Abstract
Background
Longitudinal measurement of glioma burden with MRI is the basis for treatment response assessment. In this study, we developed a deep learning algorithm that automatically segments abnormal fluid attenuated inversion recovery (FLAIR) hyperintensity and contrast-enhancing tumor, quantitating tumor volumes as well as the product of maximum bidimensional diameters according to the Response Assessment in Neuro-Oncology (RANO) criteria (AutoRANO).
Methods
Two cohorts of patients were used for this study. One consisted of 843 preoperative MRIs from 843 patients with low- or high-grade gliomas from 4 institutions and the second consisted of 713 longitudinal postoperative MRI visits from 54 patients with newly diagnosed glioblastomas (each with 2 pretreatment “baseline” MRIs) from 1 institution.
Results
The automatically generated FLAIR hyperintensity volume, contrast-enhancing tumor volume, and AutoRANO were highly repeatable for the double-baseline visits, with an intraclass correlation coefficient (ICC) of 0.986, 0.991, and 0.977, respectively, on the cohort of postoperative GBM patients. Furthermore, there was high agreement between manually and automatically measured tumor volumes, with ICC values of 0.915, 0.924, and 0.965 for preoperative FLAIR hyperintensity, postoperative FLAIR hyperintensity, and postoperative contrast-enhancing tumor volumes, respectively. Lastly, the ICCs for comparing manually and automatically derived longitudinal changes in tumor burden were 0.917, 0.966, and 0.850 for FLAIR hyperintensity volume, contrast-enhancing tumor volume, and RANO measures, respectively.
Conclusions
Our automated algorithm demonstrates potential utility for evaluating tumor burden in complex posttreatment settings, although further validation in multicenter clinical trials will be needed prior to widespread implementation.
Keywords: deep learning, glioma, longitudinal response assessment, RANO, segmentation
Key Points.
1. An algorithm that automatically segments postoperative glioblastoma was developed.
2. The calculation of the product of maximum bidimensional diameters was automated.
3. Automated measures are in agreement with human experts for changes in tumor burden.
Importance of the Study.
Longitudinal measurement of glioma burden with MRI is the basis for treatment response assessment. Experts in neuro-oncology routinely make manual estimates of tumor size based on the product of bidimensional diameters of enhancing tumor. However, this procedure is time-consuming and subject to interobserver variability. In this study, we developed an algorithm that automatically segments FLAIR hyperintensity and contrast-enhancing tumor, quantitating tumor volumes as well as the product of maximum bidimensional diameters according to the RANO criteria (AutoRANO). We show that automated volume and RANO measurements are highly repeatable and are in agreement with human experts in terms of change in tumor burden during the course of treatment. This tool may be helpful in clinical trials and clinical practice for expediting measurement of tumor burden in the evaluation of treatment response, decreasing the time expended by clinicians for manual tumor segmentation, and decreasing interobserver variability.
Gliomas are primary central nervous system tumors with variable natural histories and prognoses depending on their histologic and molecular characteristics.1 The current gold standard to determine treatment response and assess tumor progression in clinical trials is the Response Assessment in Neuro-Oncology (RANO) criteria.2 For high-grade gliomas, including glioblastomas (GBMs), radiographic response assessment is based on (i) measurement of the 2D product of maximum bidimensional diameters of contrast-enhancing tumor and (ii) qualitative evaluation of T2/fluid-attenuated inversion recovery (FLAIR) abnormal hyperintense regions.2,3 However, manual delineation of tumor boundaries can be difficult due to the infiltrative nature of gliomas and presence of heterogeneous contrast enhancement, which is particularly common during anti-angiogenic treatment. As a result, there can be substantial interrater variability in 2D measurements for both contrast-enhancing and FLAIR hyperintense tumors.4–6 Furthermore, variability in segmentation can introduce substantial variability in calculated mean values of multi-parametric magnetic resonance (MR) parameters, such as the volume transfer constant.7 Consequently, there is great interest in developing reproducible automated methods for segmentation and calculation of the product of maximum bidimensional diameters.
Although 2D linear measurements currently represent the gold standard for response assessment, volumetric measurements may capture tumor burden more accurately, particularly because gliomas are often irregularly shaped. However, volumetric response assessment has not been adopted for routine use due to the laborious efforts needed to perform tumor segmentation using existing tools and a lack of large-scale studies validating its benefit over simpler 2D approaches. A recent consensus paper on brain tumor imaging in clinical trials noted volumetric analysis as an improvement to current protocols.8 An automated segmentation tool could help facilitate the use of tumor volume as a response endpoint in clinical trials and allow integration into the clinical workflow. Rapid and reproducible tumor segmentation is also an essential step toward voxel-based quantitative assessment of single as well as multi-parametric imaging biomarkers of tumor response to treatment.9–13
With the advent of more powerful graphics processing units, deep learning has become the method of choice for automatic segmentation of medical images.14,15 At the core of deep learning is the convolutional neural network; a machine learning technique that can be trained on raw image data to predict clinical outputs of interest. Existing deep learning methods have not been developed for the postoperative setting, where the surgical cavity and brain distortion make it difficult to reliably outline the boundaries of the tumor.14,16
There are 2 key challenges to automatic tumor segmentation. The first challenge is variability in brain extraction, an image preprocessing technique that separates the brain from skull and is essential for many neuroimaging applications.17 Removing the skull from the image prevents automatic segmentation algorithms from falsely labeling non-brain regions as tumor and enables consistent intensity normalization across all patients. Many automated methods exist for brain extraction, but their generalizability is limited.18–22 Without manual correction, poor brain extraction can introduce errors into downstream automatic segmentation.23 This is particularly important in the postoperative setting due to the widely heterogeneous and variable appearance of surgical cavity, calvarium, and scalp. The second challenge is generalizability: MR intensity values vary substantially depending on the MR scanner properties (including manufacturer, scanner type, and field strength) and acquisition parameters (including echo time, repetition time, and contrast injection dose/timing) and can result in substantial differences in tumor appearance.8 Consequently, algorithms trained on limited datasets may not apply well to data acquired from different institutions, acquisition protocols, and patient populations.
In this study, we developed a fully automated pipeline for brain extraction and tumor segmentation that can be used to reliably generate abnormal FLAIR hyperintensity and contrast-enhancing tumor volumes as well as 2D bidimensional diameters according to the RANO criteria. We then validated the performance of the algorithm in both a multi-institutional preoperative patient cohort and a longitudinal postoperative patient cohort from a single institution by comparing automated measurements to manual measurements derived from experts.
Materials and Methods
Preoperative Patient Cohort
The study was conducted following approval by the Hospital of the University of Pennsylvania (HUP) and the Partners Institutional Review Boards. Glioma patients at HUP, The Cancer Imaging Archive (TCIA), Massachusetts General Hospital (MGH), and Brigham and Women’s Hospital (BWH) were retrospectively identified. The imaging study dates for HUP, MGH, and BWH ranged from 1998 to 2016. For the TCIA cohort, we identified glioma patients with preoperative MRI data from The Cancer Genome Atlas and IvyGap.24 All patients met the following criteria: (i) histopathologically confirmed grades II–IV glioma according to World Health Organization criteria (2007 or 2016 criteria, depending on whether the case occurred before or after 2016) and (ii) available preoperative MRI consisting of T2-weighted FLAIR and post-contrast T1-weighted (T1 post-contrast) images. Patients were excluded if glioma was not histopathologically confirmed, either FLAIR or T1 post-contrast imaging was unavailable, or there was excessive motion artifact on imaging. The acquisition settings of the imaging for the preoperative patient cohort are shown in Supplementary Figures 1–2. For the preoperative cases, both 2D and 3D T1-weighted images were used, depending on which were available. Three-dimensional T1-weighted imaging was available for 29% of the patients in the preoperative patient cohort.
Postoperative Patient Cohort
MRI data were acquired from 2 clinical trials at MGH that enrolled patients with newly diagnosed glioblastoma receiving standard chemoradiation (NCT00756106) or standard chemoradiation with cediranib (NCT00662506). There were 54 total patients. The Dana-Farber/Harvard Cancer Center institutional review board approved these studies. Inclusion criteria for both trials were age >18 years, post-surgical residual contrast-enhancing tumor size of ≥1 cm in one dimension, histologically confirmed diagnosis of glioblastoma, and eligibility for standard therapy after surgery. For NCT00756106, MRI was performed at the following timepoints: within 1 week of starting chemoradiation therapy (baseline visit 1), 1 day before starting chemoradiation therapy (baseline visit 2), weekly during chemoradiation, and monthly before each cycle of adjuvant temozolomide until disease progression or at least until completion of 6 cycles of adjuvant temozolomide (whichever occurred first).25 MRI timepoints for NCT00662506 were previously described by Batchelor et al.26 MRI was performed at 3.0 T (TIM Trio, Siemens Healthcare) and included FLAIR images (repetition time [TR] = 10 000 ms, echo time [TE] = 70 ms, 5 mm slice thickness, 1 mm interslice gap, 0.43 mm in-plane resolution) and both pre- and post-contrast T1-weighted images (TR = 600 ms, TE = 12 ms, 5 mm slice thickness, 1 mm interslice gap, 0.43 mm in-plane resolution).
Expert Brain Extraction, Tumor Segmentation, and RANO Measurements
Brain extraction was performed in 42 randomly selected patients from the preoperative and postoperative patient cohort by one rater (R.Y.H., neuroradiologist, 9 years experience). Manual tumor segmentations were performed on the FLAIR hyperintense areas in the preoperative patient cohort (Q.S., neuroradiologist, 5 years experience; R.Y.H.; A.B., neurosurgery resident, 5 years experience) and the FLAIR hyperintense as well as contrast-enhancing tumor areas in the postoperative patient cohort (E.R.G., neuro-oncologist, 12 years experience; M.C.P., neuroradiologist, 11 years experience), with segmentation for each patient visit performed by a single expert evaluating both the pre- and post-contrast MRIs to exclude postoperative blood products. Manual RANO bidirectional measurements as well as assessment for FLAIR progression were performed by 2 raters (E.R.G. and K.I.L., neuro-oncologist, 7 years experience) for both baseline visits, the visit with the lowest manual contrast-enhancing tumor volume, and the last patient visit from the postoperative patient cohort.27
Deep Learning–Based Brain Extraction
The 42 patients for whom expert brain mask extraction was performed were divided into training (n = 30) and testing (n = 12) sets. The neural network was trained on the training set. As a point of reference, we compared brain extraction using our deep learning algorithm with that of other commonly used automatic brain extraction methods (Hybrid Watershed Algorithm, Robust Learning-Based Brain Extraction, Brain Extraction Tool, 3dSkullStrip, and Brain Surface Extractor).17–22 All methods were applied to the T1 post-contrast images using default parameters, except for Robust Learning-Based Brain Extraction, which has no tunable parameters.
Deep Learning–Based Abnormal FLAIR Hyperintensity and Contrast-Enhancing Tumor Segmentation
The HUP, TCIA, and MGH patient preoperative cohorts were randomly divided into training and testing sets in a 4:1 ratio. The BWH patient cohort was used as an independent testing set. A single neural network model was trained for FLAIR hyperintensity segmentation in the preoperative patient cohort using only the training set. Once the model was trained, performance was assessed on the testing and independent testing sets.
The patients from the single institutional postoperative patient cohort were randomly divided into training and testing sets in a 4:1 ratio. Data were split on a patient level such that all visits for a patient were entirely in either the training or test set (Supplementary Fig. 3). Two neural network models were trained for the postoperative patient cohort: FLAIR hyperintensity segmentation and contrast-enhancing tumor segmentation. Only the training set was used during training of the model. Once trained, the performance of the model was assessed on the separate testing set.
Neural Network Architecture and Postprocessing
We utilized the 3D U-Net architecture, a neural network designed for fast and precise segmentation, for both brain extraction and tumor segmentation (Supplementary Fig. 4B).28,29 Similar to the original 2D U-Net, our architecture consists of a downsampling and an upsampling arm with residual connections between the two that concatenate feature maps at different spatial scales. The networks were designed to receive input patches from multiple channels: (i) FLAIR and T1 post-contrast images for brain extraction, (ii) FLAIR and T1 post-contrast images for FLAIR hyperintensity segmentation in the preoperative patient cohort, (iii) FLAIR, T1 pre-contrast, and T1 post-contrast images for FLAIR tumor segmentation in the postoperative patient cohort, and (iv) FLAIR, T1 pre-contrast, T1 post-contrast, and FLAIR hyperintensity region for contrast-enhancing tumor segmentation in the postoperative patient cohort. Rectified linear unit (ReLU) activation was used in all layers, with the exception of the final sigmoid output. Batch normalization was applied after each convolutional layer for regularization. We used Nestorov adaptive moment estimation to train the 3D U-Nets with an initial learning rate 10−5, minimizing a soft Dice loss function:
| (1) |
where D is Dice, p is the probability output of the neural network, g is the ground truth, and α is a constant. Our networks were implemented in DeepNeuro with Keras/Tensorflow backend.30 Each U-Net was trained on a NVIDIA Tesla P100 graphics processing unit. During training, 20% of the training set was withheld as a validation set. For brain extraction, 50 patches (64 × 64 × 8) were extracted, randomly, for each patient in the training set and 10 patches were extracted for each patient in the validation set. For tumor segmentation, 20 patches (64 × 64 × 8) were extracted from normal brain and FLAIR hyperintense regions in a 1:1 ratio for each patient in the training set, and 4 patches were extracted for each patient in the validation set. Before patches were used to train the network, they were augmented by means of sagittal flips. Augmentation increases the size of the training set while also preventing overfitting.13 The network was trained through all extracted patches until the validation loss did not improve for 10 consecutive iterations. Once the network was trained, inference was performed by gridding the MR images into patches at 8 different offsets from the uppermost corner of the image. The model then predicted probability maps for each of these patches, and voxels with predictions from multiple overlapping patches had their probabilities averaged. For prediction of the contrast-enhancing tumor regions, the output probability map from the FLAIR hyperintensity segmentation neural network was used as input instead of the manually derived FLAIR hyperintensity region.
AutoRANO Algorithm
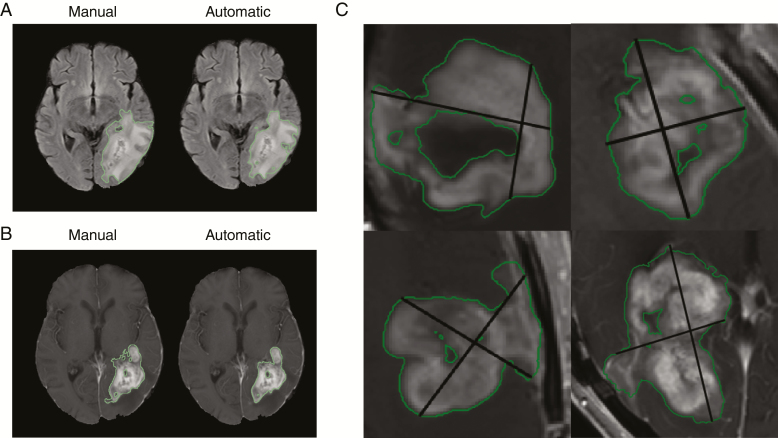
We developed an AutoRANO algorithm to automatically derive RANO measurements from our automatic deep learning–based contrast-enhancing tumor segmentations as described above. The algorithm searches for the axial slice with the largest tumor area and determines if the lesion is measurable. A measurable lesion was defined as a minimum length of both perpendicular measurements ≥12 mm (based on a threshold of 10 mm if slice thickness + gap ≤ 5 mm or a threshold of 2 × [slice thickness + gap] if slice thickness + gap > 5 mm).31 If the lesion was measurable, the product of maximum bidimensional diameters was automatically derived by first exhaustively searching for the longest diameter and then the corresponding longest perpendicular diameter. The angle between the longest diameter and the perpendicular diameter was restricted to 85–95°. If there was more than one measurable lesion on the same scan, the products of maximal bidimensional diameters were summed (for up to 5 measurable lesions).31 The AutoRANO algorithm was applied to the automatically segmented contrast-enhancing tumor regions (Fig. 1C).
Fig. 1.
Example of manual vs automatic FLAIR hyperintensity segmentation (A) and enhancing-tumor segmentation (B) for the testing set in the postoperative patient cohort. (C) Examples of AutoRANO applied to automatic enhancing segmentations on the postoperative patient cohort.
Statistical Analysis
Neural network segmentation was compared with expert segmentation by means of the Sørensen–Dice coefficient, sensitivity, and specificity, and evaluated statistically using Dunnet’s test (significance level P < 0.05). Comparison of volume and RANO measurements were assessed via either Spearman’s rank correlation coefficient (ρ [Greek letter rho]) or intraclass correlation coefficient (ICC) (significance level P < 0.05). Further details of ICC calculation are in the Supplementary Material. For the postoperative patient cohort, the nadir was defined as the minimum volume or minimum 2D linear measurements at any timepoint from baseline to last visit. For longitudinal comparison of volume and RANO measurements, the last patient visit was assessed relative to the nadir (delta measure = volume or RANO measure of the last patient visit—volume or RANO measure of the nadir).
Code and Data Availability
The codes for preprocessing, U-Net architecture, and postprocessing are publicly available at: https://github.com/QTIM-Lab/DeepNeuro. Accessed June 23, 2019.30
Results
Patient Cohorts
Our final preoperative patient cohort included 239 patients from HUP, 293 patients from TCIA, 154 patients from MGH, and 157 patients from BWH. Our final postoperative patient cohort consisted of 713 visits from 54 patients from MGH. Twenty-one patient visits were excluded due to missing MRI sequences or excessive motion artifact. Patient characteristics are shown in Supplementary Table 1.
Deep Learning–Based Brain Extraction
We compared brain extraction using our deep learning algorithm, based on the 3D U-Net architecture,29 with that of both human expert and commonly used brain extraction software packages. The mean Dice score between our algorithm and manual expert brain extraction was 0.935 (95% CI: 0.918–0.948) in the testing set (Supplementary Table 2, Supplementary Fig. 5A). Compared with other commonly used brain extraction techniques (Supplementary Table 2), our algorithm had the highest Dice score and specificity for the testing set. When the U-Net was applied to all 843 patients in the preoperative patient cohort, the mean fraction of FLAIR hyperintensity retained in the extracted brain image (defined as tumor volume remaining in the brain-extracted image divided by total tumor volume) was 0.987 (95% CI: 0.984–0.990; Supplementary Fig. 5B). When applied to the 713 patient visits in the postoperative patient cohort, the mean fraction of FLAIR hyperintensity and contrast-enhancing tumor retained in the extracted brain image was 0.996 (95% CI: 0.994–0.997; Supplementary Fig. 5C) and 0.982 (95% CI: 0.977–0.987), respectively.
Deep Learning–Based FLAIR Hyperintensity and Contrast-Enhancing Tumor Volume Segmentation
The average time for brain extraction, FLAIR hyperintensity, and contrast-enhancing tumor segmentation was 19 seconds using our trained algorithms. For the testing set of the preoperative patient cohort, the mean Dice score for FLAIR hyperintensity segmentation was 0.796 (95% CI: 0.753–0.803) (Supplementary Table 3). For the independent testing set, the mean Dice score for automatic FLAIR hyperintensity segmentation compared with expert human segmentation was 0.819 (95% CI: 0.793–0.842). Examples of FLAIR hyperintensity segmentations for the independent testing set of the preoperative patient cohort are shown in Supplementary Fig. 7. For the testing set of the postoperative patient cohort, the mean Dice score for automatic FLAIR hyperintensity segmentation compared with manual segmentation was 0.701 (95% CI: 0.670–0.731). The mean Dice score for automatic segmentation compared with manual contrast-enhancing tumor segmentation was 0.696 (95% CI: 0.660–0.728).
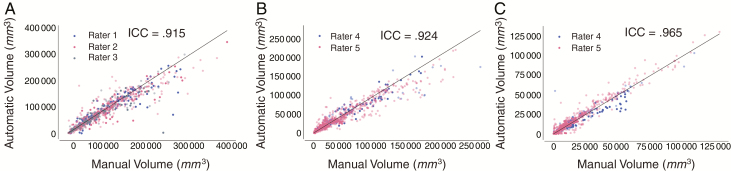
Examples of FLAIR hyperintensity and contrast-enhancing tumor segmentations for the testing set of the postoperative patient cohort are shown in Fig. 1A, B. Examples of longitudinal tracking of FLAIR hyperintensity and contrast-enhancing tumor volumes for 2 patients in the testing set are shown in Supplementary Fig. 9. The ICC for calculated FLAIR hyperintensity volumes between automatic and manual segmentation was 0.915 (P < 0.001) in the preoperative and 0.924 (P < 0.001) in the postoperative patient cohorts. The ICC for contrast-enhancing tumor volume in the postoperative patient cohort was 0.965 (P < 0.001; Fig. 3). In the rare cases when the algorithm was off, the reason was due to similarity in signal intensity between normal brain and tumor—a similar challenge for human readers (Supplementary Figures 7D and 8).
Fig. 3.
Automatically and manually derived volumes are highly correlated. Correlation between manually and automatically derived volumes for (A) FLAIR hypertintensity in the preoperative patient cohort, (B) FLAIR hyperintensity in the postoperative patient cohort, and (C) contrast-enhancing tumor in the postoperative patient cohort. Training and testing sets are shown light blue/red/gray and dark blue/red/gray, respectively. Line of identity (x = y) is shown in all plots.
Repeatability of Volume and RANO Measurements in the Postoperative Patient Cohort
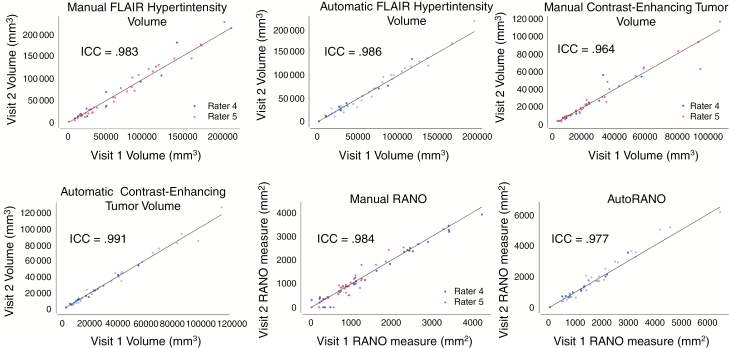
Repeatability of manual and automatic measurements was assessed by comparing measurements from the 2 baseline visits for each patient. Comparing baseline visits 1 and 2 for FLAIR hyperintensity volume, the ICC was 0.983 (P < 0.001) for manual volume measurement and 0.986 (P < 0.001) for automatic volume measurement. For contrast-enhancing tumor volume, the ICC was 0.964 (P < 0.001) for manual volume measurement and 0.991 (P < 0.001) for automatic volume measurement.
Comparing baseline visits 1 and 2 for RANO measurements, the ICC was 0.984 (P < 0.001) for manual RANO and 0.977 (P < 0.001; Fig. 2) for AutoRANO. Notably, there were 5 patients assessed by one rater who had measurable lesions on one but not the other baseline visit. Similarly, there were 3 patients assessed by the other rater who had measurable lesions on one but not the other baseline visit. By comparison, when using the AutoRANO algorithm, no patients had a discrepancy in the presence/absence of measurable lesions between the 2 baseline visits.
Fig. 2.
Volume and RANO measures are highly repeatable. Repeatability of (A) manual FLAIR hypertintensity volume, (B) automatic FLAIR hypertintensity volume, (C) manual contrast-enhancing tumor volume, (D) automatic contrast-enhancing tumor volume, (E) manual RANO, and (F) AutoRANO in the postoperative patient cohort. Training and testing sets are shown in light blue and dark blue, respectively, in B, D, and F. Line of identity (x = y) is shown in all plots.
Interrater Agreement for Manual RANO and Agreement Between Manual RANO and AutoRANO
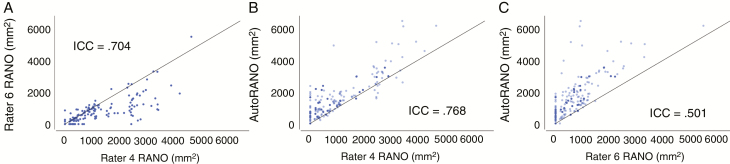
In assessing interrater agreement, the ICC for manual RANO measurements between the 2 expert raters was 0.704 (P < 0.001). In assessing rater–algorithm agreement, the ICC was 0.768 (P < 0.001) between AutoRANO and Rater 4 and 0.501 (P < 0.001; Fig. 4) between AutoRANO and Rater 6.
Fig. 4.
There was moderate interrater and manual–algorithm agreement for RANO measures. Agreement between RANO measures for (A) Rater 6 vs Rater 4, (B) AutoRANO vs Rater 4, and (C) AutoRANO vs Rater 6 in the postoperative patient cohort. Training and testing sets are light blue and dark blue, respectively, in B and C. Line of identity (x = y) is shown in all plots.
Automatic Treatment Response Assessment
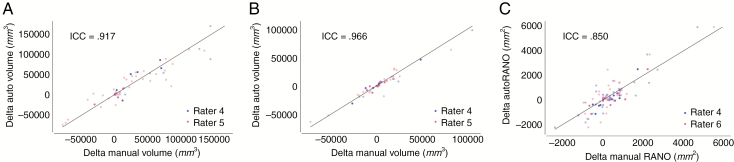
Comparisons between nadir and the last patient visit were made (delta measure = last patient visit measure—nadir measure). In assessing rater–algorithm agreement for the delta measures, the ICC between automatic and manual delta measurements were 0.917 (P < 0.001), 0.966 (P < 0.001), and 0.850 (P < 0.001) for FLAIR hyperintensity volume, contrast-enhancing tumor volume, and RANO measures, respectively (Fig. 5).
Fig. 5.
There was high agreement between manually and automatically derived longitudinal changes in volume and RANO measures. Agreement between automatic and manual delta measures for (A) FLAIR hypertintensity volume, (B) contrast-enhancing tumor volumes, and (C) RANO measure in the postoperative patient cohort. Training and testing sets are shown light blue/red and dark blue/red, respectively. Line of identity (x = y) is shown in all plots.
Correlation Between RANO Measures and Manual Volume
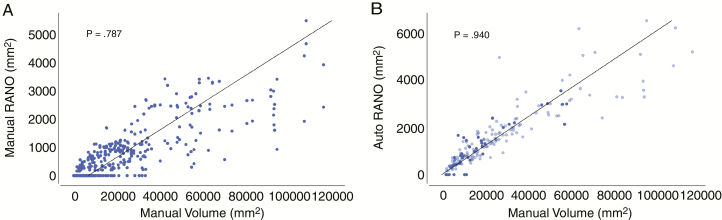
Spearman’s ρ coefficient between manual RANO measures and manual enhancing-tumor volume was 0.787 (P < 0.001). Spearman’s ρ coefficient between AutoRANO measures and manual enhancing-tumor volume was 0.940 (P < 0.001; Fig. 6). Spearman’s ρ coefficient between delta manual RANO measures and delta manual enhancing-tumor volume was 0.744 (P < 0.001). Spearman’s ρ coefficient between delta AutoRANO measures and delta manual enhancing-tumor volume was 0.832 (P < 0.001, Supplementary Fig. 11).
Fig. 6.
AutoRANO had higher agreement with manual contrast-enhancing volume than manual RANO measures. Correlation between manual contrast-enhancing volume and RANO measures for (A) manual RANO and (B) AutoRANO in the postoperative patient cohort. Training and testing sets are shown in light blue and dark blue, respectively in B. Linear fit is shown in all plots.
Discussion
In this study, we demonstrate the utility of a fully automated, deep learning–based pipeline for calculation of tumor volumes and RANO measurements. A key image preprocessing step is brain extraction, which removes non-brain tissue—a significant source of error for downstream tumor segmentation. We trained a network on expert brain-extracted images from patients who underwent heterogeneous imaging acquisition protocols and demonstrate its superiority compared with other commonly used skull-stripping methods. After brain extraction, a deep learning framework was applied for FLAIR hyperintensity and contrast-enhancing tumor volume segmentation. Even with the varied acquisition protocols, our automatic pipeline proved to be robust for segmentation in the majority of patients in our multi-institutional dataset. We further developed an algorithm for automatic calculation of RANO measurements from contrast-enhancing tumor segmentations. In addition to the preoperative setting, our algorithm demonstrated good performance in postoperative MRIs, which are particularly challenging given the frequent presence of surgical cavities and brain distortion. Furthermore, the algorithm was successfully applied in a longitudinal patient cohort including patients who had been treated with cediranib, which blunts the contrast enhancement, yielding ill-defined contrast enhancement margins that are difficult to contour. It is in these cases, particularly, that standardized segmentation is likely to be most helpful.
Based on the double baseline MRIs, both manually and automatically derived FLAIR hyperintensity volume, contrast-enhancing tumor volume, and RANO measurements were highly repeatable, showing intrarater consistency. However, there were differences in interrater consistency. The RANO measurements from the AutoRANO algorithm were, on average, larger than those of the 2 human raters. This is likely due to the fact that our AutoRANO algorithm performs an exhaustive search of the longest perpendicular diameters while a human performs this estimation by eye, which is a less accurate method. This inaccuracy is further evidenced by the fact that the average RANO measurements differed between the 2 raters. In fact, consistent with prior reports on the variability in 2D measurements,4 it is not surprising that there was substantial variability between RANO measurements between our raters. In contrast, we found high agreement between manual raters and automatic volume for both contrast-enhancing tumor and FLAIR hyperintensity. This suggests that volume measurements allow for greater consistency across raters than RANO measurements.
There was high agreement between manual and automatic measures with regard to changes in tumor burden (both contrast-enhancing and FLAIR hyperintensity) during the course of longitudinal therapy. However, there was better agreement between manual raters and automated measurements for contrast-enhancing tumor volume compared with RANO measures. Thus, automated volume measurements were superior to AutoRANO measurements due to higher concordance with manual methods.
Interestingly, AutoRANO correlated better with manual contrast-enhancing tumor volume than the manual RANO measurements. Delta AutoRANO (the difference in the bidimensional measurements between the last visit and the nadir scan) also correlated better with delta manual contrast-enhancing tumor volume than delta manual RANO measurements. This suggests that AutoRANO may be a more accurate measure of tumor burden than manual RANO measurement in addition to the advantage of being fully automated.
One point to note is that the ICC values for manual versus automatic volumes were higher than the Dice scores for manual versus automatic segmentation. This is because Dice is a measure of the spatial overlap between the ground truth and segmentations, while the ICC compares volumes without considering spatial location. Both metrics provide useful but complementary information. Dice as a measure is more sensitive to differences in segmentation along the boundary of the lesion. Thus, if manual and automatic segmentations differed along the boundary, this can compromise the dice measure which is dependent on the degree of overlap. Furthermore, Dice coefficient can be sensitive to lesion size in that a few voxel difference in the location of the boundary can substantially reduce the Dice for small lesions but not as much for large lesions. In contrast, ICC of volume is less sensitive to boundary effects. If automatic segmentation was more conservative at some boundaries and more liberal at other boundaries compared with manual segmentation, these effects would cancel out and there would still be high concordance between manual and automatic volumes. Indeed, this is the case, which is why the ICC values were higher than the Dice scores.
There are some limitations to our study. First, the expert manual volume segmentations for each patient were derived from a single rater, which limits our ability to assess interrater variability of volume segmentation. Future studies could incorporate segmentations from multiple raters for segmentation. Second, our postoperative patient cohort contained imaging from only 54 patients from a single institution. Additional studies could utilize a larger, multi-institutional cohort as well as assess performance early after surgery versus later after surgery as well as in responsive versus progressive disease. Third, our approach utilized a single neural network architecture without comparison with other approaches. Future work could explore the clinical utility of other neural network architectures as well as ensembles of neural network models.32 Furthermore, only patients with residual enhancing tumor of a certain size after surgery were enrolled in the clinical trials, which limits applicability to smaller tumors which may be harder to segment. Additionally, patient cohorts with 2D or 3D MR imaging were used in this study, as 3D MR imaging is not always available at all institutions. The utilization of only 3D MR imaging would further improve the reliability of bidirectional and volume measures.8 Lastly, the confidence of the algorithm in its segmentations could be added to our pipeline to flag segmentations that require further verification from clinicians.33 This would allow for more reliable integration into clinical workflows. Overall, our study shows that automated measures of tumor burden are highly reproducible and can reflect changes in tumor burden during the course of treatment. These automated tools could potentially be integrated in routine clinical care and imaging analyses performed as part of clinical trials and significantly enhance our accuracy in assessing treatment response.
We developed an open-source, fully automatic pipeline for brain extraction, tumor segmentation, and RANO measurements and applied it to a large, multi-institutional preoperative and postoperative glioma patient cohort. We showed that automated volume and AutoRANO measurements are highly reproducible and are in agreement with human experts in terms of change in tumor burden during the course of treatment. This tool may be helpful in clinical trials and clinical practice for expediting measurement of tumor burden in the evaluation of treatment response, decreasing clinician burden associated with manual tumor segmentation and decreasing interobserver variability. Furthermore, our study serves as a proof of concept for automated tools in the clinic with potential application to other tumor pathologies.
Funding
Research reported in this publication was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number 5T32EB1680 and by the National Cancer Institute (NCI) of the National Institutes of Health under Award Number F30CA239407 to K.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Other funding included National Institutes of Health grants R01CA129371 to T.T.B.; K23CA169021 to E.R.G.; and U01 CA154601, U24 CA180927, and U24 CA180918 to J.K-C.; the National Natural Science Foundation of China (81301988 to L.Y., 81472594/81770781 to X.L., 81671676 to W.L.), and the Shenghua Yuying Project of Central South University to L.Y.
This research was carried out in whole or in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by NIBIB, National Institutes of Health.
Supplementary Material
Acknowledgments
We would like to acknowledge the GPU computing resources provided by MGH and BWH Center for Clinical Data Science.
Conflict of interest statement.
P.Y.W. reports receiving research support from Agios, Astra Zeneca, Beigene, Eli Lily, Genentech/Roche, Kazia, Merck, MediciNova, Novartis, Oncoceutics, Sanofi-Aventis, Vascular Biogenics, and VBI Vaccines; is on the advisory board for Abbvie, Agios, Astra Zeneca, Agios, Eli Lilly, Genentech/Roche, Immunomic Therapeutics, Kayetek, Puma, Taiho, Vascular Biogenics, Deciphera, and VBI Vaccines; and is a speaker for Merck and Prime Oncology.
T.T.B. has received research support from Champions Biotechnology, AstraZeneca, Pfizer, and Millennium. He is on the advisory board for UpToDate, Inc, and is a consultant for Genomicare, Merck, NXDC, Amgen, Roche, Oxigene, Foundation Medicine, Proximagen. He has provided CME lectures or material for UpToDate, Research to Practice, Oakstone Medical Publishing, and Imedex.
B.R.R. is on the advisory board for ARIA, Butterfly, Inc, DGMIF (Daegu-Gyeongbuk Medical Innovation Foundation), QMENTA, and Subtle Medical, Inc; is a consultant for Broadview Ventures, Janssen Scientific, ECRI Institute, GlaxoSmithKline, Hyperfine Research, Inc, Peking University, Wolf Greenfield, Superconducting Systems, Inc, Robins Kaplin, LLC, Millennium Pharmaceuticals, GE Healthcare, Siemens, Quinn Emanuel Trial Lawyers, Samsung, and Shenzhen Maternity & Child Healthcare Hospital; and is a founder of BLINKAI Technologies, Inc.
J.K. is a consultant/advisory board member for Infotech, Soft. The other authors declare no competing interests.
Authorship statement.
Chang had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Chang, Beers, Bai, Arnaout, Rosen, Gerstner, Yang, Huang, Kalpathy-Cramer. Acquisition, analysis, or interpretation of data: Chang, Beers, Bai, Brown, Ly, Li, Senders, Kavouridis, Boaro, Su, Bi, Rapalino, Liao, Shen, Zhou, Xiao, Wang, Zhang, Pinho, Batchelor, Boxerman, Gerstner, Huang, Kalpathy-Cramer Drafting of the manuscript: Chang, Beers, Bai, Brown, Ly, Rapalino, Boxerman, Gerstner, Huang, Kalpathy-Cramer. Critical revision of the manuscript for important intellectual content: All authors. Supervision: Wen, Batchelor, Arnaout, Rosen, Gerstner, Yang, Huang, Kalpathy-Cramer.
References
- 1. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. . Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen PY, Macdonald DR, Reardon DA, et al. . Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology Working Group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 3. Huang RY, Rahman R, Ballman KV, et al. . The impact of T2/FLAIR evaluation per RANO criteria on response assessment of recurrent glioblastoma patients treated with bevacizumab. Clin Cancer Res. 2016;22(3):575–581. [DOI] [PubMed] [Google Scholar]
- 4. Vos MJ, Uitdehaag BM, Barkhof F, et al. . Interobserver variability in the radiological assessment of response to chemotherapy in glioma. Neurology. 2003;60(5):826–830. [DOI] [PubMed] [Google Scholar]
- 5. Boxerman JL, Zhang Z, Safriel Y, et al. . Early post-bevacizumab progression on contrast-enhanced MRI as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 Central Reader Study. Neuro Oncol. 2013;15(7):945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deeley MA, Chen A, Datteri R, et al. . Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: a multi-expert study. Phys Med Biol. 2011;56(14):4557–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barboriak DP, Zhang Z, Desai P, et al. . Interreader variability of dynamic contrast-enhanced MRI of recurrent glioblastoma: the multicenter ACRIN 6677/RTOG 0625 study. Radiology. 2018;181296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellingson BM, Bendszus M, Boxerman J, et al. ; Jumpstarting Brain Tumor Drug Development Coalition Imaging Standardization Steering Committee Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro Oncol. 2015;17(9):1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang B, Chang K, Ramkissoon S, et al. . Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19(1):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grossmann P, Narayan V, Chang K, et al. . Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smits M, van den Bent MJ. Imaging correlates of adult glioma genotypes. Radiology. 2017;284(2):316–331. [DOI] [PubMed] [Google Scholar]
- 12. Zhou H, Vallières M, Bai HX, et al. . MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19(6):862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang K, Bai HX, Zhou H, et al. . Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24(5):1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kamnitsas K, Ledig C, Newcombe VFJ, et al. . Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 15. Havaei M, Davy A, Warde-Farley D, et al. . Brain tumor segmentation with Deep Neural Networks. Med Image Anal. 2017;35:18–31. [DOI] [PubMed] [Google Scholar]
- 16. Havaei M, Davy A, Warde-Farley D, et al. . Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. [DOI] [PubMed] [Google Scholar]
- 17. Kleesiek J, Urban G, Hubert A, et al. . Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage. 2016;129:460–469. [DOI] [PubMed] [Google Scholar]
- 18. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 19. Ségonne F, Dale AM, Busa E, et al. . A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22(3):1060–1075. [DOI] [PubMed] [Google Scholar]
- 20. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. [DOI] [PubMed] [Google Scholar]
- 21. Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6(2):129–142. [DOI] [PubMed] [Google Scholar]
- 22. Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging. 2011;30(9):1617–1634. [DOI] [PubMed] [Google Scholar]
- 23.Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography. 2016;2(4):334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clark K, Vendt B, Smith K, et al. . The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ina Ly K, Vakulenko-Lagun B, Emblem KE, et al. . Probing tumor microenvironment in patients with newly diagnosed glioblastoma during chemoradiation and adjuvant temozolomide with functional MRI. Sci Rep. 2018;8(1):17062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Batchelor TT, Gerstner ER, Emblem KE, et al. . Improved tumor oxygenation and survival in glioblastoma patients who show increased blood perfusion after cediranib and chemoradiation. Proc Natl Acad Sci U S A. 2013;110(47):19059–19064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wen PY, Macdonald DR, Reardon DA, et al. . Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology Working Group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 28. Beers A, Chang K, Brown J, Gerstner E, Rosen B, Kalpathy-Cramer J. Sequential neural networks for biologically-informed glioma segmentation. In: Angelini ED, Landman BA, eds. Medical Imaging 2018: Image Processing. Vol. 10574 Houston, TX: SPIE; 2018:108. [Google Scholar]
- 29. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-net: Learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, eds. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Vol 9901 Athens, Greece: LNCS; 2016:424–432. [Google Scholar]
- 30. Beers A, Brown J, Chang K, et al. . DeepNeuro: an open-source deep learning toolbox for neuroimaging. 2018. http://arxiv.org/abs/1808.04589. Accessed August 30, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiograplc response assessment in glioblastoma clinical trials. Neurotherapeutics. 2017;14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou ZH, Wu J, Tang W. Ensembling neural networks: many could be better than all. Artif Intell. 2002;137(1–2):239–263. [Google Scholar]
- 33. Gal Y, Ghahramani Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. Proc 33rd Int Conf Int Conf Mach Learn, Vol 48. 2016:1050–1059. https://dl.acm.org/citation.cfm?id=3045502. Accessed October 23, 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The codes for preprocessing, U-Net architecture, and postprocessing are publicly available at: https://github.com/QTIM-Lab/DeepNeuro. Accessed June 23, 2019.30