serial dilutions on media lacking uracil or complete media containing 5% ethanol. (D) Wild-type strain BY4742 containing one of the tRNAs described in B and a fluorescence heat shock reporter were grown to saturation in media lacking uracil and histidine. Cells were diluted 1:20 in the same media and grown for 6 hr. Cell densities were normalized and fluorescence measured. Each point represents one biological replicate.
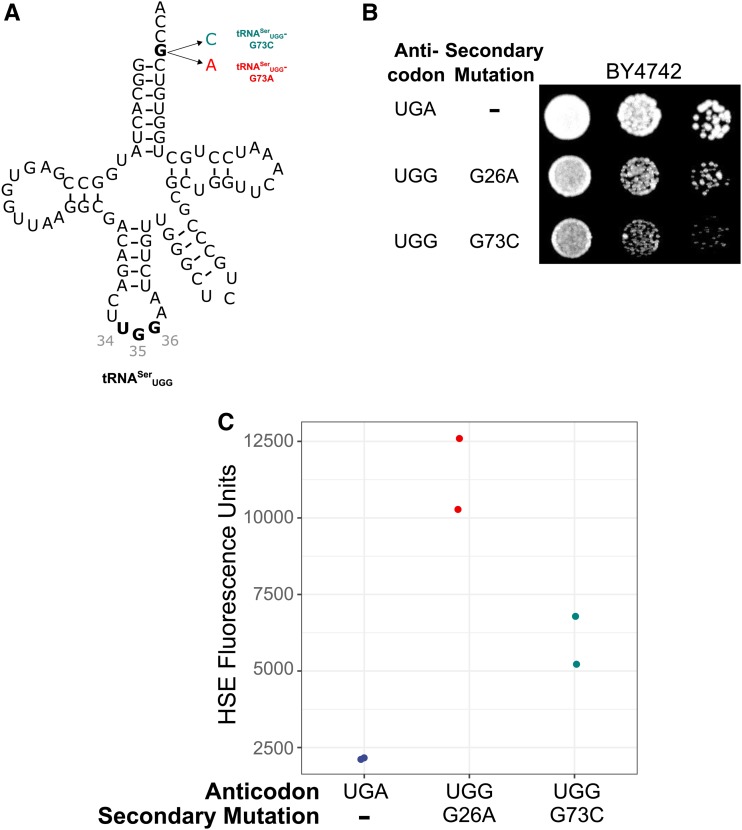
Next, we mutated the discriminator base (position 73), which plays an important role in aminoacylation of many tRNAs by making contacts with the aaRS (Hou 1997; Fukunaga and Yokoyama 2005). All tRNASer isoacceptors have G as the discriminator base. We engineered tRNASerUGG-encoding alleles with the discriminator base converted to A or C (Figure 3A) and attempted to transform these alleles on centromeric plasmids into BY4742 (Figure S4). No transformants were obtained for the 73A variant, suggesting that it is highly functional and mistranslates at a lethal level. Transformants expressing the 73C variant were obtained. Consistent with mistranslation by this variant, the transformants grew at a reduced rate relative to the control wild-type tRNASer (Figure 3B) and induced a 2.7-fold heat-shock response relative to the wild type (Figure 3C). We again note that similar to the G3:C70 variant, the extent of the heat-shock response did not correlate well with the effect of the G73C variant on growth. We were unable to transform the tRNASerUGG-G73C containing plasmid into the tti2-L187P strain, likely reflecting the combined toxicity of mistranslation with the tti2 mutation.
Figure 3.
Mutations to the discriminator base of tRNASerUGG allow mistranslation. (A) Structure of tRNASerUGG-G73C and -G73A. (B) Wild-type strain (BY4742) containing either wild-type tRNASerUGA, tRNASerUGG-G26A or tRNASerUGG-G73C were grown to saturation in media lacking uracil and spotted in 10-fold serial dilutions on media lacking uracil. (C) Wild-type strains containing one of the tRNAs described in B and a fluorescence heat shock reporter were grown in media lacking uracil and histidine. Cells were diluted 1:20 in the same media and grown for 6 hr. Cell densities were normalized and fluorescence measured. Each point represents one biological replicate.
In E. coli and Saccharomyces cerevisiae, the discriminator base for tRNASer and tRNALeu is G and A, respectively. Normanly et al. (1992) found that changing the discriminator base of a leucine tRNA from A to the G allowed E. coli tRNALeu to be charged with serine. To test this for S. cerevisiae tRNASer, we constructed tRNASer alleles with G73C and G73A mutations in the context of the wild-type UGA serine anticodon. If the tRNASerUGA variants were mischarged, they should be partially toxic in a wild-type strain and induce a heat-shock response. We did not observe toxicity in growth assays nor did these tRNAs induce a heat-shock response (Figure S5), suggesting that the discriminator base variants are not mischarged. These discriminator base variants are only toxic in the context of a mistranslating tRNASer with a noncognate anticodon.
tRNASerUGG variants randomly selected to mistranslate at different levels
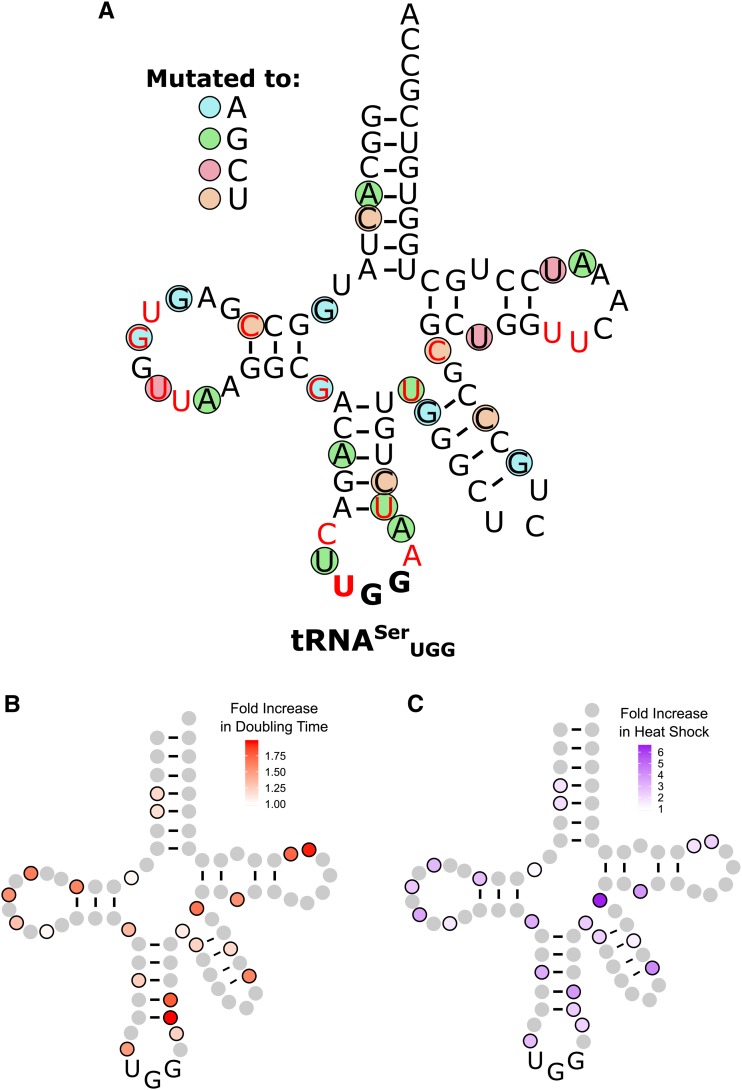
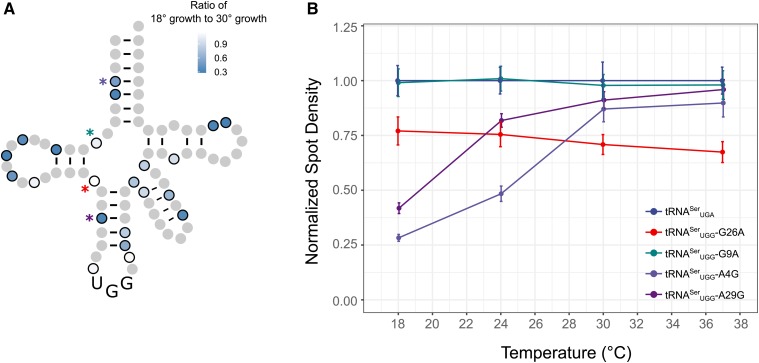
The targeted mutations in tRNASerUGG that mistranslated resulted in poor viability. To identify additional bases that, when mutated, lower tRNASerUGG mistranslation to a variety of levels in a less biased manner, we mutagenized the sup17(UGG)-containing plasmid with UV light and randomly selected mutants that suppress tti2-L187P. Plasmids that allowed growth of the tti2-L187P strain on media containing 5% ethanol were isolated, transformed back into the tti2-L187P strain to verify activity, and sequenced (suppression of tti2-L187P by the randomly selected variants is shown in Figure S6). We mapped the mutations onto the tRNASerUGG secondary structure (Figure 4A). We isolated 22 different mutations that dampen tRNASerUGG function to allow nonlethal levels of mistranslation, including mutations at G9, A20b, G26, and C40 that we had identified previously (Berg et al. 2017). Twelve mutations occur in single-stranded regions; 10 occur in stem structures. Of those occurring in stem structures, six change a Watson–Crick base pair to a G:U pair, including one in the variable arm. The three mutations that abolish base-pairing occur either at the beginning or end of a stem resulting in a shortened stem structure. tRNASerUGA contains 14 modified bases (Machnicka et al. 2013; colored red in Figure 4A). Seven of the mutations that dampen tRNASerUGG function occur at one of these modification sites.
Figure 4.
Randomly selected second-site mutations that dampen mistranslation by tRNASerUGG. (A) Base changes are indicated by the colored circles, where blue indicates a mutation to adenine, green to guanine, red to cytosine, and orange to uracil. Bases colored in red are sites of modification in tRNASer (Machnicka et al. 2013). (B) Heat map of the growth rates of strains containing mutations in tRNASerUGG that mistranslate at a level that supports viability. BY4742 containing tRNASerUGG variants were grown to saturation in media lacking uracil, diluted to an OD600 of ∼0.1 in the same media and grown for 24 hr at 30°. OD600 was measured every 15 min. Doubling time was calculated with the R package “growthcurver” (Sprouffske and Wagner 2016) and normalized to a strain containing a wild-type tRNASer. (C) Heat map of heat shock induced by tRNASerUGG derivatives. BY4742 containing different tRNASerUGG variants and a fluorescence heat shock reporter were grown to saturation in media lacking uracil in biological triplicate. Cells were diluted 1:20 in the same media and grown for 6 hr. Cell densities were normalized and fluorescence measured.
To estimate the amount of mistranslation by the randomly selected derivatives, we examined their effect on growth in a wild-type strain (Figure 4B and Table 1) and their induction of a heat-shock response (Figure 4C and Table 1). The least toxic tRNASerUGG variants, containing a C5U, G9A, A20bG, or U44G mutation, had a doubling time similar to the control strain that contained wild-type tRNASerUGA, and induced a 1.9- to 2.4-fold heat-shock response. The most toxic variants with U39G or U60C mutations increased doubling time by 2.0 and 1.7-fold, respectively, and induced a 2.4 and 1.7-fold heat-shock response.
Table 1. Mutations to tRNASerUGG allowing nontoxic levels of mistranslation.
| Mutationa | Doubling time (min) | Heat-shock induction (fold change) |
|---|---|---|
| tRNASerUGA | 65.5 | 1.0 |
| A20bG | 67.4 | 1.6 |
| G9A | 68.3 | 1.2 |
| U44G | 71.6 | 2.4 |
| C5U | 75.5 | 1.9 |
| Ce21Ub | 77.7 | 1.4 |
| A4G | 77.9 | 1.7 |
| G45A | 79.4 | 2.3 |
| A38G | 80.4 | 2.2 |
| A29G | 81.6 | 3.3 |
| U19C | 85.3 | 3.4 |
| A59G | 85.4 | 2.7 |
| G26A | 88.9 | 3.2 |
| U33G | 98.0 | 2.8 |
| U51C | 101 | 3.8 |
| G17A | 102 | 2.5 |
| C12U | 105 | 2.8 |
| Ge23Ab | 105 | 4.2 |
| G15A | 107 | 2.9 |
| C48U | 109 | 6.5 |
| U60C | 113 | 1.7 |
| C40U | 116 | 3.8 |
| U39G | 129 | 2.4 |
With the exception of the wild-type tRNASerUGA, all tRNAs have the UGG anticodon.
Refers to the extended variable arm, as numbered by Sprinzl et al. (1998).
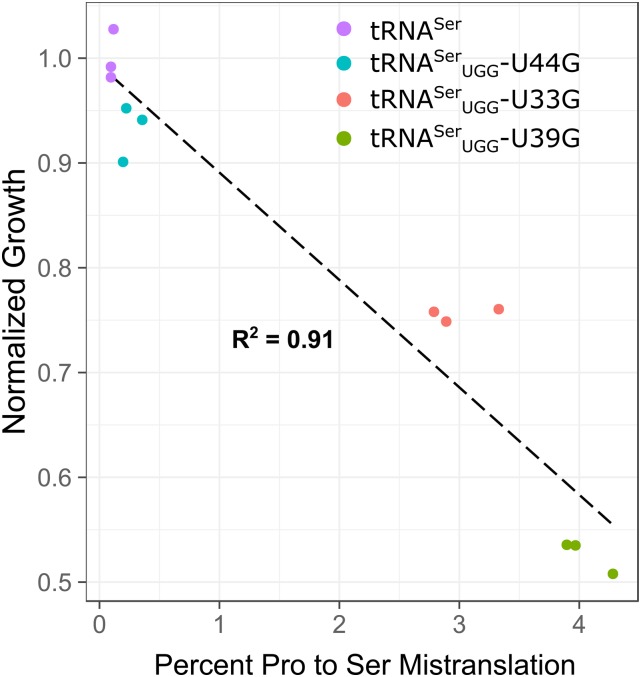
To better estimate mistranslation frequencies, we analyzed protein lysates by mass spectrometry for tRNASerUGG variants U44G, U33G, and U39G, which have ∼1-, ∼1.5-, and ∼two-fold increases in doubling time, respectively. We predicted that these variants would mistranslate at different frequencies correlating to their effect on growth. In agreement with this, the U44G variant mistranslated at 0.3%, the U33G variant mistranslated at 3.0% and the U39G variant mistranslated at 4.0% (Figure 5).
Figure 5.
Mistranslation frequency correlates with the effect of each mistranslating tRNA on growth. Whole proteome mass spectrometry was performed on wild-type strain BY4742 containing either a wild-type tRNASer or mistranslating variants tRNASerUGG-U44G, U33G, or U39G. Doubling time in minutes for each strain was determined from growth curves of each strain in media lacking uracil and normalized to the strain containing a wild-type tRNASer.
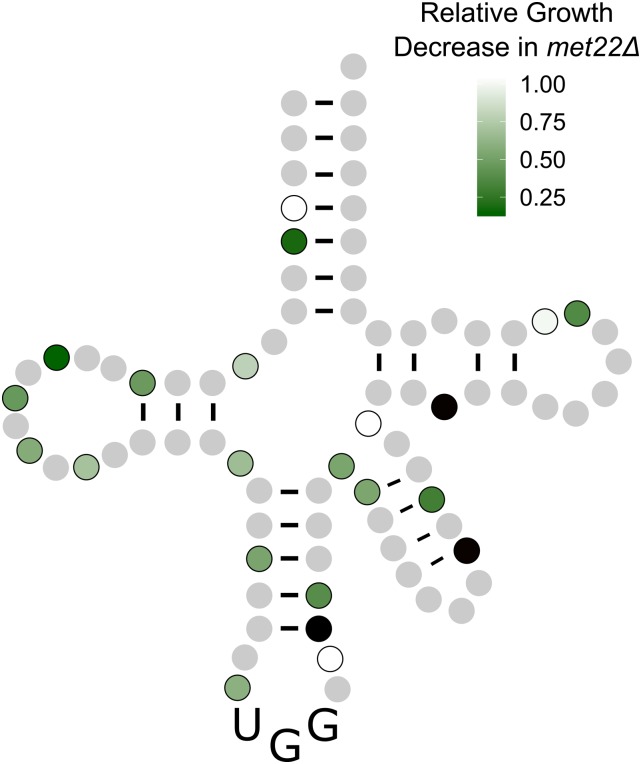
To evaluate whether the secondary mutations in the randomly selected variants reduced the stability of the tRNAs, we compared their toxicity at 18° and 30° (Figure 6A with representative temperature curves shown in Figure 6B). Seventeen of the 22 mutations became more toxic at 18°, suggesting that these mutations affect the stability or folding of the tRNA. Neither the G9A nor G26A variants were more toxic at lower temperatures (Figure 6B), suggesting that temperature induced toxicity is not due to changes in translation decoding at different temperatures. Furthermore, this result suggests that the extent of mistranslation can be regulated by growth temperature. To further evaluate if the secondary mutations affect the stability of the tRNA, we determined the toxicity of the mistranslating tRNAs in a met22Δ strain where the RTD pathway is inhibited (Dichtl et al. 1997; Chernyakov et al. 2008). If a mutation reduces tRNA function by increasing the turnover rate of the tRNA, the tRNA should be more toxic in the met22Δ strain. Eighteen of the 22 randomly selected tRNASerUGG variants were more toxic in the met22Δ strain than the wild-type strain (Figure 7 and Figure S7). Of the four variants not affected by the RTD pathway, one changed an A:U base pair in the acceptor stem to a G:U and three had mutations in loop regions. We note that of the 22 variants, all but the A38G variant showed either temperature-dependent toxicity or increased toxicity when the RTD pathway was inhibited, or both.
Figure 6.
Growth of tRNASerUGG variants at different temperatures. (A) Wild-type strains containing a mistranslating tRNASerUGG variant were grown to saturation in media lacking uracil. Optical density was normalized and cultures were diluted 1:100, spotted on media lacking uracil, and grown at either 18° for 4 days or 30° for 2 days. Pixel intensity was measured for each spot. Each value is the average of three biological replicates. (B) Five representative temperature curves from the data presented in A. Strains were grown at 18° for 4 days, 24° for 3 days, or 30° or 37° for 2 days. Each point is the average of three biological replicates and error bars indicate 1 SD.
Figure 7.
tRNASerUGG variants that are targets of the RTD pathway. Wild-type MET22 or met22Δ strains containing a mistranslating tRNASerUGG variant were grown to saturation in media lacking uracil, diluted, and spread for single colonies on media lacking uracil. Colony size was measured using ImageJ after 2 days of growth at 30° and the ratio of MET22 to met22Δ colony size determined for each mistranslating variant. Variants whose growth was not statistically different between the two strains, as determined by a Welch’s t-test, are colored white. Three variants where we could not obtain colonies in the met22Δ strain are colored black.
Mistranslating serine at other codons
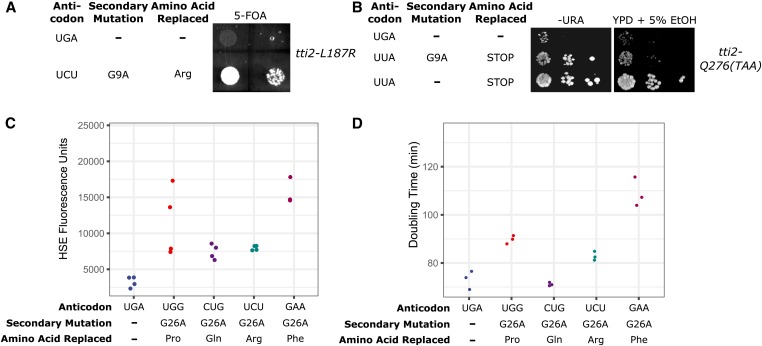
As shown by Zimmerman et al. (2018) in yeast and by Geslain et al. (2010) in mammalian cells, the ability to change the anticodon of tRNASer without affecting serylation makes it ideal to engineer mistranslation at many codons. Interestingly, Zimmerman et al. (2018) found that different amino acid substitutions are not equally toxic. To determine if secondary mutations dampen toxicity and allow mistranslation at other codons, we constructed tRNASer expressing plasmids with anticodons to decode arginine, ochre, glutamine, and phenylalanine codons. tRNASerUCU to decode arginine was examined first. No transformants were obtained when we attempted to introduce a centromeric plasmid expressing tRNASerUCU into a wild-type yeast strain (Figure S8), indicating its toxicity. To decrease mistranslation frequency and allow viable levels of serine for arginine mistranslation, we engineered a plasmid expressing tRNASerUCU with a G9A secondary mutation, which transformed into cells (Figure S8). To verify mistranslation, we transformed the plasmid expressing tRNASerUCU-G9A into a strain deleted for TTI2, but with wild-type TTI2 on a URA3 plasmid and tti2-L187R on a LEU2 centromeric plasmid (CY8607). As shown by plasmid shuffling on 5-fluoroorotic acid medium (Figure 8A), tRNASerUCU-G9A suppresses tti2-L187R, in contrast to the control wild-type tRNASerUGA. Since Tti2 with serine at position 187 supports viability (Berg et al. 2017), this result indicates that tRNASerUCU-G9A mistranslates serine for arginine.
Figure 8.
tRNASer variants mistranslate at nonserine codons. (A) tti2-L187R and either a wild-type tRNASerUGA or tRNASerUCU-G9A that mistranslates serine at arginine codons were transformed into the tti2 disruption strain CY6963 with TTI2 on a URA3 plasmid. Strains were grown in media lacking histidine and leucine and plated in 10-fold serial dilutions on 5-fluoroorotic acid containing medium to select against colonies containing wild-type TTI2. (B) Wild-type tRNASerUGA, an ochre suppressor serine tRNA (tRNASerUUA) or tRNASerUUA-G9A were transformed into a tti2 disruption strain containing tti2-Q276(TAA). Strains were spotted in 10-fold serial dilutions on media lacking uracil or on YPD containing 5% ethanol. (C) Mistranslating tRNAs induce a heat-shock response. Wild-type strain (BY4742) containing a fluorescence heat shock reporter was transformed with wild-type tRNASerUGA, tRNASerUGG-G26A, tRNASerCUG-G26A, tRNASerUCU-G26A, or tRNASerGAA-G26A. Strains were grown to saturation in selective minimal medium, diluted 1:20 in the same media, and grown for 6 hr. Cell densities were normalized and fluorescence measured. Each point represents one biological replicate. (D) Strains containing either wild-type tRNASer or mistranslating serine tRNAs from C were grown to saturation in media lacking uracil, diluted to an OD600 of ∼0.1 in the same media and grown for 24 hr. OD600 was measured every 15 min. Doubling time was calculated with the R package “growthcurver” (Sprouffske and Wagner 2016). Each point represents one biological replicate.
Previously, we identified an allele of tti2 where the glutamine codon at position 276 was mutated to an ochre stop codon (Hoffman et al. 2016). tti2-Q276TAA grows slowly on complete medium and is stress sensitive. We predict that tRNASer could be engineered to suppress ochre stop codons. As shown in Figure 8B, a wild-type tRNASer variant with an ochre anticodon (UUA) is not toxic and suppresses the tti2-Q276TAA slow growth and stress-sensitive phenotype. In this case, the tRNASerUUA lacking secondary mutations was not lethal, likely because of competition between the tRNA and release factor. The variant with the ochre anticodon and a G9A secondary mutation was less active, suppressing the slow growth of tti2-Q276TAA on complete media and only weakly restored growth on medium containing 5% ethanol.
We generated serine tRNAs with glutamine (CUG) or phenylalanine (GAA) anticodons in the context of a G26A secondary mutation, to prevent potential lethality. Since we do not have a reporter construct to detect mistranslation of glutamine or phenylalanine codons, we used heat-shock response and growth rates as proxies for mistranslation. Heat-shock response was measured in the wild-type BY4742 strain expressing wild-type tRNASerUGA or G26A variants that decode either glutamine, phenylalanine, proline, or arginine codons. All of the strains expressing a variant tRNA induced a heat-shock response, as compared to the strain containing the wild-type tRNASerUGA (Figure 8C). The effect of the tRNAs on growth is shown in Figure 8D. The serine tRNA with a glutamine anticodon did not affect growth, whereas the tRNA decoding arginine, proline, or phenylalanine significantly increased doubling time by 10, 20, and 50%, respectively (Welch’s t-test; P < 0.05), indicating that incorporating serine for different amino acids has different consequences. Our results also demonstrate that serine misincorporation at different codons can be achieved by pairing with an appropriate secondary mutation.
Discussion
tRNAs that mistranslate the genetic code have utility in a number of molecular and synthetic biology applications for both research and biotechnology. Because of its effects on the proteome, mistranslation can be toxic or even lethal, depending on the level of mistranslation and the amino acid substitution. For mistranslation to be useful in biological applications, it must be modulated to avert toxicity. Here, we identify secondary mutations within the tRNA to allow for a range of mistranslation levels for serine to proline substitution (all variants investigated are listed in Table S5). These same derivatives are applicable for substitutions at other codons.
Uses of mistranslation include expanding the genetic code to incorporate noncanonical amino acids, generating statistical proteins that have expanded activity or substrate specificity or inducing proteome wide translational errors to study the effects of amino acid substitutions on cellular functions. The range of efficiencies allows selection of the optimal variant for the application. For example, selecting the optimal tRNASer variant could allow incorporation of noncanonical amino acids or even selenocysteine in yeast, without resulting in a loss of fitness, which can lead to poor protein yields. Tuning of the mistranslation will also facilitate optimal mistranslation levels when using multiple tRNAs that substitute at different codons. The temperature-sensitive nature of the tRNAs variants we identified allows regulated mistranslation and applications such as the control of protein expression when used to suppress stop codons. For example, a stop codon could be incorporated early in the protein coding sequence of interest. In the presence of a corresponding temperature-sensitive tRNASer derivative, at low temperatures the stop codon would be read through and the protein expressed. The conditional mistranslation also allows mistranslation at levels above the lethal threshold.
Generating tRNAs that mistranslate at different levels
We found that an unbiased genetic selection was the most efficient approach to identify tRNASer bases with nonlethal levels of mistranslation. Bases altered in the targeted strategy were often essential and many of the variants did not mistranslate. By selecting for randomly induced mutations of tRNASerUGG that suppressed tti2-L187P and were therefore mistranslating at a nonlethal level, we identified 22 single nucleotide mutations that induce mistranslation, including mutations on G9, A20b, G26, and C40 that we had identified previously (Berg et al. 2017). Most of these changes occur in single-stranded regions (12 variants) or create a G:U pair (six variants), and thus they likely maintain the overall tRNA structure. The mutations that abolish base pairing occur at the beginning or end of the stem resulting in its shortening. Despite this, we conclude that most of the secondary mutations have a role in tRNA stability and/or turnover, since 21 of the variants were more susceptible to the RTD pathway (18 variants) and/or showed enhanced toxicity at low temperature (19 variants). Included in the latter were base changes that altered base pairing (A4G, C5U, C12U, A29G, U39G, C40U, G45A, Ge23A, and Ce21U). Many of these create a G:U base pair, thus supporting the idea that although thermodynamically similar to a Watson–Crick pair, the G:U pair has different structural and chemical properties that alter tRNA structure (Varani and McClain 2000).
We considered that the enhanced toxicity of the mistranslating variants at lower temperatures may be due to a temperature-dependent effect of mistranslation on cells, rather than being an effect of tRNA stability. However, neither the G9A nor G26A variant (shown in the representative temperature curves in Figure 7B), with mistranslation frequencies of <1% and ∼5%, were more toxic at lower temperatures. Their lack of increased toxicity suggests that the temperature-induced toxicity seen for other variants is a result of tRNA stability, rather than a temperature-dependent effect of mistranslation on the proteome.
The concentration of a tRNA in the cell depends upon both its synthesis and degradation. We note that six variants had base changes in the A box or B box region, required for transcription by RNA polymerase III (Hamada et al. 2001). The EufindtRNA algorithm, a tool that predicts likelihood of tRNA expression based upon a consensus sequence (Pavesi et al. 1994), predicts that all the variants will be expressed and only the G17A secondary mutation substantially decreases the Eufind score (Table S4). As these variants are all temperature sensitive, we suggest the base changes have a more pronounced effect on stability than synthesis.
Nucleotide changes that affect tRNA stability yet enable partial activity in vivo are difficult to predict. Therefore, using a genetic selection as we have done here or a screen for alleles that induce a partial heat-shock response appears to be the most efficient way to identify mistranslating tRNA variants.
Features of the sequence-function relationships of S. cerevisiae tRNASer
With the exception of mitochondrial tRNASer in metazoans (Helm et al. 2000), the long variable arm is conserved in all tRNASer molecules and is a key element in recognition by SerRS (Normanly et al. 1986, 1992; Sampson and Saks 1993; Asahara et al. 1994; Biou et al. 1994; Himeno et al. 1997; Bilokapic et al. 2006). Our results indicate the importance of both the length and sequence orientation of the variable arm for function of tRNASer in S. cerevisiae. As little as a single base-pair deletion impaired function below a detectable level in the sensitive tti2-L187P suppression assay. Similarly, reversing the orientation of the G:C base pairs in the variable arm eliminated function. Our results also confirm the essential nature of the D-arm C11:G24 base pair of tRNASerUGG, previously identified for its role in serylation in E. coli (Normanly et al. 1986, 1992; Asahara et al. 1994).
SerRS is a class II synthetase that contains a seven-sheet, antiparallel β-fold (Artymiuk et al. 1994). In this fold, motif 2 interacts with the acceptor stem (Burke et al. 2000) and models of the interaction position motif 2 close to base pairs 3:70 of the acceptor stem (Eichert et al. 2011; Berg et al. 2018). Our results indicate that base pairs 1:72 and 3:70 play partial roles in tRNASer function in S. cerevisiae, whereas base pairs 2:71 and 4:69 have lesser roles. This is consistent with a previous study, where we found that cross-species differences in the function of human and yeast tRNASer functions are the result of differences at the 3:70 position (Berg et al. 2018).
Modifications play many roles in tRNA function, from facilitating accurate decoding to maintaining the correct tRNA structure (Lorenz et al. 2017). Fourteen bases in tRNASer are modified (Machnicka et al. 2013). In our random selection for reduced function, we identified mutations at seven of these modified bases. Six were more toxic at lower temperatures and were turned over by the RTD pathway consistent with the modifications enhancing folding and protecting the tRNA from degradation at higher temperatures. Also consistent with this idea, thermophiles have more abundant and diverse tRNA modifications, including thiolations and methylations, which rigidify the tRNA structure (Watanabe et al. 1976; Kawai et al. 1992; Kowalak et al. 1994; McCloskey et al. 2001).
Cellular effects of mistranslation
The effect of the tRNASerUGG variants on doubling time varied from almost no change in growth to a twofold increase in doubling time. The change in growth correlated well with the frequencies of mistranslation as estimated by mass spectrometry for the five mistranslating tRNAs that were tested. Based on this correlation, we predict mistranslation rates as high as 8% could be achieved with a 90% decrease in fitness. This is consistent with other measures for the maximum tolerable amount of mistranslation. For example, Ruan et al. (2008) found E. coli can tolerate up to 10% mismade protein and Mohler et al. (2017) found yeast can tolerate 8% tyrosine incorporation at phenylalanine codons. We also note that based on the correlation we observed, growth rate provides a better proxy for mistranslation than level of heat-shock response.
We compared the growth characteristics of strains mistranslating at proline, arginine, glutamine, and phenylalanine codons. Of these, misincorporation at phenylalanine codons resulted in the most pronounced decrease in growth rate. A similar result was obtained by Zimmerman et al. (2018), who found that tRNASer with phenylalanine anticodons were depleted from pools of cells containing tRNASer with randomized anticodons. Zimmerman et al. (2018) discuss possible reasons for differences in toxicity noting that level of misincorporation does not correlate well with toxicity when comparing different amino acid substitutions. Reasons include whether the amino acid substitution is conservative or nonconservative, features of codon usage, and the ratio of the mistranslating tRNA to the native isodecoder. Zimmerman et al. (2018) investigated the effect of competition between a mistranslating tRNA and native cognate tRNA on toxicity, finding that overexpressing a wild-type cognate tRNA reduced the toxicity of mistranslation. The mistranslating serine derivatives generated here that mistranslate at phenylalanine, proline, arginine, or glutamine codons all compete with approximately the same number of wild-type cognate tRNAs (10 for GAA phenylalanine, 10 for UGG proline, 11 for UCU arginine, and 10 for GUC glutamine). We believe that the specific chemistry of the substitution explains the varying toxicities of mistranslation at different codons, although we recognize that the analysis is complicated by difficulties quantitating all of the parameters in cells; for example, the extent and efficiency of wobble.
Acknowledgments
We are grateful to Stan Fields and Stephanie Zimmerman for assistance on the experiments presented in this study and feedback on the manuscript. We thank Kyle Hoffman for comments on the manuscript, Dan Giguere for help computing the tRNAscan scores, and Patrick O’Donoghue for suggestions. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; grant RGPIN-2015-04394 to C.J.B.) and generous donations from Graham Wright and James Robertson to M.D.B. Mass spectrometry work was supported by a research grant from the Keck Foundation (to J.V.). M.D.B. holds an NSERC Alexander Graham Bell Canada Graduate Scholarship. B.Y.R. holds a National Science Foundation Graduate Research Fellowship (DGE-1256082).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9742577.
Communicating editor: E. Tran
Literature Cited
- Artymiuk P. J., Rice D. W., Poirrette A. R., and Willet P., 1994. A tale of two synthetases. Nat. Struct. Biol. 1: 758–760. 10.1038/nsb1194-758 [DOI] [PubMed] [Google Scholar]
- Asahara H., Himeno H., Tamura K., Nameki N., Hasegawa T. et al. , 1994. Escherichia coli Seryl-tRNA synthetase recognizes tRNASer by its characteristics tertiary structure. J. Mol. Biol. 236: 738–748. 10.1006/jmbi.1994.1186 [DOI] [PubMed] [Google Scholar]
- Bacher J. M., Waas W. F., Metzgar D., De Crécy-Lagard V., and Schimmel P., 2007. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. J. Bacteriol. 189: 6494–6496. 10.1128/JB.00622-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M. D., Hoffman K. S., Genereaux J., Mian S., Trussler R. S. et al. , 2017. Evolving mistranslating tRNAs through a phenotypically ambivalent intermediate in Saccharomyces cerevisiae. Genetics 206: 1865–1879. 10.1534/genetics.117.203232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M. D., Genereaux J., Zhu Y., Mian S., Gloor G. B. et al. , 2018. Acceptor stem differences contribute to species-specific use of yeast and human tRNASer. Genes (Basel) 9: 612. 10.3390/genes9120612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Maier T., Ahel D., Gruic-Sovulj I., Söll D. et al. , 2006. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 25: 2498–2509. 10.1038/sj.emboj.7601129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., and Cusack S., 1994. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science 263: 1404–1410. 10.1126/science.8128220 [DOI] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C. et al. , 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Yang F., Chen F., Stehlin C., Chan B. et al. , 2000. Evolutionary coadaptation of the Motif 2−acceptor stem interaction in the class II prolyl-tRNA synthetase system. Biochemistry 39: 15540–15547. 10.1021/bi001835p [DOI] [PubMed] [Google Scholar]
- Chapeville F., Lipmann F., Ehrenstein G., Weisblum B., Ray W. J. et al. , 1962. On the role of soluble ribonucleic acid in coding for amino acids. Proc. Natl. Acad. Sci. USA 48: 1086–1092. 10.1073/pnas.48.6.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I., Whipple J. M., Kotelawala L., Grayhack E. J., and Phizicky E. M., 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 22: 1369–1380. 10.1101/gad.1654308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., 1997. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast 13: 647–653. [DOI] [PubMed] [Google Scholar]
- de Duve C., 1988. Transfer RNAs: the second genetic code. Nature 333: 117–118. 10.1038/333117a0 [DOI] [PubMed] [Google Scholar]
- Dichtl B., Stevens A., and Tollervey D., 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16: 7184–7195. 10.1093/emboj/16.23.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Uhlenbeck O. C., and Browning K. S., 1999. Quantitative assessment of EF-1alpha.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 274: 666–672. 10.1074/jbc.274.2.666 [DOI] [PubMed] [Google Scholar]
- Drummond D. A., and Wilke C. O., 2009. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10: 715–724. 10.1038/nrg2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichert A., Oberthuer D., Betzel C., Geßner R., Erdmann V. A. et al. , 2011. The Seryl-tRNA synthetase/tRNA Ser acceptor stem interface is mediated via a specific network of water molecules. Biochem. Biophys. Res. Commun. 412: 532–536. 10.1016/j.bbrc.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Elias J. E., and Gygi S. P., 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4: 207–214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Eng J. K., Jahan T. A., and Hoopmann M. R., 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics 13: 22–24. 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- Francklyn C., and Schimmel P., 1989. Aminoacylation of RNA minihelices with alanine. Nature 337: 478–481. 10.1038/337478a0 [DOI] [PubMed] [Google Scholar]
- Fukunaga R., and Yokoyama S., 2005. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 12: 915–922. 10.1038/nsmb985 [DOI] [PubMed] [Google Scholar]
- Geslain R., Cubells L., Bori-Sanz T., Álvarez-Medina R., Rossell D. et al. , 2010. Chimeric tRNAs as tools to induce proteome damage and identify components of stress responses. Nucleic Acids Res. 38: e30 10.1093/nar/gkp1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Sissler M., and Florentz C., 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. 10.1093/nar/26.22.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A. C., Kordala A. J., Strack R., Wang X., Geslain R. et al. , 2016. A dual fluorescent reporter for the investigation of methionine mistranslation in live cells. RNA 22: 467–476. 10.1261/rna.054163.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustilo E. M., Vendeix F. A., and Agris P. F., 2008. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 11: 134–140. 10.1016/j.mib.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M., Huang Y., and Lowe T. M., 2001. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol. Cell. Biol. 21: 6870–6881. 10.1128/MCB.21.20.6870-6881.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Brulé H., Friede D., Giegé R., Pütz D. et al. , 2000. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379. 10.1017/S1355838200001047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., and Shimizu M., 1990. Conversion of aminoacylation specificity from tRNATyrto tRNASer in vitro. Nucleic Acids Res. 18: 6815–6819. 10.1093/nar/18.23.6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Yoshida S., Soma A., and Nishikawa K., 1997. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNA(Leu) in vitro. J. Mol. Biol. 268: 704–711. 10.1006/jmbi.1997.0991 [DOI] [PubMed] [Google Scholar]
- Hoffman K. S., Duennwald M. L., Karagiannis J., Genereaux J., McCarton A. S. et al. , 2016. Saccharomyces cerevisiae Tti2 regulates PIKK proteins and stress response. G3 (Bethesda) 6: 1649–1659. 10.1534/g3.116.029520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman K. S., Berg M. D., Shilton B. H., Brandl C. J., and O’Donoghue P., 2017. Genetic selection for mistranslation rescues a defective co-chaperone in yeast. Nucleic Acids Res. 45: 3407–3421. 10.1093/nar/gkw1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., 1997. Discriminating among the discriminator bases of tRNAs. Chem. Biol. 4: 93–96. 10.1016/S1074-5521(97)90252-0 [DOI] [PubMed] [Google Scholar]
- Jackman J. E., and Alfonzo J. D., 2013. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 4: 35–48. 10.1002/wrna.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javid B., Sorrentino F., Toosky M., Zheng W., Pinkham J. T. et al. , 2014. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. USA 111: 1132–1137. 10.1073/pnas.1317580111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G. et al. , 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18: 1227–1240. 10.1101/gad.1183804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J., 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4: 923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M. et al. , 1992. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2ʹ-hydroxyl group. Biochemistry 31: 1040–1046. 10.1021/bi00119a012 [DOI] [PubMed] [Google Scholar]
- Kowalak J. A., Dalluge J. J., McCloskey J. A., and Stetter K. O., 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33: 7869–7876. 10.1021/bi00191a014 [DOI] [PubMed] [Google Scholar]
- Kramer E. B., and Farabaugh P. J., 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96. 10.1261/rna.294907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant J. T., Berg M. D., Sze D. H., Hoffman K. S., Akinpelu I. C. et al. , 2017. Visualizing tRNA-dependent mistranslation in human cells. RNA Biol. 15: 567–575. 10.1080/15476286.2017.1379645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Kim D. G., Kim B.-G., Yang W. S., Hong J. et al. , 2014. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 127: 4234–4245. 10.1242/jcs.152470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Boniecki M. T., Jaffe J. D., Imai B. S., Yau P. M. et al. , 2011. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. USA 108: 9378–9383. 10.1073/pnas.1016460108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., and Söll D., 2010. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. USA 107: 4028–4033. 10.1073/pnas.1000315107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Reynolds N., and Ibba M., 2009. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63: 61–78. 10.1146/annurev.micro.091208.073210 [DOI] [PubMed] [Google Scholar]
- Lorenz C., Lünse C. E., and Mörl M., 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7: 35. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka M. A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M. et al. , 2013. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 41: D262–D267. 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey S. E., Moura G., Beltrão P., Almeida R., Garey J. R. et al. , 2003. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 13: 544–557. 10.1101/gr.811003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., and Foss K., 1988. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240: 793–796. 10.1126/science.2452483 [DOI] [PubMed] [Google Scholar]
- McCloskey J. A., Graham D. E., Zhou S., Crain P. F., Ibba M. et al. , 2001. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29: 4699–4706. 10.1093/nar/29.22.4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal A., Mohler K., and Ibba M., 2014. Mistranslation of the genetic code. FEBS Lett. 588: 4305–4310. 10.1016/j.febslet.2014.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler K., Aerni H.-R., Gassaway B., Ling J., Ibba M. et al. , 2017. MS-READ: quantitative measurement of amino acid incorporation. Biochim. Biophys. Acta, Gen. Subj. 1861: 3081–3088. 10.1016/j.bbagen.2017.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasvall S. J., Chen P., and Björk G. R., 2004. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNApro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 10: 1662–1673. 10.1261/rna.7106404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer N., Goodenbour J. M., David A., Dittmar K. A., Jones R. B. et al. , 2009. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462: 522–526. 10.1038/nature08576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard M., Thirup S., Clark B. F., and Nyborg J., 1996. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie 78: 921–933. 10.1016/S0300-9084(97)86714-4 [DOI] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., and Abelson J., 1986. Changing the identity of a transfer RNA. Nature 321: 213–219. 10.1038/321213a0 [DOI] [PubMed] [Google Scholar]
- Normanly J., Ollick T., and Abelson J., 1992. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl. Acad. Sci. USA 89: 5680–5684. 10.1073/pnas.89.12.5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. L. J., Poruri K., and Martinis S. A., 2014. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA 5: 461–480. 10.1002/wrna.1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes J. A., Carreto L., Simões J., Bezerra A. R., Gomes A. C. et al. , 2012. Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast. BMC Biol. 10: 55 10.1186/1741-7007-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., Conterio F., Bolchi A., Dieci G., and Ottonello S., 1994. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix anaylsis of transcriptional control regions. Nucleic Acids Res. 22: 1247–1256. 10.1093/nar/22.7.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M., and Hopper A. K., 2010. tRNA biology charges to the front. Genes Dev. 24: 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y., 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2: 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rich A., and RajBhandary U. L., 1976. Transfer RNA: molecular structure, sequence, and properties. Annu. Rev. Biochem. 45: 805–860. 10.1146/annurev.bi.45.070176.004105 [DOI] [PubMed] [Google Scholar]
- Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S. et al. , 2008. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl. Acad. Sci. USA 105: 16502–16507. 10.1073/pnas.0809179105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks M. E., and Sampson J. R., 1996. Variant minihelix RNAs reveal sequence-specific recognition of the helical tRNA(Ser) acceptor stem by E. coli seryl-tRNA synthetase. EMBO J. 15: 2843–2849. 10.1002/j.1460-2075.1996.tb00645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., and Saks M. E., 1993. Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 21: 4467–4475. 10.1093/nar/21.19.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M. A. S., Cheesman C., Costa V., Moradas-Ferreira P., and Tuite M. F., 1999. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 31: 937–947. 10.1046/j.1365-2958.1999.01233.x [DOI] [PubMed] [Google Scholar]
- Schimmel P., 2011. Mistranslation and its control by tRNA synthetases. Philos. Trans. R. Soc. B Biol. Sci. 366: 2965–2971. 10.1098/rstb.2011.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., and Jensen T. H., 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33: 501–510. 10.1016/j.tibs.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Schneider C., Anderson J. T., and Tollervey D., 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell 27: 324–331. 10.1016/j.molcel.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., and Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. H., and Pan T., 2016. Temperature dependent mistranslation in a hyperthermophile adapts proteins to lower temperatures. Nucleic Acids Res. 44: 294–303. 10.1093/nar/gkv1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. H., and Pan T., 2017. tRNA misacylation with methionine in the mouse gut microbiome in situ. Microb. Ecol. 74: 10–14. 10.1007/s00248-016-0928-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., and Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Horn C., Brown M., Ioudovitch A., and Steinberg S., 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. 10.1093/nar/26.1.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K., and Wagner A., 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17: 172 10.1186/s12859-016-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D. et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Urbonavičius J., Qian Q., Durand J. M. B., Hagervall T. G., and Björk G. R., 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20: 4863–4873. 10.1093/emboj/20.17.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., and McClain W. H., 2000. The G·× U wobble base pair. a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1: 18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Pan T., 2016. Stress response and adaptation mediated by amino acid misincorporation during protein synthesis. Adv. Nutr. 7: 773S–779S. 10.3945/an.115.010991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Shinma M., Oshima T., and Nishimura S., 1976. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72: 1137–1144. 10.1016/S0006-291X(76)80250-1 [DOI] [PubMed] [Google Scholar]
- Wei F., Suzuki T., Watanabe S., Kimura S., Kaitsuka T. et al. , 2011. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121: 3598–3608. 10.1172/JCI58056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple J. M., Lane E. A., Chernyakov I., D’Silva S., and Phizicky E. M.. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 25: 1173–1184. 10.1101/gad.2050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout E., Goodenbour J. M., Fréchin M., and Pan T., 2012. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res. 40: 10494–10506. 10.1093/nar/gks805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., and Davis R. W., 1997. Functional analysis of the yeast genome. Curr. Opin. Genet. Dev. 7: 771–776. 10.1016/S0959-437X(97)80039-1 [DOI] [PubMed] [Google Scholar]
- Woese C. R., 1965. On the evolution of the genetic code. Proc. Natl. Acad. Sci. USA 54: 1546–1552. 10.1073/pnas.54.6.1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Fan Y., and Ling J., 2014. Mechanism of oxidant-induced mistranslation by threonyl-tRNA synthetase. Nucleic Acids Res. 42: 6523–6531. 10.1093/nar/gku271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. M., Kon Y., Hauke A. C., Ruiz B. Y., Fields S. et al. , 2018. Conditional accumulation of toxic tRNAs to cause amino acid misincorporation. Nucleic Acids Res. 46: 7831–7843. 10.1093/nar/gky623 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. File S1 contains all supplemental figures and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9742577.