Abstract
Purpose of review:
In this review we highlight recent advances in the human genetics of frontotemporal dementia (FTD). In addition to providing a broad survey of genes implicated in FTD in the last several years, we also discuss variation in genes implicated in both hereditary leukodystrophies and risk for FTD (e.g., TREM2, TMEM106B, CSF1R, AARS2, NOTCH3).
Recent findings:
Over the past five years, genetic variation in approximately 50 genes has been confirmed or suggested to cause or influence risk for FTD and FTD-spectrum disorders. We first give background and discuss recent findings related to C9ORF72, GRN and MAPT, the genes most commonly implicated in FTD. We then provide a broad overview of other FTD-associated genes and go on to discuss new findings in FTD genetics in East Asian populations, including pathogenic variation in CHCHD10, which may represent a frequent cause of disease in Chinese populations. Finally, we consider recent insights gleaned from genome-wide association and genetic pleiotropy studies.
Summary:
Recent genetic discoveries highlight cellular pathways involving autophagy, the endolysosomal system and neuroinflammation, and reveal an intriguing overlap between genes that confer risk for leukodystrophy and FTD.
Keywords: frontotemporal lobar degeneration, leukodystrophy, genetics, autophagy, lysosomes, inflammation
Introduction
Frontotemporal dementia (FTD), one of the most common forms of dementia after Alzheimer’s disease (AD) in people younger than 65, encompasses a broad, clinically heterogeneous group of disorders involving pathological changes in personality, behavior and/or language due to underlying neurodegeneration of the frontal and temporal lobes of the cerebral cortex (i.e., frontotemporal lobar degeneration [FTLD]). When the primary clinical manifestations include behavioral disinhibition, apathy, loss of empathy, hyperorality, and/or compulsive behaviors, patients are likely to meet clinical criteria for diagnosis of behavioral variant FTD (bvFTD). Conversely, patients that present with difficulty understanding the meaning of words or naming people or objects are most often diagnosed with semantic variant primary progressive aphasia (svPPA); difficulties with motoric aspects of speech (e.g., word generation, pronunciation) are associated with a diagnosis of nonfluent variant PPA (nfvPPA). Individuals diagnosed with FTD-spectrum disorders often have a family history of dementia or other neurodegenerative disease (~40% of cases [1,2]), and it has been estimated that ~10–30% of FTD is inherited in an autosomal-dominant manner [1–3], a much higher proportion than that which has been found for AD (<1% [4]), highlighting the importance of genetics in the etiology of FTD.
FTLD underlying clinical diagnoses within the FTD spectrum can be characterized neuropathologically by the abnormal accumulation of the proteins TDP-43 (FTLD-TDP; ~50–60% of cases; [5,6]), tau (FTLD-tau; ~40–45% of cases [5,7]) or FET (FUS, EWS, TAF15; FTLD-FET; ~5–10% of cases [5,7]). In rare cases, clinical FTD is associated with white matter degeneration rather than pathological protein accumulation (see leukodystrophy section below). It is important to note that clinical diagnosis of a particular FTD subtype (e.g., bvFTD) may be caused by a variety of distinct neuropathologies, while a particular subtype of FTLD (e.g., FTLD-TDP) may, in different individuals, lead to distinct diagnoses within the FTD spectrum. In addition, it is important to keep in mind that perhaps 20% or more of clinically diagnosed FTD cases may arise from underlying AD or other non-FTLD pathology [5]. These issues highlight how cohort selection (e.g., clinically vs. pathologically defined) will have a marked influence on the outcome of genetic studies.
A combination of clinical observation, genetic discovery and overlapping pathology described over the last 20 years has led to the current view that FTD and the motor neuron disease, amyotrophic lateral sclerosis (ALS), have a shared etiology and can be thought to exist within a disease continuum. In this review, we (i) highlight the most important genetic contributions to disorders within the FTD and FTD-ALS spectrum, emphasizing in particular those discoveries made in the last five years; (ii) place a special emphasis on genes involved in both leukodystrophy and FTD; and (iii) discuss genes implicated as disease risk factors in East Asian populations.
MAPT, GRN and C9ORF72
Pathogenic variation in MAPT, encoding the microtubule-binding protein tau, was first linked to an autosomal-dominant form of FTD with parkinsonism over 20 years ago [8–11] and has since been estimated to account for 5–20% of familial FTD cases (see Table 1 for a summary of the major genes implicated in familial FTD; [5,7,12,13]). The large range in estimated prevalence of pathogenic variation in MAPT (and other genes) in familial FTD reflects both the malleability of the term ‘familial’ and the nature and ethnicity of the cohorts being studied. To date, over 50 pathogenic mutations in MAPT have been identified [13], but only rarely (0–2% of cases) do these mutations account for sporadic (non-familial) FTD (e.g., see [14]). Although MAPT mutations are generally thought to display complete penetrance, a recent report of the well-established p.V337M mutation (rs63750570) has described very slow disease progression in one carrier and apparent absence of disease in her 67-year-old son [15]. Relatedly, the p.G389R mutation (rs63750512) in some cases results in aggressive FTD, but it can also be found in unaffected individuals [16]. Cases such as these illustrate that even for well-characterized genes, presumed “causal” mutations may not be completely penetrant, possibly due to the modifying effects of other genetic or environmental factors. In addition, although the vast majority of pathogenic MAPT mutations are associated with bvFTD and FTD-spectrum disorders, the p.R406W mutation (rs63750424) is instead associated primarily with clinically diagnosed AD [17,18]. Curiously, however, in two independent families, patients that are homozygous for the p.R406W mutation present with bvFTD [19,20], suggesting that the dosage of this mutation can influence disease outcome.
Table 1. Major genes implicated in autosomal-dominant forms of FTD.
Genes implicated in ≥1% of familial FTD cases are listed in descending order of mutation frequency in cohorts of European ancestry. Estimated mutation frequency data are derived from sources indicated within the main text. Estimated numbers of pathogenic (including presumptive pathogenic) mutations are from the Alzheimer Disease & Frontotemporal Dementia Mutation Database (http://www.molgen.ua.ac.be/ADmutations/default.cfm?MT=1&ML=6&Page=StatPerGene) and [12,13,39].
| Gene | Chr. | Protein | Protein function | Mutation freq. in familial FTD | Mutation freq. in sporadic FTD | # Pathogenic mutationsa |
|---|---|---|---|---|---|---|
| C9ORF72 | 9p21.2 | C9ORF72 | Lysosomal homeostasis | 20–30% | 6% | b |
| GRN | 17q21.31 | Progranulin | Lysosomal homeostasis; inflammation | 5–25% | 5% | 79 |
| MAPT | 17q21.31 | Microtubule-associated protein tau | Microtubule stabilization and assembly | 5–20% | 0–2% | >50 |
| TBK1 | 12q14.2 | Serine/threonine-protein kinase TBK1 | Regulator of autophagy and inflammation | 3% | 2–4% | 28 |
| SQSTM1 | 5q35.3 | p62 (Sequestosome-1) | Selective autophagy receptor | 1–3% | 1–3% | ~20 |
| TARDBP | 1p36.22 | TAR DNA-binding protein 43 (TDP-43) | RNA processing and metabolism | 1% | 1% | 33 |
A subset of these mutations are implicated in ALS or ALS-FTD (e.g., TARDBP mutations). Pathogenicity is not established for all mutations.
Indicates variable-length hexanucleotide repeat expansions in C9ORF72.
After the discovery of pathogenic MAPT mutations linked to familial FTD with parkinsonism in 1998, several families remained who possessed neither MAPT mutations nor tau pathology but who displayed autosomal-dominant FTD linked to the same chromosomal region (17q21). In 2006, two groups independently identified null mutations in GRN, located <2 Mb centromeric of MAPT and encoding the secreted glycoprotein progranulin, as the cause of disease in these families [21,22]. Since this discovery, over 70 pathogenic mutations in GRN have been described, most of which are thought to result in loss of function via interference with GRN transcription or translation resulting in protein haploinsufficiency; missense mutations have also been described [6]. Pathogenic variation in GRN is currently estimated to account for 5–25% of familial FTD [6,13] and perhaps as much as 10% of all FTD cases [23]. TMEM106B is an important modifier of FTD risk and age at disease onset in individuals with GRN mutations [24] and will be discussed in more detail below. While the initial identification of pathogenic GRN mutations suggested an important role for microglia in FTD pathogenesis [22,25], the 2012 discovery of a homozygous, loss-of-function GRN mutation as a cause of neuronal ceroid lipofuscinosis (NCL) [26], a lysosomal storage disorder (LSD), provided additional insight into progranulin biology, and suggested that lysosomal homeostasis might represent a cellular pathway relevant to FTD pathogenesis. Moreover, the recent identification of potentially pathogenic mutations in the CTSF gene—encoding the lysosomal hydrolase, cathepsin F, mutations in which also cause NCL—in patients with FTD and early-onset AD indicates that additional NCL genes may contribute risk for FTD and related dementias [27,28]. In support of this idea, recent evidence from our laboratory suggests that rare variant enrichment in MFSD8 (CLN7) may contribute to FTLD risk [29].
Additional rare causes of FTD, ALS-FTD and complex clinical syndromes that sometimes include FTD were reported in the years following the discoveries of pathogenic variation in MAPT (including mutations in VCP [30] and CHMP2B [31]) and GRN (including mutations in TARDBP [32], FUS [33] and UBQLN2 [34]). However, as these mutations collectively account for only a small fraction (<5%) of familial FTD [7,13], it was clear by 2011 that there must be additional pathogenic mutations accounting for a larger proportion of familial FTD. Thus, the discovery that a hexanucleotide G4C2 repeat expansion intronic to C9ORF72 was a common cause of familial and sporadic forms of FTD, ALS and ALS-FTD [14,35], reported independently by two groups, was met with great excitement. It is currently estimated that pathogenic repeat expansion in C9ORF72 accounts for 20–30% of familial and 6% of sporadic FTD in cohorts of European descent [6,7,13], although the expansion appears to be much less common in East Asian patients with FTD (see below). C9ORF72 repeat expansions have also been reported in a small proportion of familial, late-onset AD cases [36]. Models for the pathogenicity of C9ORF72 expansion remain hotly debated and include haploinsufficiency; toxicity from the transcribed, expanded-repeat-containing RNA; and toxic dipeptide repeat proteins generated through non-canonical expanded-repeat translation. A detailed discussion of such mechanisms is beyond the scope of this article, but other recent reviews provide excellent descriptions (e.g., see [37]).
Additional FTD genes
SQSTM1, OPTN and TBK1
Mutations in SQSTM1, which encodes a multifunctional autophagy adaptor protein, p62, were first discovered in FTD patients in 2012 [38], and rare variants in this gene were subsequently found to increase risk for disease in a large cohort of European FTD patients [39]. Interestingly, the recent identification of compound heterozygous loss-of-function mutations in SQSTM1 as a cause of childhood-onset neurodegenerative disease [40] suggests that heterozygous mutations implicated in FTD may confer risk via haploinsufficiency. Although mutations in OPTN, encoding another autophagy adaptor protein, optineurin, have generally been associated with ALS rather than FTD, several reports have identified heterozygous mutations in ALS cases that also presented with aspects of FTD [41,42]. In addition, compound heterozygous mutations in OPTN have more recently been identified in FTLD-TDP by whole-genome sequencing [43]. Functional studies suggest that disease-associated mutations in both SQSTM1 and OPTN result in impaired macroautophagy (reviewed in [37]), strongly implicating this cellular pathway in FTD and ALS pathogenesis.
Heterozygous, loss-of-function mutations in TBK1 (encoding TANK-binding kinase 1) were recently implicated in FTD and ALS as well as pathologically confirmed cases of FTLD-TDP [43,44]. Interestingly, some pathogenic TBK1 mutations appear to impair the ability of TBK1 to bind optineurin, thus implicating these mutations in dysfunctional autophagy, similar to pathogenic SQSTM1 and OPTN mutations [44]. Findings from European FTD cohorts indicate that pathogenic TBK1 mutations may be a relatively common genetic cause of disease (accounting for 1–5% of FTD and FTD-ALS), perhaps second only to pathogenic variation in C9ORF72 and GRN [45]. TBK1 was very recently suggested to be a key regulator of inflammation in the brain by acting as a negative regulator of RIPK1 kinase activity [46], thus placing TBK1 at the crossroads of autophagy and inflammation. Analogously, PRKN, encoding the key mitophagy regulator parkin, was recently shown to regulate inflammation by preventing the accumulation of damaged mitochondria [47]. Although a well-established familial Parkinson’s disease (PD) gene, pathogenic mutations in PRKN may also be a rare cause of bvFTD in the absence of parkinsonism [48]. These findings illustrate the interconnectedness of autophagy and neuroinflammation, and indicate both may represent key pathways in multiple forms of neurodegeneration.
TIA1 and CCNF
A missense mutation in the low-complexity domain (LCD) of TIA1, encoding T-cell-restricted intracellular antigen-1, was recently identified as the potentially causal mutation in a family with ALS-FTD [49]. Further analysis by the same investigators suggested an increased burden of rare variants specifically within the LCD of TIA1 in ALS and ALS-FTD patients relative to controls. As an LCD-containing, RNA-binding protein, TIA1 shares characteristics with other well known FTD-associated proteins such as TDP-43 and FUS. However, it remains unclear whether rare variants in TIA1 indeed impart risk for ALS or ALS-FTD, as several groups have more recently failed to replicate the aforementioned findings [50,51].
Intriguingly, in Tia1 knockout mice, the phenotype of which was reported prior to the identification of rare variation in TIA1 associated with ALS-FTD, cyclin F (Ccnf) is significantly up-regulated in the central nervous system [52], and a pathogenic mutation in human CCNF was recently linked to disease in a large ALS-FTD family [53]. Follow-up analyses identified additional rare variants enriched in sporadic FTD and ALS cohorts, as well as in familial ALS and FTD-ALS cohorts [53]. Cyclin F is a component of an SCF-type E3 ubiquitin ligase, and the variants associated with FTD/ALS have been suggested to impair protein degradation via the ubiquitin-proteasome system, indicating that cyclin F, like many other FTD-associated proteins, may regulate cellular proteostasis.
Genes implicated in hereditary leukodystrophies and FTD
Hereditary leukodystrophies (also referred to as leukoencephalopathies) represent a diverse array of inherited disorders of myelin formation and maintenance. Below we describe seven genes implicated in various classes of leukodystrophy that are also associated with FTD risk. In some cases, patients harboring pathogenic mutations in these genes present clinically with aspects of FTD occurring as part of a constellation of symptoms characteristic of the underlying white matter loss (e.g., CSF1R). In another scenario, one class of variant in a given gene may modify the onset of FTD, while a distinct, pathogenic mutation in the same gene (TMEM106B) may cause a childhood-onset leukodystrophy. The genes highlighted below have diverse expression patterns among human brain cell types, including predominant expression in microglia (TREM2, TYROBP, CSF1R); neurons, astrocytes and oligodendrocytes (TMEM106B); and, potentially, endothelial cells (NOTCH3) ([54]; brainrnaseq.org ). Intriguingly, using publicly available data, we find that most genes implicated in both leukodystrophy and FTD risk show differential expression in FTLD postmortem brain (Figure 1; [55]). Using gene interaction network analysis, we found that leukodystrophy/FTD-associated genes are interconnected with genes regulating immunological function and lysosomal homeostasis, including some that are implicated in Mendelian neurodegenerative LSDs (Figure 2; [56]). Below we describe in more detail genes implicated in both leukodystrophy and FTD.
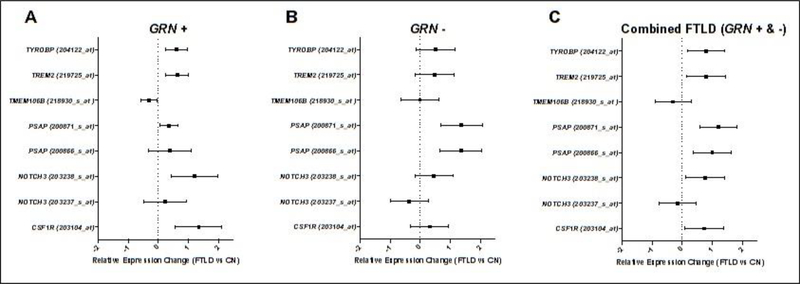
Figure 1. Leukodystrophy genes are dysregulated in pathologically proven FTLD.
Forest plots depict results from differential expression analyses in pathologically proven frontotemporal lobar degeneration (FTLD) due to progranulin mutations (GRN+; n=7) and sporadic FTLD (GRN−; n=10) compared to controls (n=11). (A) In GRN+ cases, all tested leukodystrophy genes (6/6) demonstrated dysregulated expression for at least one microarray probe (praw<0.05). (B) In contrast, the only dysregulated gene in GRN− cases was PSAP (praw<0.05). (C) In a combined analysis, all genes but TMEM106B demonstrated significant dysregulation for at least one microarray probe (praw<0.05). All analyses utilized linear regression. The main effect of diagnosis on relative expression for each gene is plotted with 95% confidence intervals. For details on the samples and gene expression measurement, see Chen-Plotkin et al., 2008 [55] and data available through GEO (GSE13162). Gene expression was estimated using an Affymetrix Human Genome U133A microarray. The forest plots depict all available microarray probes mapping to the query genes (AARS2 was not available for analysis). Probe identifiers are provided alongside each gene’s name in parentheses.
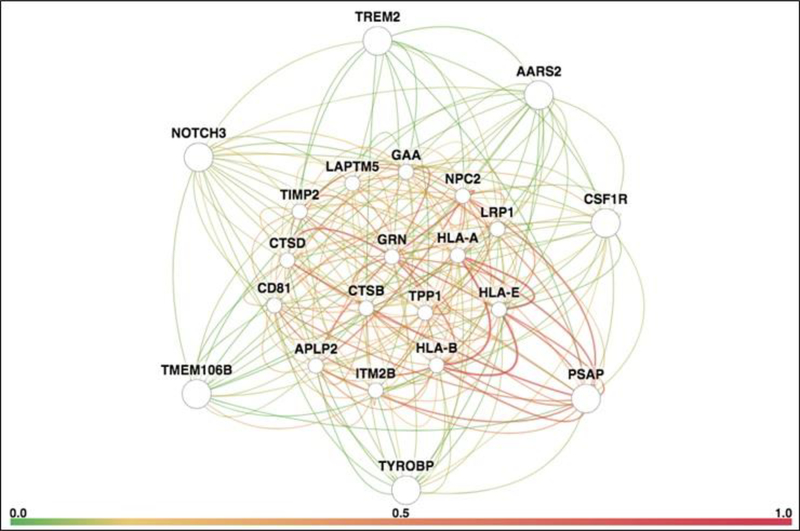
Figure 2. Gene interaction network for leukodystrophy genes (PSAP, TYROBP, TMEM106B, NOTCH3, TREM2, AARS2 and CSF1R).
The network is remarkable for extensive interactions with progranulin (GRN, center of network), genes implicated in immunological function (CD81, HLA-A, HLA-B and HLA-E), lysosomal genes (CTSB, CTSD, GAA, LAPTM5, LRP1, NPC2 and TPP1), genes implicated in neurodegenerative lysosomal storage disorders (CTSD, GRN, NPC2 and TPP1), and a gene implicated in familial dementia (ITM2B). The network diagram was generated using HumanBase, a publicly available online database and analytical pipeline hosted by the Flatiron Institute (http://www.simonsfoundation.org/flatiron/). We limited our search to high-quality, brain-specific relationships connected to the query genes through co-expression, protein interaction, or shared transcription factor binding. Query genes are located on the periphery of the network to facilitate visualization of their connections with interacting network genes. The thickness of a connection represents edge weight, or strength of ties. Connection color represents ‘evidence’ for an edge, defined as the posterior probability of a functional relationship given the brain-tissue specific connectivity dataset, minus the prior probability. Detailed descriptions of the techniques used are provided in Greene et al., 2015 [56] and at https://humanbase.readthedocs.io/en/latest/functional-networks.html.
TREM2 and TYROBP
TREM2 and TYROBP (encoding DAP12), which together encode a receptor signaling complex expressed in myeloid cells, were first linked to autosomal-recessive Nasu-Hakola disease (NHD; also known as polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy) more than 15 years ago [57,58]. Although NHD classically involves early-onset dementia and recurrent bone fractures, a number of patients with FTD-like syndromes lacking a bone phenotype (often with early-onset symptoms) have also been identified who harbor homozygous or compound heterozygous mutations in TREM2 [59–65]. Moreover, a recent meta-analysis of rare TREM2 variants found that variants p.R47H (rs75932628) and p.T96K (rs2234253) confer an ~2–3-fold increase in risk for FTD in European populations [66]; p.R47H is also a well known AD risk factor [67] and a possible risk factor for PD [68].
CSF1R
The CSF1R gene, expressed in microglia and encoding the colony stimulating factor 1 receptor, was originally linked to autosomal-dominant inheritance of adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) in 2011 [69]. More recently, heterozygous variants (including missense, splice-site, and in-frame deletion variants) in CSF1R have been identified in ALSP patients who presented clinically with aspects of bvFTD [70–72]. The clinicopathologic similarities between NHD and ALSP along with the shared microglial expression pattern of CSF1R, TREM2 and TYROBP highlight the importance of microglial function in white matter homeostasis.
TMEM106B and PSAP
Variants in TMEM106B were first linked to FTLD in 2010 [73]. Though originally identified as a risk modifier in FTLD-TDP due to GRN mutations [24,73,74] and significantly associated with circulating progranulin levels [24,74], TMEM106B was subsequently found to modify FTLD risk due to C9ORF72 expansion [75]. Interestingly, a recurrent de novo mutation in TMEM106B was recently identified in several unrelated patients of both European and Chinese descent with a form of hypomyelinating leukodystrophy [76,77].
Prosaposin, encoded by PSAP, regulates the sortilin-independent delivery of progranulin to the lysosome [78]. In addition, the PSAP locus has been shown to be an important regulator of circulating progranulin levels in humans [79]. Intriguingly, PSAP has also been implicated in hereditary sphingolipidoses including metachromatic leukodystrophy [80]. Thus, PSAP and TMEM106B collectively regulate circulating progranulin levels and can cause distinct forms of leukodystrophy. Given that TMEM106B is an established FTD risk modifier, it will be interesting to determine in the future if variants in PSAP also confer risk for FTD or act as disease modifiers.
In contrast to TMEM106B and PSAP, SORT1 (encoding sortilin) has not, to our knowledge, been implicated in leukodystrophy, but like TMEM106B and PSAP, the SORT1 locus is associated with plasma progranulin levels [81]; sortilin also affects the intracellular trafficking of progranulin [82]. Intriguingly, rare variants in SORT1 were recently reported to be enriched in FTD in multiple large, independent case–control cohorts [83].
AARS2
Compound heterozygous mutations in AARS2, which encodes mitochondrial alanyl-tRNA synthetase 2, have been identified in a leukodystrophy syndrome that includes features of cognitive decline, frontal lobe dysfunction and, in women, ovarian failure [84]. Pathogenic mutations in AARS2 appear to be a common cause of disease in patients with symptoms characteristic of ALSP but who do not harbor CSF1R mutations (see above) [85]. Recent case reports implicate pathogenic mutations in AARS2 in leukodystrophy in both Asian [86,87] and European populations [88], as well as bvFTD in Asian populations [72].
NOTCH3
NOTCH3 was originally linked to cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) in 1996 [89]. More recent findings suggest that pathogenic NOTCH3 mutations may, in rare cases, be identified in patients that present primarily with aspects of bvFTD [90], ALS-FTD [91] or AD [92].
FTD genetics in East Asian populations
C9ORF72
As mentioned above, recent genetic analyses have indicated that hexanucleotide expansion intronic to C9ORF72 is rare in Chinese FTD patients. For example, in a cohort of 82 sporadic FTD patients in which putatively pathogenic mutations were identified in MAPT and CHCHD10, no C9ORF72 expansions were identified [93]. In another recent study, a small number of FTD-associated genes were sequenced in 38 FTD patients of Han Chinese ethnicity and, although presumptively pathogenic mutations were identified in MAPT, GRN and VCP, no C9ORF72 expansions were identified [94]. Similarly, analysis of 52 Han Chinese FTD patients found mutations in MAPT and GRN but no C9ORF72 expansions [95]. On the other hand, a cohort of 128 Chinese patients with ALS and/or FTD identified a single FTD patient with a C9ORF72 expansion [96], while separate studies detected the hexanucleotide expansion at frequencies of <1% in large cohorts of Han Chinese ALS patients [97,98]. Importantly, the rare Chinese ALS patients that do harbor C9ORF72 expansions also possess C9ORF72 risk haplotypes similar to the 20-SNP haplotype identified in European expansion carriers, providing support for the idea that C9ORF72 expansion may be derived from a single European founder [96,98,99].
CHCHD10
CHCHD10 encodes a mitochondrial protein reported to localize to cristae junctions, and represents the first reported link between FTD and a mitochondria-localized protein [100]. In contrast to the relative scarcity of C9ORF72 hexanucleotide expansions in the Chinese FTD population, pathogenic mutations in CHCHD10 may be quite common, accounting for ~8% of FTD cases in the first-reported Chinese cohort [101]. On the other hand, in a cohort of 82 sporadic FTD cases from the Shanghai area, only a single missense variant was identified [93]. Beyond Chinese patients, putatively pathogenic CHCHD10 mutations have been estimated to account for ~1–3% of cases in European populations [13], suggesting that variation in this gene may account for a greater proportion of FTD cases in East Asian populations relative to European populations. However, more studies are needed for a reliable estimate of pathogenic CHCHD10 mutation prevalence in Chinese and other East Asian FTD populations.
TBK1
Although pathogenic mutations in TBK1 appear to be relatively common in European FTD cohorts (see above), they appear to be rare in the limited studies available on East Asian populations. To date only a single sporadic ALS-FTD patient of Han Chinese descent has been reported to harbor a putatively pathogenic TBK1 mutation; in this study the reported frequency of TBK1 mutations in a cohort of 207 ALS and ALS-FTD patients was 0.5% [102].
ANXA11
ANXA11, encoding the calcium- and membrane-binding protein annexin A11, was first linked to ALS in 2017 [103]. Rare variants in ANXA11 have recently been implicated in a relatively high proportion of Chinese cases of familial (~6%) and sporadic (~2%) ALS, and were detected in 1 of 12 patients with ALS-FTD in the same study [104]. Because of its recent association with ALS-FTD, it is unclear how prevalent pathogenic variation in ANXA11 is in FTD-spectrum disorders in either European or East Asian populations.
Genome-wide association studies and genetic modifiers
Although FTD is a clinically and neuropathologically heterogeneous disease, genome-wide association studies (GWAS) have implicated common variants in or near multiple genes as risk factors in FTD patients of European ancestry. The largest GWAS to date included over 3,500 patients clinically diagnosed with an FTD-spectrum disorder, and identified three SNPs in the HLA locus associated with increased risk of FTD [105]. This region includes three genes with important immune system functions: BTNL2, HLA-DRA and HLA-DRB5. A sub-analysis in this study also identified two SNPs near RAB38 and CTSC, both of which have roles in lysosomal biology, suggestive of an association with risk specifically for bvFTD. Subsequently, a separate study utilizing summary statistics from the 2014 Ferrari et al. GWAS found gene-wide aggregate common variation associated with different clinical subtypes of FTD. This study identified AD-associated genes TOMM40 and APOE as risk factors for bvFTD, while ARHGAP35 and SERPINA1 were associated with nfvPPA risk [106]. Although findings from these studies require replication in independent cohorts, the identified novel genetic risk factors are consistent with results from rare variant studies implicating immune and lysosomal dysfunction in the pathobiology of FTD.
Other recent GWAS have focused on identifying risk factors for progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD), both of which are tauopathies that fall within the FTLD spectrum. The first study included over 2,000 clinically and/or pathologically diagnosed PSP patients of European ancestry, and found SNPs associated with disease risk in STX6, EIF2AK3 and MOBP [107]. This study also identified an expression quantitative trait locus (eQTL) SNP in MAPT associated with PSP risk that influences brain expression of MAPT, and confirmed previously observed associations between the H1 allele of the H1/H2 MAPT haplotype and increased PSP risk. Two subsequent independent studies identified additional genetic risk factors for PSP by jointly analyzing the original PSP case–control cohort with additional PSP cases and controls. These studies confirmed the original findings and identified SNPs near RUNX2 [108], DUSP10 [109] and SLCO1A2 [108,109] as PSP risk factors. Finally, a small GWAS confirmed the H1 MAPT haplotype as a CBD risk factor [110]. In addition, in this study the MOBP SNP previously associated with PSP also showed suggestive association with CBD risk. These findings suggest that beyond their clinical and neuropathological similarities, PSP and CBD may share genetic risk factors in addition to the H1 MAPT haplotype.
Beyond identifying novel genetic risk factors, GWAS have also identified several modifiers of risk and clinical characteristics such as age at onset for FTD-spectrum disorders. For example, a study confirming the association between SNPs in TMEM106B and FTLD-TDP risk in both GRN mutation carrier and non-carrier sporadic cases also identified GFRA2 as a modifier of disease risk [111]. Similarly, genetic modifiers of disease course have been identified in FTLD caused by C9ORF72 repeat expansion. A recent study analyzing variants in CpG islands that affect DNA methylation status found an association between a SNP near non-coding RNA gene LOC101929163 and C6ORF10 and later disease onset in C9ORF72 repeat expansion carriers as well as non-carrier patients with FTD and/or ALS [112]. In addition, this SNP was shown to be an eQTL for both the above non-coding RNA and HLA-DRB1 in the brain [112]. Taken together, these findings highlight the importance of genetic modifiers even in the context of mutations that cause autosomal-dominant FTLD.
Genetic pleiotropy studies
Pleiotropy studies leverage how a single gene may influence multiple distinct traits, making it possible to identify shared genetic risk across multiple biologically related diseases. Using this approach to jointly analyze summary statistics from FTD, AD and PD GWAS, SNPs nominally associated with AD and PD were up to 140- and 120-fold enriched, respectively, in FTD-associated SNPs; SNPs near HLA-DRA, HLA-DRB5, MAPT (tagging the H1 haplotype), SCARB2 and SLC2A13 (near LRRK2) were identified as risk factors for FTD and PD [113]. This study also provided further evidence that variation in the APOE region confers risk for FTD and AD, consistent with the recent finding that ApoE4 exacerbates tau-mediated neurodegeneration [114]. In line with previous findings, another recent study identified novel overlapping genetic risk factors shared between PSP and CBD, including SNPs in or near CXCR4, EGFR and GLDC, and confirmed MAPT and MOBP as risk factors for both disorders [115]. Interestingly, CXCR4 was also identified as a shared risk factor for PSP and PD in a separate study [116]. The widely perceived shared etiology between FTD and ALS was further supported by a recent study of ALS and several FTD-spectrum disorders that identified 29 shared risk loci, 22 of which are novel risk loci for ALS [117]. This study found that SNPs nominally associated with FTLD-TDP were up to 300-fold enriched in ALS-associated SNPs. In addition, a SNP tagging the MAPT H1 haplotype was identified as a risk factor for ALS; this study also provided evidence that BNIP1, which encodes a pro-apoptotic protein with an important role in mitophagy, represents an ALS risk gene. Beyond novel genetic risk factors shared between neurodegenerative disorders, pleiotropy has also been used to characterize the overlapping genetic architecture of FTD-spectrum and immune-mediated disorders. This approach identified up to 270-fold enrichment in FTD-associated SNPs for SNPs nominally associated with rheumatoid arthritis. Specifically, variants in or near AGPAT1, BTNL2, GPSM3, HLA-DQA2, HLA-DQB1, HLA-DRA, PAQR8, TRIM15 and TRIM26 in the HLA region; as well as those in or near the MAPT H1 haplotype, TNS3, TWISTNB, CR590356, SLC2A13 and DCC were associated with risk for FTD and several immune system disorders [118]. Although limited to patient populations of European ancestry, these studies suggest that FTD and other neurodegenerative disorders share considerable genetic risk with disorders mediated by immune dysfunction.
Summary and conclusions
Recent research into the human genetics of FTD has highlighted the central roles of genes involved in autophagy and endolysosomal homeostasis. Beyond this, work in mouse models suggests that cellular regulators of autophagy such as TBK1 and parkin are also crucial regulators of the inflammatory response in vivo. These results dovetail nicely with other recent findings in FTD genetics indicating that a substantial amount of genetic risk for FTD derives from genes encoding regulators of the immune response.
In this review we highlighted accumulating evidence that several genes implicated in inherited leukodystrophies may also confer risk for FTD. Our gene expression analyses (Figure 1) indicate that many of these joint leukodystrophy/FTD genes show dysregulated expression in the brain in GRN+ FTD, and possibly in FTD more generally. In addition, our gene interaction network (Figure 2) suggests that joint leukodystrophy/FTD genes exist in a network of genes regulating immune processes and endolysosomal homeostasis. Importantly, some genes in the latter class are established causes of Mendelian neurodegenerative LSDs. Collectively, the findings from the last five years of human genetics point to the intersection between intracellular protein clearance (autophagy and lysosomal degradation) and neuroinflammation, as well as shared genetic risk not only between FTD and immune-mediated disorders but also between FTD and inherited leukodystrophies.
Finally, we have emphasized recent genetic studies of Chinese patients with FTD and/or ALS, which have indicated that pathogenic C9ORF72 expansion appears to be less common in this population, while potentially pathogenic variation in CHCHD10 may be more common than in European populations. These findings highlight the limitations of most FTD genetics studies conducted to date, which have primarily focused on populations of European ancestry. It will therefore be crucial for future studies to focus on non-European FTD cohorts to increase our understanding of the genetic architecture of FTD in diverse populations. In addition, it will be essential to clarify the link between intracellular degradation and the inflammatory response, to understand why these pathways confer risk for FTD and other forms of neurodegeneration, and to identify novel targets and pathways for therapeutic intervention.
Acknowledgements
We thank Lin Yuan (UCSF) for her helpful reading of the manuscript. Primary research support in the Yokoyama lab is provided by the Rainwater Charitable Foundation, the Bluefield Project to Cure FTD, the Association for Frontotemporal Degeneration Susan Marcus Memorial Fund Clinical Research Grant, the Larry L. Hillblom Foundation (2016-A-005-SUP), the National Institute on Aging (K01 AG049152), and the John Douglas French Alzheimer’s Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest
Daniel W. Sirkis, Ethan G. Geier, Luke W. Bonham, Celeste M. Karch and Jennifer S. Yokoyama each declare no potential conflicts of interest.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
• Papers of particular interest
- 1.Goldman JS, Farmer JM, Wood EM, Johnson JK, Boxer A, Neuhaus J, et al. Comparison of family histories in FTLD subtypes and related tauopathies. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2005;65:1817–9. [DOI] [PubMed] [Google Scholar]
- 2.Rohrer JD, Guerreiro R, Vandrovcova J, Uphill J, Reiman D, Beck J, et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2009;73:1451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seelaar H, Kamphorst W, Rosso SM, Azmani A, Masdjedi R, de Koning I, et al. Distinct genetic forms of frontotemporal dementia. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2008;71:1220–6. [DOI] [PubMed] [Google Scholar]
- 4.Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016;12:733–48. [DOI] [PubMed] [Google Scholar]
- 5.Mackenzie IRA, Neumann M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem. Wiley/Blackwell (10.1111); 2016;138 Suppl 1:54–70. [DOI] [PubMed] [Google Scholar]
- 6.Van Mossevelde S, Engelborghs S, van der Zee J, Van Broeckhoven C. Genotype-phenotype links in frontotemporal lobar degeneration. Nat Rev Neurol. Nature Publishing Group; 2018;14:363–78. [DOI] [PubMed] [Google Scholar]
- 7.Pottier C, Ravenscroft TA, Sanchez-Contreras M, Rademakers R. Genetics of FTLD: overview and what else we can expect from genetic studies. J Neurochem. Wiley/Blackwell (10.1111); 2016;138 Suppl 1:32–53. [DOI] [PubMed] [Google Scholar]
- 8.Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol Wiley-Blackwell; 1998;43:815–25. [DOI] [PubMed] [Google Scholar]
- 9.Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc Natl Acad Sci USA. 1998;95:13103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. Nature Publishing Group; 1998;393:702–5. [DOI] [PubMed] [Google Scholar]
- 11.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 1998;95:7737–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD. Exp Cell Res. 2014;325:27–37. [DOI] [PubMed] [Google Scholar]
- 13.Deleon J, Miller BL. Frontotemporal dementia. Handb Clin Neurol. 2018;148:409–30. [DOI] [PubMed] [Google Scholar]
- 14.Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domoto-Reilly K, Davis MY, Keene CD, Bird TD. Unusually long duration and delayed penetrance in a family with FTD and mutation in MAPT (V337M) Tsuang DW, Bird TD, editors. Am. J. Med. Genet. B Neuropsychiatr. Genet Wiley-Blackwell; 2017;174:70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Chen K, Li X, Xiao S. Rapidly Progressive Frontotemporal Dementia Associated with MAPT Mutation G389R. J Alzheimers Dis. IOS Press; 2017;55:777–85. [DOI] [PubMed] [Google Scholar]
- 17.Ygland E, van Westen D, Englund E, Rademakers R, Wszolek ZK, Nilsson K, et al. Slowly progressive dementia caused by MAPT R406W mutations: longitudinal report on a new kindred and systematic review. Alzheimers Res Ther. BioMed Central; 2018;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carney RM, Kohli MA, Kunkle BW, Naj AC, Gilbert JR, Zuchner S, et al. Parkinsonism and distinct dementia patterns in a family with the MAPT R406W mutation. Alzheimers Dement. 2014;10:360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behnam M, Ghorbani F, Shin J-H, Kim D-S, Jang H, Nouri N, et al. Homozygous MAPT R406W mutation causing FTDP phenotype: A unique instance of a unique mutation. Gene. 2015;570:150–2. [DOI] [PubMed] [Google Scholar]
- 20.Ng ASL, Sias AC, Pressman PS, Fong JC, Karydas AM, Zanto TP, et al. Young-onset frontotemporal dementia in a homozygous tau R406W mutation carrier. Ann Clin Transl Neurol. Wiley-Blackwell; 2015;2:1124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. [DOI] [PubMed] [Google Scholar]
- 22.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. [DOI] [PubMed] [Google Scholar]
- 23.Wauters E, Van Mossevelde S, van der Zee J, Cruts M, Van Broeckhoven C. Modifiers of GRN-Associated Frontotemporal Lobar Degeneration. Trends in molecular medicine. 2017;23:962–79. [DOI] [PubMed] [Google Scholar]
- 24.Cruchaga C, Graff C, Chiang H-H, Wang J, Hinrichs AL, Spiegel N, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. American Medical Association; 2011;68:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JSK, Shears S, et al. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann. Neurol. Wiley-Blackwell; 2006;60:314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Zee J, Mariën P, Crols R, Van Mossevelde S, Dillen L, Perrone F, et al. Mutated CTSF in adult-onset neuronal ceroid lipofuscinosis and FTD. Neurol Genet. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2016;2:e102.• Provides evidence that additional genes involved in neuronal ceroid lipofuscinosis may confer risk for FTD.
- 28.Bras J, Djaldetti R, Alves AM, Mead S, Darwent L, Lleo A, et al. Exome sequencing in a consanguineous family clinically diagnosed with early-onset Alzheimer’s disease identifies a homozygous CTSF mutation. Neurobiol. Aging 2016;46:236.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geier EG, Bourdenx M, Storm NJ, Cochran JN, Sirkis DW, Hwang J-H, et al. Rare variants in the neuronal ceroid lipofuscinosis gene MFSD8 are candidate risk factors for frontotemporal dementia. Acta Neuropathol. Springer Berlin Heidelberg; 2018;526:1–18.• Provides evidence that additional genes involved in neuronal ceroid lipofuscinosis may confer risk for FTD.
- 30.Watts GDJ, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. Nature Publishing Group; 2004;36:377–81. [DOI] [PubMed] [Google Scholar]
- 31.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8. [DOI] [PubMed] [Google Scholar]
- 32.Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann. Neurol Wiley-Blackwell; 2009;65:470–3. [DOI] [PubMed] [Google Scholar]
- 33.Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2010;74:366–71. [DOI] [PubMed] [Google Scholar]
- 34.Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harms M, Benitez BA, Cairns N, Cooper B, Cooper P, Mayo K, et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. American Medical Association; 2013;70:736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao F-B, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. EMBO Press; 2017;36:2931–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubino E, Rainero I, Chiò A, Rogaeva E, Galimberti D, Fenoglio P, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2012;79:1556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Zee J, Van Langenhove T, Kovacs GG, Dillen L, Deschamps W, Engelborghs S, et al. Rare mutations in SQSTM1 modify susceptibility to frontotemporal lobar degeneration. Acta Neuropathol. Springer Berlin Heidelberg; 2014;128:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haack TB, Ignatius E, Calvo-Garrido J, Iuso A, Isohanni P, Maffezzini C, et al. Absence of the Autophagy Adaptor SQSTM1/p62 Causes Childhood-Onset Neurodegeneration with Ataxia, Dystonia, and Gaze Palsy. Am J Hum Genet. 2016;99:735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito H, Nakamura M, Komure O, Ayaki T, Wate R, Maruyama H, et al. Clinicopathologic study on an ALS family with a heterozygous E478G optineurin mutation. Acta Neuropathol. Springer-Verlag; 2011;122:223–9. [DOI] [PubMed] [Google Scholar]
- 42.Czell D, Andersen PM, Neuwirth C, Morita M, Weber M. Progressive aphasia as the presenting symptom in a patient with amyotrophic lateral sclerosis with a novel mutation in the OPTN gene. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14:138–40. [DOI] [PubMed] [Google Scholar]
- 43.Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. Springer Berlin Heidelberg; 2015;130:77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. Nature Publishing Group; 2015;18:631–6. [DOI] [PubMed] [Google Scholar]
- 45.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Philtjens S, Heeman B, et al. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2015;85:2116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D, Jin T, Zhu H, Chen H, Ofengeim D, Zou C, et al. TBK1 Suppresses RIPK1-Driven Apoptosis and Inflammation during Development and in Aging. Cell. 2018;174:1477–1491.e19.• Provides evidence that a regulator of autophagy influences the inflammatory response.
- 47.Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. Nature Publishing Group; 2018;561:258–62.• Provides evidence that regulators of mitophagy influence the inflammatory response.
- 48.Zimmermann M, Wilke C, Schulte C, Hoffmann J, Klopfer J, Reimold M, et al. Biallelic Parkin (PARK2) mutations can cause a bvFTD phenotype without clinically relevant parkinsonism. Parkinsonism Relat. Disord. 2018. [DOI] [PubMed] [Google Scholar]
- 49.Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, et al. TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron. 2017;95:808–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Spek RA, van Rheenen W, Pulit SL, Kenna KP, Ticozzi N, Kooyman M, et al. Reconsidering the causality of TIA1 mutations in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baradaran-Heravi Y, Dillen L, Nguyen HP, Van Mossevelde S, Baets J, De Jonghe P, et al. No supportive evidence for TIA1 gene mutations in a European cohort of ALS-FTD spectrum patients. Neurobiol. Aging 2018;69:293.e9–293.e11. [DOI] [PubMed] [Google Scholar]
- 52.Heck MV, Azizov M, Stehning T, Walter M, Kedersha N, Auburger G. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics. Springer Berlin Heidelberg; 2014;15:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams KL, Topp S, Yang S, Smith B, Fifita JA, Warraich ST, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. Nature Publishing Group; 2016;7:11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen-Plotkin AS, Geser F, Plotkin JB, Clark CM, Kwong LK, Yuan W, et al. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17:1349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. Nature Publishing Group; 2015;47:569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paloneva J, Kestilä M, Wu J, Salminen A, Böhling T, Ruotsalainen V, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet. 2000;25:357–61. [DOI] [PubMed] [Google Scholar]
- 58.Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chouery E, Delague V, Bergougnoux A, Koussa S, Serre J-L, Mégarbané A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum. Mutat 2008;29:E194–204. [DOI] [PubMed] [Google Scholar]
- 60.Guerreiro R, Bilgic B, Guven G, Bras J, Rohrer J, Lohmann E, et al. Novel compound heterozygous mutation in TREM2 found in a Turkish frontotemporal dementia-like family. Neurobiol. Aging 2013;34:2890.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerreiro RJ, Lohmann E, Brás JM, Gibbs JR, Rohrer JD, Gurunlian N, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013;70:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Ber I, De Septenville A, Guerreiro R, Bras J, Camuzat A, Caroppo P, et al. Homozygous TREM2 mutation in a family with atypical frontotemporal dementia. Neurobiol. Aging 2014;35:2419.e23–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng ASL, Tan YJ, Yi Z, Tandiono M, Chew E, Dominguez J, et al. Targeted exome sequencing reveals homozygous TREM2 R47C mutation presenting with behavioral variant frontotemporal dementia without bone involvement. Neurobiol. Aging 2018;68:160.e15–9. [DOI] [PubMed] [Google Scholar]
- 64.Peplonska B, Berdynski M, Mandecka M, Barczak A, Kuzma-Kozakiewicz M, Barcikowska M, et al. TREM2 variants in neurodegenerative disorders in the Polish population. Homozygosity and compound heterozygosity in FTD patients. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19:407–12. [DOI] [PubMed] [Google Scholar]
- 65.Redaelli V, Salsano E, Colleoni L, Corbetta P, Tringali G, Del Sole A, et al. Frontotemporal Dementia and Chorea Associated with a Compound Heterozygous TREM2 Mutation. J Alzheimers Dis. IOS Press; 2018;63:195–201. [DOI] [PubMed] [Google Scholar]
- 66.Su W-H, Shi Z-H, Liu S-L, Wang X-D, Liu S, Ji Y. The rs75932628 and rs2234253 polymorphisms of the TREM2 gene were associated with susceptibility to frontotemporal lobar degeneration in Caucasian populations. Ann. Hum. Genet Wiley/Blackwell (10.1111); 2018;82:177–85. [DOI] [PubMed] [Google Scholar]
- 67.Carmona S, Zahs K, Wu E, Dakin K, Bras J, Guerreiro R. The role of TREM2 in Alzheimer’s disease and other neurodegenerative disorders. Lancet Neurol. 2018;17:721–30. [DOI] [PubMed] [Google Scholar]
- 68.Benitez BA, Cruchaga C, United States–Spain Parkinson’s Disease Research Group. TREM2 and neurodegenerative disease. N. Engl. J. Med 2013;369:1567–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. Nature Publishing Group; 2011;44:200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gore E, Manley A, Dees D, Appleby BS, Lerner AJ. A young-onset frontal dementia with dramatic calcifications due to a novel CSF1R mutation. Neurocase. 2016;22:257–62. [DOI] [PubMed] [Google Scholar]
- 71.Kawakami I, Iseki E, Kasanuki K, Minegishi M, Sato K, Hino H, et al. A family with hereditary diffuse leukoencephalopathy with spheroids caused by a novel c.2442+2T>C mutation in the CSF1R gene. J. Neurol. Sci 2016;367:349–55. [DOI] [PubMed] [Google Scholar]
- 72.Kim E-J, Kim Y-E, Jang J-H, Cho E-H, Na DL, Seo SW, et al. Analysis of frontotemporal dementia, amyotrophic lateral sclerosis, and other dementia-related genes in 107 Korean patients with frontotemporal dementia. Neurobiol. Aging 2018. [DOI] [PubMed] [Google Scholar]
- 73.Van Deerlin VM, Sleiman PMA, Martinez-Lage M, Chen-Plotkin A, Wang L-S, Graff-Radford NR, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. Nature Publishing Group; 2010;42:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2011;76:467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Blitterswijk M, Mullen B, Nicholson AM, Bieniek KF, Heckman MG, Baker MC, et al. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simons C, Dyment D, Bent SJ, Crawford J, D’Hooghe M, Kohlschütter A, et al. A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain. 2017;140:3105–11.• An important FTD risk modifier is implicated in leukodystrophy.
- 77.Yan H, Kubisiak T, Ji H, Xiao J, Wang J, Burmeister M. The recurrent mutation in TMEM106B also causes hypomyelinating leukodystrophy in China and is a CpG hotspot. Brain. 2018;141:e36.• An important FTD risk modifier is implicated in leukodystrophy.
- 78.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. Rockefeller University Press; 2015;210:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholson AM, Finch NA, Almeida M, Perkerson RB, van Blitterswijk M, Wojtas A, et al. Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun. Nature Publishing Group; 2016;7:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Motta M, Tatti M, Furlan F, Celato A, Di Fruscio G, Polo G, et al. Clinical, biochemical and molecular characterization of prosaposin deficiency Clin. Genet 8 ed. Wiley/Blackwell (10.1111); 2016;90:220–9. [DOI] [PubMed] [Google Scholar]
- 81.Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, et al. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87:890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu F, Padukkavidana T, Vægter CB, Brady OA, Zheng Y, Mackenzie IR, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Philtjens S, Van Mossevelde S, van der Zee J, Wauters E, Dillen L, Vandenbulcke M, et al. Rare nonsynonymous variants in SORT1 are associated with increased risk for frontotemporal dementia. Neurobiol. Aging 2018;66:181.e3–181.e10. [DOI] [PubMed] [Google Scholar]
- 84.Dallabona C, Diodato D, Kevelam SH, Haack TB, Wong L-J, Salomons GS, et al. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2014;82:2063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lynch DS, Zhang WJ, Lakshmanan R, Kinsella JA, Uzun GA, Karbay M, et al. Analysis of Mutations in AARS2 in a Series of CSF1R-Negative Patients With Adult-Onset Leukoencephalopathy With Axonal Spheroids and Pigmented Glia. JAMA Neurol. American Medical Association; 2016;73:1433–9. [DOI] [PubMed] [Google Scholar]
- 86.Lee J-M, Yang H-J, Kwon J-H, Kim W-J, Kim S-Y, Lee E-M, et al. Two Korean siblings with recently described ovarioleukodystrophy related to AARS2 mutations. Eur. J. Neurol Wiley/Blackwell (10.1111); 2017;24:e21–2. [DOI] [PubMed] [Google Scholar]
- 87.Hamatani M, Jingami N, Tsurusaki Y, Shimada S, Shimojima K, Asada-Utsugi M, et al. The first Japanese case of leukodystrophy with ovarian failure arising from novel compound heterozygous AARS2 mutations. J. Hum. Genet Nature Publishing Group; 2016;61:899–902. [DOI] [PubMed] [Google Scholar]
- 88.Taglia I, Di Donato I, Bianchi S, Cerase A, Monti L, Marconi R, et al. AARS2-related ovarioleukodystrophy: Clinical and neuroimaging features of three new cases. Acta Neurol. Scand Wiley/Blackwell (10.1111); 2018;42:S27. [DOI] [PubMed] [Google Scholar]
- 89.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. Nature Publishing Group; 1996;383:707–10. [DOI] [PubMed] [Google Scholar]
- 90.Alexander SK, Brown JM, Graham A, Nestor PJ. CADASIL presenting with a behavioural variant frontotemporal dementia phenotype. J Clin Neurosci. 2014;21:165–7. [DOI] [PubMed] [Google Scholar]
- 91.Kim H-J, Kim HY, Paek WK, Park A, Young Park M, Ki CS, et al. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration in association with CADASIL. Neurologist. 2012;18:92–5. [DOI] [PubMed] [Google Scholar]
- 92.Guerreiro RJ, Lohmann E, Kinsella E, Brás JM, Luu N, Gurunlian N, et al. Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer’s disease. Neurobiol. Aging 2012;33:1008.e17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Che X-Q, Zhao Q-H, Huang Y, Li X, Ren R-J, Chen S-D, et al. Genetic Features of MAPT, GRN, C9orf72 and CHCHD10 Gene Mutations in Chinese Patients with Frontotemporal Dementia. Curr Alzheimer Res. 2017;14:1102–8. [DOI] [PubMed] [Google Scholar]
- 94.Shi Z, Liu S, Xiang L, Wang Y, Liu M, Liu S, et al. Frontotemporal dementia-related gene mutations in clinical dementia patients from a Chinese population. J. Hum. Genet Nature Publishing Group; 2016;61:1003–8. [DOI] [PubMed] [Google Scholar]
- 95.Tang M, Gu X, Wei J, Jiao B, Zhou L, Zhou Y, et al. Analyses MAPT, GRN, and C9orf72 mutations in Chinese patients with frontotemporal dementia. Neurobiol. Aging 2016;46:235.e11–5. [DOI] [PubMed] [Google Scholar]
- 96.Jiao B, Tang B, Liu X, Yan X, Zhou L, Yang Y, et al. Identification of C9orf72 repeat expansions in patients with amyotrophic lateral sclerosis and frontotemporal dementia in mainland China. Neurobiol. Aging 2014;35:936.e19–22. [DOI] [PubMed] [Google Scholar]
- 97.Liu F, Liu Q, Lu CX, Cui B, Guo XN, Wang RR, et al. Identification of a novel loss-of-function C9orf72 splice site mutation in a patient with amyotrophic lateral sclerosis. Neurobiol. Aging 2016;47:219.e1–219.e5. [DOI] [PubMed] [Google Scholar]
- 98.Chen Y, Lin Z, Chen X, Cao B, Wei Q, Ou R, et al. Large C9orf72 repeat expansions are seen in Chinese patients with sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 2016;38:217.e15–217.e22. [DOI] [PubMed] [Google Scholar]
- 99.Mok K, Traynor BJ, Schymick J, Tienari PJ, Laaksovirta H, Peuralinna T, et al. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol. Aging 2012;33:209.e3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiao B, Xiao T, Hou L, Gu X, Zhou Y, Zhou L, et al. High prevalence of CHCHD10 mutation in patients with frontotemporal dementia from China. Brain. 2016;139:e21.• Indicates that pathogenic CHCHD10 mutations may be a common cause of FTD in Chinese populations.
- 102.Tsai P-C, Liu Y-C, Lin K-P, Liu Y-T, Liao Y-C, Hsiao C-T, et al. Mutational analysis of TBK1 in Taiwanese patients with amyotrophic lateral sclerosis. Neurobiol. Aging 2016;40:191.e11–6. [DOI] [PubMed] [Google Scholar]
- 103.Smith BN, Topp SD, Fallini C, Shibata H, Chen H-J, Troakes C, et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci Transl Med. American Association for the Advancement of Science; 2017;9:eaad9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang K, Liu Q, Liu K, Shen D, Tai H, Shu S, et al. ANXA11 mutations prevail in Chinese ALS patients with and without cognitive dementia. Neurol Genet. Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology; 2018;4:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JBJ, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mishra A, Ferrari R, Heutink P, Hardy J, Pijnenburg Y, Posthuma D, et al. Gene-based association studies report genetic links for clinical subtypes of frontotemporal dementia. Brain. 2017;140:1437–46. [DOI] [PubMed] [Google Scholar]
- 107.Höglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang L-S, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. Nature Publishing Group; 2011;43:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen JA, Chen Z, Won H, Huang AY, Lowe JK, Wojta K, et al. Joint genome-wide association study of progressive supranuclear palsy identifies novel susceptibility loci and genetic correlation to neurodegenerative diseases. Mol Neurodegener. BioMed Central; 2018;13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sanchez-Contreras MY, Kouri N, Cook CN, Serie DJ, Heckman MG, Finch NA, et al. Replication of progressive supranuclear palsy genome-wide association study identifies SLCO1A2 and DUSP10 as new susceptibility loci. Mol Neurodegener. BioMed Central; 2018;13:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. Nature Publishing Group; 2015;6:7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pottier C, Zhou X, Perkerson RB, Baker M, Jenkins GD, Serie DJ, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 2018;17:548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang M, Ferrari R, Tartaglia MC, Keith J, Surace EI, Wolf U, et al. A C6orf10/LOC101929163 locus is associated with age of onset in C9orf72 carriers. Brain. 2018;141:2895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ferrari R, Wang Y, Vandrovcova J, Guelfi S, Witeolar A, Karch CM, et al. Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer’s and Parkinson’s diseases. J. Neurol. Neurosurg. Psychiatr 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. Nature Publishing Group; 2017;549:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yokoyama JS, Karch CM, Fan CC, Bonham LW, Kouri N, Ross OA, et al. Shared genetic risk between corticobasal degeneration, progressive supranuclear palsy, and frontotemporal dementia. Acta Neuropathol. Springer Berlin Heidelberg; 2017;133:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bonham LW, Karch CM, Fan CC, Tan C, Geier EG, Wang Y, et al. CXCR4 involvement in neurodegenerative diseases. Transl Psychiatry. Nature Publishing Group; 2018;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Karch CM, Wen N, Fan CC, Yokoyama JS, Kouri N, Ross OA, et al. Selective Genetic Overlap Between Amyotrophic Lateral Sclerosis and Diseases of the Frontotemporal Dementia Spectrum. JAMA Neurol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Broce I, Karch CM, Wen N, Fan CC, Wang Y, Hong Tan C, et al. Immune-related genetic enrichment in frontotemporal dementia: An analysis of genome-wide association studies. PLoS Med. Public Library of Science; 2018;15:e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]