Summary
ASPP (apoptosis-stimulating proteins of p53) proteins bind PP-1c (protein phosphatase 1) and regulate p53 impacting cancer cell growth and apoptosis. Here we determine the crystal structure of the oncogenic ASPP protein, iASPP, bound to PP-1c. The structure reveals a 1:1 complex that relies on interactions of the iASPP SILK and RVxF motifs with PP-1c, plus interactions of the PP-1c PxxPxR motif with the iASPP SH3 domain. Small angle X-ray scattering analyses suggest the crystal structure undergoes slow interconversion with more extended conformations in solution. We show that iASPP, and the tumour suppressor ASPP2, enhance the catalytic activity of PP-1c against the small molecule substrate, pNPP as well as p53. The combined results suggest that PxxPxR binding to iASSP SH3 domain is critical for complex formation, and that the modular ASPP-PP-1c interface provides dynamic flexibility that enables functional binding and dephosphorylation of p53 and other diverse protein substrates.
Keywords: iASPP, ASPP2, PP-1c, p53, ANK repeats, RVxF motif, dephosphorylation, SH3
Introduction
The tumour suppressor p53 is regulated by a complex network of interacting proteins and post-translational modifications (Bieging et al., 2014). A key family of p53 regulatory proteins is the ASPP family (apoptosis-stimulating proteins of p53), which has three members: the highly homologous ASPP1 and ASPP2, and the inhibitory iASPP protein (Trigiante and Lu, 2006). ASPP1 and ASPP2 have been shown to bind the p53 DNA binding domain and potentiate the pro-apoptotic transcriptional activity of p53 (Mori et al., 2000; Samuels-Lev et al., 2001; Sgroi et al., 1999). In contrast, iASPP binds to p53 and inhibits pro-apoptotic p53 activity. Given their roles in regulating p53 activity, it is not surprising that ASPP1 or ASPP2 levels are decreased in several human cancers and this is associated with the transition to metastases in breast cancer (Agirre et al., 2006; Li et al., 2012; Liu et al., 2010; Liu et al., 2005; Liu et al., 2004; Lossos et al., 2002; Mori et al., 2004; Mori et al., 2000; Samuels-Lev et al., 2001; Sgroi et al., 1999; Zhao et al., 2010). The iASPP protein is overexpressed in multiple cancers, typically in those that express wild-type p53 (Bergamaschi et al., 2003; Cao et al., 2013; Li et al., 2011; Li et al., 2012; Lin et al., 2011; Liu et al., 2010; Lu et al., 2010; Pinto et al., 2010; Zhang et al., 2011; Zhang et al., 2005). In addition, increased ASPP1/2 expression is correlated with enhanced sensitivity of tumour cells to cytotoxic chemotherapy and radiation, while enhanced iASPP expression leads to resistance (Ao et al., 2001; Bergamaschi et al., 2003; Cao et al., 2013; Jia et al., 2014; Li et al., 2012; Liu et al., 2009; Mori et al., 2000; Wang et al., 2012; Zhang et al., 2005).
All three ASPP proteins share a conserved C-terminal domain consisting of a proline rich region, followed by four ankyrin (ANK) repeats and a SH3 domain that binds p53. While the N-terminal regions of ASPP1/2 and iASPP are poorly conserved and are thought to be mainly unstructured, studies have shown that this region is regulated by multiple post-translational modifications and protein-protein interactions (Aylon et al., 2010; Godin-Heymann et al., 2013; Lu et al., 2013; Trigiante and Lu, 2006). The ANK/SH3 domain of ASPP1/2 binds to the DNA binding domain (DBD) of p53 (Gorina and Pavletich, 1996; Patel et al., 2008) and the majority of cancer-associated p53 mutations that inhibit the ability of p53 to bind to DNA also block its interaction with ASPP1/2 (Samuels-Lev et al., 2001). While iASPP has a similar ANK/SH3 domain, it is not thought to bind tightly to the p53 DBD. Instead, proline-rich sequences in other regions of p53 are thought to bind to the iASPP SH3 domain (Ahn et al., 2009; Bergamaschi et al., 2006). Not only is the ANK/SH3 region critical for interactions with p53, but it has also been shown to interact with the serine/threonine protein phosphatase-1c (PP-1c) (Helps et al., 1995; Llanos et al., 2011; Skene-Arnold et al., 2013), fueling speculation that the ASPP proteins may regulate the phosphorylation status of p53.
Together with Protein Phosphatase-2A (PP-2A), PP-1c handles most Ser/Thr dephosphorylation in human cells (Shi, 2009) and gains its specificity through selective interactions with hundreds of regulatory proteins that target PP-1c to specific phosphorylated substrates (Bollen et al., 2010; Brautigan and Shenolikar, 2018). A growing body of structural studies have indicated some conserved themes in PP-1c regulatory proteins. Many of these proteins are largely unstructured, and utilize extended coil structures to wrap around PP-1c to contact various targeting surfaces (Bollen et al., 2010; Choy et al., 2012). Not only are these interactions thought to bind the regulatory protein to PP-1c, but they can also block potential substrate binding grooves on the PP-1c surface to modulate substrate specificity. The most common PP-1c binding motif identified is the tetrapeptide RVxF motif ([R/K]-[V/I/L]-X-[F/W], where X can be any amino acid except proline), and other secondary motifs have been identified such as the MyPhone, SILK ([S/G]-I-L-K), and Φ motifs (Bollen et al., 2010; Choy et al., 2012). In addition to these short peptide motifs, ANK domains have also been shown to bind PP-1c, as in the PP-1c regulatory factor, MYPT1 (Terrak et al., 2004). MYPT1 also binds to PP-1c via MyPhone and RVXF motifs N-terminal to its ANK repeats.
The fact that all three ASPP proteins contain RVxF-like motifs immediately N-terminal to their ANK domains suggested that they might bind PP-1c in a manner similar to MYPT. Building on this work, we recently showed that purified ASPP1, ASPP2 and iASPP can all form 1:1 stoichiometric complexes with PP-1c that critically involve the unstructured C-terminal tail of PP-1c (Skene-Arnold et al., 2013). Here we determine the crystal structure of iASPP bound to PP-1c, which reveals that the interactions between the two proteins are stabilized by several distinct contact surfaces. Small angle X-ray scattering however indicates that the complex accesses additional conformational states in solution that likely involve the release of one or more of the peptide contacts. From the combined results, we propose a structure-based model for how the modular nature of the ASPP-PP-1c contact surface provides dynamic flexibility that enables p53 binding and dephosphorylation.
RESULTS
Overview of the crystal structure of iASPP608–828 bound to PP-1cα
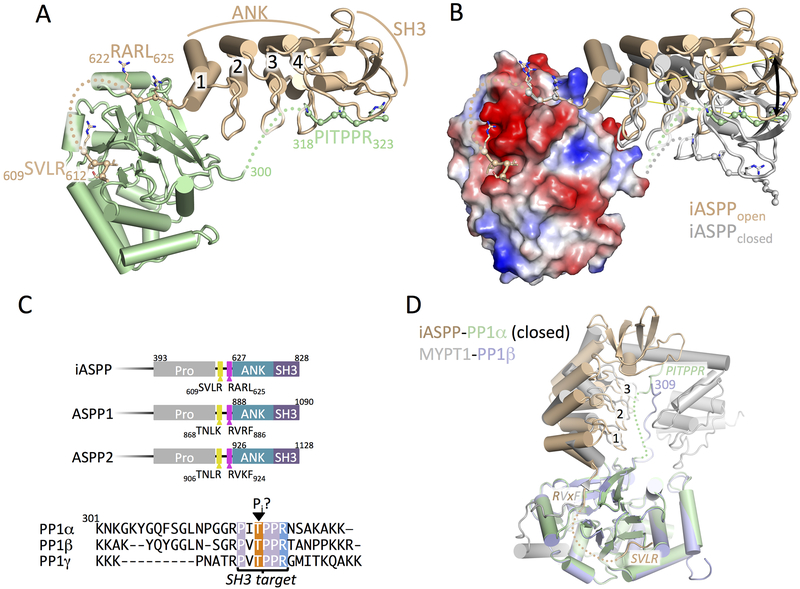
To understand the structural basis for interactions between iASPP and PP-1c, we crystallized and determined the structure of the minimal PP-1c-binding iASPP fragment, iASPP608–828 (Skene-Arnold et al., 2013), bound to PP-1cα at 3.41 Å resolution (Figure 1, Table 1, STAR Methods). iASPP residues 627–828 constitute the folded core of iASPP, and is composed of four N-terminal ankyrin (ANK) repeats and a C-terminal SH3 domain that we find are essentially identical to the structures of this region in free iASPP (Robinson et al., 2008) as well as ASPP2 (Gorina and Pavletich, 1996). The iASPP N-terminal ANK repeat docks against a shallow groove on PP-1c and the N-terminal peptide iASPP608–626 extends away from this contact surface, making two additional interactions with the PP-1c catalytic core (Figure 1A and B). The first contact involves residues 622RARL625, which plays the role of an RVxF motif (Figure S1A) as previously predicted (Skene-Arnold et al., 2013). N-terminal to this sequence the electron density becomes poor and tracks away from PP-1c, however, additional density is observed packed into an adjacent groove on PP-1c groove that can bind SILK-like motifs. We suggest this density represents the N-terminal-most region, 609SVLR612, which may play the role of a SILK motif (Figure S1B). Notably, the conservation of the 609SVLR612 and 622RARL625 sequences in the ASPP protein family suggest that similar interactions may stabilize complexes with PP-1c in the other ASPP proteins (Figure 1C). The final major contact between PP-1c and iASPP involves the SH3 domain of iASPP and a region of left handed helical density that we have modeled as the 318PxxPxR323 motif near the C-terminus of the flexible PP-1c tail (Figure S1C). This motif is conserved in all PP-1c isoforms, although the length of the tail linking the PxxPxR motif to the catalytic domain varies between isoforms (Figure 1C).
Figure 1.
Crystal structure of iASPP608–828 bound to PP-1cα.
(A) Overview of the structure of iASPP608–828 bound to PP-1cα. iASPP is brown, PP-1cα is in green. Peptide segments involved in key protein-protein contacts are illustrated with Cα spheres and the four ANK repeats are numbered. Regions of polypeptide chain not visible in the electron density are indicated by dotted lines.
(B) Comparison of the two iASPP608–828-PP-1cα complexes in the asymmetric unit, aligned on PP-1cα, with the second iASPP in the closed conformation colored grey and an electrostatic charge surface displayed for PP-1cα. The ~22° angular difference in the orientation of the two iASPPs relative to PP-1cα is indicated.
(C) Top panel, comparison of the primary structures of the C-terminal regions of iASPP, ASPP1 and ASPP2 with key domains and motifs indicated. Bottom panel, sequence alignment of the C-terminal unstructured tails of the three PP-1cα isoforms.
(D) Structural comparison of the closed complex of iASPP-PP-1cα with the structure of MYPT1-PP-1β, aligned on PP-1cα with the different chains colored as indicated in the figure. Crystallographic statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Resolution range1 | 48.87 - 3.41 (3.53 - 3.41) |
| Space group | P41212 |
| Unit cell | 135.46 135.46 165.39 90 90 90 |
| Number of total/unique reflections1 | 422341 (39508)/ 21564 (2031) |
| Completeness (%)1 | 99.46 (95.94) |
| Mean I/sigma(I) 1 | 11.80 (2.51) |
| Wilson B-factor | 92.17 |
| Rmerge1 | 0.296 (1.73) |
| Rmeas1 | 0.304 (1.77) |
| Rpim1 | 0.0679 (0.399) |
| CC1/21 | 0.997 (0.675) |
| CC*1 | 0.999 (0.898) |
| Refinement | |
| Reflections for refinement/Reflections for Rfree1 | 21546 (2031)/ 1112 (97) |
| Rwork/Rfree1 | 0.1882 (0.2573)/ 0.2415 (0.2844) |
| Number of non-hydrogen atoms | 7972 |
| Number of protein residues | 1010 |
| RMS(bonds) | 0.004 |
| RMS(angles) | 0.68 |
| Ramachandran favored/allowed/outliers (%) | 95.08/4.72/0.20 |
| Rotamer outliers (%) | 1.17 |
| Clashscore | 3.95 |
| Average B-factor | 103.12 |
| Number of TLS groups | 18 |
Values in brackets are for highest resolution bin
Two distinct iASPP-PP-1c complexes are observed in the crystallographic asymmetric unit. The overall structures are similar, however a structural alignment of the PP-1c catalytic domains of two complexes (Figure 1B) reveals conformational differences between the binding orientation of the iASPP ANK-SH3 domain relative to PP-1c. The difference between the two complexes likely reflects the conformational flexibility of the complex in solution, as the alignment shows that iASPP pivots on the N-terminal ANK repeat such that there is an ~20° difference in the angular orientation of iASPP with respect to PP-1c between the two complexes. The MYPT family of PP-1c regulatory subunits also use an RVxF motif N-terminal to ANK repeats to bind PP-1c (Grassie et al., 2011; Ito et al., 2004) and the overall orientation of the more compact or closed iASPP-PP-1c complex resembles the PP-1c binding orientation of the MYPT1 regulatory subunit (Figure 1D) (Terrak et al., 2004).
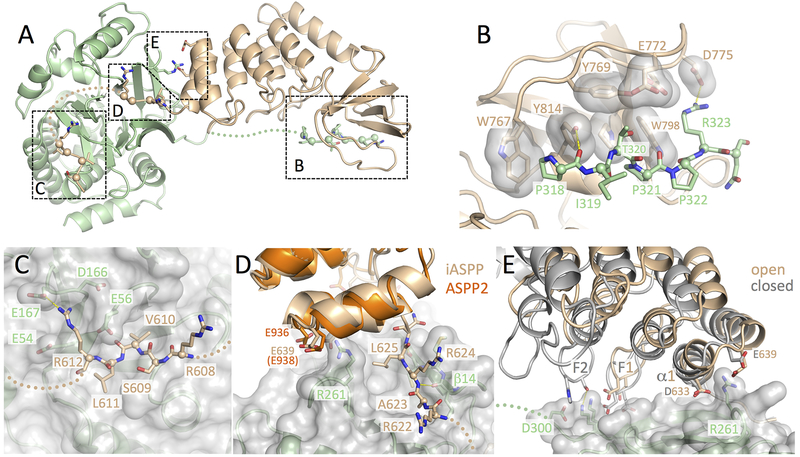
Four distinct contact surfaces stabilize the iASPP-PP-1c complex
Our structure shows that a large ~3100 Å2 solvent accessible surface area is buried upon formation of the iASPP-PP-1c complex, explaining the tight binding affinity of the complex (26 nM (Skene-Arnold et al., 2013)). The two proteins do not interact through a single contiguous binding surface; however, they instead contact one another through four distinct interfaces (Figure 2A). The largest of these interfaces is formed between the PxxPxR motif of PP-1c and the SH3 domain of iASPP, which buries ~990 Å2 of the solvent accessible surface (Figure 2B, Figure S1C). The PP-1c C-terminal PxxPxR motif adopts a polyproline helical structure similar to the classical interactions observed between SH3 domains and class II polyproline peptide targets (Feng et al., 1994; Lim et al., 1994). Within the iASPP SH3 domain, Trp767, Tyr814, Pro811 and Trp798 form the base of the peptide binding cleft and contact PP-1c residues Pro318 and Pro321, which protrude from the PxxPxR motif. Arg323 of PP-1c packs against a cluster of negatively charged residues from the iASPP SH3 RT loop (Glu772, Asp775, Glu776). The importance of the PxxPxR-SH3 contact is consistent with the finding that deletion of the PP-1c tail abrogates interactions with ASPP proteins in pull-down experiments (Skene-Arnold et al., 2013). The PxxPxR motif also contains a conserved Thr320 (Figure 1C), which packs into a pocket formed by Tyr769, Trp798, Pro811, Tyr814, and Glu772 (Figure 2B). Thr320 is a known cdk2-cyclin A phosphorylation site that is phosphorylated during mitosis and is associated with PP-1c auto-inhibition (Berndt et al., 1997; Dohadwala et al., 1994). Based on our iASPP-PP-1c crystal structure, phosphorylation of PP-1c Thr320 would sterically interfere with the packing of this residue into the ASPP SH3 binding pocket and would also introduce electrostatic repulsions with iASPP Glu772. Indeed, we have previously shown that a phospho-mimetic mutant of PP-1c (PP-1cγ T311D) destabilizes PP-1c-ASPP interactions (Skene-Arnold et al., 2013).
Figure 2.
Four distinct contact surfaces stabilize the iASPP-PP-1c complex.
(A) Overview of the iASPP608–828-PP-1cα structure with the four intermolecular contact surfaces boxed and labeled corresponding to the subsequent panels.
(B) Details of the SH3-PxxPxR motif interaction. Key residues from the iASPP SH3 domain that interact with the PxxPxR motif are shown as sticks with transparent surfaces. The PxxPxR peptide is shown in sticks with Cα spheres displayed.
(C) Details of the SILK-PP-1c interaction. A transparent surface is shown for PP-1c with key side chains shown as sticks. The SILK peptide is shown in sticks with Cα spheres displayed.
(D) Details of the RARL-PP-1c interaction and ANK-PP-1c contact. The RARL peptide is shown in sticks with Cα spheres displayed, while PP-1c is displayed with a transparent surface and key contact side chains as sticks. The ASPP2 structure (PDB ID: 1YCS) is shown aligned on the iASPP structure with conserved acidic residues in the first ANK repeat shown as sticks.
(E) Interactions between the iASPP ANK repeats and PP-1c. Shown are the two iASPP-PP-1c complexes aligned on PP-1c. The fingers from the first two ankyrin repeats are labeled F1 and F2 and residues that make intermolecular contacts are shown as sticks. Electron density for each of these contact surfaces is shown in Figure S1.
The second largest contact surface involves the iASPP 609SVLR612 motif that packs into a depression on the PP-1c catalytic domain and buries ~790 Å2 of solvent accessible surface area (Figure 2C, Figure S1B). This interaction is essentially identical to the interaction of the “SILK” motif of inhibitor 2 with PP-1c (Hurley et al., 2007). Val610 and Leu611 pack into hydrophobic pockets in PP-1c while Arg612 makes electrostatic contact with an extensive negatively charged PP-1c surface composed of Glu167, Asp166, Glu56, and Glu54.
The next largest contact surface involves iASPP RVxF-like motif (622RARL625), which buries ~600 Å2 of solvent accessible surface area (Figure 2D, Figure S1C). As observed in other RVXF motif interactions (Bollen et al., 2010), the second and fourth residues of the motif (Ala623 and Leu625) form a key part of the interface by binding to hydrophobic pockets on PP-1c. Further anchoring the interaction are main chain hydrogen bonds between the iASPP RARL motif and PP-1c β14. Mutation of Leu625 to Ala significantly weakens the interaction with PP-1c in pull-down experiments (Skene-Arnold et al., 2013). In addition, deletion of both the SILK and RARL motifs (iASPP626–828), decreases but does not completely abolish iASPP-PP-1c interactions, suggesting that these contact sites may not be as important as the PxxPxR-SH3 interaction (Skene-Arnold et al., 2013).
The final point of interaction is between the iASPP ANK repeats and PP-1c catalytic domain (Figure 2D and E). The interaction is not as extensive as the other contact surfaces and only buries a maximum of ~340 Å2 solvent accessible surface area in the closed complex, less in the more open form. The interaction pivots on α1 of the first ANK motif and involves electrostatic interactions between PP-1c Arg261 and a conserved negatively charged surface encompassing the C-terminus of α1 and N-terminus of α2 (Figure 2D and E). The pivoting of iASPP allows additional contacts between the fingers of the first two iASPP ANK repeats (F1, F2) and the PP-1c catalytic domains (Figure 2E). These contacts likely limit the degree of closure of the complex. While the surface area buried in this interaction is modest, mutation of Arg261 to Ser has been shown to significantly reduce ASPP-PP-1c affinity in pull-down assays (Skene-Arnold et al., 2013).
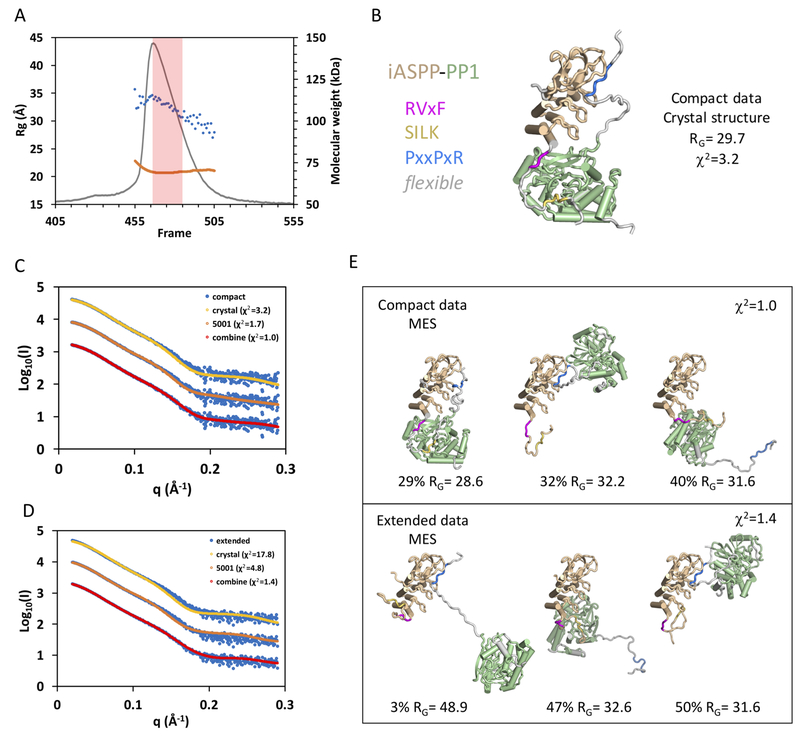
Flexibility of ASPP-PP-1c complexes revealed by SEC-SAXS
To characterize the structure and flexibility of ASPP-PP-1c complexes in solution by SAXS (Rambo and Tainer, 2013), we applied SAXS to ASPP-PP-1c complexes separated by in-line size exclusion chromatography (SEC-SAXS; Figure 3). In this method, purified iASPP608–828-PP-1cα or ASPP2905–1128-PP-1cα was run on size exclusion chromatography in-line with SAXS and multi-angle laser light scattering (MALS). For both samples, the complexes eluted as a single asymmetric peak (Figure 3A, Figure S2). MALS analysis of this peak suggested a small amount of aggregation near the leading edge of the peak, however the mass across the majority of the peak corresponded closely to the expected mass of the heterodimer (ASPP2905–1128-PP-1cα MW (theoretical) = 62.4 kDa; MW (MALLS) = 69.6 ± 0.4 kDa). Automated Guinier calculation of the radius of gyration (RG) across the peak revealed a significant decrease in the RG from ~35 Å at the peak, to ~25–30 Å in the tail (Figure 3A). Taken together, these data suggest that heterodimeric iASPP-PP-1c adopts a mixture of different conformational states that slowly interconvert over the time course of the chromatography. Similar results were obtained with ASPP2-PP-1c complexes (Figure S2).
Figure 3.
Conformational flexibility of iASPP-PP-1c revealed by SAXS.
(A) SEC-SAXS chromatogram with raw scattering trace in grey, the blue dots give the RG values calculated by Guinier approximation for each frame, and the orange trace gives the MW as determined by MALS.
(B) MES modeling using the 5011 library (based on the open form of the iASPP-PP-1c crystal structure). The single model is shown with color coding as shown on the left.
(C) and (D) Fit of the compact (C) and extended (D) data sets to calculated scattering from the MES models derived from the 5011 library (crystal), the 5001 library, and the combined libraries.
(E) Three model ensembles derived from the combined libraries for the compact data set (top panel) and the extended data set (bottom panel).
Additional details and a comparable analysis of ASPP2-PP-1c and iASPP(621-828)-PP-1c are given in Figures S2, S3, S4 and S5 and Tables S1 and S2.
To further analyze the structure and dynamics of these conformational states, we used the program RAW (Hopkins et al., 2017) to deconvolute the scattering data into two separate sets representing compact (RG = 31.7 Å; Dmax = 113 Å) and extended (RG = 34.5 Å; Dmax = 121 Å) conformational states (Figure S2). To address whether this scattering data could be explained by the iASPP-PP-1c crystal structure, we used a minimal ensemble search (MES) of diverse conformational libraries of ASPP-PP-1c (Pelikan et al (2009); see Star Methods and Table S1 for details of libraries used and the MES procedure). The compact data subset could be fit with a model derived from the crystal structure (χ2 = 3.2; library “crystal”, Figure 3B, C, Table S1), and the quality of the fit could be improved by allowing flexibility in the docking of the ASPP ANK/SH3 domain against PP-1c (χ2 = 1.7; library 5001, Figure 3C, Table S1). The extended data set, however, was only poorly approximated by models from these libraries (Figure 3D, Table S1), suggesting that more extended models would be required to account for this scattering. To allow further flexibility, we generated model libraries in which one or more of the RVxF, SILK or PxxPxR peptides were released from their respective binding partners. MES analysis using these model libraries for the compact data subsets gave ensembles that contained models resembling the crystal structure as well as more extended conformations (Figure 3C, E, Table S1). MES analyses of the extended data subsets in contrast yielded ensembles dominated by more extended structures that do not resemble the crystal structure (Figure 3E). Taken together, this analysis indicates that the ASPP-PP-1c complexes adopt conformations in solution that can sample the compact conformations observed in the crystal structure. However, these compact conformations interconvert with more extended conformations that may involve release of one or more of the RVxF, SILK or PxxPxR contacts. Very similar results were obtained with ASPP2-PP-1c, suggesting that ASPP2-PP-1c may exhibit similar conformational flexibility in solution (Figure S2, S3, Table S2). We next tested the role of the SILK motif in the conformational dynamics of the iASPP-PP-1c complex. We expressed and purified a PP-1c complex containing iASPP(621–828) that lacks the SILK motif and analyzed this complex by SEC-SAXS. Similar to the SILK-containing complexes, this complex eluted as a single peak containing the heterodimer as assessed by MALS (Figure S4A). Evaluation of the RG across the peak revealed that this sample is in slow conformational equilibrium between extended states near the peak to more compact states in the tail. Deconvolution of the data and minimal ensemble modeling suggests that the compact forms dominating in the tail of the peak correspond to the crystal structure, while the extended conformations that dominate near the peak maximum may correspond to states in which either the RVxF or PxxPxR motifs have released. Together, these results indicate that while ASPP-PP-1c complexes can sample compact conformations that resemble the iASPP-PP-1c crystal structure, in solution they exhibit conformational flexibility and populate more highly extended forms that likely involve dynamic disengagement of one or more of the discrete contact surfaces.
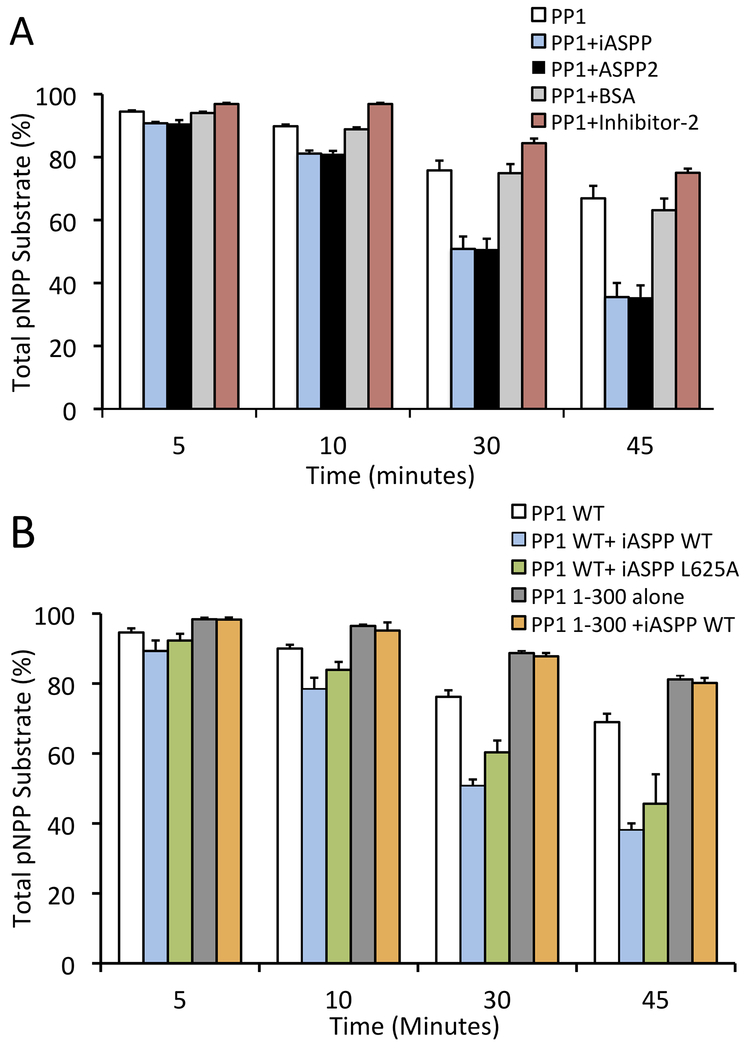
ASPP proteins modulate PP-1c catalytic activity
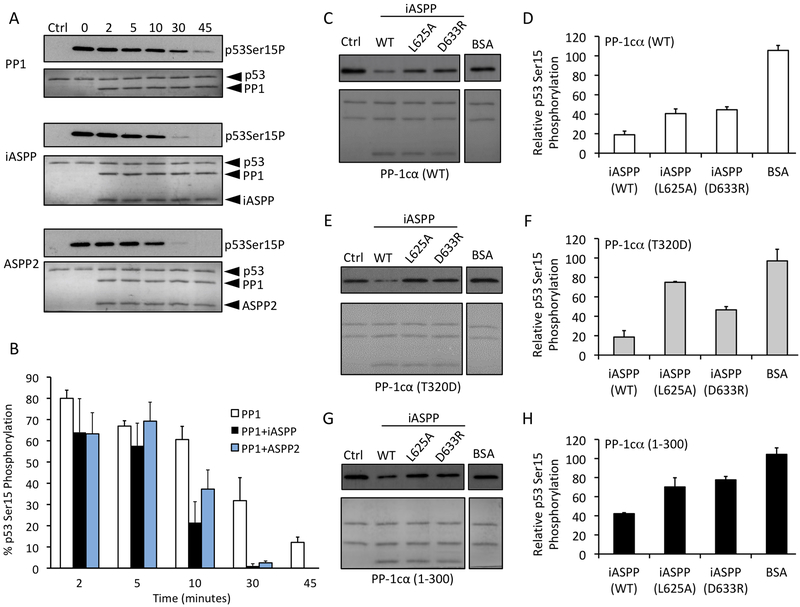
To test the ability of ASPP proteins to regulate PP-1c catalytic activity, we first tested the effect of the ASPPs on the activity of PP-1cα towards the chemical PP-1c substrate, para-nitrophenylphosphate (pNPP) (Figure 4A). pNPP is a small molecule substrate that interacts directly with the PP-1c active site and has been used as a valuable research tool for investigating the effects on the PP-1c active site upon the binding to regulatory proteins and toxins (Choy et al., 2014; Ragusa et al., 2010; Shopik et al., 2013). Both iASPP and ASPP2 were found to increase the activity of PP-1c compared to either a BSA control or the PP-1c inhibitory protein, Inhibitor-2 (Figure 4A). Deletion of the PP-1cα C-terminal tail (PP-1cα 1–300), which is critical for specific interactions with iASPP, effectively blocked the ability of iASPP to enhance PP-1cα dephosphorylation of pNPP, while mutation of the iASPP RVxF motif (L625A) had little effect on the stimulatory activity of iASPP (Fig 4B). Taken together, these data indicate that the specific interactions between ASPP proteins and PP-1c enhance the core catalytic activity of PP-1c.
Figure 4.
iASPP and ASPP2 enhance the activity of PP-1c towards pNPP and inhibit PP-1cα towards Phosphorylase a. (A) PP-1cα colorimetric assay using para-nitrophenylphosphate (pNPP) as a substrate for PP-1cα. PP-1cα was incubated alone or in the presence of iASPP608–828, ASPP2905–1128, bovine serum albumin (BSA) or Inhibitor-2. Graph depicts the total amount of pNPP substrate left in each sample at each time point. Error bars indicate s.d of 3 replicates. (B) PP-1c colorimetric pNPP assay comparing the activity of PP-1cα WT or PP-1cα1-300 alone, or in the presence of WT iASPP608–828, or iASPP608–828 L625A mutant. Error bars indicate s.d of 3 replicates.
We next addressed the effect of the ASPP proteins on p53 dephosphorylation by PP-1c. We set up an in vitro system using purified recombinant proteins to monitor site-specific dephosphorylation of p53 (STAR Methods). We chose to monitor the dephosphorylation of p53 on Ser15, a phosphorylation site known to be involved in the regulation of p53 activity and previously shown to be dephosphorylated by PP-1cα both in vitro and in vivo (Fiscella et al., 1993; Li et al., 2006). DNA-dependent protein kinase (DNA-PK), a Ser/Thr kinase that is activated upon binding to double-stranded DNA during the cellular response to DNA damage, was used to phosphorylate p53 on Ser15, as described previously (Lees-Miller et al., 1992; Soubeyrand et al., 2004). Supporting and extending previous results (Li et al., 2006), we found that purified PP-1cα alone could dephosphorylate p53 phospho-Ser15 over a 45 min time course under our reaction conditions (Figure 5A and B). The addition of an equivalent amount of iASPP608–828 or ASPP2905–1128 markedly increased the rate of dephosphorylation.
Figure 5.
iASPP and ASPP2 enhance the dephosphorylation of p53 Ser-15.
(A) Western blot analysis of in vitro dephosphorylation reactions of DNA-PK phosphorylated p532–293 (blotting for phosho-Ser-15) incubated with PP-1cα alone (top panel) or in the presence of iASPP608–828 (middle panel) or ASPP2905–1128 (lower panel) for 0 to 45 min. The lanes labeled “ctrl” contain a sample of p53 where DNA-PK was inhibited with LY294002 for the duration of the experiment. The lanes labeled “0” contain no PP1 or ASPP-PP1.
(B) Quantification of the amount of phosphorylated p53 Ser-15 in each sample relative to the total phospho-Ser-15 in control. Error bars indicate s.d. of 4 replicates.
(C-H) Western blot analyses of in vitro dephosphorylation reactions of DNA-PK phosphorylated p53 (blotting for phospho-Ser-15 p53) by PP-1cα WT (C and D), PP-1cα T320D (E and F), or PP-1cα1–300 (G and H) in the presence of iASPP WT, iASPP L625A, or iASPP D633R, with results quantitated as in (B). Error bars indicate s.d. of 3 replicates.
In order to identify the molecular interactions between iASPP and PP-1cα that are important for the ASPP-mediated dephosphorylation of p53, we compared the effect of wild-type and mutant iASPP and PP-1cα proteins on the dephosphorylation of p53 Ser15 (Figure 5C–H). To test the importance of the iASPP RVxF-like motif (622RARL625), we tested the effect of the iASPP L625A mutant on p53 dephosphorylation. This mutation resulted in a moderate reduction p53 Ser15 dephosphorylation, compared to WT iASPP (Figure 5C and D). We also tested the importance of the iASPP ANK-PP-1c pivot interaction by creating an iASPP D663R mutant, which reverses the charge of a key residue forming an electrostatic contact with residue Arg261 of PP-1cα (Figure 2E). This mutation exhibited a similar defect in p53 dephosphorylation as iASPP L625A (Figure 5C and D). We next tested the effects of two mutations in the PP-1cα C-terminal tail that is essential for binding to the SH3 domain of ASPP proteins: PP-1cα1–300, which lacks the entire C-terminal tail, and PP-1cα T320D, a mutant that mimics phosphorylation on Thr320 that has been associated with PP-1c autoinhibition (Berndt et al., 1997; Dohadwala et al., 1994; Skene-Arnold et al., 2013). Despite the fact that these mutations have been shown to destabilize iASPP-PP-1c interactions (Skene-Arnold et al., 2013), mutation of PP-1cα T320D had no apparent effect on iASPP-mediated dephosphorylation of p53 and the PP-1cα1–300 mutant only marginally decreased the dephosphorylation of p53 Ser-15 (Figure 5E and F). Combining either of the PP-1cα tail mutations with either of the iASPP mutations further decreased but did not abolish the iASPP effect (Figure 5C–H). Taken together, these data indicate that ASPP proteins can facilitate dephosphorylation of a critical phosphosite in p53 in a manner that relies on the integrity of multiple contacts between ASPP and PP-1cα.
Differential inhibition of p53 DNA binding by ASPP and ASPP-PP-1c complexes
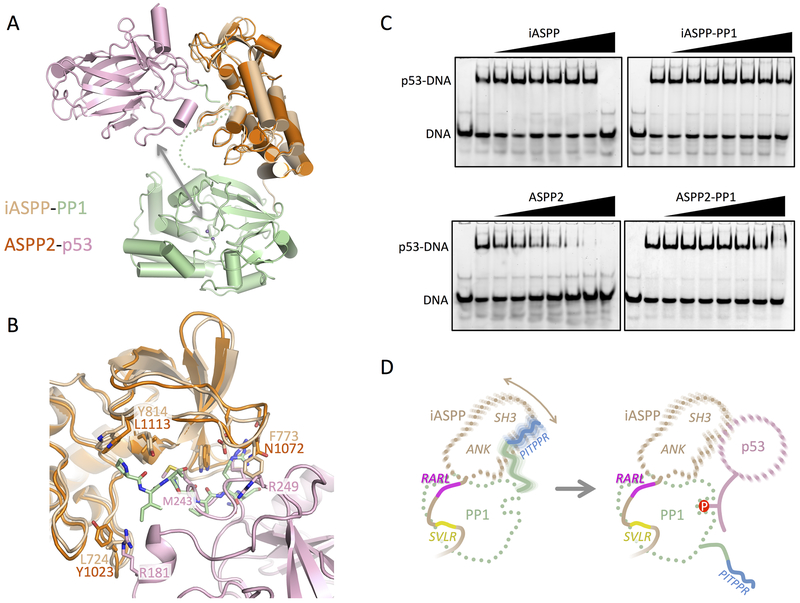
ASPP2 has long been known to bind the p53 DNA binding domain (DBD) through interactions between the ASPP2 SH3 domain and the DNA binding loops of p53 (Gorina and Pavletich, 1996; Patel et al., 2008). The ability of the ASPP proteins to specifically bind p53 suggests that the ASPP proteins could act as a targeting protein, bringing p53 to the vicinity of PP-1c for dephosphorylation (Figure 6A). While the striking similarity of iASPP to ASPP2 suggests that similar interactions may occur between iASPP and p53 DBD, other studies have indicated that the poly-proline regions in p53 may also bind the iASPP SH3 domain (Ahn et al., 2009; Bergamaschi et al., 2006). Alignment of iASPP-PP-1c on the ASPP2-p53 structure reveals that while the overall p53 binding surface is similar, limited substitutions in the binding interface might lead to different binding properties of the two ASPP proteins (Figure 6B). The fact that the p53 DBD can potentially bind either DNA or the ASPP SH3 presents the possibility that these interactions could be mutually exclusive.
Figure 6.
ASPP-PP-1c-p53 interactions and a model for the targeted dephosphorylation of p53 by ASPP-PP-1c.
(A) Interactions between ASPP-PP-1cα complexes and p53. Shown is a structural overlay of the closed iASPP-PP-1cα complex and the ASPP2-p53 DBD complex (PDB ID: 1YCS), aligned on their ASPP components. The arrow indicates the proximity of the p53 DBD to the PP-1c active site catalytic metal ions (purple spheres).
(B) Details of the superposition of the iASPP-PP-1c and ASPP2-p53 DBD complex. Shown is the ASPP SH3 domain and C-terminal ANK finger with the p53 DBD and PP-1c PxxPxR peptide shown. Key residues involved in intermolecular contacts are shown as sticks.
(C) EMSA analysis probing the ability of ASPP or ASPP- PP-1cα complexes to inhibit p53 DBD binding to a PUMA p53 DNA site. Each lane contains 80 nM PUMA. In each panel, lanes 2–9 contain 800 nM p53 DBD, and a gradient of ASPP protein or ASPP- PP-1cα complex at 0, 0.8, 1.6, 3.2, 6.4, 12.8, 25.6, and 51.2 μM.
(D) Model for how a dynamic ASPP-PP-1c complex could bind and target p53 for dephosphorylation. Left panel – the iASPP-PP-1c complex is stabilized by discrete interactions involving the SVLR, RARL and PITPPR peptide motifs, resulting in a flexible complex that allows motion of the iASPP ANK/SH3 domain relative to the PP-1c catalytic domain. Right panel – dynamic release of the PP-1c PITPPR motif allows binding of p53 to the ANK/SH3 domain and dephosphosphorylation.
To directly test this idea, we used an electrophoretic mobility shift assay to examine the effects of either free ASPP or ASPP-PP-1c on the binding of the p53 DBD to a consensus p53 DNA recognition sequence (Figure 6C). The results show that the p53-DNA complex is largely resistant to challenge by iASPP, and competition is only observed at the highest iASPP concentration. In contrast, ASPP2 competes p53 off the DNA at much lower concentrations, suggesting that ASPP2 SH3 binds more tightly to the p53 DBD than iASPP. Complex formation between either ASPP protein and PP-1c abrogates the p53 inhibitory effect of the ASPP proteins, consistent with the idea that the ASPP SH3 peptide binding groove is critical for both p53 and PP-1c binding.
DISCUSSION
Here we present a structural model for interactions between iASPP and PP-1c. Our model suggests that interactions between these proteins are stabilized by multiple discrete contacts that together constitute a large binding interface that can explain the high affinity of this complex (KD ~ 26 nM) (Skene-Arnold et al., 2013). These include the docking of the PxxPxR motif of the PP-1c tail onto the ASPP SH3 domain, docking of iASPP SILK-like and RVxF-like motifs onto the PP-1c catalytic domain, as well as docking of the N-terminal iASPP ANK motif against the PP-1c catalytic domain. The high degree of sequence conservation within the ASPP family suggest that ASPP1/2 may bind PP-1c in a very similar manner, and indeed SAXS analysis indicates that the structure and dynamics of the ASPP2-PP-1c complex is similar to iASPP-PP-1c in solution (Figure S2., S3). While this manuscript was under review, Bertran et al. published a paper reporting on the crystal structure of the ASPP2-PP-1c complex (Bertran et al., 2019). The structure is very similar to the open form of iASPP-PP-1c, except that the ASPP2 construct used by Bertran lacks the SILK motif. These authors went on to test the binding affinities of the three ASPP variants with the three PP-1c variants. They discovered that while all pair-wise complexes are possible, the binding affinities of different pairs can vary by almost 100-fold. ASPP2 showed a significant preference for PP-1cα, and this selectivity could be attributed to interactions between charged residues immediately C-terminal to the PP-1cα PxxPxR motif and the ASPP SH3 domain (Bertran et al., 2019).
The interactions of the RVxF and ANK repeats of iASPP with PP-1c are reminiscent of the interactions with PP-1c of the myosin binding protein and PP-1cβ regulatory subunit, MYPT1 (Terrak et al., 2004) (Figure 1D). The orientation of the N-terminal ANK repeats of MYPT1 are similar to the more closed conformation of the iASPP-PP-1c complex in which the fingers of the two N-terminal-most ANK repeats contact the PP-1c catalytic domain. MYPT1, however, does not contain an SH3 domain but contains additional C-terminal ANK repeats that wrap around the PP-1c C-terminal tail, potentially explaining the specificity of MYPT1 for PP-1cβ (Terrak et al., 2004) (Figure 1D).
Intriguingly, analysis of either iASPP-PP-1c or ASPP2-PP-1c by SEC-SAXS indicates a surprising degree of conformational heterogeneity in both of these complexes (Figures 3, S3, S5). Our analysis and structural modeling based on these data suggests that the conformation represented by the iASPP-PP-1c crystal structure is in slow exchange with more extended conformations under physiological solution conditions. Notably, the more extended conformations cannot be fit by simply allowing rigid body movement of the PP-1c catalytic domain relative to the iASPP ANK/SH3 domain (Figure 3D). However, our analysis suggests that these more extended states could be accessed through the release of one or more of the contacts involving the discrete peptide-domain interactions such as the PxxPxR-SH3 interaction, or the interactions between the SILK or RVxF motifs with the PP-1c catalytic domain. Each of these individual peptide-domain interactions are expected to be of relatively low affinity compared to the overall high affinity of the complex, and are therefore likely to be microscopically dynamic. Indeed, SAXS analysis has indicated that a similar dynamic release of SILK and RVxF motifs are involved in the formation of a heterotrimeric complex between spinophilin, inhibitor 2 and PP-1c (Dancheck et al., 2011). Despite the fact that both spinophilin and inhibitor 2 have been shown to bind to PP-1c primarily via a RVxF motif (Hurley et al., 2007; Ragusa et al., 2010), within the heterodimeric complex, only the RVxF motif from spinophilin was bound to PP-1c.
The work here also has implications for the functional role of the ASPP proteins in the regulation of PP-1c activity. We found that ASPP proteins can activate PP-1c catalytic activity against the small molecule substrate pNPP as well as the physiologically relevant Ser15 phosphosite in p53 (Figure 4, 5). The ASPP proteins, like the MYPT proteins have been implicated as PP-1c targeting proteins that might act to recruit specific substrates to PP-1c for dephosphorylation (Helps et al., 1995; Llanos et al., 2011; Skene-Arnold et al., 2013). ASPP proteins are not only known to specifically bind to p53 using their ANK-SH3 domain (Helps et al., 1995; Llanos et al., 2011; Skene-Arnold et al., 2013), but they also can bind to a variety of other potential targets. These include NF-kB p65 (Yang et al., 1999a; Yang et al., 1999b), Yes-associated protein YAP (Espanel and Sudol, 2001), and Bcl-2 (Katz et al., 2008; Naumovski and Cleary, 1996), many of which may also be regulated by PP-1c (Mui et al., 2015; Vigneron et al., 2010). While there is no high resolution structural information for these other interactions, it is likely that the SH3 peptide binding groove is involved in at least some of these complexes.
Together with our previous binding studies (Skene-Arnold et al., 2013), our crystal structure demonstrates that the PxxPxR motif in the PP-1c tail is critical for ASPP-PP-1c complex formation. This interaction however occludes the SH3 surface that is critical for binding to the p53 DNA binding domain, and indeed we show that the binding of PP-1c to ASPP reduces the ability of ASPP proteins to inhibit p53 DNA binding (Figure 6). This finding then raises the question as to how ASPP proteins could play a targeting role if their primary target binding surface, the SH3 domain, is occluded by PP-1c. Our combined results support the possible dynamic release of the SH3 domain by the PxxPxR peptide, as suggested by our SAXS analysis. This dynamic interface plasticity might free up the SH3 domain to bind protein targets for dephosphorylation by PP-1c (Figure 6D). The additional flexibility afforded by the transient dissociation of the PxxPxR-SH3 interaction may furthermore facilitate dephosphorylation of diverse phosphosites.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Mark Glover (mark.glover@ualberta.ca)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Protein phosphatase-1cα, iASPP608–828, iASPP621–828, ASPP2905–1128 and p532–293 were expressed and purified for biochemical studies as previously described (Skene-Arnold et al., 2013). Mutagenesis of PP-1cα T320D, iASPP L625A, and iASPP D633R was carried out using QuikChange site-directed mutagenesis (Stratagene).
METHOD DETAILS
Purification of ASPP-PP-1c for crystallization and small angle X-ray scattering.
iASPP608–828 bacterial pellets from 4 L culture were resuspended in 100 mL lysis Buffer A (25 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol) containing protease inhibitor cocktail (Pierce Halt). Cells were lysed with three passes through an Emulsiflex-C3 high pressure homogenizer (Avestin). Lysates were clarified by centrifugation at 13000g for 30 minutes at 4 °C. The iASPP supernatant was loaded onto a 4 mL pre-equilibrated Ni-NTA sepharose column, followed by washing with 150 mL wash buffer 1 (25 mM Tris, pH 8.0, 500 mM NaCl, 10 mM imidazole). iASPP-bound Ni-NTA beads were incubated with PP-1cα at 4oC for 1 hour. The supernatant was removed, then the beads were washed with 150 mL wash buffer 1 and 100 mL wash buffer 2 (25 mM Tris, pH 8.0, 500 mM NaCl, 20 mM imidazole). The iASPP His-tag was removed by on-column digestion with 300 µL 6 mg/mL GST-3C protease in digestion buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 2 mM DTT) at 4°C overnight. The cleaved iASPP-PP-1cα complex was further purified by Q sepharose chromatography with Buffer A (40 mM Tris, pH 8.0, 20 mM NaCl, 1 mM DTT) and Buffer B (40 mM Tris, pH 8.0, 1 M NaCl, 1 mM DTT). iASPP-PP1cα was finally purified by size exclusion chromatography using a Superdex 75 16/60 column with Buffer S (25 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT). The protein complex was exchanged into crystallization Buffer C (25 mM Tris, pH 8.0, 100 mM NaCl, 1 mM TCEP, 5% glycerol), concentrated to 4 mg/mL. iASPP621–828 and ASPP2905–1128 were purified similarly.
Crystallization and structure determination of iASPP-PP-1cα.
Crystals of iASPP608–828-PP-1cα complex were grown by sitting drop vapor diffusion with a reservoir solution containing 12–15% PEG6000, 0.1 M sodium citrate, pH 5.3 and were improved by seeding by hanging vapor diffusion at room temperature.
For data crystallographic data collection, iASPP608–828-PP-1cα crystals were flash frozen in liquid nitrogen with a cryoprotectant (reservoir solution with 30% glycerol). Diffraction data were collected on beamline 12.3.1 at the Advanced Light Source and indexed, integrated and scaled using HKL2000. Two data sets were collected, exposing different parts of a single crystal. An automated combinatorial algorithm was used to select the range of frames to maximize data redundancy while excluding frames suffering from radiation damage. Molecular replacement was carried out using PHASER, placing two copies of human PP-1 catalytic subunit (search model PDB ID 3EGG, chain A) and two copies of human iASPP607–822 (PDB ID 2VGE). Refinement in PHENIX utilized automated non-crystallographic symmetry and reference model restraints throughout with the latter being removed for the final refinement round. The PxxPPR, RARL and SILK motif regions were sequentially built toward the end of refinement when the difference density had improved sufficiently to be confident of placement. Careful analysis of binding modes in homologous PxxPPR, RARL and SILK motif complex structures were used to guide sequence register and orientation.
Size exclusion chromatography – small angle X-ray scattering (SEC-SAXS) analysis of ASPP-PP-1c complexes.
SEC-SAXS and multi-angle laser light scattering (SEC-MALLS) was performed on iASPP608–828-PP-1c, iASPP621–828 and ASPP2905–1128–PP-1c complexes at beamline 12.3.1 at the Advanced Light Source (ALS). iASPP-PP-1c and ASPP2-PP-1c samples at either 10 mg/mL and 15 mg/mL were loaded onto a Shodex KW-803 size exclusion column (Shodex) in 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT buffer running at 0.5 mL/min. The column eluant passed through a splitter and was simultaneously analyzed for SAXS on the beamline using a PILATUS3 X 2M detector and MALS using a DAWN detector (Wyatt Technology). MALS data was analyzed using ASTRA VI software (Wyatt Technology) to provide MW information across the eluted peak. SAXS data was analyzed with RAW (Hopkins et al., 2017) to derive RG values for individual 3 sec. frames across the peak using the Guinier approximation.
RAW was used for deconvolution of iASPP-PP1c and ASPP2-PP1c data into separate compact and extended subsets. The range of data selected for the deconvolution extended from the peak to ½ maximal peak height in the tail, a region where MALLS indicated the sample only contained ASPP-PP-1c heterodimer and not higher order aggregates. Each of these datasets was further processed using SCATTER (https://bl1231.als.lbl.gov/scatter/) to determine RG by Guinier analysis for data truncated to q x RG < 1.3 and Dmax by P(r)-distribution plot.
SAXS-based modeling of ASPP-PP-1c structure and dynamics using a minimal ensemble search (MES).
BILBOMD was used to generate model libraries for MES analysis (Pelikan et al., 2009). The starting PDB file for BILBOMD was based on the crystal structure with the flexible regions of peptide not visualized in the crystal structure modeled using SWISS-MODEL (https://swissmodel.expasy.org) and ALossMod-FoXS (https://modbase.compbio.ucsf.edu/allosmod-foxs/). Several model sets with different levels of flexibility were created. The first library (the 5011 library) was designed to represent the open crystal form. In this library, the iASPP ANK-SH3 domain, the PP-1c catalytic domain, as well as the RVxF, SILK and PxxPxR peptides of the open crystal form were treated as a single rigid body, while the peptide linkers and tails that were not observed in the crystallographic electron density were defined as flexible. Library 5013 is analogous to 5011, and uses instead the closed conformation observed crystallographically. Library 5001 allowed additional flexibility by treating the iASPP ANK-SH3 domain and the PP-1c catalytic domain as independent rigid bodies. While these libraries could model the compact datasets generated by RAW, the extended data sets were not well fit by models from these libraries. To allow further flexibility to model the extended ASPP-PP-1c structures observed in solution, we created several additional model libraries in which one or more of the RVxF, SILK or PxxPxR peptides were released from their respective binding partners for iASPP608–828 and ASPP905–1128. These model libraries provided much larger degrees of flexibility as expressed by the range of RG and Dmax for the models in these libraries. In the 5002 library, the PxxPxR peptide was released from its interaction with the iASPP SH3, while the RVxF and SILK interactions were maintained. In the 5003 library, the SILK interaction was released, while the RVxF and PxxPxR contacts were maintained. In the 5004 library, the RVxF interaction was released, while the SILK and PxxPxR contacts were maintained. In the final libraries, two of the peptide contacts were released, leaving only a single peptide interaction to tether the iASPP and PP-1c domains. In library 5005, the two proteins were tethered by the SILK interaction, in library 5006, the tether was provided by the RVxF, while in library 5007 the PxxPxR provided the tethering interaction. We also generated library for iASPP621–828-PP-1c which lacks SILK motif on iASPP. Library 7001 for iASPP621–828-PP-1c were created with restraints corresponding to library 5001. In the library 7002, the PxxPxR peptide was released from its interaction with the iASPP SH3, while the RVxF interactions were fixed. In the 7003 library, the RVxF contacts with the PP-1c were released, while the PxxPxR peptide interaction was maintained. MES was performed on each of the individual model sets, as well as on all of the model sets combined as previously described (Aceytuno et al., 2017; Hodge et al., 2016; Pelikan et al., 2009). Details of the model libraries and the MES results for each of the model pools are given in Supplementary Table 1 for iASPP608–828-PP-1c and Supplementary Table 2 for ASPP2905–1128-PP-1c.
pNPP Phosphatase Activity Assay (Zhuo et al., 1993).
Colorimetric phosphatase activity assays using para-nitrophenylphosphate (pNPP) were carried out as follows. ASPP2905–1128, WT iASPP608–828, iASPP608–828 L625A mutant, Bovine Serum Albumin, and inhibitor-2 proteins were diluted in Buffer L (50 mM Tris-HCl, pH 8.0, 75 mM KCl, 10 mM NaCl, 10 mM MgCl2, 0.2 mM EGTA, 1 mM MnCl2 and 0.2 % β-mercaptoethanol) to a final concentration of 500 nM. PP-1cα WT and PP-1cα 1–300 was diluted in buffer L to 333 nM and added together with each regulatory protein in a 96 well plate. The samples were then incubated for 10 min and reaction was initiated by addition of pNPP (1.2 mM final concentration). Reaction was monitored by absorbance at 405 nm over a 45 min. time course at 37 °C. The percentage of total pNPP substrate was calculated from the raw absorbance of each reaction at a given time point (Abx) and the total absorbance after ~5 hrs (Abtotal) as follows: [(Abtotal – Abx) / Abtotal ]. Each experiment was carried out three times in duplicate and the standard deviation is shown as error bars.
In Vitro p53 Dephosphorylation Assay by Protein Phosphatase-1cα.
Purified p532–293 (0.5 µg, 940 nM final concentration) was phosphorylated with 75 U of DNA-PK (Promega) in DNA-PK kinase buffer (50 mM Tris-HCl, pH 8.0, 75 mM KCl, 10 mM NaCl, 10 mM MgCl2, 1 MnCl2 0.2 mM EGTA, 2 mM DTT, 0.5 mM ATP, 10 µg/mL sonicated calf thymus DNA, 0.025% Tween-20) for 90 minutes at 30°C and stopped with 1 mM LY294002 (Calbiochem). PP-1cα, either alone or combined with ASPP, was pre-incubated at 30°C for 15 minutes in DNA-PK kinase buffer and dephosphorylation was initiated by addition of phosphorylated p53 in a 10 µL reaction volume (final concentrations of 470 nM p532–293, 330 nM PP-1cα, 330 nM PP-1cα (T320D), 360 nM PP-1cα1–300, 520 nM iASPP608–828, or 490 nM ASPP2905–1128). The dephosphorylation reaction was stopped using 2X SDS-PAGE sample buffer.
Dephosphorylation was detected and quantitated by western blotting. p53 samples were separated by 12% SDS-PAGE and transferred onto nitrocellulose using a Mini-Blot Module (Biorad). Nitrocellulose blots were first stained with Memcode Reversible Protein Stain kit (Pierce Scientific) and then scanned to analyze total amount of protein in each dephosphorylation reaction. After removing the reversible stain, blots were blocked using 5 % milk in TBST buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1 % Tween-20) for 1 hr at room temperature. Membranes were then incubated at 4 °C overnight (or for 1 hour at room temperature) with antibody specific for p53 phosphorylated on Ser-15 (Cell Signaling), diluted to 1:15000, in 5 % milk in TBST buffer. Membranes were washed with TBST and further incubated with a horseradish peroxidase-conjugated immunoglobin G secondary antibody (Cell Signaling) for 1 hr at room temperature. Blots were developed for 1 min in enhanced chemiluminescence solution (Clarity™ Western ECL substrate, Biorad) and exposed to Fuji Medical X-Ray Film (Fujifilm). Densitometry for total p53 (stained with Memcode Reversible stain) and phospho-p53 (western blot) were quantified using Image Studio Light or ImageJ software. The amount of phospho-p53 (either p53 phospho-Ser-15) was normalized to the total p53 protein present in each sample. Data was evaluated for outliers by calculating the interquartile range (IQR) and data points were deemed an outlier if they were greater than 1.5 times the IQR. Data was subjected to a one-way analysis of variance (ANOVA) and a post hoc Tukey’s test for individual multiple comparison (p <0.05).
Electrophoretic mobility shift assay.
Complementary nucleotides, PUMA forward and reverse strands (Star method table), were annealed in annealing buffer (10 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA) at 95°C for 2 min and slowly cooled to 23oC. Double stranded DNA (final concentration 80 nM) was incubated with p53 DBD (residues 94–292, final concentration 800 nM) on ice for 30 minutes. ASPP or ASPP-PP1c complex were subsequently added at the concentrations indicated (Figure 6) and incubated on ice for 30 minutes. Binding reactions were performed in 40 mM Tris pH 7.5, 150 mM NaCl, 3% glycerol. Samples were separated by electrophoresis on 6% (wt/vol) native polyacrylamide gels in TBE at 4°C. Gels were visualized by a Typhoon scanner (GE HealthCare).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-p53 (Ser15) Antibody | Cell Signaling | Cat# 9284 |
| Anti-rabbit IgG, HRP Antibody | Cell Signaling | Cat# 7074 |
| Bacterial and Virus Strains | ||
| Rosetta (DE3) pLysS | Novagen | Cat# 70956 |
| C41 (DE3) | Lucigen | Cat# 60442 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Para-nitrophenyl phosphate (pNPP) | Sigma | Cat# P4744 |
| LY294002, PI3-kinase inhibitor | Sigma (Calbiochem) | Cat# 440202 |
| Clarity Western ECL substrate | Biorad | Cat # 1705060 |
| Critical Commercial Assays | ||
| QuikChange II site-directed mutagenesis kit | Stratagene | Cat# 200521 |
| DNA-Dependent Protein Kinase (DNA-PK) | Promega | Cat# V5811 |
| Reversible Protein Stain Kit | Thermo Scientific/Pierce | Cat# 24585 |
| Deposited Data | ||
| Crystal structure, iASPP (608-828)-PP-1cα | This paper | 6DCX |
| Small angle X-ray scattering data, iASPP(608-828)-PP-1cα, compact | This paper | Draft ID 1666 |
| Small angle X-ray scattering data, iASPP(608-828)-PP-1cα, extended | This paper | Draft ID 1667 |
| Small angle X-ray scattering data, His-tagged iASPP(621-828)-PP-1cα, compact | This paper | Draft ID 1657 |
| Small angle X-ray scattering data, His-tagged iASPP(621-828)-PP-1cα, extended | This paper | Draft ID1651 |
| Small angle X-ray scattering data, ASPP2(905-1128)-PP-1cα, compact | This paper | Draft ID1671 |
| Small angle X-ray scattering data, ASPP2(905-1128)-PP-1cα, extended | This paper | Draft ID1672 |
| Oligonucleotides | ||
| PUMA forward strand5’-/56FAM/CGCGCCTGCAAGTCCTGACTTGTCCGCGGC - 3’ | Integrated DNA Technologies | N/A |
| PUMA reverse strand5’ - GCCGCGGACAAGTCAGGACTTGCAGGCGCG -3’ | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Human His(6)-iASPP (608-828) in pET47b | (Skene-Arnold et al., 2013) | N/A |
| Human His(6)-iASPP (621-828) in pET47b | This paper | N/A |
| Human His(6)-iASPP (905-1128) in pET47b | (Skene-Arnold et al., 2013) | N/A |
| Human PP-1cα (1-330) in pKK233-3 vector | (Skene-Arnold et al., 2013) | N/A |
| Human p53(2-293) in pET28A | (Skene-Arnold et al., 2013) | N/A |
| Human p53(94-292) in pGEX-6p-1 | This paper | N/A |
| Software and Algorithms | ||
| PHENIX suite | (Adams et al., 2010) | https://www.phenix-online.org/ |
| HKL2000 | (Otwinowski and Minor, 1997) | http://www.hkl-xray.com/ |
| COOT | (Emsley et al., 2010) | https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot |
| ATSAS suite | (Franke et al., 2017) | https://www.emblhamburg.de/biosaxs/software.html |
| SWISS-MODEL | (Guex et al., 2009; Waterhouse et al., 2018) | https://swissmodel.expasy.org |
| AllosMod-FoXS | (Schneidman-Duhovny et al., 2010; Weinkam et al., 2012) | https://modbase.compbio.ucsf.edu/allosmod-foxs/ |
| RAW | (Hopkins et al., 2017) | https://bioxtas-raw.readthedocs.io/en/latest/ |
| BILBOMD | (Pelikan et al., 2009) | https://bl1231.als.lbl.gov/bilbomd |
| FOXS | (Schneidman-Duhovny et al., 2013, 2016) | https://modbase.compbio.ucsf.edu/foxs/ |
| MES | (Pelikan et al., 2009) | https://bl1231.als.lbl.gov/saxs_protocols/mes.php |
| SCATTER | Bioisis | http://www.bioisis.net/tutorial/9 |
| Image studio lite | LI-COR Biosciences | https://www.licor.com/bio/image-studio-lite/ |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/index.html |
QUANTIFICATION AND STATISTICAL ANALYSES
Crystal structure determination.
Diffraction data statistics were calculated with HKL2000 and crystallographic refinement statistics were calculated in PHENIX and are reported in Table 1.
SAXS analyses.
Goodness of fit (χ2) of individual models to experimental scattering data was calculated using FoXS and these values are reported in Figures 3, S2, S3, S4 and Tables S1 and S2.
pNPP Phosphatase Activity Assay.
The percentage of total pNPP substrate was calculated from the raw absorbance of each reaction at a given time point (Abx) and the total absorbance after ~5 hrs (Abtotal) as follows: [(Abtotal – Abx) / Abtotal ]. Each experiment was carried out three times in duplicate and the standard deviation is shown as error bars. Results are reported in Figure 4.
In Vitro p53 Dephosphorylation Assay by Protein Phosphatase-1cα.
Densitometry for total p53 (stained with Memcode Reversible stain) and phospho-p53 (western blot) were quantified using Image Studio Light or ImageJ software. The amount of phospho-p53 (either p53 phospho-Ser-15) was normalized to the total p53 protein present in each sample. Data was evaluated for outliers by calculating the interquartile range (IQR) and data points were deemed an outlier if they were greater than 1.5 times the IQR. Data was subjected to a one-way analysis of variance (ANOVA) and a post hoc Tukey’s test for individual multiple comparison (p <0.05). Results are reported in Figure 5.
DATA AND CODE AVAILABILITY
The coordinates and the structure factors for the iASPP-PP-1c complex are available in the PDB with accession code 6DCX. All small angle X-ray scattering data have been deposited in the small angle scattering biological data bank (https://www.sasbdb.org).
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Classen and the staff at the Advanced Light Source beamline 12.3.1 for assistance with synchrotron data collection. This work was funded by grants from the Canadian Cancer Society Research Institute to J.N.M.G. and C.F.B.H. (#703775), NIH to J.N.M.G., M.H. and J.A.T. (PO1CA092584), and Natural Sciences and Engineering Council of Canada to J.N.M.G. (2016–05163). J.A.T. is also supported by NIH R35 CA22043, a Robert A. Welch Chemistry Chair, and the Cancer Prevention and Research Institute of Texas. This research used resources of the ALS SIBYLS facilities, which are a DOE Office of Science User Facility supported under the Integrated Diffraction Analysis Technologies (IDAT) program
Footnotes
DATA AVAILABILITY
The coordinates and the structure factors for the iASPP-PP-1c complex has been deposited in the PDB with accession code 6DCX. All small angle X-ray scattering data have been deposited in the small angle scattering biological data bank (https://www.sasbdb.org). Other data are available from corresponding authors upon reasonable request.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Aceytuno RD, Piett CG, Havali-Shahriari Z, Edwards RA, Rey M, Ye R, Javed F, Fang S, Mani R, Weinfeld M, et al. (2017). Structural and functional characterization of the PNKP-XRCC4-LigIV DNA repair complex. Nucleic Acids Res 45, 6238–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X, Roman-Gomez J, Jimenez-Velasco A, Garate L, Montiel-Duarte C, Navarro G, Vazquez I, Zalacain M, Calasanz MJ, Heiniger A, et al. (2006). ASPP1, a common activator of TP53, is inactivated by aberrant methylation of its promoter in acute lymphoblastic leukemia. Oncogene 25, 1862–1870. [DOI] [PubMed] [Google Scholar]
- Ahn J, Byeon IJ, Byeon CH, and Gronenborn AM (2009). Insight into the structural basis of pro- and antiapoptotic p53 modulation by ASPP proteins. The Journal of biological chemistry 284, 13812–13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y, Rohde LH, and Naumovski L (2001). p53-interacting protein 53BP2 inhibits clonogenic survival and sensitizes cells to doxorubicin but not paclitaxel-induced apoptosis. Oncogene 20, 2720–2725. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, and Oren M (2010). The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev 24, 2420–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, O’Neil NJ, Trigiante G, Crook T, Hsieh JK, O’Connor DJ, Zhong S, Campargue I, Tomlinson ML, et al. (2003). iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet 33, 162–167. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. (2006). iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet 38, 1133–1141. [DOI] [PubMed] [Google Scholar]
- Berndt N, Dohadwala M, and Liu CW (1997). Constitutively active protein phosphatase 1alpha causes Rb-dependent G1 arrest in human cancer cells. Curr Biol 7, 375–386. [DOI] [PubMed] [Google Scholar]
- Bertran MT, Mouilleron S, Zhou Y, Bajaj R, Uliana F, Kumar GS, van Drogen A, Lee R, Banerjee JJ, Hauri S, et al. (2019). ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail. Nat Commun 10, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, and Attardi LD (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M, Peti W, Ragusa MJ, and Beullens M (2010). The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci 35, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigan DL, and Shenolikar S (2018). Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu Rev Biochem 87, 921–964. [DOI] [PubMed] [Google Scholar]
- Cao L, Huang Q, He J, Lu J, and Xiong Y (2013). Elevated expression of iASPP correlates with poor prognosis and chemoresistance/radioresistance in FIGO Ib1-IIa squamous cell cervical cancer. Cell and tissue research 352, 361–369. [DOI] [PubMed] [Google Scholar]
- Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, et al. (2014). Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc Natl Acad Sci U S A 111, 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Page R, and Peti W (2012). Regulation of protein phosphatase 1 by intrinsically disordered proteins. Biochem Soc Trans 40, 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancheck B, Ragusa MJ, Allaire M, Nairn AC, Page R, and Peti W (2011). Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry 50, 1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala M, da Cruz e Silva EF, Hall FL, Williams RT, Carbonaro-Hall DA, Nairn AC, Greengard P, and Berndt N (1994). Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc Natl Acad Sci U S A 91, 6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta crystallographica Section D, Biological crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espanel X, and Sudol M (2001). Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem 276, 14514–14523. [DOI] [PubMed] [Google Scholar]
- Feng S, Chen JK, Yu H, Simon JA, and Schreiber SL (1994). Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science 266, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Fiscella M, Ullrich SJ, Zambrano N, Shields MT, Lin D, Lees-Miller SP, Anderson CW, Mercer WE, and Appella E (1993). Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene 8, 1519–1528. [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, et al. (2017). ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. Journal of applied crystallography 50, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin-Heymann N, Wang Y, Slee E, and Lu X (2013). Phosphorylation of ASPP2 by RAS/MAPK pathway is critical for its full pro-apoptotic function. PLoS One 8, e82022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorina S, and Pavletich NP (1996). Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Grassie ME, Moffat LD, Walsh MP, and MacDonald JA (2011). The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Arch Biochem Biophys 510, 147–159. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, and Schwede T (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 Suppl 1, S162–173. [DOI] [PubMed] [Google Scholar]
- Helps NR, Barker HM, Elledge SJ, and Cohen PT (1995). Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett 377, 295–300. [DOI] [PubMed] [Google Scholar]
- Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, and Glover JN (2016). RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment. J Biol Chem 291, 9396–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, and Skou S (2017). BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. Journal of applied crystallography 50, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, and DePaoli-Roach AA (2007). Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem 282, 28874–28883. [DOI] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, and Hartshorne DJ (2004). Myosin phosphatase: structure, regulation and function. Mol Cell Biochem 259, 197–209. [DOI] [PubMed] [Google Scholar]
- Jia Y, Peng L, Rao Q, Xing H, Huai L, Yu P, Chen Y, Wang C, Wang M, Mi Y, et al. (2014). Oncogene iASPP enhances self-renewal of hematopoietic stem cells and facilitates their resistance to chemotherapy and irradiation. FASEB J 28, 2816–2827. [DOI] [PubMed] [Google Scholar]
- Katz C, Benyamini H, Rotem S, Lebendiker M, Danieli T, Iosub A, Refaely H, Dines M, Bronner V, Bravman T, et al. (2008). Molecular basis of the interaction between the antiapoptotic Bcl-2 family proteins and the proapoptotic protein ASPP2. Proc Natl Acad Sci U S A 105, 12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, and Anderson CW (1992). Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol 12, 5041–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Liu JP, Schmid PC, Schlosser R, Feng H, Liu WB, Yan Q, Gong L, Sun SM, Deng M, et al. (2006). Protein serine/threonine phosphatase-1 dephosphorylates p53 at Ser-15 and Ser-37 to modulate its transcriptional and apoptotic activities. Oncogene 25, 3006–3022. [DOI] [PubMed] [Google Scholar]
- Li G, Wang R, Gao J, Deng K, Wei J, and Wei Y (2011). RNA interference-mediated silencing of iASPP induces cell proliferation inhibition and G0/G1 cell cycle arrest in U251 human glioblastoma cells. Molecular and cellular biochemistry 350, 193–200. [DOI] [PubMed] [Google Scholar]
- Li S, Shi G, Yuan H, Zhou T, Zhang Q, Zhu H, and Wang X (2012). Abnormal expression pattern of the ASPP family of proteins in human non-small cell lung cancer and regulatory functions on apoptosis through p53 by iASPP. Oncology reports 28, 133–140. [DOI] [PubMed] [Google Scholar]
- Lim WA, Richards FM, and Fox RO (1994). Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Lin BL, Xie DY, Xie SB, Xie JQ, Zhang XH, Zhang YF, and Gao ZL (2011). Down-regulation of iASPP in human hepatocellular carcinoma cells inhibits cell proliferation and tumor growth. Neoplasma 58, 205–210. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang M, Diao S, Rao Q, Zhang X, Xing H, and Wang J (2009). siRNA-mediated down-regulation of iASPP promotes apoptosis induced by etoposide and daunorubicin in leukemia cells expressing wild-type p53. Leukemia research 33, 1243–1248. [DOI] [PubMed] [Google Scholar]
- Liu WK, Jiang XY, Ren JK, and Zhang ZX (2010). Expression pattern of the ASPP family members in endometrial endometrioid adenocarcinoma. Onkologie 33, 500–503. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Lu X, Zhang Y, Zhong S, Gu SZ, Zhang XB, Yang X, and Xin HM (2005). Downregulated mRNA expression of ASPP and the hypermethylation of the 5’-untranslated region in cancer cell lines retaining wild-type p53. FEBS letters 579, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Zhang Y, Zhang XB, and Yang X (2004). Abnormal mRNA expression of ASPP members in leukemia cell lines. Leukemia 18, 880. [DOI] [PubMed] [Google Scholar]
- Llanos S, Royer C, Lu M, Bergamaschi D, Lee WH, and Lu X (2011). Inhibitory member of the apoptosis-stimulating proteins of the p53 family (iASPP) interacts with protein phosphatase 1 via a noncanonical binding motif. J Biol Chem 286, 43039–43044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos IS, Natkunam Y, Levy R, and Lopez CD (2002). Apoptosis stimulating protein of p53 (ASPP2) expression differs in diffuse large B-cell and follicular center lymphoma: correlation with clinical outcome. Leukemia & lymphoma 43, 2309–2317. [DOI] [PubMed] [Google Scholar]
- Lu B, Guo H, Zhao J, Wang C, Wu G, Pang M, Tong X, Bu F, Liang A, Hou S, et al. (2010). Increased expression of iASPP, regulated by hepatitis B virus X protein-mediated NF-kappaB activation, in hepatocellular carcinoma. Gastroenterology 139, 2183–2194 e2185. [DOI] [PubMed] [Google Scholar]
- Lu M, Breyssens H, Salter V, Zhong S, Hu Y, Baer C, Ratnayaka I, Sullivan A, Brown NR, Endicott J, et al. (2013). Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer cell 23, 618–633. [DOI] [PubMed] [Google Scholar]
- Mori S, Ito G, Usami N, Yoshioka H, Ueda Y, Kodama Y, Takahashi M, Fong KM, Shimokata K, and Sekido Y (2004). p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer 100, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Mori T, Okamoto H, Takahashi N, Ueda R, and Okamoto T (2000). Aberrant overexpression of 53BP2 mRNA in lung cancer cell lines. FEBS Lett 465, 124–128. [DOI] [PubMed] [Google Scholar]
- Mui MZ, Zhou Y, Blanchette P, Chughtai N, Knight JF, Gruosso T, Papadakis AI, Huang S, Park M, Gingras AC, et al. (2015). The Human Adenovirus Type 5 E4orf4 Protein Targets Two Phosphatase Regulators of the Hippo Signaling Pathway. J Virol 89, 8855–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L, and Cleary ML (1996). The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol 16, 3884–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Patel S, George R, Autore F, Fraternali F, Ladbury JE, and Nikolova PV (2008). Molecular interactions of ASPP1 and ASPP2 with the p53 protein family and the apoptotic promoters PUMA and Bax. Nucleic Acids Res 36, 5139–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, and Hammel M (2009). Structure and Flexibility within proteins as identified through small angle X-ray scattering. General Physiology and Biophysics 28, 174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto EM, Musolino NR, Cescato VA, Soares IC, Wakamatsu A, de Oliveira E, Salgado LR, and Bronstein MD (2010). iASPP: a novel protein involved in pituitary tumorigenesis? Frontiers of hormone research 38, 70–76. [DOI] [PubMed] [Google Scholar]
- Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, and Peti W (2010). Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nature structural & molecular biology 17, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, and Tainer JA (2013). Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual review of biophysics 42, 415–441. [DOI] [PubMed] [Google Scholar]
- Robinson RA, Lu X, Jones EY, and Siebold C (2008). Biochemical and structural studies of ASPP proteins reveal differential binding to p53, p63, and p73. Structure 16, 259–268. [DOI] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, and Lu X (2001). ASPP proteins specifically stimulate the apoptotic function of p53. Molecular cell 8, 781–794. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, and Sali A (2010). FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38, W540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2013). Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J 105, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2016). FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res 44, W424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR Jr., and Elkahloun AG (1999). In vivo gene expression profile analysis of human breast cancer progression. Cancer Res 59, 5656–5661. [PubMed] [Google Scholar]
- Shi Y (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. [DOI] [PubMed] [Google Scholar]
- Shopik MJ, Li L, Luu HA, Obeidat M, Holmes CF, and Ballermann BJ (2013). Multi-directional function of the protein phosphatase 1 regulatory subunit TIMAP. Biochem Biophys Res Commun 435, 567–573. [DOI] [PubMed] [Google Scholar]
- Skene-Arnold TD, Luu HA, Uhrig RG, De Wever V, Nimick M, Maynes J, Fong A, James MN, Trinkle-Mulcahy L, Moorhead GB, et al. (2013). Molecular mechanisms underlying the interaction of protein phosphatase-1c with ASPP proteins. The Biochemical journal 449, 649–659. [DOI] [PubMed] [Google Scholar]
- Soubeyrand S, Schild-Poulter C, and Hache RJ (2004). Structured DNA promotes phosphorylation of p53 by DNA-dependent protein kinase at serine 9 and threonine 18. Eur J Biochem 271, 3776–3784. [DOI] [PubMed] [Google Scholar]
- Terrak M, Kerff F, Langsetmo K, Tao T, and Dominguez R (2004). Structural basis of protein phosphatase 1 regulation. Nature 429, 780–784. [DOI] [PubMed] [Google Scholar]
- Trigiante G, and Lu X (2006). ASPP [corrected] and cancer. Nat Rev Cancer 6, 217–226. [DOI] [PubMed] [Google Scholar]
- Vigneron AM, Ludwig RL, and Vousden KH (2010). Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev 24, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xing H, Tian Z, Peng L, Li Y, Tang K, Rao Q, Wang M, and Wang J (2012). iASPPsv antagonizes apoptosis induced by chemotherapeutic agents in MCF-7 cells and mouse thymocytes. Biochem Biophys Res Commun 424, 414–420. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkam P, Pons J, and Sali A (2012). Structure-based model of allostery predicts coupling between distant sites. Proc Natl Acad Sci U S A 109, 4875–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JP, Hori M, Sanda T, and Okamoto T (1999a). Identification of a novel inhibitor of nuclear factor-kappaB, RelA-associated inhibitor. J Biol Chem 274, 15662–15670. [DOI] [PubMed] [Google Scholar]
- Yang JP, Hori M, Takahashi N, Kawabe T, Kato H, and Okamoto T (1999b). NF-kappaB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene 18, 5177–5186. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xiao HJ, Chen J, Tao X, and Cai LH (2011). Inhibitory member of the apoptosis-stimulating protein of p53 (ASPP) family promotes growth and tumorigenesis in human p53-deficient prostate cancer cells. Prostate cancer and prostatic diseases 14, 219–224. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang M, Zhou C, Chen S, and Wang J (2005). The expression of iASPP in acute leukemias. Leukemia research 29, 179–183. [DOI] [PubMed] [Google Scholar]
- Zhao J, Wu G, Bu F, Lu B, Liang A, Cao L, Tong X, Lu X, Wu M, and Guo Y (2010). Epigenetic silence of ankyrin-repeat-containing, SH3-domain-containing, and proline-rich-region- containing protein 1 (ASPP1) and ASPP2 genes promotes tumor growth in hepatitis B virus-positive hepatocellular carcinoma. Hepatology 51, 142–153. [DOI] [PubMed] [Google Scholar]
- Zhuo S, Clemens JC, Hakes DJ, Barford D, and Dixon JE (1993). Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. The Journal of biological chemistry 268, 17754–17761. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and the structure factors for the iASPP-PP-1c complex are available in the PDB with accession code 6DCX. All small angle X-ray scattering data have been deposited in the small angle scattering biological data bank (https://www.sasbdb.org).