The small ovary (sov) locus was identified in a female sterile screen, yet its molecular identity and function remained a mystery for decades. In the present work, Benner et al. molecularly map...
Keywords: sov zinc-finger, heterochromatin, gene expression, HP1a, oogenesis, position-effect variegation
Abstract
Heterochromatin-mediated repression is essential for controlling the expression of transposons and for coordinated cell type-specific gene regulation. The small ovary (sov) locus was identified in a screen for female-sterile mutations in Drosophila melanogaster, and mutants show dramatic ovarian morphogenesis defects. We show that the null sov phenotype is lethal and map the locus to the uncharacterized gene CG14438, which encodes a nuclear zinc-finger protein that colocalizes with the essential Heterochromatin Protein 1 (HP1a). We demonstrate Sov functions to repress inappropriate gene expression in the ovary, silence transposons, and suppress position-effect variegation in the eye, suggesting a central role in heterochromatin stabilization.
While gene activation by specific transcription factors is important during development, coordinated repression is also an essential process (Beisel and Paro 2011). Gene silencing represents an effective method to ensure that genes and transposons are not inappropriately activated. Gene repression can be carried out on a regional basis to inactivate large blocks of the genome by the formation of heterochromatin (Elgin and Reuter 2013). Heterochromatization relies on the dense, higher-order packing of nucleosomes, which compete for DNA binding with transcription factors (Jenuwein and Allis 2001; Kouzarides 2007; Lorch and Kornberg 2017). Once heterochromatin forms, it is maintained by a set of largely conserved proteins.
In Drosophila, heterochromatin formation occurs early during embryogenesis, where it is targeted to large blocks of repetitive DNA sequences (Elgin and Reuter 2013), including both mobile transposons and immobile mutated derivatives (Vermaak and Malik 2009). Active suppression through condensation into heterochromatin prevents the mobilization of transposons. Nevertheless, some regulated transcription of heterochromatic sequences is required for normal cellular functions. For example, the telomeres of Drosophila are maintained by the transcription and transposition of mobile elements from heterochromatic sites (Mason et al. 2008), and actively expressed histone and ribosomal RNA genes are located within heterochromatic regions (Yasuhara and Wakimoto 2006). Thus, heterochromatin serves many distinct functions and must be tightly regulated to coordinate gene expression.
The spreading of heterochromatin results in an interesting phenotype in Drosophila, whereby genes near a heterochromatin–euchromatin boundary may become repressed (Elgin and Reuter 2013). For example, when white+ (w+) transgenes are located at such a boundary, the eye can have a mottled appearance where some ommatidia express pigment while others do not. This phenomenon is known as position-effect variegation (PEV). Mutations that suppress PEV {i.e., suppressors of variegation [Su(var)]} identify genes that promote heterochromatin formation. Indeed, many of the genes required for heterochromatin function were identified in PEV modifier screens (Reuter et al. 1986; Eissenberg et al. 1990; Brower-Toland et al. 2009). Reducing heterochromatization by Su(var) mutations results in derepression of gene expression at the edges of heterochromatin blocks, suggesting that the boundaries between repressed and active chromatin expand and contract (Reuter and Spierer 1992; Weiler and Wakimoto 1995). For example, the highly conserved Heterochromatin Protein 1a (HP1a), encoded by Su(var)205, is critical for heterochromatin formation and function (Ebert et al. 2006); mutants show strong suppression of variegation (James and Elgin 1986; Eissenberg et al. 1990; Clark and Elgin 1992). Similarly, HP1a is also required to repress the expression of transposons (Vermaak and Malik 2009). HP1a may also be required for preventing the misexpression of large cohorts of genes, such as those normally expressed in specialized cell types (Greil et al. 2003; Figueiredo et al. 2012). Indicative of its central role in heterochromatization, HP1a associates with newly formed heterochromatin in the Drosophila embryo and is thought to be found in all cells throughout the life of the animal (James et al. 1989).
Both the formation and relaxation of heterochromatin depend primarily on histone modifications. Heterochromatin is thought to be nucleated by HP1a binding to di- and trimethylated Histone 3 Lys9 (H3K9me) and spread by multimerization of HP1a over blocks of chromatin (Nakayama et al. 2001; Canzio et al. 2011). In Drosophila, the methyltransferases Eggless (Egg)/SETDB1 and Su(var)3–9 catalyze the addition of methyl groups at H3K9 (Clough et al. 2007; Yoon et al. 2008), but there may be additional ways to bind HP1a to chromatin. Work in Drosophila shows that destruction of the H3K9me site recognized by HP1a (via a K9 to R9 mutation) reduces, but does not abolish, HP1a binding and heterochromatin formation (Penke et al. 2016). Moreover, HP1a binding does not always occur in regions with high H3K9me or methyltransferase activity (Figueiredo et al. 2012). These findings raise the possibility that HP1a may bind to chromatin independently of H3K9me.
Originally described over 40 years ago (Mohler 1977), the small ovary (sov) locus is associated with a range of mutant phenotypes including disorganized ovarioles, egg chamber fusions, undifferentiated tumors, and ovarian degeneration (Wayne et al. 1995). In the present work, we define the molecular identity and function of Sov. We demonstrate that sov encodes an unusually long C2H2 zinc-finger (ZnF) nuclear protein. Interestingly, Sov is in a complex with HP1a (Alekseyenko et al. 2014), suggesting that it could be important for heterochromatin function. Indeed, we find that loss-of-function sov mutations result in strong dominant suppression of PEV in the eye, similar to HP1a (Elgin and Reuter 2013). Moreover, we show that Sov and HP1a colocalize in the interphase nucleus in blastoderm embryos. Additionally, our RNA-sequencing (RNA-Seq) analysis indicates that sov activity is required in the ovary to repress the expression of large gene batteries that are normally expressed in nonovarian tissues. We further demonstrate that Sov represses transposons, including those that are required for telomere maintenance. Collectively, these data indicate that Sov is a novel repressor of gene expression involved in many, if not all, of the major functions of HP1a in heterochromatization.
Materials and Methods
We have adopted the FlyBase-recommended resources table, which includes all genetic, biological, cell biology, genomics, manufactured reagents, and algorithmic resources used in this study (Supplemental Material, Table S1).
Flies and genetics
The sov locus was defined by three X-linked, female-sterile mutations including sov2, which was mapped to ∼19 cM (Mohler 1977; Mohler and Carroll 1984) and refined to cytological region 6BD (Wayne et al. 1995). We used existing and four custom-made deletions (Df(1)BSC276, BSC285, BSC286, and BSC297) to map sov to the four-gene CG14438–shf interval (Figure 1). sov mutations complemented shf2, but not P{SUPor-P}CG14438KG00226 or P{GawB}NP6070, suggesting that CG14438 or CR43496 was sov, which we confirmed by generating Df(1)sov (X:6756569..6756668;6770708) from FLP recombination between P{XP}CG14438d07849 and PBac{RB}e03842 (Parks et al. 2004; Cook et al. 2012) to remove only those two genes.
Figure 1.
sov is CG14438. (A) Genes of the genomic interval X:6710000–6810000 (Gramates et al. 2017). (B) Deficiency (Df) and Duplication (Dp) mapping. Noncomplementing (sov–, red) and complementing (sov+, green) alleles, and rearrangements, are shown. (C) Schematic of the CG14438 (black) and CR43496 (purple) genes. Transcription start sites (bent arrows), introns (thin lines), noncoding regions (medium lines), and coding regions (thick lines) are shown. Transposon insertions (red triangles), GFP tag insertion in rescuing transgene (green triangle), point mutations (red lines), the region targeted by the short hairpin RNA interference transgene (base-paired), and Sov protein features are shown. ChrX, X chromosome; NLS, nuclear localization sequence.
We used the dominant female-sterile technique for germline clones (Chou and Perrimon 1996). Because ovoD1 egg chambers do not support development to vitellogenic stages of oogenesis, we scored for the presence of vitellogenic eggs in sov mutant germline clones. Test chromosomes that were free of linked lethal mutations by male viability (sov2) or rescue by Dp(1;3)DC486 [sovEA42 and Df(1)sov] were recombined with P{ry+t7.2 = neoFRT}19A, and verified by complementation tests and PCR. We confirmed P{ry+t7.2 = neoFRT}19A functionality by crossing to P{w+mC = GMR-hid}SS1, y1 w* P{ry+t7.2 = neoFRT}19A; P{w+m*=GAL4-ey.H}SS5, P{w+mC = UAS-FLP.D}JD2 and scoring for large eye size. We crossed females with FRT chromosomes to P{w+mC = ovoD1-18}P4.1, P{ry+t7.2 = hsFLP}12, y1 w1118 sn3 P{ry+t7.2 = neoFRT}19A/Y males for 24 hr of egg laying at 25°. We heat shocked for 1 hr at 37° on days 2 and 3. We dissected females (5 days posteclosion) to score for ovoD1 or vitellogenic morphology.
We generated a PBac{GFP-sov} transgene sufficient to rescue the sov mutant phenotype. The encoded N-terminal fusion protein uses the Sov initiation codon, followed immediately by a multitag sequence [3xFLAG, Tobacco Etch Virus (TEV) protease site, streptavidin tag II (StrepII), superfolder GFP (sfGFP), fluorescein arsenical helix binder (FLAsH) tetracysteine tag, and flexible 4xGlyGlySer (GGS) linker] and the full Sov coding sequence. The transgene contains 3040 nt of regulatory sequences upstream of the distal transcriptional start site of sov and 2692 nt downstream of the 3′ untranslated region. See the supplement for FASTA files of the genomic and complementary DNA GFP-sov sequences. The GFP-sov construct derives from P[acman] BAC clone CH322-191E24 (X:6753282–6773405) (Venken et al. 2009) grown in the SW102 strain (Warming et al. 2005). In step one of the construction, we integrated the positive/negative marker CP6-RpsL/Kan (CP6 promoter with a bicistronic cassette encoding the RpsL followed by the Kan), PCR-amplified with primers N-CG14438-CP6-RN-F and -R between the first two codons of sov, and selected (15 μg/ml Kanamycin). We integrated at the galK operon in DH10B bacteria using mini-λ-mediated recombineering (Court et al. 2003). We amplified DH10B::CP6-RpsL/Kan DNA using primers N-CG14438-CP6-RN-F and -R. Correct events were identified by PCR, as well as resistance (15 µg/ml kanamycin) and sensitivity (250 µg/ml streptomycin). In step two, we replaced the selection markers with a multitag sequence (Venken et al. 2011), tailored for N-terminal tagging (N-tag) and counterselected (250 µg/ml streptomycin). The N-tag (3xFLAG, TEV protease site, StrepII, superfolder GFP, FLAsH tetracysteine tag, and flexible 4xGlyGlySer (GGS) linker) was Drosophila codon optimized in an R6Kγ plasmid. We transformed the plasmid into EPI300 for copy number amplification. We confirmed correct events by PCR and Sanger DNA sequencing. Tagged P[acman] BAC clone DNA was injected into y1 M{vas-int.Dm}ZH-2A w*; PBac{y+-attP-3B}VK00033 embryos, resulting in w1118; PBac{y+mDint2 w+mC = GFP-sov}VK00033. All strains were maintained using standard laboratory conditions.
Western blotting
We dissected ovaries from 10 well-fed 0–2-day-old females into PBS and lysed them in 300 μl 0.1% PBST (PBS + 0.1% Tween 20), 80 μl of 5 × SDS loading dye was added to the total protein extract, and the samples were boiled at 95° for 10 min. Next, 30 μl of extract per sample was loaded into a commercial 7.5% polyacrylamide gel and run until the large bands of a high-molecular weight protein standard ladder were fully separated. Gels were blotted onto a 0.2-μm nitrocellulose membrane by the wet transfer method, using Tris/glycine transfer buffer containing 0.02% SDS and 10% MeOH under 150 mA for 3 hr. Primary antibodies used were: rabbit anti-GFP (1:2000, A11122; Fisher Scientific, Pittsburgh, PA), mouse anti-FLAG (1:5000, F1804; Sigma [Sigma Chemical], St. Louis, MO), and mouse anti-β-Tubulin [1:1000, Developmental Studies Hybridoma Bank (DSHB) E7]. Secondary antibodies used were: HRP-conjugated goat anti-mouse (1:5000, 31430; Fisher Scientific) and HRP-conjugated goat anti-rabbit (1:5000, 31460; Fisher Scientific).
Microscopy
We fixed ovaries in 4 or 5% EM-grade paraformaldehyde in PBS containing 0.1 or 0.3% Triton X-100 (PBTX) for 10–15 min, washed 3 × 15 min in PBTX, and blocked for > 30 min in 2% normal goat serum, and 0.5–1% bovine serum albumin in PBS with 0.1% Tween 20 or 0.1% Triton X-100. Antibodies and DAPI were diluted into blocking buffer. We incubated in primary antibodies overnight at 4° and secondaries for 2–3 hr at room temperature. The following primary antibodies were used: rabbit anti-Vasa (1:10,000, gift from Ruth Lehmann), rat anti-Vasa (1:10, DSHB), mouse anti-αSpectrin (αSpec; 1:200, DSHB), mouse anti-Orb (1:10, DSHB orb 4H8), guinea pig anti-Traffic jam (Tj) (1:1500, gift from Mark Van Doren, (Jemc et al. 2012), mouse anti-GFP (1:200, DSHB DSHB-GFP-4C9), and rabbit anti-GFP (1:700–1:1000, A11122; Fisher Scientific). Secondary antibodies and stains used were: Alexa-fluor 488, 568, or 647 goat anti-mouse, goat anti-rat, goat anti-guinea pig, and goat anti-rabbit (1:500; Molecular Probes, Eugene, OR), DAPI (1:1000, D1306; Invitrogen, Carlsbad, CA), phalloidin (1:500, A12380; Invitrogen), and Alexa-fluor 633-conjugated wheat germ agglutinin (WGA) (1:1000, W21404; Fisher Scientific).
Embryos (1–2 hr) were prepared for live imaging in halocarbon oil (Lerit et al. 2015). We imaged ovaries and embryos using a Nikon (Garden City, NY) Ti-E system, or Zeiss ([Carl Zeiss], Thornwood, NY) LSM 780 microscope and eyes with a Nikon SMZ.
Image analysis
In this study, tumors were defined as cysts enveloped by follicle cells containing at least six spectrosome dots, as indicated by αSpec immunostaining. Egg chamber fusion events were defined as a single cyst enveloped by a monolayer of follicle cells containing > 15 nurse cells and > 1 oocyte, but no spectrosome dots. Germaria were counted from randomly sampled dispersed ovarioles isolated from ≥ 10 individual females and replicated in at least two independent experiments; representative data from a single experiment are reported in the text.
Images were assembled using ImageJ (National Institutes of Health) and Photoshop (Adobe) software to crop regions of interest, adjust brightness and contrast, separate or merge channels, and generate maximum-intensity projections as noted.
For image intensity quantification of nuclear staining in embryos, line scans were converted to a –3- to 3-μm scale such that the 0.0 position marked the peak HP1a fluorescence intensity. The fluorescence intensities of red fluorescent protein (RFP)-HP1a and GFP-Sov were individually normalized to set their respective maxima to 100.
DNA-Seq
Genomic DNA was extracted from 30 whole flies per genotype (Huang et al. 2009); (Sambrook and Russell 2006) to prepare DNA-sequencing (DNA-Seq) libraries (Nextera DNA Library Preparation Kit). We used 50-bp, single-end sequencing (Illumina HiSeq 2500, CASAVA base calling). Sequence data are available at the Sequence Read Archive (SRA) (SRP14438). We mapped DNA-Seq reads to the FlyBase r6.16 genome with Hisat2 (–k 1 –no-spliced-alignment)(Kim et al. 2015). We used mpileup and bcftools commands from SAMtools within the genomic region X:6756000–6771000 (Li et al. 2009; Li 2011) for variant calling and SnpEff to determine the nature of variants in sov mutants (Cingolani et al. 2012).
RNA-Seq
Stranded PolyA+ RNA-Seq libraries from sov (maternal allele listed first: sovEA42/sov2, sovML150/sov2, Df(1)sov/sov2, c587-GAL4 > sovRNAi, tj-GAL4 > sovRNAi, da-GAL4 > sovRNAi, nos-GAL4 > sovRNAi) and control ovaries (maternal allele listed first: sovEA42/w1118, sovML150/w1118, Df(1)sov/w1118, sov2/w1118, w1118/sov2, w1118/w1118, c587-GAL4 > mCherryRNAi, tj-GAL4 > mCherryRNAi, da-GAL4 > mCherryRNAi, nos-GAL4 > mCherryRNAi) were created using a standard laboratory protocol (Lee et al. 2016b) and are available at the Gene Expression Omnibus (GEO) (GSE113977). We collected only ovaries containing distinguishable germaria and/or egg chambers for these experiments. We extracted total RNA (RNeasy Mini Kit, QIAGEN, Valencia, CA) in biological triplicates from 15 ovaries (4–5 days posteclosion) and used 200 ng with 10 pg of External RNA Controls Consortium (ERCC) spike-in control RNAs (pools 78A or 78B) for libraries (Jiang et al. 2011; Zook et al. 2012; Lee et al. 2016a; Pine et al. 2016). We used 50-bp, single-end sequencing as above. Tissue expression analyses are from GEO accession GSE99574 (Yang et al. 2018), a resource for comparing gene expression patterns.
We mapped RNA-Seq reads to FlyBase r6.21 with Hisat2 (–k 1 –rna-strandness R –dta) (Kim et al. 2015). We determined read counts for each attribute of the FlyBase r6.21 GTF file (with ERCC and transposable element sequences) with HTSeq-count (Anders et al. 2015). Transposon sequences were from the University of California Santa Cruz (UCSC) Genome Browser RepeatMasker track (Smit et al. 2013–2015; Casper et al. 2018).
We conducted differential expression analysis with DESeq2 (pAdjustMethod = “fdr”) (Love et al. 2014). We removed genes with read counts less than one and read counts for transposable elements with more than one location were summed for the DESeq2 analysis. Df(1)sov/sov2 replicate 3 and sov2/w1118 replicate 1 failed. For sov mutant vs. control DESeq2 analysis, all sov mutants were compared to all wild-type controls. For tissue types, we compared each sexed tissue to each sexed whole organism. We used reads per kilobase per million reads for gene-level expression. FlyBase identifiers of genes found to be expressed according to either “high” or “very high” levels in 0–2-hr-old embryos according to Graveley et al. (2011) were batch downloaded from flybase.org RNA-Seq profiles (Table S3). Raw read FASTA files of RNA-Seq data for HP1a, SETDB1, and WDE germline knockdown (Smolko et al. 2018; GSE109850) were downloaded from the SRA (SRP131778) and analyzed according to the above parameters.
For heatmaps, we calculated Euclidean distance and performed hierarchical cluster analysis (agglomeration method = Ward) on the respective columns. The rows for gene and tissue heatmaps were k-means clustered (k = 5 for gene and k = 7 for tissue heatmaps), and values were mean-subtracted scaled across genotypes. The rows for the transposon heatmap were ordered according to element and values were mean-subtracted scaled across genotypes.
For read density tracks, replicate raw read files were combined. Bedgraph files were created with bedtools genomecov (Quinlan and Hall 2010) visualized on the UCSC genome browser (Kent et al. 2002). Tracks were scaled by the number of reads divided by total reads per million.
We used PANTHER to perform gene ontology (GO) term enrichment analysis (Ashburner et al. 2000; Mi et al. 2017; The Gene Ontology Consortium 2017). We report significantly enriched GO terms as above but with the PANTHER GO-Slim Biological Process annotation data set (Table 3).
Table 3. GO term enrichment analysis of derepressed genes in sov mutants.
| Enriched GO terms | Fold enrichment | P-value |
|---|---|---|
| Neuromuscular synaptic transmission (GO:0007274) | 4.29 | 1.61E-02 |
| Synaptic transmission (GO:0007268) | 3.42 | 1.83E-12 |
| Neuron–neuron synaptic transmission (GO:0007270) | 3.24 | 3.17E-03 |
| Cyclic nucleotide metabolic process (GO:0009187) | 3.16 | 6.46E-03 |
| Cell–cell signaling (GO:0007267) | 3.16 | 1.48E-12 |
| System process (GO:0003008) | 2.27 | 2.35E-13 |
| Single-multicellular organism process (GO:0044707) | 2.15 | 2.23E-14 |
| Multicellular organismal process (GO:0032501) | 2.1 | 8.09E-14 |
| Neurological system process (GO:0050877) | 2.08 | 5.49E-09 |
| Cell surface receptor signaling pathway (GO:0007166) | 1.82 | 6.25E-03 |
| Cell communication (GO:0007154) | 1.71 | 6.31E-08 |
| Signal transduction (GO:0007165) | 1.64 | 4.65E-05 |
Enriched GO terms in sov mutants. Terms in bold are significantly enriched GO terms from genes within group 1 cluster analysis (Figure 5B). GO, gene ontology.
Statistical analysis
Data were plotted and statistical analysis was performed (Microsoft Excel and GraphPad Prism). Data were subjected to the D’Agnostino and Pearson normality test, followed by a Student’s two-tailed t-test or a Mann–Whitney U-test. We used Fisher’s exact test and Bonferroni correction on the GO biological process complete annotation data set.
Data availability
Strains and plasmids, and their sources, are detailed in Table S1. All sequencing data are available at public repositories: DNA-Seq data are available at the SRA (SRP14438) and RNA-Seq data are available at the GEO (GSE113977 and GSE99574). Supplementary tables are available at Figshare: Table S1, FlyBase ART file contains all experimental materials used in the study; Table S2, Differentially expressed genes in sov mutants vs. controls; Table S3, gene list of high or very high levels in 0–2-hr-old embryos according to Graveley et al. (2011); and Table S4, common and differentially expressed genes following germline knockdown of sov vs. HP1a, SETDB1, and WDE. A supplementary text file includes FASTA sequences for the GFP-sov transgene. Supplemental material available at Figshare: https://figshare.com/articles/Genetics_09_13_2019/9828002.
Results
The essential gene small ovary corresponds to CG14438
To refine the previous mapping of sov (Mohler 1977; Mohler and Carroll 1984; Wayne et al. 1995), we complementation tested sov mutations with preexisting and custom-generated deficiencies, duplications, and transposon insertions (Figure 1, A and B). Both female-sterile and lethal sov alleles mapped to the two-gene deletion Df(1)sov, showing that sov corresponds to either the protein-coding CG14438 gene or the intronic, noncoding CR43496 gene. Rescue of female sterility and/or lethality of sov2, sovEA42, and Df(1)sov with Dp(1;3)sn13a1, Dp(1;3)DC486, and PBac{GFP-sov} (GFP-sov) confirmed our mapping and did not replicate previous duplication-rescue results, suggesting that sovEA42 disrupts adjacent female-sterile and lethal loci (Table 1) (Wayne et al. 1995). We conclude that female sterility and lethality map to the same location.
Table 1. Complementation of sov alleles.
| sovNP6070 | sovKG00226 | sovML150 | sovEA42 | sov2 | Df(1)sov | |
|---|---|---|---|---|---|---|
| sovNP6070 | Fs | Fertile | Fs | Fs | Fertile | Fs |
| sovKG00226 | Fertile | Fertile | Fs | Fs | Fertile | Fs |
| sovML150 | Lethal | Lethal | Lethal | Lethal | Lethal | Lethal |
| sovEA42 | Lethal | Lethal | Lethal | Lethal | Lethal | Lethal |
| sov2 | Fertile | Fertile | Fs | Fs | Fs | Fs |
| Df(1)sov | Lethal | Lethal | Lethal | Lethal | Lethal | Lethal |
| GFP-sov+ | N.D. | N.D. | N.D. | Rescue | Rescue | Rescue |
| Dp(1;3)DC486 | N.D. | N.D. | N.D. | Rescue | Rescue | Rescue |
| Dp(1;3)sn13A1 | N.D. | N.D. | Rescue | Rescue | Rescue | Rescue |
Maternally contributed chromosome listed on top. Fs denotes female sterile. Fertile denotes female fertility. N.D. denotes no data.
To further map sov, we sequenced CG14438 and CR43496 to determine if DNA lesions in these genes exist on the chromosomes carrying sov mutations. While CR43496 contained three polymorphisms relative to the genomic reference strain, none were specific to sov mutant chromosomes. In contrast, we found disruptive mutations in CG14438 (Figure 1C and Table 2), which encodes an unusually long, 3313-residue protein with 21 C2H2 ZnFs, multiple nuclear localization sequence (NLS) motifs, and coiled-coil regions (Figure 1C). The lethal allele sovEA42 has a nonsense mutation (G to A at position 6,764,462) in the CG14438 open reading frame that is predicted to truncate the Sov protein before the ZnF domains. The lethal sovML150 allele has a missense mutation that results in a glutamine to glutamate substitution within a predicted coiled-coil domain (C to G at position 6,763,888). While this is a conservative substitution and glutamate residues are common in coiled-coil domains, glutamine to glutamate substitutions can be disruptive as seen in the coiled-coil region of the σ transcription factor (Hsieh et al. 1994). We found a frameshift insertion (T at position 6,769,742) located toward the end of CG14438 in the female-sterile allele sov2 that encodes 30 novel residues followed by a stop codon within the C-terminal ZnF and removes a predicted terminal NLS. Two P-element insertions in the promoter region are weaker female-sterile alleles. Although the locus was named for the female sterility phenotypes of partial loss-of-function alleles, such as sov2, the phenotype of the more severely disrupted sovML150, sovEA42, and Df(1)sov mutations is lethality (Table 1). We conclude that CG14438 encodes sov and is an essential gene.
Table 2. Allele-specific sov sequences.
| Allele | Lesion location | Location relative to first TSS | Change | Codon change |
|---|---|---|---|---|
| sovNP6070 | 6757638 | 1,316 | Deletion and insertion | None |
| sovKG00226 | 6759438 | 3,116 | Insertion | None |
| sovML150 | 6763888 | 7,566 | C > G | Gln1246Glu |
| sovEA42 | 6764462 | 8,140 | G > A | Trp1437Stop |
| sov2 | 6769742 | 13,420 | Insert T | Frameshift |
| Df(1)sov | 6756569-6770708 | — | Deletion | Deletes ORF |
sov transcript expression and Sov protein localization
To determine where sov is expressed and if it encodes multiple isoforms, we analyzed its expression in adult tissues by RNA-Seq. We noted that while sov employs two promoters and two transcription start sites, sov transcripts are broadly expressed in multiple tissues as a single messenger RNA (mRNA) isoform with highest expression in ovaries (Figure 2A). The modENCODE (Graveley et al. 2011; Brown et al. 2014) and FlyAtlas (Robinson et al. 2013; Leader et al. 2018) reference sets show similar enrichments in the ovaries and early embryos due to maternal deposition. These expression patterns suggest that sov is required broadly, but is particularly important for the ovary.
Figure 2.
Sov is widely expressed and localizes to the soma and germline during oogenesis. (A) RNA expression tracks by tissue type from female (red) or male (blue) adults. (B) Cartoon of Drosophila egg chamber development showing germarium regions I–III and young egg chamber with cell types labeled. (C) Western blot analysis of WT (genotype: y1w67c23) and GFP-sov ovarian lysates probed for expression of tagged Sov protein using anti-FLAG and anti-GFP antibodies. βTub antibodies were used as a loading control. A specific band of ∼450 kDa is detected in GFP-sov extracts, corresponding to the predicted 446.6-kDa molecular weight of 3xFLAG-GFP-Sov. (D–F’) One-day-post eclosion WT germaria, visualized for GFP-Sov (green). All images show single optical sections and anterior is to the left. Dashed boxes highlight inset regions, magnified below. GFP-Sov is enriched in germarium region I (dashed line) with high expression in GSCs (arrows). (D) Sov contrasted with anti-αSpec to label spectrosomes (magenta) and anti-Vas to label germline (red). (E) Sov contrasted with anti-αSpec to label spectrosomes (red) and anti-Tj to label the soma (magenta). (F) Open arrowheads highlight localization of Sov within follicle cell nuclei outlined with WGA. (F’) Sov localizes within nurse cell and follicle cell nuclei in mature egg chambers. Bars, 10 μm, insets 5 μm. GSC, germline stem cell; reprod., reproductive; αSpec, αSpectrin; Tj, Traffic jam; Tub, Tubulin; Vas, Vasa; WGA, wheat germ agglutinin; WT, wild-type.
Given the ovarian enrichment of sov RNA, we examined the distribution of Sov protein in developing egg chambers (Figure 2B). For these studies, we employed our genetically functional GFP-sov transgene, which contains ∼3 kb of upstream and downstream regulatory sequences (Table 1; see Materials and Methods). The GFP-sov fusion contains multiple N-terminal tags, including FLAG and sfGFP, and it has a predicted molecular weight of ∼450 kDa. Using two different antibodies, we assayed ovarian extracts by western blot and confirmed that GFP-sov generates a protein product of the expected molecular size (Figure 2C). This protein product is specific to GFP-sov lysates, as it is missing in wild-type controls.
Using GFP antibodies to monitor the localization of Sov together with antibodies recognizing Vasa to label the germline, we observed nuclear localization of Sov surrounded by perinuclear Vasa in the germline cells within region 1 of the germarium (dashed line, Figure 2D and insets), with the highest levels evident within the germline stem cells (GSCs) (arrows; Figure 2, D–F). Compared to region 1, levels of Sov appear diminished in other regions of the germarium. Nonetheless, we noted nuclear enrichment of Sov in several Tj-positive cells, including follicle stem cells (Figure 2E, insets). We confirmed nuclear localization of Sov by using fluorescently conjugated WGA to label the nuclear envelope, where Sov was found within follicle cells and follicle stem cell nuclei (arrowheads, Figure 2, F and F’). In addition, we noted that Sov localized within the nuclei of nurse cells and follicle cells in later-stage egg chambers (Figure 2F’). These data indicate that Sov localizes to the nucleus in both germline and somatic cells.
sov is required in the soma and germline for oogenesis
Prior characterization of sov described pleiotropic female sterility phenotypes, including the complete absence of one or more ovaries, ovarian tumors of germ cells undergoing complete cytokinesis, egg chambers with > 16 nuclei, arrested egg chamber development in vitellogenic stages, and egg chamber degeneration (Wayne et al. 1995). We revisited the phenotypic profile of sov by examining transheterozygous females of sov2 with classic and new sov alleles (Figure 3). While we recapitulated the female sterility phenotypes, we note these phenotypes are variable within individuals. For example, a female could be missing one ovary, with the contralateral ovary showing any combination of the egg chamber phenotypes. This variability extended to within-brood effects. For example, sov females eclosing first (from mothers allowed 48 hr to lay eggs) showed a weaker phenotype relative to those eclosing on subsequent days. In addition, females eclosing later in the brood showed frequent partial tergite deletions.
Figure 3.
Sov permits normal oogenesis. (A–E) Maximum-intensity projections of germaria (0–2 day posteclosion) stained with anti-Vas (green), anti-Tj (magenta), αSpec (red), and DAPI (blue) in the noted genotypes, with excess dot spectrosomes (arrows) and empty ovarioles (asterisks) shown. (F–I) Quantification of ovarian differentiation phenotypes. sov2 represents the paternal allele in all genotypes. (F and H) Quantification of the number of dot spectrosomes per germarium from 0 to 2- or 5 to 6-day post eclosion females, respectively. (G and I) Quantification of the number of developing egg chambers per ovariole in 0–2- or 5–6-day post eclosion females, respectively. For quantification of dot spectrosomes within 0–2-day germaria, N = 28 sov2/+, N = 35 sov2/sovNP6070, N = 55 sov2/sovEA42, N = 80 sov2/sovML150, and N = 136 sov2/Df(1)sov. For quantification of the number of egg chambers within 0–2-day ovarioles, N = 26 sov2/+, N = 35 sov2/sovNP6070, N = 55 sov2/sovEA42, N = 76 sov2/sovML150, and N = 138 sov2/Df(1)sov. For quantification of dot spectrosomes within 5–6-day germaria, N = 51 sov2/+, N = 37 sov2/sovNP6070, N = 36 sov2/sovEA42, N = 64 sov2/sovML150, and N = 189 sov2/Df(1)sov. For quantification of the number of egg chambers within 5–6 day ovarioles, N = 41 sov2/+, N = 27 sov2/sovNP6070, N = 36 sov2/sovEA42, N = 62 sov2/sovML150, and N = 189 sov2/Df(1)sov. Data shown are from a single representative experiment, and the experiment was repeated twice with similar results. Bars, 10 μm. αSpec, αSpectrin; Tj, Traffic jam; Vas, Vasa.
To examine the phenotypic consequences of sov depletion in greater detail, we focused our analysis on the germarium, where sov expression is particularly strong (Figure 2). The germaria of heterozygous sov females appear normal (Figure 3A). Similarly, germaria from sov2/sovNP6070 females show no gross morphological defects (Figure 3B). In contrast, sovEA42, sovML150, and Df(1)sov mutants often show an ovarian tumor phenotype, with a greater than expected number of germ cells with dot spectrosomes (arrows, Figure 3, C–E), the cytoskeletal organelle normally associates with GSCs and their early cystoblast daughters (Deng and Lin 1997). The ovaries from sovML150 and Df(1)sov mutants consisted of degenerate tissue characterized by small, nub-like ovaries comprising dysmorphic germaria (Figure 3D), ovarian tumors, and empty ovarioles (asterisks, Figure 3E). To assay the incidence of ovarian stem cell-like tumors, we quantified the number of dot spectrosomes within sov germaria using αSpec antibodies (Eliazer et al. 2014). Normally, germaria contain one dot spectrosome in each of the two or three GSCs (Figure 2B). Germaria from sov2/+ and sov2/sovNP6070 females contain the expected number of dot spectrosomes (Figure 3F). In contrast, a significant proportion of germaria from sovEA42, sovML150, and Df(1)sov mutants contained at least six dot spectrosomes (Figure 3F). Similarly, strong sov alleles were associated with a reduction in the number of developing egg chambers within a given ovariole (Figure 3G). For example, the null allele Df(1)sov produced degenerate ovarian tissue consisting largely of empty ovarioles (Figure 3E, asterisks and Figure 3G). Similar results were observed in germaria from 5–6-day-old females (Figure 3, H and I). These findings are consistent with GSC or cystoblast hyperproliferation, and/or failed differentiation.
Previous work suggested that sov activity is solely required in the soma to permit germline development (Wayne et al. 1995). To examine cell type-specific requirements for sov, we used tj-GAL4 to knock down sov using the upstream activation sequence short hairpin P{TRiP.HMC04875} (Figure 1C) construct (sovRNAi) or mCherryRNAi controls in somatic escort and follicle cells, or nanos-GAL4 (nos-GAL4) for RNA interference (RNAi) in the germline. Relative to the controls, the tj > sovRNAi germaria (Figure 4, A and B) were abnormal, showing ovarian tumor phenotypes similar to sov mutants. Germaria were often filled with germline cells with dot spectrosomes (arrows, Figure 4B) and the follicle cells encroached anteriorly. Similar results were also observed using other somatic drivers (c587-GAL4 and da-GAL4). This demonstrates a clear somatic requirement for Sov. Interestingly, germline knockdown via nos>sovRNAi permitted the production of egg chambers representing all 14 morphological stages of oogenesis, which appeared phenotypically normal (data not shown); however, eggs from these females did not develop, suggesting that sov expression in the germline is required maternally to support embryogenesis.
Figure 4.
Sov is required in the germline and soma. Immunofluorescence for the indicated probes in the noted genotypes. (A and B) Single optical sections of control (mCherryRNAi) and sovRNAi expressed in germaria (4–5 days post eclosion) using tj > GAL4 and stained with anti-Vas (blue), -Tj (green), -αSpec (red), and DAPI (white) with dot spectrosomes (arrows). (C) Cartoon of dominant female-sterile germline clonal analysis technique. Recombination occurs between homologs at FRT sequences only in the presence of HS-induced FLP expression (Chou and Perrimon 1996). (D) Quantification of vitellogenic egg production from sov germline mutant clones. HS was used (+) to induce expression of FLP, but was omitted (–, no HS) in controls. (E–G) Maximum intensity projections of egg chambers (1 day posteclosion) stained with anti-Orb (green), phalloidin (red), and DAPI (blue). Orb+ cells shown (*). Bars, (A–D) 20 μm, (G–I) 10 μm. HS, heat shock; RNAi, RNA interference; αSpec, αSpectrin; Tj, Traffic jam; Vas, Vasa.
Our results showing that germline knockdown of sov results in maternal-effect lethality contradict prior work reporting the production of viable embryos from sov germline clones (Wayne et al. 1995). However, in that study, only the weaker sov alleles were examined. To reassess sov function in the female germline, we performed germline clonal analysis to assay the production of vitellogenic egg chambers in dissected ovaries (Figure 4C, see Materials and Methods) across an allelic series of sov mutations. In the absence of heat-shock/clone induction (negative control), none of the sov/ovoD1 genotypes resulted in clones giving rise to eggs. In the positive control, we observed germline clones of sov+ following heat-shock induction of mitotic clones in sov+/ovoD1. Clones of the weak sov2 allele yielded eggs that developed into viable progeny, in agreement with previous work (Wayne et al. 1995). In contrast, germline clones of the strong sov alleles sovEA42 and Df(1)sov failed to produce vitellogenic egg chambers or mature eggs (Figure 4D). The failure to recover sovEA42 and Df(1)sov germline clones was not due to spontaneous lethal or female-sterile mutations. Alleles were backcrossed and/or selected for viability in the presence of Dp(1:3)DC486, and subsequently maintained as attached-X stocks once the alleles were recombined with FRT sites. Most importantly, we did recover sovEA42 and Df(1)sov germline clones in the presence of the rescuing Dp(1:3)DC486 duplication (data not shown). These data indicate that sov is required for germline development into the vitellogenic stages and imply that the apparent strict somatic dependence of sov observed by Wayne et al. (1995) was likely the result of using weak female-sterile alleles in their analysis, which we also observed in the recovery of sov2 germline clones. The sov germline phenotype in the clonal analysis was also more extreme than the germline RNAi phenotype, suggesting that our RNAi experiments only partially reduced sov expression. Taken together, our data support a role for Sov in both the germline and the soma, consistent with recent reports (Jankovics et al. 2018).
While many of the sov phenotypes result in aberrant germline development, we also observed what appeared to be defective follicle encapsulation of germline cysts following sov depletion. To examine this more closely, we used the oocyte-specific expression of Oo18 RNA-binding protein (Orb) (Lantz et al. 1994) to count the number of oocytes per cyst. Orb specifies the future single oocyte at the posterior of the egg chamber in control egg chambers (Figure 4E). Somatic depletion of sov resulted in examples of egg chambers with either too many oocytes (Figure 4F) or no oocytes (Figure 4G). In a wild-type 16-cell germline cyst, one of the two cells that has four ring canals becomes the oocyte, and this feature was used to determine if extra germline divisions had occurred in cysts or if multiple cysts were enveloped by the follicle cells. We saw that the egg chambers with multiple Orb+ cells always had > 16 germ cells, and in the representative example shown all three Orb+ cells had four ring canals, indicating that egg chamber fusion had occurred (Figure 4F). We conclude that defective follicle encapsulation contributes, in part, to the irregular morphology and abortive development of sov mutants.
Sov represses gene expression in the ovary
Our data show that Sov localizes to the nucleus and is required for oogenesis. To test if sov regulates gene expression, we performed transcriptome profiling using triplicated PolyA+ RNA-Seq analyses of ovaries from sov mutant females, females with ovary-specific knockdown of sov using sovRNAi, and several control females (17 genotypes, 50 samples in total, see Materials and Methods). The gene expression profiles of ovaries from sterile females were markedly different from controls, primarily due to derepression in the mutants (Figure 5A). Differential gene expression was strikingly asymmetric, as genes were overexpressed in sov mutants and sov knockdowns generated with a variety of GAL4 drivers more often than they were downregulated.
Figure 5.
sov functions as a repressor of gene expression. (A) Heatmap of all expressed genes in all genotypes assessed in this study. k-means cluster analysis (k = 5) was performed on gene RPKMs. The black bar demarks sterile vs. fertile phenotypes. (B) Heatmap of differentially expressed genes in sov mutants. k-means cluster analysis (k = 5) was performed on genes and clustered into groups 1–5. Group 1 (dotted box) represents genes that were derepressed in sov mutants and somatic knockdown of sov. Group 2 (solid box) represents genes that were derepressed in sov mutants and germline knockdown of sov. Values are mean-subtracted ratios scaled across genotypes (red = higher and blue = lower). Groups 3 and 4 represent genes showing allele-specific derepression. Group 5 shows genes that were repressed in sov mutants. The black bar demarks sterile vs. fertile phenotypes. (C) Tissue-biased expression in wild-type tissues for genes derepressed in sov mutant ovaries. Heatmap from mean-subtracted ratios scaled across tissues (red = higher and blue = lower). RPKM, reads per kilobase per million reads; NRC FC, normalized read count fold change.
To robustly identify which genes are repressed by Sov+, we used replicated samples from all females with three mutant genotypes (sovEA42/sov2, sovML150/sov2, and Df(1)sov/sov2) using the different homozygous and heterozygous genotypes in a nested analysis, and calculated a locus-level change in gene expression. There were 6091 genes expressed at significantly different levels in sov mutant ovaries relative to the heterozygous controls [false discover rate (FDR) padj < 0.05]. Due to the large number of differentially expressed genes, we reduced the number of genes in our analysis using a highly conservative fourfold differential expression cut off. Among genes showing differences in expression (FDR padj < 0.05) in this nested analysis, we found 1661 genes with more than fourfold increased expression in mutants, while there were only 172 genes with more than fourfold decreased expression (Table S2).
To determine what types of genes are differentially expressed in sov mutants, we performed a k-means cluster analysis (genes and genotypes) on this subset of 1833 genes (1,661 + 172) and 17 genotypes to generate five groups of genes, and neatly split sterile and fertile genotypes (Figure 5B). We also performed GO term analysis (Ashburner et al. 2000; Mi et al. 2017; The Gene Ontology Consortium 2017) (Table 3). As the largest cluster, group 1 contained genes that were derepressed in sov mutants and with somatic sov RNAi (Figure 5B, dotted line). GO terms of group 1 genes were enriched for cell signaling, including neuronal communication (Table 3, shown in bold), and thus might represent a somatic-specific function of sov to repress inappropriate signaling pathways detrimental to somatic–germline communication. Similar upregulation of neuronal genes was previously described in the ovary following loss of other transcriptional repressors (Soshnev et al. 2013). Group 2 contained a small subset of genes that seemed to be selectively derepressed with germline knockdown of sov (Figure 5B, solid line), based on RNAi driven by nos-GAL4. Groups 3 and 4 highlight genes that are significantly derepressed in sov mutants; however, the magnitude of derepression seems to be allele-specific, suggesting that differences in the genetic background may also contribute to these gene expression changes (Figure 5B). There was no significant GO term enrichment in groups 2–4. Group 5 represents genes with decreased expression in sov mutants (Figure 5B). This group contained only a few significant GO terms that were predominantly oogenic in nature, as expected given the general lack of mature eggs. For example, there was poor expression of the chorion genes that are required to build the eggshell (Orr-Weaver 1991).
It is formally possible that mRNAs expressed in wild-type late-stage egg chambers in our RNA-Seq analysis could skew the differential expression analysis in comparison to sov mutant ovaries, which lack these stages. To address this concern, we calculated the log2 fold change values of genes that are considered to have high (1980 genes) or very high (742 genes) expression in 0–2-hr-old embryos according to FlyBase RNA-Seq expression profiles (Table S3) (Graveley et al. 2011). Genes in the high and very high categories had a mean log2 fold change of −0.14 and −0.23, respectively, in sov mutant ovaries. Conservatively correcting the log2 fold change values of all genes in our analysis by subtracting the very high category’s −0.23 log2 fold change resulted in 1512 genes with more than fourfold increased expression in mutants and 199 genes with more than fourfold decreased expression. Thus, this correction resulted in only a minor effect on the trend we identified. We also removed the genes with high expression in 0–2-hr-old embryos and recalculated differential expression. Again, this had no overt effect on the outcome. We observed 1625 or 1590 genes with increased expression in sov mutants even with the very high or high genes removed, respectively. We conclude that the differences in gene expression observed in sov mutants are not due to the absence of advanced oogenic stages. Rather, our expression analyses are most consistent with a role for Sov in the widespread repression of gene expression.
Our gene expression analysis showcases the aberrant derepression genes in response to sov loss. To explore where these derepressed genes are normally expressed, we examined their expression in other female tissues and in testes in a set of quadruplicated RNA-Seq experiments in wild-type flies (Figure 5C). Many of the genes derepressed in sov mutant ovaries were highly expressed in other tissues. Notably, we found that these depressed genes were largely uncharacteristic of ovary or testes expression, and were more similar to that of head and thorax tissue expression. We conclude that sov functions broadly to repress gene expression, preventing the misexpression of genes normally expressed elsewhere.
Comparison to previous HP1a expression data in ovaries
The repressive role of sov might require other chromatin-associated factors that also repress gene expression in the ovary. We compared our RNA-Seq data to data generated from germline knockdown of HP1a, SETDB1, and WDE in a common pipeline and we found limited overlap (Table S4). When germ cells have a male identity in a female soma, this results in ovarian tumors (Oliver 2002; Murray et al. 2010). The dot spectrosome phenotype in sov ovaries could be due to sex transformation. In addition to a general role, it has been suggested that heterochromatin factors might play a critical role in determining germline sexual identity (Smolko et al. 2018). SETDB1 has been reported to silence the male-determining phf7 gene to allow for female germline sexual identity. Therefore, we specifically asked if sov alters phf7 expression, both overall and at the level of sex-specific promoter choice. Loss of sov was associated with modestly altered expression of phf7 (sov mutants: padj = 0.02 and log2 fold change = –0.21, and sov germline RNAi: padj = 0.98 and log2 fold change = 0.08), but sov was not required for male-specific promoter repression (Figure 5D). Taken together, these data do not support a role for sov in germline sex determination, but they do support the importance of sov as a general repressive factor.
Sov represses transposon expression
Our data strongly support a general role of Sov in regulating gene expression. Recent work described a role for Sov function in the regulation of transposon expression (Czech et al. 2013). Indeed, our RNA-Seq analysis identified a dramatic and coherent elevation of transposon expression in all sov mutant genotypes; no cases of underexpression were observed (Figure 6A and Table S2). For example, when we used the weak female-sterile sov2 allele in trans to strong alleles (sovEA42/sov2, sovML150/sov2, and Df(1)sov/sov2), 117 out of 138 transposable elements were expressed at significantly higher levels (FDR padj < 0.05). Using the same conservative fourfold cutoff as before, we found that 91 transposable elements had more than fourfold increased expression (FDR padj < 0.05) in sov mutants, while none had more than fourfold decreased expression. These data confirm that Sov is required to suppress transposon expression.
Figure 6.
Transposon expression in sov mutants. (A) Number of transposons with a greater (red) or less than (blue) fourfold change in gene expression (FDR padj value < 0.05) in sov mutants or RNAi knockdown when compared to controls. (B) Transposable element expression in sov mutants (sterile) and controls (fertile). Heatmap from mean-subtracted reads (in RPKM; red = higher and blue = lower) scaled for each transposable element (rows) across genotypes (columns). The black bar demarks sterile vs. fertile phenotypes. Transposon classes for DNA, non-LTR (Jockey), telomeric repeat (dashed box), and LTR (Gypsy, Copia, and PAO) are indicated. FDR, false discovery rate; rel. relative; RNAi, RNA interference; RPKM, reads per kilobase per million reads.
There is tremendous diversity in transposon types in Drosophila (McCullers and Steiniger 2017). Broadly, there are retrotransposons that utilize reverse transcription and DNA elements, such as P-elements, that excise and reinsert. Retrotransposons can be further divided into those with long terminal repeats (LTRs)—such as Gypsy, Copia, and PAO—and those lacking LTRs, such as Jockey. While many transposons are considered to be detrimental when active, the non-LTR HeT-A, TAHRE, and TART transposons are the structural basis of Drosophila telomeres (Mason et al. 2008). All of these classes of transposable elements were derepressed in sov mutants and in flies where sovRNAi was driven by the ubiquitously expressed da-GAL4 driver (Figure 6B). Interestingly, somatic knockdown of sov in the cells encasing the germ cells using c587-GAL4 or tj-GAL4 resulted in derepression of most of the transposon classes, including the Gypsy and Copia classes, which are Drosophila retroviruses that develop in somatic cells and are exported to the developing germline (Yoshioka et al. 1990). Thus, wild-type sov may be important to protect the germline from infection.
Germline knockdown of sov driven by nos-GAL4 resulted in significant derepression of 20 transposable elements (Figure 6B and Table S2). Among these, we found that there was a greater magnitude of derepression of the transposons required for normal telomere function. HeT-A, TAHRE, and TART, which are important for telomere function, had a mean log2 fold change of 7.8 with germline knockdown of sov, while the other 17 transposable elements had a mean log2 fold change of 3.9. This indicates that telomere classes of transposons were more sensitive to loss of sov than other classes of transposons in the germline. These results are strikingly similar to the derepression of telomere transposons following germline-specific knockdown of HP1a (Teo et al. 2018). Comparative analysis between our data set and RNA-Seq reported for HP1a, SETDB1, or WDE (Smolko et al. 2018) highlights overlapping derepression of HeT-A and several other transposons (Table S4). These findings suggest that the general role of sov in silencing transposon expression includes both transposons necessary for normal cellular functions, exemplified by the telomeric transposons, and transposons with no known beneficial cellular roles. Taken together, our RNA-Seq analyses define Sov as a negative regulator of gene expression, of which transposons represent one class of Sov targets.
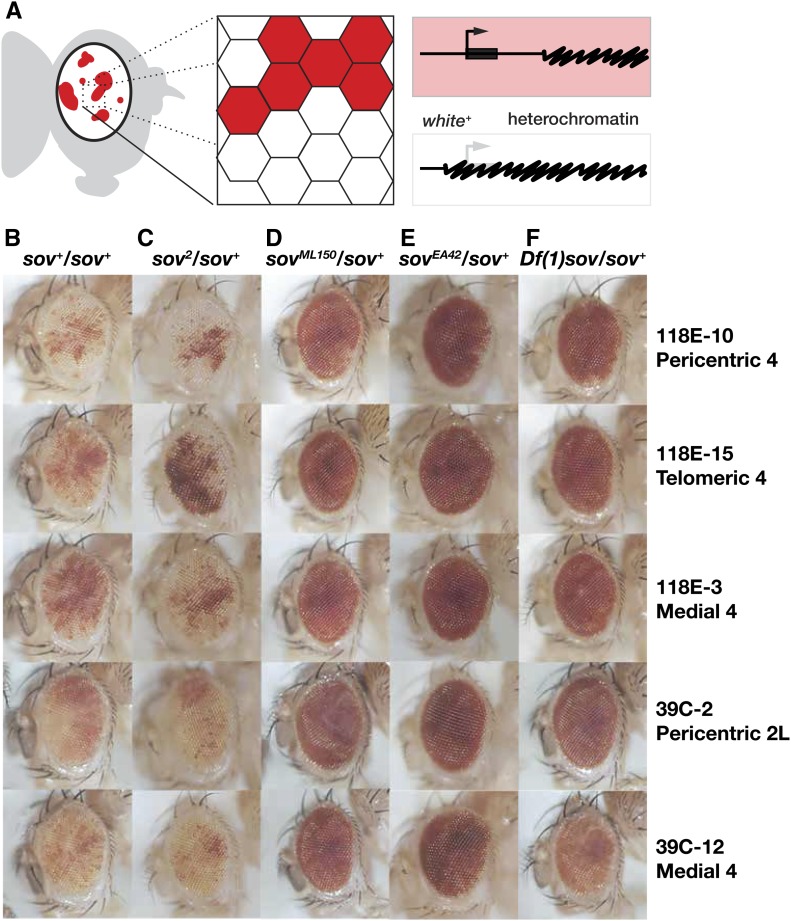
Sov is a suppressor of PEV
The ovary may be particularly susceptible to heterochromatin defects, as mutations in several members of the HP1a complex result in female sterility (Clough et al. 2007, 2014; Yoon et al. 2008; Teo et al. 2018). Loss of the H3K9 methyltransferase SETDB1/egg (Clough et al. 2007, 2014; Wang et al. 2011) or the H3K4 demethylase encoded by lysine-specific demethylase 1 (LSD1) (Di Stefano et al. 2007; Rudolph et al. 2007; Eliazer et al. 2011, 2014) results in degenerate phenotypes in the ovary, yet these factors function widely to regulate gene expression. We reasoned that Sov may similarly function to regulate gene expression outside of the ovary.
HP1a was first characterized in Drosophila as a suppressor of PEV (Clark and Elgin 1992). To test the hypothesis that sov negatively regulates gene expression by promoting heterochromatin formation, we examined the effects of sov mutations on PEV in the eye, where patches of w+ (red pigmented) and w– (unpigmented) eye facets are easily observed (Figure 7A). If, like HP1a, Sov represses gene expression by promoting heterochromatin formation (Eissenberg et al. 1990), then sov mutations should suppress PEV, which is scored as increased eye pigmentation. We obtained five variegating w+ transgene insertions associated with either the heterochromatic pericentric region of chromosome arm 2L or spread along the length of heterochromatin-rich chromosome 4. In control animals, these insertions show characteristic eye variegation patterns (Figure 7B). Consistent with the allelic strengths seen in previous experiments, the weak female-sterile sov2 allele did not suppress PEV (Figure 7C), but the stronger lethal mutations sovML150, sovEA42, and Df(1)sov dominantly suppressed PEV (Figure 7, D–F). These data establish a role for Sov in heterochromatin function and demonstrate that Sov is capable of regulating gene expression outside of oogenic contexts.
Figure 7.
Sov is a dominant suppressor of position-effect variegation. (A) Cartoon of position-effect variegation in the eye. Expression of the white gene (bent arrow, thick bar, red) can be silenced (white) by proximal heterochromatin (squiggled) spreading. (B–F) Eyes from adults of the indicated genotypes (columns) with variegated expression of P{hsp26-pt-T} transgenes inserted into the indicated chromosomal positions (rows).
Sov colocalizes with HP1a during heterochromatin formation
Our loss-of-function studies allow us to draw several parallels between Sov and the core heterochromatin factor HP1a. Both proteins localize to the nucleus, are required to suppress transposons, and function more broadly as general repressors of gene expression. Moreover, recent work in Drosophila S2 cells suggests that Sov complexes with HP1a (Alekseyenko et al. 2014), hinting that Sov and HP1a may coordinate the functions of heterochromatin.
To determine if Sov colocalizes with HP1a in the nucleus, we examined HP1a-RFP and GFP-Sov localization in 1–2-hr-old live embryos. During Drosophila embryogenesis, nuclei undergo rapid synchronous nuclear divisions prior to cellularization at cleavage division/nuclear cycle (NC) 14 (Foe and Alberts 1983). HP1a and Sov colocalized in the nuclei of all NC 10–12 embryos examined, including those that were monitored and quantified by live imaging (Figure 8A; N = 7 embryos). During interphase, prophase, and nuclear envelope breakdown, both HP1a and Sov were nuclear (Figure 8A). In prophase, HP1a and Sov colocalized, and were enriched in regions of condensed DNA (Figure 8A, 4:30). Whereas low levels of HP1a decorated DNA throughout nuclear division, Sov was depleted during mitosis (Figure 8A, 10:00 and 11:10). However, upon reentry into interphase, Sov localization to nuclei resumed and was coincident with HP1a (Figure 8A, 14:30).
Figure 8.
Sov colocalizes with HP1a. Stills from live imaging of embryos expressing GFP-Sov and RFP-HP1a. (A) Localization of GFP-Sov and RFP-HP1a (rows) in an embryo progressing from NC 10 to 11, with cell cycle stages (columns) and time (min:sec) shown. (B) NC 14 embryo. Boxed regions are magnified in insets below. An HP1a subnuclear domain is shown (arrows). (B’) Single optical section containing peak HP1a fluorescence of inset from (B). Dashed line indicates region used for histogram analysis. (A and B) Images show maximum projections through 1.5-μm volume and were captured at 1F/30 sec. Bar, 10 μm; insets, 5 μm. (C) Histogram of HP1a and Sov fluorescence intensity measured in (B’). Fluorescence levels (arbitrary units) normalized to the peak fluorescence intensity for each channel and the distance (μm) to peak HP1a signal. Half-maximum HP1a fluorescence is shaded (yellow). a.u., arbitrary units; NC, nuclear cycle; NEB, nuclear envelope breakdown; RFP, red fluorescent protein.
Formation of heterochromatin is contemporaneous with or slightly proceeds HP1a apical subnuclear localization in NC 14 (Rudolph et al. 2007; Yuan and O’Farrell 2016). At this stage, we observed a strong colocalization of HP1a and Sov. Measuring the distribution of HP1a and Sov (Figure 8, B and B’) revealed high levels of colocalization within HP1a subnuclear domains (Figure 8C, shaded region). These data support the idea that HP1a and Sov assemble into a complex in the nucleus. Moreover, our live imaging reveals that Sov localizes to HP1a-positive foci around the time heterochromatin first forms. These findings support the possibility that Sov functionally coordinates heterochromatin stabilization and/or maintenance together with HP1a.
Discussion
Sov is a novel heterochromatin-associated protein
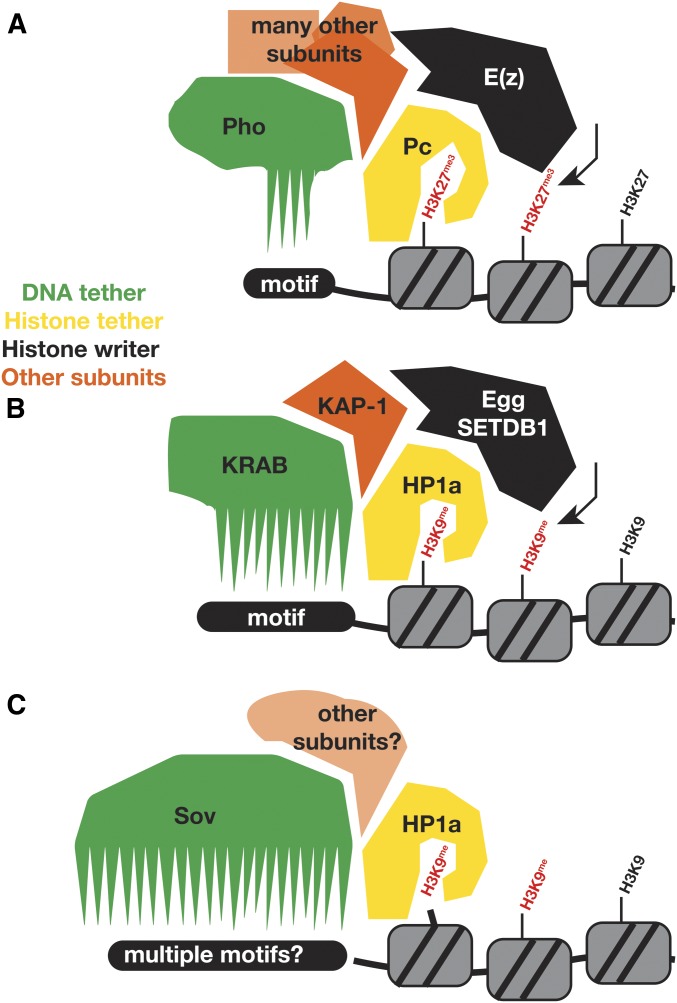
Gene repression is often stable over extended timescales. Emerging themes suggest that multiple members of protein complexes, rather than a single key component, maintain a stabilized chromatin state. For example, complexes formed by proteins of the repressive Polycomb group (PcG) provide a repressive epigenetic memory function (Kassis et al. 2017). To create a stable epigenetic state, PcG complexes have subunits that modify histones and bind these modifications (Figure 9A). Chromatin binding of PcG complexes also involves DNA-binding proteins, such as the YY1-like ZnF protein Pleiohomeotic (Pho), which further reinforce localization (Brown et al. 2003). The DNA-anchoring proteins within PcG complexes are at least partially redundant with the histone-binding components (Brown et al. 2003), suggesting that localization is robust due to multiple independent localization mechanisms. Such partially redundant components functionally contribute to stable gene repression.
Figure 9.
Working model of sov function. (A) Polycomb complexes (Pc) in flies and (B) Krüppel associated box (KRAB) complexes in mammals use a DNA-binding protein (Pho and KRAB; green) that binds sequence motifs in addition to histone code readers (Pc and HP1a; yellow), which bind to modified histones (red). These complexes also bear enzymes that create the histone marks [E(z) and Egg or SETDB1; black]. They have other proteins as well (such as KAP-1; orange). Like KRAB proteins, Sov is in complex with HP1a (yellow). We propose that it binds to DNA to provide another mechanism for tethering to chromosomes, in addition to HP1a binding to methylated H3K9. This partially redundant mechanism contributes to the stabilization of a repressed chromatin state.
The repressive chromatin protein HP1a may coordinate with other repressive proteins (Figure 9B). The Krüppel associated box (KRAB) family members use the KRAB-Associated Protein (KAP1) adapter protein to associate with HP1a and the SETDB1 methylase that modifies histones to enable HP1a binding. Although KRAB proteins have not been identified in Drosophila, the bonus (bon) gene encodes a KAP1 homolog (Beckstead et al. 2005). Drosophila also has a SETDB1 encoded by egg, and loss of egg results in ovarian phenotypes reminiscent of those observed in sov loss-of-function females (Clough et al. 2007, 2014; Wang et al. 2011). The single, very long, and ZnF-rich Sov protein may play a DNA- or RNA-binding role to help tether HP1a to chromatin (Figure 9C). If Sov uses subsets of fingers to bind sequences, it could localize to many locations. As in the case of Pho in the PcG complexes, Sov might contribute to heterochromatin stability rather than its formation, as we did not observe gross delocalization of HP1a following sov knockdown via RNAi, similar to recent findings (Jankovics et al. 2018). Consistent with this idea, nuclear colocalization of Sov is cell cycle-dependent whereas HP1a decorates DNA within cycling embryos constitutively. Perhaps recruitment of Sov to DNA serves to stabilize HP1a-containing complexes.
Repression by Sov promotes germline differentiation
The ovary may be particularly sensitive to loss of sov and other HP1a complex members. For example, the activities of both the H3K9 methyltransferase SETDB1, encoded by egg (Clough et al. 2007, 2014; Wang et al. 2011), and the H3K4 demethylase encoded by LSD1 (Di Stefano et al. 2007; Rudolph et al. 2007; Eliazer et al. 2011, 2014) are required in the somatic cells of the germarium for GSC maintenance, normal patterns of differentiation, and germline development. Dynamic changes in chromatin landscapes within germline and somatic cells are critical for oogenesis, and contribute to the regulated gene expression required for tissue development (McConnell et al. 2012; Soshnev et al. 2013; Barton et al. 2016; Börner et al. 2016; Peng et al. 2016; Li et al. 2017). Genome-wide profiling hints at a progression from open chromatin in stem cells to a more closed state during differentiation (Chen and Dent 2014). We predict that disrupting this progressive repression in the ovary through loss of sov or other heterochromatin factors contributes to stem cell hyperproliferation, and defective oogenesis.
The sov female sterility phenotype is complex and somewhat variable, but weak sov alleles have more severe consequences in somatic cells than germ cells. Stronger lethal alleles show a germline-dependent block in egg chamber development, based on our failure to recover germline clones of sovEA42 and Df(1)sov. We propose that the pleiotropy of sov mutations may be attributed to a single mechanism wherein Sov helps control facultative heterochromatin stabilization required to repress ectopic gene expression and tissue-inappropriate responses in the ovary, and elsewhere in the organism. For example, sov has a clear impact on PEV in the eye.
Sov represses transposons
We observed dramatic derepression of transposons in sov mutants. Consistent with our observations, Czech et al. (2013) reported a role for sov in transposon repression in a genome-wide RNAi screen. That work raised the possibility that transposon repression by Sov occurs via the Piwi-interacting RNA (piRNA) pathway (Brennecke et al. 2007; Yin and Lin 2007; Teixeira et al. 2017). However, genes involved more strictly with transposable element regulation, such as Piwi, do not have strong dosage effects on PEV (Gu and Elgin 2013), while sov (this study) and HP1a/Su(var)205 do (James and Elgin 1986; Eissenberg et al. 1990; Clark and Elgin 1992). Furthermore, amorphic sov alleles are embryonic lethal, whereas piRNA pathway mutants are viable with maternal-effect lethal phenotypes. These distinctions suggest that Sov has a more general role in heterochromatin, rather than a function restricted to transposon repression. Germline knockdown of HP1a results in a strong derepression of telomeric transposons (Teo et al. 2018), just like that found with germline knockdown of Sov. These key differences between the sov and piwi phenotypes imply a general role of sov with HP1a and heterochromatin, rather than a specific role in the piRNA pathway and oogenesis as recently reported (Jankovics et al. 2018).
Conclusions
The sov locus is required for the same major functions in PEV, viability, transposon repression, and gene repression as the HP1a locus. Further, HP1a and Sov colocalize in the nucleus. These data support the idea that Sov encodes a novel and essential protein that is generally repressive, which may act by stabilizing heterochromatin by facilitating HP1 functions.
Acknowledgments
We thank BACPAC Resources for plasmids; Norbert Perrimon for the sovEA42 stock; Don Court, Neal Copeland, and Nancy Jenkins for the recombineering strain; Sally Elgin for the PEV stocks; Astrid Haase, Nasser Rusan, David Katz, and members of the Oliver and Lerit laboratories for stimulating discussions; and particularly Stacy Christensen and Kimberley Cook for generating and characterizing the sov deletions. Stocks obtained from the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) grant P40 OD-018537) and Drosophila Genomics and Genetic Resources at the Kyoto Institute of Technology were used in this study. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the NIH and maintained at the Department of Biology, University of Iowa, Iowa City, IA 52242. Sequencing was performed by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Genomics Core, under the direction of Harold Smith. Genetic and genomic information was obtained from FlyBase (U41 HG-000739). This work utilized the computational resources of the NIH High-Performance Computing Biowulf cluster (http://hpc.nih.gov). This research was supported in part by the Intramural Research Program of the NIH NIDDK (awarded to B.O.). D.A.L. and E.A.C. were supported by NIH grant 5K22 HL-126922 (D.A.L.). L.B. was supported by the NIH Graduate Partners Program. K.J.T.V. was supported by Baylor College of Medicine, the Albert and Margaret Alkek Foundation, the McNair Medical Institute at The Robert and Janice McNair Foundation, the March of Dimes Foundation (#1-FY14-315), the Foundation for Angelman Syndrome Therapeutics (FT2016-002), the Cancer Prevention and Research Institute of Texas (R1313), and NIH grants 1R21 HG-006726, 1R21 GM-110190, 1R21 OD-022981, and R01 GM-109938). C.W. and K.R.C. were supported by the Office of the NIH Director, the National Institute of General Medical Sciences, and the NICHD (P40 OD-018537). K.R.C. was supported by the National Center for Resource (R24RR014106) and the National Science Foundation (DBI-9816125).
Author contributions: L.B., E.A.C., K.R.C., B.O., and D.A.L. designed the project and analyzed data. L.B., K.R.C., C.W., and K.J.T.V. performed genetics. L.B. and H.Y. performed genomics. J.F. performed western blotting. L.B., E.A.C., C.W., and D.A.L. performed imaging. L.B., B.O., and D.A.L. wrote the manuscript. All authors reviewed data and provided feedback on the manuscript.
Footnotes
Supplemental material available at Figshare: https://figshare.com/articles/Genetics_09_13_2019/9828002.
Communicating editor: P. Geyer
Literature Cited
- Alekseyenko A. A., Gorchakov A. A., Zee B. M., Fuchs S. M., Kharchenko P. V. et al. , 2014. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28: 1445–1460. 10.1101/gad.241950.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L. J., Lovander K. E., Pinto B. S., and Geyer P. K., 2016. Drosophila male and female germline stem cell niches require the nuclear lamina protein Otefin. Dev. Biol. 415: 75–86. 10.1016/j.ydbio.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead R. B., Ner S. S., Hales K. G., Grigliatti T. A., Baker B. S. et al. , 2005. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics 169: 783–794. 10.1534/genetics.104.037085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C., and Paro R., 2011. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12: 123–135. 10.1038/nrg2932 [DOI] [PubMed] [Google Scholar]
- Börner K., Jain D., Vazquez-Pianzola P., Vengadasalam S., Steffen N. et al. , 2016. A role for tuned levels of nucleosome remodeler subunit ACF1 during Drosophila oogenesis. Dev. Biol. 411: 217–230. 10.1016/j.ydbio.2016.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M. et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Riddle N. C., Jiang H., Huisinga K. L., and Elgin S. C. R., 2009. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181: 1303–1319. 10.1534/genetics.108.100271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H. et al. , 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Fritsch C., Mueller J., and Kassis J. A., 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130: 285–294. 10.1242/dev.00204 [DOI] [PubMed] [Google Scholar]
- Canzio D., Chang E. Y., Shankar S., Kuchenbecker K. M., Simon M. D. et al. , 2011. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41: 67–81. 10.1016/j.molcel.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J., Zweig A. S., Villarreal C., Tyner C., Speir M. L. et al. , 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46: D762–D769. 10.1093/nar/gkx1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., and Dent S. Y., 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 15: 93–106. 10.1038/nrg3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. B., and Perrimon N., 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T. et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. F., and Elgin S. C., 1992. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 20: 6067–6074. 10.1093/nar/20.22.6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Moon W., Wang S., Smith K., and Hazelrigg T., 2007. Histone methylation is required for oogenesis in Drosophila. Development 134: 157–165. 10.1242/dev.02698 [DOI] [PubMed] [Google Scholar]
- Clough E., Tedeschi T., and Hazelrigg T., 2014. Epigenetic regulation of oogenesis and germ stem cell maintenance by the Drosophila histone methyltransferase Eggless/dSetDB1. Dev. Biol. 388: 181–191. 10.1016/j.ydbio.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R. K., Christensen S. J., Deal J. A., Coburn R. A., Deal M. E. et al. , 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13: R21 10.1186/gb-2012-13-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D. L., Swaminathan S., Yu D., Wilson H., Baker T. et al. , 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315: 63–69. 10.1016/S0378-1119(03)00728-5 [DOI] [PubMed] [Google Scholar]
- Czech B., Preall J. B., McGinn J., and Hannon G. J., 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell 50: 749–761. 10.1016/j.molcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., and Lin H., 1997. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 189: 79–94. 10.1006/dbio.1997.8669 [DOI] [PubMed] [Google Scholar]
- Di Stefano L., Ji J.-Y., Moon N.-S., Herr A., and Dyson N., 2007. Mutation of Drosophila Lsd1 disrupts H3–K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 17: 808–812. 10.1016/j.cub.2007.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A., Lein S., Schotta G., and Reuter G., 2006. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14: 377–392. 10.1007/s10577-006-1066-1 [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V. et al. , 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. 10.1073/pnas.87.24.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. R., and Reuter G., 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S., Shalaby N. A., and Buszczak M., 2011. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 108: 7064–7069. 10.1073/pnas.1015874108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S., Palacios V., Wang Z., Kollipara R. K., Kittler R. et al. , 2014. Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLoS Genet. 10: e1004200 10.1371/journal.pgen.1004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo M. L. A., Philip P., Stenberg P., and Larsson J., 2012. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet. 8: e1003061 10.1371/journal.pgen.1003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., and Alberts B. M., 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61: 31–70. [DOI] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J.-M., Antonazzo G. et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M. et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F., van der Kraan I., Delrow J., Smothers J. F., de Wit E. et al. , 2003. Distinct HP1 and Su(var)3–9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 17: 2825–2838. 10.1101/gad.281503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T., and Elgin S. C. R., 2013. Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS Genet. 9: e1003780 10.1371/journal.pgen.1003780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M., Tintut Y., and Gralla J. D., 1994. Functional roles for the glutamines within the glutamine-rich region of the transcription factor sigma 54. J. Biol. Chem. 269: 373–378. [PubMed] [Google Scholar]
- Huang A. M., E. J. Rehm, and G. M. Rubin, 2009 Quick preparation of genomic DNA from Drosophila Cold Spring Harb. Protoc. 2009: pdb.prot5198. doi: 10.1101/pdb.prot5198 10.1101/pdb.prot5198 [DOI] [PubMed]
- James T. C., and Elgin S. C., 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862–3872. 10.1128/MCB.6.11.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. C., Eissenberg J. C., Craig C., Dietrich V., Hobson A. et al. , 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50: 170–180. [PubMed] [Google Scholar]
- Jankovics F., Bence M., Sinka R., Faragó A., Bodai L. et al. , 2018. Drosophila small ovary gene is required for transposon silencing and heterochromatin organization, and ensures germline stem cell maintenance and differentiation. Development 145: dev170639. 10.1242/dev.170639 [DOI] [PubMed] [Google Scholar]
- Jemc J. C., Milutinovich A. B., Weyers J. J., Takeda Y., and Van Doren M., 2012. Raw Functions through JNK signaling and cadherin-based adhesion to regulate Drosophila gonad morphogenesis. Dev. Biol. 367: 114–125. 10.1016/j.ydbio.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., and Allis C. D., 2001. Translating the histone code. Science 293: 1074–1080. 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Jiang L., Schlesinger F., Davis C. A., Zhang Y., Li R. et al. , 2011. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21: 1543–1551. 10.1101/gr.121095.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Kennison J. A., and Tamkun J. W., 2017. Polycomb and trithorax group genes in Drosophila. Genetics 206: 1699–1725. 10.1534/genetics.115.185116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H. et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang J. S., Horabin J. I., Bopp D., and Schedl P., 1994. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8: 598–613. 10.1101/gad.8.5.598 [DOI] [PubMed] [Google Scholar]
- Leader D. P., Krause S. A., Pandit A., Davies S. A., and Dow J. A. T., 2018. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46: D809–D815. 10.1093/nar/gkx976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Pine P. S., McDaniel J., Salit M., and Oliver B., 2016a External RNA controls Consortium beta version update. J Genomics 4: 19–22. 10.7150/jgen.16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Cho D.-Y., Whitworth C., Eisman R., Phelps M. et al. , 2016b Effects of gene dose, chromatin, and network topology on expression in Drosophila melanogaster. PLoS Genet. 12: e1006295 10.1371/journal.pgen.1006295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D. A., Jordan H. A., Poulton J. S., Fagerstrom C. J., Galletta B. J. et al. , 2015. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210: 79–97. 10.1083/jcb.201503117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Seidel C. W., Szerszen L. T., Lange J. J., Workman J. L. et al. , 2017. Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev. 31: 1588–1600. 10.1101/gad.300988.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., and Kornberg R. D., 2017. Chromatin-remodeling for transcription. Q. Rev. Biophys. 50: e5 10.1017/S003358351700004X [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Frydrychova R. C., and Biessmann H., 2008. Drosophila telomeres: an exception providing new insights. Bioessays 30: 25–37. 10.1002/bies.20688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell K. H., Dixon M., and Calvi B. R., 2012. The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development 139: 3880–3890. 10.1242/dev.083576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers T. J., and Steiniger M., 2017. Transposable elements in Drosophila. Mob. Genet. Elements 7: 1–18. 10.1080/2159256X.2017.1318201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C. et al. , 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler J. D., 1977. Developmental genetics of the Drosophila egg. I. Identification of 59 sex-linked cistrons with maternal effects on embryonic development. Genetics 85: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler D., and Carroll A., 1984. Report of Dawson Mohler and Andrea Carroll. Drosoph. Inf. Serv. 60: 236–241. [Google Scholar]
- Murray S. M., Yang S. Y., and Van Doren M., 2010. Germ cell sex determination: a collaboration between soma and germline. Curr. Opin. Cell Biol. 22: 722–729. 10.1016/j.ceb.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D., and Grewal S. I., 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- Oliver B., 2002. Genetic control of germline sexual dimorphism in Drosophila, pp. 1–60 in International Review of Cytology, edited by Jeon K. W., Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., 1991. Drosophila chorion genes: cracking the eggshell’s secrets. Bioessays 13: 97–105. 10.1002/bies.950130302 [DOI] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R. et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Peng J. C., Valouev A., Liu N., and Lin H., 2016. Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat. Genet. 48: 283–291. 10.1038/ng.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke T. J., McKay D. J., Strahl B. D., Matera A. G., and Duronio R. J., 2016. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev. 30: 1866–1880. 10.1101/gad.286278.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine P. S., Munro S. A., Parsons J. R., McDaniel J., Lucas A. B. et al. , 2016. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol. 16: 54 10.1186/s12896-016-0281-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., and Spierer P., 1992. Position effect variegation and chromatin proteins. Bioessays 14: 605–612. 10.1002/bies.950140907 [DOI] [PubMed] [Google Scholar]
- Reuter G., Dorn R., Wustmann G., Friede B., and Rauh G., 1986. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202: 481–487. 10.1007/BF00333281 [DOI] [Google Scholar]
- Robinson S. W., Herzyk P., Dow J. A. T., and Leader D. P., 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 41: D744–D750. 10.1093/nar/gks1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T., Yonezawa M., Lein S., Heidrich K., Kubicek S. et al. , 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26: 103–115. 10.1016/j.molcel.2007.02.025 [DOI] [PubMed] [Google Scholar]
- Sambrook J., and Russell D. W., 2006. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006: pdb.prot4455. 10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- Smit A., Hubley R., and Green P., 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org/faq.html
- Smolko A.E., Shapiro-Kulnane L., and Salz H. K., 2018 . The H3K9 methyltransferase SETDB1 maintains female identity in Drosophila germ cells. Nat Commun. 9: 4155 10.1038/s41467-018-06697-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., Baxley R. M., Manak J. R., Tan K., and Geyer P. K., 2013. The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 140: 3613–3623. 10.1242/dev.094953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F. K., Okuniewska M., Malone C. D., Coux R.-X., Rio D. C. et al. , 2017. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552: 268–272. 10.1038/nature25018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo R. Y. W., Anand A., Sridhar V., Okamura K., and Kai T., 2018. Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila. Nat. Commun. 9: 1735 10.1038/s41467-018-03908-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2017. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45: D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Carlson J. W., Schulze K. L., Pan H., He Y. et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Schulze K. L., Haelterman N. A., Pan H., He Y. et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D., and Malik H. S., 2009. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43: 467–492. 10.1146/annurev-genet-102108-134802 [DOI] [PubMed] [Google Scholar]
- Wang X., Pan L., Wang S., Zhou J., McDowell W. et al. , 2011. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet. 7: e1002426 10.1371/journal.pgen.1002426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., and Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne S., Liggett K., Pettus J., and Nagoshi R. N., 1995. Genetic characterization of small ovaries, a gene required in the soma for the development of the Drosophila ovary and the female germline. Genetics 139: 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K. S., and Wakimoto B. T., 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. 10.1146/annurev.ge.29.120195.003045 [DOI] [PubMed] [Google Scholar]
- Yang H., Jaime M., Polihronakis M., Kanegawa K., Markow T. et al. , 2018. Re-annotation of eight Drosophila genomes. Life Sci Alliance 1: e201800156 10.26508/lsa.201800156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara J. C., and Wakimoto B. T., 2006. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 22: 330–338. 10.1016/j.tig.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Yin H., and Lin H., 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308. 10.1038/nature06263 [DOI] [PubMed] [Google Scholar]
- Yoon J., Lee K.-S., Park J. S., Yu K., Paik S.-G. et al. , 2008. dSETDB1 and SU(VAR)3–9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS One 3: e2234 10.1371/journal.pone.0002234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Honma H., Zushi M., Kondo S., Togashi S. et al. , 1990. Virus-like particle formation of Drosophila copia through autocatalytic processing. EMBO J. 9: 535–541. 10.1002/j.1460-2075.1990.tb08140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., and O’Farrell P. H., 2016. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 30: 579–593. 10.1101/gad.272237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook J. M., Samarov D., McDaniel J., Sen S. K., and Salit M., 2012. Synthetic spike-in standards improve run-specific systematic error analysis for DNA and RNA sequencing. PLoS One 7: e41356 10.1371/journal.pone.0041356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids, and their sources, are detailed in Table S1. All sequencing data are available at public repositories: DNA-Seq data are available at the SRA (SRP14438) and RNA-Seq data are available at the GEO (GSE113977 and GSE99574). Supplementary tables are available at Figshare: Table S1, FlyBase ART file contains all experimental materials used in the study; Table S2, Differentially expressed genes in sov mutants vs. controls; Table S3, gene list of high or very high levels in 0–2-hr-old embryos according to Graveley et al. (2011); and Table S4, common and differentially expressed genes following germline knockdown of sov vs. HP1a, SETDB1, and WDE. A supplementary text file includes FASTA sequences for the GFP-sov transgene. Supplemental material available at Figshare: https://figshare.com/articles/Genetics_09_13_2019/9828002.