Abstract
Objective
We investigated the relationship of the transcriptional tumor immune microenvironment with prognosis of patients with locally advanced lung adenocarcinoma (LUAD).
Materials and methods
A targeted RNA-Seq approach was used to measure the abundance of 395 immune-related transcripts of 24 formalin-fixed paraffin embedded (FFPE) tumor specimens from our institution and transcription data of 85 matched LUAD samples from The Cancer Genome Atlas (TCGA). Gene set variation analysis (GSVA) was used to identify gene sets related to prognosis, and the microenvironment cell-population (MCP)-counter method was used to quantify infiltrated immune cells. Survival analysis with the log rank test was used to determine the relationships of different immune-related transcripts with prognosis. Cox proportional hazards models were also used to identify risk factors associated with poor prognosis.
Results
Among our patients, GSVA and the log rank test demonstrated that enrichment of the antigen processing pathway (P = 0.01) correlated with a favorable prognosis. MCP-counter and survival analysis demonstrated that greater CD8 T cell infiltration correlated with a favorable prognosis (P = 0.05), but greater infiltration of neutrophils (P = 0.014) and NK cells (P = 0.015) correlated with poor prognoses. Cox hazard analysis showed that greater infiltration of neutrophils was an independent risk factor for poor prognosis. These results were consistent with LUAD data from TCGA.
Conclusion
When integrated with computational bioinformatics methods, targeted RNA-Seq from FFPE specimens provides profiles of the tumor immune microenvironment that have prognostic value for patients with locally advanced LUAD.
Keywords: NSCLC, lung adenocarcinoma, RNA-Seq, tumor immune microenvironment, transcriptomic markers
Introduction
Lung cancer is one of the most common malignancies worldwide, and lung adenocarcinoma (LUAD) accounts for about 60% of all lung cancer cases.1–4 The five-year survival rate of LUAD is about 50% for early-stage disease, but only about 20% for locally advanced disease.1,5 The presence of lymphatic metastasis is associated with a poor prognosis, a higher rate of local regional relapse, and distant metastasis.1,6 However, there is heterogeneity in the prognosis of patients with locally advanced LUAD.7,8 Some patients experience relapse or distant metastasis soon after initial treatment, but others experience long-term disease-free survival (DFS). The reason for these differences is unclear.9
A tumor is a complex ecosystem consisting of various types of cells and cell components.10,11 Researchers have increasingly recognized tumor-infiltrating lymphocytes (TILs) as cells that can profoundly affect the course of tumor progression and the therapeutic response.12–14 Most of these previous studies investigated the composition of TILs using immunohistochemistry (IHC), immunofluorescence, and flow cytometry, but these methods can only provide simultaneous examination of a limited number of immune cells. Thus, little is known about how all these cell components together affect tumor progression and alter the tumor during the course of therapy.
Several recent computational bioinformatic methods quantified immune infiltrates based on transcriptomics data.15 The ability to simultaneously quantify multiple cell types within a tissue specimen is critical for determining the function of immune infiltrates within a tumor, and may also improve our understanding of the mechanisms underlying different therapeutic responses and allow stratification of patients with different prognoses.16,17 Previous studies that characterized transcription within the tumor immune microenvironment have used whole transcriptome sequencing data.15,18,19 However, this method requires many fresh tumor specimens, which are not always available, and the cost is currently too high for routine use in clinical practice.
In the present study, we used targeted RNA-Seq to measure the abundance of 395 immune transcripts from formalin-fixed paraffin embedded (FFPE) LUAD specimens to characterize the transcriptome profile of the tumor immune microenvironment. Although this approach only examined 395 immune-related genes, it provided more deep sequencing, and had superior sensitivity to bulk whole transcriptome sequencing.20,21 Our purpose was to demonstrate the combined use of computational bioinformatic methods with a targeted RNA-Seq assay to profile the tumor immune microenvironment and to stratify patients with LUAD according to prognosis.
Materials And Methods
Patient Enrollment
This study was approved by the Local Ethics Committee of Xiangya Hospital of Central South University (Changsha, China). All patients provided written informed consent prior to participation. Patients who met the following criteria were enrolled: receipt of surgery as initial treatment, but no preoperative therapy; confirmation of postoperative pathology as locally advanced LUAD (N1–2) with pathological stage of IIB–IIIA; refusal of postoperative adjuvant therapy; receipt of surgery in our department from December 2014 to December 2016; no other malignant tumors; attainment of the clinical endpoint (local lymph node recurrence or distant metastasis); no previous myocardial infarction, unstable angina, or stroke; no systemic immune disease.
The clinical data of all 24 patients were collected, and FFPE specimens were retrieved from the tissue archive of Xiangya Hospital. Sections from the FFPE blocks (stained with hematoxylin and eosin) were used to confirm the presence of tumor tissue by a qualified pathologist. Only FFPE specimens with 10% or more viable tumor cells were used for RNA extraction.
RNA Extraction, RNA-Seq Library Preparation And Sequencing
The truXTRAC™ FFPE RNA Kit (Covaris, Inc., Woburn, MA) was used for extraction of RNA from FFPE sections. Thus, RNA was extracted from FFPE lysates, purified, eluted, and quantified using the Quant-iT RNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA).
The Ion AmpliSeq™ targeted sequencing system (Thermo Fisher Scientific, Waltham, MA) was used for preparation of RNA-Seq libraries. The assay was designed to quantify the expression of 395 immune-related genes, including 10 house-keeping (HK) genes as controls (Table S1). Following reverse transcription into cDNA, targets of the 395 genes were amplified, and barcode adapters were ligated to the partially digested amplicons. After purification and normalization, equimolar libraries were pooled. Following enrichment and template preparation using the Ion Chef™ system (Thermo Fisher Scientific), 200-bp sequencing was performed on the Ion Torrent S5 system (Thermo Fisher Scientific) to obtain mapped reads (about 2- to 3-million per sample).
Next Generation Sequencing (NGS), Quality Control And Gene Expression Normalization
Sequencing data were analyzed using Ion Torrent Suite software version 5.2.0 (Thermo Fisher Scientific) for reference mapping and base calling. Quality control was conducted using acceptance criteria, and gene expression normalization was performed using HK gene background-subtracted reads as a control, as described by Conroy et al.21 Finally, the normalized gene expression, presented as normalized reads per million (nRPM), were log2-transformed.21
Heatmap-Based Hierarchal Clustering And Gene Set Variation Analysis (GSVA)
Patients were divided into two groups — favorable prognosis or poor prognosis — based on median time of DFS. The “limma” package in R (version 3.4.1) was used to identify differentially expressed genes (DEGs) in the two groups. A heatmap of DEGs was created with z-score normalization for each gene, in which the rows listed genes, and the columns listed patients. The targeted 395 genes are annotated as 36 classes according to signal/function pathway (Table S1). The “GSVA” package in R was then used to analyze and cluster DEGs according to 36 signal/function pathways for the two groups.
Profiling Of Immune Infiltrates Using MCP-Counter And TCGA LUAD Dataset
The “MCPcounter” package in R was used for analysis of microenvironment cell populations (MCPs) and quantification of immune cells from transcriptomic data. This method estimates the abundance of different immune cells based on specific molecular markers, as described by Becht et al.19
Normalized level 3 gene expression data and annotations of LUAD samples from The Cancer Genome Atlas (TCGA) in projects that were not currently embargoed were retrieved from the Genomic Data Commons (GDC) portal. Finally, transcriptional data from 85 patients with locally advanced LUAD (Stage IIb–IIIa, N1–2) and available DFS information were analyzed to explore the relationship of the tumor immune microenvironment with prognosis.
Statistical Analysis
All statistical tests were performed using R (version 3.4.1). The Wilcoxon rank-sum test was used for hierarchical data, and the chi-square test or Fisher’s exact test for binary data. To analyze the impacts of gene set enrichment score and immune infiltrates on clinical outcomes, Kaplan-Meier curves were used with the log rank test to analyze DFS, in which the median score was the cutoff value. Cox proportional hazards models for DFS were also used to identify the association of prognosis with infiltration by different types of immune cells. A p-value of 0.05 or less was regarded as significant.
Results
Correlation Of Clinical Data And Prognosis
We analyzed transcriptional data of 395 immune-related genes from FFPE specimens of 24 patients from our institution who had locally advanced LUAD (Table S2). We divided these patients into two groups — favorable prognosis or poor prognosis — using the median time (16 months) of DFS as the cutoff (Table 1). These two groups had no significant differences in any major clinical parameters. Survival analysis with the log rank test (data not shown) showed no significant correlation of clinical outcomes with major clinical parameters, including sex (P = 0.47), age (P = 0.30), T stage (P = 0.13), N stage (P = 0.35), or degree of differentiation (P = 0.44).
Table 1.
Baseline Characteristics Of Patients With Advanced Lung Adenocarcinoma From Xiangya Hospital
| Characteristic | Favorable Prognosis (n=12) | Poor Prognosis (n=12) | P value |
|---|---|---|---|
| Age (years) | |||
| ≥60 | 4 | 8 | >0.05 |
| <60 | 8 | 4 | |
| Sex | |||
| Male | 8 | 8 | >0.05 |
| Female | 4 | 4 | |
| T Stage | |||
| T1 | 6 | 6 | >0.05 |
| T2 | 4 | 4 | |
| T3 | 2 | 2 | |
| N Stage | |||
| N1 | 3 | 1 | >0.05 |
| N2 | 9 | 11 | |
| Differentiation | |||
| High | 3 | 4 | >0.05 |
| Medium | 7 | 2 | |
| Low | 2 | 6 |
Analysis Of DEGs And Function Analysis Using Gene Set Variation Analysis (GSVA)
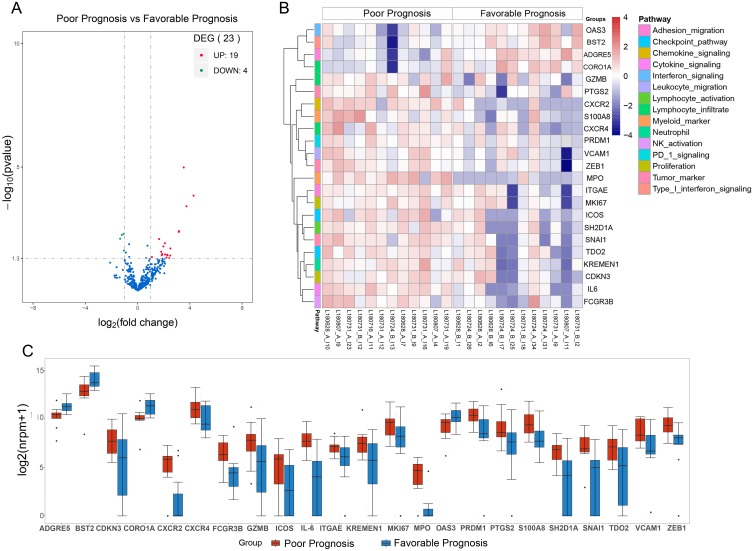
A comparison of the two groups indicated the presence of 23 DEGs (filter criteria: fold change ≥2, P < 0.05; Figure 1A and B). Nineteen genes had significantly higher expression in patients with favorable prognoses, and 4 genes had significantly higher expression in patients with poor prognoses (Figure 1C).
Figure 1.
(A) Volcano plot of the expression of immune-related genes in patients with favorable rather than poor prognoses. (B) Heatmap of DEGs in patients with favorable and poor prognoses. (C) Boxplots of DEGs in patients with favorable and poor prognoses (center line: median, box: second and third quartiles, bars: 95% CI, points: outliers).
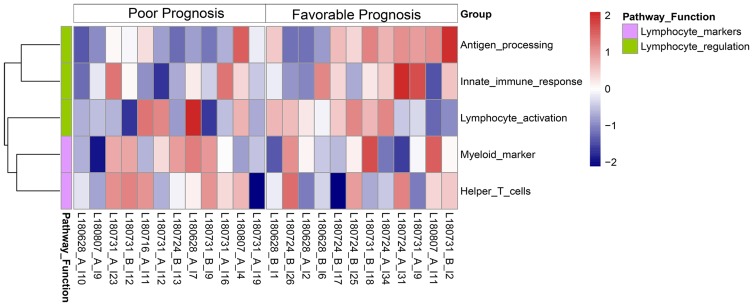
Gene set variation analysis (GSVA) demonstrated the two groups had significant differences in 5 pathways (Padj < 0.05): antigen processing, innate immune response, lymphocyte activation, myeloid markers, and helper T cells (Figure 2). Furthermore, survival analysis with the log rank test demonstrated that greater enrichment of gene sets that function in the antigen processing pathway (P = 0.01) and lymphocyte activation pathway (P = 0.043) correlated with a favorable prognosis (Figure S1).
Figure 2.
Heat map of GSVA enrichment scores of immune-related pathways in patients with favorable and poor prognoses.
Profiles Of Tumor Immune Infiltrates And Impact On Clinical Outcome: Xiangya Hospital LUAD Data
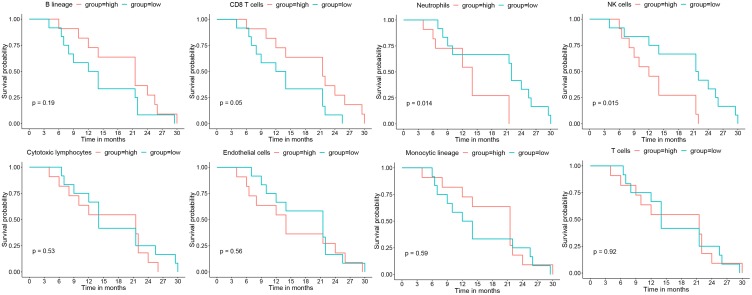
Analysis of data from our institution demonstrated that greater infiltration of CD8 T cells correlated with longer DFS (long-rank test: P = 0.05), but greater infiltration of neutrophils (P = 0.015) and NK cells (P = 0.015) correlated with shorter DFS (Figure 3). However, there were no significant associations of DFS time with infiltration of cytotoxic lymphocytes, endothelial cells, cells with monocytic lineage, or T cells. Univariate and multivariate Cox hazard analysis also showed that infiltration of neutrophils was an independent risk factor for poor prognosis (adjusted hazard ratio [aHR] = 1.67, 95% confidence interval [CI] = 1.09 to 2.55, P = 0.019; Table 2).
Figure 3.
Kaplan-Meier survival analyses of the relationships of different immunological infiltrates with DFS, in which the median value of each MCP-counter score of immune infiltrates was used as the cutoff (Xiangya Hospital LUAD data, n = 24).
Table 2.
Univariate And Multivariate Cox Hazard Analysis Of The Association Of Different Tumor Immunological Infiltrates With PFS (Xiangya Hospital Data, N = 24)
| Cell Type | Univariate Cox Hazard Analysis | Multivariate Cox Hazard Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | aHR | 95% CI | P | |
| T cells | 1 | 0.68–1.48 | 0.996 | NA | NA | NA |
| CD8 T cells | 0.78 | 0.48–1.25 | 0.298 | NA | NA | NA |
| Cytotoxic lymphocytes | 1.12 | 0.72–1.76 | 0.612 | NA | NA | NA |
| NK cells | 1.51 | 0.96–2.39 | 0.076 | NA | NA | NA |
| B lineage | 1 | 0.63–1.6 | 0.992 | NA | NA | NA |
| Monocytic lineage | 1.01 | 0.67–1.51 | 0.98 | NA | NA | NA |
| Neutrophils | 1.72 | 1.12–2.65 | 0.014 | 1.67 | 1.09–2.55 | 0.019 |
| Endothelial cells | 0.91 | 0.61–1.38 | 0.668 | NA | NA | NA |
Abbreviations: HR, hazard ratio; CI95, 95% confidence interval; NA, non-available.
Profiles Of Tumor Immune Infiltrates And Impact On Clinical Outcome: TCGA LUAD Data
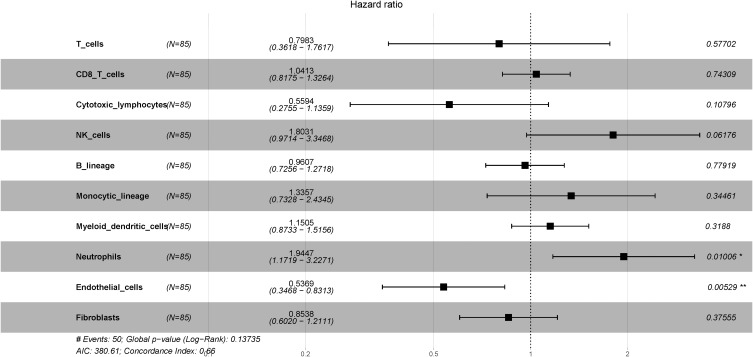
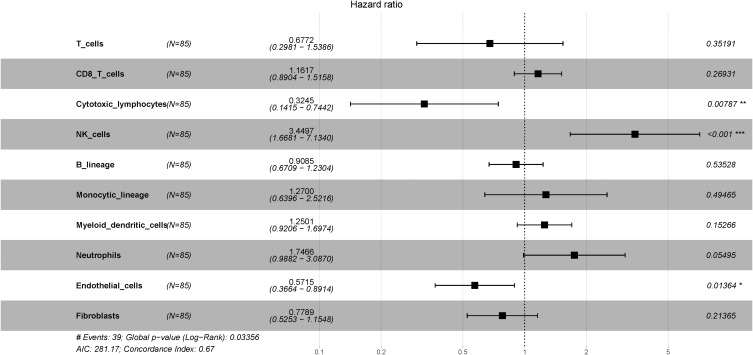
We also used LUAD data from TCGA with MCP-counter estimates to examine 85 different patients with LUAD (Stage IIb–IIIa, N1–2, with available DFS data). A Cox proportional hazard ratio analysis indicated greater infiltration of neutrophils was associated with shorter DFS (P = 0.01), but the presence of more endothelial cells was associated with slower disease progression (P = 0.005; Figure 4). Next, we determined the correlation between immune cell infiltration and overall survival (OS) in the same cohort (Figure 5). The results indicated that infiltration of NK cells (P < 0.001) and neutrophils (P = 0.055) negatively correlated with OS; on the other hand, cytotoxic lymphocytes (P = 0.008) and endothelial cells (P = 0.014) positively correlated with OS. These observations were largely in accordance with the data from our institution.
Figure 4.
Forest plot showing multivariate Cox regression analysis of the effect of different tumor immunological infiltrates on DFS (TCGA LUAD data, n = 85). *P<0.05; **P<0.01.
Figure 5.
Forest plot showing multivariate Cox regression analysis of the effect of different tumor immunological infiltrates on OS (TCGA LUAD data, n = 85). *P<0.05; **P<0.01; ***P<0.001.
Discussion
The targeted RNA-Seq assay used in this study is more sensitive than bulk whole transcriptome data20,21 and requires very little starting material. Thus, RNA-Seq can be used with FFPE specimens, even though formalin can cause RNA degradation. Paluch et al20 first developed this assay in 2017 to examine a cohort of 13 patients with ovarian cancer. They found that targeted RNA-Seq data from FFPE specimens were highly correlated with identical samples that were freshly frozen from the same tumors. These researchers also found that the expression data from FFPE specimens measured by RNA-Seq were highly concordant with data measured by quantitative RT-PCR and IHC of the same specimens.20 Intriguingly, a recent study used the targeted RNA-Seq approach (Thermo Fisher Scientific) on FFPE samples to explore the tumor microenvironment and identify potential responders to immune therapy.22
In 2016, Becht et al19 first described MCP-counter, a method used to quantify tumor-infiltrating immune cells based on a stringent set of marker genes. These researchers performed an in vitro RNA mixture validation experiment and found that MCP-counter score had a strong correlation with the known proportions of RNAs in these mixtures. Moreover, their in vivo experiments indicated that MCP-counter score had a high correlation with IHC data used to determine the densities of different cells. Thus, our combined use of the targeted RNA-Seq assay and the MCP-counter method is a valid approach for comparison of tumor samples based on tumor infiltrated immune cells and transcriptome data.19
Our results indicated that greater infiltration of CD8 T cells correlated with better prognosis in patients with LUAD. Previous studies also reported that greater infiltration of CD8+ T cells into tumors was associated with slower progression in melanoma,23 ovarian cancer,24 colorectal cancer,25 and breast cancer.26 Furthermore, greater CD8+ T cell infiltration of tumors can predict response to standard chemotherapy27 and to immune checkpoint blockade therapy, such as anti–CTLA-4 therapy28 or anti–PD-1 therapy.29 Therefore, it is crucial to measure the extent of CD8+ T cell infiltration of solid tumors.30
Our GSVA analysis also demonstrated that enrichment of the antigen processing pathway (P = 0.01), and possibly the lymphocyte activation pathway (P = 0.08), correlated with better prognosis (Figure S1). The antigen processing pathway prepares antigens by converting them into MHC-I bound peptides for presentation to T lymphocytes.31 Activation of cytotoxic T lymphocytes is an antigen-specific process that requires an interaction of the TCR-CD3 complex with a processed tumor antigen-derived peptide bound to an MHC class I molecule, as well as CD8 and CD28.32 These two pathways interact, and both are critical for an effective immune response against tumors. Only the activated CD8+ T lymphocytes can recognize and kill tumor cells that harbor neoantigens.30,33 Thus, the higher enrichment scores in the antigen processing pathway and lymphocyte activation pathway that we observed indicate a stronger adaptive immune response, protective T cell mounting, and therefore a more favorable prognosis.
We found similarities of our LUAD patients (Figure 3) with LUAD patients in TCGA in terms of NK cell infiltration (Figure 5), in that abundant NK cell infiltration correlated with poor prognosis. A study by Gordon et al showed that continuous exposure of NK cells to tumors reduced their cytotoxic activity, and that excessive proliferation of NK cells contributed to their dysfunction.34 Several mechanisms may influence NK cell activity within tumor tissues. Platonova et al showed that intratumoral NK cells have strongly down-regulated activating receptors, and this impairs their cytotoxic effects.35 Additionally, Carrega et al found that non-small cell lung cancer (NSCLC) samples had more CD56hiCD16-NK cells than normal lung tissues, leading to expression of inhibitory killer cell immunoglobulin-like receptors and suppression of the activity of NK cells within tumor tissue.36 Furthermore, these researchers found that NK cells were mainly retained in the stroma and had little direct contact with NSCLC cells.36
Our results showed that stronger neutrophil infiltration into the tumor was associated with a shorter DFS (P = 0.015), and our Cox proportional hazard analysis showed that greater tumor infiltration by neutrophils was an independent risk factor for poor prognosis. The results were similar in the TCGA LUAD cohort (Figures 4 and 5). Previous studies also found that greater infiltration of neutrophils into tumors predicts poor prognosis in other solid tumors, such as lung cancer,37 breast cancer,38 gastric cancer,39 and esophageal cancer.40 There is also evidence that the presence of neutrophils can directly enhance tumor growth. In particular, a major neutrophil protein, neutrophil elastase (NE), can enter cancer cells via clathrin-coated pit-mediated endocytosis, leading to degradation of the insulin receptor substrate 1 (IRS-1), which generally binds to phosphatidylinositol 3-kinase (PI-3K). Following degradation of IRS-1, tumor cells have increased availability of PI-3K as an energy source, leading to enhanced proliferation.37,41 Another possibility is that neutrophils suppress anti-tumor immune responses by suppressing cytotoxic T-cell function and proliferation. Under such circumstances, these neutrophils are regarded as granulocytic-myeloid-derived suppressor cells, which function in immunosuppression.42
One of the drawbacks of the present study is that the sample size was small, and we only analyzed patients from a single institution. Studies with larger samples and from multiple institutions are needed to verify our results. Second, we only examined the abundance of 395 immune-related transcripts, so this could not be considered a thorough transcriptional immune profile of LUAD. A comprehensive profile of the whole transcriptome for analysis of immune infiltrations of fresh frozen tissues would be a valuable step forward.
In conclusion, our study of patients with LUAD indicated that greater tumor infiltration by CD8 T cells correlated with a favorable prognosis, but greater tumor infiltration by neutrophils or NK cells correlated with a poor prognosis. Thus, our results demonstrate that targeted RNA-Seq of immune-related genes from FFPE specimens, when integrated with computational bioinformatic methods, can effectively profile the tumor immune microenvironment. This method thus provides a fundamental understanding of the versatile role of immune infiltrates within tumors, allows stratification of patients with different prognoses, and is especially useful when bulk tumor tissues are unavailable.
Acknowledgments
The present study was funded by the Human Provincial Science and Technology Department (2017JJ2345) and the National Natural Science Foundation of China (No. 81673516).
Ethical Approval
This study has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rami-Porta R, Bolejack V, Crowley J, et al. The IASLC lung cancer staging project: proposals for the revisions of the T descriptors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10(7):990–1003. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3.Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer. 2015;121(Suppl 17):3080–3088. [DOI] [PubMed] [Google Scholar]
- 4.Li YX, Le Wang J, Gao M, Tang H, Gui R, Fu YF. Celecoxib-erlotinib combination delays growth and inhibits angiogenesis in EGFR-mutated lung cancer. Am J Cancer Res. 2016;6(7):1494. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Peng X, Zhou Y, Xia K, Zhuang W. Comparing the benefits of chemoradiotherapy and chemotherapy for resectable stage III A/N2 non-small cell lung cancer: a meta-analysis. World J Surg Oncol. 2018;16(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (Eighth) Edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. [DOI] [PubMed] [Google Scholar]
- 7.Zeng X, Li J, Peng L, et al. Economic outcomes of maintenance gefitinib for locally advanced/metastatic non-small-cell lung cancer with unknown EGFR mutations: a semi-Markov model analysis. PLoS One. 2014;9(2):e88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Lei J, Wang Q, Li J, Wu L. Clinical characteristics of patients with non-small cell lung cancers harboring anaplastic lymphoma kinase rearrangements and primary lung adenocarcinoma harboring epidermal growth factor receptor mutations. Genet Mol Res. 2015;14(4):12973–12983. [DOI] [PubMed] [Google Scholar]
- 9.Shukla S, Evans JR, Malik R, et al. Development of a RNA-seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2017;109(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. [DOI] [PubMed] [Google Scholar]
- 11.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. [DOI] [PubMed] [Google Scholar]
- 12.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becht E, Giraldo NA, Germain C, et al. Immune contexture, immunoscore, and malignant cell molecular subgroups for prognostic and theranostic classifications of cancers. Adv Immunol. 2016;130:95–190. [DOI] [PubMed] [Google Scholar]
- 14.Angelova M, Mlecnik B, Vasaturo A, et al. Evolution of metastases in space and time under immune selection. Cell. 2018;175(3):751–765.e716. [DOI] [PubMed] [Google Scholar]
- 15.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Z, Chen J, Cheng JN, et al. Tumor microenvironment properties are associated with low CD68-positive cell infiltration and favorable disease-free survival in EGFR-mutant lung adenocarcinoma. Clin Lung Cancer. 2018;19(5):e551–e558. [DOI] [PubMed] [Google Scholar]
- 18.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paluch BE, Glenn ST, Conroy JM, et al. Robust detection of immune transcripts in FFPE samples using targeted RNA sequencing. Oncotarget. 2017;8(2):3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conroy JM, Pabla S, Glenn ST, et al. Analytical validation of a next-generation sequencing assay to monitor immune responses in solid tumors. J Mol Diagn. 2018;20(1):95–109. [DOI] [PubMed] [Google Scholar]
- 22.Morrison C, Pabla S, Conroy JM, et al. Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden. J Immunother Cancer. 2018;6(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahbar M, Naraghi ZS, Mardanpour M, Mardanpour N. Tumor-infiltrating CD8+ lymphocytes effect on clinical outcome of muco-cutaneous melanoma. Indian J Dermatol. 2015;60(2):212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato E, Olson SH, Ahn J, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci. 2005;102(51):18538–18543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 26.Mahmoud SM, Paish EC, Powe DG, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29(15):1949–1955. doi: 10.1200/JCO.2010.30.5037 [DOI] [PubMed] [Google Scholar]
- 27.Seo AN, Lee HJ, Kim EJ, et al. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br J Cancer. 2013;109(10):2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashidian M, Ingram JR, Dougan M, et al. Predicting the response to CTLA-4 blockade by longitudinal noninvasive monitoring of CD8 T cells. J Exp Med. 2017. doi: 10.1084/jem.20161950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen P-L, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Gruosso T, Zuo D, et al. Infiltration of CD8(+) T cells into tumor cell clusters in triple-negative breast cancer. Proc Natl Acad Sci U S A. 2019;116:3678–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Lostao L, Anel A, Pardo J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res. 2015;21(22):5047–5056. [DOI] [PubMed] [Google Scholar]
- 33.Thommen DS, Koelzer VH, Herzig P, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon SM, Chaix J, Rupp LJ, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36(1):55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platonova S, Cherfils-Vicini J, Damotte D, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. [DOI] [PubMed] [Google Scholar]
- 36.Carrega P, Morandi B, Costa R, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells.. Cancer. 2008;112(4):863–875. [DOI] [PubMed] [Google Scholar]
- 37.Houghton AM, Rzymkiewicz DM, Ji H, et al. Neutrophil elastase–mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 2010;16(2):219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawa M, Osada S, Morimitsu K, et al. Growth effect of neutrophil elastase on breast cancer: favorable action of sivelestat and application to anti-HER2 therapy. Anticancer Res. 2012;32(1):13–19. [PubMed] [Google Scholar]
- 39.Wada Y, Yoshida K, Hihara J, et al. Sivelestat, a specific neutrophil elastase inhibitor, suppresses the growth of gastric carcinoma cells by preventing the release of transforming growth factor‐α. Cancer Sci. 2006;97(10):1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada Y, Yoshida K, Tsutani Y, et al. Neutrophil elastase induces cell proliferation and migration by the release of TGF-α, PDGF and VEGF in esophageal cell lines. Oncol Rep. 2007;17(1):161–167. [PubMed] [Google Scholar]
- 41.Sato T, Takahashi S, Mizumoto T, et al. Neutrophil elastase and cancer. Surg Oncol. 2006;15(4):217–222. [DOI] [PubMed] [Google Scholar]
- 42.Treffers LW, Hiemstra IH, Kuijpers TW, van Den Berg TK, Matlung HL. Neutrophils in cancer. Immunol Rev. 2016;273(1):312–328. [DOI] [PubMed] [Google Scholar]