Homologous recombination is induced to high levels in meiosis and is clustered at hotspots that regulate its frequency and distribution in the genome. By studying five different classes of DNA sequence-dependent recombination hotspots in the fission yeast...
Keywords: meiosis, genetic recombination, nucleosome, chromatin remodeling, epigenetics
Abstract
In meiosis, multiple different DNA sequence motifs help to position homologous recombination at hotspots in the genome. How do the seemingly disparate cis-acting regulatory modules each promote locally the activity of the basal recombination machinery? We defined molecular mechanisms of action for five different hotspot-activating DNA motifs (M26, CCAAT, Oligo-C, 4095, 4156) located independently at the same site within the ade6 locus of the fission yeast Schizosaccharomyces pombe. Each motif promoted meiotic recombination (i.e., is active) within this context, and this activity required the respective binding proteins (transcription factors Atf1, Pcr1, Php2, Php3, Php5, Rst2). High-resolution analyses of chromatin structure by nucleosome scanning assays revealed that each motif triggers the displacement of nucleosomes surrounding the hotspot motif in meiosis. This chromatin remodeling required the respective sequence-specific binding proteins, was constitutive for two motifs, and was enhanced meiotically for three others. Hotspot activity of each motif strongly required the ATP-dependent chromatin remodeling enzyme Snf22 (Snf2/Swi2), with lesser dependence on Gcn5, Mst2, and Hrp3. These findings support a model in which most meiotic recombination hotspots are positioned by the binding of transcription factors to their respective DNA sites. The functional redundancy of multiple, sequence-specific protein–DNA complexes converges upon shared chromatin remodeling pathways that help provide the basal recombination machinery (Spo11/Rec12 complex) access to its DNA substrates within chromatin.
MEIOSIS couples one round of DNA replication, high-frequency recombination between homologous chromosomes (homologs), and two rounds of chromosome segregation to produce haploid meiotic products. In most eukaryotes, recombination is required for the faithful segregation of homologs at the first meiotic division and it generates genetic diversity upon which natural selection can act (Székvölgyi and Nicolas 2009; Handel and Schimenti 2010).
The broadly conserved catalytic subunit of the basal meiotic recombination machinery, Spo11 (Rec12 in fission yeast), introduces DNA double-strand breaks (DSBs) that initiate, and are required for, meiotic recombination throughout the genome (Keeney et al. 1997; Gerton et al. 2000; Sharif et al. 2002; Buhler et al. 2007; Cromie et al. 2007; Kan et al. 2011). While DSB-initiated recombination can occur anywhere along chromosomes, it is clustered preferentially at hotspots that regulate its frequency and positioning in the genome (Smukowski and Noor 2011; Dluzewska et al. 2018; Tock and Henderson 2018). As is the case for transcription, cis-acting regulatory elements (transcription factor binding sites) help to localize the activity of the basal recombination machinery at its preferred sites of action (Wahls and Davidson 2010, 2012).
Allele-specific (i.e., cis-acting) regulation of meiotic recombination was first reported for the ade6-M26 hotspot of fission yeast (Gutz 1971). A single base pair substitution in the ade6 locus created serendipitously a 7-bp DNA sequence motif (the M26 DNA site, Table 1) that is essential for hotspot activity (Schuchert et al. 1991). Binding of the heterodimeric transcription factor Atf1-Pcr1 (originally called Mts1-Mts2) (Wahls and Smith 1994) to the M26 DNA site increases recombination ∼20-fold, relative to control alleles of ade6 that lack the M26 DNA site (Wahls and Smith 1994; Kon et al. 1997, 1998; Gao et al. 2008). The Atf1-Pcr1-M26 protein–DNA complex triggers chromatin remodeling in meiosis (Yamada et al. 2004; Hirota et al. 2007) and stimulates the catalysis of recombination-initiating DSBs by Rec12/Spo11 (Steiner et al. 2002). The hotspot-specific chromatin remodeling precedes the formation of DSBs and does not require Rec12 (Hirota et al. 2007), suggesting that chromatin remodeling functions upstream of (i.e., likely promotes) the formation of DSBs at this hotspot.
Table 1. Hotspot-activating DNA sequence motifs and their binding proteins.
| Motif name | Motif sequencea | Binding proteins |
|---|---|---|
| M26 | 5′-ATGACGT-3′ | Atf1-Pcr1 |
| CCAAT | 5′-CCAATCA-3′ | Php2-Php3-Php5 |
| Oligo-C | 5′-CCCCGCAC -3′ | Rst2 |
| 4095 | 5′-GGTCTRGACC-3′ | Unknown |
| 4156 | 5′-TCGGCCGA-3′ | Unknown |
Experimentally defined core sequences that are sufficient to promote recombination locally; R = A or G. The base pair substitutions used to generate these DNA binding sites in the ade6 gene and the DNA sequences of all other ade6 alleles used in this study are provided in Table S2.
A subset of naturally occurring and artificially created M26 DNA sites located elsewhere in the genome, within both coding and noncoding regions, promote recombination locally (Virgin et al. 1995; Steiner and Smith 2005). Interestingly, around three-quarters of the M26 sites in the genome are not recombinogenic (Wahls and Davidson 2010), even though most of them are occupied by the Atf1-Pcr1 heterodimer (Kon et al. 1998; Eshaghi et al. 2010; Woolcock et al. 2012). Additional factors, such as nearby, cis-linked, DNA sequence elements (Zahn-Zabal et al. 1995), and, potentially, structural features such as chromatin loop-axis domains (Miyoshi et al. 2012; Yamada et al. 2018), are required for Atf1-Pcr1-M26 complexes to promote recombination locally. Such “context-variable penetrance” (Wahls and Davidson 2012) also occurs for other known and inferred cis-acting regulatory factors of fission yeast and other species, including other recombinogenic DNA sequences (Mieczkowski et al. 2006; Myers et al. 2008; Steiner et al. 2011), histone PTMs (Brick et al. 2012; Yamada et al. 2013), open chromatin (Berchowitz et al. 2009; de Castro et al. 2011), and long noncoding RNAs (Wahls et al. 2008). Nevertheless, the subset of naturally occurring M26 DNA sites that do promote recombination are implicated to help position ∼20% of all recombination in the fission yeast genome (Wahls and Davidson 2010).
A large, gain-of-function screen identified numerous short DNA sequences that promote meiotic recombination in fission yeast, and M26 was among the consensus sequence motifs discovered, validating the approach (Steiner et al. 2009). Four additional, newly discovered consensus motifs (CCAAT, Oligo-C, 4095, and 4156) were subsequently refined functionally at single-nucleotide resolution by systematic base pair substitution analyses (Table 1) (Steiner et al. 2009, 2011; Foulis et al. 2018). As with M26-promoted recombination (Wahls and Smith 1994; Kon et al. 1997, 1998; Gao et al. 2008), the CCAAT and Oligo-C motifs are bound by transcription factors (Table 1) that are essential for hotspot activity (Steiner et al. 2009, 2011; Foulis et al. 2018). Notably, the proteins that bind to and activate these specific, sequence-dependent hotspots have no significant impact on the activation of heterologous sequence-dependent hotspots to which they do not bind (Steiner et al. 2009, 2011). Proteins that bind to DNA motifs 4095 and 4156 have not yet been identified. Nearly 200 additional hotspot-activating DNA sequences that were identified in the screen share no obvious homology with each other or with the defined motifs. Thus, ∼200 (and potentially more) short, distinct DNA sequence elements, and, by inference, their sequence-specific binding proteins, help to position recombination at hotspots in fission yeast—and, together, they have the potential to regulate all hotspots in the genome (Steiner et al. 2009, 2011; Wahls and Davidson 2010, 2012).
The DNA sequence-dependent regulation of meiotic recombination hotspots has also been reported for budding yeast, mice, and humans, and has been implicated by association in many other species. In budding yeast, deletions and insertions that contain transcription factor binding sites ablate and create hotspots, respectively, and the corresponding binding proteins are required for hotspot activity (White et al. 1991, 1993; Fan et al. 1995). The different protein–DNA complexes can function redundantly at the same locus, which can explain (Wahls and Davidson 2012) why ablating a transcription factor does not necessarily abolish hotspot activity near all of its binding sites (Mieczkowski et al. 2006; Zhu and Keeney 2015). Like the regulatory protein–DNA complexes of fission yeast and mammals, those of budding yeast display context-variable penetrance (Mieczkowski et al. 2006; Zhu and Keeney 2015). Nevertheless, genome-wide, ∼52% of DSB hotspots in budding yeast colocalize with DNA binding sites for 77 transcription factors (Pan et al. 2011). Extrapolation for the estimated 140–250 transcription factors (Hughes and de Boer 2013) suggests that transcription factor binding sites could account for the regulated positioning of most, if not all, hotspots in this organism.
Among the five well-characterized regulatory elements of fission yeast, M26 DNA sites and Atf1-Pcr1 heterodimer contribute the most to recombination across the genome (Steiner et al. 2009, 2011; Wahls and Davidson 2010; Foulis et al. 2018). The homologous recombination activation (HRA) domain resides in Atf1 (Gao et al. 2008). The Atf1 ortholog Sko1 of budding yeast also seems to be quite recombinogenic, based on the very high frequency with which DSBs are directed to Sko1 binding sites (Pan et al. 2011). Other sequence-specific binding proteins proven to activate hotspots in fission yeast (Php2, Php3, Php5, Rst2) (Steiner et al. 2009, 2011) also have orthologs in budding yeast (Hap2, Hap3, Hap5, Adr1), and, in each case, DSBs are directed preferentially to their binding sites (Pan et al. 2011). Moreover, four of the regulatory DNA sequences discovered in fission yeast also promote recombination (i.e., generate hotspots) when placed at a test locus in budding yeast (Steiner and Steiner 2012). Thus, the positioning of meiotic recombination hotspots by specific DNA sites and their binding proteins (transcription factors) is conserved between two species that are as evolutionarily distant from each other as either species is from human beings (Sipiczki 2000). Data from other taxa are also consistent with broad conservation of cis-acting regulatory mechanisms. For example, DNA sequence motifs implicated in helping position meiotic recombination in honeybees (Mougel et al. 2014) match, or are similar to, respectively, the CCAAT and Oligo-C motifs that are known to activate hotspots in fission yeast (Steiner et al. 2009, 2011).
How do seemingly disparate DNA sequences and their binding proteins each promote locally the activity of the basal recombination machinery? Here, we report that each of five distinct, well-defined, sequence-dependent recombination hotspots of fission yeast (Table 1) employs a common mechanism of function. They each trigger the remodeling of chromatin structure in meiosis, and, in each case, the ATP-dependent chromatin remodeling enzyme Snf22 (Snf2/Swi2) is a key effector of high frequency recombination.
Materials and Methods
Fission yeast husbandry
Genotypes of S. pombe strains used in this study are listed in Supplemental Material, Table S1 and the DNA sequences of all ade6 alleles used are provided in Table S2. We use standard fission yeast genetic nomenclature (Kohli 1987) for wild-type genes (e.g., ade6), variant alleles (e.g., ade6-M210), and deletions (e.g., pcr1-D1), although such deletions are designated with deltas in the text for clarity (e.g., pcr1Δ). Strains were constructed using standard genetic methods, and were cultured in rich media or minimal media supplemented with specific nutrients and/or G418 at 100 µg per ml (Gutz et al. 1974; Forsburg and Rhind 2006).
Analyses of meiotic recombination
Methods to measure rates of intragenic meiotic recombination were as described (Kon et al. 1997, 1998), are depicted schematically in Figure 1A, and are summarized here. Heterothallic, haploid strains with different ade6 alleles (see Table S2 for their DNA sequences) were crossed, and haploid meiotic products (spores) were plated on minimal media that contains or lacks adenine. The titer of Ade+ recombinant spore colonies was divided by the titer of all viable spore colonies to yield the recombinant frequency from each cross. Recombinant frequencies (mean ± SD from three independent biological replicates) are plotted in the figures; primary data values from each cross are reported in Table S4 and Table S5.
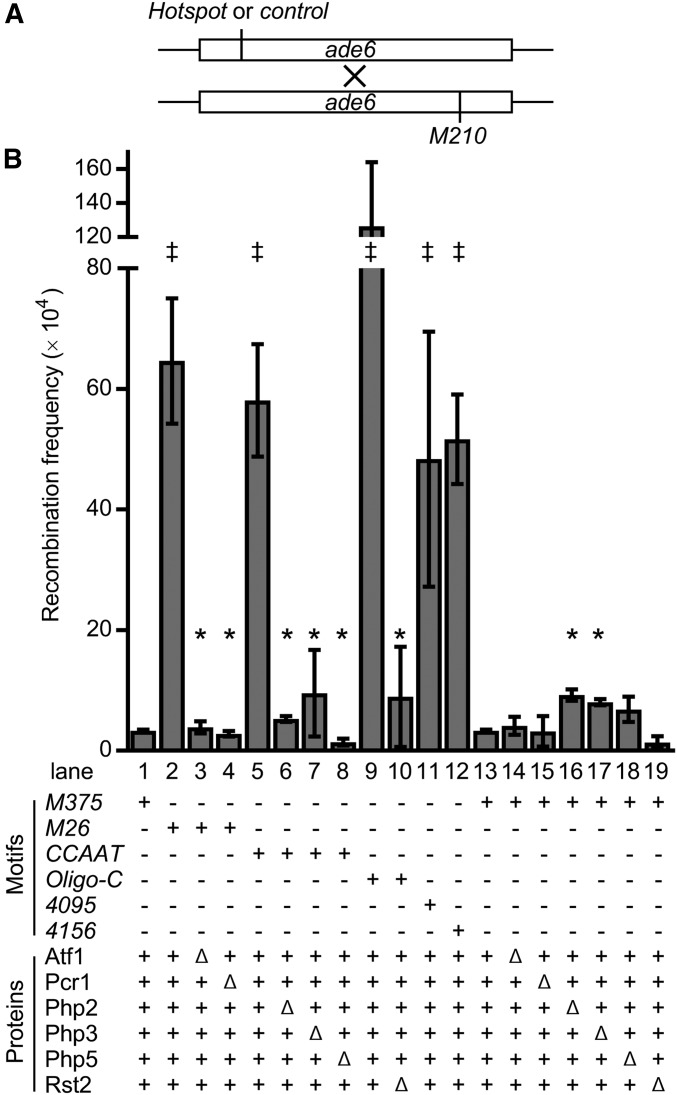
Figure 1.
DNA sequence-specific, binding protein-dependent activation of meiotic recombination hotspots. (A) Assay for meiotic recombination. Diagram shows ade6 ORF (boxes) and relative positions of alleles used. Haploid cells harboring a basal recombination control allele (M375) or alleles that contain a hotspot DNA sequence motif (M26, CCAAT, Oligo-C, 4095, and 4156) were crossed to a strain with a tester allele (M210) and haploid meiotic products were scored for frequencies of ade6+ recombinants. (B) Recombinant frequencies from basal control and hotspot crosses in the presence or absence of proteins that bind to the hotspot DNA sequence motifs. Data are mean ± SD from three biological replicates; statistically significant differences (P ≤ 0.05% from t-test) are shown for hotspot vs. control (‡) and hotspot lacking its binding proteins vs. with its binding proteins (*). Note that hotspot activation requires both the DNA sequence motif and the protein(s) that bind to that motif, and that ablating these binding proteins does not reduce significantly rates of basal recombination at the control M375. The DNA sequences and precise locations of alleles are provided in Table S2; primary data values are provided in Table S4.
Induction of meiosis
The induction of synchronous meiosis by thermal inactivation of Pat1-114ts and the monitoring of meiotic progression were as described (Storey et al. 2018), although a different procedure was used to synchronize cells in G0 (G1) phase of the cell cycle prior to inducing meiosis. The cells were grown to midlog phase (A595 = 0.5) at 25° in 167 ml of EMM2 minimal medium containing 1% glucose and 3.75 g/l glutamate as the nitrogen source. Cells were harvested by centrifugation (2500 × g for 5 min), washed with ddH2O, inoculated into 500 ml of EMM2 media that lacked glutamate, and incubated at 25° for 16 hr to synchronize them in G0 phase. Glutamate was added (to 1.0 g/l), cultures were allowed to recover from starvation at 25° for 15 min, and then brought rapidly to 35° (by swirling the flasks in a hot water bath). Cultures were incubated at 35° and samples were collected at the desired time points.
Mapping of chromatin structure
Our methods for the preparation of mononucleosomes are based on procedures optimized for fission yeast (Lantermann et al. 2009, 2010) and that we refined empirically for cells in meiosis. Cell cultures were treated with 0.5% formaldehyde for 20 min to crosslink and stabilize proteins and DNA within chromatin, then the crosslinking reactions were quenched by the addition of glycine to 125 mM. After 10 min, cells were harvested by centrifugation (2500 × g for 5 min at 4°), washed with ddH2O, collected by centrifugation, and stored as frozen cell pellets at −20° until processed further.
Each preparation of spheroplasts employed formaldehyde-treated cells from 250 ml of culture (∼1.3 × 109 cells). Cells were thawed on ice and resuspended in 6.7 ml of preincubation buffer (20 mM citric acid, 20 mM Na2HPO4, 40 mM EDTA, pH 8.0) supplemented with 14 µl of β-mercaptoethanol (BME). Cells were collected by centrifugation at 4°, then resuspended in 3.3 ml of spheroplast buffer (1 M Sorbitol, 50 mM Tris-HCl, pH 7.4) supplemented with 2.3 µl of BME and 50 µl of yeast lytic enzyme (100 mg/ml; MP Biomedicals, Santa Anna, CA). Reactions were incubated at 32° for the amount of time determined empirically within each experiment (usually between 30 and 60 min) required to convert essentially all cells to spheroplasts (as judged by light microscopy to monitor changes in cell shape and birefringence, as well as the susceptibility of cells to lysis when exposed to 0.5% SDS). The spheroplasts were collected by centrifugation, were washed with 3.3 ml of the spheroplast buffer without lytic enzyme, and were collected by centrifugation. The spherophasts were then resuspended in 2.5 ml of nuclease reaction buffer (1 M Sorbitol, 50 mM NaCl, 10 mM Tris-HCl pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 0.75% NP-40) supplemented with 0.5 µl of BME.
For the preparation of mononucleosomes, one-third of each 2.5 ml spheroplast suspension (830 µl) was placed into a tube to serve as an intact-DNA control; two-thirds of each suspension (1660 µl) was placed into another tube for digestion with micrococcal nuclease (MNase). To each sample for digestion we added 3 µl of MNase (0.59 units per microliter; Sigma, Saint Louis, MO), then the no MNase (intact-DNA control) and MNase (nucleosome) samples were incubated in parallel at 37°. After 20 min of incubation, half of the MNase sample (830 µl) was processed, and, at 40 min, the other half of the MNase sample (830 µl) and the intact-DNA control (no MNase) sample (830 µl) were each processed. (Although we titrated the amount and time of MNase digestion for each separate batch of reagents and biological samples to ensure optimal results, we still found that bracketing of reaction times within each experiment was required to ensure the likelihood of obtaining a high proportion of mononucleosomes.) To each tube (830 µl) we added 110 µl of stop buffer (5% SDS, 100 mM EDTA) and 100 µl proteinase K (10 mg/ml; Sigma). The samples were then incubated at 65° for ∼16 hr to reverse crosslinks and digest cellular proteins. Samples of DNA were isolated from intact chromatin and from nucleosome preparations by phenol/chloroform extractions and ethanol precipitation. Both the high MW (undigested) DNA and the mononucleosome-sized (digested) DNA molecules were then isolated by preparative electrophoresis on 2% agarose gels. The DNAs were eluted using Freeze “N” Squeeze DNA gel extraction spin columns (Bio-Rad, Hercules, CA), were recovered by precipitation with isopropanol, were resuspended in 50 µl of TE buffer (10 mM Tris/HCl pH 8.0, 1 mM EDTA), and aliquots were stored at −20° until used as templates for PCR.
Our approach for the PCR-based mapping of chromatin structure follows those described previously (Sansó et al. 2011; Infante et al. 2012; García et al. 2014; Small et al. 2014). Samples of DNA obtained from MNase-digested chromatin were analyzed by real-time, quantitative PCR (qPCR) using All in One qPCR Master Mix (GeneCopeia, Rockville, MD) and the PCR primers listed in Table S3. These primer pairs were based on published global maps of nucleosome occupancy in the fission yeast genome (Lantermann et al. 2009, 2010; Soriano et al. 2013), and were designed to generate 15 overlapping PCR amplicons that cover a 1.2 kbp region of ade6 (see Figure 2). The qPCR reactions were carried out using a CFX96 Real Time System (Bio-Rad). Each qPCR reaction (20 µl) contained 0.5 µl of template and 500 nM of forward and reverse primers. Thermocycler parameters were: one cycle at 95° for 10 min; followed by 40 cycles of 95° for 10 sec, 62° for 20 sec, and 72° for 15 sec. In each experiment specificity was confirmed by melting point analyses. For each amplicon, mononucleosomal DNA enrichment was calculated using the ∆∆Ct method (Schmittgen and Livak 2008), with ∆∆Ct = [(Ctpp − Ctref)No Mnase − (Ct pp − Ctref)+MNase], where pp refers to the primer pair (1–15), and ref is the smc5 reference primer pair, in both undigested (No MNase) and digested (+MNase) conditions. Data values for each amplicon under each experimental condition are provided in Table S6. In the figures, each data point (the average from three independent biological replicates) and its SD was plotted at the midpoint of the corresponding amplicon (relative to its position along the X-axis). In each case, these positional coordinates were plotted relative to the first nucleotide of the ade6 start codon (designated as nucleotide position +1). The data points were connected by smoothed curves to represent the inferred positions of nucleosomes, as is commonly done in nucleosome scanning assays that employ a qPCR-based readout [e.g., (Sansó et al. 2011; Infante et al. 2012; Garcia et al. 2014; Small et al. 2014]).
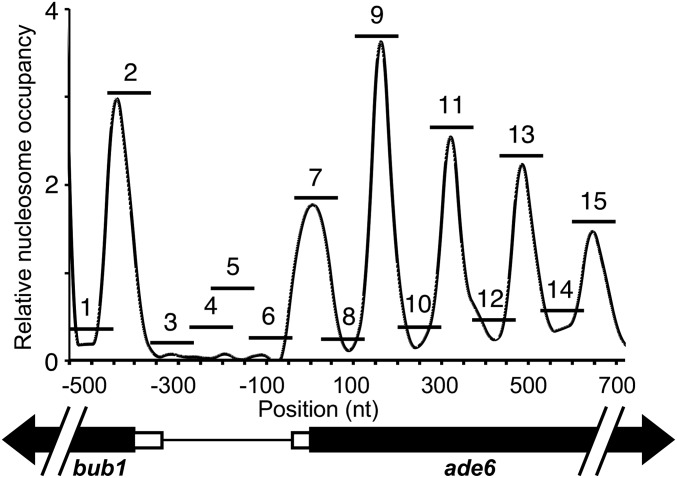
Figure 2.
Nucleosome occupancy at ade6 and design of PCR amplicons used to map chromatin structure. Graph displays our plot of published data on nucleosome occupancy in a pat1-114 strain background (like that used in our study) prior to entering meiosis (Soriano et al. 2013). A diagram of the 1.2 kb region of interest is below, indicating the 5′ UTRs (open boxes) and coding regions (black arrows) of the bub1 and ade6 genes. Relative nucleosome occupancy (GEO study GSE41773, sample GSM1024000, from single-end sequencing of DNA molecules after MNase digestion of chromatin) was plotted with the A of the ade6 start codon designated as nucleotide (nt) position +1. The horizontal bars numbered 1 through 15 indicate the positions of the overlapping PCR amplicons designed for, and used in, this study. See Table S3 for the DNA sequences and coordinates of the PCR primer pairs.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Yeast strains and other materials generated by this study are available upon request. All data supporting the conclusions of this study are available within the paper and its supplemental material file. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9745103.
Results
Factors that help to position meiotic recombination at hotspots, such as sequence-specific protein–DNA complexes and histone PTMs, each display context-variable penetrance (Wahls and Davidson 2012). Thus to elucidate whether multiple, cis-acting, regulatory factors function through a common mechanism, it is necessary to study them within the same chromosomal context. We therefore analyzed the functions of five different hotspot-activating DNA sequence motifs (M26, CCAAT, Oligo-C, 4095, 4156; Table 1) (Schuchert et al. 1991; Wahls and Smith 1994; Steiner et al. 2009, 2011; Foulis et al. 2018) located independently at the same site within the ade6 locus of fission yeast (Figure 1A). In each case, the hotspot alleles were generated by base pair substitutions that create the respective DNA sequence motifs (Table S2), thus maintaining the overall structure and spacing of the locus. The basal recombination control allele (M375) and the hotspot alleles were each located near the 5′ end of the ade6 coding region, mapping within the position of the second nucleosome (+2 nucleosome). Placing the hotspot motifs within a phased nucleosome array allowed us to monitor their impacts on a well-defined, well-organized, chromatin structure.
Discrete protein–DNA complexes function redundantly to promote meiotic recombination
Haploid strains harboring the basal recombination control (M375) or hotspot DNA sequence motifs (M26, CCAAT, Oligo-C, 4095, 4156) within ade6 are each auxotrophic for adenine. These were crossed to a strain with a distal tester allele (M210) that is likewise an adenine auxotroph, and the resulting haploid meiotic products were scored for the frequency Ade+ (recombinant) spore colonies (Figure 1A). In each case, the hotspot crosses yielded substantially higher recombinant frequencies than the control cross (Figure 1B). For example, the presence of the M26 DNA sequence motif stimulated recombination ∼20-fold relative to the M375 control (compare lane 2 to lane 1). This hotspot ratio ranged from 15-fold (for motif 4095) to 38-fold (for the Oligo-C motif), demonstrating that each motif is proficient for promoting recombination (i.e., is active) in this chromosomal context.
Sequence-specific DNA binding proteins have been identified for three of the five motifs (Wahls and Smith 1994; Kon et al. 1997; Steiner et al. 2009, 2011). Analyses of crosses using null mutants revealed that the stimulation of meiotic recombination by each DNA sequence motif strictly required the proteins that bind to that motif (Figure 1B). Ablating either subunit of the M26-binding heterodimer Atf1-Pcr1 abolished hotspot activity of M26 (compare lanes 3 and 4 to lane 2); removal of the Php2, Php3 or Php5 subunits of the CCAAT box-binding complex abolished hotspot activity of CCAAT (compare lanes 6–8 to lane 5); and Rst2 was essential for hotspot activity of Oligo-C (compare lane 10 to lane 9). Notably, these null mutant strains were still proficient for meiotic recombination, but not hotspot activity, and yielded recombinant frequencies that were indistinguishable statistically from that of the basal recombination control in wild-type cells (M375, lane 1). Furthermore, while ablating these hotspot binding proteins abolished the stimulation of recombination conferred by their respective DNA sequence motifs, the protein deletions did not reduce recombination of the basal recombination control allele, M375 (compare lanes 14–19 to lane 13). We conclude that the basal recombination machinery (including its catalytic subunit, Rec12) is intact in the respective null mutants. Moreover, because Rec12 (Spo11) and its active site tyrosine are essential for meiotic recombination throughout the fission yeast genome (Sharif et al. 2002), including at the ade6 locus (Kan et al. 2011), the hotspot-specific effects indicate that the protein–DNA complexes function specifically to promote the activity of the basal recombination machinery at hotspots. This interpretation is consistent with the fact that the Atf1-Pcr1-M26 protein–DNA complex promotes the formation of Rec12-catalyzed, recombination-initiating DSBs in its vicinity (Steiner et al. 2002).
These findings provided independent confirmation, using a different configuration of test crosses, that multiple different DNA sequence motifs and their binding proteins (transcription factors) each position meiotic recombination at hotspots in fission yeast (Wahls and Smith 1994; Kon et al. 1997; Steiner et al. 2009, 2011). Moreover, the fact that each cis-acting regulatory module is active when placed within the identical chromosomal context allowed us to compare directly whether the different regulatory modules each function through common downstream mechanisms (next three sections).
Functionally redundant hotspot-activating DNA sequence motifs each induce the remodeling of chromatin structure
It was reported previously, based on Southern blotting, that the ade6-M26 hotspot allele undergoes chromatin remodeling in meiosis (Yamada et al. 2004; Hirota et al. 2007). We adopted a well-established MNase and PCR-based approach (Sansó et al. 2011; Infante et al. 2012; Garcia et al. 2014; Small et al. 2014) to map chromatin structure of the ade6 locus at single-nucleosome resolution. Published data on genome-wide nucleosome occupancy (Soriano et al. 2013) were used to design 15 pairs of PCR primers that generate tiling, overlapping, amplicons spanning the interval from bub1 to ade6 that contains the hotspot DNA sequence motifs (Figure 2, see Table S3 for primer sequences and coordinates). These amplicons were positioned to measure signal intensity within nucleosomes, between nucleosomes, and within a nucleosome-depleted region (NDR).
Cells were treated with formaldehyde to cross-link and stabilize chromatin, then chromatin was digested with MNase, which cleaves DNA preferentially in the linker region between nucleosomes and in NDRs (Lantermann et al. 2009, 2010). Following reversal of cross-links, mononucleosomal DNA was purified and used as a template for quantitative, real-time PCR (qPCR). For each ade6 amplicon, we determined a mononulceosomal DNA enrichment value by comparing signals from untreated (intact) and MNase-treated (mononucleosomal) samples, with internal normalization to an unrelated region of chromosome I (at smc5) whose occupancy does not change in meiosis (de Castro et al. 2011; Soriano et al. 2013). As additional internal controls, we included the shared promoter region between ade6 and bub1 and the +1 nucleosome of bub1, which is transcribed divergently from ade6 (Figure 2). Our results with this qPCR-based assay (described below) are consistent with the overall structure of chromatin and the discrete phasing of nucleosomes encompassing the bub1-ade6 interval, as defined previously using the same MNase sensitivity assay, but with DNA microarray and deep-sequencing readouts of DNA abundance (Lantermann et al. 2009, 2010; de Castro et al. 2011; Soriano et al. 2013).
In our figures, we plotted nucleosome positions relative to the start codon of ade6. For the sake of reference, the 5′ UTR of the ade6 transcript is short (46 nucleotides) (Wood et al. 2012; Thodberg et al. 2019). Except for the differences noted below, all maps conformed to the canonical chromatin structure of fission yeast genes (Lantermann et al. 2009, 2010). Nucleosomes were depleted from the shared promoter region between bub1 and ade6; each gene had a well-positioned nucleosome at its transcription start site (denoted in the figures as nucleosome +1 for ade6 and -1 for bub1); and nucleosomes were discretely phased within the ade6 coding region (Figure 3, Figure 4, and Figure S1).
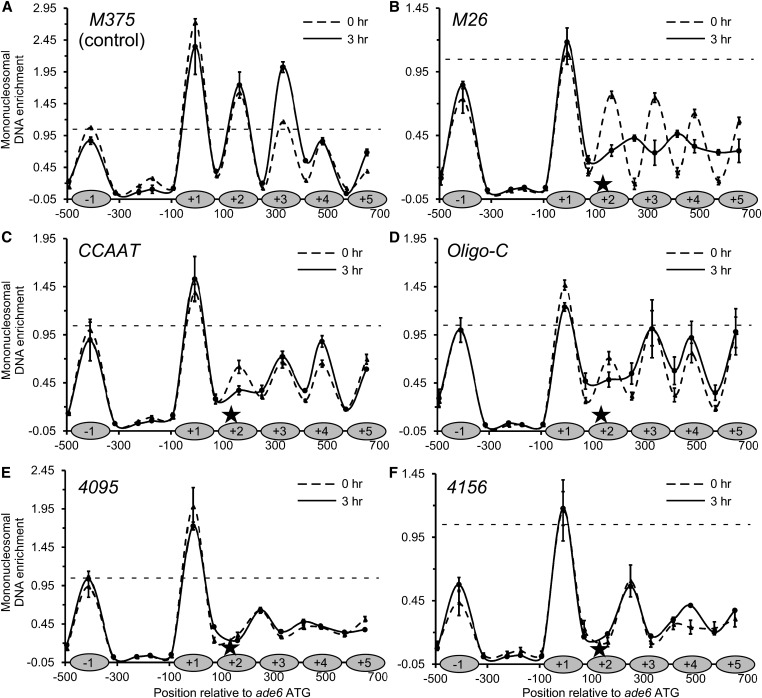
Figure 3.
Multiple hotspot-activating DNA sequence motifs each trigger the remodeling of chromatin structure. Plots show chromatin structures at ade6 before meiosis (0 hr) and during meiosis (3 hr). Data are mean ± SD from three biological replicates, with internal normalization to a nucleosome in smc5. (A) Basal recombination control allele (M375). Shaded ovals on the X-axis represent protection of DNA by nucleosomes. (B–F) Recombination hotspot alleles (M26, CCAAT, Oligo-C, 4095 and 4156). The positions of the hotspot-activating DNA sequence motifs (stars) and nucleosomes in the basal recombination control [from (A)] are shown on the X-axis of each panel for the sake of comparison. Note differences in scale for Y-axes (horizontal dashed line at Y = 1.0 is included in each panel for visual reference). Tabular data used to generate this figure are provided in Table S5; color-coded plots of superimposed data for different hotspots vs. each other and the control are provided in Figure S1.
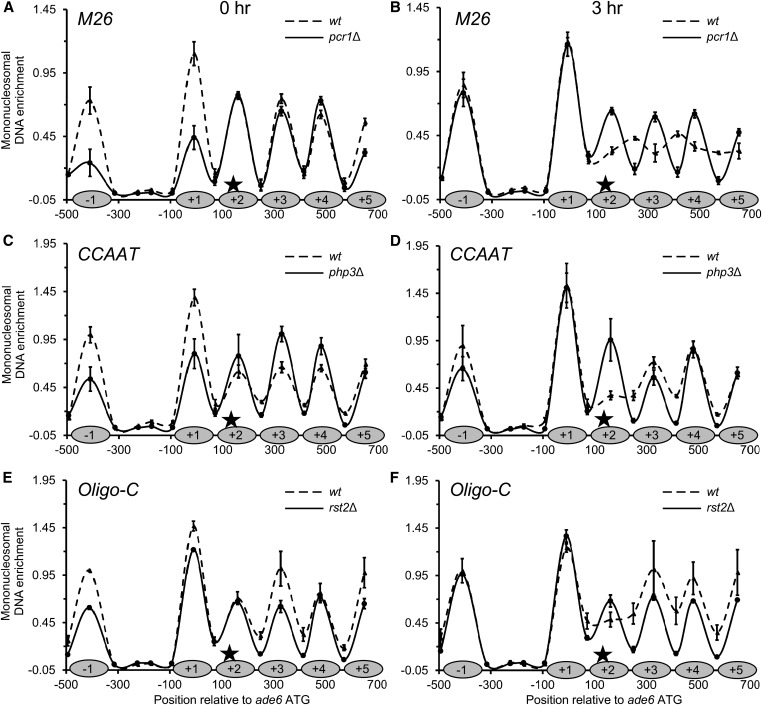
Figure 4.
DNA sequence-specific binding proteins regulate chromatin remodeling at sequence-dependent hotspots. Chromatin structures of hotspot alleles before (0 hr; A, C, and E) and during meiosis (3 hr; B, D, and F) in wild-type cells and in cells lacking the respective DNA binding proteins. Data are mean ± SD from three biological replicates, with internal normalization to a nucleosome in smc5. The positions of nucleosomes in the basal recombination control [from (A) of Figure 3] are depicted on each X-axis (shaded ovals) for the sake of comparison. Note that ablating the hotspot-activating binding proteins restores a more normal phasing of nucleosomes and that this effect is most pronounced at 3 hr of meiosis. Tabular data used to generate this figure are provided in Table S5.
We sought to define dynamic changes, if any, in chromatin structure during meiosis. We were particularly interested in changes that are present just before, and are thus a potential prerequisite for, recombination-initiating DSBs catalyzed by Rec12 (Spo11) (Cromie et al. 2007; Kan et al. 2011). To do so, we took advantage of the fact that one can induce highly synchronous meiosis in large cultures of fission yeast by thermal inactivation of the Pat1-114ts repressor of meiosis (Wahls and Smith 1994; Storey et al. 2018). We focused our comparative analyses of chromatin structure, in nine different genetic backgrounds, at time points before meiosis (0 hr) and during meiosis (3 hr), with the latter time point being just before maximal induction of DSBs.
Chromatin structure of basal control M375:
Except for a difference in the amplitude of signal for nucleosome +3 of ade6, the overall structure and phasing of nucleosomes for ade6-M375 before and during meiosis each superimposed well (Figure 3A). We conclude that meiosis does not trigger any substantial changes in the organization of chromatin at the basal recombination control. The amplitude and positioning for the +1 nucleosome of bub1 (denoted in the figures as the −1 nucleosome), the low signals within the NDR of the intergenic promoter region between bub1 and ade6, and the amplitude and positioning of nucleosomes within ade6 each provided controls for the mapping of chromatin structures of ade6 alleles that harbor hotspot DNA sequence motifs.
Chromatin structure of hotspot M26:
For this and other hotspot alleles, the position of the hotspot-activating DNA sequence motif is displayed on the X-axis (star), and the inferred positions of nucleosomes as detected in the basal recombination control, M375, are provided for comparison (shaded ovals). From cells bearing the ade6-M26 hotspot, the amplitude of signals for the −1 (bub1) nucleosome and for the intergenic NDR region, before and during meiosis, were like those of the control ade6-M375 (compare Figure 3B to Figure 3A; note difference in scales of Y-axes). We infer that the M26 hotspot motif, which maps within the +2 nucleosome of ade6, does not affect substantially chromatin structure outside of the ade6 transcription unit in which it resides.
Before meiosis, the chromatin structure of the ade6 coding region harboring the hotspot allele M26 (Figure 3B, 0 hr) was similar to that of the basal recombination control, ade6-M375 (Figure 3A). In each case, nucleosomes were well defined and discretely phased within the ade6 transcription unit. However, there were significant reductions in peak heights for nucleosomes within the ade6-M26 coding region (Figure 3B) relative to the control (Figure 3A). This can also be seen within each data set by comparing relative peak heights within ade6 to that of the −1 nucleosome of bub1, and by superimposing data sets plotted at the same scale (Figure S1). Meiosis triggered substantial further reorganization of chromatin at ade6-M26 (Figure 3B, 3 hr). DNA normally protected from MNase digestion by nucleosome +2 (where the M26 site resides) and downstream nucleosomes became even more accessible to digestion, DNA that is normally in the linker regions became more protected, and there was evidence for the repositioning of nucleosomes (new, minor peaks at +2.5 and +3.5). We conclude that the M26 hotspot motif regulates meiotically enhanced displacement of nucleosomes, which is consistent with its induction of a more open chromatin structure, as determined previously by Southern blotting (Yamada et al. 2004).
Our observation that there were some M26 hotspot-specific changes in chromatin structure before meiosis (0 hr) was made using cells that had been synchronized in G0 (G1) phase of the cell cycle, which is necessary to induce synchronous meiosis. To see if such changes were induced by the synchronization process (nitrogen starvation), we analyzed chromatin structures in cells that were growing vegetatively in rich medium (asynchronous, log-phase, mitotic cultures). In these samples, the chromatin structure of the hotspot ade6-M26 was very similar to that of the basal recombination control, ade6-M375 (Figure S1). Thus, nitrogen starvation triggers incipient chromatin remodeling at this hotspot before meiosis (0 hr), and there are substantial, meiosis-dependent changes thereafter (3 hr) (Figure 3B).
Chromatin structure of hotspot CCAAT:
For the CCAAT hotspot motif, the chromatin structures at the −1 (bub1) nucleosome and intergenic NDR region were like those of the basal control before and during meiosis (Figure 3C vs. Figure 3A). Thus, the CCAAT DNA sequence motif, like the M26 motif, does not seem to affect chromatin structure outside of the ade6 transcription unit in which it resides.
Before meiosis, the phasing of nucleosomes at ade6-CCAAT (Figure 3C, 0 hr) was similar to that of the basal recombination control (Figure 3A). However, there was substantially less protection of DNA within nucleosomes at and flanking the hotspot motif, indicating that the CCAAT motif triggers changes in chromatin structure that do not require meiotic factors. These changes surrounding the CCAAT motif in ade6 are attributable to the process of synchronizing cells in G0 (G1) by nitrogen starvation because the chromatin pattern in vegetative cells was like that of the basal recombination control, ade6-M375 (Figure S1). Meiosis triggered an additional reduction in the protection of DNA within the +2 nucleosome in which the CCAAT motif resides (Figure 3C, 3 hr). We conclude that the CCAAT motif, like the M26 motif, induces chromatin remodeling that encompasses the nucleosome in which it resides and nearby nucleosomes.
Chromatin structure of hotspot Oligo-C:
Results for this hotspot motif were similar to those for M26 and CCAAT. Before and during meiosis, there was no evidence of chromatin changes at the −1 (bub1) nucleosome and the intergenic NDR (Figure 3D). Before meiosis, there were significant reductions in DNA protection by nucleosomes in the ade6 coding region, relative to the M375 control. This property, of changes in chromatin structure preceding meiosis, is shared with the CCAAT hotspot (Figure 3C) and the M26 hotspot (Figure 3B). Meiosis triggered further, Oligo-C-dependent remodeling of chromatin structure in its vicinity (Figure 3D), which is also a property shared by each of these three motifs. We conclude that the Oligo-C DNA sequence motif, like the other recombination-promoting DNA sequence motifs, regulates changes in local chromatin structure.
Chromatin structure of hotspot 4095:
Like the hotspot-activating DNA sequence motifs described above, the 4095 motif had no detectable effect on chromatin structure at the −1 nucleosome and within the NDR, but triggered chromatin remodeling within ade6 (Figure 3E). The pattern closely resembles that of M26 in meiosis, with substantial eviction of the +2 and downstream nucleosomes, accompanied by the inferred repositioning of nucleosomes extending to the right of where the 4095 motif resides (new, minor peaks at +2.5 and +3.5). The chromatin maps from before (0 hr) and during meiosis (3 hr) were essentially identical. Analyses of chromatin in vegetative cells revealed the same, 4095-dependent changes in chromatin structure (Figure S1), suggesting that this hotspot DNA sequence motif induces remodeling constitutively.
Chromatin structure of hotspot 4156:
The timing, constellation, and magnitude of chromatin changes elicited by the 4156 motif (Figure 3F), including the remodeling of chromatin in vegetative cells (Figure S1), were like those for 4095. One difference, relative to all of the other hotspot-activating motifs and the basal recombination control, is that there was significantly reduced protection of DNA within the −1 (bub1) nucleosome (Figure 3F), although this was not observed in vegetative cells (Figure S1). This suggests that 4156-regulated changes in chromatin structure might spread more broadly during meiosis than those triggered by the four other DNA sequence motifs.
In summary, five different meiotic recombination hotspot-activating DNA sequence motifs each regulate the remodeling of chromatin structure locally (Figure 3 and Figure S1). In each case, the chromatin is extensively modified at 3 hr of meiosis, shortly before maximal induction of recombination-initiating DSBs by the catalytic subunit (Rec12/Spo11) of the basal recombination machinery (Cromie et al. 2007; Kan et al. 2011). For two of the motifs (4095, 4156), the changes are present at both meiotic time points and even in vegetative cells, indicating constitutive disruption of the canonical nucleosome array. For three of the motifs (M26, CCAAT, Oligo-C), remodeling is induced by nitrogen starvation before meiosis (0 hr) and is further enhanced by meiosis (3 hr). For one of these, M26, the most substantial changes occur during meiosis. These findings have important implications for molecular mechanisms by which meiotic recombination is positioned throughout the genome (see Discussion).
Hotspot-activating proteins regulate hotspot-specific chromatin remodeling
We next sought to determine whether the sequence-specific, hotspot binding/activating proteins regulate the sequence-dependent chromatin remodeling. We confirmed the ability of null mutant strains to undergo meiosis in a pat1-114ts genetic background, setting aside the strains in which we could not induce synchronous meiosis (atf1Δ and php5Δ). The results of chromatin mapping for the three relevant hotspot motifs (M26, CCAAT and Oligo-C), before and during meiosis, and in the presence or absence of their cognate DNA binding proteins, are shown in Figure 4.
Effects of Pcr1 protein on chromatin remodeling for hotspot M26:
In a wild-type background, M26 motif-dependent chromatin remodeling was maximal at 3 hr, with substantial lateral displacement of nucleosomes (Figure 3B and Figure 4B). Under these conditions, the removal of the M26 binding protein Pcr1 restored the phasing of nucleosome to one like that of the basal recombination control, ade6-M375 (Figure 4B and Figure 3A, respectively). Pcr1 was required for the increased sensitivity of M26 hotspot DNA to MNase within canonical nucleosome positions +2 to +5, for the increased protection of DNA between canonical nucleosome positions +1 to +5, and for the apparent repositioning of nucleosomes (new, minor peaks at +2.5 and +3.5).
Effects of Php3 protein on chromatin remodeling for hotspot CCAAT:
In a wild-type background, CCAAT motif-dependent chromatin remodeling was observed before, and was enhanced further during, meiosis (Figure 3C and Figure 4, C and D). The removal of the CCAAT binding protein Php3 restored the chromatin structure of the CCAAT hotspot to one similar to that of the basal recombination control. This is most evident during meiosis (Figure 4D, 3 hr). Php3 was also required for the displacement of the +2 nucleosome, and for the increased protection of DNA between the +2 to +5 nucleosomes at the hotspot.
Effects of Rst2 protein on chromatin remodeling for hotspot Oligo-C:
In a wild-type background, the Oligo-C motif-dependent chromatin remodeling was observed upon nitrogen starvation and was enhanced further by meiosis (Figure 3D and Figure 4, E and F). As was the case for the preceding two hotspot motifs and their binding proteins, removing the Oligo-C binding protein Rst2 restored a more normal phasing of nucleosomes, and this was most evident in meiosis (Figure 4F). Rst2 was required for the meiotically enhanced, Oligo-C-dependent displacement of the +2 nucleosome, and for the increased protection of DNA between nucleosome positions +1 to +5.
In summary, multiple different DNA sequence-specific, hotspot-activating proteins (transcription factors) each regulate hotspot-specific chromatin remodeling at their DNA binding sites. These changes involve both the nucleosome that contains the hotspot DNA sequence motif and nearby nucleosomes.
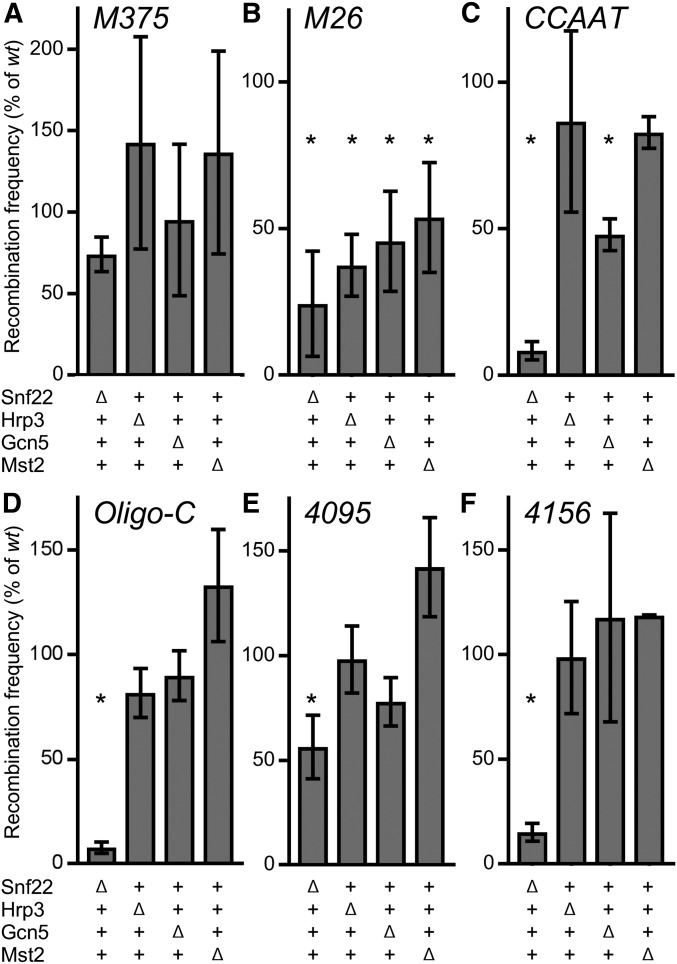
Chromatin remodeling enzyme Snf22 (Snf2/Swi2) is a major effector of sequence-dependent recombination hotspots
Our experimental system—in which different cis-acting regulatory modules are each analyzed within the identical chromosomal context—also allows one to compare directly whether, and the extent to which, various chromatin modifying enzymes regulate diverse classes of DNA sequence-dependent hotspots. There is a plethora of chromatin modifiers, and our choice of which ones to analyze combinatorially (using null mutants and hotspot DNA sequence motifs) was guided by prior findings. For this study, we focused on enzymes already known to be required for efficient chromatin remodeling and for high-frequency recombination at the ade6-M26 hotspot (Yamada et al. 2004, 2013; Hirota et al. 2007, 2008). These are the ATP-dependent DNA helicase/chromatin remodeling enzymes Snf22 (of the SWI/SNF family) and Hrp3 (CHD1 family), the SAGA complex histone acetyltransferase catalytic subunit Gcn5, and the Mst2 acetyltransferase of the Mst2/NuA4 complex. We examined the roles of these enzymes in recombination at the five different hotspots (M26, CCAAT, Oligo-C, 4095, 4156) and the basal recombination control (M375) (Figure 5).
Figure 5.
ATP-dependent chromatin remodeling enzyme Snf22 (Snf2/Swi2) is a key regulator of multiple sequence-dependent recombination hotspots. Recombinant frequencies for crosses with basal recombination control [(A) M375] and hotspot DNA sequence motifs [(B–F) M26, CCAAT, Oligo-C, 4095, and 4156, respectively] were determined as in Figure 1A. Frequencies for the indicated null mutants are expressed as percent relative to frequencies of wild-type cells from crosses conducted in parallel. Data are mean ± SD from three biological replicates; statistically significant differences at P ≤ 0.05% (*) from t-test are based on recombinant frequencies in both wild type and mutant; note differences in scales of Y-axes. Tabular data used to generate this figure are provided in Table S5.
In null mutants lacking Snf22, Hrp3, Gcn5, or Mst2, the recombinant frequencies for the basal recombination control (M375) were indistinguishable statistically (at a threshold of P ≤ 0.05) from that of wild-type cells (Figure 5A). This indicates that all essential components of the basal recombination machinery are present and functional in the null mutants.
For the M26 hotspot, recombinant frequencies were significantly lower in snf22Δ, hrp3Δ, gcn5Δ, and mst2Δ mutants than in wild-type cells (Figure 5B). Thus, each of the respective chromatin modifying enzymes contributes to high-frequency recombination at ade6-M26, with Snf22 being the most important (Snf22 > Hrp3 > Gcn5 > Mst2). Since ablating these proteins did not affect significantly basal recombination (Figure 5A), we conclude that each of these chromatin-modifying enzymes contributes specifically to activation of the M26 hotspot. These findings are consistent with those reported previously (Yamada et al. 2004, 2013; Hirota et al. 2007, 2008), providing independent confirmation of results, and serving as a good comparator for additional experiments.
In the snf22Δ mutant background, recombination rates were strongly reduced, relative to wild-type cells, for all five different hotspot-activating DNA sequence motifs (Figure 5, B–F). We conclude that diverse hotspot-regulating protein-DNA complexes share an effector mechanism that is mediated by the ATP-dependent chromatin remodeling enzyme Snf22.
Interestingly, while recombination at the M26 hotspot was attenuated significantly in the hrp3Δ and mst2Δ mutants, in these mutants there were no significant reductions in recombination for the CCAAT, Oligo-C, 4095 and 4156 hotspots (Figure 5). Thus, the Hrp3 and Mst2 proteins are required only for activation of one of the five DNA sequence-dependent hotspots, M26. Similarly, deletion of gcn5 did not affect significantly recombination at Oligo-C, 4095, and 4156, although Gcn5 contributed to high-frequency recombination at M26 and CCAAT (Figure 5).
In summary, the ATP-dependent chromatin remodeling enzyme Snf22 is a crucial mediator of hotspot activation for all five different DNA sequenced-dependent meiotic recombination hotspots analyzed; whereas Gcn5, Hrp3, and Mst2 contribute to the activation of only one or two of the five different classes of hotspots.
Discussion
Diverse cis-acting regulators employ common mechanism for hotspot activation
The original, evidence-based hypothesis that “Discrete [DNA] sites and their binding proteins could account for the observed regulation of recombination both along the chromosome and during the life cycle” (Wahls and Smith 1994) has been supported by subsequent findings in organisms ranging from fungi to mammals. The hundreds of short, distinct DNA sequence elements shown experimentally to activate meiotic recombination hotspots in fission yeast (Schuchert et al. 1991; Wahls and Smith 1994; Kon et al. 1997; Steiner et al. 2009, 2011; Foulis et al. 2018), and those implicated by association in budding yeast [our interpretation of primary data reported by Pan et al. (2011)], could account for the positioning of essentially all meiotic recombination hotspots in these two highly diverged species, and, by extension, in others (Steiner et al. 2009, 2011; Wahls and Davidson 2010, 2012). This study revealed shared molecular mechanisms by which the functionally redundant, cis-acting regulatory elements control recombination locally.
We report that multiple, distinct, recombination hotspot-activating DNA sequence motifs and their binding proteins, where identified (Table 1), each promote recombination by remodeling chromatin structure (Figure 3, Figure 4, Figure 5, and Figure S1). In each case, the hotspot-specific changes in chromatin encompass the nucleosome that contains the regulatory DNA motif, extend to nearby nucleosomes, and precede the time at which the catalytic subunit of the basal recombination machinery (Rec12/Spo11) catalyzes the formation of recombination-initiating DSBs (Sharif et al. 2002; Cromie et al. 2007; Kan et al. 2011). Our finding that chromatin remodeling at five different hotspots precedes recombination is consistent with the observation that meiotically induced chromatin remodeling at the ade6-M26 hotspot occurs normally in null mutants lacking Rec12 (Spo11) (Hirota et al. 2007), even though Rec12 and its active site tyrosine are essential for recombination throughout the fission yeast genome (Sharif et al. 2002; DeWall et al. 2005; Kan et al. 2011). It thus seems clear that neither Rec12 nor Rec12-dependent recombination intermediates contribute appreciably to the hotspot-specific chromatin remodeling that we observed. These findings support a unifying model in which transcription factor-induced changes in chromatin structure help to position the catalytic activity of the basal recombination machinery at hotspots.
Recombinationally poised epigenetic states
Our mapping of chromatin structure using the highly sensitive, qPCR-based readout revealed increased accessibility of DNA within nucleosomes at the M26 hotspot before meiosis, relative to the basal recombination control (compare Figure 3B to Figure 3A, see also Figure S1), which had escaped detection by the Southern blotting approach employed previously (Yamada et al. 2004, 2013). The finding of some chromatin remodeling before meiosis make sense given that the Atf1-Pcr1 heterodimer binds to M26 DNA sites in the genome of vegetative cells as well as cells in meiosis (Kon et al. 1998; Eshaghi et al. 2010; Woolcock et al. 2012), promotes the acetylation of histone H3 surrounding the hotspot before meiosis (Yamada et al. 2004, 2013), and in vegetative cells affects the chromatin structure of stress-responsive genes whose transcription it regulates (Garcia et al. 2014).
We also observed hotspot motif-dependent changes in chromatin structure, relative to the basal recombination control, prior to meiosis for all other DNA sequence-dependent hotspots (Figure 3 and Figure S1). We speculate that changes in such poised epigenetic states might contribute to the modulation of hotspot positioning or strength that are induced by differences in mating type (Parvanov et al. 2008), auxotrophies and nutritional states (Abdullah and Borts 2001; Cotton et al. 2009), temperature (Fan et al. 1995; Zhang et al. 2017), and prior freezing (Stahl et al. 2016). The fact that transcription factors of fission yeast and budding yeast respond to such intracellular and environmental cues, and that they are rate-limiting for hotspot recombination at their DNA binding sites (there is a protein dose-dependent response) (White et al. 1991; Kon et al. 1997), supports this idea.
Global positioning of hotspots by transcription factor-dependent chromatin remodeling
The global distribution of fission yeast DSB hotspots has been mapped (Cromie et al. 2007), and analyses of those data revealed that hotspots are directed preferentially to transcription factor binding sites (Wahls and Davidson 2010). Correspondingly, ∼80% of hotspots cluster preferentially at intergenic, promoter-containing regions (Wahls et al. 2008; Fowler et al. 2014). About 20% of DSB hotspots reside within protein-coding genes (Cromie et al. 2007) and occur preferentially in the vicinity of transcription factor binding sites therein (Wahls and Davidson 2010). In every case tested by measuring frequencies of recombination-initiating DSBs or rates of recombination in the presence or absence of candidate regulatory DNA sequences, hotspot activity requires specific DNA sites and their binding proteins [e.g., Figure 1 and (Schuchert et al. 1991; Virgin et al. 1995; Kon et al. 1997; Steiner and Smith 2005; Steiner et al. 2009, 2011; Foulis et al. 2018)]. Hence, the hotspot-specific, transcription factor-dependent chromatin remodeling mechanisms that we discovered (Figure 3, Figure 4, Figure 5 and Figure S1) are germane to previously reported, genome-wide associations between chromatin accessibility and hotspot positions (de Castro et al. 2011; Yamada et al. 2013).
We discovered that hotspot-activating DNA sequence motifs can trigger chromatin remodeling before meiosis (as well as during meiosis), and, in some cases, even do so constitutively in vegetative cells (Figure 3 and Figure S1). This striking finding can explain the observation that ∼80% of hotspots colocalize with NDRs that are already present before meiosis and the other 20% are associated with meiotically induced NDRs (de Castro et al. 2011). Moreover, our finding that specific chromatin remodeling enzymes are required for the activation of all five different classes of DNA sequence-dependent hotspots (Figure 5) reinforces the conclusion that, in each case, the pathway mechanism proceeds sequentially through transcription factor binding, chromatin remodeling, and stimulation of the basal meiotic recombination machinery.
Not simply “windows of opportunity”
Our assays of chromatin structure, like those in other studies, involve population-average measurements that infer the distribution of nucleosomes based on the accessibility of DNA to MNase. The interpretation of some changes, like the lateral displacement of nucleosomes (e.g., Figure 3F), is straightforward. Other changes, such as reduced peak height of normally phased nucleosomes (e.g., Figure 3B, 0 hr), might be due to the removal of nucleosomes in a subset of cells, or by looser wrapping of DNA around nucleosomes, or some combination of the two. For example, histone PTMs (such the acetylation of lysine residues) can affect how tightly DNA is wrapped around nucleosomes, and, consequently, sensitivity of that DNA to MNase (Zhao et al. 2014; Lorzadeh et al. 2016; Jha et al. 2017; Kurup et al. 2019). Notably, at least 23 different combinations of histone PTMs are enriched preferentially around the ade6-M26 hotspot at one or more time points of meiosis, relative to basal recombination control (Storey et al. 2018). The relevant point here is that DNA sequence-dependent hotspots trigger many changes in the underlying constituents of chromatin, and the MNase sensitivity assay reveals only their aggregate impact on the accessibility of DNA within chromatin. For these and the following reasons, we posit that the types of chromatin remodeling induced by the hotspot-activating, sequence-specific binding proteins (not simply “windows of opportunity” associated with NDRs) position recombination at hotspots.
Although most hotspots in the fission yeast genome colocalize with NDRs, the vast majority of NDRs (>90%) do not have an associated hotspot (de Castro et al. 2011). Moreover, the NDRs that are associated with hotspots typically have reduced (not fully depleted) nucleosome occupancy (although they are still referred to as NDRs in the literature) (de Castro et al. 2011). For example, based on the distribution of a core histone (H3), there is on average only about a 40% reduction of nucleosome occupancy at hotspot centers vs. average occupancy genome-wide (Yamada et al. 2013). Together, the various association studies have revealed the following, statistically significant trends: the hotspot peaks tend to localize just 5′ of transcription start sites (i.e., in promoter regions) (Wahls et al. 2008; Fowler et al. 2014); they tend to cluster around transcription factor binding sites (Wahls and Davidson 2010); and they tend to coincide with reduced (but not eliminated) nucleosome occupancy (de Castro et al. 2011; Yamada et al. 2013).
Each of these associations can also be explained by our data on underlying mechanisms. The hotspot-activating protein-DNA complexes (which in every case defined so far involve transcription factors) each trigger a reduction in the protection of DNA by nucleosomes, without necessarily abolishing the phasing of nucleosomes (Figure 3, B–D). In a subset of cases, there is also evidence for the lateral displacement of nucleosomes, at least in meiosis (Figure 3, B, E, and F). But, in all cases, it seems clear that there is only partial, incomplete hotspot-specific eviction of nucleosomes. In short, the magnitude of hotspot-specific changes that we defined at single-nucleosome resolution, as well as their spatial and temporal patterns at each hotspot analyzed, provide an underlying molecular basis for the genome-wide associations between transcription factor binding sites, changes in chromatin structure and hotspot positions. Together, the findings indicate that transcription factor-specific changes in chromatin—not simply increased accessibility of DNA within NDRs—help to position recombination at hotspots.
Transcription factors regulate recombination via chromatin modifying enzymes
The remarkably high multiplicity, short lengths, functional redundancy, and context variable penetrance of hotspot-regulating DNA sequences explains why computational analyses have low predictive power for the regulation of recombination by—and can even fail to identify—discrete DNA sequences that are demonstrably recombinogenic (Wahls and Davidson 2012). Similar considerations apply for the multitude of potential downstream effectors, such as chromatin modifying enzymes and histone codes (Storey et al. 2018). We reasoned that direct measurements of recombination, for each hotspot vs. basal recombination control analyzed under identical conditions, provides the best way to determine whether and the extent to which chromatin remodeling factors regulate diverse classes of recombination hotspots. Positive and negative results of this powerful, well-controlled bioassay are each informative, as exemplified by the following.
The histone acetyltransferases Gcn5 and Mst2, which are required for full activity of M26-class hotspots (Yamada et al. 2004, 2013; Hirota et al. 2007, 2008), had no significant impact on recombination rates at three and four other classes of DNA sequence-dependent hotspots, respectively (Figure 5). This is concordant with the finding that there is only modest association between H3K9ac and essentially no association between H3K14ac with recombination hotspots across the genome, and that mutating the H3K9 acceptor residue has only a very weak impact on the activity of hotspots overall (Yamada et al. 2013). Similarly, our finding that the ATP-dependent chromatin remodeling enzyme Hrp3 contributes only to the activation of recombination at M26, with no significant impact on recombination for CCAAT, Oligo-C, 4095 and 4156 (Figure 5), suggests that Hrp3 is a supporting player, rather than a central effector of hotspot activation.
In contrast, the ATP-dependent chromatin remodeling enzyme Snf22 was strongly required for the stimulation of recombination by all five different classes of sequence-dependent hotspots tested (Figure 5). Snf22 is known to be recruited directly to M26 hotspots, based on chromatin affinity purification of hotspot vs. basal recombination control (Storey et al. 2018), and is required for both hotspot-specific chromatin remodeling (Yamada et al. 2004) and for high-frequency recombination (Figure 5). Together, these findings suggest that transcription factor-regulated remodeling of chromatin structure by Snf22 helps to localize the catalytic activity of the basal recombination machinery at diverse classes of hotspots. Other chromatin remodeling enzymes and histone PTMs that are recruited to hotspots (Storey et al. 2018) could be similarly tested with the comparative bioassay. Those that are required for the activation of diverse classes of DNA sequence-dependent hotspots, such as Snf22 (Figure 5), will reveal the subset of chromatin remodelers and histone codes with the greatest impact on the cis-acting regulation of recombination, helping to define how they affect the distribution of recombination hotspots genome-wide. Ultimately, as a long-term goal, it would be interesting to test whether the functionally important remodeling enzymes uncovered by the comparative bioassay are recruited to, and regulate via chromatin remodeling, hotspots throughout the genome.
Conclusions
Our findings support a model in which the binding of transcription factors to their respective DNA sites helps to position most, if not all, meiotic recombination hotspots of fission yeast. The functional redundancy of multiple, sequence-specific protein–DNA complexes converges upon shared chromatin remodeling pathways that help provide the basal recombination machinery (Spo11/Rec12 complex) access to its DNA substrates within chromatin. This mechanism might be conserved in other eukaryotes, including the subset of metazoans that superimpose regulation by another sequence-specific DNA binding protein, Prdm9.
Acknowledgments
We thank Giulia Baldini, Robert Eoff, Galina Glazko, and Alan Tackett for discussions of the study. This work was supported by National Institutes of Health grant number GM081766 to W.P.W.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9745103.
Communicating editor: M. Freitag
Literature Cited
- Abdullah M. F., and Borts R. H., 2001. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 14524–14529. 10.1073/pnas.201529598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Hanlon S. E., Lieb J. D., and Copenhaver G. P., 2009. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 19: 2245–2257. 10.1101/gr.096297.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., and Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. 10.1038/nature11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C., Borde V., and Lichten M., 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5: e324 [corrigenda: PLoS Biol. 6 (2008)]. 10.1371/journal.pbio.0050324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton V. E., Hoffmann E. R., Abdullah M. F., and Borts R. H., 2009. Interaction of genetic and environmental factors in Saccharomyces cerevisiae meiosis: the devil is in the details. Methods Mol. Biol. 557: 3–20. 10.1007/978-1-59745-527-5_1 [DOI] [PubMed] [Google Scholar]
- Cromie G. A., Hyppa R. W., Cam H. P., Farah J. A., Grewal S. I. et al. , 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 3: e141 10.1371/journal.pgen.0030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E., Soriano I., Marin L., Serrano R., Quintales L. et al. , 2011. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 31: 124–137. 10.1038/emboj.2011.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWall K. M., Davidson M. K., Sharif W. D., Wiley C. A., and Wahls W. P., 2005. A DNA binding motif of meiotic recombinase Rec12 (Spo11) defined by essential glycine-202, and persistence of Rec12 protein after completion of recombination. Gene 356: 77–84. 10.1016/j.gene.2005.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzewska J., Szymanska M., and Ziolkowski P. A., 2018. Where to cross over? Defining crossover sites in plants. Front. Genet. 9: 609 10.3389/fgene.2018.00609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M., Lee J. H., Zhu L., Poon S. Y., Li J. et al. , 2010. Genomic binding profiling of the fission yeast stress-activated MAPK Sty1 and the bZIP transcriptional activator Atf1 in response to H2O2. PLoS One 5: e11620 10.1371/journal.pone.0011620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Xu F., and Petes T. D., 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15: 1679–1688. 10.1128/MCB.15.3.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., and Rhind N., 2006. Basic methods for fission yeast. Yeast 23: 173–183. 10.1002/yea.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulis S. J., Fowler K. R., and Steiner W. W., 2018. Sequence requirement of the ade6–4095 meiotic recombination hotspot in Schizosaccharomyces pombe. Genetica 146: 65–74. 10.1007/s10709-017-9997-3 [DOI] [PubMed] [Google Scholar]
- Fowler K. R., Sasaki M., Milman N., Keeney S., and Smith G. R., 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24: 1650–1664. 10.1101/gr.172122.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Davidson M. K., and Wahls W. P., 2008. Distinct regions of ATF/CREB proteins Atf1 and Pcr1 control recombination hotspot ade6–M26 and the osmotic stress response. Nucleic Acids Res. 36: 2838–2851. 10.1093/nar/gkn037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P., Paulo E., Gao J., Wahls W. P., Ayté J. et al. , 2014. Binding of the transcription factor Atf1 to promoters serves as a barrier to phase nucleosome arrays and avoid cryptic transcription. Nucleic Acids Res. 42: 10351–10359. 10.1093/nar/gku704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O. et al. , 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. 10.1073/pnas.97.21.11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69: 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., Heslot H., Leupold U., and Loprieno N., 1974. Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by King R. C. Plenum Press, New York. [Google Scholar]
- Handel M. A., and Schimenti J. C., 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11: 124–136. 10.1038/nrg2723 [DOI] [PubMed] [Google Scholar]
- Hirota K., Steiner W. W., Shibata T., and Ohta K., 2007. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot. Cell 6: 2072–2080. 10.1128/EC.00246-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Mizuno K., Shibata T., and Ohta K., 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6–M26. Mol. Biol. Cell 19: 1162–1173. 10.1091/mbc.e07-04-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., and de Boer C. G., 2013. Mapping yeast transcriptional networks. Genetics 195: 9–36. 10.1534/genetics.113.153262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante J. J., Law G. L., and Young E. T., 2012. Analysis of nucleosome positioning using a nucleosome-scanning assay. Methods Mol. Biol. 833: 63–87. 10.1007/978-1-61779-477-3_5 [DOI] [PubMed] [Google Scholar]
- Jha P. K., Khan M. I., Mishra A., Das P., and Sinha K. K., 2017. HAT2 mediates histone H4K4 acetylation and affects micrococcal nuclease sensitivity of chromatin in Leishmania donovani. PLoS One 12: e0177372 10.1371/journal.pone.0177372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan F., Davidson M. K., and Wahls W. P., 2011. Meiotic recombination protein Rec12: functional conservation, crossover homeostasis and early crossover/non-crossover decision. Nucleic Acids Res. 39: 1460–1472. 10.1093/nar/gkq993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., and Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kohli J., 1987. Genetic nomenclature and gene list of the fission yeast Schizosaccharomyces pombe. Curr. Genet. 11: 575–589. 10.1007/BF00393919 [DOI] [PubMed] [Google Scholar]
- Kon N., Krawchuk M. D., Warren B. G., Smith G. R., and Wahls W. P., 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94: 13765–13770. 10.1073/pnas.94.25.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N., Schroeder S. C., Krawchuk M. D., and Wahls W. P., 1998. Regulation of the Mts1-Mts2-dependent ade6–M26 meiotic recombination hotspot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol. Cell. Biol. 18: 7575–7583. 10.1128/MCB.18.12.7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup J. T., Campeanu I. J., and Kidder B. L., 2019. Contribution of H3K4 demethylase KDM5B to nucleosome organization in embryonic stem cells revealed by micrococcal nuclease sequencing. Epigenetics Chromatin 12: 20 10.1186/s13072-019-0266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann A., Stralfors A., Fagerstrom-Billai F., Korber P., and Ekwall K., 2009. Genome-wide mapping of nucleosome positions in Schizosaccharomyces pombe. Methods 48: 218–225. 10.1016/j.ymeth.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Lantermann A. B., Straub T., Stralfors A., Yuan G. C., Ekwall K. et al. , 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17: 251–257. 10.1038/nsmb.1741 [DOI] [PubMed] [Google Scholar]
- Lorzadeh A., Bilenky M., Hammond C., Knapp D., Li L. et al. , 2016. Nucleosome density ChIP-seq identifies distinct chromatin modification signatures associated with MNase accessibility. Cell Rep. 17: 2112–2124. 10.1016/j.celrep.2016.10.055 [DOI] [PubMed] [Google Scholar]
- Mieczkowski P. A., Dominska M., Buck M. J., Gerton J. L., Lieb J. D. et al. , 2006. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1014–1027. 10.1128/MCB.26.3.1014-1027.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Ito M., Kugou K., Yamada S., Furuichi M. et al. , 2012. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47: 722–733. 10.1016/j.molcel.2012.06.023 [DOI] [PubMed] [Google Scholar]
- Mougel F., Poursat M. A., Beaume N., Vautrin D., and Solignac M., 2014. High-resolution linkage map for two honeybee chromosomes: the hotspot quest. Mol. Genet. Genomics 289: 11–24. 10.1007/s00438-013-0784-2 [DOI] [PubMed] [Google Scholar]
- Myers S., Freeman C., Auton A., Donnelly P., and McVean G., 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40: 1124–1129. 10.1038/ng.213 [DOI] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G. et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. 10.1016/j.cell.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E., Kohli J., and Ludin K., 2008. The mating-type-related bias of gene conversion in Schizosaccharomyces pombe. Genetics 180: 1859–1868. 10.1534/genetics.108.093005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansó M., Vargas-Perez I., Quintales L., Antequera F., Ayte J. et al. , 2011. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 39: 6369–6379. 10.1093/nar/gkr255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., and Livak K. J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schuchert P., Langsford M., Kaslin E., and Kohli J., 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10: 2157–2163. 10.1002/j.1460-2075.1991.tb07750.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif W. D., Glick G. G., Davidson M. K., and Wahls W. P., 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1: 1 10.1186/1475-9268-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M., 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1: REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. C., Xi L., Wang J. P., Widom J., and Licht J. D., 2014. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc. Natl. Acad. Sci. USA 111: E2462–E2471. 10.1073/pnas.1400517111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski C. S., and Noor M. A., 2011. Recombination rate variation in closely related species. Heredity 107: 496–508. 10.1038/hdy.2011.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano I., Quintales L., and Antequera F., 2013. Clustered regulatory elements at nucleosome-depleted regions punctuate a constant nucleosomal landscape in Schizosaccharomyces pombe. BMC Genomics 14: 813 10.1186/1471-2164-14-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., Rehan M. B., Foss H. M., and Borts R. H., 2016. Apparent epigenetic meiotic double-strand-break disparity in Saccharomyces cerevisiae: a meta-analysis. Genetics 204: 129–137. 10.1534/genetics.116.191635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., and Smith G. R., 2005. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol. Cell. Biol. 25: 9054–9062. 10.1128/MCB.25.20.9054-9062.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., and Steiner E. M., 2012. Fission yeast hotspot sequence motifs are also active in budding yeast. PLoS One 7: e53090 10.1371/journal.pone.0053090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., Schreckhise R. W., and Smith G. R., 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9: 847–855. 10.1016/S1097-2765(02)00489-6 [DOI] [PubMed] [Google Scholar]
- Steiner W. W., Steiner E. M., Girvin A. R., and Plewik L. E., 2009. Novel nucleotide sequence motifs that produce hotspots of meiotic recombination in Schizosaccharomyces pombe. Genetics 182: 459–469. 10.1534/genetics.109.101253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., Davidow P. A., and Bagshaw A. T., 2011. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics 187: 385–396. 10.1534/genetics.110.124636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey A. J., Wang H. P., Protacio R. U., Davidson M. K., Tackett A. J. et al. , 2018. Chromatin-mediated regulators of meiotic recombination revealed by proteomics of a recombination hotspot. Epigenetics Chromatin 11: 64 10.1186/s13072-018-0233-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székvölgyi L., and Nicolas A., 2009. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 277: 571–589. 10.1111/j.1742-4658.2009.07502.x [DOI] [PubMed] [Google Scholar]
- Thodberg M., Thieffry A., Bornholdt J., Boyd M., Holmberg C. et al. , 2019. Comprehensive profiling of the fission yeast transcription start site activity during stress and media response. Nucleic Acids Res. 47: 1671–1691. 10.1093/nar/gky1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock A. J., and Henderson I. R., 2018. Hotspots for initiation of meiotic recombination. Front. Genet. 9: 521 10.3389/fgene.2018.00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin J. B., Metzger J., and Smith G. R., 1995. Active and inactive transplacement of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 141: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., and Davidson M. K., 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 26: 202–208. 10.1016/j.tig.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., and Davidson M. K., 2012. New paradigms for conserved, multifactorial, cis-acting regulation of meiotic recombination. Nucleic Acids Res. 40: 9983–9989. 10.1093/nar/gks761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., and Smith G. R., 1994. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 8: 1693–1702. 10.1101/gad.8.14.1693 [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Siegel E. R., and Davidson M. K., 2008. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS One 3: e2887 10.1371/journal.pone.0002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Wierdl M., Detloff P., and Petes T. D., 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl. Acad. Sci. USA 88: 9755–9759. 10.1073/pnas.88.21.9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Dominska M., and Petes T. D., 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 6621–6625. 10.1073/pnas.90.14.6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V., Harris M. A., McDowall M. D., Rutherford K., Vaughan B. W. et al. , 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40: D695–D699. 10.1093/nar/gkr853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock K. J., Stunnenberg R., Gaidatzis D., Hotz H. R., Emmerth S. et al. , 2012. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 26: 683–692. 10.1101/gad.186866.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Ohta K., and Yamada T., 2013. Acetylated histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 41: 3504–3517. 10.1093/nar/gkt049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Kugou K., Ding D. Q., Fujita Y., Hiraoka Y. et al. , 2018. The conserved histone variant H2A.Z illuminates meiotic recombination initiation. Curr. Genet. 64: 1015–1019. 10.1007/s00294-018-0825-9 [DOI] [PubMed] [Google Scholar]
- Yamada T., Mizuno K., Hirota K., Kon N., Wahls W. P. et al. , 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23: 1792–1803. 10.1038/sj.emboj.7600138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn-Zabal M., Lehmann E., and Kohli J., 1995. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics 140: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wu X. C., Zheng D. Q., and Petes T. D., 2017. Effects of temperature on the meiotic recombination landscape of the yeast Saccharomyces cerevisiae. MBio 8: pii: e02099-17. 10.1128/mBio.02099-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wang P., Yan S., Gao F., Li H. et al. , 2014. Promoter-associated histone acetylation is involved in the osmotic stress-induced transcriptional regulation of the maize ZmDREB2A gene. Physiol. Plant. 151: 459–467. 10.1111/ppl.12136 [DOI] [PubMed] [Google Scholar]
- Zhu X., and Keeney S., 2015. High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201: 525–542. 10.1534/genetics.115.178293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Yeast strains and other materials generated by this study are available upon request. All data supporting the conclusions of this study are available within the paper and its supplemental material file. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9745103.