Abstract
Background
Glioblastoma (GBM) represents an aggressive cancer type with a median survival of only 14 months. With fewer than 5% of patients surviving 5 years, comprehensive profiling of these rare patients could elucidate prognostic biomarkers that may confer better patient outcomes. We utilized multiple molecular approaches to characterize the largest patient cohort of isocitrate dehydrogenase (IDH)–wildtype GBM long-term survivors (LTS) to date.
Methods
Retrospective analysis was performed on 49 archived formalin-fixed paraffin embedded tumor specimens from patients diagnosed with GBM at the Mayo Clinic between December 1995 and September 2013. These patient samples were subdivided into 2 groups based on survival (12 LTS, 37 short-term survivors [STS]) and subsequently examined by mutation sequencing, copy number analysis, methylation profiling, and gene expression.
Results
Of the 49 patients analyzed in this study, LTS were younger at diagnosis (P = 0.016), more likely to be female (P = 0.048), and MGMT promoter methylated (UniD, P = 0.01). IDH-wildtype STS and LTS demonstrated classic GBM mutations and copy number changes. Pathway analysis of differentially expressed genes showed LTS enrichment for sphingomyelin metabolism, which has been linked to decreased GBM growth, invasion, and angiogenesis. STS were enriched for DNA repair and cell cycle control networks.
Conclusions
While our findings largely report remarkable similarity between these LTS and more typical STS, unique attributes were observed in regard to altered gene expression and pathway enrichment. These attributes may be valuable prognostic markers and are worth further examination. Importantly, this study also underscores the limitations of existing biomarkers and classification methods in predicting patient prognosis.
Keywords: copy number alterations, glioblastoma, methylation, mutation analysis, RNA sequencing
Key Points.
Genomic profiles of 12 LTS and 37 STS IDH-wildtype GBMs were established.
Enrichment for DNA repair and cell cycle pathways was observed in patients with short survival.
Sphingomyelin-related pathways were enriched in LTS.
Importance of the Study.
Glioblastoma is a lethal cancer for which durable therapeutic responses are lacking. Identifying the genetic features that promote resistance will enable the development of therapeutic strategies that address the limitations of current standards of care. This report compares patients with short survival times with those who lived more than 5 years following diagnosis to broaden our understanding of molecular features associated with prognosis and outcome. To date, these features have been limited to the O6-methylguanine-DNA methyltransferase gene, IDH, and the telomerase reverse transcriptase gene. Comprehensive characterization of IDH-wildtype patients revealed remarkable similarity at the level of mutations, copy number alterations, and methylation profiling; however, global gene expression data demonstrated a propensity to upregulate DNA repair and cell cycle pathways in GBM patients with lower survival rates. Conversely, sphingomyelin-related pathways were enriched in long-term survivors. Evaluating the role of such pathways in tumor growth and therapeutic sensitivity is warranted if these targets are to be valuable in GBM.
Glioblastoma (GBM) is the most commonly diagnosed primary malignant brain tumor in adults and overall prognosis remains poor for a majority of patients. It remains unclear how genomic events that occur in GBM impact therapeutic efficacy and survival. Prior to 2005, the standard of care was confined to surgical resection and radiation therapy. A pivotal phase III trial demonstrated that the addition of concurrent and adjuvant temozolomide (TMZ) to a standard radiation schedule (hereafter denoted the Stupp regimen) produced better overall survival (OS) than radiation alone in newly diagnosed GBM patients (95% CI: 13.2–16.8 mo vs 11.2–13.0 mo).1 Results from the subsequent phase III EF-14 trial found that the addition of tumor treating fields to TMZ not only provided longer median OS than TMZ alone (20.9 mo vs 16.0 mo; 95% CI: 0.53–0.76) but also extended 5-year survival rates from 5% to 13%.2 However, beyond these treatments, there has been minimal progress in extending survival for patients with GBM.
O6-methylguanine-DNA methyltransferase (MGMT) methylation status has been shown to be associated with improved survival and TMZ sensitivity, and as a result, MGMT testing became a part of routine clinical practice for newly diagnosed GBM.3 Beyond MGMT methylation, there are several other important prognostic biomarkers. Mutations in isocitrate dehydrogenase (IDH) 1 and 2 occur in up to 10% of primary GBMs and are associated with substantially longer OS (median 3.8 y vs 1.1 y).4 However, IDH mutations are typically observed in younger patients and are more indicative of secondary GBMs.4 This observation prompted the World Health Organization (WHO) to modify its existing central nervous system tumor classification scheme to distinguish IDH-wildtype from IDH-mutant GBM.5 Other molecular characteristics, such as promoter-specific telomerase reverse transcriptase (TERT) mutations, have also been observed in GBM more frequently than other tumor types.6 The presence of TERT promoter mutations has been associated with poorer OS in GBM.7 Collectively, MGMT, IDH, and TERT represent the few known prognostic markers associated with patient outcome in GBM.
Large-scale transcriptomic characterization has been used in an attempt to establish clinically relevant GBM subtypes.8,9 While the most recent analyses by The Cancer Genome Atlas (TCGA) recognize 3 mRNA subtypes, significant survival differences across subtypes were not observed.9 Similarly, epigenetic analysis has revealed hypermethylation of a subset of loci, more commonly referred to as glioma cytosine-phosphate-guanine island methylator phenotype (G-CIMP), which is strongly associated with IDH status, younger age, and prolonged survival.10,11
While molecular characterization has provided a better understanding of the molecular basis of GBM, it remains a clinical challenge to identify patients who will have long-term survival.8,12,13 To further understand the molecular features that are associated with long-term survival in GBM patients, we performed transcriptomic, genetic, and epigenetic profiling on tissue samples from IDH-wildtype GBM patients as classified by the updated 2016 WHO criteria.
Materials and Methods
Patients
This single-institution study was approved by the Mayo Clinic institutional review board (protocol #14-002252). Long-term survivors (LTS) were selected for inclusion if they had an initial diagnosis of GBM between December 1995 and October 2008, were at least 18 years of age at diagnosis, and survived at least 5 years. Central pathology review was performed by the Mayo Clinic Department of Pathology using the 2016 WHO criteria. Patients in the cohort of short-term survivors (STS) were diagnosed with GBM between May 2001 and September 2013 and later classified as IDH-wildtype GBM. All patients were required to have survived more than 6 months and less than 12 (TERT-mutant) or 24 months (TERT-wildtype).
Nucleic Acid Isolation
Ten micron section slides were cut from formalin fixed, paraffin embedded (FFPE) specimen blocks on to uncharged slides. Immediately after completion, representative hematoxylin and eosin (H&E) slides for each sample were reviewed by a certified neuropathologist to identify tumor tissue. All normal tissue was discarded and RNA was extracted from the remaining tumor tissue using the Qiagen AllPrep RNA/DNA FFPE kit according to the manufacturer’s instructions. Supplementary Table 1 reports the sample size for each genomic technology reported in this study.
Quantitative Reverse Transcription Polymerase Chain Reaction
Quantitative reverse transcription PCR was performed as previously reported.14 For more information, see the Supplementary Methods.
RNA Sequencing
A total of 1 µg of RNA was extracted from FFPE tumor samples and used to construct stranded cDNA libraries using the TruSeq RNA Access preparation protocol (Illumina). Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using a HiSeq 2500 instrument. Each run was a 76 basepair paired-end with an 8-base index barcode read. Data were analyzed using the Broad Picard Pipeline, which includes de-complexing and data aggregation.
High-throughput sequencing raw count data were processed with DESeq2.15 Sample read counts were normalized by size factor that was calculated using the median-of-ratios method in DESeq2.15 Median absolute deviation (MAD) of normalized counts was calculated for each gene across all samples. Hierarchical clustering was applied using the 200 genes with the largest MAD value (distance = Euclidean, clustering = complete).
For differential gene expression analysis, FASTQ formatted raw files for each sample were mapped and aligned in reference to hg19. Further processing of the RNA sequencing data was done using Map-Rseq, a Mayo developed bioinformatic pipeline.16 For more details, see the Supplementary Methods.
Immune Scoring Using ESTIMATE
See the Supplementary Methods.17
Illumina MethylationEPIC Array
The Infinium MethylationEPIC Beadchip was used to evaluate methylation in 31 IDH-wildtype GBM patient tumors (8 LTS, 23 STS). For details, see the Supplementary Methods.
OncoScan
Somatic copy number variation (CNV) and copy neutral loss of heterozygosity (cnLOH) were detected using OncoScan (Affymetrix) run in accordance with the manufacturer’s instructions. CNV and cnLOH were manually analyzed using Chromosome Analysis Suite 2.1 (Affymetrix). CNV burden, overall and separately for each chromosome, was calculated for each subject by summing the total number of CNV alterations, and comparisons were made between LTS and STS subjects using a Kruskal–Wallis test.
Statistical Analysis
Patient characteristics were compared across LTS and STS groups using Kruskal–Wallis and chi-square tests as appropriate. Cumulative survival probabilities were estimated using the Kaplan–Meier method. Differential probe methylation was compared using the Wilcoxon non-parametric test.
Results
A total of 1164 patients were identified in the Mayo Clinic Rochester archives with a primary diagnosis of GBM between April 1989 and September 2013. Of the 1164 patients, 57 (4.9%) survived at least 5 years. H&E-stained slides were available for 40 patients (3.4%). Following central review by a neuropathologist, 22 had sufficient tissue for molecular analysis (1.9%): 12 IDH-wildtype tumors (1.0%) and 10 IDH-mutant tumors (0.9%) (Supplementary Fig. 1). Since IDH mutation confers improved survival, the focus herein was to identify molecular features associated with LTS in IDH-wildtype GBM. These tumors were also stratified by TERT promoter status to determine the influence of these mutations on the molecular profile of a given tumor.
The STS cohort accurately represented the broader population of 1164 patients with GBM diagnoses at Mayo Clinic (Supplementary Fig. 2). Clinical data from both LTS and STS are shown in Table 1. LTS were younger at diagnosis (median 48 vs 60 y, P = 0.016) and more likely to be female (58% vs 27%, P = 0.048). Differences in tumor location (P = 0.14), extent of resection (P = 0.67), and tumor size (P = 0.99) were not observed between LTS and STS; the incidence of TERT mutations and the distribution of gene expression subtype were also similar (P = 0.35 and P = 0.28, respectively). Year of diagnosis (P < 0.001) was significantly different between LTS and STS. Seven LTS were diagnosed prior to 2005 when TMZ became the standard of care, while only one STS patient was from that era.
Table 1.
Patient demographics
| LTS (N = 12) | STS (N = 37) | P-value | |
|---|---|---|---|
| Age, y | 0.016a | ||
| Mean (SD) | 48 (15) | 60 (9) | |
| Median | 50 | 61 | |
| Q1, Q3 | 42, 58 | 53, 66 | |
| Range | 19–70 | 39–80 | |
| Sex | 0.048b | ||
| F | 7 (58%) | 10 (27%) | |
| M | 5 (42%) | 27 (73%) | |
| Resection | 0.67b | ||
| Gross total | 7 (58%) | 19 (51%) | |
| Subtotal | 5 (42%) | 18 (49%) | |
| Tumor size, cm | 0.99a | ||
| Mean (SD) | 4.4 (1.8) | 4.6 (1.4) | |
| Median | 5.0 | 4.3 | |
| Q1, Q3 | 3.3, 5.8 | 3.8, 5.6 | |
| Range | 0.8–6.8 | 2.3–7.7 | |
| Not available | 1 | 2 | |
| Location | 0.14a | ||
| Frontal | 6 (50%) | 15 (41%) | |
| Occipital | 0 | 1 (3%) | |
| Parietal | 1 (8%) | 7 (19%) | |
| Temporal | 1 (8%) | 11 (30%) | |
| Includes >1 lobe | 4 (33%) | 3 (8%) | |
| Molecular features | |||
| TERT mutations | 9 (75%) | 32 (87%) | 0.35b |
| ATRX mutations | 0 | 2 (5%) | 0.41b |
| Gene expression subtype 9 | 0.28b | ||
| Mesenchymal | 1 (13%) | 9 (39%) | |
| Classical | 6 (75%) | 10 (44%) | |
| Proneural | 1 (13%) | 4 (17%) | |
| Not available | 4 | 14 | |
| G-CIMP status | 1.00b | ||
| Negative | 8 (100%) | 23 (100%) | |
| Positive | 0 | 0 | |
| Not available | 4 | 14 | |
| Diagnosis | <0.001b | ||
| Pre-2005 | 7 (58%) | 1 (3%) | |
| Post-2005 | 5 (42%) | 36 (97%) | |
| Treatment | 0.04b | ||
| Radiation with concurrent/adjuvant chemotherapy* | 6 (50%) | 31 (84%) | |
| Radiation with concurrent chemotherapy | 3 (25%) | 2 (5%) | |
| Radiation with adjuvant chemotherapy | 2 (17%) | 0 | |
| Radiation only | 0 | 1 (3%) | |
| Chemotherapy only | 0 | 1 (3%) | |
| Unknown | 1 | 2 |
*5 LTS were on Stupp. In only 1 of these patients was tumor diagnosed prior to 2005. aKruskal–Wallis rank sum test; bPearson’s chi-squared test.
To further confirm that our patient cohorts represent the broader GBM population, DNA sequencing was performed using a custom gene panel (Supplementary Table 2) that targets the coding regions of 50 genes commonly altered in central nervous system tumors (n = 49, 12 LTS and 37 STS). Results are shown in Supplementary Fig. 3. TERT was the most frequently mutated gene (75% LTS vs 92% STS), followed by phosphatase and tensin homolog (PTEN) (50% LTS vs 35% STS) and tumor protein 53 (TP53) (33% LTS vs 24% STS). As expected, TERT and alpha thalassemia/mental retardation syndrome X-linked (ATRX) mutations were mutually exclusive with only TERT-wildtype STS possessing pathogenic ATRX mutations (50%). Pathogenic epidermal growth factor receptor (EGFR) mutations occurred at similar rates to those reported by TCGA (25% LTS vs 27% STS vs 21% TCGA GBMs). Collectively, these mutation profiles are consistent with conventional GBMs.
While virtually all patients received radiation therapy, chemotherapy was more varied (Table 1, Supplementary Table 3). STS were more likely to be prescribed longer durations of chemotherapy, with 84% receiving treatment both during and after radiation compared with only 50% of LTS. The remaining LTS were largely enrolled in chemotherapy regimens limited to radiation or the adjuvant setting (concurrent with radiation 25% vs 5% STS; adjuvant 17% vs 0% STS). These treatment differences were shown to be significant (P = 0.04). More specifically, STS were exclusively enrolled into TMZ-based regimens, while LTS were just as likely to receive nitrosourea compounds as TMZ (36% vs 42%, respectively). While almost 75% of LTS did not receive the current standard (Stupp) regimen, LTS still achieved extended survival. Collectively, these data suggest that the choice of therapeutic intervention alone cannot explain the survival achieved by the LTS cohort.
Copy Number Variation
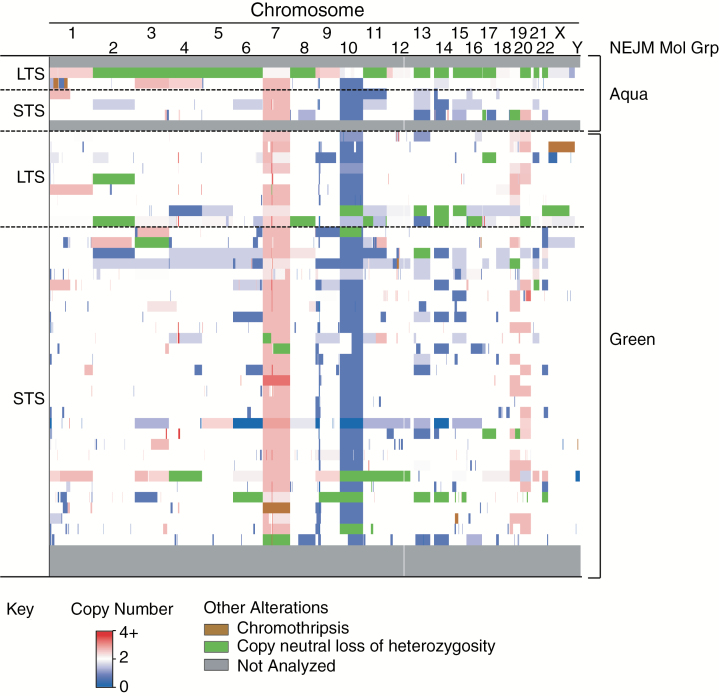
Assessment of genome-wide copy number changes in 44 IDH-wildtype GBM (11 LTS, 33 STS) demonstrated classic GBM hallmarks, including chromosome 7 gains (82% LTS vs 82% STS), EGFR amplification (64% LTS vs 67% STS), chromosome 10 loss (73% LTS vs 79% STS), and cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) homozygous loss (55% LTS vs 67% STS) (Fig. 1). Overall genome-wide CNV burden was similar across groups (24.6 LTS vs 21.5 STS, P = 0.71) except for chromosome 11 (1.18 STS vs 0.18 LTS, P = 0.017) (Supplementary Tables 4 and 5). CNVs varied in both size and location, which supports the idea that few, if any, genes were universally affected across patients.
Figure 1.
Copy number profile of IDH-wildtype GBM. Copy number alteration data were grouped by survival and TERT status. Copy number variations and copy neutral loss of heterozygosity calls were identified by manual examination of OncoScan array data using Chromosome Analysis Suite.
RNA Sequencing
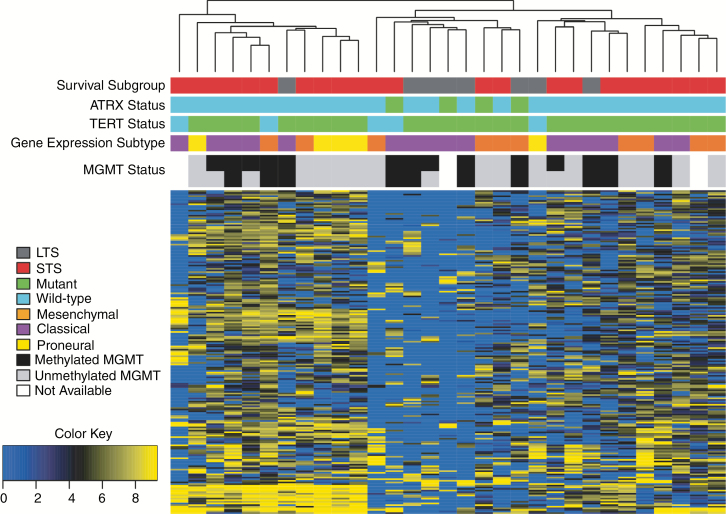
Global RNA expression patterns were assessed in 31 IDH-wildtype GBMs (8 LTS, 23 STS). Unsupervised hierarchal clustering was performed on the 200 most variable genes, and no discernible expression patterns related to survival group, gene expression subtype, or MGMT methylation were observed (Fig. 2). Differentially expressed gene (DEG) analysis of 26 TERT-mutated, IDH-wildtype GBMs (7 LTS, 19 STS) revealed 106 DEGs between LTS and STS (Supplementary Fig. 4a, Supplementary Table 6). Genes upregulated in STS included those previously linked to GBM growth (CEACAM1, MUC4) and poorer prognosis (ALDH1A3, RAB38, and MIR335).18–23 Of these candidate DEGs, CD300LF and RAB38 were upregulated in STS and significantly associated with OS for all GBMs in TCGA (P = 0.0068 and P = 0.0066, respectively). No additional DEGs were observed when this analysis was expanded to include the 5 IDH-wildtype GBMs that were also TERT-wildtype (Supplementary Fig. 4b, Supplementary Table 7). Quality control metrics are provided in Supplementary Table 8. Analysis of the database of TCGA (13 LTS, 90 STS) validated only a single DEG (FGF9); however, only 6 of these LTS had IDH-wildtype GBMs.
Fig. 2.
Unsupervised hierarchal clustering of gene expression in all IDH-wildtype GBM. Following removal of 4 poor quality samples, raw counts were normalized using the R package DEseq2 and log2(norm_count + 1) transformed. Median absolute deviation (MAD) was used to select the 200 most variant genes across all samples and the hclust() function (R package, v3.3) with complete distance method was applied. Note: MGMT status was predicted using STP27 (top row) and UniD (bottom row) models.
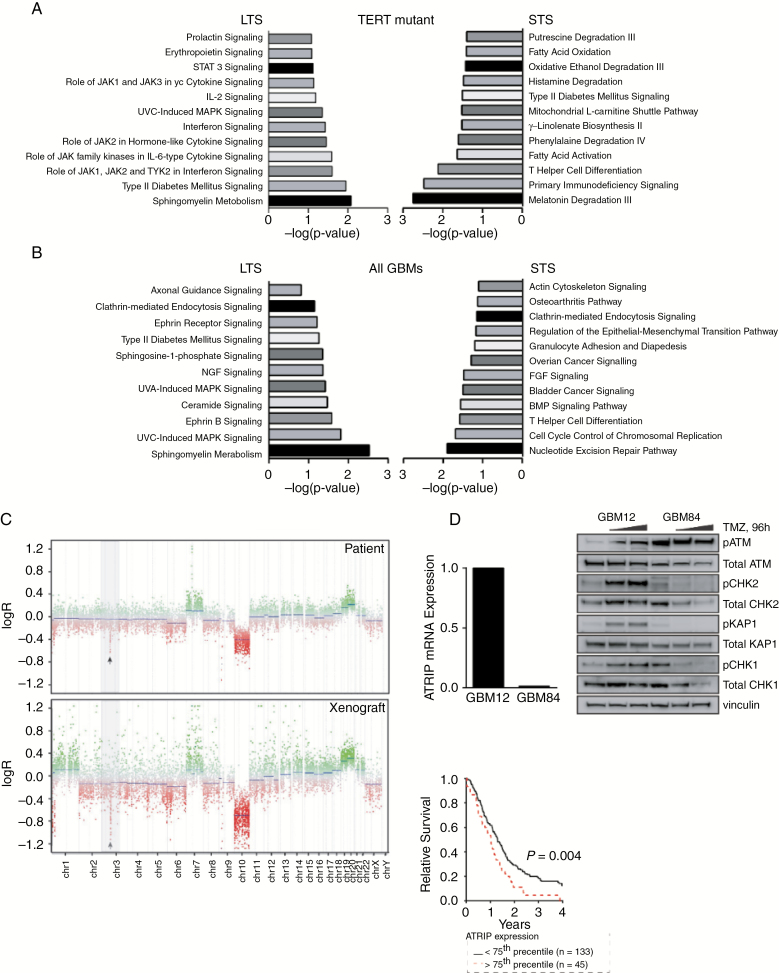
Ingenuity pathway analysis (IPA) of the 106 candidate DEGs in the 26 TERT-mutated, IDH-wildtype GBMs demonstrated distinct enrichment for sphingomyelin metabolism in the LTS group, while Janus family kinases in interferon and cytokine signaling were also highlighted (Fig. 3A, left). Genes in the STS group predominantly function in T helper cell differentiation and multiple protein degradation pathways (Fig. 3A, right). Further examination of immune cell signatures failed to detect differences in stromal and immune infiltration between STS and LTS (Supplementary Fig. 5).
Fig. 3.
DNA repair defects can improve OS in GBM. Differentially expressed genes upregulated in LTS or STS were uploaded into IPA and core analysis of expression was performed for (A) IDH-wildtype GBMs with TERT promoter mutations and (B) all IDH-wildtype GBMs. (C, D) One LTS and derivative PDX (GBM84) demonstrated an ATRIP deficiency and profound DNA repair signaling defects following TMZ dosing compared with another MGMT methylated PDX (GBM12). (E) Analysis of REMBRANDT expression data showed that decreased ATRIP expression correlated with better survival in GBM patients.
Enrichment of sphingomyelin metabolism in LTS was also observed following IPA analysis of all IDH-wildtype GBMs (8 LTS, 23 STS). This pathway, in addition to ceramide signaling, represented 2 of the top 4 hits (Fig. 3B, left). Biological functions and disease annotations illustrated enrichment for brain cancer susceptibility, fibroblast necroptosis, and apoptosis of cortical astrocytes. Conversely, nucleotide excision repair (NER), cell cycle control, and T helper cell differentiation were top canonical pathways observed in DEGs within STS (Fig. 3B, right). NER represents the most versatile DNA repair system in humans, and while not classically viewed as a mediator of TMZ lesion repair, can assist in the repair of these lesions.24 These contributions could facilitate resistance, as NER inhibition enhances sensitivity to alkylating agents in multiple myeloma.25 Along similar lines, an LTS patient from our study had a single copy deletion of the ataxia telangiectasia and Rad3-related (ATR) interacting protein (ATRIP) locus on chromosome 3 and associated hypermethylation of the remaining allele; this patient achieved more than 60 months progression-free survival following TMZ-based chemoradiation. The Mayo patient-derived xenograft (PDX) from this patient’s primary tumor (GBM84) demonstrated profound defects in TMZ-induced DNA damage signaling (Fig. 3C, D). Validation using the Repository of Molecular Brain Neoplasia Data (REMBRANDT) database associated decreased ATRIP expression with better survival (Fig. 3E). These data suggest that defects in DNA repair proficiency or activation of pathways implicated in reduced growth or pro-death may contribute to long-term survival in GBM.
Methylation
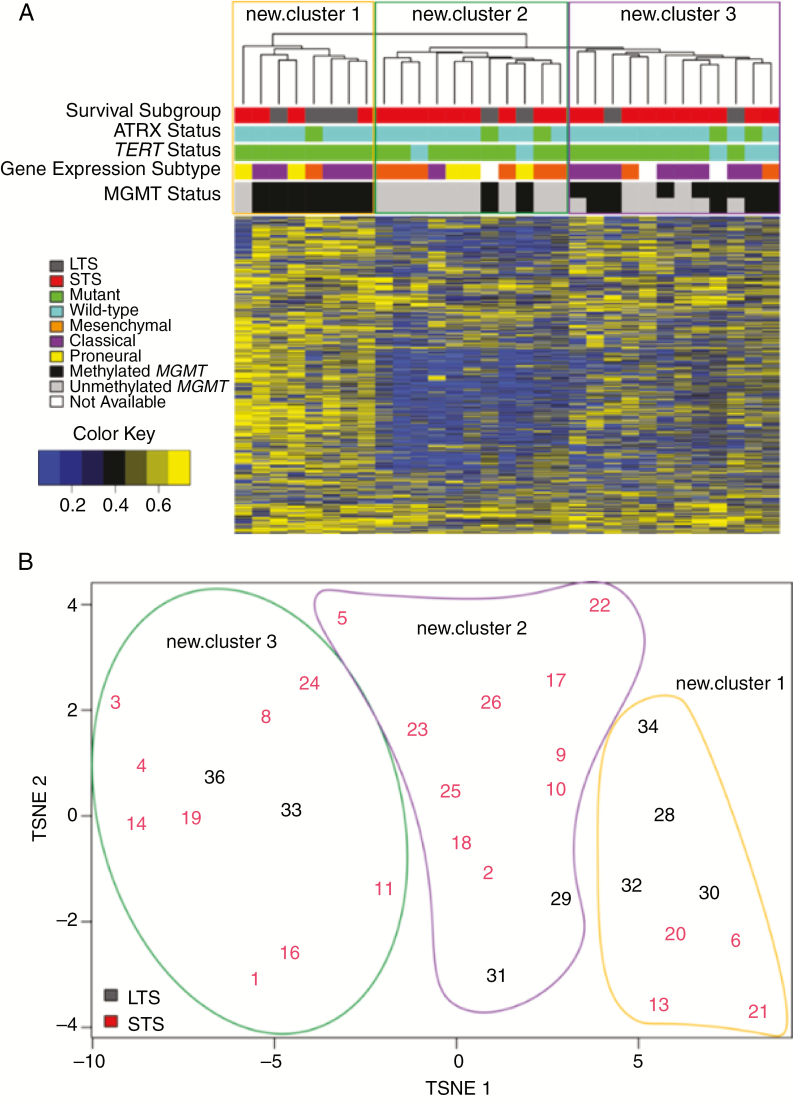
Global methylation patterns were assessed in 31 IDH-wildtype GBMs (8 LTS, 23 STS) using the Infinium MethylationEPIC 850K methylation platform (Fig. 4). MGMT methylation was more common in LTS versus STS (100% vs 44% using STP27,26P = 0.005; 88% vs 30% using UniD,27P = 0.005), which confirms previous studies. Unsupervised hierarchal clustering did not identify methylation patterns that universally related to survival, ATRX or TERT status, gene expression subtype, or MGMT status (Supplementary Fig. 6, Supplementary Table 9).
Fig. 4.
(A) Unsupervised hierarchal clustering and (B) t-distributed stochastic neighbor embedding (t-SNE) plots of top 2000 methylation probes with the highest MAD value in all IDH-wildtype GBM. MAD values were calculated for each probe across all samples. The top 2000 were selected and the complete distance method was applied. Beta values are shown in (A). Randomly assigned sample identifiers are plotted and associated by t-SNE analysis in (B).
Note: MGMT status was predicted using STP27 (top row) and UniD (bottom row) models.
Patient Follow-up
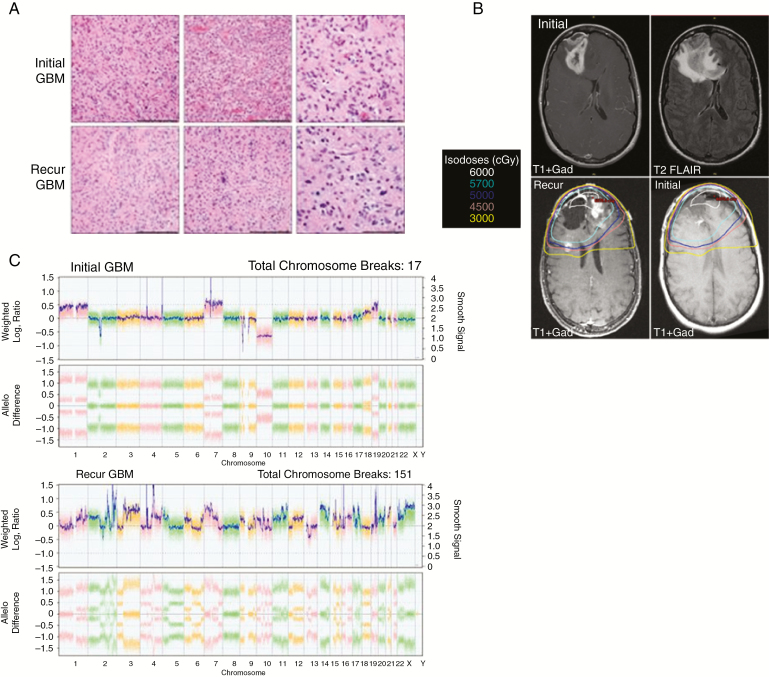
We identified one LTS with a recurrence defined as radiation-induced GBM (Fig. 5). After a gross total resection (Fig. 5A, top row) and chemoradiation, the patient remained disease free for almost 15 years before a presumed biopsy-confirmed recurrence (Fig. 5A, bottom row) within the previous intermediate-dose radiation field (Fig. 5B). Comparative mutation and copy number analysis revealed that the initial GBM had characteristic molecular features of GBM, including but not limited to TERT promoter mutation, EGFR amplification, and CDKN2A deletion. However, the recurrent tumor had a dramatically different molecular profile: TERT-wildtype, platelet derived growth factor receptor A (PDGFRA) amplification and mutation, and no EGFR amplification (Fig. 5C). The tumor had highly complex chromosomal alterations, with a vast number of intrachromosomal breakpoints, a feature more typical of radiation-induced malignancies than primary GBM. Taken together, these data suggest that this event was a radiation-induced GBM and not a local recurrence of the original tumor.
Fig. 5.
LTS patient with radiation-induced GBM. (A) One LTS was initially diagnosed with IDH-wildtype, TERT-mutant GBM (top row, H&E at 20 and 40x magnification). Scale bars reflect 200 µm and 100 µm, respectively. Almost 15 years later, the patient developed an IDH-wildtype, TERT-wildtype GBM (bottom row, H&E at 20 and 40x, respectively). (B) Preoperative MRI scans of the initial GBM (top row) highlight lesions in the right frontal lobe. The recurrence (bottom left) arose within the prior radiation field (dosimetry shown in bottom row) that received between 3000 and 6000 cGy. (C) Chromosomal analysis revealed a highly complex profile in the recurrent GBM relative to the initial tumor.
Discussion
GBM is a highly aggressive, essentially incurable malignancy with 5-year survival rates of 5–13%. Previously published studies have associated the following characteristics with long-term survival: female sex, younger age at diagnosis, higher Karnofsky performance scores, MGMT methylation, IDH mutations, gross total resection with adjuvant chemotherapy, increased immunity and genomic instability, differential retinoic acid, and methylation patterns.28–36 Many of these previous studies analyzed GBM as a single entity without distinction between fundamentally different IDH-wildtype and IDH-mutant GBMs. Because IDH-mutant GBM has been associated with longer survival, we focused on the molecular profiling of IDH-wildtype GBM to understand why some patients experience long-term survival.
Among IDH-wildtype GBM, significant differences between LTS and STS were limited to age at diagnosis, sex, and year of diagnosis. Classic GBM hallmarks including TERT, PTEN, TP53, and EGFR alterations were observed in both groups. Patients were prescribed similar initial treatment regimens regardless of survival, although more STS received the Stupp regimen (87% vs 27% LTS, respectively). After publication of the results of the European Organisation for Research and Treatment of Cancer/National Cancer Institute of Canada trial in 2005, TMZ was approved and adopted into widespread clinical use,1 and this difference in use of TMZ therapy between STS and LTS likely reflects an unintentional bias toward identifying more recently treated STS for inclusion in this study.
MGMT promoter methylation is mechanistically linked to increased TMZ sensitivity and is associated with superior survival in patients treated with the Stupp regimen. Of the 8 LTS samples that received epigenetic profiling, virtually all were MGMT methylated and treated with TMZ-containing regimens; however, only 38% (3 of 8) received the Stupp regimen. Conversely, ~35% of STS were MGMT methylated, which is similar to the prevalence of MGMT methylation across all GBM, and yet these patients garnered little to no survival benefit from TMZ therapy. While rare clones with mismatch repair (MMR) mutations could have been present at initial diagnosis to promote rapid emergence of TMZ resistance, our clinical experience suggests that MMR-deficient tumors are associated with a hypermutator phenotype which was not observed in our 50-gene panel. These data highlight uncertainties about current prognostic and predictive markers beyond MGMT promoter methylation status.
Genome-wide copy number analysis was used to quantify the degree of DNA genomic variability across groups. Differences were observed on chromosome 11, with STS possessing higher copy number burden than LTS. This result validates a previously published study reporting that CNVs on chromosome 11 are associated with reduced survival times in patients with brain tumors.37 This collection of work highlights the potential importance of chromosome 11 aberrations in GBM development. Our study suggests that chromosome 11 alterations can impact patient survival, although further work is needed to validate this finding and elucidate specific gene(s) that may be driving this phenomenon.
Analysis of DEGs showed preferential engagement of sphingomyelin-related metabolic pathways in LTS. Attempts to perform TCGA validation were subject to limited sample availability, with only 13 LTS who fit our survival criteria. Closer examination revealed molecular discrepancies between these TCGA LTS and those from our study. Only 6 were IDH-wildtype, while 2 patients also possessed 1p/19q codeletion, which represents a misclassification of these cases as GBM. The TERT status of these LTS was also unknown. These differences, in conjunction with small sample sizes, limit the value of TCGA-based validation. Alternatively, we performed a literature-based validation to ascertain if any of the networks that were differentially expressed in our patient cohorts had previously been implicated in GBM tumor biology. DEGs in our LTS cohort preferentially enriched in sphingomyelin-related pathways, which have notable connections to GBM growth, invasion, and angiogenesis.38 Activation of ceramide, the basic structural unit of sphingolipids, can produce antitumor effects in response to radiotherapy and a range of chemotherapeutics.39,40 Upregulated sphingomyelin signaling may explain why our LTS cohort demonstrated extreme survival despite only 50% receiving TMZ-based therapy. Conversely, the STS group demonstrated preferential enrichment for DNA repair and cell cycle control mechanisms. Tumors with the capability to coordinate cell cycle entry with the repair of therapy-induced damage could manifest diminished treatment benefits and increased likelihood of disease progression. By extension, pathways that regulate such processes could be relevant chemo- or radiosensitizing drug targets. Consistent with this idea, defects in ATR signaling following TMZ treatment and markedly suppressed ATRIP expression were demonstrated in a PDX model from an LTS. ATR is critically important for recovery from replication stress induced by TMZ, and there is significant interest in developing small-molecule kinase inhibitors of ATR signaling as chemosensitizing agents.41 Taken together, these data suggest that survival in some GBM patients may be linked, at least in part, to upregulated sphingomyelin signaling and/or defects in DNA repair signaling.
Radiation-induced malignancies are a well-known complication of cranial irradiation that typically manifest in patients with low-grade or highly curable malignancies that have extended survivals.42 Similar to our patient, a recent study of radiation-induced gliomas reported a relatively unique molecular signature which lacked TERT promoter mutations, which are typically seen in more than 90% of primary GBM and PDGFRA amplification and/or mutation, in addition to a significant increase in intrachromosomal breaks.43 Consistent with this molecular phenotype, the recurrent GBM in our study was TERT-wildtype, possessed both mutant and amplified PDGFRA, and had a highly altered chromosomal profile. While anecdotal, this case highlights the importance of obtaining tumor tissue for molecular analysis at the time of tumor recurrence in LTS patients if subsequent molecularly targeted therapies are contemplated.
In summary, long-term survival for patients with IDH-wildtype GBM is rare, and these tumors share classic genomic features with GBM from patients with more typical short survival. At the gene expression level, there is a suggestion that pathways involved in sphingomyelin signaling, cell cycle control, and DNA damage response may be potential mediators of extended survival in some GBM. In the future, these data highlight potential therapeutic strategies that should be explored as potential adjuvants to conventional chemoradiation.
Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award R01CA184320 to J.N.S.
Conflict of interest statement.
The authors declare no potential conflicts of interest.
Authorship statement:
The original idea was developed by J.E.P., J.N.S., and R.B.J.
Review of the electronic medical records was conducted by D.M.B., R.S.Y., D.R.
Sample collection and preparation was done by F.B.M., D.H.L., I.J.P., C.G., D.M.B., A.C.M., A.A.C.
Benchwork was completed by D.M.B., S.K.G, G.J.K., T.M.K., J.S.V., and E.G.B.
Data analysis was performed by D.M.B., J.Y., P.A.D., T.M.K., M.L.K., R.A.V., H.S., C.M.I., B.R.K., and J.E.P.
Informatics analysis strategy was developed by J.Y, E.P.S., J.E.P., and P.A.D.
Tables and figures were created by D.M.B., R.A.V., T.M.K., K.L.K., P.A.D., R.B.J., J.E.P., and J.N.S.
The manuscript was drafted by D.M.B., J.N.S., and J.E.P.
All authors read and approved the final manuscript.
Supplementary Material
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Taillibert S, Kanner A, et al. . Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hegi ME, Diserens AC, Godard S, et al. . Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 4. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 6. Killela PJ, Reitman ZJ, Jiao Y, et al. . TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Killela PJ, Pirozzi CJ, Healy P, et al. . Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Q, Hu B, Hu X, et al. . Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56 e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turcan S, Rohle D, Goenka A, et al. . IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marko NF, Toms SA, Barnett GH, Weil R. Genomic expression patterns distinguish long-term from short-term glioblastoma survivors: a preliminary feasibility study. Genomics. 2008;91(5):395–406. [DOI] [PubMed] [Google Scholar]
- 14. Kitange GJ, Carlson BL, Schroeder MA, et al. . Expression of CD74 in high grade gliomas: a potential role in temozolomide resistance. J Neurooncol. 2010;100(2):177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalari KR, Nair AA, Bhavsar JD, et al. . MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshihara K, Shahmoradgoli M, Martínez E, et al. . Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneko S, Nakatani Y, Takezaki T, et al. . Ceacam1L modulates STAT3 signaling to control the proliferation of glioblastoma-initiating cells. Cancer Res. 2015;75(19):4224–4234. [DOI] [PubMed] [Google Scholar]
- 19. Li W, Wu C, Yao Y, et al. . MUC4 modulates human glioblastoma cell proliferation and invasion by upregulating EGFR expression. Neurosci Lett. 2014;566:82–87. [DOI] [PubMed] [Google Scholar]
- 20. Zhang W, Liu Y, Hu H, et al. . ALDH1A3: a marker of mesenchymal phenotype in gliomas associated with cell invasion. PLoS One. 2015;10(11):e0142856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H, Jiang C. RAB38 confers a poor prognosis, associated with malignant progression and subtype preference in glioma. Oncol Rep. 2013;30(5):2350–2356. [DOI] [PubMed] [Google Scholar]
- 22. Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47(1):131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang J, Sun X, Wang W, et al. . Tumor microRNA-335 expression is associated with poor prognosis in human glioma. Med Oncol. 2012;29(5):3472–3477. [DOI] [PubMed] [Google Scholar]
- 24. Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010;2010:543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szalat R, Samur MK, Fulciniti M, et al. . Nucleotide excision repair is a potential therapeutic target in multiple myeloma. Leukemia. 2018;32(1):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van den Bent MJ, Erdem-Eraslan L, Idbaih A, et al. . MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19(19):5513–5522. [DOI] [PubMed] [Google Scholar]
- 27. Yang J, Wang Q, Long L, Ezhilarasan R, Sulman EP. UniD: unified and integrated diagnostic pipeline for malignant gliomas based on DNA methylation data [abstract]. Proceedings of the American Association for Cancer Research Annual Meeting 2017; 2017 Apr 1–5; Washington, DC Philadelphia (PA): AACR;. Vol 77. Cancer Res2017:Abstract nr 3348. [Google Scholar]
- 28. Gerber NK, Goenka A, Turcan S, et al. . Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol. 2014;16(9):1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez R, Schackert G, Yaya-Tur R, Rojas-Marcos I, Herman JG, Esteller M. Frequent hypermethylation of the DNA repair gene MGMT in long-term survivors of glioblastoma multiforme. J Neurooncol. 2007;83(1):91–93. [DOI] [PubMed] [Google Scholar]
- 30. Peng S, Dhruv H, Armstrong B, et al. . Integrated genomic analysis of survival outliers in glioblastoma. Neuro Oncol. 2017;19(6):833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shinawi T, Hill VK, Krex D, et al. . DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics. 2013;8(2):149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barbus S, Tews B, Karra D, et al. . Differential retinoic acid signaling in tumors of long- and short-term glioblastoma survivors. J Natl Cancer Inst. 2011;103(7):598–606. [DOI] [PubMed] [Google Scholar]
- 33. Donson AM, Birks DK, Schittone SA, et al. . Increased immune gene expression and immune cell infiltration in high-grade astrocytoma distinguish long-term from short-term survivors. J Immunol. 2012;189(4):1920–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reifenberger G, Weber RG, Riehmer V, et al. ; German Glioma Network Molecular characterization of long-term survivors of glioblastoma using genome- and transcriptome-wide profiling. Int J Cancer. 2014;135(8):1822–1831. [DOI] [PubMed] [Google Scholar]
- 35. Scott JN, Rewcastle NB, Brasher PM, et al. . Which glioblastoma multiforme patient will become a long-term survivor? A population-based study. Ann Neurol. 1999;46(2):183–188. [PubMed] [Google Scholar]
- 36. Yang W, Warrington NM, Taylor SJ, et al. . Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med. 2019;11(473). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sintupisut N, Liu PL, Yeang CH. An integrative characterization of recurrent molecular aberrations in glioblastoma genomes. Nucleic Acids Res. 2013;41(19):8803–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. [DOI] [PubMed] [Google Scholar]
- 39. Mehta S, Blackinton D, Omar I, et al. . Combined cytotoxic action of paclitaxel and ceramide against the human Tu138 head and neck squamous carcinoma cell line. Cancer Chemother Pharmacol. 2000;46(2):85–92. [DOI] [PubMed] [Google Scholar]
- 40. Myrick D, Blackinton D, Klostergaard J, et al. . Paclitaxel-induced apoptosis in Jurkat, a leukemic T cell line, is enhanced by ceramide. Leuk Res. 1999;23(6):569–578. [DOI] [PubMed] [Google Scholar]
- 41. Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat Rev Cancer. 2018;18(9):586–595. [DOI] [PubMed] [Google Scholar]
- 42. Kim J, Jackman JG, Woodring S, et al. . Second primary cancers in long-term survivors of glioblastoma. Neurooncol Pract. 2019:1–6. doi:10.1093/nop/npz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. López GY, Van Ziffle J, Onodera C, et al. . The genetic landscape of gliomas arising after therapeutic radiation. Acta Neuropathol. 2019;137(1):139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.