Abstract
Average daily gain (ADG) and daily dry matter intake (DMI) are key determinants of beef industry profitability. These traits together with metabolic body weight (MWT) are combined as component traits to calculate residual feed intake (RFI), a common measure of feed efficiency in beef cattle. Recently, there have been significant efforts towards molecular genetic characterization of RFI through transcriptomic studies in different breeds and tissues. However, molecular mechanisms of RFI component traits still remain predominately unexplored. Therefore, in the current study, we investigated the hepatic transcriptomic profiles and their associations with ADG, DMI, and MWT in Angus, Charolais, and Kinsella Composite (KC) populations through global RNAseq analyses. In each population and for each trait, 12 steers with extreme phenotypes (n = 6 low and n = 6 high) were analyzed for differential gene expression. These animals were from 20 beef steers of each Angus, Charolais, and KC breed population that were initially selected for a transcriptome study of RFI. At a false discovery rate <0.05 and fold change >1.5, we identified 123, 102, and 78 differentially expressed (DE) genes between high- and low-ADG animals of Angus, Charolais, and KC populations, respectively. For DMI, 108, 180, and 156 DE genes were identified between high- and low-DMI from Angus, Charolais, and KC populations, respectively, while for MWT, 80, 82, and 84 genes were differentially expressed between high- and low-MWT animals in Angus, Charolais, and KC populations, respectively. The identified DE genes were largely breed specific (81.7% for ADG, 82.7% for DMI, and 83% for MWT), but were largely involved in the same biological functions across the breeds. Among the most enriched biological functions included metabolism of major nutrients (lipids, carbohydrates, amino acids, vitamins, and minerals), small molecule biochemistry, cellular movement, cell morphology, and cell-to-cell signaling and interaction. Notably, we identified multiple DE genes that are involved in cholesterol biosynthesis, and immune response pathways for the 3 studied traits. Thus, our findings present potential molecular genetic mechanisms and candidate genes that influence feed intake, growth, and MWT of beef cattle.
Keywords: average daily gain, beef cattle, daily dry matter intake, liver transcriptome profiling, metabolic body weight
Introduction
Animal growth rate and feed intake are very important traits to the beef industry as they both directly and indirectly affect the productivity, and thus profitability of the industry. To finish beef cattle for meat production, feedlot operators maintain their animals in the feedlot and incur costs such as labor, management, veterinary, feed, and feeding-related costs, with the latter accounting for up to 75% of the total production costs (Ahola and Hill, 2012). It is therefore of great interest to beef producers to raise faster growing animals with minimal or reduced daily feed consumption to optimize productivity of production systems profits (Hill and Ahola, 2012). It has been reported that growth rate measured as average daily gain (ADG) and feed intake measured as daily dry matter intake (DMI) are moderately to highly heritable traits, with estimated heritability of 0.35 to 0.59 (Schenkel et al., 2004; Nkrumah et al., 2007; Mao et al., 2013). Additionally, ADG and DMI together with metabolic body weight (MWT) are key component traits used in the calculation of animal feed efficiency termed as residual feed intake (RFI) in beef cattle (Koch et al., 1963). Furthermore, ADG and DMI are also important traits that can be included in beef cattle genetic selection and breeding programs to improve the efficiency of beef production (Amer et al., 2001). In recent years, several transcriptome studies on different tissues including the liver have been performed to identify molecular mechanisms of feed efficiency traits including RFI in different beef cattle breeds or populations (Chen et al., 2011; Al-Husseini et al., 2014; Alexandre et al., 2015; Paradis et al., 2015; Tizioto et al., 2015, 2016; Kong et al., 2016; Weber et al., 2016; Khansefid et al., 2017; Mukiibi et al., 2018). In our recent transcriptome study involving the liver tissue of beef steers from 3 breeds including Angus, Charolais, and Kinsella Composite (KC), we identified 253 genes as associated with RFI, of which up to 85% were breed specific (Mukiibi et al., 2018). In addition, lipid metabolism was found as a key metabolic function associated with RFI across the 3 studied populations (Mukiibi et al., 2018), and lipid synthesis and accumulation were predicted to be downregulated in the liver tissue of more efficient animals across the 3 breeds (Mukiibi et al., 2018). With respect to ADG and DMI, several tissues especially of the digestive system including the rumen (Kern et al., 2016; Reynolds et al., 2017), duodenum (Lindholm-Perry et al., 2016a), jejunum (Lindholm-Perry et al., 2016a; Foote et al., 2017), and ileum (Lindholm-Perry et al., 2016a) have been investigated to identify genes associated with body weight gain and feed intake in beef cattle. Genes associated to ADG and DMI have also been identified in the mesenteric adipose (Lindholm-Perry et al., 2017), spleen (Lindholm-Perry et al., 2016b), skeletal muscle (Keel et al., 2018), and liver (Tizioto et al., 2015; Zarek et al., 2017). However, each of these studies used a single breed or a breed population from their specific management and environmental conditions, making it difficult to directly infer common differentially expressed (DE) genes and biological functions associated with ADG and DMI across various beef cattle breeds. Therefore, in the current study, we analyzed RNAseq data of liver tissues of steers from our previous study for RFI (Mukiibi et al., 2018), with the aim to identify DE genes and metabolic or biological functions that underlie ADG, DMI, and MWT phenotypic differences of the animals from 3 Canadian beef populations including Angus, Charolais, and KC that were born, raised, and managed on the same ranch.
Materials and Methods
Animal Populations and Management
Populations and management of the animals used in this study have been described in our previous study by Mukiibi et al. (2018). Briefly, the animals were managed under the Canadian Council of Animal Care (CCAC) guidelines on the care and use of farm animals in research, teaching, and testing (CCAC, 2009), and the experimental procedures were approved by the University of Alberta Animal Care and Use Committee (AUP00000777). Beef steers from 3 beef cattle herds including purebred Angus, Charolais, as well as the KC population were used in this study. All animals were born, raised, and managed similarly at the Roy Berg Kinsella Research Ranch, University of Alberta, Canada. The purebred Angus and Charolais cows were bred by artificial insemination (AI) followed by natural service bulls and their pedigree information was maintained by the Canadian Angus or Charolais Association, respectively. The KC herd descended from crosses between Angus, Charolais, or Alberta Hybrid bulls and the University of Alberta’s hybrid dam line that was generated by crossing composite cattle lines of multiple beef breeds as described by Goonewardene et al. (2003). Commercial crossbred bulls have also been used in the KC herd since 2012. The animals used in this study were born during the months of April and May in 2014 and were castrated right after birth. The steer calves remained with their dams over the summer and grazed on a mixed tame grass pasture, then weaned at approximately 6 mo of age. They were transitioned to a backgrounding diet composed of 80% barley silage, 17% barley grain, and 3% rumensin pellet supplement, and then fed set-up diets with gradually decreasing barley silage and increasing barley grain proportions for 3 wk prior to introducing them to the finishing diet of 75% barley grain, 20% barley silage, and 5% rumensin pellet supplement (as fed basis).
GrowSafe Feedlot Test, Phenotype Measurement, and Calculations
In 2015, 50 Angus, 48 Charolais, and 158 KC steers were measured for individual feed intake between April and August using the GrowSafe system (GrowSafe Systems Ltd., Airdrie, AB, Canada). During the test period, animals were fed a finishing diet as described above. Mao et al. (2013) have previously described the process of measuring the individual animals’ daily feed intake using the GrowSafe automated system in details. Briefly, the DMI value of each steer was calculated by averaging the daily feed intake measurements of the animal recorded over the feedlot test period (70 to 73 d). The daily feed intake measurements were then standardized to 12 MJ ME/kg of dry matter based on the energy content of the diet. Initial body weight and ADG for each animal were obtained from a linear regression of serial body weight (BW) measurements that were recorded on 2 consecutive days at the beginning, at approximately 14-d intervals during the feedlot test, and on 2 consecutive days at the end of test. MWT was calculated as midpoint BW0.75, where midpoint BW was computed as the sum of initial BW of the animal and the product of its ADG multiplied by half the number of days under the feedlot test. The RFI value of each animal was computed as the difference between DMI of the animal and the expected feed intake of the animal based on the animal’s ADG and MWT. The expected feed intake of an individual animal was obtained from a linear regression of DMI on ADG and MWT of each breed population. Within each breed, the feedlot tested animals were initially sorted by their RFI phenotypes, and consequently 20 animals including 10 with the lowest RFI phenotypes and 10 with the highest RFI phenotypes were selected for the differential gene expression study of RFI as described in the previous study (Mukiibi et al., 2018). In the current study, we analyzed the RNAseq data with respect to RFI’s component traits ADG, DMI, and MWT.
Liver Tissue Collection
At the end of the feedlot test, all animals from each of the 3 breeds were slaughtered at Agriculture and Agri-Food Canada (AAFC) Lacombe Research and Development Centre (Lacombe, AB, Canada) between July and September of 2015. Steers were rendered fit for slaughter at a back fat thickness of ≥8 mm as predicted from a final ultrasound back fat measurement that was performed between the 12th and 13th ribs at the end of the GrowSafe feedlot test using an Aloka 500 diagnostic Realtime ultrasound machine with a 17-cm, 3.5-Mhz linear array transducer (Overseas Monitor Corporation Ltd., Richmond BC, Canada). In preparation for slaughter, steers of a single breed were randomly assigned to one of slaughter batches of 12 animals each. The animals had access to both feed and water until transportation for slaughter. Each batch was slaughtered on a day within a 7- to 14-d period at the same abattoir with the same procedure. The animal age at slaughter was recorded and the 3 beef steer populations had average slaughter ages of 494 ± 3, 518 ± 4, and 457 ± 4 d for Angus, Charolais, and KC, respectively. The liver sample of each animal was collected immediately after slaughter and the tissue was dissected from approximately the same location on the right lobe with the fibrous capsule removed. Samples were separately bagged, labeled, and were immediately flash frozen in liquid nitrogen. Subsequently, the liver samples were transported to the laboratory on dry ice within 6 h, and then stored at −80 °C until RNA extraction.
RNA Isolation and Purification
From the frozen liver samples, a total of 60 samples (20 from each breed) were selected for total RNA extraction based on their RFI values (i.e., 10 steers with high and 10 with low RFI values) as described previously (Mukiibi et al., 2018). The frozen liver tissue of each steer was pulverized into fine powder in liquid nitrogen with a pre-chilled mortar and pestle on dry ice. Total RNA was then extracted from 10 mg of the pulverized tissue using a Qiagen RNeasy Plus Universal Kit (Qiagen, Toronto, ON, Canada) and further purified using a Zymo RNA Clean and Concentrator (Zymo, Irvine, CA). RNA was quantified using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and was deemed acceptable if its absorbance (A260/280) was between 1.8 and 2.0. RNA integrity was confirmed using a TapeStation-Agilent instrument (Agilent Technologies, Mississauga, ON, Canada), and the RNA integrity number values for all samples were higher than 8.
cDNA Library Preparation and Sequencing
Preparation of cDNA libraries and subsequent next-generation sequencing of each of the 60 libraries were also described previously (Mukiibi et al., 2018). Construction of cDNA libraries and sequencing were performed at the Clinical Genomics Centre (Toronto, ON, Canada) using the Illumina TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA), where mRNA was purified and enriched from 1 µg of each of the total RNA samples using oligo-dT attached magnetic beads, and then fragmented through elevated heating to produce mRNA fragments of length 120 to 200 bp and a median of 150 bp. Thereafter, the first strand of the cDNA was synthesized using SuperScript II Reverse Transcriptase enzyme (Thermo Fisher Scientific, San Jose, CA) and the second strand was synthesized using the DNA Polymerase I and RNase H enzymes (Illumina). The cDNA libraries were validated using gel electrophoresis to confirm that the fragment size was 150 bp (on average) and concentration was on average 25 ng/µL per sample. Unique oligonucleotide adapters were added to the cDNA of each sample to allow for multiplexing. Of the prepared cDNA sample libraries, 48 (all Angus, all KC, and 8 Charolais) samples were single end sequenced (100 bp) under the high output run mode of the Illumina Hiseq 2500 System on 8 flow cell lanes. The other 12 Charolais samples were sequenced under the rapid run mode of the same sequencing equipment. All sequence and phenotype data used in this study have been submitted to the Gene Expression Omnibus (GEO) repository under the accession number GSE107477.
RNAseq Data Bioinformatic Analyses
Raw sequence data for each sample was assessed for sequencing quality using FASTQC software (version 0.11.5) with default parameters (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed in July 2017) (Andrews, 2010). Tophat2 (version 2.1.1) RNA-seq mapper was used to align and map the reads to the bovine reference genome UMD3.1 using default single-end read alignment parameters (Kim et al., 2013). Reads that were uniquely aligned to each gene annotated in the bovine gene transfer format (GTF) file (ftp://ftp.ensembl.org/pub/release-89/gtf/bos_taurus/Bos_taurus.UMD3.1.89.gtf.gz, accessed in July 2017) were counted using HTSeq-count package (Anders et al., 2015) with default parameters to generate read count tables that were further used for differential gene expression statistical analyses.
Differential Gene Expression Analyses
The 20 animals with the RNAseq data from each of the 3 cattle populations were sorted independently by their ADG, DMI, or MWT phenotypic values. Subsequently, out of the 20 samples, 12 samples within each breed with extreme phenotypic values (n = 6 high and n = 6 low) for each trait were then analyzed for differential gene expression. The gene count tables generated by HTSeq-count, the bovine gene features annotation file (Release 89) downloaded from Ensembl Biomart, and the sample information file were used for differential gene expression statistical analyses using the R Bioconductor package edgeR (Robinson et al., 2010; Anders et al., 2013). Genes within each breed that had less than 1 count per million (CPM) of mapped reads in at least 6 samples (half of the analyzed samples) were filtered out from the analyses as proposed by Anders et al. (2013). Counts of the remaining genes were normalized using the trimmed mean M values (TMM) method (Robinson and Oshlack, 2010), to account for the technical variations between samples due to RNA extraction, cDNA library construction, and differences in library sequencing depths of genes (Robinson and Oshlack, 2010). The normalized counts were modeled using a generalized linear model, and the differential gene expression between high- and low-phenotype groups of each trait within a breed was tested using a likelihood ratio test under assumption of a negative binomial distribution with the trait group as a fixed effect. For Charolais, sequencing run mode, either rapid or high output, was also included in the model to account for the difference in the sequencing modes. Genes were considered significantly DE between the trait groups at a threshold of Benjamin–Hochberg’s false discovery rate (FDR) <0.05 and fold change (FC) of >1.5.
Functional Enrichment Analysis
Functional enrichment analysis of the DE genes for each trait within a breed was performed using Ingenuity Pathway Analysis (IPA) software (Redwood City, CA; www.qiagen.com/ingenuity), with Ensembl bovine gene IDs and log2FC of the DE genes as input data. To increase the mapping rate to the IPA database, the DE bovine genes that did not map to the IPA database had their Ensembl IDs replaced by their respective human orthologue Ensembl gene IDs. The core analysis in IPA was performed on the mapped genes to identify significantly enriched biological functions, canonical pathways, and upstream regulators. Molecular and cellular functions (biological functions), canonical pathways, and upstream regulators were considered significantly enriched if the overlap comparison test (Fisher’s exact test) between the input DE gene list and the IPA Knowledge base database for that given biological function had a P-value less than 0.05. Activation or deactivation of a specific enriched metabolic process, pathway or gene expression regulator was defined by the Z-score (Krämer et al., 2014) that was calculated based on the log2FCs of the overlapping DE genes involved in a process or canonical pathway, where a negative or a positive score indicated deactivation or activation of a process, respectively.
Results
Phenotypic Differences Between Animal Groups
For ADG, the steer groups of high-ADG and low-ADG within all the 3 studied breeds were significantly different from each other at P <0.0042 with Bonferroni correction of 12 multiple tests at α <0.05 (Supplementary Table S1). There were no significant differences between high- and low-ADG steers groups across breeds for all the other phenotypic traits except for final ultrasound rib eye area for the KC steers.
For DMI, steers from low-DMI group significantly consumed less feed per day as compared to those from the high-DMI group within each breed (P < 0.0042) (Supplementary Table S2). As expected, low-DMI steers had significantly lower RFI than high-DMI animals in Charolais and KC (P < 0.0042), and for Angus, low-DMI steers also had lower RFI than their high-DMI counterparts although the difference did not reach the significance level of P < 0.0042. When the steer groups in each breed were compared for the other production phenotypes, no significant difference was observed between the high- and low-DMI animals except for MWT, for which the low-DMI steers showed lower MWT than the high-DMI steers and the difference reached the significance level of P <0.0042 for the KC population.
For MWT, our results showed that animals in the high-MWT group within each of the studied populations had significantly (P < 0.0042) higher MWT than those in the low-MWT group (Supplementary Table S3). It is observed that animals with high MWTs on average also had significantly (P < 0.0042) higher hot carcass weight (HCW) than those with lower MWT. For Angus, animals with low-MWT consumed significantly less feed per day as compared to the high-MWT animals. All the other phenotypes were not significantly different between the MWT groups across the 3 breeds.
Sequencing and Alignment Quality Assessment
High-throughput sequencing on average generated more than 32, 40, and 29 million raw single-end sequence reads per cDNA library from the Angus, Charolais, and KC animals respectively (Table 1). FASTQC sequence data quality assessment results showed that the sequence reads were of high quality with the reads having an average length of 101 bp and an average Phred quality score of more than 36. For alignment to the bovine reference genome, we obtained a high unique feature alignment of approximately 87% reads per sample (Table 1).
Table 1.
Averages of the sequencing quality and alignment assessment parameters for Angus, Charolais, and Kinsella Composite (KC) animals
| Angus (SD1) | Charolais (SD1) | KC (SD1) | |
|---|---|---|---|
| Total number of reads | 32,419,572 (2,527,134) | 40,796,790 (8,826,642) | 29,571,035 (5,730,204) |
| Uniquely aligned reads | 28,388,072 (2,394,131) | 35,584,367 (8,224,313) | 25,680,361 (4,964,214) |
| Average Phred Score | 35.6 (0.23) | 35.6 (0.60) | 37.8 (0.18) |
| Uniquely aligned reads (%) | 87.5 (1.12) | 87.0 (2.01) | 86.9 (0.84) |
1SD = standard deviation.
Differential Gene Expression Between ADG Divergent Steer Groups
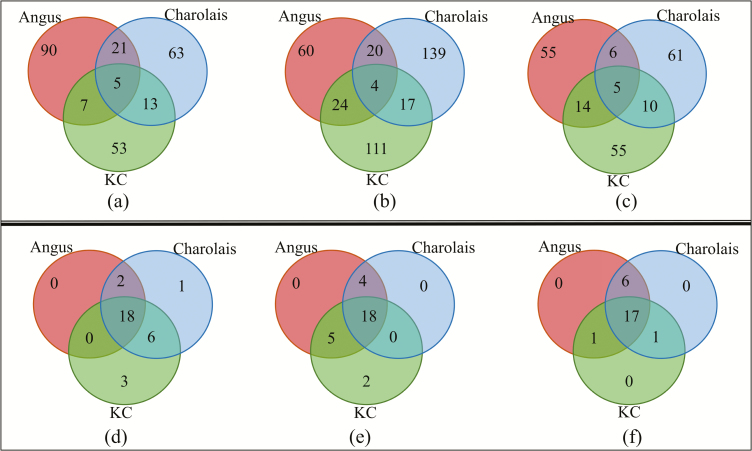
Of the 24,616 annotated bovine genes, 11,849, 11,923, and 11,809 were found to be of adequate expression level (i.e., CPM in at least 6 samples > 1) for differential liver gene expression analyses in ADG divergent steers from Angus, Charolais, and KC populations, respectively. For Angus, 123 DE genes were identified between the ADG divergent steers, of which 74 genes were upregulated and 49 genes downregulated in fast-growing (high-ADG group) animals. For Charolais, we identified 102 DE genes for ADG with 39 and 63 DE genes that were up- and downregulated, respectively, in the high-ADG steers. For KC, 78 genes showed significant DE between the high- and low-ADG steers, with 23 and 55 of these genes, respectively, up- and downregulated in the high-ADG steers. Based on FDR, the 40 topmost significantly DE genes which code for characterized proteins for each breed are presented in Table 2. The full list of all DE genes identified as associated with ADG for each breed are provided in the Supplementary File 1. Most of the DE genes (81.7%) were breed specific; however, a sizable number of DE genes were shared at least between 2 breeds (Fig. 1a). Five DE genes including SLC17A9, CXCL3, IFI27, JSP.1, and ENSBTAG00000003492 were shared among 3 breeds, with SLC17A9, CXCL3, and IFI27 showing consistent direction of expression in fast-growing steers across the 3 studied populations as presented in Supplementary Figure S1.
Table 2.
Top 40 significantly (by FDR) differentially expressed genes of characterized proteins between high- and low-ADG steers from Angus, Charolais, and Kinsella Composite populations
| Angus | Charolais | Kinsella Composite | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | log2FC1 | FDR2 | Gene | Log2FC1 | FDR2 | Gene | log2FC1 | FDR2 |
| TMEM45A | −3.87 | 4.69E-76 | TMEM45A | −1.81 | 2.08E-19 | IFI27 | −2.69 | 9.87E-39 |
| SERPINA3 | 3.32 | 1.74E-55 | HOPX | −1.63 | 5.59E-16 | LPIN1 | −2.54 | 1.48E-34 |
| GPX3 | 2.94 | 1.75E-53 | AKR1B15 | 1.59 | 1.66E-14 | IFI6 | −1.67 | 5.27E-15 |
| AKR1B15 | 2.68 | 2.33E-38 | TNC | −1.73 | 2.20E-13 | SERPINA3 | 1.56 | 1.01E-14 |
| GPNMB | 1.85 | 3.79E-21 | HLA-DQB1 | −1.86 | 6.68E-13 | ISG15 | −1.59 | 8.76E-14 |
| HP | 2.43 | 8.42E-18 | GPC3 | −1.64 | 3.17E-12 | HBB | −1.49 | 8.54E-13 |
| S100A2 | 1.58 | 1.09E-15 | KEL | 1.53 | 1.19E-11 | HERC6 | −1.44 | 2.45E-12 |
| SERPINA3 | 1.95 | 7.74E-15 | DDO | −1.90 | 2.07E-10 | GNMT | −1.42 | 2.57E-12 |
| HOPX | 1.43 | 1.32E-14 | GPX3 | −2.20 | 2.72E-10 | GADD45G | −1.35 | 6.14E-11 |
| IFI6 | −1.47 | 1.96E-13 | SERPINA3 | 2.45 | 3.14E-10 | SLC5A8 | 1.75 | 9.76E-10 |
| UGT2B7 | −1.35 | 4.93E-13 | AC108941.2 | −1.55 | 3.09E-09 | CES1 | 1.24 | 5.26E-09 |
| IFI27 | −1.42 | 2.42E-12 | IGLV2-18 | −1.60 | 3.17E-08 | SERPINI2 | 1.29 | 3.15E-08 |
| GPC3 | −1.37 | 1.83E-11 | CYP2B6 | 1.29 | 4.48E-08 | CYP7A1 | 1.44 | 4.51E-08 |
| HMGCS1 | −1.30 | 2.50E-10 | SLC25A45 | 1.80 | 1.42E-07 | TSKU | −1.24 | 1.43E-07 |
| SECTM1 | 1.19 | 4.95E-10 | SERPINA3 | −1.61 | 1.97E-07 | IFIT1 | −1.38 | 1.43E-07 |
| SULT2A1 | 1.18 | 7.65E-10 | IGLV2-18 | −1.44 | 3.27E-07 | UHRF1 | −1.60 | 2.15E-07 |
| SPIDR | 1.20 | 7.83E-10 | SCD | −1.26 | 3.28E-07 | BOLA-DQA5 | −1.29 | 2.72E-06 |
| IGHG1 | 1.17 | 2.32E-08 | S100A10 | −1.14 | 4.06E-07 | NOCT | −1.10 | 3.39E-06 |
| ECEL1 | 1.10 | 5.27E-08 | STS | −1.12 | 7.69E-07 | WFDC2 | −1.03 | 4.29E-06 |
| JAKMIP2 | 1.44 | 9.89E-08 | UHRF1 | −1.47 | 1.48E-06 | ZNF385B | 1.19 | 4.29E-06 |
| CYP51A1 | −1.12 | 1.57E-07 | CCDC80 | −1.41 | 4.63E-06 | MX2 | −1.26 | 6.20E-06 |
| AIF1L | 1.02 | 2.62E-07 | SLC13A2 | −1.21 | 5.92E-06 | RSAD2 | −1.01 | 6.91E-06 |
| AKR1C1 | 1.22 | 2.91E-07 | EGR1 | −1.33 | 6.60E-06 | GSTM2 | −1.07 | 9.32E-06 |
| CCL24 | 1.35 | 4.38E-07 | CRYAB | −1.04 | 7.08E-06 | C12orf45 | −1.01 | 1.15E-05 |
| DLK1 | −1.28 | 4.54E-07 | HIST1H2BI | −0.99 | 7.11E-06 | IGFBP1 | 0.97 | 1.50E-05 |
| VCAM1 | −1.28 | 2.21E-06 | GNMT | −0.90 | 3.75E-05 | EXTL1 | 0.96 | 2.94E-05 |
| MT1G | 1.07 | 6.21E-06 | EPCAM | −1.05 | 8.16E-05 | ALAS1 | −1.00 | 3.46E-05 |
| SQLE | −0.99 | 1.05E-05 | IFI27 | −0.90 | 9.13E-05 | STS | −0.95 | 4.15E-05 |
| IL1R2 | 1.09 | 2.69E-05 | ACSS2 | −0.88 | 1.24E-04 | PLEKHG6 | 1.11 | 1.36E-04 |
| SERPINE1 | 0.88 | 4.03E-05 | SLC17A9 | 0.92 | 1.31E-04 | LURAP1L | −1.02 | 1.65E-04 |
| SLC13A2 | 0.91 | 5.28E-05 | SCD | −1.29 | 1.37E-04 | PRAP1 | −0.93 | 1.92E-04 |
| SERPINI2 | −1.21 | 5.40E-05 | THNSL2 | 0.91 | 2.19E-04 | SCD | −0.95 | 2.95E-04 |
| TNFRSF10A | −1.21 | 7.39E-05 | RCL1 | −0.88 | 2.30E-04 | FKBP5 | −0.89 | 5.62E-04 |
| RAP1GAP | 0.96 | 7.48E-05 | MID1IP1 | −0.85 | 2.42E-04 | TAT | −1.02 | 6.56E-04 |
| OXER1 | −0.84 | 8.35E-05 | MBOAT2 | −1.24 | 2.42E-04 | FGF21 | 0.95 | 6.85E-04 |
| SLC1A2 | −0.88 | 1.25E-04 | SOAT2 | 1.30 | 3.09E-04 | ACE2 | −0.84 | 7.03E-04 |
| MAMDC2 | −1.08 | 1.25E-04 | FOXA3 | −0.84 | 3.13E-04 | MX1 | −0.87 | 7.98E-04 |
| DENND2A | 0.82 | 1.50E-04 | MAMDC2 | −1.09 | 4.79E-04 | IFI44L | −0.83 | 8.80E-04 |
| ROS1 | −1.09 | 1.83E-04 | REC8 | −0.84 | 7.97E-04 | WFS1 | 0.81 | 1.24E-03 |
| CLBA1 | 1.15 | 2.55E-04 | ISG15 | −0.78 | 9.07E-04 | ALOX15B | 1.15 | 1.64E-03 |
1log2FC = log2(fold change of a gene in high-average daily gain [ADG] animals with reference to low-ADG animals).
2FDR = false discovery rate adjusted P-value.
Figure 1.
Venn diagrams showing differentially expressed genes overlap among Angus, Charolais, and Kinsella Composite (KC) for (a) average daily gain (ADG), (b) daily dry matter intake (DMI), and (c) metabolic body weight (MWT). Venn diagrams showing significant enriched biological functions overlap among breeds for (d) ADG, (e) DMI, and (f) MWT.
Differential Gene Expression Between DMI Divergent Steer Groups
For DMI, 11,871, 11,961, and 11,793 genes were expressed sufficiently for differential gene expression in Angus, Charolais, and KC steers, respectively. For Angus, we identified 108 DE genes, with 57 genes upregulated and 51 genes downregulated in the low-DMI steers. Among the Charolais steers, 180 genes (120 upregulated and 60 downregulated in the low-DMI animals) were differentially expressed. For KC, 156 genes (107 upregulated and 49 downregulated in the low-DMI steers) were differentially expressed. The 40 most significant protein-coding DE genes by FDR are presented in Table 3, and all identified DE genes associated with DMI for each breed are provided in the Supplementary File 2. Also for DMI, most (82.7%) of the identified DE genes were breed specific, with only 4 DE genes including IFI27, ENSBTAG00000003492, ENSBTAG00000024700, and ENSBTAG00000047029 common among the 3 studied breeds, and a considerable number of DE genes (17 to 24 DE genes) were uniquely shared between breed pairs as shown in Fig. 1b. However, none of the common DE genes showed consistent expression direction across the 3 breeds (Supplementary Figure S2).
Table 3.
Top 40 significantly (by FDR) differentially expressed genes of characterized proteins between high- and low-DMI steers from Angus, Charolais, and Kinsella Composite populations
| Angus | Charolais | Kinsella Composite | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | log2FC1 | FDR2 | Gene | log2FC1 | FDR2 | Gene | log2FC1 | FDR2 |
| IFIT1 | −2.49 | 1.38E-34 | SLC22A2 | 4.47 | 1.92E-47 | IFI27 | 3.13 | 9.17E-50 |
| GPX3 | −2.38 | 1.42E-30 | REC8 | −2.61 | 1.22E-37 | CXCL9 | 2.82 | 2.83E-44 |
| GPNMB | −2.23 | 4.16E-27 | EGR1 | 2.38 | 3.46E-31 | GBP3 | 2.99 | 3.15E-41 |
| HBB | 2.97 | 3.93E-26 | IGLC1 | 2.48 | 3.46E-31 | IFI6 | 2.24 | 1.09E-27 |
| SERPINA3 | −1.90 | 1.24E-20 | IGHG1 | 2.31 | 3.95E-28 | CYP2B6 | −2.17 | 1.62E-27 |
| ISG15 | −1.83 | 1.04E-17 | SERPINA3 | 2.47 | 3.92E-26 | HERC6 | 2.07 | 1.07E-25 |
| SFRP2 | 1.72 | 9.15E-17 | CCDC80 | 2.25 | 1.56E-23 | IFIT1 | 2.35 | 3.23E-25 |
| HERC6 | −1.44 | 1.18E-11 | SFRP1 | −1.79 | 1.00E-18 | ISG15 | 2.03 | 3.28E-23 |
| DDO | −1.63 | 3.99E-10 | GPX3 | 2.08 | 6.20E-17 | CXCL10 | 2.22 | 1.06E-22 |
| FKBP5 | 1.37 | 7.10E-10 | BOLA-DQB | −2.11 | 5.26E-16 | TMEM45A | −2.84 | 1.63E-22 |
| RSAD2 | −1.35 | 1.01E-09 | CLDN15 | 1.61 | 4.46E-15 | MX2 | 2.07 | 4.80E-20 |
| SDS | 1.36 | 1.82E-09 | ABCG8 | 1.54 | 2.98E-14 | AK4 | 1.79 | 2.11E-19 |
| APOA4 | 1.29 | 6.27E-09 | CES1 | −1.52 | 2.20E-12 | CD274 | 2.44 | 7.13E-16 |
| MBOAT2 | −1.28 | 7.41E-09 | S100A10 | 1.41 | 1.49E-11 | SERPINA3 | −1.60 | 2.60E-15 |
| CDHR5 | −1.25 | 7.65E-09 | CYP11A1 | 1.41 | 2.03E-11 | AKR1B15 | −1.75 | 7.10E-14 |
| MX1 | −1.26 | 1.44E-08 | NNAT | 1.63 | 8.45E-11 | OAS1 | 1.44 | 1.49E-12 |
| IL20RA | −1.32 | 4.02E-08 | FGF21 | 1.43 | 6.40E-10 | GBP7 | 1.71 | 2.55E-11 |
| SLC2A5 | 1.28 | 1.40E-07 | AC108941.2 | 1.50 | 8.82E-10 | RSAD2 | 1.36 | 2.55E-11 |
| STEAP4 | 1.19 | 2.94E-07 | CYR61 | 1.32 | 1.15E-09 | MKI67 | 1.45 | 4.35E-11 |
| GNMT | 1.10 | 1.18E-06 | PRAP1 | 1.26 | 1.73E-09 | PSMB9 | 1.46 | 7.64E-11 |
| MYOM1 | 1.21 | 2.78E-06 | CUX2 | 1.41 | 2.73E-09 | KYAT1 | −1.21 | 1.03E-08 |
| GPC3 | 1.06 | 7.85E-06 | CARNS1 | 1.27 | 3.43E-09 | IFI44L | 1.15 | 1.01E-07 |
| SERPINA3 | −1.31 | 1.98E-05 | TNC | 1.32 | 4.57E-09 | RTP4 | 1.17 | 1.01E-07 |
| HP | −1.52 | 2.27E-05 | SLC7A2 | −1.26 | 6.17E-09 | ATP6V1C2 | −1.66 | 2.12E-07 |
| LPIN1 | 1.03 | 2.27E-05 | HMGCS1 | −1.26 | 2.55E-08 | RBP5 | −1.12 | 3.20E-07 |
| SECTM1 | 1.02 | 2.28E-05 | IL1R2 | 1.48 | 3.82E-08 | GBP3 | 1.31 | 5.23E-07 |
| SCD | −0.99 | 3.23E-05 | LPIN1 | −1.23 | 4.91E-08 | HAPLN3 | −1.14 | 9.08E-07 |
| NR1D1 | −0.96 | 3.32E-05 | CDH17 | 1.13 | 1.82E-07 | CTGF | 1.49 | 1.29E-06 |
| CREM | 0.96 | 3.57E-05 | HP | 1.61 | 3.99E-07 | PSMB8 | 1.04 | 2.77E-06 |
| PYROXD2 | 0.98 | 4.35E-05 | IGLV2-18 | 1.29 | 6.43E-07 | TAP1 | 1.02 | 4.52E-06 |
| RTP4 | −0.93 | 8.43E-05 | ABCG5 | 1.16 | 6.55E-07 | FOXS1 | 1.45 | 7.19E-06 |
| SCD | −0.95 | 9.57E-05 | SLC4A4 | −1.08 | 9.88E-07 | PIM1 | −1.00 | 8.79E-06 |
| RNF125 | 0.96 | 1.20E-04 | IFI27 | −1.13 | 1.19E-06 | WFS1 | −0.99 | 9.48E-06 |
| IFI44L | −0.91 | 2.21E-04 | CLDN4 | 1.47 | 2.00E-06 | IFIT2 | 1.41 | 1.53E-05 |
| NOCT | 0.92 | 2.59E-04 | FOS | 1.21 | 2.91E-06 | NLRC5 | 1.05 | 2.76E-05 |
| CYP7A1 | −0.88 | 3.43E-04 | HOOK1 | −1.04 | 2.91E-06 | UBA7 | 0.97 | 2.77E-05 |
| AKR1B15 | −0.94 | 4.51E-04 | SQLE | −1.09 | 3.37E-06 | RAB20 | −0.96 | 3.20E-05 |
| DDIT4 | 0.91 | 4.80E-04 | STRIP2 | −1.03 | 3.92E-06 | CITED4 | 0.96 | 3.20E-05 |
| SPTB | −0.83 | 9.02E-04 | IGLC1 | 1.15 | 4.94E-06 | RRM2 | 1.36 | 3.20E-05 |
| CKAP4 | −0.83 | 9.42E-04 | DLK1 | 1.19 | 5.02E-06 | IL20RA | 1.13 | 3.33E-05 |
1log2FC = log2(fold change of a gene in low-daily dry matter intake [DMI] animals with reference to high-DMI animals).
2FDR = false discovery rate adjusted P-value.
Differential Gene Expression Between MWT Divergent Steer Groups
For MWT, 11,843, 11,908, and 11,774 genes were adequately expressed and hence were considered for analysis for the Angus, Charolais, and KC steers, respectively. Of these expressed genes, 80 (46 upregulated and 34 downregulated in the low-MWT steers), 82 (61 upregulated and 21 downregulated in the low-MWT steers), and 84 (44 upregulated and 40 downregulated in the low-MWT steers) genes were differentially expressed in Angus, Charolais, and KC steers, respectively. The 40 most significant DE genes (coding for characterized proteins) by FDR are presented in Table 4 and all identified DE genes associated with MWT in each of the studied breeds are provided in the Supplementary File 3. Comparison of the identified DE genes across breeds showed a similar trend as for ADG and DMI with most of the DE genes (83%) being breed specific and only 5 DE genes including MT1E, CTGF, PRAP1, TMEM45A, and CYP2B6 were identified as common across the 3 breeds (Fig. 1c). Two of these shared genes (i.e., MT1E and CTGF) showed consistent expression across the 3 populations with MT1E (ENSBTAG00000038706) being downregulated and CTGF upregulated in the low-MWT animals as shown in Supplementary Figure S3.
Table 4.
Top 40 significantly (by FDR) differentially expressed genes of characterized proteins between high- and low-MWT steers from Angus, Charolais, and Kinsella Composite populations
| Angus | Charolais | Kinsella Composite | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | logFC1 | FDR2 | Gene | logFC1 | FDR2 | Gene | logFC1 | FDR2 |
| GPX3 | −2.55 | 5.66E-37 | SERPINA3 | 4.94 | 1.74E-99 | TMEM45A | −5.23 | 2.12E-119 |
| IFI27 | 2.50 | 1.14E-35 | CYP2B6 | −1.57 | 2.13E-17 | HOPX | −1.76 | 1.53E-17 |
| SERPINA3 | −2.45 | 9.25E-32 | BOLA-DQB | −1.83 | 1.04E-13 | SERPINA3 | −1.73 | 1.04E-16 |
| IFI6 | 1.98 | 5.95E-22 | IGLC1 | −1.44 | 5.06E-13 | IFI27 | 1.64 | 3.16E-15 |
| SERPINA3 | −2.51 | 4.61E-21 | TMEM45A | −1.25 | 2.49E-11 | IFI6 | 1.54 | 5.51E-14 |
| GPNMB | −1.57 | 1.91E-14 | ECEL1 | 1.17 | 6.28E-10 | GPX3 | −1.36 | 3.81E-11 |
| SERPINI2 | 1.73 | 5.62E-13 | AC108941.2 | 1.16 | 6.19E-08 | CYP2B6 | −1.36 | 1.02E-10 |
| HBB | 1.99 | 5.62E-13 | KEL | 1.20 | 6.19E-08 | FBLN2 | −1.36 | 6.92E-09 |
| AKR1B15 | −1.58 | 1.39E-12 | FBLN2 | 1.17 | 6.19E-08 | WFS1 | −1.21 | 1.34E-08 |
| AC108941.2 | 1.37 | 1.84E-11 | HOPX | −1.02 | 2.74E-07 | CTGF | 1.53 | 5.43E-08 |
| SDS | 1.37 | 5.82E-11 | FGF21 | −1.12 | 7.30E-07 | HBB | 1.32 | 1.72E-07 |
| AIF1L | −1.27 | 6.18E-10 | S100A10 | 0.97 | 2.04E-06 | CXCL9 | 1.29 | 5.13E-07 |
| TMEM45A | 1.26 | 9.39E-09 | PLA2G2D | 1.41 | 2.12E-06 | UGT2B7 | −1.08 | 7.53E-07 |
| SULT2A1 | −1.16 | 5.72E-08 | PLTP | 0.92 | 7.78E-06 | CYP3A7-CYP3A51P | −1.19 | 7.53E-07 |
| CKAP4 | −1.10 | 2.23E-07 | IL1B | 1.11 | 8.46E-06 | ATP5MGL | −1.47 | 9.00E-07 |
| RAP1GAP | −1.22 | 1.55E-06 | KCTD12 | 1.02 | 1.93E-05 | PIM1 | −1.06 | 1.74E-06 |
| NUF2 | 1.44 | 1.55E-06 | CSF2RB | 0.93 | 4.06E-05 | IGHG1 | 1.06 | 2.99E-06 |
| TNC | 1.08 | 1.55E-06 | SFRP1 | −0.87 | 4.19E-05 | ZNF385B | −1.21 | 4.85E-06 |
| CDHR5 | −1.00 | 5.21E-06 | MARCO | 0.85 | 6.56E-05 | IGLC1 | 1.39 | 9.42E-06 |
| SLC13A2 | −1.11 | 6.80E-06 | PRAP1 | −1.00 | 8.72E-05 | IGLC1 | 1.00 | 2.61E-05 |
| ROS1 | 1.32 | 9.40E-06 | FAM47E | −1.01 | 1.05E-04 | BoLA-DQB1 | 0.97 | 3.59E-05 |
| HP | −1.57 | 1.81E-05 | HLA-DQB1 | 1.05 | 1.25E-04 | CES1 | 0.94 | 4.26E-05 |
| AK4 | 0.92 | 6.05E-05 | CSF1R | 0.83 | 1.84E-04 | SERPINI2 | 1.08 | 4.26E-05 |
| DENND2A | −0.89 | 1.12E-04 | UCP2 | 0.80 | 3.12E-04 | ASIP | 1.00 | 9.88E-05 |
| SFRP2 | 0.89 | 1.16E-04 | ADGRE1 | 0.79 | 3.31E-04 | UGT2B7 | 0.91 | 1.57E-04 |
| SDCBP2 | 1.20 | 2.19E-04 | PTN | 0.91 | 3.31E-04 | ACE2 | 0.88 | 1.79E-04 |
| CYP2B6 | 0.88 | 4.01E-04 | IGHA1 | −0.89 | 5.98E-04 | HP | −1.34 | 1.89E-04 |
| PLCD4 | −0.89 | 4.20E-04 | SLC13A2 | 1.02 | 1.05E-03 | AKR1C1 | 1.01 | 2.44E-04 |
| FAM13A | 0.99 | 4.81E-04 | PDK4 | 0.79 | 1.15E-03 | REEP5 | −0.86 | 3.59E-04 |
| GNMT | 0.82 | 6.79E-04 | PTGS1 | 0.75 | 1.23E-03 | TGM2 | 0.85 | 4.61E-04 |
| CFH | 0.95 | 7.04E-04 | FADS1 | 0.91 | 1.25E-03 | CDH11 | −0.99 | 5.44E-04 |
| CYR61 | 0.94 | 1.04E-03 | SLC7A5 | 1.00 | 1.46E-03 | PRAP1 | 0.85 | 5.49E-04 |
| ABHD6 | 0.85 | 1.44E-03 | SOAT2 | 0.86 | 1.94E-03 | DDO | 0.99 | 6.82E-04 |
| HMGCR | −0.83 | 1.44E-03 | PPP1R3C | −0.74 | 2.00E-03 | CARNS1 | 0.88 | 1.00E-03 |
| GPC3 | 0.81 | 1.76E-03 | IGHG1 | −0.71 | 2.79E-03 | SLCO4A1 | −0.95 | 1.46E-03 |
| BICC1 | −0.96 | 2.27E-03 | LPIN1 | 0.72 | 2.96E-03 | RAB20 | −0.81 | 1.62E-03 |
| IGFBP2 | −0.80 | 2.49E-03 | IGLV2-18 | −0.95 | 2.96E-03 | RFLNA | −0.83 | 1.74E-03 |
| ISG15 | 0.76 | 3.08E-03 | RASL10A | 0.99 | 3.33E-03 | SDS | −0.81 | 1.91E-03 |
| PRAP1 | 0.79 | 3.30E-03 | TMEM176B | 0.71 | 4.09E-03 | ECEL1 | 0.88 | 2.09E-03 |
| DLK1 | 0.97 | 3.49E-03 | SMPDL3B | 0.81 | 4.75E-03 | CLDN15 | 0.78 | 2.09E-03 |
1log2FC = log2(fold change of a gene in low metabolic body weight [MWT] animals with reference to high-MWT animals).
2FDR = false discovery rate adjusted P-value.
Gene Expression Across Traits Within Breed
DE genes identified to be associated with ADG, DMI, and MWT were compared with the DE genes for RFI as reported by Mukiibi et al. (2018) within each studied population as shown in Supplementary Figure S4. Within each breed, the DE genes were largely trait specific with only 4 (HP, ENSBTAG00000047029, SERPINA3, and IFI27), 1 (ENSBTAG00000048094), and 2 (ENSBTAG00000022590 and ENSBTAG00000003492) DE genes shared across 4 traits in Angus, Charolais, and KC, respectively. However, there was some reasonable number of genes shared between pairs of the traits. For example, 46 genes were common between ADG and MWT, 31 genes shared between ADG and DMI, and 39 genes shared between ADG and RFI in Angus, Charolais, and KC steers, respectively.
Functional Enrichment for ADG-Associated DE Genes
For ADG, a total of 120, 102, and 78 DE genes were mapped to the IPA knowledgebase database for Angus, Charolais, and KC, respectively. These mapped DE genes were significantly (P < 0.05) involved in 20 molecular and cellular functions for Angus, 27 for Charolais, and 27 for KC. Of all the identified molecular and cellular functions, 18 (60%) were common to all the 3 breeds as shown in Fig. 1d. The most significantly enriched functions included cellular movement, lipid metabolism, small molecule biochemistry, vitamin and mineral metabolism, cell-to-cell signaling and interaction, molecular transport, amino acid metabolism, and carbohydrate metabolism (Supplementary Figures S5–S7). It is worth noting that lipid metabolism and small molecule biochemistry functions were among the top 5 enriched biological functions across the 3 breeds.
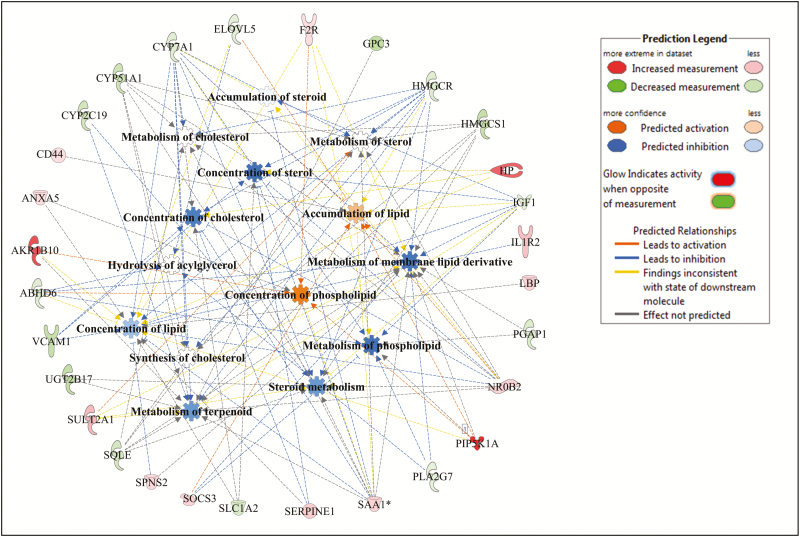
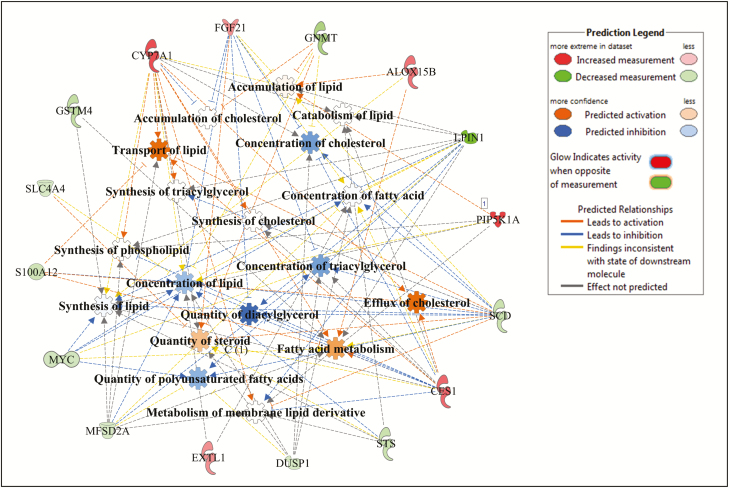
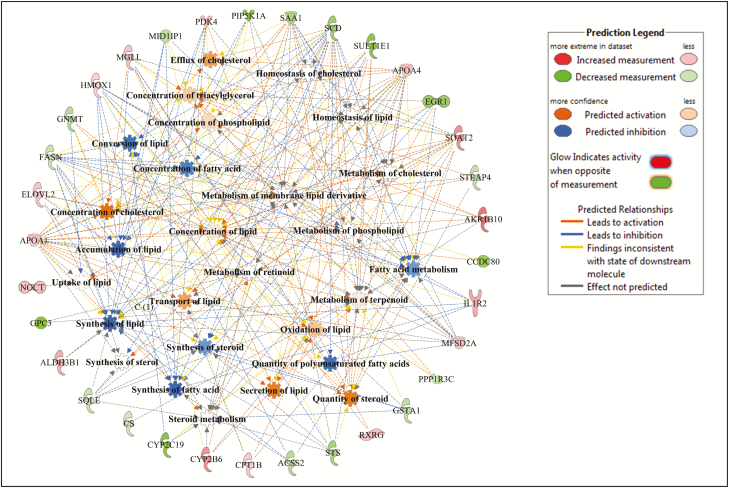
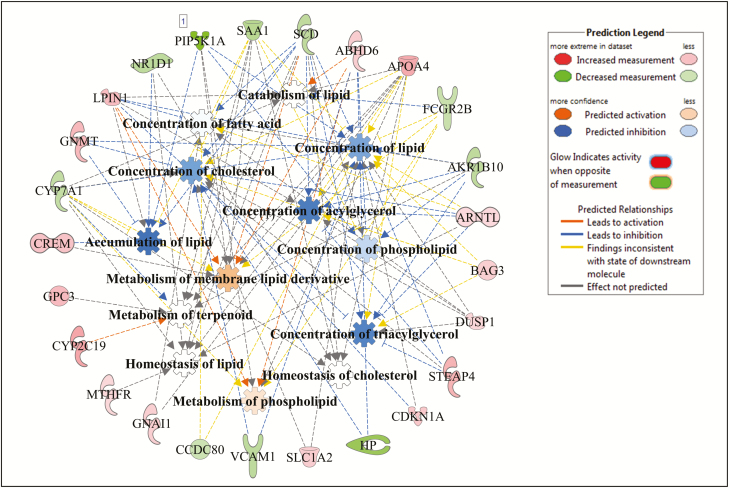
Within lipid metabolism, several metabolic processes related to lipid accumulation, lipid synthesis, lipid oxidation, and lipid transport were identified as enriched by the DE genes as shown in Figs. 2–4 for Angus, Charolais, and KC, respectively. Synthesis of lipids (e.g., steroids, fatty acids, and acylglycerol) was predicted to be downregulated (negative Z-scores) in the liver tissue of high-ADG animals from Charolais and KC steers (Figs. 3 and 4). Accumulation of lipid was predicted to be downregulated in Charolais (Fig. 3), while upregulated in Angus and KC high-ADG steers, as shown in Figs. 2 and 4, respectively. Transport of lipid and fatty acid oxidation were predicted as upregulated in both Charolais and KC high-ADG steers. Some of the key DE genes associated with lipid metabolism identified in the current study include CYP7A1, IGF1, SAA1, HMGCR, and NROB2 for Angus, SCD, FASN, APOA1, APOA4, SAA1, PDK4, and HMOX1 for Charolais, and SCD, LPIN1, FGF21, CYP7A1, and CES1 for KC. Lists of all DE genes involved in each of the 5 topmost enriched functions within each breed are provided in Supplementary Table S4.
Figure 2.
Lipid metabolism gene and molecular processes interaction network within lipid metabolism function as associated to average daily gain (ADG), and predicted activation or inhibition in high-ADG Angus steers.
Figure 4.
Lipid metabolism gene and molecular processes interaction network within lipid metabolism function as associated to average daily gain (ADG), and predicted activation or inhibition in high-ADG Kinsella Composite (KC) steers.
Figure 3.
Lipid metabolism gene and molecular processes interaction network within lipid metabolism function as associated to average daily gain (ADG), and predicted activation or inhibition in high-ADG Charolais steers.
Other than being among the topmost enriched molecular and cellular functions for KC, amino acid and carbohydrate metabolism were also enriched for both Angus and Charolais with important enriched underlying processes. In relation to amino acid metabolism, some of the enriched metabolic processes included transport of amino acids, synthesis of amino acids, and catabolism of amino acids. Top amino acid metabolism-related processes for each breed and the DE genes involved in these processes, activation/deactivation score, and overlap test P-values are presented in Supplementary Table S5. For carbohydrate metabolism, glucose uptake, and carbohydrate synthesis (gluconeogenesis), carbohydrate oxidation and transport were among the enriched metabolic processes as shown in Supplementary Table S6.
IPA also revealed several interesting enriched activated or deactivated pathways for the identified DE genes in relation to growth rate in the 3 studied breed populations, with the topmost enriched pathways for each breed shown in Table 5. In Angus, superpathway of cholesterol biosynthesis was the most significantly (P = 1.35E-05) enriched pathway involving 4 of the DE genes (SQLE, HMGCR, HMGCS1, and CYP51A1) and was predicted to be inactivated in the high-ADG steers with a Z-score of −2.00 (Table 5). Liver X receptor (LXR)/retinoid X receptor (RXR) and PXR/RXR activation pathways were the most significant pathways involving 7 (IL1R2, SCD, RXRG, APOA1, APOA4, FASN, and SAA1) and 5 (GSTM1, SCD, CYP7A1, IGFBP1, and ALAS1) DE genes for Charolais and KC, respectively (Table 5). Additionally, IPA identified several upstream gene expression regulators and their predicted activation or deactivation level in the liver tissue of the high-ADG animals across the 3 studied breeds. SREBF1 is a transcription factor that was predicted as the most significant (P = 9.41E-11) expression regulator in Angus and was shown to regulate expression of 14 (AK4, CYP51A1, CYP7A1, GPNMB, GPX3, HMGCR, HMGCS1, IFI30, IL1R2, NR0B2, OAT, SERPINA3, SERPINE1, and SQLE) of the identified DE genes in this breed (Supplementary Table S7A). For Charolais, the P450 oxidoreductase (POR) enzyme was the most significant (P = 1.86E-12) regulator, regulating expression of 13 DE genes (ACTG1, APOA4, CSAD, CYP2B6, ELOVL2, GADD45B, HMOX1, NOCT, PDK4, SCD, SDS, SERPINA3, and SQLE) (Supplementary Table S7A). For KC, interferon-beta (IFN-β) was the most significant (P = 5.63E-14) upstream regulator, predicted to regulate 12 DE genes (DUSP1, HLA-B, IFI44, IFI6, ISG15, MX1, MX2, MYC, OAS1, RSAD2, SLC16A6, and USP18) and to be inactivated in the high-ADG animals with a Z-score of −3.08 (Supplementary Table S7A).
Table 5.
Top enriched canonical pathways associated with growth rate, feed intake, and metabolic body weight in Angus, Charolais, and Kinsella Composite (KC) animals
| Trait_breed1 | Ingenuity Canonical Pathways | P-value | Ratio | Z−score2 | Molecules |
|---|---|---|---|---|---|
| ADG_ Angus | Superpathway of Cholesterol Biosynthesis | 1.35E-05 | 0.14 | −2.00 | SQLE, HMGCR, HMGCS1, CYP51A1 |
| Nicotine Degradation II | 2.40E-05 | 0.08 | −2.24 | UGT2B17, FMO2, INMT, CYP51A1, CYP2C19 | |
| LPS/IL-1 Mediated Inhibition of RXR Function | 2.40E-05 | 0.04 | 2.24 | IL1R2, NR0B2, FMO2, CYP7A1, LBP, HMGCS1, SULT2A1, CYP2C19 | |
| Acute Phase Response Signaling | 4.17E-05 | 0.04 | 0.82 | SOCS3, HP, SAA1, SOCS2, SERPINA3, LBP, SERPINE1 | |
| LXR/RXR Activation | 4.37E-05 | 0.05 | −1.00 | IL1R2, SAA1, CYP7A1, LBP, HMGCR, CYP51A1 | |
| ADG_Charolais | LXR/RXR Activation | 1.23E-06 | 0.06 | 0.45 | IL1R2, SCD, RXRG, APOA1, APOA4, FASN, SAA1 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 6.61E-05 | 0.03 | 0.00 | IL1R2, SULT1E1, CPT1B, ALDH3B1, GSTA1, CYP2B6, CYP2C19 | |
| PXR/RXR Activation | 2.09E-04 | 0.06 | NC | SCD, GSTA1, CYP2B6, CYP2C19 | |
| FXR/RXR Activation | 2.63E-04 | 0.04 | NC | APOA1, APOA4, FASN, SAA1, FOXA3 | |
| Glycine Betaine Degradation | 8.71E-04 | 0.20 | NC | SDS, SHMT2 | |
| ADG_KC | PXR/RXR Activation | 3.39E-06 | 0.08 | NC | GSTM1, SCD, CYP7A1, IGFBP1, ALAS1 |
| Interferon Signaling | 7.76E-06 | 0.11 | −2.00 | OAS1, MX1, IFI6, ISG15 | |
| LPS/IL-1 Mediated Inhibition of RXR Function | 1.38E-04 | 0.03 | GSTM1, IL36G, GSTM4, CYP7A1, ALAS1, SOD3 | ||
| 2-amino-3-carboxymuconate Semialdehyde Degradation to Glutaryl-CoA | 3.55E-03 | 1.00 | NC | ACMSD | |
| 4-hydroxybenzoate Biosynthesis | 3.55E-03 | 1.00 | NC | TAT | |
| DMI_ Angus | Pyrimidine Ribonucleotides Interconversion | 1.20E-03 | 0.07 | NC | NUDT5, CMPK2, AK8 |
| Pyrimidine Ribonucleotides De Novo Biosynthesis | 1.38E-03 | 0.07 | NC | NUDT5, CMPK2, AK8 | |
| LXR/RXR Activation | 2.69E-03 | 0.03 | NC | SCD, APOA4, SAA1, CYP7A1 | |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 3.39E-03 | 0.05 | NC | DHX58, IFIT2, ISG15 | |
| GADD45 Signaling | 3.63E-03 | 0.11 | NC | GADD45B, CDKN1A | |
| DMI_Charolais | LPS/IL-1 Mediated Inhibition of RXR Function | 2.63E-10 | 0.07 | 0.33 | IL1R2, ABCG8, GSTM1, ABCG5, SULT1E1, JUN, SULT1C4, NR0B2, CYP7A1, SLC27A6, IL1B, ALDH3B1, HMGCS1, ABCA1, CYP2C19 |
| Superpathway of Cholesterol Biosynthesis | 2.34E-06 | 0.18 | −2.24 | SQLE, PMVK, IDI1, HMGCR, HMGCS1 | |
| Mevalonate Pathway I | 2.45E-06 | 0.31 | −2.00 | PMVK, IDI1, HMGCR, HMGCS1 | |
| LXR/RXR Activation | 5.13E-06 | 0.07 | −1.34 | IL1R2, ABCG8, ABCG5, SAA1, CYP7A1, IL1B, HMGCR, ABCA1 | |
| Superpathway of Geranylgeranyl diphosphate Biosynthesis I (via Mevalonate) | 7.94E-06 | 0.24 | −2.00 | PMVK, IDI1, HMGCR, HMGCS1 | |
| DMI_KC | Interferon Signaling | 1.00E-10 | 0.22 | 2.83 | OAS1, IFI6, PSMB8, STAT1, TAP1, IRF1, IFITM1, ISG15 |
| Antigen Presentation Pathway | 6.31E-09 | 0.18 | NC | PSMB9, NLRC5, HLA-B, HLA-DQB1, PSMB8, HLA-DQA2, TAP1 | |
| Th1 Pathway | 3.80E-07 | 0.07 | 1.41 | NFIL3, CD3E, HLA-B, CD274, HLA-DQB1, HLA-DQA2, STAT1, CD3D, IRF1 | |
| Th1 and Th2 Activation Pathway | 5.25E-06 | 0.05 | NC | NFIL3, CD3E, HLA-B, CD274, HLA-DQB1, HLA-DQA2, STAT1, CD3D, IRF1 | |
| PKCθ Signaling in T Lymphocytes | 1.82E-05 | 0.05 | 2.45 | CACNA1I, RAC2, CACNG1, CD3E, HLA-B, HLA-DQB1, CD3D, LCP2 | |
| MWT_Angus | LPS/IL-1 Mediated Inhibition of RXR Function | 1.38E-05 | 0.03 | −2.00 | IL1R2, SULT1E1, CYP2B6, LBP, HMGCS1, SULT2A1, CYP2C19 |
| Melatonin Degradation I | 8.13E-05 | 0.06 | 1.00 | SULT1E1, CYP2B6, SULT2A1, CYP2C19 | |
| Superpathway of Melatonin Degradation | 1.10E-04 | 0.06 | 1.00 | SULT1E1, CYP2B6, SULT2A1, CYP2C19 | |
| IGF-1 Signaling | 6.46E-04 | 0.04 | NC | CTGF, IGF1, CYR61, IGFBP2 | |
| Mevalonate Pathway I | 9.33E-04 | 0.15 | NC | HMGCR, HMGCS1 | |
| MWT_Charolais | Neuroinflammation Signaling Pathway | 1.45E-04 | 0.02 | 1.89 | HMOX1, VCAM1, HLA-B, IL1B, HLA-DQB1, CSF1R, GRIN3A |
| Prostanoid Biosynthesis | 4.68E-04 | 0.22 | NC | PTGS1, TBXAS1 | |
| Granulocyte Adhesion and Diapedesis | 5.37E-04 | 0.03 | NC | VCAM1, SELL, IL1B, MMP11, SDC3 | |
| Graft-versus-Host Disease Signaling | 7.24E-04 | 0.06 | NC | HLA-B, IL1B, HLA-DQB1 | |
| Altered T-Cell and B-Cell Signaling in Rheumatoid Arthritis | 4.47E-03 | 0.03 | NC | HLA-B, IL1B, HLA-DQB1 | |
| MWT_KC | Antigen Presentation Pathway | 3.39E-04 | 0.08 | NC | HLA-B, HLA-DQB1, HLA-DQA2 |
| Pathogenesis of Multiple Sclerosis | 4.47E-04 | 0.22 | NC | CXCL10, CXCL9 | |
| Nicotine Degradation III | 1.07E-03 | 0.05 | NC | UGT2B17, CYP2B6, CYP2C19 | |
| Th1 Pathway | 1.41E-03 | 0.03 | NC | SOCS3, HLA-B, HLA-DQB1, HLA-DQA2 | |
| Melatonin Degradation I | 1.62E-03 | 0.05 | NC | UGT2B17, CYP2B6, CYP2C19 |
1ADG = average daily gain; DMI = average daily dry matter intake; MWT = metabolic body weight.
2The Z-score indicates activation (+) or inactivation (−) of the process in high-ADG, low-DMI, or low-MWT steers. NC = not calculated.
Functional Enrichment for DMI-Associated DE Genes
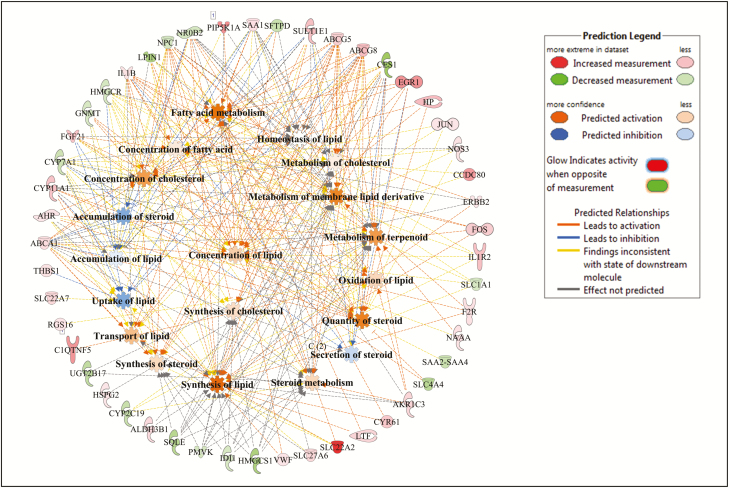
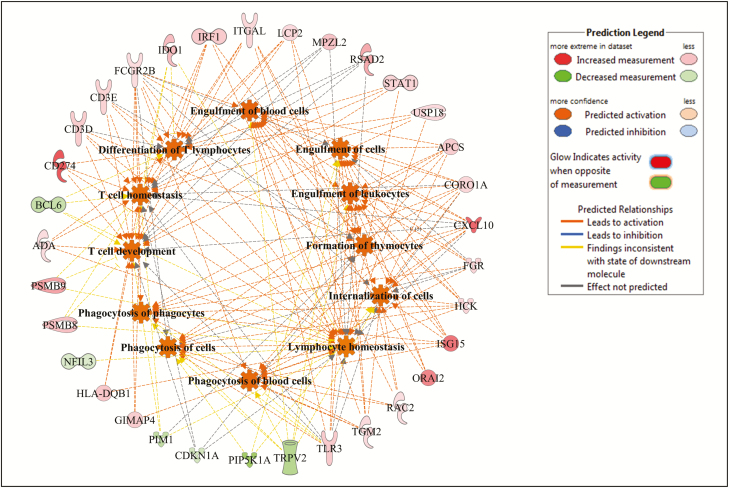
For DMI, 107, 177, and 155 DE genes were mapped to the IPA database for Angus, Charolais, and KC, respectively, and we identified 27, 22, and 25 significantly enriched biological functions for Angus, Charolais, and KC respectively, with 18 of them (62%) common to all breeds (Fig. 1e). The top enriched functions associated with DMI included lipid metabolism, molecular transport, small molecule biochemistry, cell death and survival, carbohydrate metabolism, vitamin and mineral metabolism, cellular movement, cellular function and maintenance, cell-to-cell signaling and interaction, and cellular development (Supplementary Figures S8–S10). The genes involved in the top enriched molecular and cellular functions associated with DMI in each breed are provided in Supplementary Table S4. Lipid metabolism was among the top enriched molecular and cellular functions in Angus and Charolais. For KC, all the top enriched functions we identified were related to functionality of cells, with cellular function and maintenance being the most significantly enriched function. Within lipid metabolism for Angus, 30 DE genes were involved in several lipid-related metabolic processes including concentration and accumulation of multiple lipids (cholesterol, phospholipids, triacylglycerol, and acylglycerol), and catabolism of lipid as shown in Fig. 5. Accumulation of lipid and concentration of lipids such as cholesterols and triacyl glycerides were predicted to be downregulated in the low-DMI steers, whereas metabolism of membrane lipid derivative and quantity of polyunsaturated fatty acids were predicted to be upregulated. Some key DE genes involved in the metabolism of lipids in Angus included SCD, ARNTL, LIPN1, APOA4, and ABHD6. For Charolais, 47 DE genes were identified as involved in different lipid metabolism processes. Some of the processes including uptake of lipid, accumulation of lipid, and uptake of cholesterol were predicted to be downregulated in the liver tissue of low-feed intake animals as shown in Fig. 6. However, other processes such as synthesis of lipid, synthesis of cholesterol, transport of lipid, and fatty acid metabolism were predicted to be upregulated in the same animals as shown in Fig. 6. Some of the major DE genes related to lipid metabolism identified in Charolais included ABCA1, ABCG5, ABCG8, CYP7A1, NROB2, NPC1, CES1, SAA1, IL1B, and SULT1E1. For DMI in KC, 51 DE genes were identified as involved in cellular function and maintenance, and these genes are mainly involved in a number of immune-related functions such as proliferation of T lymphocytes, T-cell development, phagocytosis of cells, and T-cell homeostasis which were predicted to be upregulated in liver tissue of low-DMI animals as shown in Fig. 7.
Figure 5.
Lipid metabolism gene and molecular processes interaction network within lipid metabolism function as associated to daily dry matter intake (DMI), and predicted activation or inhibition in low DMI in Angus steers.
Figure 6.
Lipid metabolism gene and molecular processes interaction network within lipid metabolism function as associated to daily dry matter intake (DMI), and predicted activation or inhibition in low-DMI Charolais steers.
Figure 7.
Cellular function and maintenance gene and molecular processes interaction network as associated to daily dry matter intake (DMI), and predicted activation or inhibition in low-DMI Kinsella Composite (KC) steers.
Pyrimidine ribonucleotides interconversion was identified as the most significantly (P = 1.20E-03) enriched pathway for Angus with 3 (NUDT5, CMPK2, and AK8) DE genes involved in this pathway (Table 5). For Charolais, lipopolysaccharide (LPS)/interleukin-1 (IL-1)-mediated inhibition of RXR function was the most significant (P = 2.63E-10) pathway, with 15 of the DE genes (IL1R2, ABCG8, GSTM1, ABCG5, SULT1E1, JUN, SULT1C4, NR0B2, CYP7A1, SLC27A6, IL1B, ALDH3B1, HMGCS1, ABCA1, CYP2C19) involved in this pathway and it was predicted to be relatively activated (Z-score = 0.33) in the low-feed intake steers (Table 5). For KC, we identified IFN signaling pathway as the most significant (P = 1.00E-10) pathway for this breed involving 8 of the identified DE genes (all upregulated) as shown in Supplementary Figure S11, and it was predicted to be activated (Z-score = 2.83) in the low-DMI animals (Table 5). All the top enriched canonical pathways associated with DMI for each breed are presented in Table 5. Besides canonical pathways, IPA also predicted the top gene expression regulators and their activation/deactivation state as associated with feed intake for each of the breeds. IFN-α cytokine group was predicted as the most significant gene expression regulator in Angus and KC. It was predicted to be inactivated (Z-score = −0.63) in Angus and activated in KC (Z-score = 3.86) in low-DMI animals. For Charolais, FGF19 growth factor was the most significant (P = 2.30E-16) regulator involved in the regulation of 15 DE genes. Top 5 enriched upstream gene expression regulators and their predicted activation or deactivation state in the low-DMI steers from the 3 studied populations are presented in Supplementary Table S7B.
Functional Enrichment for MWT-Associated DE Genes
For MWT, 80, 81, and 83 DE genes from Angus, Charolais, and KC, respectively, were mapped to the IPA database. These genes significantly enriched 24, 24, and 19 molecular and cellular functions for Angus, Charolais, and KC, respectively, with 17 of the enriched functions in common (68%) across breeds (Fig. 1f). The major functions that were identified as associated with MWT included lipid metabolism, amino acid metabolism, small molecule biochemistry, vitamin and mineral metabolism, molecular transport, cell morphology, cellular movement, cell-to-cell signaling and interaction, cell death and survival, and drug metabolism. The genes involved in these major molecular and cellular functions associated with MWT in each breed are provided in Supplementary Table S4. As for ADG and DMI, lipid metabolism and small molecule metabolism were among the top functions for both Angus and Charolais. Topmost (by P-value) enriched processes within amino acid metabolism and lipid metabolism for Angus, lipid metabolism and cellular movement for Charolais, and cell death and survival, and cellular movement for KC are presented in Supplementary Table S8. IPA additionally identified several significantly enriched canonical pathways associated with MWT in each of the breeds, and the top enriched canonical pathways are presented in Table 5. LPS/IL-1-mediated inhibition of RXR function was the most significant (P = 1.38E-05) pathway for Angus and was predicted to be inactivated (Z-score = −2.00) in the low-MWT steers. For Charolais, neuroinflammation signaling pathway was identified as the most significant (P = 1.45E-04) pathway and predicted to be activated (Z-score = 1.89) in the low-MWT steers (Table 5). For KC, antigen presentation pathway was the most significant (P = 3.39E-04) involving 3 of the identified DE genes (HLA-B, HLA-DQB1, and HLA-DQA2) (Table 5). For the upstream gene expression regulatory factors, the ligand-dependent nuclear receptor RORA was identified as the most significant (P = 5.29E-07) expression regulator in Angus and was shown to affect expression of 8 of the DE genes including CCL24, CYP2B6, HMGCR, IGF1, ITPR1, SLC13A2, SULT1E1, and SULT2A1. Albeit, cytokines IFNG and IL6 were identified as the most significant gene expression regulators in Charolais and KC steers, respectively. IFNG was predicted to be activated (Z-score = 1.22), whereas IL6 was predicted to be inactivated (Z-score = −1.27) in the low-MWT steers of the respective breeds. The top enriched gene expression regulators associated with MWT for each breed are presented in Supplementary Table S7C.
Discussion
In the current study, we employed RNAseq analyses of liver tissue from 60 steers of 3 Canadian beef cattle populations (Angus, Charolais, and KC) to study gene expression difference between the high- and low-phenotype steer groups for 3 feed efficiency-related traits ADG, DMI, and MWT. To maximize the phenotypic divergence between 2 animal groups for the particular trait under investigation, we sorted the 20 steers of each of the 3 breeds and selected the 6 highest and 6 lowest steers for differential gene expression gene analyses. Each trait under investigation showed significant differences between the high- and low-extreme-phenotype animal groups (Supplementary Tables S1–S3). It is worth noting that with the aim of minimizing environmental differences between the studied animals, they were raised on the same experimental farm and were managed similarly. In addition, the 2 extreme groups of each population did not differ significantly in their age when the liver samples were collected (Supplementary Table S1–S3). Although the 2 extreme groups of the target trait also exhibited significant differences in a few of other production traits due to their biological correlations between the traits, the strongest divergence for ADG, DMI, or MWT provided suitable contrast of animal groups for differential gene expression analysis for each of the trait under investigation.
Our results showed a great diversity in terms of DE genes between breeds for the same trait (Fig. 1a–c). For example, of the 252, 375, and 206 DE genes associated with ADG, DMI, and MWT, only 1% to 2% of them were shared among all 3 breeds, while 81.7% to 83% were breed specific, and 3% to 8% are uniquely common between 2 breeds for a trait. This diversity of differential gene expression implies that probably these traits are largely controlled by different genes in different breeds. A similar trend of predominantly breed-specific differential gene expression profiles across breeds in the same animal populations (including animals used in the current study) has been previously observed and reported for RFI by Mukiibi et al. (2018). These breed-specific DE gene results concur with the identification of quantitative trait loci for feed efficiency traits by Saatchi et al. (2014), in which quantitative trail loci associated with RFI, ADG, DMI, and MWT are largely breed specific (Saatchi et al., 2014). When DE genes among the traits were compared within a breed, our results showed a relatively moderate number of gene overlap (12 – 29%) between MWT and ADG or DMI. This could be an indication of shared genetic mechanisms underlying these traits, which supports the moderate to high genetic correlations between MWT and ADG or DMI reported in beef cattle (Crowley et al., 2010; Mao et al., 2013). In the liver tissue of Nellore cattle, Tizioto et al. (2015) also reported DE gene overlap of 14% or 20% between RFI and ADG or DMI, respectively. In addition, IFI27 which was among the 7 DE genes associated with RFI, ADG, and DMI in the Nellore cattle steers (Tizioto et al., 2015) was also identified as one of the 4 genes common to all the 4 traits in the current study for the Angus population. IFI27 codes for IFN-α-inducible protein 27 and plays roles in the innate immune response (Dill et al., 2014), indicating the potential importance of immune-related genes in influencing feed efficiency and the related traits in beef cattle.
Although a low DE gene overlap for each trait was observed among the breeds, we identified significant overlap of the biological functions (60% to 68%) affecting the traits across the 3 studied breeds. These results suggest that despite the high diversity in terms of genes controlling these traits in each breed, the biological processes influencing the traits across the breeds or biological types are largely the same. Some of the major cellular and molecular functions identified as associated with ADG or DMI or MWT included lipid metabolism, molecular transport, carbohydrate metabolism, amino acid metabolism, vitamin metabolism, molecular transport, cell-to-cell signaling and interaction, cell morphology, cell death and survival, cellular movement, and immune-related functions. Given the central roles played by the liver in nutrient metabolism and distribution in the body (Jeremy et al., 2002; Rui, 2011), and its function as a major immunological organ in the body (Parker and Picut, 2005; Racanelli and Rehermann, 2006), we will further discuss the associations of lipid metabolism, amino acid and carbohydrate metabolism, and immunological functions with ADG, DMI, and MWT in the following sections.
Association of Lipid Metabolism With Growth Rate, Feed Intake, and Metabolic Body Weight
Lipid metabolism was identified in this study as an important biological function influencing growth rate, feed intake, and metabolic weight in beef cattle. These results are consistent with reports from other similar studies that have identified genes involved in lipid metabolism for feed efficiency (Chen et al., 2011; Alexandre et al., 2015; Tizioto et al., 2015; Weber et al., 2016), growth rate (Kern et al., 2016; Lindholm-Perry et al., 2016a; Foote et al., 2017), and feed intake (Lindholm-Perry et al., 2016a) in beef cattle. Some of the highly represented lipid metabolic functions identified in our study as associated with ADG or DMI or MWT included synthesis of lipid, synthesis of cholesterol, accumulation of lipid, oxidation of fatty acids, and cellular transport of lipid. With respect to growth, our results suggest that fast-growing animals tend to have reduced synthesis of lipid. This is demonstrated by the IPA results that showed synthesis of lipids involving 19 DE genes in Charolais and 10 DE genes in KC was predicted to be downregulated in the fast-growing animals (Figs. 3 and 4). Additionally, in Angus, 14 lipogenic genes (AKR1C3, IL1R2, SOCS3, NR0B2, F2R, IGF1, ELOVL5, CYP7A1, ABHD6, HMGCR, PGAP1, SQLE, CYP51A1, and HMGCS1) were identified as associated with ADG, of which 8 genes (IGF1, ELOVL5, CYP7A1, ABHD6, HMGCR, PGAP1, SQLE, CYP51A1, and HMGCS1) were downregulated in the liver tissue of high-ADG animals (Fig. 2). The difference in lipid synthesis between fast- and slow-growing animals could indicate differences in energy utilization efficiency, where the fast-growing animals optimize their energy towards protein deposition and less towards fat synthesis and deposition. Indeed, fast-growing animals tend to deposit less fat and produce more lean meat than the slow-growing animals (Mitchell, 2007). In our previous study, we identified 253 DE genes of liver for RFI in the same 3 breed populations (Mukiibi et al., 2018), of which 14 DE genes including HMGCR, STS, IGF1, APOA1, LPIN1, MFSD2A, GSTM4, ELOVL2, SCD, PDK4, CES1, SQLE, NR0B2, and IGFBP2 were common to DE genes involved in lipid synthesis for ADG, DMI, and MWT from this study. These genes probably contribute to the variation in feed efficiency and its component traits through modulation of lipid synthesis. Additionally, associations of expression of lipogenic genes and the other lipid metabolic processes such as accumulation, transport, oxidation, and uptake of lipid with ADG, DMI, and MWT could in part explain the reported genetic relationships between these traits and carcass or meat fat content and fat composition traits in beef cattle (Lancaster et al., 2009; Inoue et al., 2011; Zhang et al., 2017). Therefore, the DE genes associated with lipid metabolism should be further investigated to identify causative DNA markers for the feed efficiency traits in beef cattle.
Notably, of the lipogenic DE genes identified in this study, several of them are involved in cholesterol metabolism. The superpathway of cholesterol biosynthesis was identified among the top significantly enriched pathways for ADG in Angus where it was predicted to be downregulated in the high-ADG steers, and for DMI in Charolais where it was predicted to be downregulated in the low-DMI steers. Four of the DE genes identified as associated with ADG in Angus (SQLE, HMGCR, HMGCS1, and CYP51A1) are involved in the cholesterol biosynthesis pathway and were all downregulated in the high-ADG animals. For DMI in Charolais, 5 DE genes including SQLE, PMVK, IDI1, HMGCR, and HMGCS1 are involved in the cholesterol biosynthesis pathway, and they were all downregulated in the low-DMI steers. Interestingly, HMGCS1, HMGCR, PMVK, SQLE, and IDI1 are key enzymes catalyzing important steps in cholesterol biosynthesis (Brown and Sharpe, 2016). For example, HMGCS1 codes for the 3-hydroxy-3-methylglutaryl-CoA synthase 1 that characterizes the condensation of acetoacetyl-CoA and acetyl-CoA to 3-hydroxy-3-methylglutaryl-CoA, an initial reaction in cholesterol biosynthesis (Brown and Sharpe, 2016). HMGCR encodes for 3-hydroxy-3-methylglutaryl-CoA reductase, an enzyme that characterizes the reduction of 3-hydroxy-3-methylglutaryl-CoA to mevalonic acid, a rate-limiting step in cholesterol synthesis (Brown and Sharpe, 2016). Gene SQLE codes for squalene monooxygenase, which is an enzyme that oxidizes the first oxygenation step in cholesterol/sterol biosynthesis and is considered a rate-limiting enzyme in this process (Brown and Sharpe, 2016). LXRs and RXRs, which are heterodimer nuclear receptors that regulate cholesterol metabolism through regulation of cholestrogenic enzymes, and carriers (Sharpe and Brown, 2013; Hong and Tontonoz, 2014) were identified to be associated with ADG, DMI, and MWT. Additionally, SREBF1 transcription regulator was identified as an enriched regulator in this study (for ADG and MWT in Angus) and predicted to regulate several genes including cholesterol biosynthesis genes. SREBF1 codes for sterol regulatory element-binding protein 1, which is a key (together with SREBF2) expression regulator of genes involved in cholesterol biosynthesis (Hua et al., 1995). These results present interesting revelations on the potential correlations of beef production traits such as growth rate with cholesterol content in beef as the liver is also a major site for cholesterol biosynthesis in the animal body (Bell, 1981).The associations of cholesterol biosynthesis with growth and feed intake is an interesting revelation from our results as they imply that selection of fast-growing animals or low feed intake could result in the production of beef with low cholesterol content, a dietary health concern of many beef consumers. Consistent with our findings, association of cholesterol metabolism with feed efficiency was reported by Karisa et al. (2014), and lower blood cholesterol content has been observed in more efficient beef animals as compared to inefficient animals (Alexandre et al., 2015; Bourgon et al., 2017). Also, downregulation of HMGCR and SQLE in the liver tissue of more feed efficient animals has been previously reported in crossbred steers (Mukiibi et al., 2018).
Association of Amino Acid and Carbohydrate Metabolism With Growth, Feed Intake, and Metabolic Body Weight
Amino acid metabolism was among the top enriched cellular and molecular functions associated with ADG in Charolais and KC animals, while carbohydrate metabolism was one of the top enriched functions in KC. With respect to amino acid metabolism in Charolais, processes such as metabolism of serine family amino acids (involving genes CSAD, SDS, and SHMT2), synthesis of amino acid (involving genes ARG1, CNDP2, CS, and SHMT2), and transport of arginine (involving genes ARG1 and SLC3A1) were strongly enriched. For KC, processes such as catabolism of amino acids, synthesis of l-proline, metabolism of essential amino acids, and others (shown in Supplementary Table S5) were enriched for KC with DE genes ARG1 and AASS being involved in most of these processes. ARG1 and AASS code for critical enzymes in amino acid metabolism. ARG1 codes for arginase enzyme which catalyzes conversion of arginine to urea and orthenine in the urea cycle (Morris Jr, 2002), whereas AASS codes for alpha-aminoadipate-semialdehyde synthase, a bifunctional enzyme that catalyzes a 2-step conversion of lysine to alpha-aminoadipic semialdehyde in the lysine degradation pathway (Sacksteder et al., 2000). For Angus, amino acid metabolism was not among the most significant functions associated with ADG; however, it was significantly enriched by mainly amino acid transport DE genes including SLC16A10, SLC1A2, SLC25A15, and SLC3A1 as presented in Supplementary Table S5. Amino acid metabolism also showed a strong association with MWT in Angus steers with numerous enriched amino acid metabolic processes such as uptake of cystine (Supplementary Table S5). For DMI, amino acid metabolism was not among the top enriched biological functions in any of the 3 breeds; however, it was enriched in Angus and Charolais by 10 and 17 DE genes, respectively. Although in the current study, we did not identify large numbers of DE genes involved in amino acid metabolism, the few genes we identified are involved in key hepatic amino acid metabolic process such as catabolism and synthesis of amino acids. Through these processes, the liver modulates nitrogenous compounds content in the body (Reynolds, 1992).
As the liver is a major source of glucose in the body of a ruminant animal (Nafikov and Beitz, 2007), it is of interest to note that carbohydrate metabolism was among the most significantly enriched functions in Angus as well as enriched in Charolais and KC with respect to ADG. Key processes including oxidation of carbohydrate, synthesis of carbohydrate, glycogenolysis, intake of glucose, and gluconeogenesis were identified as associated with growth rate in the studied animals (Supplementary Table S6). In agreement with our results, Foote et al. (2017) have reported carbohydrate and amino acid metabolism associations with beef cattle growth and feed intake in the jejunum tissue. Studying the rumen epithelial tissue transcriptome of crossbred steers with divergent feed intake and growth phenotypes, Kern et al. (2016) reported that carbohydrate metabolism was associated with feed intake and body weight gain. Besides feed intake and body weight gain, hepatic carbohydrate, and amino acid metabolism have also been identified to be associated with feed efficiency in beef cattle (Chen et al., 2011; Mukiibi et al., 2018). These observations suggest that animals divergent for growth or feed intake might inherently possess differences in their carbohydrate (majorly glucose) synthesis or utilization for energy production.
Association of Immunological Functions With Growth, Feed Intake, and Metabolic Body Weight
In the current study, we identified immune function-related genes that were differentially expressed in the liver tissue of animals with divergent growth rate or feed intake or MWT phenotypes. For example, acute phase response signaling was among the top enriched canonical pathway for Angus steers with divergent growth rate phenotypes involving SOCS3, HP, SAA1, SOCS2, SERPINA3, LBP, and SERPINE1, and it was predicted as upregulated in the fast-growing animals. Within the composite breed KC, steers with divergent DMI had a large number of DE immune-related genes, and they were involved in multiple immune function processes such as engulfment of cells by macrophages, T-cell homeostasis, T-cell development, and differentiation of T lymphocytes, which were predicted to be upregulated in the liver tissue of low-feed intake KC. Additionally, IFN signaling pathway (involving OAS1, IFI6, PSMB8, STAT1, TAP1, IRF1, IFITM1, and ISG15), Th1 pathway (involving NFIL3, CD3E, HLA-B, CD274, HLA-DQB1, HLA-DQA2, STAT1, CD3D, and IRF1), and PKCθ signaling in T lymphocytes (involving CACNA1I, RAC2, CACNG1, CD3E, HLA-B, HLA-DQB1, CD3D, and LCP2) were among the top enriched pathways associated with feed intake in KC and also predicted to be activated in KC steers with lower feed intake (Table 5). Since the liver is a major organ to process absorbed materials from the gastrointestinal tract including microbes and toxins, it plays an important role in defending the body against invading pathogens through phagocytosis by the Kupfer cells or killing the infected cells through lysis and inducing apoptosis by natural killer cells and natural killer T cells (Nakamoto and Kanai, 2014). Immune genes from similar gene clusters as those identified in this study have also been reported to be associated to feed intake or gain in the rumen (Kern et al., 2016; Reynolds et al., 2017), duodenum (Lindholm-Perry et al., 2016a), jejunum (Lindholm-Perry et al., 2016a), and ileum (Lindholm-Perry et al., 2016a) in beef cattle. With respect to hepatic transcriptome studies, associations of feed efficiency and hepatic immune response in beef cattle have been reported by Alexandre et al. (2015) and Paradis et al. (2015). Paradis et al. (2015) identified 5 immune genes (HBB, MX1, ISG5, HERC6, and IF44) associated with feed efficiency in crossbred heifers. Similarly, in our study, MX1, ISG5, HERC6, and IF44 were also identified as associated either with ADG, DMI, or MWT in KC steers, whereas for Charolais steers, MX1, ISG5, and IF44 were either associated with DMI or MWT. These and several other genes we identified in this study are regulated by INF-α and INF-β signaling as shown by canonical pathways and upstream regulator results and are hence involved in innate immune function against invading pathogens (Stetson and Medzhitov, 2006; Boxx and Cheng, 2016). Our results and those reported by other similar studies in beef cattle indicate possible immunological adaptations to the feedlot challenges by some of the animals, which probably have implications on animal’s feed intake, growth, and feed efficiency.
Conclusions
We identified a total of 252, 375, and 206 protein-coding genes associated with growth rate, feed intake, and MWT of beef cattle, respectively, through hepatic transcriptome sequence data analyses. The majority of the identified DE genes for the traits were breed specific. However, most of the enriched biological functions are common across the 3 breeds. Functional enrichment showed that the identified DE genes were involved in multiple cellular and molecular functions that mainly include metabolism of lipids, carbohydrates, amino acids, vitamins and minerals, small molecule biochemistry, cellular movement, cell morphology, and cell-to-cell signaling and interaction. Further identification of pathways and upstream gene expression regulators revealed strong associations of both cholesterol biosynthesis and immune-related functions with growth, feed intake, and MWT. The DE genes and major biological functions associated with growth, feed intake, and MWT advance our understanding of genetic mechanisms that regulate feed intake, growth, and feed efficiency in beef cattle with respective to various breeds/breed populations, which will also help with identification of causative DNA polymorphisms for the traits and lead to designing of better genetic and genomic selection and breeding programs to improve the traits.
Supplementary Data
Supplementary data are available at Journal of Animal Science online.
Supplementary Figure S1. Expression profile (log2(fold change)) in high-ADG steers of the 5 differentially expressed (DE) genes common across all 3 breeds.
Supplementary Figure S2. Expression profile (log2(fold change)) in low-DMI steers of the 4 differentially expressed (DE) genes common across all 3 breeds.
Supplementary Figure S3. Expression profile (log2(fold change)) in low-MWT steers of the 4 differentially expressed (DE) genes common across all 3 breeds.
Supplementary Figure S4. Venn diagram showing overlap between DE gene of ADG, DMI, MWT, and RFI from Mukiibi et al. (2018), within Angus, Charolais, and Kinsella Composite populations.
Supplementary Figure S5. Enriched molecular and cellular functions associated with ADG in Angus steers.
Supplementary Figure S6. Enriched molecular and cellular functions associated with ADG in Charolais steers.
Supplementary Figure S7. Enriched molecular and cellular functions associated with ADG in KC steers.
Supplementary Figure S8. Enriched molecular and cellular functions associated with DMI in Angus steers.
Supplementary Figure S9. Enriched molecular and cellular functions associated with DMI in Charolais steers.
Supplementary Figure S10. Enriched molecular and cellular functions associated with DMI in KC steers.
Supplementary Figure S11. Interferon Signaling Canonical pathway associated with DMI in KC animals.
Supplementary Figure S12. Enriched molecular and cellular functions associated with MWT in Angus steers.
Supplementary Figure S13. Enriched molecular and cellular functions associated with MWT in Charolais steers.
Supplementary Figure S14. Enriched molecular and cellular functions associated with MWT in KC steers.
Supplementary Table S1. Differences of ADG and other performance traits between groups of high- (n = 6) and low-ADG steers (n = 6) of the 3 breeds.
Supplementary Table S2. Differences of DMI and other performance traits between groups of high- (n = 6) and low-DMI steers (n = 6) of the 3 breeds.
Supplementary Table S3. Differences of MWT and other performance traits between groups of high- (n = 6) and low-MWT steers (n = 6) of the 3 breeds.
Supplementary Table S4. Top 5 enriched functions and the DE genes involved in the functions for ADG, DMI, and MWT within Angus, Charolais, and KC populations.
Supplementary Table S5. Enriched biological processes within amino acid metabolism biological functions associated with ADG in Angus, Charolais, and KC steers.
Supplementary Table S6. Enriched biological processes within carbohydrate metabolism biological functions associated with ADG in Angus, Charolais, and KC steers.
Supplementary Table S7. Predicted activated and deactivated gene expression regulators associated with ADG, DMI, and MWT in Angus, Charolais, and KC.
Supplementary Table S8. Enriched biological processes within the topmost enriched molecular and cellular functions associated with MWT in Angus, Charolais, and KC.
Supplementary File 1. All DE genes between high- and low-ADG steers from Angus, Charolais, and KC populations.
Supplementary File 2. All DE genes between high- and low-DMI steers from Angus, Charolais, and KC populations.
Supplementary File 3. All DE genes between high- and low-MWT steers from Angus, Charolais, and KC populations.
Footnotes
This work was supported by Alberta Livestock and Meat Agency (ALMA)/Alberta Agriculture and Forestry (AAF) (Project 2014F047R) and in part by the Genome Alberta (Project GAB-A3GP37). R.M. acknowledges financial support from University of Alberta-Teagasc (Walsh Fellowship). This research had been enabled by the use of computing resources provided by Compute/Calcul Canada.
Literature Cited
- Ahola J. K., and Hill R. A.. . 2012. Input factors affecting profitability: a changing paradigm and a challenging time. In: Hill R. A., editor, Feed efficiency in the beef industry. Wiley-Blackwell, Ames, IA, p. 10. [Google Scholar]
- Alexandre P. A., Kogelman L. J., Santana M. H., Passarelli D., Pulz L. H., Fantinato-Neto P., Silva P. L., Leme P. R., Strefezzi R. F., Coutinho L. L.. . 2015. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics 16:1073. doi: 10.1186/s12864-015-2292-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Husseini W., Gondro C., Quinn K., Herd R. M., Gibson J. P., and Chen Y.. . 2014. Expression of candidate genes for residual feed intake in Angus cattle. Anim. Genet. 45:12–19. doi: 10.1111/age.12092 [DOI] [PubMed] [Google Scholar]
- Amer P. R., Simm G., Keane M. G., Diskin M. G., and Wick-ham B. W.. . 2001. Breeding objective for beef cattle in Ireland. Livest. Prod. Sci. 67:223–239. [Google Scholar]
- Anders S., McCarthy D. J., Chen Y., Okoniewski M., Smyth G. K., Huber W., and Robinson M. D.. . 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8:1765–1786. doi: 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W.. . 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed 7 June 2019).
- Bell A. W. 1981. Lipid metabolism in liver and selected tissues and in the whole body of ruminant animals. In: Christie W. W., editor, Lipid metabolism in ruminant animals. Pergamon Press, Amsterdam, the Netherlands: p. 363–410. doi: 10.1016/B978-0-08-023789-3.50012-X [DOI] [Google Scholar]
- Bourgon S. L, de Amorim M. D., Miller S. P., and Montanholi Y. R.. . 2017. Associations of blood parameters with age, feed efficiency and sampling routine in young beef bulls. Livest. Sci. 195:27–37. doi: 10.1016/j.livsci.2016.11.003 [DOI] [Google Scholar]
- Boxx G. M., and Cheng G.. . 2016. The roles of type I interferon in bacterial infection. Cell Host Microbe 19:760–769. doi: 10.1016/j.chom.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., and Sharpe L. J.. . 2016. Cholesterol synthesis. In: Ridgway N. D. and McLeod R. S., editors, biochemistry of lipids, lipoproteins and membranes. Elsevier, Amsterdam, the Netherlands: p. 327–358. doi: 10.1016/B978-0-444-63438-2.00011-0 [DOI] [Google Scholar]
- CCAC 2009. Guidelines on the care and use of farm animals in research, teaching and testing Canadian Council on Animal Care, Ottawa, ON, Canada: https://www.ccac.ca/Documents/Standards/Guidelines/Farm_Animals.pdf (Accessed 7 June 2019.) [Google Scholar]
- Chen Y., Gondro C., Quinn K., Herd R. M., Parnell P. F., and Vanselow B.. . 2011. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim. Genet. 42:475–490. doi: 10.1111/j.1365-2052.2011.02182.x [DOI] [PubMed] [Google Scholar]
- Crowley J. J., McGee M., Kenny D. A., Crews D. H. Jr, Evans R. D., and Berry D. P.. . 2010. Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance-tested beef bulls. J. Anim. Sci. 88:885–894. doi: 10.2527/jas.2009-1852 [DOI] [PubMed] [Google Scholar]
- Dill M. T., Makowska Z., Trincucci G., Gruber A. J., Vogt J. E., Filipowicz M., Calabrese D., Krol I., Lau D. T., Terracciano L.. . 2014. Pegylated IFN-α regulates hepatic gene expression through transient Jak/STAT activation. J. Clin. Invest. 124:1568–1581. doi: 10.1172/JCI70408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote A. P., Keel B. N., Zarek C. M., and Lindholm-Perry A. K.. . 2017. Beef steers with average dry matter intake and divergent average daily gain have altered gene expression in the jejunum. J. Anim. Sci. 95:4430–4439. doi:org/10.2527/jas2017.1804 [DOI] [PubMed] [Google Scholar]
- Goonewardene L. A., Wang Z., Price M., Yang R.-C., Berg R. T., and Makarechian M.. . 2003. Effect of udder type and calving assistance on weaning traits of beef and dairy× beef calves. Livest. Prod. Sci. 81:47–56. doi: 10.1016/S0301-6226(02)00194-X [DOI] [Google Scholar]
- Hill R. A., and Ahola J. K.. . 2012. Feed efficiency interactions with other traits: growth and product quality. In: Hill R. A., editor, Feed efficiency in the beef industry. Wiley-Blackwell, Ames, IA: p. 148. [Google Scholar]
- Hong C., and Tontonoz P.. . 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug. Discov. 13:433–44. doi: 10.1038/nrd4280 [DOI] [PubMed] [Google Scholar]
- Hua X., Wu J., Goldstein J. L., Brown M. S., and Hobbs H. H.. . 1995. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics 25:667–673. doi: 10.1016/0888-7543(95)80009-b [DOI] [PubMed] [Google Scholar]
- Inoue K., Kobayashi M., Shoji N., and Kato K.. . 2011. Genetic parameters for fatty acid composition and feed efficiency traits in Japanese Black cattle. Animal 5:987–994. doi: 10.1017/S1751731111000012 [DOI] [PubMed] [Google Scholar]
- Jeremy M. B., John L. T., and Lubert S.. . 2002. Each organ has a unique metabolic profile. In: Moran S., editor, Biochemistry. W. H. Freeman and Company, New York: pp. 1259 https://www.ncbi.nlm.nih.gov/books/NBK22436/ (Accessed 7 June 2019). [Google Scholar]
- Karisa B., Moore S., and Plastow G.. . 2014. Analysis of biological networks and biological pathways associated with residual feed intake in beef cattle. Anim. Sci. J. 85:374–387. doi: 10.1111/asj.12159 [DOI] [PubMed] [Google Scholar]
- Keel B. N., Zarek C. M., Keele J. W., Kuehn L. A., Snelling W. M., Oliver W. T., Freetly H. C., and Lindholm-Perry A. K.. . 2018. RNA-Seq meta-analysis identifies genes in skeletal muscle associated with gain and intake across a multi-season study of crossbred beef steers. BMC Genomics 19:430. doi: 10.1186/s12864-018-4769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern R. J., Lindholm-Perry A. K., Freetly H. C., Snelling W. M., Kern J. W., Keele J. W., Miles J. R., Foote A. P., Oliver W. T., Kuehn L. A., and Ludden P. A.. . 2016. Transcriptome differences in the rumen of beef steers with variation in feed intake and gain. Gene 586:12–26. doi: 10.1016/j.gene.2016.03.034 [DOI] [PubMed] [Google Scholar]
- Khansefid M., Millen C. A., Chen Y., Pryce J. E., Chamberlain A. J., Vander Jagt C. J., Gondro C., and Goddard M. E.. . 2017. Gene expression analysis of blood, liver, and muscle in cattle divergently selected for high and low residual feed intake. J. Anim. Sci. 95:4764–4775. doi: 10.2527/jas2016.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L.. . 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. M., Swiger L. A., Chambers D., and Gregory K. E.. . 1963. Efficiency of feed use in beef cattle. J. Anim. Sci. 22:486–494. doi: 10.2527/jas1963.222486x [DOI] [Google Scholar]
- Kong R. S., Liang G., Chen Y., Stothard P., and Guan l. e. L.. . 2016. Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genomics 17:592. doi: 10.1186/s12864-016-2935-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J. Jr, and Tugendreich S.. . 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30:523–530. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster P. A., Carstens G. E., Crews D. H. Jr, Welsh T. H. Jr, Forbes T. D., Forrest D. W., Tedeschi L. O., Randel R. D., and Rouquette F. M.. . 2009. Phenotypic and genetic relationships of residual feed intake with performance and ultrasound carcass traits in Brangus heifers. J. Anim. Sci. 87:3887–3896. doi: 10.2527/jas.2009-2041 [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Butler A. R., Kern R. J., Hill R., Kuehn L. A., Wells J. E., Oliver W. T., Hales K. E., Foote A. P., and Freetly H. C.. . 2016a. Differential gene expression in the duodenum, jejunum and ileum among crossbred beef steers with divergent gain and feed intake phenotypes. Anim. Genet. 47:408–427. doi: 10.1111/age.12440 [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Cunningham H. C., Kuehn L. A., Vallet J. L., Keele J. W., Foote A. P., Cammack K. M., and Freetly H. C.. . 2017. Relationships between the genes expressed in the mesenteric adipose tissue of beef cattle and feed intake and gain. Anim. Genet. 48:386–394. doi: 10.1111/age.12565 [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Kern R. J., Keel B. N., Snelling W. M., Kuehn L. A., and Freetly H. C.. . 2016b. Profile of the spleen transcriptome in beef steers with variation in gain and feed intake. Front. Genet. 7:127. doi: 10.3389/fgene.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F., Chen L., Vinsky M., Okine E., Wang Z., Basarab J., Crews D. H. Jr, and Li C.. . 2013. Phenotypic and genetic relationships of feed efficiency with growth performance, ultrasound, and carcass merit traits in Angus and Charolais steers. J. Anim. Sci. 91:2067–2076. doi: 10.2527/jas.2012-5470 [DOI] [PubMed] [Google Scholar]
- Mitchell A. D. 2007. Impact of research with cattle, pigs, and sheep on nutritional concepts: body composition and growth. J. Nutr. 137:711–714. doi: 10.1093/jn/137.3.711 [DOI] [PubMed] [Google Scholar]
- Morris S. M., Jr 2002. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 22:87–105. doi: 10.1146/annurev.nutr.22.110801.140547 [DOI] [PubMed] [Google Scholar]
- Mukiibi R., Vinsky M., Keogh K. A., Fitzsimmons C., Stothard P., Waters S. M., and Li C.. . 2018. Transcriptome analyses reveal reduced hepatic lipid synthesis and accumulation in more feed efficient beef cattle. Sci. Rep. 8:7303. doi: 10.1038/s41598-018-25605-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafikov R. A., and Beitz D. C.. . 2007. Carbohydrate and lipid metabolism in farm animals. J. Nutr. 137:702–705. doi: 10.1093/jn/137.3.702 [DOI] [PubMed] [Google Scholar]
- Nakamoto N., and Kanai T.. . 2014. Role of toll-like receptors in immune activation and tolerance in the liver. Front. Immunol. 5:221. doi: 10.3389/fimmu.2014.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkrumah J., Basarab J., Wang Z., Li C., Price M., Okine E., Crews D., and Moore S.. . 2007. Genetic and phenotypic relationships of feed intake and measures of efficiency with growth and carcass merit of beef cattle. J. Anim. Sci. 2007. 85:2711–2720. doi: 10.2527/jas.2006-767 [DOI] [PubMed] [Google Scholar]
- Paradis F., Yue S., Grant J. R., Stothard P., Basarab J. A., and Fitzsimmons C.. . 2015. Transcriptomic analysis by RNA sequencing reveals that hepatic interferon-induced genes may be associated with feed efficiency in beef heifers. J. Anim. Sci. 93:3331–3341. doi: 10.2527/jas.2015-8975 [DOI] [PubMed] [Google Scholar]
- Parker G. A., and Picut C. A.. . 2005. Liver immunobiology. Toxicol. Pathol. 33:52–62. doi: 10.1080/01926230590522365 [DOI] [PubMed] [Google Scholar]
- Racanelli V., and Rehermann B.. . 2006. The liver as an immunological organ. Hepatology 43(2 Suppl. 1):S54–S62. doi: 10.1002/hep.21060 [DOI] [PubMed] [Google Scholar]
- Reynolds C. K. 1992. Metabolism of nitrogenous compounds by ruminant liver. J. Nutr. 122(Suppl. 3):850–854. doi: 10.1093/jn/122.suppl_3.850 [DOI] [PubMed] [Google Scholar]
- Reynolds J. G., Foote A. P., Freetly H. C., Oliver W. T., and Lindholm-Perry A. K.. . 2017. Relationships between inflammation- and immunity-related transcript abundance in the rumen and jejunum of beef steers with divergent average daily gain. Anim. Genet. 48:447–449. doi: 10.1111/age.12546 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K.. . 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., and Oshlack A.. . 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. Energy metabolism in the liver. 2011. Compr. Physiol. 4: 177–197. doi: 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Beever J. E., Decker J. E., Faulkner D. B., Freetly H. C., Hansen S. L., Yampara-Iquise H., Johnson K. A., Kachman S. D., Kerley M. S., Kim J., Loy D. D., Marques E., Neibergs H. L., Pollak E. J., Schnabel R. D., Seabury C. M., Shike D. W., Snelling W. M., Spangler M. L., Weaber R. L., Garrick D. J., and Taylor J. F.. . 2014. QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies. BMC Genomics 15:1004. doi: 10.1186/1471-2164-15-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder K. A., Biery B. J., Morrell J. C., Goodman B. K., Geisbrecht B. V., Cox R. P., Gould S. J., and Geraghty M. T.. . 2000. Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am. J. Hum. Genet. 66:1736–1743. doi: 10.1086/302919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel F., Miller S., and Wilton J.. . 2004. Genetic parameters and breed differences for feed efficiency, growth, and body composition traits of young beef bulls. Can. J. Anim. Sci. 84:177–186. doi: 10.4141/A03-085 [DOI] [Google Scholar]
- Sharpe L. J., and Brown A. J.. . 2013. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288:18707–18715. doi: 10.1074/jbc.R113.479808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D. B., and Medzhitov R.. . 2006. Type I interferons in host defense. Immunity 25:373–381. doi: 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Tizioto P. C., Coutinho L. L., Decker J. E., Schnabel R. D., Rosa K. O., Oliveira P. S., Souza M. M., Mourão G. B., Tullio R. R., Chaves A. S., Lanna D. P., Zerlotini-Neto A., Mudadu M. A., Taylor J. F., and Regitano L. C.. . 2015. Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes. BMC Genomics 16:242. doi: 10.1186/s12864-015-1464-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizioto P. C., Coutinho L. L., Oliveira P. S., Cesar A. S., Diniz W. J., Lima A. O., Rocha M. I., Decker J. E., Schnabel R. D., Mourão G. B., Tullio R. R., Zerlotini A., Taylor J. F., and Regitano L. C.. . 2016. Gene expression differences in longissimus muscle of Nelore steers genetically divergent for residual feed intake. Sci. Rep. 6:39493. doi: 10.1038/srep39493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. L., Welly B. T., Van Eenennaam A. L., Young A. E., Porto-Neto L. R., Reverter A., and Rincon G.. . 2016. Identification of gene networks for residual feed intake in angus cattle using genomic prediction and RNA-seq. PLoS ONE 11:e0152274. doi: 10.1371/journal.pone.0152274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarek C. M., Lindholm-Perry A. K., Kuehn L. A., and Freetly H. C.. . 2017. Differential expression of genes related to gain and intake in the liver of beef cattle. BMC Res. Notes 10:1. doi: 10.1186/s13104-016-2345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ekine-Dzivenu C., Vinsky M., Basarab J. A., Aalhus J. L., Dugan M. E. R., and Li C.. . 2017. Phenotypic and genetic relationships of residual feed intake measures and their component traits with fatty acid composition in subcutaneous adipose of beef cattle. J. Anim. Sci. 95(7):2813–2824. doi: 10.2527/jas.2017.1451 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.