Abstract
Arginine methylation plays crucial roles in many cellular functions including signal transduction, RNA transcription, and regulation of gene expression. Protein arginine methyl-transferase 8 (PRMT8), a unique brain-specific protein, is localized to the plasma membrane. However, the detailed molecular mechanisms underlying PRMT8 plasma membrane targeting remain unclear. Here, we demonstrate that the N-terminal 20 amino acids of PRMT8 are sufficient for plasma membrane localization and that oligomerization enhances membrane localization. The basic amino acids, combined with myristoylation within the N-terminal 20 amino acids of PRMT8, are critical for plasma membrane targeting. We also found that substituting Gly-2 with Ala [PRMT8(G2A)] or Cys-9 with Ser [PRMT8(C9S)] induces the formation of punctate structures in the cytosol or patch-like plasma membrane localization, respectively. Impairment of PRMT8 oligomerization/dimerization by C-terminal deletion induces PRMT8 mis-localization to the mitochondria, prevents the formation of punctate structures by PRMT8(G2A), and inhibits PRMT8(C9S) patch-like plasma membrane localization. Overall, these results suggest that oligomerization/dimerization plays several roles in inducing the efficient and specific plasma membrane localization of PRMT8.
Keywords: Dimerization, Myristoylation, Oligomerization, Plasma membrane, PRMT8
INTRODUCTION
Post-translational modifications of proteins are important for their functional diversity. Methylation of arginine residues within proteins is associated with several important cellular functions such as signal transduction, DNA repair, RNA transcription, and regulation of gene expression (1–4). In human cells, protein arginine methyl transferases (PRMTs) are classified as type I, II, or III enzymes (1). Types I and II catalyze the formation of monomethylarginine as an intermediate. The type I enzymes PRMT1, PRMT2, PRMT3, CARM1/PRMT4, PRMT6, and PRMT8 promote the formation of asymmetric dimethylarginine, whereas the type II enzyme PRMT5 facilitates symmetric dimethylarginine formation and the type III enzyme PRMT7 only monomethylates guanidinium groups (5).
Among the PRMT proteins, the type I PRMT8 is a unique brain-specific member and is the only PRMT that localizes to the plasma membrane (6, 7). This enzyme has a unique N-terminal extended region that is involved in plasma membrane localization via myristoylation, and is suggested to regulate PRMT8 enzymatic activity (7, 8). It is a multifunctional protein with arginine methyl transferase and phospholipase D activities (9), localizes to both presynaptic and postsynaptic sites, and plays multiple roles in the brain, including in Purkinje cell morphology, perineuronal net formation in the visual cortex, fear learning in the hippocampus, and neuroprotection against age-related increases in cellular stress (9–12). Type I PRMTs may undergo oligomerization/dimerization through an interaction between the dimerization arm projecting off the β-barrel and the Rossman fold of another subunit (13). Similarly, PRMT8 can form homo- or heterodimers with PRMT1 but not with PRMT3, PRMT4, or PRMT6. Although oligomerization plays key roles in PRMT8 plasma membrane targeting and enzyme activity (7, 14), the detailed molecular mechanisms involved in PRMT8 plasma membrane targeting remain unclear.
In this study, we investigated the detailed molecular mechanisms of PRMT8 plasma membrane targeting in HEK293T cells and neurons. We found that the N-terminal 20 amino acids of PRMT8 are sufficient for targeting this protein to the plasma membrane, and the combination of myristoylation and N-terminal basic amino acids is important for PRMT8 plasma membrane localization, both in HEK293T cells and in neurons. Furthermore, PRMT8 oligomerization/dimerization can enhance its plasma membrane localization.
RESULTS AND DISCUSSION
Mapping the minimal PRMT8 membrane-targeting domains
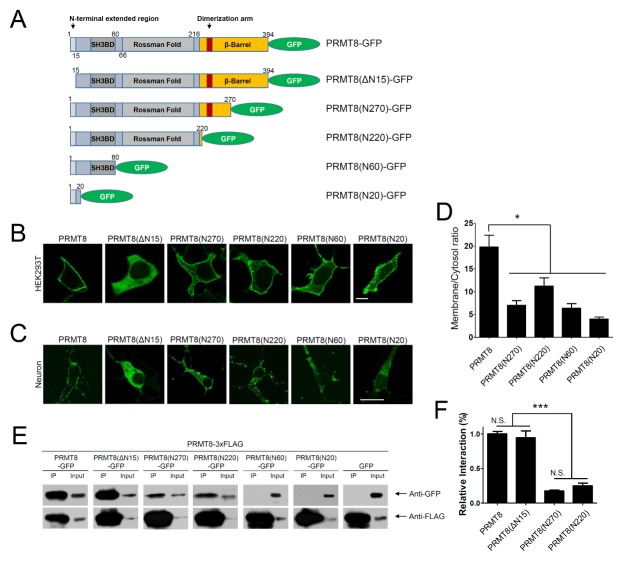
The PRMT8 enzyme is a unique PRMT that is expressed in the brain and localizes specifically to the plasma membrane for proper functioning (7). To understand the cellular mechanisms of PRMT8 targeting to the plasma membrane, we generated a GFP-fused, full-length PRMT8 (PRMT8-GFP) (Fig. 1A) and expressed this recombinant protein in HEK293T cells and in cultured cortical neurons (Fig. 1B and C). As shown in Fig. 1B and 1C, PRMT8-GFP localized to the plasma membrane of HEK293T cells and cultured cortical neurons.
Fig. 1.
Plasma membrane targeting of PRMT8-GFP. (A) Schematic diagram of PRMT8 wild-type (PRMT8-GFP) and serial mutants. (B, C) Cellular localization of PRMT8 serial deletion mutants. PRMT8-GFP, PRMT8(ΔN15)-GFP, PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, and PRMT8(N20)-GFP localized to the plasma membrane in HEK293T cells (B) and in cultured cortical neurons (C). Scale bar, 20 μm. SH3BD, SH3-binding domain. (D) Quantification of the ratio between the fluorescent intensity at the plasma membrane and in the cytosol of cells expressing the PRMT8 constructs in HEK293T cells. *P < 0.001, one-way ANOVA; F = 12.96, Tukey’s post-hoc test. Values are presented as means ± SEM. Scale bar, 20 μm. (E) Oligomerization/dimerization of PRMT8. PRMT8-3×FLAG was co-expressed with PRMT8-GFP, PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, or GFP in HEK293T cells. The data shown represent the results from three independent experiments. 1% of total lysate was used as input. (F) Quantification of the relative interaction of PRMT8-3xFLAG to PRMT8, PRMT8(ΔN15), PRMT8(N270), and PRMT8(N220)-GFP. ***P < 0.0001, one-way ANOVA; F = 69.36, Tukey’s post-hoc test. Values are presented as means ± SEM. N. S., not significant.
To examine whether the unique N-terminal extended region of PRMT8 is involved in plasma membrane targeting, we deleted the N-terminal extended region from the full-length PRMT8 to generate PRMT8(ΔN15)-GFP (Fig. 1A) and expressed this mutant in HEK293T cells and in cultured cortical neurons. As shown in Fig. 1B and 1C, PRMT8(ΔN15)-GFP localized to the cytosol in HEK293T cells and cultured cortical neurons, indicating that the N-terminal extended region is involved in plasma membrane targeting of PRMT8.
Next, we generated four serial PRMT8 C-terminal deletion mutants: PRMT8(N270)-GFP, carrying the dimerization arm but only a partial β-barrel domain; PRMT8(N220)-GFP that excludes the β-barrel domain; PRMT8(N60)-GFP containing the N-terminal extended region and an SH3-binding domain (SH3BD); and PRMT(N20)-GFP containing the N-terminal extended region (Fig. 1A). These constructs were then expressed in HEK293T cells (Fig. 1B) and in cultured cortical neurons (Fig. 1C). As shown in Fig. 1B, PRMT8(N270)-GFP, PRMT8 (N220)-GFP, PRMT8(N60)-GFP, and PRMT(N20)-GFP showed some localization to the plasma membrane of HEK293T cells. However, unlike PRMT8-GFP, the PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, and PRMT8(N20)-GFP signals also exhibited significant cytosolic and intracellular localization in HEK293T cells. In cultured cortical neurons, PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, and PRMT8(N20)-GFP similarly localized to the plasma membrane (Fig. 1C). We then quantified the plasma membrane localization efficiency of PRMT8 and its deletion mutants by measuring the ratio of plasma membrane versus cytoplasmic fluorescent intensity in HEK293T cells using ImageJ. As shown in Fig. 1D, the PRMT8-GFP membrane-cytoplasmic ratio was significantly higher than that of PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, or PRMT8(N20)-GFP. These results suggest that the full-length PRMT8 is more efficiently localized to the plasma membrane than the other deletion mutants.
It has previously been shown that PRMT8 can oligomerize/dimerize (7, 14). To further confirm this, we generated a PRMT8-3×FLAG construct and co-expressed it with the PRMT8-GFP serial deletion mutants, as shown in Fig. 1A, and performed FLAG co-immunoprecipitation (co-IP). We then quantified the relative interaction of PRMT8-3×FLAG to PRMT8, PRMT8(ΔN15), PRMT8(N270), and PRMT8(N220)-GFP by dividing the signal intensities obtained from the immunoprecipitated protein by the partial intensities in the whole cell lysate using ImageJ (Fig. 1F). As shown in Fig. 1E, we found that PRMT8-3×FLAG could bind PRMT8-GFP and PRMT8 (ΔN15)-GFP, but not PRMT8(N60)-GFP or PRMT8(N20)-GFP. As also shown in Fig. 1F, PRMT8(N270)-GFP and PRMT8 (N220)-GFP showed a weaker association with PRMT8-3×FLAG than PRMT8-GFP or PRMT8(ΔN15)-GFP (Fig. 1F). This suggests that PRMT8(N270)-GFP and PRMT8(N220)-GFP, but not PRMT8(N60)-GFP or PRMT8(N20)-GFP, may contain the Rossman fold that could facilitate binding to the β-barrel domain of PRMT8-3×FLAG, and may explain why PRMT8(N270)-GFP and PRMT8(N220)-GFP could bind relatively weakly to PRMT8-3×FLAG. Taken together, our results suggest that full-length PRMT8 is required for oligomerization/dimerization.
In addition, our results also suggest that the N-terminal 20 amino acids of PRMT8 can still localize to the plasma membrane and that full-length PRMT8, containing both the β-barrel and the Rossman fold, is more efficiently localized to the plasma membrane than the other deletion mutants (Fig. 1D). It is therefore plausible that oligomerization/dimerization may enhance PRMT8 plasma membrane localization. Similar to our results, it has been reported that oligomerization is necessary for proper PRMT8 plasma membrane localization (14) since oligomer-disrupted PRMT8 mutants were shown to be localized diffusely in the cytosol (14). However, our data showed that the N-terminal 20 amino acids of PRMT8 are sufficient for plasma membrane targeting, which can be further enhanced by PRMT8 oligomerization (Fig. 1). Similarly, the Aplysia Sec7 protein (ApSec7) was shown to localize to the plasma membrane through its PH domains, and ApSec7 plasma membrane targeting was enhanced by homodimerization via its coiled-coil (CC) domains (15). In addition, oligomerization of ApPDE4 short- and long-forms were reported to enhance its plasma membrane targeting via interaction of two upstream conserved regions (UCR1 and UCR2) (16). Collectively, our results suggest that PRMT8 localizes to the plasma membrane via its unique N-terminal region and that membrane localization is enhanced by its oligomerization that occurs through interactions between the dimerization arm projecting off the β-barrel and the Rossman fold of another subunit.
Basic amino acids within the N-terminus of PRMT8 are involved in plasma membrane localization
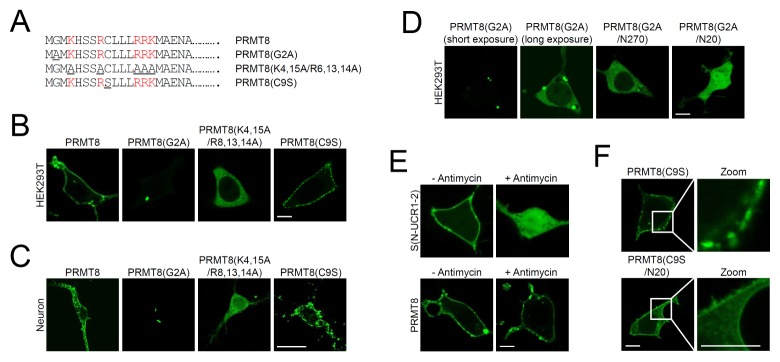
We undertook a detailed examination of the role of the PRMT8 N-terminal extended region in membrane targeting. It has been reported that myristoylation of PRMT8 is important for its plasma membrane localization (7). To verify the role of myristoylation in PRMT8 cellular localization, we changed Gly-2 to Ala in PRMT8 to obtain PRMT8(G2A)-GFP (Fig. 2A). As shown in Fig. 2B and 2C, unlike PRMT8-GFP that localized to the plasma membrane, PRMT8(G2A)-GFP expression showed only weak and highly punctate expression in the cytosol of HEK293T cells and cortical neurons. To examine whether the formation of these punctate structures is mediated by oligomerization/dimerization, we generated the PRMT8(G2A/N270)-GFP and PRMT8(G2A/N20)-GFP constructs. As shown in Fig. 2D, PRMT8(G2A)-GFP generated highly punctate structures, but PRMT8(G2A/N270)-GFP and PRMT8(G2A/N20)-GFP were observed mostly in the cytosol and in both cytosol and nucleus in HEK293T cells, respectively. These results suggest that disruption of PRMT8(G2A)-GFP oligomerization/dimerization reduces punctate structure formation and results in diffuse cytosolic localization.
Fig. 2.
Characterization of the PRMT8 plasma membrane-targeting domains. (A) Schematic diagrams of point mutations within the N-terminal 20 amino acids of PRMT8. Mutated amino acids are underlined. Basic amino acids are colored in red. (B, C) Cellular localization of PRMT8-GFP, PRMT8(G2A)-GFP, and PRMT8(K4,15A/R6,13,14A)-GFP in HEK293T cells (B) and in cultured cortical neurons (C) Scale bar, 20 μm. (D) Cellular localization of PRMT8(G2A)-GFP, PRMT8(G2A/N270)-GFP, and PRMT8(G2A/N20)-GFP in HEK293T cells. Scale bar, 20 μm. (E) Effects of phosphoinositide depletion on the plasma membrane localization of PRMT8-GFP and Aplysia PDE4 short-form, S(N-UCR1/2)-GFP after antimycin treatment. Images were acquired before and after treatment with 10 μM antimycin for 40 min. Scale bar, 20 μm. (F) Cellular localization of PRMT8(C9S)-GFP and PRMT8 (C9S/N20)-GFP in HEK293T cells. Scale bar, 20 μm.
To narrow down the basic amino acids within the N-terminal 20 amino acids of PRMT8 that are important for membrane targeting, we replaced five basic amino acids (Lys-4, Arg-8, Arg-13, Arg-14, and Lys-15) in PRMT8 with Ala [PRMT8 (K4,15A,R8,13,14A)-GFP]. As shown in Fig. 2B and 2C, PRMT8(K4,15A,R8,13,14A)-GFP localized to the cytosol of HEK293T cells and neurons, showing that five basic PRMT8 N-terminal amino acids are critical for its targeting to the plasma membrane in cells. Therefore, N-terminal basic amino acids combined with myristoylation play key roles in PRMT8 plasma membrane targeting. The myristoylated alanine-rich C kinase substrate (MARCKS) is similarly localized to the plasma membrane through hydrophobic insertion of its myristate chain into the membrane and electrostatic interaction of its clustered basic amino acids with negatively charged phospholipids, including phaophatidylserine and phosphoinositides, in the plasma membrane (17, 18).
The cytoplasmic layer of the plasma membrane is the most negatively charged membrane in cells owing to the presence of phosphoinositol-4-phosphate (PI4P), phosphoinositol-4,5-diphosphate [PI(4,5)P2], and phosphoinositol-4,5-triphosphate [PI(3,4,5)P3] in the inner leaflet of the membrane (19, 20). Given that the basic N-terminal amino acids of PRMT8 play crucial roles in plasma membrane targeting, this process may be mediated through electrostatic interactions with the negative charges within the cytoplasmic surface of the membrane. To examine this hypothesis, we depleted cellular phosphoinositides like PI4P, PI(4,5)P2, and PI(3,4,5)P3 by incubating cells with Antimycin, an ATP synthesis inhibitor. As a control, we examined the localization of the Aplysia phosphodiesterase 4 (ApPDE4) short-form [ApPDE4 S(N-UCR1-2)-mRFP], known to be targeted to the plasma membrane through electrostatic interaction (16) upon Antimycin treatment. Following Antimycin treatment, ApPDE4 S(N-UCR1-2)-mRFP localized to the cytoplasm, whereas the plasma membrane targeting of PRMT8-GFP was unchanged (Fig. 2E), suggesting that the electrostatic interaction generated by phosphoinositides is not critical for PRMT8-GFP plasma membrane localization.
We also examined the role of the cysteine residue in PRMT8 plasma membrane localization by mutating the cysteine residue to serine, generating PRMT8(C9S)-GFP. As shown in Fig. 2B and C, PRMT8(C9S)-GFP localized to the plasma membrane, and the results were comparable to those observed for PRMT8-GFP in HEK293T cells and cortical neurons. While PRMT8(C9S)-GFP localized to the plasma membrane in a patch-like manner, PRMT8-GFP showed continuous localization in the plasma membrane of cultured cortical neurons (Fig. 2C). These results suggest that the cysteine residue within the PRMT8 N-terminus may have played a small role in plasma membrane targeting, but was required for proper plasma membrane localization in neurons. To examine whether patch-like plasma membrane targeting is mediated by oligomerization/dimerization, we generated the PRMT8(C9S/N20)-GFP construct. As shown in Fig. 2F, PRMT8(C9S)-GFP localized to the plasma membrane in a patch-like manner, but PRMT8(C9S/N20)-GFP showed continuous localization in the plasma membrane. These results suggest that disruption of PRMT8(C9S)-GFP oligomerization/dimerization inhibits patch-like membrane localization.
Oligomerization/dimerization inhibits mis-targeting of PRMT8 to mitochondria
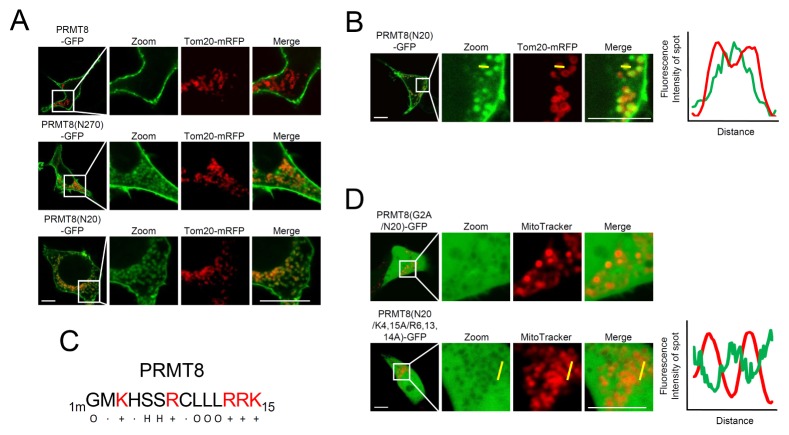
As shown in Fig. 1B and C, we found that PRMT8(N270)-GFP, PRMT8(N220)-GFP, PRMT8(N60)-GFP, and PRMT8(N20)-GFP exhibited significant intracellular localization in HEK293T cells and cortical neurons, unlike PRMT8-GFP. To investigate intracellular localization, PRMT8-GFP, PRMT8(N270)-GFP, or PRMT8(N20)-GFP were co-expressed with Tom20-mRFP, a mitochondrial outer membrane (MOM) marker in HEK293T cells. As shown in Fig. 3A, PRMT8(N270)-GFP and PRMT8(N20)-GFP, but not PRMT8-GFP, co-localized with Tom20-mRFP, indicating their targeting to the mitochondria. However, Tom20-mRFP localized to MOM, while PRMT8(N20)-GFP localized to the inner mitochondrial space, suggesting that PRMT8(N20)-GFP was targeted to the mitochondrial matrix (Fig. 3B).
Fig. 3.
Mitochondria mis-targeting of PRMT8 mutants. (A) Mitochondrial localization of PRMT8 mutants. PRMT8-GFP, PRMT8(N270)-GFP, and PRMT8(N20)-GFP co-localized with Tom20-mRFP, a mitochondria marker, in HEK293T cells. Scale bar, 20 μm. (B) Mitochondrial luminal localization of PRMT8(N20)-GFP in HEK293T cells. PRMT8(N20)-GFP co-localized with Tom20-mRFP, a mitochondrial outer membrane marker. The yellow line in the confocal fluorescence images indicates the paths along which the fluorescence intensities of the corresponding images were plotted to the bottom [red line: Tom20-mRFP, green line: PRMT8(N20)-GFP]. Scale bar, 20 μm. (C) N-terminal 15 amino acid sequences of PRMT8 within the amino acids. +, basic amino acids; H, hydroxylated amino acids; O, hydrophobic amino acids; mG, myristoylated Gly. (D) Cellular localization of PRMT8(N20/G2A)-GFP or PRMT8(N20/K4,15A,R6,13,14A)-GFP. Mitochondria are stained with MitoTracker, a mitochondria marker. The yellow line in the confocal fluorescence images indicates the paths along which the fluorescence intensities of the corresponding images were plotted to the bottom [red line: MitoTracker; green line: PRMT8(N20/K4,15A,R6,13,14A)-GFP]. Scale bar, 20 μm.
Mitochondrial presequences are enriched in scattered positively charged amino acids, including Arg and Lys, and hydroxylated and hydrophobic amino acids like Ser and Leu, respectively (20, 21). Scattered positively charged amino acids (K4, R8, R13, R14, and K15) and hydroxylated (S6 and S7) and hydrophobic amino acids (mG2 [myristoylated Gly], L10, L11, and L12) are similarly present within the PRMT8 N-terminal 15 amino acids (Fig. 3C), and may explain the mis-targeting of PRMT(N20)-GFP to the mitochondrial matrix. To test this possibility, we generated the PRMT8(G2A/N20)-GFP and PRMT8(K3,15A,R7,13,14A/N20)-GFP constructs and expressed them in HEK293T Cells. To examine mitochondria localization, we treated cells with MitoTracker, a cell membrane-permeable mitochondrial marker dye. As shown in Fig. 3D, PRMT8(G2A/N20)-GFP and PRMT8(K3,15A,R7,13,14A/N20)-GFP were localized to the cytoplasm and nucleus in a diffuse manner. However, neither PRMT8(G2A/N20)-GFP nor PRMT8 (K3,15A,R7,13,14A/N20)-GFP localized to mitochondria (Fig. 3D). Overall, these results indicate that myristoylated Gly and basic amino acids within the PRMT8 N-terminus are involved in PRMT8 mis-targeting to mitochondria, a process that is inhibited by PRMT8 oligomerization/dimerization.
The PRMT8 protein has critical roles in numerous physiological functions, including in the brain and stem cells, in a variety of animals (9, 10, 12, 22, 23). In zebrafish, for example, PRMT8 has an extra N-terminal sequence, and shows brain-specific expression that is critical for the development of the zebrafish brain. However, N-terminal deletion of PRMT8 failed to complement prmt8 morphants (22). In pluripotent human embryonic stem cells (hESCs), PRMT8 interacts directly with p85, a regulatory subunit of PI3K in the plasma membrane, inducing the accumulation of phosphoinositol 3-phosphate and consequently high AKT activity, that leads to increased maintenance of hESC pluripotency (23). Therefore, it will be interesting to examine the effects on physiological functions of mistargeting PRMT8.
MATERIALS AND METHODS
DNA constructs
To identify the PRMT8 protein in mice, PRMT8(Δ15) was cloned by reverse-transcription polymerase chain reaction (RT-PCR) from mouse cDNA using nested PCR with a specific primer set, namely, PRMT8(Δ15)-RT-S, PRMT8(Δ15)-RT-A1, and PRMT8 (Δ15)-RT-A2. To generate PRMT8(Δ15)-GFP, the region encoding PRMT8(Δ15) was amplified by PCR with the PRMT8 (Δ15)-HindIII-S and PRMT8-KpnI-A primer set and inserted between the HindIII and KpnI sites of the pGFP-N3 vector. The region encoding PRMT8 was obtained by extension PCR with the PRMT8-Ex-HindIII–S2, PRMT8-Ex–S1, and PRMT8-KpnI-A primer set using PRMT8(Δ15)-GFP as a template, and subcloned into the HindIII-Kpn1-digested pGFP-N3, pmRFP-N3, or pCDNA3.1-3×FLAG vectors (16, 24). To generate PRMT8 serial deletion mutants, regions encoding PRMT8(N270), PRMT8 (N220), PRMT8(N60), and PRMT8(N20) were generated by PCR using primers PRMT8(N270)-KpnI-A, PRMT8(N220)-KpnI-A, PRMT8(N60)-KpnI-A, and PRMT8 (N20)-KpnI-A, respectively; the amplified fragments were then inserted separately between the HindIII and KpnI sites of the pGFP-N3 vector (Table 1). Mutagenesis of PRMT8 was performed by PCR using the PRMT8(G2A)-HindIII-S, PRMT8(C9S)-HindIII-S, PRMT8(5A)-EX-HindIII-S2, and PRMT8(5A)-EX-S1 primers, and the amplified fragments were separately subcloned into the pGFP-N3 vector between the HindIII and KpnI sites (Table 1).
Table 1.
Primer sequences used for the experiments
| Name | Primer sequence |
|---|---|
| PRMT8(Δ15)-RT-S | 5′-AGGAAAATGGCGGAGAATG-3′ |
| PRMT8(Δ15)-RT-A1 | 5′-GCAGCATGCAAAACTTGAAA-3′ |
| PRMT8(Δ15)-RT-A2 | 5′-CAAGGCAGAGTGGAAAGCTC-3′ |
| PRMT8(Δ15)-HindIII-S | 5′-CGCCCAAGCTTGCCACCATGGCGGAGAATGCAGTC-3′ |
| PRMT8-KpnI-A | 5′-GACGGTA CCACGCATTTTGTAGTCATT-3′ |
| PRMT8-Ex-HindIII–S2 | 5′-CCCAAGCTTGCCACCATGGGCATGAAACACTCCTCCCGCTGCCTGCTC-3′ |
| PRMT8-Ex–S1 | 5′-TCCTCCCGCTGCCTGCTCCTCCGGAGGAAAATGGCGGAGAATGCAGTC-3′ |
| PRMT8(N270)-KpnI-A | 5′-GACGGTACCGGTCACCACTTGCTTTGG-3′ |
| PRMT8(N220)-KpnI-A | 5′-GACGGTACCAGCTGCCCGGTCTGGAAA-3′ |
| PRMT8(N60)-KpnI-A | 5′-GACGGTACCCTTGCCCCGTCCAGGGCAGCT-3′ |
| PRMT8(N20)-KpnI-A: | 5′-GACGGTACCTGCATTCTCCGCCATTTT-3′ |
| PRMT8(G2A)-HindIII-S | 5′-CGCCCAAGCTTGCCACCATGGCCATGAAACACTCC-3′ |
| PRMT8(C9S)-HindIII-S | 5′-CGCCCAAGCTTG CCACCATGGGCATGAAACACTCCTCCCGCTCCCTG-3′ |
| PRMT8(5A)-EX-HindIII-S2 | 5′-CGCCCAAGCTTGCCACCATGGGCATGGCGCACTCCTCCGCGTGCCTGCT CCTC-3′ |
| PRMT8(5A)-EX-S1 | 5′-TCCGCGTGCCTGCTCCTCGCGGCGGCGATGGCGGAGAATGCAGTCGAA-3′ |
Cell culture, transfection, confocal microscopy, and drug treatment
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and penicillin/streptomycin in a humidified atmosphere with 5% (v/v) CO2 at 37°C. The cells were seeded in the sticky-Slide 8-well system (Catalog #: 80828; Ibidi, Martinsried, Germany) to obtain 40–60% confluent cells on the day of imaging. The cells were transfected with the DNA constructs using calcium phosphate or Lipofectamine® 2000 (Life Technologies, Carlsbad, CA, USA) 24–26 h before imaging. The relative amount of each construct was empirically determined based on the relative expression of each construct combination. Cells were observed under an inverted Zeiss LSM-700 scanning laser confocal microscope operated by ZEN software (Carl Zeiss, Oberkochen, Germany). The laser lines for excitation and the emission wavelengths for the fluorochromes were 488 with 508–543 nm for GFP and 561 with 578–649 nm for mRFP, respectively. Appropriate GFP (500–550 nm) and mRFP (575–625 nm) emission filters were used during the sequential imaging of each fluorescent protein. Most images were taken with live cells. Rapamycin was obtained from Sigma-Aldrich (Catalog #: R8781; St. Louis, MO, USA). MitoTracker® Red was purchased from Invitrogen (Catalog #: qM7512; Thermo Fisher Scientific, Waltham, MA, USA). ATP was depleted by incubating cells with 200 nM Antimycin A (Sigma-Aldrich, St. Louis, MO, USA) in PBS for at least 40 min, as previously described (16). All the treatments and assays were performed at 37°C unless otherwise indicated.
Immunoprecipitation
Immunoprecipitation were performed as previously described (25–27). Briefly, for transient transfections, HEK293T cells were plated at a density of 5–7 × 105 cells/well in six-well plates and cultured for 24 h. The cells were transfected with DNA constructs using calcium phosphate (Clontech laboratories, Mountain View, CA, USA) and incubated for 24 h. For FLAG immunoprecipitation, the transfected HEK293T cells were washed twice with PBS and lysed with a buffer containing 1% Triton X-100, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), and a protease inhibitor cocktail (Roche, Basel, Basel-Stadt, Switzerland). The cell lysates were incubated with 50 μl (bead volume) of mouse anti-FLAG M2 antibody-conjugated beads (Sigma-Aldrich, St. Louis, MO, USA) at 4°C overnight. The beads were subsequently washed three times with lysis buffer. The immunoprecipitates were eluted by adding 2 μg/ml of 3× FLAG peptides and analyzed by Coomassie blue staining.
ACKNOWLEDGEMENTS
J.A.L was supported by the Basic Science Research Program through the NRF (NRF-2017R1D1A3B03030972), by the National Honor Scientist Program of Korea, the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (2017M3A9G7073521) and Hannam Research Program (2019). D.J.J was supported by the Basic Science Research Program through the NRF (NRF-2018R1D1A1B07048822).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Blanc RS, Richard S. Arginine Methylation: The Coming of Age. Molecular cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morales Y, Caceres T, May K, Hevel JM. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs) Arch Biochem Biophys. 2016;590:138–152. doi: 10.1016/j.abb.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 4.Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y, Maity R, Whitelegg JP, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem. 2013;288:37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kousaka A, Mori Y, Koyama Y, Taneda T, Miyata S, Tohyama M. The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience. 2009;163:1146–1157. doi: 10.1016/j.neuroscience.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Sayegh J, Daniel J, Clarke S, Bedford M. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 8.Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J Biol Chem. 2007;282:36444–36453. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- 9.Kim JD, Park K, Ishida J, et al. PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci Adv. 2015;1:e1500615. doi: 10.1126/sciadv.1500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penney J, Seo J, Kritskiy O, Elmsaouri S. Loss of Protein Arginine Methyltransferase 8 Alters Synapse Composition and Function, Resulting in Behavioral Defects. J Neurosci. 2017;37:8655–8666. doi: 10.1523/JNEUROSCI.0591-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee PK, Goh WW, Sng JC. Network-based characterization of the synaptic proteome reveals that removal of epigenetic regulator Prmt8 restricts proteins associated with synaptic maturation. J Neurochem. 2017;140:613–62. doi: 10.1111/jnc.13921. [DOI] [PubMed] [Google Scholar]
- 12.Simandi Z, Paje K, Karolyi K, Sieler T. Arginine Methyltransferase PRMT8 Provides Cellular Stress Tolerance in Aging Motoneurons. J Neurosci. 2018;38:7683–7700. doi: 10.1523/JNEUROSCI.3389-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schapira M, Ferreira de Freitas R. Structural biology and chemistry of protein arginine methyltransferases. Medchemcomm. 2014;5:1779–178. doi: 10.1039/C4MD00269E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toma-Fukai S, Kim JD, Park KE, et al. Novel helical assembly in arginine methyltransferase 8. J Mol Biol. 2016;428:1197–1208. doi: 10.1016/j.jmb.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Jun YW, Lee SH, Shim J, et al. Dual roles of the N-terminal coiled-coil domain of an Aplysia sec7 protein: homodimer formation and nuclear export. J Neurochem. 2016;139:1102–1112. doi: 10.1111/jnc.13875. [DOI] [PubMed] [Google Scholar]
- 16.Kim KH, Jun YW, Park Y, et al. Intracellular membrane association of the Aplysia cAMP phosphodiesterase long and short forms via different targeting mechanisms. J Biol Chem. 2014;289:25797–25811. doi: 10.1074/jbc.M114.572222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/S0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 18.Swierczynski SL, Blackshear PJ. Myristoylation-dependent and electrostatic interactions exert independent effects on the membrane association of the myristoylated alanine-rich protein kinase C substrate protein in intact cells. J Biol Chem. 1996;271:23424–23430. doi: 10.1074/jbc.271.38.23424. [DOI] [PubMed] [Google Scholar]
- 19.Jang DJ, Lee JA. The roles of phosphoinositides in mammalian autophagy. Arch Pharm Res. 2016;39:1129–36. doi: 10.1007/s12272-016-0777-x. [DOI] [PubMed] [Google Scholar]
- 20.Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 21.Pfanner N. Protein sorting: recognizing mitochondrial presequences. Curr Biol. 2000;10:R412–415. doi: 10.1016/S0960-9822(00)00507-8. [DOI] [PubMed] [Google Scholar]
- 22.Lin YL, Tsai YJ, Liu YF, et al. The critical role of protein arginine methyltransferase prmt8 in zebrafish embryonic and neural development is non-redundant with its paralogue prmt1. PLoS One. 2013;8:e55221. doi: 10.1371/journal.pone.0055221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong HC, Park SJ, Choi JJ, et al. PRMT8 Controls the Pluripotency and Mesodermal Fate of Human Embryonic Stem Cells By Enhancing the PI3K/AKT/SOX2 Axis. Stem Cells. 2017;35:2037–2049. doi: 10.1002/stem.2642. [DOI] [PubMed] [Google Scholar]
- 24.Lee YK, Jun YW, Choi HE, et al. Development of LC3/GABARAP sensors containing a LIR and a hydrophobic domain to monitor autophagy. EMBO J. 2017;36:1100–1116. doi: 10.15252/embj.201696315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin CH, Ryu S, Kim HH. hnRNPK-regulated PTOV1-AS1 modulates heme oxygenase-1 expression via miR-1207-5p. BMB Rep. 2017;50:220–225. doi: 10.5483/BMBRep.2017.50.4.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang E, Lee HR, Lee GH, et al. Bach2 represses the AP-1-driven induction of interleukin-2 gene transcription in CD4 T cells. BMB Rep. 2017;50:472–477. doi: 10.5483/BMBRep.2017.50.9.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang HS, Choi MH, Kim HA. 29-kDa FN-f inhibited autophagy through modulating localization of HMGB1 in human articular chondrocytes. BMB Rep. 2018;51:508–513. doi: 10.5483/BMBRep.2018.51.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]