Abstract
Sequential metabolic enzymes have long been hypothesized to form multienzyme metabolic complexes to regulate metabolic flux in cells. Although in vitro biochemistry has not been much fruitful to support the hypothesis, advanced biophysical technologies have successfully resurrected the hypothesis with compelling experimental evidence. As biochemistry has always evolved along with technological advancement over the century (e.g. recombinant protein expression, site-directed mutagenesis, advanced spectroscopic and structural biology techniques, etc), there has been growing interests in advanced imaging-based biophysical methods to explore enzymes inside living cells. In this work, we describe how we visualize two phase-separated biomolecular condensates of multienzyme metabolic complexes that are associated with de novo purine biosynthesis and glucose metabolism in living human cells and how imaging-based data are quantitatively analyzed to advance our knowledge of enzymes and their assemblies in living cells. Therefore, we envision that the framework we describe here would be the starting point to investigate other metabolic enzymes and their assemblies in various cell types with an unprecedented potential to comprehend enzymes and their network in native habitats.
Keywords: Purinosome, Glucosome, Metabolon, Metabolic Complex, Phase-separated Biomolecular Condensate, Fluorescence Microscopy, Live-cell Imaging, High-Content Analysis, Single Cell Analysis
Introduction
Discoveries of multienzyme metabolic assemblies in live cells have revived the importance of metabolic enzymes and their complexes as fundamental metabolic units that not only regulate metabolic pathways but also communicate with other cellular processes in living organisms (Schmitt and An, 2017; Srere, 1987). Without a doubt, conventional in vitro biochemical approaches have significantly contributed to our atomic-level understanding of enzyme structure, catalysis and mechanism. However, their intrinsic experimental prerequisite that one must purify the enzymes and should reconstitute their active environment in test tubes has substantially limited to our understanding of their functional interplays with other enzymes in native habitats. Furthermore, alternative functional contributions of metabolic enzymes to a cell, that are commonly referred as moonlighting functions, have been overlooked due to the unknown nature of our knowledge at the time of in vitro experimental designs. Even after novel biological roles of metabolic enzymes are uncovered by in vivo physiological and/or genetic methods, transient nature of their participation in given biology has restricted to expand our understanding of the enzymes and their interactions with other biological processes in live cells. Therefore, to address scientific gaps between in vitro biochemistry and biology, there have been great strides in the field of cellular biochemistry to understand how metabolic enzymes interact with each other to regulate the pathways and interplay with other cellular processes in living cells.
Over the last decade, we have contributed to such endeavors by focusing on the enzymes in de novo purine biosynthesis and glucose metabolism in human cells with strong emphasis on fluorescence live-cell techniques (An et al., 2008; Kohnhorst et al., 2017; Schmitt and An, 2017). Multienzyme metabolic assemblies that are associated with glycolysis have been identified in various tissues/species, including but not limited to protists, yeast, mammalian erythrocytes and human cancer cells (Schmitt and An, 2017). Particularly, we have discovered namely the “glucosome” in which rate-determining enzymes in not only glycolysis but also gluconeogenesis are spatially and temporally organized together to regulate glucose flux at subcellular levels in human cancer cells (Jeon et al., 2018; Kohnhorst et al., 2017). Meanwhile, a multienzyme metabolic assembly that is involved in human de novo purine biosynthesis, so-called the “purinosome,” has been also discovered as an intracellular metabolic assembly in living cells (An et al., 2008; Pedley and Benkovic, 2017). Therefore, we describe here technical details with which we have successfully visualized the intracellular metabolic assemblies (a.k.a. metabolons or metabolic condensates), purinosome and glucosome, in live cells and further provide quantitative biophysical techniques by which we have advanced our understanding of functional metabolic condensates at subcellular levels.
2. Visualization of Metabolic Condensates in Living Human Cells: Purinosome and Glucosome
The enzymes in purinosomes and glucosomes are assembled into biomolecular condensates where the enzymes transiently and reversibly interact with each other in space and time. Not surprisingly, dynamics of such metabolic condensates are highly dependent on environmental and growth conditions of living cells at given metabolic stages. Here, we describe our best practice of how we visualize the metabolic condensates in human cancer cells, HeLa (ATCC CCL-2) and Hs578T (ATCC HTB-126) cells (An et al., 2008; Kohnhorst et al., 2017; Kyoung et al., 2015).
All the materials described here and below are recommended to warm up at room temperature for a few hours before use. The following two items have been used to visualize of the metabolic condensates in HeLa and Hs578T cells (Figure 1).
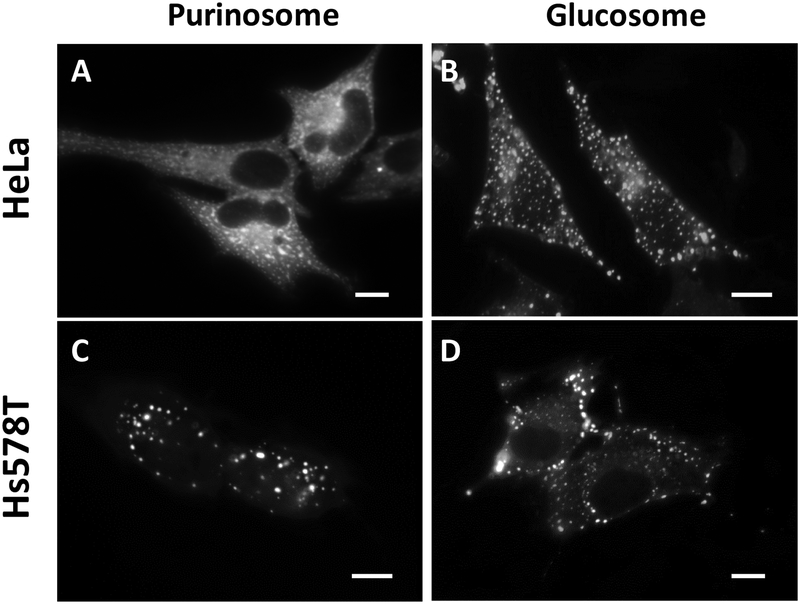
Figure 1. Representative images of metabolic condensates, the purinosome and the glucosome, in HeLa and Hs578T cells.
(A and C) Purinosomes are visualized using hFGAMS-mEGFP. (B and D) Glucosomes are visualized using hPFKL-mEGFP. Scale bars, 10 um.
Buffered Saline Solution (BSS) is used for washing and imaging: 20 mM HEPES (pH 7.4), 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2 and 5.6 mM glucose. BSS should be sterile filtered and can be stored at 4 °C for ~6 months. Bovine serum albumin (1 mg/ml) can be alternatively supplemented for long-term time-lapse imaging if it is desired.
Dialyzed fetal bovine serum (FBS) was prepared in advance before mammalian cell culture. We dialyze regular FBS (Atlanta Biological - Cat# S11550, or Sigma Aldrich - Cat# F2442) against 0.9% NaCl solution at 4 °C for ~ 48 hours and exchange the solution with fresh saline solution every 8–10 hours.
2.1. Mammalian Cell Culture: HeLa and Hs578T Cells
Thaw and plate a vial of cells (~ 1–2 × 106 cells) in tissue culture flask containing standard growth medium (a.k.a. nutrient-rich medium). MEM (Mediatech, Cat# 10–010-CV), 10% FBS and 50 ug/ml gentamicin solutions are used for HeLa cells while DMEM (HyClone, Cat# SH30022.01), 1 mM sodium pyruvate, 10% FBS and 50 ug/ml gentamicin solutions are prepared for Hs578T cells.
Maintain cells in the nutrient-rich medium for about 2 weeks by passing cells every 3–4 days.
Then, seed the 2-week-old cells into a tissue culture flask containing a nutrient-depleted medium (i.e. RPMI 1640 (Mediatech, Cat# 10–040-CV), 10% dialyzed FBS and 50 ug/ml gentamicin solution). After passing cells every 3–4 days for another 2 weeks, prepare samples for live-cell imaging every passage if desired.
2.2. Fluorescence Live-Cell Microscopy
We have used both live and fixed cells for wide-field, confocal and lattice light sheets microscopy. However, due to our focus on real-time dynamics of the metabolic condensates in live cells, we do not describe fixation protocols in details for the metabolic condensates, but commonly used protocols are available in literature (An et al., 2008; Kohnhorst et al., 2017).
Day-1: Sample Dish Preparation
-
1
Seed ~ 2 × 105 cells per glass-bottomed 35 mm Petri dish (MatTek, Cat# P35G-1.5–14-C) in 2 mL of growth medium without antibiotics. For lattice light sheets microscopy (LLSM), multiple 5 mm round coverslips (Warner Instruments, Cat# 64–0700) are placed on a 35 mm Petri dish (MatTEK) prior to seeding cells. Cells should be 70–80% confluent at the time of transfection in Day-2. Therefore, optimal seeding conditions need to be determined for each cell line. In addition, cells should be distributed as evenly as possible in your sample dishes. Heterogeneous distribution negatively affects transfection efficiency. More than 80% populated areas would show significantly poor transfection efficiency. Less than 40% populated cells also experience dramatic cytotoxicity during transfection.
-
2
Incubate the dishes at 37 °C in a CO2 incubator (5% CO2, 95% humidity) for 20–24 hours.
Day-2: Transfection
-
3Prepare transfection complexes as follows:
- Dilute a plasmid (0.8 ug) in 50 uL of Opti-MEM-I Reduced Serum Medium Without Serum (Opti-MEM-I; Gibco, Cat#11058). Mix gently and incubate for 5 min at room temperature. Note that cells’ viability appears to be dependent on a plasmid of choice containing your protein of interest being expressed. Optimal conditions for the amount of a plasmid and the volume of the Opti-MEM-I need to be extensively determined for every plasmid.
- In parallel, dilute 1.5 uL of Lipofectamine 2000 (ThermoFisher Scientific, Cat# 11668019) in 50 uL of Opti-MEM-I. Mix gently and incubate for 5 min at room temperature. Note that the ratio of a plasmid to Lipofectamine 2000 is one of the critical elements to be intensively optimized for every plasmid for each cell line.
- Combine the diluted plasmid into the diluted Lipofectamine 2000 solutions. Mix gently and incubate for 30 min at room temperature.
- Pipet 900 uL of Opti-MEM-I into the tube containing transfection complexes. Mix gently.
-
4During the 30 min incubation at STEP-3c, prepare cell dishes as follows:
- Take out the sample dishes from a CO2 incubator.
- Rinse the dishes with 2 mL of Opti-MEM-I.
- Pipet 1 mL of Opti-MEM-I per dish.
-
5
Add entire transfection complexes (about 1 mL; STEP-3) dropwise to the dish containing cells. Mix gently by rocking the dish back and forth.
-
6
Incubate cells at 37 °C in a CO2 incubator (95% humidity) for 5 hours.
-
7
Exchange the medium with fresh growth medium without antibiotics.
-
8
Incubate cells at 37 °C in a CO2 incubator (95% humidity) for additional 18–24 hours.
Day-3: Wide-Field or Confocal Microscopic Imaging
-
9
Rinse the sample dishes with 2 mL of BSS solution at room temperature.
-
10
Add 2 mL of BSS solution and incubate for at least 10 min at room temperature.
-
11
Repeat twice STEP-10.
-
12
Equilibrate the dishes at room temperature 1–2 hours before imaging.
-
13
Cells are imaged at ambient temperature (~25 °C) with a 60 × 1.45 NA objective (Nikon CFI Plan Apo TIRF) using a Photometrics CoolSnap EZ monochrome CCD camera on a Nikon Eclipse Ti inverted C2 confocal microscope. Wide-field imaging are carried out using the following filter sets from Chroma Technology; mEGFP detection by a set of Z488/10-HC cleanup, HC TIRF Dichroic and 525/50-HC emission filter; and mCherry detection by a set of Z561/10-HC cleanup, HC TIRF Dichroic and 600/50-HC emission filter. Confocal Imaging are carried out using JDSU argon ion 488 nm laser for mEGFP detection and Coherent Sapphire 561 nm laser for mCherry detection via a 488/561/640 dichroic mirror with 525/50 and 600/50 emission filters.
Day-3: Lattice Light Sheets Microscopy (LLSM) Imaging
Unlike wide-field and confocal imaging, LLSM imaging requires optical calibration described below prior to imaging. Note that our in-house LLSM was constructed as previously reported (Chen et al., 2014).
-
ACalibration of Lattice Light Sheet Illumination
-
9Fill the sample chamber of the LLSM with 7 mL of a mixture of 500 nM fluorescein (ThermoFisher Scientific, Cat# F1300) and 500 nM sulforhodamine B (Invitrogen, Cat# S1307) in the imaging buffer (i.e. BSS) at room temperature.
-
10Manually adjust the position of an illumination objective lens to make rough focus of the illumination plane. Then, further align the illumination plane by adjusting neighboring mirrors and stages. Finally, fine-tune the z-scanning piezo of an imaging objective lens to ensure that the plane of illumination is well overlaid on the imaging plane. Note that this is achieved using a Bessel beam illumination and scanning the z-galvo mirror while ensuring that both ends of the Bessel beam focus and defocus coincidentally.
-
11Additionally, adjust neighboring mirrors of objective lenses to align the Bessel beam to be perpendicular to the x-scanning galvo. In this way, the illumination plane of the lattice light sheet is aligned to the detection plane and thus dithered orthogonally to the detection direction.
-
12After imaging the Bessel beam and lattice light sheets, thoroughly clean the sample chamber to remove the fluorescein and sulforhodamine B solution.
-
13Rinse multiple times to clean the chamber using sterile water followed by several washes with the BSS imaging buffer.
-
14Immobilize 200 nm blue/green/orange/dark-red TetraSpek™ microspheres (Invitrogen, Ref# T7280) on a clean 5 mm round coverslip.
-
15Place the 5 mm round coverslip on the sample chamber and image an isolated bead to obtain a 3-dimensional point spread function (3D PSF) of the LLSM for the day of imaging. This is done by paired scanning of the z-galvo mirror and the z-piezo imaging objective lens stage.
-
9
-
BCalibration of Dual-Color Imaging Capability
-
16To calibrate the spatial aberration of dual-color images, TetraSpek™ microspheres are embedded in polyacrylamide gel on the surface of a 5 mm round coverslip.
-
17Perform dual-color imaging of multi-color beads that are distributed in 3D space (Figure 2A).
-
18Bead images of red and green channels are corrected to have overlaid images in 3D space within a sub-voxel range. Briefly, correction factors are calculated using an average distance in each axis between beads in red and green channels. The correction factors are then employed to define and apply the vector translation to the images by the imtranslate syntax of MatLab.
-
16
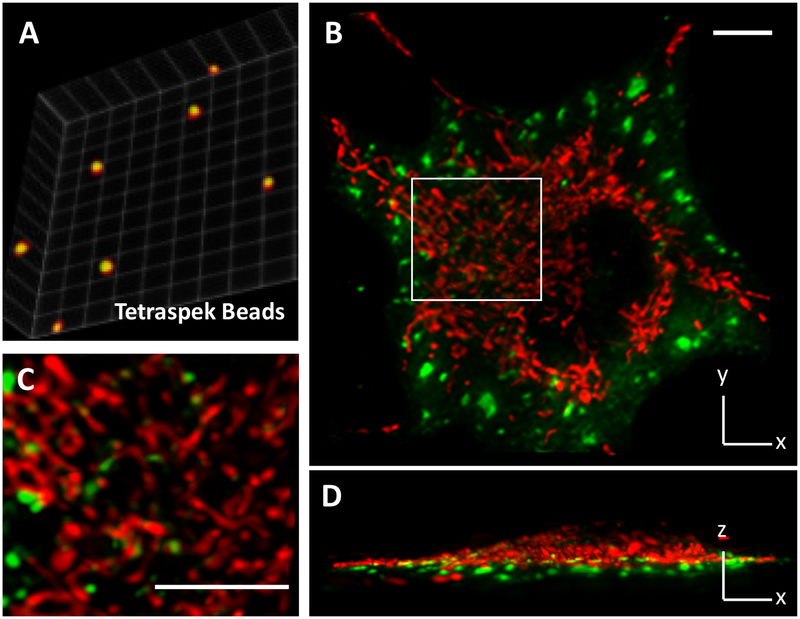
Figure 2. Dual-color LLSM imaging of metabolic condensates in live cells.
(A) Dual color calibration with TetraSpek™ microspheres (200 nm in diameter) that are embedded in polyacrylamide gel. The centroids of red and green images are overlapped with subvoxel precision. Calibration factors are applied to dual-color data. (B, C and D) A representative image shows the spatial distribution of glucosomes (green, hPFKL-mEGFP) and mitochondria (red, MitoTracker Red CMXRos, Cat# M7512) in a Hs578T cell. The indicated region of interest (a white box) is enlarged for clarification. Scale bars, 10 um.
Note that STEPS 16 and 17 can be replaced by imaging TetraSpek™ microspheres with cells as long as the microspheres do not interfere with data collection. Details are added in STEP-21 below.
-
3LLSM Imaging
- Clean and fill the sample chamber of the LLSM with fresh 7 mL of BSS solution at room temperature.
- Remove a 5 mm round coverslip containing cells from the MatTEK dish, place it into a new dish with sterile tweezers, and rinse it with 2 mL of BSS solution.
- Repeat twice STEP-19.
- If it is desired, add TetraSpek™ microspheres as one drop directly over the 5 mm round coverslip in the BSS solution and leave undisturbed for ~10 min. It is recommended to evaluate a batch of TetraSpek™ microspheres in advance to result in approximately 10 beads within an imaging region of interest.
- Submerge the 5 mm round coverslip containing cells (and beads) in the sample chamber of the LLSM.
- Equilibrate the 5 mm round coverslip in the BSS solution at room temperature for ~ 1 hour prior to imaging.
- Cells are imaged at ambient temperature (~25 °C) with a 25 × 1.1 NA objective (Nikon CFI Apo LWD) using a sCMOS camera (Orca Flash 4.0 v2 camera Hamamatsu) with a quad-band pass filter (FF01–446/523/600/677–25 Semrock) (Figure 2B). Note that we recommend using hex-patterned phase masks on the spatial light modulator in the LLSM if the spatial resolution is a limited factor. Otherwise, square-patterned phase masks provide relatively higher illumination power resulting in better signal to noise ratios.
2.3. Data Acquisition
Take differential interference contrast (DIC) images to count the total number of cells from the sample dish.
Take fluorescent images from the identical field of views in the DIC images to count cells expressing fluorescence.
Count at least 100 cells per dish. Note that mitotic cells and/or dead cells identified as rounded cells are not counted for analysis.
Repeat at least 2–3 dishes per day. Also, repeat at least 3 times in different days. During the image acquisition, we further recommend using at least two different batches of cells starting all over from a frozen cell stock.
Transfection efficiency is calculated by counting fluorescent cells over the total number of cells. Clustering efficiency is calculated by counting cells displaying purinosomes or glucosomes over the number of transfected cells. The procedure above consistently yields the clustering efficiency of ~ 40–60% for purinosomes or 50–80% for glucosomes in nutrient-depleted medium. Note that lower clustering efficiencies have been also seen based on the quality of seeded cells in sample dishes (e.g. cell density, cell distribution, etc) and the applications of less optimal transfection conditions, different cell lines, different batches of serum, different transfection reagents, etc.
Purinosome-Positive Cells
Human formylglycinamidine ribonucleotide synthase fused with a monomeric form of enhanced green fluorescence protein (hFGAMS-mEGFP) has been used to visualize purinosomes in living human cells (An et al., 2008; Kyoung et al., 2015). However, we have recently demonstrated that hFGAMS form its own self-assemblies to downregulatie the pathway (Schmitt et al., 2016) whereas hFGAMS is also part of the three-enzyme core complex (Schmitt et al., 2018). To distinguish which hFGAMS granules are being regulated in given systems, at least two additional enzymes that are involved in de novo purine biosynthesis are strongly recommended to be evaluated in the same conditions; one of two enzymes catalyzing the first half of de novo purine biosynthesis (i.e. phosphoribosyl pyrophosphate amidotransferase (hPPAT-mEGFP) and a trifunctional enzyme containing the activities of glycinamide ribonucleotide synthetase, glycinamide ribonucleotide transformylase and aminoimidazole ribonucleotide synthetase (hTrifGART-GFP)), and another one among three enzymes catalyzing the second half of de novo purine biosynthesis (i.e. a bifunctional enzyme with the activities of carboxyaminoimidazole ribonucleotide synthetase and succinylaminoimidazolecarboxamide ribonucleotide synthetase (hPAICS-mEGFP), adenylosuccinate lyase (hASL-mEGFP) and a bifunctional enzyme with the activities of aminoimidazolecarboxamide ribonucleotide transformylase and inosine monophosphate cyclohydrolase (GFP-hATIC)). In this way, one can properly evaluate their metabolic condensates with appropriate negative and positive controls. Note that the plasmids expressing these tagged enzymes are available from Addgene.com.
Glucosome-Positive Cells
Human liver-type phosphofructokinase fused with mEGFP (hPFKL-mEGFP) has been used to visualize glucosomes in living human cells (Jeon et al., 2018; Kohnhorst et al., 2017). When we collect glucosome-positive cells under fluorescence microscopy, we have categorized them into three subclasses; namely small (< 0.1 um2), medium (0.1 – 3 um2), and large (3 – 8 um2) sized granules (Kohnhorst et al., 2017). Granules that are smaller than an optical diffraction limit are categorized as small-sized cluster (< 0.1 um2). Medium-sized granules are then defined in sizes between 0.1 to 3 um2 because granules that are larger than 3 um2 are not detected in non-cancerous cells (i.e. Hs578Bst) (Kohnhorst et al., 2017). However, any granules larger than 8 um2 are considered as aggregations. The described sizes are based on wide-field imaging. It is also worthwhile to mention that when cells are defined to show certain sized clusters, ‘smaller’ sized clusters are also displayed in the given cell.
3. High-Content Image Analysis of Single Cells
3.1. 2D Granule Size Analysis
Granule size analysis is carried out using the ImageJ processing software (National Institutes of Health, NIH).
Unless raw images show one cell, raw images are edited to isolate in-focus, single, whole cells from the image. Cropping the original image and, in some instances, manually outlining the cell to entirely remove surrounding pixel intensity information are acceptable as long as neither affect the original pixel information of the image.
- The raw but cropped images are then processed through ImageJ using its built-in module, so-called Robust Automatic Threshold Selection (RATS).
- Scale images according to the pixel size of the microscope (e.g. 0.12 μm/pixel) before the RATS segmentation tool.
- Run the RATS module with default parameters for RATS (i.e., noise threshold = 25, λ factor = 3).
- Run the Inverse Look-up Table function to generate a mask of the original image that only displays fluorescent clusters.
- Apply the Particle Analysis module to this masked image to attain both the number and area of fluorescent clusters within the image.
- Repeat the process for all subsequent cell images.
- Evaluates the original cell images against the masked image to eliminate data in which more than one cluster was counted as a single particle.
3.2. 2D Single-Cell Fluorescence Intensity Analysis
Either Nikon Elements imaging software (Nikon) or the ImageJ processing software (NIH) can be used to compare fluorescence intensities between fluorescence-positive cells as long as they are captured by the same light source at fixed power with the same exposure time. Here, we provide a protocol using Nikon Elements imaging software.
Outline single cells as described above in STEP-1 of Section 3.1.
The raw but cropped images are subjected to Nikon imaging software, with which we defined the boundary of cells to quantify total fluorescent intensities and the size of cells.
Mean fluorescent intensities are calculated by dividing the total fluorescent intensities of whole cells with the area of the cells.
The mean or total fluorescence intensities are graphed with the number of granules per cell or the average size of granules per cell that we obtain from the Granule Size Analysis (Section 3.1).
3.3. 3D Granule Volume and Intensity Analysis
LLSM images are deconvolved using the experimentally obtained PSF on the day of imaging (STEP-15, Section 2.2) (Gao et al., 2014). Afterward, the images are processed with either the Allen Cell Structure Segmenter (Chen et al., 2018) or ImageJ (NIH). Binary object maps are generated using the Allen Cell Structure Segmenter and overlaid onto the original data to mask objects that retain their original intensities. The detected objects are processed using the ImageJ 3D Objects Counter plugin (Bolte and Cordelieres, 2006). From the 3D Objects Counter Analysis tool, both the volume and the integrated density are used to analyze granule sizes in single cells. Note that the experimental PSF and the comparison between deconvolved and non-deconvolved 488 nm bead scans are used to determine a minimal filter size of 17 voxels for the volume unless otherwise noted. The final deconvolved voxel dimensions are 107 nm × 107 nm × 107 nm in the respective XYZ dimensions in our home-built LLSM instrument.
4. Quantitative Analysis of Metabolic Condensates in Single Cells
4.1. Fluorescence Recovery After Photobleaching (FRAP)
The metabolic condensates for de novo purine biosynthesis and glucose metabolism have been also subjected to FRAP measurement from single cells expressing mEGFP-tagged enzymes (Kohnhorst et al., 2017; Kyoung et al., 2015). Specific regions of interest in a cell (i.e. part of or whole granules) are selected and photobleached with JDSU argon ion 488 nm laser (50 mW) at 20–75% power for 0.5 s. The power of the laser and the exposure time of photobleaching are optimized prior to collect a data set for analysis. In addition, we select three regions of interest per cell and at least three reference regions of interest per cell during the data acquisition. All the regions of interest are recommended to be selected in different parts of the cell and as far as possible from each other. At least 10 images are obtained before bleaching, and subsequent images are acquired every 0.5 s for at least 50 s. Since every image we collect during FRAP experiments subsequently suffers from additional photobleaching, which would negatively impact the degree of diffusion of the target enzymes, the reference regions of interest are used to correct the additional photobleaching effect that occurs during data acquisition. Each corrected fluorescence recovery is then fitted by one of the following equations,
and/or
where τ1/2 is a diffusion time constant and t is the time (s). Apparent diffusion coefficients (Dapp) were then calculated by the following equation,
where re is a radius (μm) of a photobleached area, which we experimentally determine for data analysis.
4.4. Intracellular Fluorescence Resonance Energy Transfer (FRET)
To measure intracellular FRET from live cells, we have applied a so-called acceptor photobleaching method (Kohnhorst et al., 2017). Experimentally, mEGFP- and mCherry-tagged enzymes are dually transfected into cells. Co-clustering cells are subjected to measurement of their FRET signals under a confocal mode. Here, we detect increased emission of the donors’ signals upon the acceptors’ photobleaching due to the loss of FRET. Coherent Sapphire 561 nm laser is applied at 40–50% power for 0.5 s to bleach acceptor molecules at the regions of interest. At least 10 images are obtained before the acceptor bleaching, and subsequent images are acquired every 0.5 s for at least 50 s. After the photobleaching of the donor molecules is corrected in each data point, the temporal increase of the emission of the mEGFP-tagged donor molecules from the same areas is graphed to reveal their direct interaction in live cells. In our system, dual-color confocal imaging is achieved via a 488/561/640 dichroic mirror with 525/50 and 600/50 emission filters and photomultipliers. Sufficient spectral overlap between the donor emission and the acceptor excitation is required for FRET measurement.
5. Discussion
5.1. Advantages of the Intrinsically Fluorescent Protein Tag over Disadvantages
To quantitatively analyze real-time spatial dynamics of metabolic enzymes and their interplays with other cellular processes in live cells, we need to ensure at least two factors in designing fluorescence live-cell imaging. First, it is imperative to specifically label only the enzymes of interest in live cells. Accordingly, we have genetically labeled the target enzymes with intrinsically fluorescent proteins in live cells. In this way, we can rule out fluorescence signals from non-specifically labeled proteins or free fluorophores. Second, high photostability and high signal-to-noise ratios of the fluorescent probes of interest are essential for time-lapse observation of the labeled enzymes in live cells. Evidently, fluorescence signals from the mEGFP-tagged enzymes in the described cell culture conditions are strong enough for us to investigate their dynamics in live cells (Kohnhorst et al., 2017; Kyoung et al., 2015).
Importantly, we have not encountered potential caveats of using mEGFP-tagged enzymes in transfected cells; which are often referred to as overexpression-mediated and/or GFP-mediated artifacts. Briefly, we have introduced site-directed mutations to the gene of EGFP, which has shown to prevent so-called EGFP-mediated aggregates in cells (Landgraf et al., 2012; Zacharias et al., 2002). We also track several hundreds of single cells displaying varying degrees of fluorescence intensities, which reflects the varying ranges of protein expression levels. Single cell analyses described above support that our results are expression level-independent phenomena (Kohnhorst et al., 2017). Furthermore, we have shown as negative controls that many mEGFP-tagged metabolic enzymes do not form spatially resolved granules in HeLa and Hs578T cells (An et al., 2008; Kohnhorst et al., 2017; Kyoung et al., 2015). Most importantly, the assembly and disassembly of purinosomes and glucosomes are reversibly regulated in live cells, strongly supporting they are not technique-driven artifacts. Collectively, it is apparent that we do not observe the overexpression and/or mEGFP-mediated aggregation artifacts at least in HeLa and Hs578T cells under the described imaging conditions.
5.2. Alternative Live-Cell Approach with Fluorescent Tags
Nevertheless, the size of fluorescent protein tags cannot be ignored particularly when tagged enzymes are studied for protein-protein interactions or macromolecular complexation. Although various fluorescent tags have been developed, it is unfortunate that there is barely an option if cytoplasmic enzymes are being imaged under fluorescence live-cell imaging, except in our opinions for the tetracysteine (TC) tag.
Briefly, the TC tag, which is composed of only six amino acids (e.g., N-CCPGCC-C), can be fluorescently labeled with small biarsenical compounds (Crivat and Taraska, 2012). To evaluate the specificity of TC-mediated fluorescent staining in HeLa and Hs578T cells (Kohnhorst et al., 2017), we transfect cells with a dually tagged enzyme with TC and mEGFP (e.g. hPFKL-TC-mEGFP). We then incubate the cells with a biarsenical resorufin-derivative agent, ReAsH-EDT2 (ThermoFisher Scientific, Cat# T34562), to visualize the PKFL granules through the TC tag. Clearly, colocalization of ReAsH (in the red channel) and mEGFP (in the green channel) signals supports the specific labeling of ReAsH to PFKL-TC-mEGFP in HeLa and Hs578T cells (Kohnhorst et al., 2017). Afterward, TC-tagged enzymes without fluorescent proteins have been applied to visualize glucosome granules in our live-cell platform in the presence of FlAsH (ThermoFisher Scientific, Cat# T34561) or ReAsH. Therefore, we establish that the TC-tag can visualize metabolic condensates in the cytoplasm of live cells with the minimum steric interference.
5.3. Complementary Approaches without Fluorescent Tags
We and others have also performed immunocytochemistry against endogenous hFGAMS or hPFKL in various cell lines (An et al., 2008; Baresova et al., 2012; Kohnhorst et al., 2017). In our work, cells are fixed with freshly prepared 3% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 10% normal donkey serum (Jackson ImmunoResearch Lab). The cells are then incubated with a rabbit polyclonal anti-FGAMS/PFK antibody and a Cy3-conjugated goat anti-rabbit IgG (H+L) (Jackson ImmunoResearch Lab). Controls for non-specificity and autofluorescence include the fixed cells incubated with primary only, secondary only, and neither antibody. It is important to note here that specificity of primary antibody is strongly recommended to be evaluated before carrying out immunocytochemistry. Conventional western blot analysis or immunostaining of cells expressing mEGFP-tagged target enzyme are acceptable for antibody validation. Alternatively, in situ proximity ligation assays using oligonucleotide-modified antibodies have also been developed to visualize endogenous proteins and their interactions in mammalian cells (Klaesson et al., 2018; Soderberg et al., 2006). Nevertheless, one may now compare their live or fixed cell images with the immunestained images available at the Human Protein Atlas database (https://www.proteinatlas.org/) (Thul et al., 2017).
6. Conclusion
Fluorescence live-cell microscopy has allowed us to discover multienzyme metabolic assemblies that are functionally associated with de novo purine biosynthesis and glucose metabolism in living human cells. We believe that we have now walked into a new territory where metabolic enzymes, their interactions and their complexes are associated with other cellular processes (e.g. signaling pathways and the cell cycle progression). Indeed, new information we and others have obtained so far has advanced our understanding of metabolic assemblies and their functional roles in living cells. However, we are certainly far away from extracting comprehensive information from imaging-based data and thus it is important to correlate the imaging-based knowledge with the information we have obtained from ensemble assays. We believe that the described strategies in this work would serve as good entry points for cellular biochemistry but cannot be limited to future investigation of enzymes or proteins in living cells. Therefore, various single-cell techniques should be innovatively employed to explore the new territory of living cells in our endeavors to comprehend metabolic enzymes and their network in cells.
Acknowledgments
We would like to thank Dr. Eric Betzig for sharing the design of LLSM. We also thank our current and previous group members for their contributions to the optimized protocols. This work was funded in part by UMBC and NIH (R01GM125981, R03CA219609, R25GM55036 and T32GM066706)
References
- An S, Kumar R, Sheets ED, and Benkovic SJ (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science, 320, 103–106. [DOI] [PubMed] [Google Scholar]
- Baresova V, Skopova V, Sikora J, Patterson D, Sovova J, Zikanova M, et al. (2012). Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum Mol Genet, 21, 1534–1543. [DOI] [PubMed] [Google Scholar]
- Bolte S, and Cordelieres FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc, 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science, 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ding L, Viana MP, Hendershott MC, Yang R, Mueller IA, et al. (2018). The Allen Cell Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images. bioRxiv, 10.1101/491035. [DOI] [Google Scholar]
- Crivat G, and Taraska JW (2012). Imaging proteins inside cells with fluorescent tags. Trends Biotechnol, 30, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Shao L, Chen BC, and Betzig E (2014). 3D live fluorescence imaging of cellular dynamics using Bessel beam plane illumination microscopy. Nature protocols, 9, 1083–1101. [DOI] [PubMed] [Google Scholar]
- Jeon M, Kang HW, and An S (2018). A Mathematical Model for Enzyme Clustering in Glucose Metabolism. Sci Rep, 8, 2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaesson A, Grannas K, Ebai T, Heldin J, Koos B, Leino M, et al. (2018). Improved efficiency of in situ protein analysis by proximity ligation using UnFold probes. Sci Rep, 8, 5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnhorst CL, Kyoung M, Jeon M, Schmitt DL, Kennedy EL, Ramirez J, et al. (2017). Identification of a multienzyme complex for glucose metabolism in living cells. J Biol Chem, 292, 9191–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyoung M, Russell SJ, Kohnhorst CL, Esemoto NN, and An S (2015). Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry, 54, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Okumus B, Chien P, Baker TA, and Paulsson J (2012). Segregation of molecules at cell division reveals native protein localization. Nature methods, 9, 480–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley AM, and Benkovic SJ (2017). A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci, 42, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt DL, and An S (2017). Spatial Organization of Metabolic Enzyme Complexes in Cells. Biochemistry, 56, 3184–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt DL, Cheng YJ, Park J, and An S (2016). Sequestration-Mediated Downregulation of de Novo Purine Biosynthesis by AMPK. ACS Chem Biol, 11, 1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt DL, Sundaram A, Jeon M, Luu BT, and An S (2018). Spatial alterations of De Novo purine biosynthetic enzymes by Akt-independent PDK1 signaling pathways. PLoS One, 13, e0195989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. (2006). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods, 3, 995–1000. [DOI] [PubMed] [Google Scholar]
- Srere PA (1987). Complexes of sequential metabolic enzymes. Annu Rev Biochem, 56, 89–124. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. (2017). A subcellular map of the human proteome. Science, 356. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, and Tsien RY (2002). Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science, 296, 913–916. [DOI] [PubMed] [Google Scholar]