Abstract
Purpose
The purpose of this study was to extend the current understanding of endogenous lysine-specific demethylase 1 (LSD1) expression spatially and temporally in the retina. Toward that end, we determined the localization and levels of LSD1 and its substrates H3K4me1 and H3K4me2 (H3K4me1/2) within the murine eye.
Methods
Immunofluorescent microscopy for LSD1, H3K4me1, and H3K4me2 was conducted on murine formalin-fixed paraffin-embedded eye sections across development in addition to Western immunoblotting to assess localization and protein levels.
Results
Retinal LSD1 protein levels were highest at postnatal day 7 (P7), whereas its substrates H3K4me1 and H3K4me2 had equally high levels at P2 and P14. Concentrations of all three proteins gradually decreased over developmental time until reaching a basement level of ∼60% of maximum at P36. LSD1 and H3K4me1/2 were expressed uniformly in all retinal progenitor cells. By P36, there was variability in LSD1 expression in the ganglion cell layer, uniform expression in the inner nuclear layer, and dichotomous expression between photoreceptors in the outer nuclear layer. This contrasted with H3K4me1/2 expression, which remained uniform. Additionally, LSD1 was widely expressed in the lens, cornea, and retinal pigment epithelium.
Conclusions
Consistent with its known role in neuronal differentiation, LSD1 is highly and uniformly expressed throughout all retinal progenitor cells. Variability in LSD1 expression, particularly in photoreceptors, may be indicative of their unique transcriptomes and epigenetic patterns of rods and cones. Murine rod nuclei exhibit LSD1 expression in a ring or shell, rather than throughout the nucleus, consistent with their unique inverted chromatin organization. LSD1 has substantial expression throughout adulthood, especially in cone nuclei. By providing insight into endogenous LSD1 expression, our current findings could directly inform future studies to determine the exact role of Lsd1 in the development and maintenance of specific structures and cell types within the eye.
Keywords: LSD1, epigenetics, histone demethylation, heterochromatin, euchromatin, rod and cone photoreceptors, retinal development, ocular development
Epigenetic modifications, such as DNA methylation and histone modifications, including acetylation, methylation, and phosphorylation, are dynamic and can change due to environmental influence.1,2 These dynamic changes contribute to the regulation of gene expression without altering the DNA sequence itself. Aberrant epigenetic alterations have been implicated in the development of ocular disorders, such as uveal melanoma, age-related macular degeneration, and glaucoma through the dysregulation of key biological processes, such as cell proliferation, oxidative stress, angiogenesis, and inflammation.3
Lysine-specific demethylase 1 (Lsd1) (Online Mendelian Inheritance in Man [OMIM] #609132), also known as Kdm1a and Aof2, was the first histone demethylase to be discovered. This protein acts specifically to demethylate the active mark H3K4 mono- and dimethyl (H3K4me1 and H3K4me2) and the repressive mark H3K9 mono- and dimethyl (H3K9me1 and H3K9me2) depending on its associated complex; thus, LSD1 can switch between a transcriptional repressor and activator.4,5 Inhibitors of LSD1 and its complex members have been explored for anticancer therapies and have shown promising results in clinical trials.6–8
Lsd1 and its downstream targets are involved in a wide range of biological functions, including embryonic development,9 neurogenesis,10,11 tumor-cell growth and metastasis,12,13 stress-induced emotional behaviors,14 and maternal reprogramming at fertilization.15 Three patients with de novo missense mutations in Lsd1 display numerous clinical symptoms, including ocular defects such as blue sclera, exotropia, and strabismus.16,17 In addition, patients with mutations in related epigenetic proteins, including KMT2D (OMIM #602113) or KDM6A (OMIM #300128), are often diagnosed with Kabuki syndrome. Kabuki syndrome 1 and 2 (OMIM #147920 and OMIM #300867, respectively) are characterized by intellectual disability and distinctive craniofacial features, and recently, a patient with a suspected deleterious mutation in Lsd1 exhibited Kabuki-like clinical features.17
Within the central nervous system, Lsd1 is involved in terminal differentiation of neurons. Inducible deletion of Lsd1 in adult mice lead to paralysis and hippocampal and cortex cell death as well as associated learning and memory problems.18 This may be, in part, facilitated through interactions in both the brain and retina between LSD1 and TLX, also known as NR2E1 (OMIM #603849), a master regulator of neural stem cell maintenance and neurogenesis.19,20
Despite the retina being a component of the central nervous system, little is known about the role of Lsd1 in ocular development or maintenance. Recently, Popova and colleagues21 found that Lsd1 is highly expressed in late progenitor retinal cells as they become postmitotic and begin to differentiate and that inhibition of LSD1 blocks the differentiation of the retinoblast into rod photoreceptors. Tsutsumi et al.22 found potential neuroprotective effects of an LSD1 inhibitor that may protect retinal ganglion cells (RGCs), which may have implications in glaucoma. These studies have examined the effects of LSD1 inhibition in the retina, and we aimed to extend the current understanding of endogenous LSD1 expression spatially and temporally and compare and contrast our work with theirs.
In this study, we evaluated the protein levels and localization of Lsd1 and its associated substrates H3K4me1 and H3K4me2 within the developing murine eye. Additionally, we looked at LSD1 expression within the adult human retina. Such “mapping” of Lsd1 could provide useful and necessary information for subsequent studies in the important field of epigenetic changes in retinal development and retinal diseases. We hypothesized that due to its role in neuron terminal differentiation, initiation of Lsd1 expression induces terminal differentiation in at least some retinal progenitor cells (RPCs). We also hypothesized that LSD1 would not be needed after retinal cells have terminally differentiated; thus, LSD1 levels would likely dramatically decrease. Testing these hypotheses are the goal of future experiments.
Methods
Animal Studies
Mouse housing, experiments, and handling were approved by the Emory University Institutional Animal Care and Use Committee, and the studies were conducted in adherence with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and followed the guidance and principles of the Association for Assessment and Accreditation of Laboratory Animal Care. C57BL/6J (wild type [WT]) and Thy1-YFPH mice were maintained on a 12-hour light/dark cycle at 23°C, and standard mouse chow (Lab Diet 5001; PMI Nutrition Inc., LLC, Brentwood, MO, USA) and water were provided ad libitum. The mice were managed and housed by Emory University Division of Animal Resources. Adult mice were euthanized using CO2 gas asphyxiation for 5 minutes, followed by cervical dislocation. Weanling pups (aged <postnatal day 21 [P21]) were sacrificed using decapitation. Animals ranged in age from P2 to P330.
Human Studies
Within the records of the L.F. Montgomery Laboratory at the Emory Eye Center, enucleation specimens ranging from January 1940 to August 2017 were identified that contained intact retinas. Many of these samples were enucleated due to a diagnosis of retinoblastoma. For the purposes of this study, only samples that contained parts of the retina with normal morphology and three nuclear layers were included for further LSD1 expression analysis. These studies were determined to be exempt by the Emory Institutional Review Board in full compliance with the tenets of the Declaration of Helsinki and Association for Research in Vision and Ophthalmology guidelines.
Immunofluorescence
All antibodies are listed in the Table. Unless otherwise noted, one to three drops of 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain plus fluorshield (#F6057; Sigma, St. Louis, MO, USA) was applied on top of each immunofluorescence slide (both sections and flatmounts), and a coverslip (#22; #152250; Thermo Fisher, Waltham, MA, USA) was placed on top of the slide. The edges of the coverslip were sealed with nail polish and allowed to dry overnight in darkness at room temperature before imaging.
Table.
List of Antibodies Used in These Experiments
| Antibody |
Antibody Type |
Company and Catalog Number |
Concentration |
Application |
| Rabbit anti-LSD1 | Primary antibody, monoclonal | Abcam, ab129195 | [1:250] | Sections + Western blotting + retina flatmount + RPE flatmount |
| Goat anti-short wavelength cone opsin | Primary antibody, polyclonal | Santa Cruz, sc-14363 | [1:250] | Sections |
| Rabbit anti-H3K4me1 | Primary antibody, polyclonal | Abcam, ab205256 | [1:1000] | Western blotting |
| Rabbit anti-H3K4me2 | Primary antibody, polyclonal | Abcam, ab7766 | [1:1000] | Western blotting |
| Rabbit anti-H3 | Primary antibody, polyclonal | Abcam, ab1791 | [1:1000] | Western blotting |
| Mouse anti-beta-actin | Primary antibody, monoclonal | Sigma Aldrich, A5441 | [1:1000] | Western blotting |
| Goat anti-GFP | Primary antibody, polyclonal | Novus Biologicals, NB100-170 | [1:750] | Retina flatmount |
| Rat anti-ZO1 | Primary antibody, monoclonal | EMD Millipore, MABt11 | [1:100] | RPE flatmount |
| Donkey anti-rabbit AF488 | Secondary antibody, polyclonal | Jackson Immunological, 711-545-152 | [1:1000] | Sections + retina flatmount + RPE flatmount |
| Donkey anti-goat redX AF568 | Secondary antibody, polyclonal | Jackson Immunological, 705-295-147 | [1:1000] | Sections + retina flatmount |
| Goat anti-rat AF 568 | Secondary antibody, polyclonal | Life Technologies, A10042 | [1:1000] | RPE flatmount |
| Goat anti-mouse IgG-HRP | Secondary antibody | Abcam, ab7068 | [1:5000] | Western blotting |
| Mouse anti-rabbit IgG-HRP | Secondary antibody | Santa Cruz, sc-2357 | [1:5000] | Western blotting |
Sections
Murine eyes were enucleated and placed in zinc + formaldehyde (Z-fix; #SKU622; Anatech Ltd., Battle Creek, MI, USA) for fixation for 1 hour at room temperature. Afterward, the eyes were washed three times in 1× phosphate-buffered saline (PBS) (46-013-CM; Corning, Corning, NY, USA), dehydrated using increasing percentages of ethanol and xylene, and embedded in paraffin. Five-micron-thick sections were cut using a microtome, placed on superfrost glass micro slides (#48311-703; VWR, Radnor, PA, USA), and allowed to air dry vertically at ambient room temperature overnight. The slides were then incubated in fresh xylenes for 8 minutes, 5 minutes, and 2 minutes before being washed in decreasing ethanol percentages for 2 minutes each (100%, 90%, 80% 70%, 60%, and 50%) and then twice in 1× PBS for 2 minutes each. Afterward, the slides were heated for 30 minutes in citrate buffer (10 mM sodium citrate, 0.05% (vol/vol) Tween 20 [pH 6.0]) in a 95°C water bath for antigen retrieval. Coplin jars with the slides were removed from the water bath and cooled to room temperature in a beaker filled with distilled room temperature water for 15 minutes. Slides were washed one final time for 5 minutes in 1× PBS before being placed in a humid chamber. A boundary was drawn around each individual retinal section using a pap pen (#195505; Research Products International, Mt. Prospect, IL, USA). Slides were incubated in 1× Powerblock solution (#NC9495720; Fisher Scientific, Hampton, NH, USA) for 1 hour. Primary antibodies were diluted using 1× Powerblock and 1× PBS (dilutions indicated in the Table). Sections were incubated in primary antibody overnight at 4°C in a humidified chamber. The next day, slides were washed three times in 1× PBS for 5 minutes each and then incubated in secondary antibodies for 2 hours at room temperature (in a sealed humidified black plastic box). Slides were again washed three times in 1× PBS for 5 minutes each.
Retina Flatmounts
Murine eyes from P30 Thy1-YFPH mice were enucleated and placed in prechilled 10 mL of 97% methanol (#34860; Sigma Aldrich) + 3% glacial acetic acid solution for 4 days at −80°C.23 These mice endogenously express yellow fluorescent protein under the control of the Thy1 promoter, a RGC marker,24 in a random subset of 3% to 5% of total RGCs.25,26 Prior to dissection, eyes were left at room temperature for 3 hours. Dissection was conducted as described in Boatright et al.27 with one adjustment; the neural retina was not removed and remained intact and attached to the retinal pigment epithelium (RPE). After dissections were complete, samples were washed for 5 minutes in 1× PBS (46-013-CM; Corning) with 0.1% vol/vol Triton X-100 (Sigma) (0.1% PBS-TX). Samples were then blocked in 1× Powerblock solution for 1 hour and then incubated overnight at 4°C with primary antibody. Primary antibodies were diluted to the appropriate concentrations (see Table) using 1× PBS and 1× Powerblock. The following day, samples were washed 5 times with a 0.1% PBS-TX solution and then incubated with secondary antibodies (see Table) at room temperature for 1 to 2 hours. Samples were then washed three times in 0.1% PBS-TX, once in 0.01% PBS-TX, and once in 1× PBS.
RPE Flatmounts
Murine eyes were enucleated and placed in zinc + formaldehyde for fixation for 10 minutes at room temperature. Afterward, the eyes were washed three times in 1× PBS (46-013-CM; Corning) and stored at 4°C for 1 to 2 hours before dissection. RPE flatmount dissections were prepared following Boatright et al.27 Following dissection, RPE flatmounts were individually transferred into a well created by attaching a silicone gasket (#GBL665104-25EA; Sigma Aldrich) to a SuperFlost Plus microscope glass slide (#12-550-15; Fisher Scientific). Flatmounts were incubated in 300 μL of blocking buffer (1% [wt/vol] bovine serum albumin [catalog #BP9703-100] and 0.1% [vol/vol] Triton X-100 (Sigma) in Hanks' balanced salt solution [catalog #MT21023CV; Fisher Scientific]) for 1 hour at room temperature in a humidified chamber. Primary antibodies (see Table) were diluted and preblocked in the blocking buffer for 1 hour prior to being applied to the flatmounts. Blocking buffer was aspirated from the flatmounts, and the flatmounts were incubated overnight at room temperature in primary antibodies. The next day, primary antibodies were aspirated and the flatmounts were rinsed five times for 2 minutes each with wash buffer (Hanks' balanced salt solution and 0.1% vol/vol Triton X-100). Secondary antibodies (see Table) were diluted and preblocked in blocking buffer for 1 hour at room temperature before being applied to the flatmounts overnight at room temperature. The next day, flatmounts were rinsed five times for 2 minutes with the wash buffer. Afterward, the gasket was removed from the glass slide and the flatmounts were mounted with Fluoromount-G (catalog #0100-01; SouthernBiotech, Birmingham, AL, USA) and covered with a 22 × 40-mm coverslip (#152250; Thermo Fisher).
Confocal Microscopy
Imaging of the retina sections was performed using a Nikon C1 confocal imaging system with solid-state laser excitation at 488 and 568 nm. Confocal images were stitched together using Adobe Photoshop CS2.
Western Blots
Sample Preparation
Retinas were dissected from each eye separately and placed in a round-bottom 0.5-ml screwcap tube with both retinas from the same animal pooled in one tube (catalog #022363344; Eppendorf, Hamburg, Germany). Homogenization buffer was prepared from 10-mL radioimmunoprecipitation assay buffer, one tablet of protease inhibitor (compete mini protein inhibitor catalog #118361530001), and one tablet of phosphatase inhibitor (PhosSTOP EASypack #04906845001; Roche, Basel, Switzerland). A total of 75 μL of homogenization buffer was added to each pair of retinas along with one 4.7-mm ferric bead in a screw cap tube. The homogenizer was a Qiagen (Hilden, Germany) TissueLyser LT.24 The retinas were macerated at 50 oscillations per second for 3 minutes at 4°C. Afterward, the ferric bead was removed using a magnet, and samples were centrifuged at 4°C for 3 minutes (10,000g) to remove particulate debris. Supernatants were collected and transferred to fresh 500-μL Eppendorf tubes and stored at −80°C until further use.
Protein Concentration Determination
The overall protein concentration of samples was determined using a bicinchoninic acid (BCA) assay.28 A 1:10 dilution of the supernatant was prepared with homogenization buffer (described above) for quantification. A total of 10 μL of each diluted sample was pipetted in an individual well of a 96-well cell culture plate (#165306; Thermo Scientific) in triplicate along with standards from a Pierce BCA Protein Assay Kit (catalog #23227; ThermoFisher) prepared according to the manufacturer's instructions. A total of 200 μL of BCA working reagent, also prepared according to the manufacturer's instructions, was added to each well, and the entire culture plate was incubated at 37°C for 30 minutes before absorbance at 562 nm was measured using a Synergy H1 Hybrid Plate Reader (BioTek, Winooski, VT, USA).
SDS-PAGE
The sample supernatants were adjusted to a protein concentration of 0.8 mg/mL in 200 μL by using 100 μL of 2× Laemelli buffer29 prepared with 2-mercaptoethanol (#21985023; ThermoFisher) (50 μL of 14.3 M 2-mercaptoethanol and 950 μL 2× Laemelli buffer29) and cold homogenization buffer (described above) and were stored at −20°C up to a week following preparation. Immediately before, electrophoresis samples were heated for 5 minutes at 95°C in a thermocycler.
Running/Staining Gel
A total 30 μL of each sample was loaded into individual lanes onto a precast Criterion gel (TGX Stain Free Gel 4%–15%; catalog #567-1083; BioRad, Hercules, CA, USA) as well as 10 μL of ladder (catalog #1610376; BioRad) and run at 100 V for 90 minutes. Samples were then transferred for 7 minutes onto a polyvinylidene difluoride blotting membrane30 by using the Trans-blot turbo pack (catalog #170-4157; BioRad) and Trans-blot Turbo Transfer System (BioRad). The gel was checked pre- and posttransfer to assure that a good transfer of proteins from the gel to the membrane was achieved.
Prior to primary antibody incubation, membranes were blocked for 2 hours at room temperature with 5% (wt/vol) instant nonfat dry milk (catalog #A614-1005; Quality Biological, Gaithersburg, MA, USA) in Tris-buffered saline (#1706435; Bio-Rad) with 0.1% (vol/vol) Tween 20 (BP337-100; Fisher Scientific) (TBST). Primary antibodies (see Table) were diluted with 5% milk in TBST, and membranes were incubated overnight on a 4°C shaker. The membrane was washed three times for 5 minutes each using 0.1% TBST. Horseradish peroxidase (HRP)-conjugated secondary antibodies (see Table) were diluted with 5% milk in TBST, and membranes were incubated 1 to 2 hours at room temperature on a shaker. The membrane was washed three times for 5 minutes each using 0.1% TBST. A total of 10 mL of Luminata Crescendo Western HRP substrate (catalog #WBLUR0500; EMD Millipore, Burlington, MA, USA) was applied to the membrane for 5 minutes. The membrane was imaged in chemiluminescence mode using the MP ChemiDoc Imaging System (Bio-Rad). Exposure times varied from 30 to 180 seconds. In order to re-probe the same membrane with multiple antibodies, after imaging, 10 mL of Restore Western blot stripping buffer (catalog #21059; Thermo Scientific) was applied to the blot for 10 minutes; the blot was washed for 5 minutes using 0.1% TBST, then blocked with 5% milk (wt/vol) in TBST, and incubated with the appropriate primary and secondary antibody as described above.
Western Blot Data Analysis
Densitometry was conducted on the Western blot images using PhotoShop CS6. All protein levels were normalized to (divided by) their respective loading control levels (either beta Actin or H3). Mouse retina samples ranged from P2 to P330 for 11 different time points (P2, P4, P5, P6, P7, P10, P14, P21, P36, P121, and P330), and two independent sets of samples were run. In order to determine possible statistical differences in LSD1, H3K4me1, and H3K4me2 across different ages, retina samples ranged from P2 to P36 for five different time points (P2, P7, P14, P21, and P36) and three independent samples were run for each time point.
Statistical Analysis
Statistical analysis was conducted using Prism 7 for Mac OS X Version 7 (GraphPad Software, Inc., La Jolla, CA, USA). All data are summarized as mean ± SD, and individual statistical tests are listed in figure legends. P values of <0.05 were considered to be statistically significant. Each sample group member is an independent mouse.
Results
Strong Expression of LSD1 in Murine Retinas Throughout Development
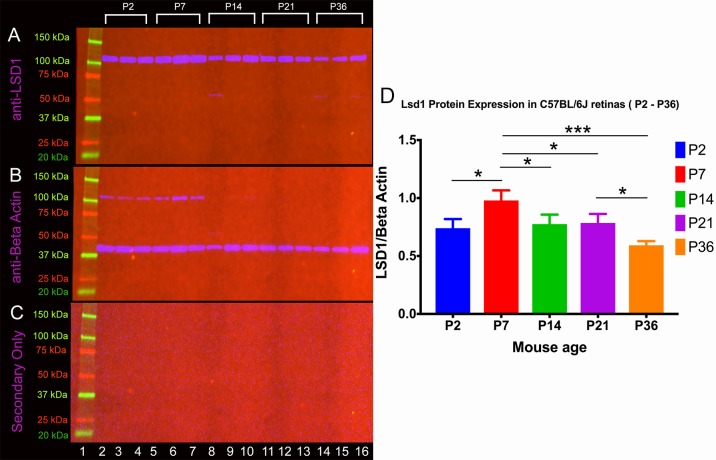
LSD1 is highly expressed in late-stage RPCs; however, it is not known whether this expression is maintained after development is completed.19 In order to resolve spatial and temporal expression of LSD1 in more detail, we performed Western blot analysis from C57Bl/6J animals starting at P2 during development through maturation at P330 (Supplementary Fig. S1). We probed for LSD1 (Supplementary Fig. S1A), and a secondary-only control was used to identify any nonspecific antibody binding (Supplementary Fig. S1B). Single bands were detected when probing for LSD1, and no bands were detected in the secondary-only control blots. Two independent Western blots were conducted using independent sets of samples (all from different mice), and quantification of the results for both experiments are shown in Supplementary Figure S1C. In general, LSD1 levels are higher at younger ages, which correspond to the retinoblast stage, and steadily decreased over development.
To determine if there was a significant reduction in LSD1 protein levels as the retina matures, we conducted Western blotting on a subset of developmental time points (P2, P7, P14, P21, and P36) by using three independent samples (Fig. 1). We probed for LSD1 (Fig. 1A), beta-actin served as a loading control (Fig. 1B), and a secondary-only control determined any nonspecific antibody binding. We detected a single band for LSD1 (expected size, 107 kDa), a single strong band for beta-actin (expected size, 42 kDa) with a faint band at 107 kDa that likely corresponds to residual LSD1 antibody that remained after the stripping process, and no band in the secondary-only control. The Western blot results were quantified using densitometry, and LSD1 was normalized to beta-actin for each individual sample. This subset of samples allowed for better resolution of changes in LSD1 protein levels during retinal development. LSD1 is present at P2, and then its expression significantly increases and peaks by P7, which corroborates results from Popova et al.21 LSD1 then decreases significantly from P7 to P21 until reaching a final “basement” level at P36 (Fig. 1D). This basement level is a plateau corresponding to ∼60% of the maximum LSD1 levels observed at P7. A two-way ANOVA shows a statistically significant difference between LSD1 protein levels between P2 and P7, P7 and P14, P7 and P21, P7 and P36, and P21 and P36, and a full list of the statistical tests and their results are listed in Supplementary Table S1. Thus, although LSD1 levels decrease after terminal differentiation is complete, it remains present in the retina throughout the lifetime of the mouse.
Figure 1.
LSD1 protein levels peaks at P7. Western blot analysis was conducted on C57BL/6J mouse retina samples at five different time points (P2, P7, P14, P21, and P36) in triplicate. Samples were probed with an anti-LSD1 antibody (single band, expected size, 107 kDa) (A), anti-beta-actin (single band, expected size, 43 kDa) served as a loading control (B), and secondary-only controls detected no nonspecific antibody binding (C). (D) Quantification of results was achieved using densitometry, and LSD1 levels were normalized to beta-actin. A two-way ANOVA with Tukey's multiple comparison test was conducted between the mean expression level in all possible pair combinations. There is a statistically significant decrease in LSD1 protein levels between P2 and P7, P7 and P14, P7 and P21, P7 and P36, and P21 and P36. A full list of comparisons and P values is listed in Supplementary Table S1. *P < 0.05, **P < 0.01, ***P < 0.001.
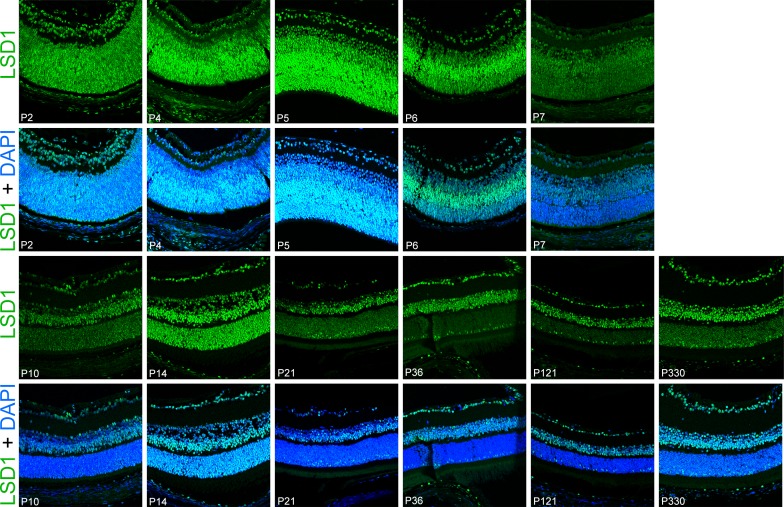
In order to determine the localization of LSD1 during and after retinal development, we performed immunofluorescence staining for LSD1 in retinal sections from C57BL/6J animals at the same time points used in Supplementary Figure S1 (P2–P330). Throughout the lifetime of the mouse, we observed LSD1 expression solely within the nuclei of cells, which is consistent with its role in demethylating histone proteins (Fig. 2).31,32 We observed high expression of LSD1 throughout the developing retinoblast from P2 to P14. At P21 (weaning age), LSD1 expression remained high; however, the expression pattern began to lose uniformity from one nucleus to the next in a given cell type. At P36, when retinal maturation is complete, the three nuclear layers showed variation in LSD1 expression from layer to layer and among different cells within a layer. Within the retinal ganglion cell layer (GCL), LSD1 had variable expression among different RGCs and displaced amacrine cells, as will be explored later. Within the inner nuclear layer (INL), which consists of amacrine, bipolar, and horizontal cells, there was uniform staining throughout each nucleus regardless of cell type. However, as will be further explained below, in the outer nuclear layer (ONL), there was a distinctly different expression pattern among the photoreceptor cells. This variation of LSD1 expression among different cell types of the GCL and ONL was maintained from P36 until P330. Thus, both the levels and cellular/histological expression patterns of LSD1 changed throughout the lifetime of the mouse, although this does not necessarily equate to altered enzymatic activity.
Figure 2.
LSD1 detected in all retinal cells starting at P2 until P330. Immunofluorescence staining of C57BL/6J mouse retinas for LSD1 alone (green) (top row of images) and LSD1 (green) + DAPI nuclear stain (blue) (bottom row of images). Images were taken using a 40× objective lens on a confocal microscope. LSD1 expression was uniformly present in all cells in the retinoblast starting at P2, and this uniform expression was consistent until the retina fully matured at P21. From P36 until P330, LSD1 was present in all three nuclear layers; however, the expression pattern was variable among different retinal subtypes.
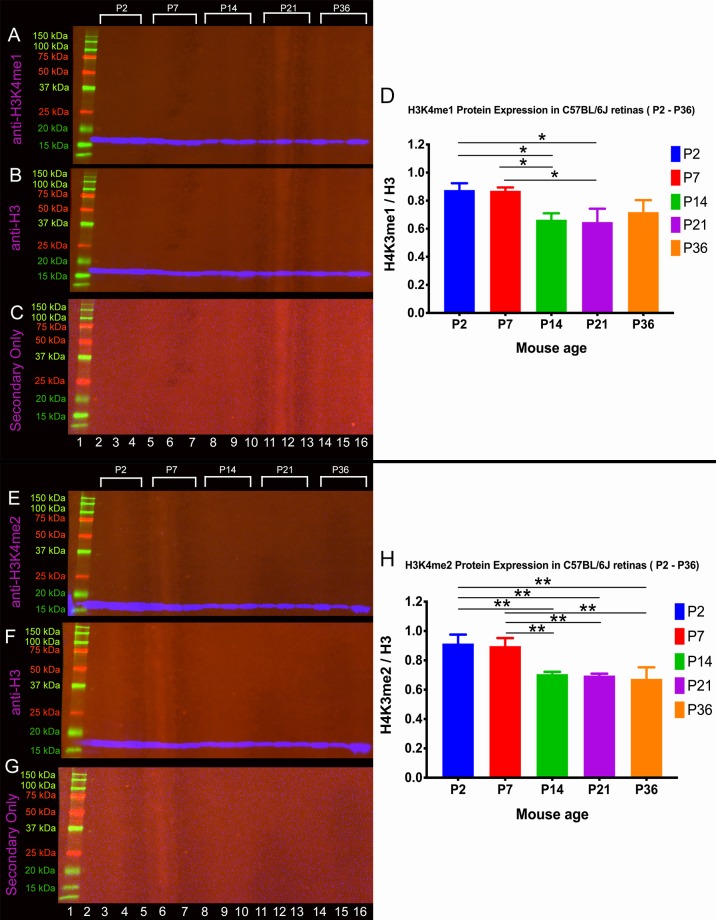
LSD1 demethylates histone modifications H3K4me1/2 and H3K9me1/2. Like LSD1, it was unclear whether H3K4me1 or H3K4me2 protein levels were changing or not. We conducted Western blotting for H3K4me1 and H3K4me2 (Fig. 3) on the same subset of developmental time points (P2, P7, P14, P21, and P36) as LSD1 in triplicate to determine if there was a significant reduction in either substrate as the retina matures. Figure 3A and 3E show H3K4me1 and H3K4me2, respectively, and H3 served as an internal loading control for each sample (Figs. 3B and 3F). Unlike LSD1, which peaked at P7, both H3K4me1 and H3K4me2 have equally high levels of expression at P2 and P7, and expression significantly decreases as the retina fully matures. A two-way ANOVA shows a statistically significant difference for H3K4me1 protein levels between P2 and P14, P2 and P21, P7 and P14, and P14 and P21 (Fig. 3D), and a full list of the statistical tests and their results are listed in Supplementary Table S2. For H3K4me2, a two-way ANOVA shows a statistically significant difference between P2 and P14, P2 and P21, P2 and P36, P7 and P14, P7 and P21, and P7 and P36. A full list of the statistical tests results for H3K4me2 is listed in Supplementary Table S3. Although both substrate levels decreased after terminal differentiation is complete, they are expressed in the retina throughout the lifetime of the mouse. Changes in H3K4me1 and H3K4me2 are unlikely to be solely influenced by changes in LSD1 levels. Histone methyltransferases, such as SET1, can have direct impacts, whereas epigenetic remodelers, such as the CHD family, can influence the epigenome as a whole by disrupting, moving, and exchanging nucleosomes.33,34 Those additional epigenetic proteins are likely to be active as the progenitor retinal cells differentiate into their respective terminal cell types, thus, leading to the observed changes in H3K4me1 and H3K4me2 levels seen in Figure 3.
Figure 3.
LSD1 substrates H3K4me1 and H3K4me2 peak at P2 and significantly decrease across retinal developmental time. Western blot analysis was conducted on C57BL/6J mouse retina samples at five different time points (P2, P7, P14, P21, and P36) in triplicate. Samples were probed with an anti-H3K4me1 (A) or anti-H3K4me2 antibody (E) (single band, expected size, 18 kDa), and an anti-H3 antibody (B, F) (single band, expected size, 18 kDa) served as a loading control. Quantification of results was achieved using densitometry, and H3K4me1/H3K4me2 levels were normalized to H3. A two-way ANOVA with Tukey's multiple comparison test was conducted between the mean expression level in all possible pair combinations. For H3K4me1, there is a statistically significant decrease between P2 and P14, P2 and P21, P7 and P14, and P7 and P21 (D). For H3K4me2, there is a statistically significant decrease between P2 and P14, P2 and P21, P2 and P36, P7 and P14, P7 and P21, and P7 and P36 (H). A full list of comparisons and P values are listed in Supplementary Table S2 and Supplementary Table S3. *P < 0.05, **P < 0.01, ***P < 0.001.
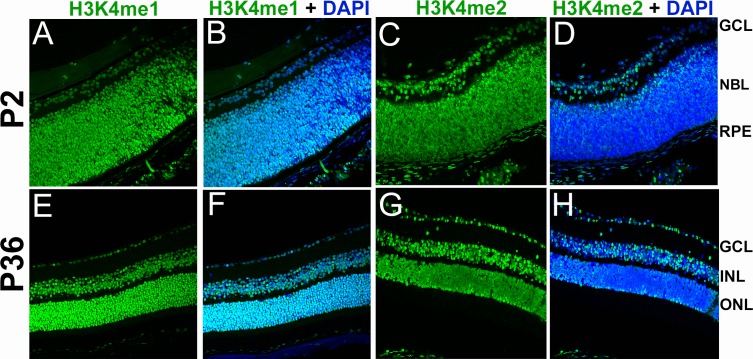
As seen in Figure 2, LSD1 localization changes over retinal developmental time. To determine whether similar changes are seen in H3K4me1 and H3K4me2, we performed immunofluorescence at P2 and P36 for these histone modifications (Fig. 4). Both H3K4me1 and H3K4me2 are uniformly highly expressed throughout all RPCs (P2) (Figs. 4A and 4C show H3K4me1 and H3K4me2 alone; Figs. 4B and 4D show H3K4me1 + DAPI nuclear stain and H3K4me2 + DAPI nuclear stain) and all differentiated retinal subtypes (P36) (Figs. 4E and 4G show H3K4me1 and H3K4me2 alone; Figs. 4F and 4H show H3K4me1 + DAPI nuclear stain and H3K4me2 + DAPI nuclear stain). Thus, in contrast to LSD1, both H3K4me1 and H3K4me2 expression remain consistent in all cells across developmental time.
Figure 4.
LSD1 substrates H3K4me1 and H3K4me2 are expressed throughout the retinoblast and mature retina. Immunofluorescence staining of C57BL/6J mouse retinas at P2 and P36 for H3K4me1 and H3K4me2 (green) with a DAPI nuclear stain (blue). Images were taken using a 40× objective lens on a confocal microscope. At P2, H3K4me1 (A, B) and H3K4me2 (C, D) were expressed uniformly in the retinoblast throughout all RPCs. At P36 in the mature retina H3K4me1 (E, F) and H3K4me2 (G, H) maintain uniform expression in all retinal cell subtypes. GCL, ganglion cell layer; NBL, neuroblastic layer; RPE, retinal pigment epithelial layer; INL, inner nuclear layer; ONL, outer nuclear layer.
Differential Expression of LSD1 in Rod and Cone Photoreceptors
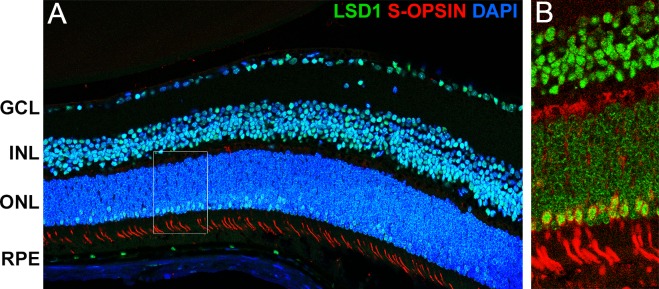
Within the ONL, only a small subset of cells have LSD1 expression levels that appear to be similar to the expression seen in either the GCL or INL; the majority of the cells appear to display relatively less expression. Because cone photoreceptors make up only 3% of the total cells within the ONL,35 we hypothesized that these relatively high-expressing LSD1 cells within the ONL are cone photoreceptors. Retinal sections from P36 C57BL/6J mice were stained for LSD1 and short-wavelength cone opsin (S-OPSIN), a marker for cone photoreceptors. Cells in the photoreceptor layer exhibiting high levels of LSD1 immunosignal also exhibited S-OPSIN immunosignal, whereas cells in the photoreceptor layer that exhibited only modest levels of LSD1 immunosignal were not stained for S-OPSIN (Fig. 5), suggesting that LSD1 is expressed in cone, but not rod, photoreceptors. This difference is even more striking after three-dimensional rendering using Imaris software (Fig. 6) (Imaris, version 9.2; Bitplane USA, Concord, MA, USA). Interestingly, the Imaris images also reveal a difference in the pattern of LSD1 expression between the two types of photoreceptors. Cone photoreceptors have a uniform staining pattern within the nucleus similar to cells in the GCL and INL. However, rod photoreceptors displayed a “ring-like” or “shell-like” staining pattern.
Figure 5.
High levels of LSD1 in cone photoreceptors but not rods. Immunofluorescence staining of P36 C57BL/6J mouse retinas for LSD1 (green), short wavelength cone opsin (red), and DAPI nuclear stain (blue). The 40× merged image taken with a confocal microscope showed LSD1 expression in all three nuclear layers and short wavelength cone opsin expression in cone photoreceptor outer segments (A). The 60× image shows perfect correlation between cells with high LSD1 expression (green) along the outer edge of the ONL and the cone opsin (red) (B).
Figure 6.
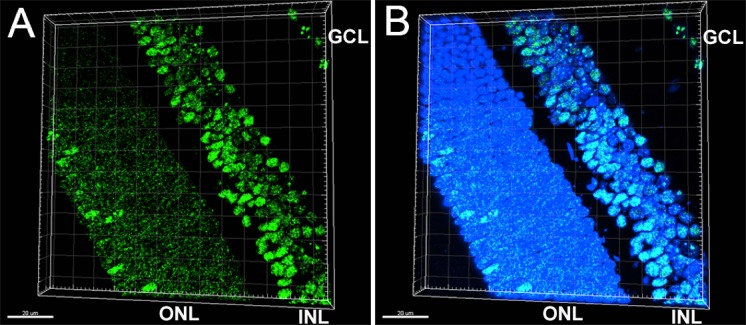
Three-dimensional rendering in Imaris of P36 C57BL/6J mouse retina section showing LSD1 ring-like staining pattern in rod photoreceptors. The 40× confocal images rendered in Imaris software show LSD1 (green) (A) and LSD1 + DAPI nuclear stain (blue) (B). Within the ONL, there is a distinct staining pattern difference between the two photoreceptors subtypes. Rod photoreceptors show a “ring-like” or “shell-like” staining pattern where LSD1 expression is located in the periphery of the nucleus. Cone photoreceptors show a uniform staining pattern which mimics the LSD1 expression found in cell types located in the INL and GCL.
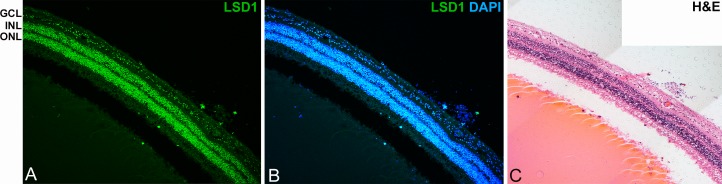
In contrast to the variation in LSD1 expression within the mature murine retina, a mature human retina showed uniform nuclear LSD1 expression in the ONL (Fig. 7). Qualitatively, there appears to be a general trend whereby all nuclei have roughly the same level of LSD1. Although the murine retina showed variability within the RGCs and photoreceptors, the human retina does not.
Figure 7.
LSD1 is uniformly expressed in all retinal cells in a normal human retina. The 20× confocal images show LSD1 expression alone (green) (A) and LSD1 (green) + DAPI nuclear stain (blue) (B). Hematoxylin and eosin (H&E) staining of the same eye (C) highlights location of cell nuclei and the intact morphology of the retinal layers. In contrast to the variable expression seen among retinal cell types in the murine eye, LSD1 is expressed at relatively uniform and equal levels throughout the entire human retina.
Variable Levels of LSD1 Within the GCL
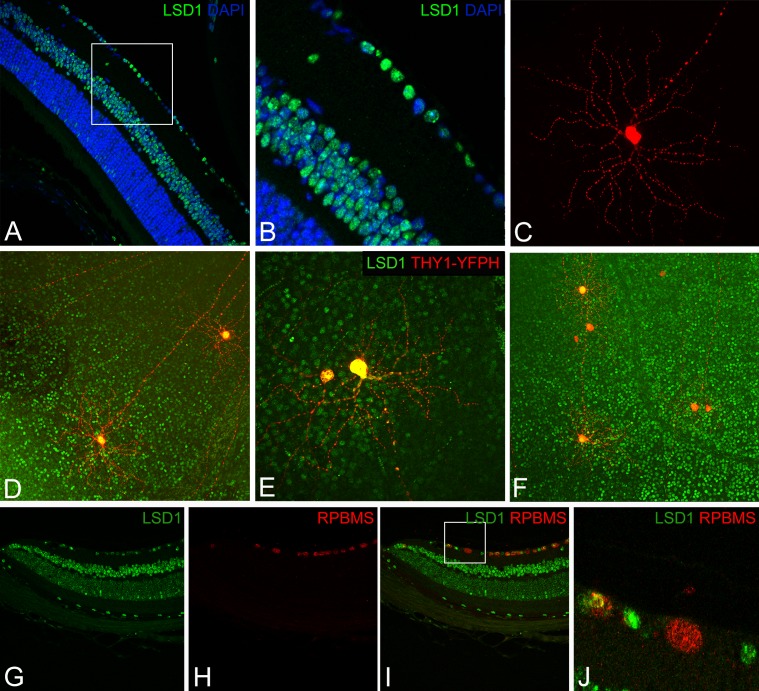
In addition to the variable expression of LSD1 observed among photoreceptor cells in the ONL, LSD1 also showed variable expression among individual cells in the GCL at P36. In Figure 8A, the GCL shows obvious differences in LSD1 levels between adjacent cells; these differences are highlighted in Figure 8B. There are two major classes of retinal cells found in the GCL: RGCs and displaced amacrine cells.36,37 To determine whether LSD1 expression was isolated to RGCs or not, we stained whole-retina flatmounts from transgenic mice expressing yellow fluorescent protein under the control of the pan-RGC marker Thy1 with LSD1.25 Using this technique, only 3% to 5% of RGCs are labeled with yellow fluorescent protein, which allows for visualization of the entire dendritic arborization of individual RGCs as well as a single axon projecting to the optic nerve head (Fig. 8C).26 In Figure 8D, two RGCs showed fairly equal LSD1 expression, as indicated by the amount of yellow color within their nuclei, which represents the overlap between the LSD1 fluorescence in the 488-nm confocal microscope channel and the THY1 fluorescence in the 568-nm channel. Figure 8E depicts two adjacent RGCs with varying levels of LSD1 expression. The left RGC had relatively less LSD1 than the right RGC. Figure 8F shows numerous RGCs with relatively high, intermediate, and low LSD1 levels within the same field of view. Supplementary Figure S2 shows LSD1 alone, THY1-YFPH alone, and LSD1 + THY1-YFPH merged for the images shown in Figures 8C to 8F. Because the THY1-YFPH mice only label 3% to 5% of the RGC population within a retina, it cannot determine whether LSD1 is expressed in displaced amacrine cells or not. For this, we colabeled LSD1 with a pan-RGC marker (RPBMS) in P90 C57Bl/6J retinal sections.38 Figures 8G to 8J show a peripheral retinal section labeled with LSD1 alone (Fig. 8G), RPBMS alone (Fig. 8H), and a merged view (Fig. 8I). A magnified area in Figure 8J shows four individual cells in the GCL. The two cells on the left are RGCs that express LSD1, as indicated by the overlap of 488-nm and 568-nm fluorescent signal. The third cell from the left only has RPBMS expression, indicating that this is a RGC that does not express LSD1. The cell on the right side only has LSD1 expression but not RPBMS, indicating that this is a displaced amacrine cell that expresses LSD1. For further clarification, Supplementary Figure S3 contains two different areas of P90 C57Bl/6J retina (peripheral and central) with individual channels for DAPI, LSD1, and RPBMS, as well as merged views of LSD1 + RPBMS and LSD1 + RPBMS + DAPI (Supplementary Figs. S3A–S3J). Additionally, we have included a displaced amacrine cell marker HPC-1 (syntaxin)39 colabeled with LSD1 with individual panels for DAPI, LSD1, and HPC-1 as well as merged views of LSD1 + HPC-1 and LSD1 + HPC-1 + DAPI (Supplementary Figs. S3K–S3O). Overall, these results indicate that LSD1 has variable expression in RGCs and displaced amacrine cells located in the GCL.
Figure 8.
LSD1 expression varies among different RGCs in the murine retina. Immunofluorescence staining of LSD1 (green) + DAPI nuclear stain (blue) in murine retinal section (A). Magnified image of GCL shows wide variability in the expression of LSD1 in adjacent RGCs (B). Thy1-YFPH-expressing RGCs stained with anti-GFP antibody (red) shows the dendrite pattern and axon projection of an individual RGC (C). Colocalization of LSD1 (green) within THY1-YFPH-expressing RGCs (red) in retina flatmounts show variability in relatively high, medium, and low LSD1 expression among adjacent RGCs (D–F). Colocalization of LSD1 (green) with a pan-RGC marker (RPBMS) shows that both RGCs and displaced amacrine cells in the GCL can express LSD1 (G–J).
LSD1 Expression in Ocular Structures Outside the Retina
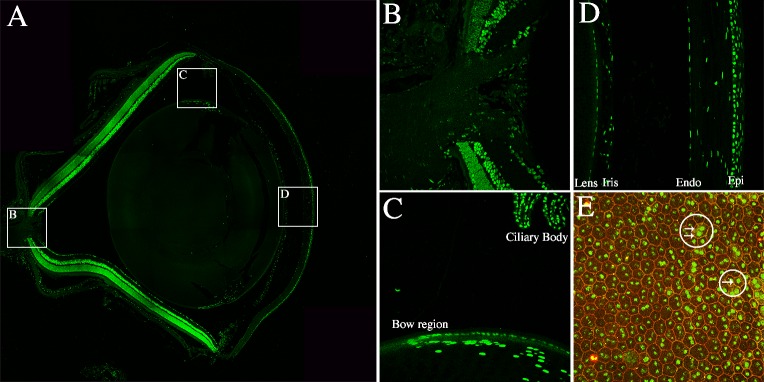
Although the focus of our study was on the expression of LSD1 within the retina, we observed LSD1 expression throughout the entire mature murine eye (Fig. 9A). At P36, LSD1 is expressed in almost all ocular structures, including the cornea, lens, retina, and RPE. In all cases, LSD1 was found only in nuclei, based on costaining of LSD1 with DAPI. At the optic nerve head, LSD1 is expressed in the central retina (Fig. 9B). In the lens, LSD1 was highly expressed in lens cells near the equatorial region and followed a gradient as the cells round the bow region (Fig. 9C). This region contains anterior epithelial cells that are beginning to differentiate and elongate into fiber cells, which will eventually lose their nuclei as they complete differentiation.40 LSD1 levels are very low to nonexistent in the anterior epithelium. The levels of LSD1 increase in these epithelial cells at the equator, and levels reach a maximum in cells that are undergoing elongation as they morph into fiber cells. In cornea, the central corneal epithelium had higher LSD1 expression than the endothelium and stroma, and LSD1 is expressed in the iris (Fig. 9D). Figure 9E shows a 40× RPE flatmount image stained for LSD1 in green and ZO-1, a protein associated with tight junctions, in red to outline the hexagonal borders of the RPE cells. Two white circles with arrows highlight different RPE cells that contain either one or two nuclei. Roughly half of RPE cells are mononuclear and half are binuclear, and LSD1 is expressed in all RPE cell nuclei.41 We did not observe a difference in the level of LSD1 comparing one nucleus to the other in binucleate RPE cells. We also did not observe any differences in LSD1 levels in nuclei located more centrally compared to those in the far periphery of the RPE layer.
Figure 9.
LSD1 is expressed throughout the murine eye. Immunofluorescence of LSD1 (green) and DAPI (blue) in P36 C57BL/6J retinal sections and RPE flatmount (A–E). The 20× confocal image of an entire P36 murine eye stained with LSD1 (green) (A) showed LSD1 expression throughout many ocular structures. White boxes indicate areas from which 40× images were taken for B–D to focus on various ocular structures outside of the retina including the optic nerve (B), lens (C), and cornea (D). In C the bow region of the lens and ciliary body are labeled, and in D the lens, iris, corneal endothelium, and corneal epithelium are labeled. A 40× image from an RPE flatmount showed LSD1 nuclear expression in green and ZO-1 in red (E) to outline all RPE cell borders. Two white circles with arrows highlight different RPE cells that contain either one or two nuclei.
Discussion
The purpose of this study was to extend the current understanding of endogenous LSD1 expression spatially and temporally in the retina. The premise of this study is that Lsd1 is known to play a major role in neuronal maturation and plasticity, specifically through a neuron-specific Lsd1 isoform, neuroLsd1 (nLsd1).42 The retina is comprised of numerous neuronal cell types; however, the role of Lsd1 in differentiating RPCs into committed neurons is largely unknown. In the developing retina, RPCs are actively undergoing mitosis during retinogenesis. As these cells start to commit to various cell lineages and undergo differentiation, gene expression patterns among different cells start to diverge. A small percentage of these changes in gene expression can be accounted for by changes in DNA methylation; however, the vast majority are due to changes in chromatin states via histone modifications.33,43 Differences in chromatin state allow for various retinal cell types to be present at distinct levels.33,44
Lsd1 serves as a key regulator of neural stem cell proliferation via its interactions with the transcription factor TLX,25 which establishes the undifferentiated and self-renewable state of neural stem cells.11,19 During embryonic stages, the mouse retina develops by first generating ganglion cells followed by the development of cones, horizontal cells, and most of the amacrine cells. Postnatally, bipolar cells, Müller glia, the remaining amacrine cells, and rod photoreceptors develop, although there is considerable overlap in the production of different retinal cells types at any specific time point.45–47 Our original hypothesis was that Lsd1 is critical for the transition between the retinal progenitor state and terminal differentiation. Although it is correlative, our data are consistent with this idea because LSD1 protein levels are highest at P7, when all RPCs showed uniform expression, until P21. Although high LSD1 expression is temporally consistent with terminal differentiation of various cell types, expression does not end abruptly in any coincidence with the end of mitotic cell division, differentiation, or synaptic maturation (Fig. 1). By P36, the different mature retinal neurons displayed various levels of LSD1 based on their unique subtypes, and LSD1 levels reached a basement maintenance level (Supplementary Fig. S1) at about 60% of maximum, which is maintained until the oldest age that we tested, P330. These expression pattern changes are also observed in localization changes that occur across developmental time (Fig. 2). To speculate, this maintenance level may be necessary to maintain the unique chromatin state and gene expression patterns that are distinct between the seven retinal cell subtypes, namely, rod photoreceptors, cone photoreceptors and bipolar, amacrine, horizontal, ganglion, and glial cells, and to maintain necessary biological functions for proper visual function.
Rod photoreceptor cells have relatively fewer regions of chromatin accessibility, and in general, the rod photoreceptor chromatin in nocturnal animals have an inverse architecture compared to those in diurnal animals.48 Nocturnal animals have chromatin in which the central part of the nucleus is heterochromatin and the surrounding peripheral areas are euchromatin.49 This inversion occurs through remodeling of the conventional nuclear architecture (euchromatin in the center and heterochromatin in the periphery), due to the lack of the inner nuclear membrane protein lamin B50 during postnatal differentiation of rod cells.51 Popova et al.21 demonstrated that pharmacological inhibition of LSD1 stops the development of rod photoreceptors. Our data show that the rod-specific changes of LSD1 nuclear partitioning occurred between P21 and P36, well after mouse retinas become functional by electroretinogram signal detection. Nocturnal animals see at light intensities that are a million times lower than those available during the day. Thus, the inverted architecture may optimize light transmission and reduce the scattering of light in the ONL.51,52 All other retinal cells within nocturnal animals have the conventional architecture, which may explain the difference in LSD1 expression patterns between rod and cone photoreceptors51 (Fig. 5). We observe relatively high expression in cone photoreceptors and relatively low expression in rods. Additionally, the “ring-like” staining pattern in the rods matches the peripheral location of euchromatin (Fig. 6). The inverted rod chromatin organization is only present in nocturnal animals. In diurnal animals, such as humans, the chromatin architecture between rod and cone photoreceptors are identical, which is consistent with the uniform LSD1 expression pattern we observed (Fig. 7).51 This may be the underlying reason why there is no distinction between cone and rod photoreceptor LSD1 staining in humans.
LSD1 acts upon histone modifications associated with active transcription, such as H3K4 mono- and dimethylation (H3K4me1 and H3K4me2), and these modifications should be located within the nuclear periphery of mouse rod photoreceptors.53 Furthermore, rod photoreceptor-specific genes accumulate H3K4me2 during retina development in the promoter and gene body.54 In contrast, H3K4 mono- and dimethylation are located in the central nucleus of cells in the inner parts of the retina.53 Yet, our immunofluorescence data show H3K4me1 and H3K4me2 located uniformly throughout all cells in the retinoblast and mature retina (Fig. 4). This contrast between the localization of LSD1 in the mature retina and its substrates H3K4me1 and H3K4me2 may be the result of LSD1 having specific demethylase activity on particular histone protein substrates, whereas H3K4me1 and H3K4me2 levels are influenced by a number of different epigenetic writers and erasers. Although the localization of the substrates in the mature retina differ from LSD1 localization, all three proteins peak in the retinoblast and decrease across retinal developmental time until basement levels at P36, indicating that they are still needed postdevelopment (Fig. 3).
Unlike the photoreceptors, which show two distinct expression patterns between the two subtypes (rods and cones), we observed variability in LSD1 expression among different RGCs in the mature murine retina (Supplementary Fig. S2) and in different cells within the ganglion cell layer, including displaced amacrine cells (Fig. 8; Supplementary Fig. S3). RGCs are classically categorized using morphologic, physiologic, and gene expression profiles. Their soma are located in the GCL with axons that project through the optic nerve to the brain, and ∼40 subtypes have been identified in the mouse.55–57 Given the numerous RGCs subtypes and the highly variable LSD1 expression, it can be hypothesized that LSD1 may play a role in the development of different types of RGCs. However, although this hypothesis warrants further investigation, it is outside the scope of the current study.
In addition to its role in neuron differentiation, Lsd1 also plays a major role in the overall differentiation of embryonic stem cells. Homozygous Lsd1 null mouse embryos are inviable and arrest in their development by embryonic day 7.5.4 Within the zygote, Lsd1 expression first appears during the morula stage, and by postimplantation embryos, expression becomes ubiquitous. Our original hypothesis was that Lsd1 expression would be relatively low and/or absent in structures outside of the adult retina; however, our results show strong LSD1 expression in all major ocular structures, including optic nerve, lens, cornea, and RPE (Fig. 9; Supplementary Fig. S4).
Despite extensive research on Lsd1 within the brain, relatively less research has been conducted on its role in the eye. Here, we sought to expand upon and contribute to previous research characterizing the importance of Lsd1 in the eye. Given the obvious differences in expression between rod and cone photoreceptors as well as the subtle differences between various RGCs, our work highlights the unique role of Lsd1 in the development of individual retinal subtypes. Additionally, despite the well-established role of Lsd1 in neuron development, the ubiquitous expression throughout the eye raises interesting questions about its role in epithelial tissues, such as cornea or RPE. Future work should investigate the basic mechanisms of how global or cell-type specific genetic or pharmacologic inhibition of Lsd1, either during or after ocular development, affects the formation of normal retinal subtypes and ocular tissues.
Lastly, from a translational perspective, inhibition of other epigenetic proteins, such as histone deacetylases, has been shown to have neuroprotective properties in retinal degenerative disorders, such as retinitis pigmentosa.58–60 Additional studies have looked at the inhibition of histone methylation, specifically H3K27me3, and its role in delaying the onset of retinal degeneration.61 LSD1 inhibitors have long been studied for their potential therapeutic abilities in relation to oncology,7,8 and clinical trials for both acute myeloid leukemia and small cell lung cancer are currently underway. Within the visual system, Tsutsumi et al.22 found potential neuroprotective effects of an LSD1 inhibitor in protecting RGCs, which may have implications in glaucoma. Thus, future research is needed to determine the exact role(s) that Lsd1 plays in ocular development in order to determine its potential as a therapeutic target for retinal diseases or in treating eye tumors.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health Grants R01EY028450, R01EY021592, P30EY006360, F31EY028855, R01EY028859, T32EY07092, and T32GM008490; the Abraham and Phyllis Katz Foundation; Veterans Affairs Rehabilitation Research & Development I01RX002806 and I21RX001924; Veterans Affairs Rehabilitation Research & Development C9246C (Atlanta Veterans Administration Center for Excellence in Vision and Neurocognitive Rehabilitation); and an unrestricted grant to the Department of Ophthalmology at Emory University from Research to Prevent Blindness, Inc.
Disclosure: S. Ferdous, None; H.E. Grossniklaus, None; J.H. Boatright, None; J.M. Nickerson, None
References
- 1.Fahrner J, Bjornsson H. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2015;510:84–91. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver VF, van Bysterveldt KA, Merbs SL. Medical Epigenetics. London: Academic Press; 2016. Epigenetics in ocular medicine; pp. 391–412. In: Tollefsbol TO, ed. [Google Scholar]
- 4.Wang J, Scully K, Zhu X, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 5.Laurent B, Ruitu L, Murn J, et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalin JH, Wu M, Gomez AV, et al. Targeting the CoREST complex with dual histone deacetylase and demethylase inhibitors. Nat Commun. 2018;9:53. doi: 10.1038/s41467-017-02242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y-C, Ma J, Wang Z, et al. A systematic review of histone lysine-specific demethylase 1 and its inhibitors. Med Res Rev. 2015;35:1032–1071. doi: 10.1002/med.21350. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Hevi S, Kurash JK, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wu Q, Yang P, et al. LSD1 co-repressor Rcor2 orchestrates neurogenesis in the developing mouse brain. Nat Commun. 2016;7:10481. doi: 10.1038/ncomms10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun G, Alzayady K, Stewart R, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Zhao Y, Wang L, et al. LSD1 promotes S-phase entry and tumorigenesis via chromatin co-occupation with E2F1 and selective H3K9 demethylation. Oncogene. 2018;37:534–543. doi: 10.1038/onc.2017.353. [DOI] [PubMed] [Google Scholar]
- 13.Cho HS, Suzuki T, Dohmae N, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 14.Rusconi F, Grillo B, Ponzoni L, et al. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A. 2016;113:3651–3656. doi: 10.1073/pnas.1511974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasson JA, Simon AK, Myrick DA, et al. Maternally provided LSD1/KDM1A enables the maternal-to-zygotic transition and prevents defects that manifest postnatally. Elife. 2016;5:e08848. doi: 10.7554/eLife.08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong JX, Yu J, Lorentzen P, et al. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med. 2016;18:788–795. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tunovic S, Barkovich J, Sherr EH, Slavotinek AM. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am J Med Genet A. 2014;164:1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- 18.Christopher MA, Myrick DA, Barwick BG, et al. LSD1 protects against hippocampal and cortical neurodegeneration. Nat Commun. 2017;8:805. doi: 10.1038/s41467-017-00922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam MM, Zhang C-L. TLX: a master regulator for neural stem cell maintenance and neurogenesis. Biochim Biophys Acta. 2015;1849:210–216. doi: 10.1016/j.bbagrm.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol. 2008;28:3995–4003. doi: 10.1128/MCB.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popova EY, Pinzon-Guzman C, Salzberg AC, Zhang SS-M, Barnstable CJ. LSD1-mediated demethylation of H3K4me2 is required for the transition from late progenitor to differentiated mouse rod photoreceptor. Mol Neurobiol. 2016;53:4563–4581. doi: 10.1007/s12035-015-9395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutsumi T, Iwao K, Hayashi H, et al. Potential neuroprotective effects of an LSD1 inhibitor in retinal ganglion cells via p38 MAPK activity. Invest Opthalmol Vis Sci. 2016;57:6461–6473. doi: 10.1167/iovs.16-19494. [DOI] [PubMed] [Google Scholar]
- 23.Sun N, Shibata B, Hess JF, Fitzgerald PG. An alternative means of retaining ocular structure and improving immunoreactivity for light microscopy studies. Mol Vis. 2015;21:428–442. [PMC free article] [PubMed] [Google Scholar]
- 24.Gooding SW, Chrenek MA, Ferdous S, Nickerson JM, Boatright JH. Set screw homogenization of murine ocular tissue, including the whole eye. Mol Vis. 2018;24:690–699. [PMC free article] [PubMed] [Google Scholar]
- 25.Barnstable CJ, Dräger UC. Thy-1 antigen: a ganglion cell specific marker in rodent retina. Neuroscience. 1984;11:847–855. doi: 10.1016/0306-4522(84)90195-7. [DOI] [PubMed] [Google Scholar]
- 26.Feng G, Mellor RH, Bernstein M, et al. Imaging neuronal sets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Boatright JH, Dalal N, Chrenek MA, et al. Methodologies for analysis of patterning in the mouse RPE sheet. Mol Vis. 2018;24:690–699. [PMC free article] [PubMed] [Google Scholar]
- 28.Smith P, Krohn R, Hermanson G, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;85:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Jin Y, Kim TY, Kim MS, Kim MA, Park SH, Jang YK. Nuclear import of human histone lysine-specific demethylase LSD1. J Biochem. 2014;156:305–313. doi: 10.1093/jb/mvu042. [DOI] [PubMed] [Google Scholar]
- 33.Aldiri I, Xu B, Wang L, et al. The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron. 2017;94:550–568. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- 36.Jeon C-J, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlamp CL, Montgomery AD, Mac Nair CE, Schuart C, Willmer DJ, Nickells RW. Evaluation of the percentage of ganglion cells in the ganglion cell layer of the rodent retina. Mol Vis. 2013;19:1387–1396. [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez AR. de Sevilla Muller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol. 2014;522:1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnstable CJ, Hofstein R, Akagawa K. A marker of early amacrine cell development in rat retina. Dev Brain Res. 1985;20:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- 40.Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodenstein L, Sidman RL. Growth and development of the mouse retinal pigment epithelium. Dev Biol. 1986;121:192–204. doi: 10.1016/0012-1606(87)90152-7. [DOI] [PubMed] [Google Scholar]
- 42.Rusconi F, Grillo B, Toffolo E, Mattevi A, Battaglioli E. NeuroLSD1: splicing-generated epigenetic enhancer of neuroplasticity. Trends Neurosci. 2016;40:1–11. doi: 10.1016/j.tins.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Hartl D, Krebs AR, Jüttner J, Roska B, Schübeler D. Cis-regulatory landscapes of four cell types of the retina. Nucleic Acids Res. 2017;45:11607–11621. doi: 10.1093/nar/gkx923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young R. Cell proliferation during postnatal development of the retina in the mouse. Dev Brain Res. 1985;21:229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- 46.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 47.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Mo A, Luo C, Davis FP, et al. Epigenomic landscapes of retinal rods and cones. Elife. 2016;5:1–29. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falk M, Feodorova Y, Naumova N, et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019;570:395–399. doi: 10.1038/s41586-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solovei I, Wang AS, Thanisch K, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Solovei I, Kreysing M, Lanctôt C, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 52.Sterling P. How retinal circuits optimize the transfer of visual information. In: Calupa LM, Werner JS, editors. The Visual Neurosciences. Cambridge, MA: MIT Press;; 2004. pp. 234–239. [Google Scholar]
- 53.Eberhart A, Feodorova Y, Song C, et al. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosom Res. 2013;21:535–554. doi: 10.1007/s10577-013-9375-7. [DOI] [PubMed] [Google Scholar]
- 54.Popova EY, Xu X, DeWan AT, et al. Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3. 2012;7:e46867. doi: 10.1371/journal.pone.0046867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levick WR. Receptive fields and trigger features of ganglion cells in the visual streak of the rabbit's retina. J Physiol. 1967;188:285–307. doi: 10.1113/jphysiol.1967.sp008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- 57.Bae JA, Mu S, Kim JS, et al. Digital museum of retinal ganglion cells with dense anatomy and physiology. Cell. 2018;173:1293–1306. doi: 10.1016/j.cell.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Dai X, Qi Y, He Y, Du W, Pang JJ. Histone deacetylases inhibitors in the treatment of retinal degenerative diseases: overview and perspectives. J Ophthalmol. 2015;2015:250812. doi: 10.1155/2015/250812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sancho-Pelluz J, Alavi MV, Sahaboglu A, et al. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death Dis. 2010;1:e24. doi: 10.1038/cddis.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sancho-Pelluz J, Paquet-Durand F. HDAC inhibition prevents Rd1 mouse photoreceptor degeneration. Adv Exp Med Biol. 2012;723:107–113. doi: 10.1007/978-1-4614-0631-0_15. [DOI] [PubMed] [Google Scholar]
- 61.Zheng S, Xiao L, Liu Y, et al. DZNep inhibits H3K27me3 deposition and delays retinal degeneration in the rd1 mice article. Cell Death Dis. 2018;9:310. doi: 10.1038/s41419-018-0349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.